Abstract
Microfluidics is an enabling technology for both cell biology and chemical analysis. We combine these attributes with a microfluidic device for on-line solid-phase extraction (SPE) and mass spectrometry (MS) analysis of secreted metabolites from living cells in culture on the chip. The device was constructed with polydimethylsiloxane (PDMS) and contains a reversibly-sealed chamber for perfusing cells. A multilayer design allowed a series of valves to control an on-chip 7.5 μL injection loop downstream of the cell chamber with operation similar to a 6-port valve. The valve collects sample and then diverts it to a packed SPE bed that was connected in-line to treat samples prior to MS analysis. The valve allows samples to be collected and injected onto the SPE bed while preventing exposure of cells to added back-pressure from the SPE bed and organic solvents needed to elute collected chemicals. Here, cultured murine 3T3-L1 adipocytes were loaded into the cell chamber and non-esterified fatty acids (NEFAs) that were secreted by the cells were monitored by SPE-MS at 30 min intervals. The limit of detection for a palmitoleic acid standard was 1.4 μM. Due to the multiplexed detection capabilities of MS, a variety of NEFAs were detected. Upon stimulation with isoproterenol and forskolin, secretion of select NEFAs was elevated an average of 1.5-fold compared to basal levels. Despite the 30 min delay between sample injections, this device is a step towards a miniaturized system that allows automated monitoring and identification of a variety of molecules in the extracellular environment.
Keywords: Microfluidic, Integration, Automation, Mass Spectrometry, Adipocyte, Solid-phase Extraction
Introduction
Innovative microfluidic technologies have been pivotal in the modern progression of analytical chemistry research by allowing miniaturization, integration, and automation, often with performance enhancements over larger scale instruments. Many examples of performing chemical assays or separations on microfluidic chips with impressive sensitivity and throughput have been reported [1, 2]. Microfluidics has also been enabling for cellular biology. Cells can be cultured on chips after loading into chambers by flow and other methods [3, 4]. The fluidic environment allows cell culture conditions to be exquisitely controlled and better match in vivo conditions [4–6]. Integrating chemical analysis with culture allows cell function to be assayed under different conditions [7, 8]. Such studies can be critical for studying cell physiology. In this work, we describe an approach to integrate living cell perfusion on a microfluidic chip with electrospray ionization mass spectrometry (ESI-MS) to analyze secretions from the cells.
Culturing and monitoring cells on microfluidic chips has many advantages over conventional techniques. The inherent small size of such devices reduces the number of cells required [9, 10]. Constant perfusion of culture media on chip allows replenishment of essential nutrients and removal of waste products, which creates a more physiologically relevant environment and provides the capability to change conditions for controlled experiments [5, 11, 12]. It is relatively straightforward to monitor cell function using fluorescence microscopy on chips [13–16]. Perfusate can also be analyzed by other analytical techniques to measure cell secretions and other effects of the cells on the chemical environment. Manually sampling perfusate from chips for detection of peptide or metabolite release from cells has been reported [17–20]; however, manual sampling of perfusate and off-line assay fails to fully to take advantage of the integration and automation potential of microfluidic chips.
Several studies have reported integration of on-line analysis of perfusates to monitor cellular secretions on chips [21, 22]. Immunoassay [23–25] or enzyme assay [26–28] with fluorescent detection, capillary electrophoresis with electrochemical detection [29], and enzyme assay with chemiluminescent detection [30] have all been used. These methods give temporal resolution and selectivity in the measurement providing excellent insight into cell function. For example, rapid electrophoresis assays have been used to monitor insulin secretion in real time from single islets of Langerhans providing information on oscillations of secretion [31]. A commonality of these methods, however, is that they are specific for a single (or small number) of compounds and new assays must be designed for each target analyte.
To develop a more versatile approach to chemical monitoring of cells on chips, we have investigated coupling MS to cell culture chips. MS is a powerful, sensitive, and widely available analytical technique. MS also provides high selectivity, the ability to detect multiple compounds in single scans, and direct detection of compounds without labels. ESI-MS is compatible with continuous flow systems and can be coupled to microfluidic chips [32–38]. All of these properties make ESI-MS well-suited as a general tool for analyzing cell perfusates. Combining the many advantages of on-chip cell perfusion and MS detection suggests potential for a powerful approach to study cell secretion dynamics.
Although ESI-MS is well suited for this application, a significant impediment is that biological samples often contain high concentrations of salts and other species that can cause ion signal suppression in MS [39]. Sample “clean-up” methods like solid phase extraction (SPE) are often used to remove interfering species prior to ESI-MS. SPE can be implemented during off-line sample preparation prior to analysis (e.g., ZipTip [18]) or in an on-line format [33, 39–41]. In SPE, samples are loaded onto a packed bed of sorbent. The bed is rinsed to remove poorly retained species, and then a solvent is passed through the bed to elute analytes. Hydrophobic sorbents are often used to retain analytes and allow removal of salts.
In previous work, microfluidic cell perfusion has been coupled to on-chip SPE and ESI-MS analysis [42, 43]. These devices were applied to monitor vitamin E metabolites secreted from lung epithelial cells [42] and glutamate release from neuronal cells [43]. This seminal work illustrated the potential for such approaches; however, in these reports, the components are on separate devices so that the steps of SPE required repeated and manual disconnection and reconnection of tubing to the chips. In addition, the cells are in direct fluidic connection to the SPE packing and therefore are exposed to higher backpressures generated by the bed. This higher (and changing) pressure is potentially detrimental to cell viability because high shear stress can cause changes in normal cell morphology and function [10]. Furthermore, increased shear can also cause detachment of adhered cells from the surface, which is especially undesirable for buoyant adipocytes.
PDMS valves have demonstrated great versatility for on-chip fluid handling, from automating fluid flow over select cell chambers [44] to transporting cells via peristaltic pumping [45]. Pneumatically-actuated valves are integrated on PDMS chips through multilayer assembly [46]. In such devices, a ‘control’ channel is overlaid across a shallow fluidic channel. When gas pressure is applied through the control channel, the fluid channel is constricted via deformation of the elastomeric PDMS to block flow. To create an automated, on-line microfluidic cell perfusion system with ESI-MS analysis, we have developed a PDMS chip that integrates a cell chamber with an injection loop consisting of a series of pneumatically-actuated valves (which essentially functions like a conventional 6-port valve). The injection loop allows automated collection of perfusate and then injection onto an in-line SPE-ESI-MS system. The injection system fluidically isolates cells from the SPE packing to prevent exposing cells to changing pressure.
Cultured adipocytes are used as a model cell system in this study. Adipose tissue is integral to maintaining systemic energy balance by storing and releasing lipids based on physiological requirements. Adipocytes secrete non-esterified fatty acids (NEFAs) through regulated lipolysis [47, 48]. Adipocytes represent a particularly challenging cell type because they are fragile and buoyant, making them susceptible to changes in pressure or shear. In previous reports, NEFAs secreted from perfused adipocytes have been measured on PDMS chips with enzyme assays and laser-induced fluorescence [26, 28]. These enzyme assays only allow total NEFA measurement; MS is a more versatile detection technique, allowing detection of specific NEFAs. With this chip-MS method, the change in specific NEFAs released during lipolysis stimulation could be determined, better indicating the composition of circulating NEFAs due to lipolysis than with previous methods.
Experimental Section
Reagents
LC-grade acetonitrile (ACN) and water, isoproterenol hydrochloride, forskolin, palmitoleic acid standard and fatty acid free bovine serum albumin (BSA) were purchased from Sigma-Aldrich (St. Louis, MO). Ammonium hydroxide was obtained from Fisher Scientific (Pittsburgh, PA). Cell culture reagents and Hank’s buffered salt solution (HBSS, Cat. No. 14175) were received from Life Technologies (Carlsbad, CA).
PDMS Chip Fabrication
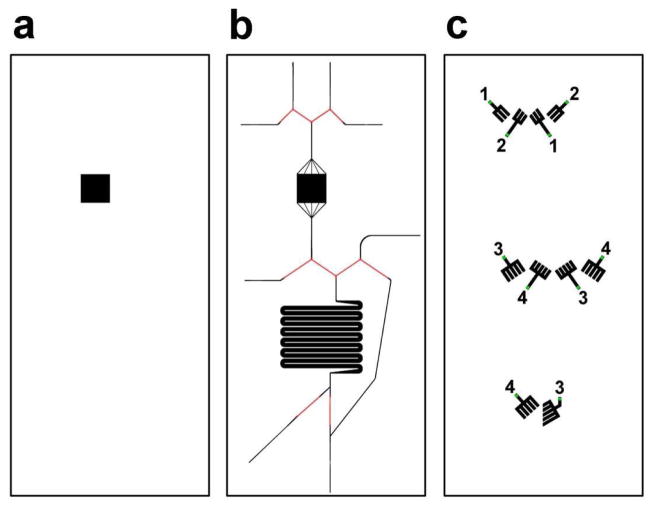
Chip fabrication used procedures similar to those described before [28]. The injection loop chip was composed of two separate pieces and all layers were formed from 10:1 base:curing agent PDMS (RTV 615, Curbell Plastics, Livonia, MI). Three Si wafers with raised features were made using SU-8 2075 photoresist (MicroChem, Newton, MA). Wafer A contained the lower cell chamber and was 250 μm thick (mask shown in Figure 1a). Wafer B consisted of the fluidic channels with SU-8 features 60 μm thick and AZ 9260 (Capitol Scientific, Austin, TX) features 12 μm thick (Figure 1b). Wafer C contained the SU-8 control channel features that were 60 μm thick (Figure 1c). PDMS was poured over wafer A and manually spread to create a layer ~1 mm thick. PDMS was spun on wafer B to a height of ~70 μm. A thick layer (~1 cm) of PDMS was poured over wafer C. Additionally, PDMS was spun on a blank wafer to a height of ~170 μm. All wafers were baked at 80 °C for 1 hr. The PDMS molded from wafer C was peeled off the wafer and holes were punched through the PDMS at the green points indicated on Figure 1c. This layer was irreversibly bonded to the PDMS spun on wafer B, feature side down, using corona discharge. The bonded layers were peeled away from wafer B and were then bonded, feature side down, to the PDMS spun on the blank wafer. The 3 layers (forming the top part of the chip) were peeled away and the bottom layer of PDMS was cut away around the cell chamber dimensions. A 4.8 mm × 4.8 mm glass coverslip (No. 1, Fisher Scientific) was bonded to the top of the cell chamber. Capillary tubing was used to move fluid in and out of the chip. One side of 50 μm inner diameter (ID)/150 μm outer diameter (OD) capillaries (Polymicro Technologies, Phoenix, AZ) were rounded with fine sand paper and inserted into the sides of the chip. The other end of the capillary was sheathed with 180/360 capillary and the two capillaries were then permanently secured using cyanoacrylate glue. Stainless steel tubing, (23 G, 1 cm long) sheathed halfway with Tygon Tubing (1/16 in. OD, 0.5 mm ID, IDEX, Lake Forest, IL), was inserted into the aforementioned holes made in the PDMS layer taken from wafer C. The PDMS from wafer A was peeled away and bonded to a glass slide, feature side up, forming the bottom part of the chip.
Figure 1.
PDMS mold layers. Each box indicates a different fabricated layer; (a) lower cell chamber, (b) fluidic layer, and (c) valve control layer. The numbers in (c) indicate the valves that are operated together on the same solenoid valve.
Adipocyte Culture
Murine 3T3-L1 adipocytes were cultured to day 11–13 post-induction as previously described [28]. Briefly, glass coverslips (No. 1, Fisher Scientific) were cut to 4.8 mm × 4.8 mm and placed in the bottom of 6-well plates. Preadipocytes were plated over the coverslips at a density of 100,000 cells/mL. Two days after the cells reached confluence, adipogenesis was induced with 500 μM isobutylmethylxanthine, 1 μM dexamethasone, and 1 μg/mL insulin in adipogenic media. Adipogenic media consisted of 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, 2 mM L-glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin in Dulbecco’s eagle medium (Cat. No. 11965, Life Technologies). 2–3 days after induction treatment, the media was replaced with adipogenic media containing 1 μg/mL insulin. Every 2–3 days following, fresh adipogenic media was added.
Chip Assembly and Operation
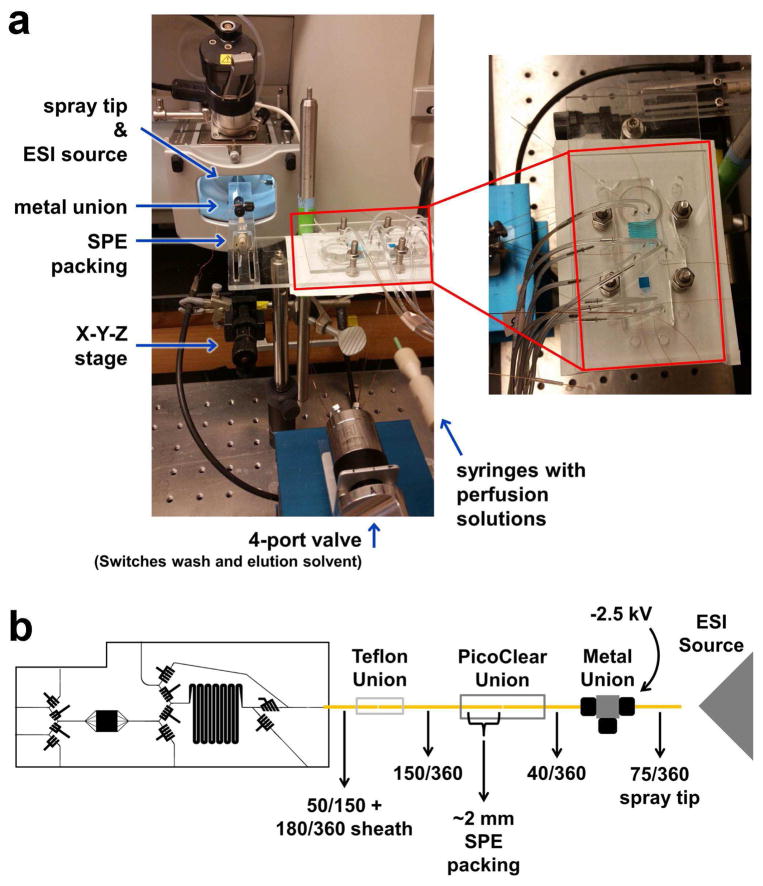
Prior to each use, the top portion of the PDMS chip was degassed for 30 min and the channels and Tygon tubing were primed with water. A coverslip with adhered adipocytes was removed from culture, rinsed with basal perfusion buffer (0.1% BSA and 25 mM glucose in HBSS) and transferred to the lower cell chamber. An additional 10 μL basal perfusion buffer was added to the top of the cells and the 2 chip pieces were sealed together, using the outline of the cell chamber in both portions for alignment. During calibration experiments, the same assembly procedure was used, but with a blank coverslip placed in the lower cell chamber. All air and excess liquid was removed between the layers. To ensure a leak-free operation, the assembled chip was compressed in a plastic frame. One side of the compression frame had holes drilled with the same spacing as the holes punched in the PDMS control channel layer. Both pieces had holes drilled around the chip with dimensions that allowed adding bolts to compress the plastic. The valve Tygon tubing was threaded through the compression frame and the chip was lightly tightened in the frame. A LabView 12.0 (National Instruments, Austin, TX) script was written to operate a series of solenoid valves (LHDA1211111H, The Lee Co., Westbrook, CT), which controlled flow from an N2 tank to the Tygon tubing in the chip, similar to that described previously [49]. The assembled chip in the compression frame is shown in Figure 2a.
Figure 2.
(a) Image of injection loop chip (filled with blue food dye for visualization) in compression frame, and connection to infusion syringes, the SPE column and ESI components. (b) A more detailed schematic of the chip connections.
Chip-to-MS Connection
A suspension of 20 μm fully porous C18 particles (Alltima, Grace Davison, Deerfield, IL) was made in methanol (~2 mg/mL). Polished ends of 150/360 and 40/360 capillary were connected with a PicoClear union (New Objective, Woburn, MA). The particle slurry was pushed into the 150/360 capillary using a syringe until the bed length was ~1 mm long. The outlet of the injection loop chip was connected to the 150/360 capillary with a Teflon union as shown in the schematic in Figure 2b. The other end of the 40/360 capillary was inserted into a metal tee which was used to apply voltage for electrospray. A spray tip (75/360 capillary with a 15 μm tip, New Objective) was inserted in the opposite end of the metal tee (the third port was plugged with an optical fiber). The entire assembly was mounted on a platform, built on an x-y-z stage for easier alignment with the MS source (shown in Figure 2a).
Mass Spectrometry Detection and Analysis
A triple quadrupole (QQQ) mass spectrometer (TSQ Quantum Ultra, Thermo-Scientific, Waltham, MA) was used for detection of the NEFA species. The platform supporting the chip and connections was inserted through the door of the ion source and the spray tip was positioned ~8 mm from the ion funnel. The spray voltage was kept off during the SPE loading phase, but was set at −2.5 kV during the wash and elution steps. The ion source capillary was maintained at a temperature of 250 °C and full scan mode over m/z 200–320 with a scan rate of 2 Hz was used for recording. Peak areas of the detected NEFAs were measured with a mass extraction range of ± 0.5 Da from the theoretical [M−H]− ion. In some cases, high resolution time-of-flight MS was also used for analysis (see Electronic Supplementary Material, ESM).
Results and Discussion
Chip Design & Operation
This work builds upon a previous chip that was reported to measure NEFA secretion from on-chip adipocyte perfusion [28]. In that work, cell perfusate was mixed on-chip with enzyme assay reagents in a continuous flow system to allow real-time monitoring of NEFA release; however, that method did not allow identification of individual NEFAs. With this new PDMS chip (Figure 2), it is possible to perfuse cells and directly monitor secretion of NEFAs via SPE-ESI-MS. The integration of an on-chip injection loop enables automated fraction collection and coupling to a SPE-ESI capillary without exposing cells to changes in pressure due to flow through the packed bed. A square cell chamber design is used on this chip as previously modeled and demonstrated [28]. Multiple chamber inlets were used and positioned so that the buffer flow velocity is constant across the width of the channel, as opposed to a parabolic shape, as previously demonstrated [28]. (The flow velocity is constant across the width of the channel; however, a parabolic flow profile would still be apparent along the height of the channel). This arrangement of inlet channels improved the temporal response of the system. The cell chamber was operated at ambient temperature because these cells are stable for several hours at room temperature. Approximately 25,000 3T3-L1 adipocytes can be loaded into the cell chamber.
Four valves were constructed upstream of the cell chamber to automate selection of cell perfusate solution (Figure 3). One inlet upstream of the cell chamber was infused with basal buffer and the other with 20 μM isoproterenol/10 μM forskolin with 0.1% DMSO in basal buffer. (During calibration, different concentrations of standards were infused through the inlets). The valves operated so that as one solution flowed to the cell chamber, the other flowed to waste. Actuating the valves changed the solution flowing over the cells. These solutions had a flow rate of 0.75 μL/min, which was therefore the flow rate perfused over the cells. Selecting the flow rate requires consideration of several tradeoffs. A high flow rate would be advantageous for temporal response by decreasing the time for each round of injections onto the SPE-MS; however, excessively high flow rate may damage or dislodge cells, overly dilute analytes, and cause valves to fail.
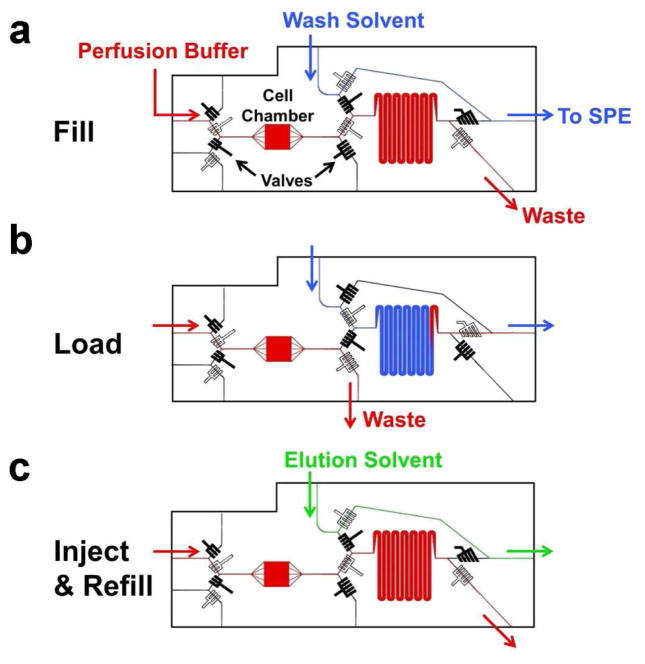
Figure 3.
Operation modes for the injection loop chip. Closed valves are black and open valves are white. (a) Loop fills with perfusate from cell chamber (red) and flows to a waste outlet, while the wash solvent (blue) bypasses the loop and is directed towards the SPE bed. (b) The cell perfusate is flushed out of the loop and onto the SPE bed by the wash solvent. Flow from the cell chamber is directed towards a waste outlet upstream of the loop. (c) Valves are in the same mode as (a), but wash solvent is replaced with elution solvent (green). Elution solvent bypasses the loop and is flowed onto the SPE bed, while the loop fills with cell perfusate again.
Downstream of the cells, the direction of flow was controlled through the switching of 6 multilayer valves (see Figure 3), which operated independently of the 4 valves upstream of the cell chamber. These valves controlled sample collection and SPE. In the first step of chip operation, cell perfusate from the cell chamber was directed into a serpentine loop while the wash solvent (10% ACN/90% H2O with 15 mM NH4OH) bypassed the loop and flowed directly into the SPE column as illustrated in Figure 3a. Once the loop was filled, the valves were switched and the sample was pumped onto the SPE bed while cell perfusate was directed to waste as shown in Figure 3b. The valves were set in this position for 12 min to fully load sample and allow rinse of the SPE column with wash solvent. Once the sample was fully loaded onto the SPE bed, the valves were switched back to their original position. The wash solvent bypassed the loop and was pumped onto the SPE bed for an additional 5 minutes to ensure all the salts were removed from the SPE packing. The wash solvent was replaced with the high organic elution solvent (75% ACN/25% H2O with 15 mM NH4OH) via an external 4-port valve, shown in Figure 2a, to elute the NEFAs for detection (Figure 3c). During the third step, the sample loop was filled with cell perfusate in preparation for the next fraction analysis. Using this procedure, 3 assays were performed with cells under basal conditions, and another 3 with cells stimulated with the isoproterenol/forskolin mixture (each iteration taking a total of 30 min). In these experiments, the NEFAs were desorbed with an isocratic method so that all eluted to the MS at the same time. Implementing a gradient elution protocol could allow chromatographic separation of the NEFAs and improve their detection.
Overall, this system operated like a conventional HPLC injection valve coupled with a short SPE bed. Integration of the loop allowed fluidic isolation of the cells from the downstream elements that could generate back pressures (SPE packing, spray tip). The serpentine pattern reduced the footprint of the chip, while providing enough channel length to collect adequate sample for MS injection. The injection loop was designed to hold 7.5 μL of sample, so with the flow rate used, it took 10 min to fill or empty the loop. The size of this channel could be reduced in future generations of the chip to improve temporal response, as long as the detection system had adequate sensitivity to detect the analytes.
Initial experiments revealed that back pressure of the SPE bed caused control channels to fail. To prevent this, the 6 valves that operated the injection loop were modified so that control channels for each valve were branched to 5 channels. We found that this branching allowed complete channel closure under conditions with downstream backpressure (the upstream valves were only branched to 3 channels as they were not exposed to any significant back pressure). Using inlet flow rates of 0.75 μL/min, the valves remain functional (i.e., completely sealed the lower fluid channels) up to back pressures of ~7 psi at the chip outlet. A video showing the operation of the multilayer valves is available in the ESM. This pressure limit, while sufficient for SPE as demonstrated here, constitutes a limit of this system for using longer columns or smaller particles.
The SPE bed was made with 20 μm particles packed into a 150 μm ID capillary. These large particles were used to reduce the flow resistance so that the backpressure of the SPE bed was lower than the maximum pressure at which the valves functioned. A 40 μm ID capillary at the exit of the SPE bed served to hold the particles in the capillary by the keystone effect [50]. A ~1 mm long SPE bed packed in this manner creates about 2.5 psi backpressure at a flow rate of 0.75 μL/min. We also explored using glass fiber frits to hold the particles in place [51]; however, we found that the back pressure was too high to operate the valves at effective flow rates.
The spray tip size was another consideration taken into account for this system. Smaller tips create more efficient spray, but they were prone to clogs and increased the back pressure. A capillary that had a 75 μm ID, pulled to 15 μm at the tip, provided sufficient spray (with low back pressure) and was able to be used for routine experiments without clogging.
Chip Characterization
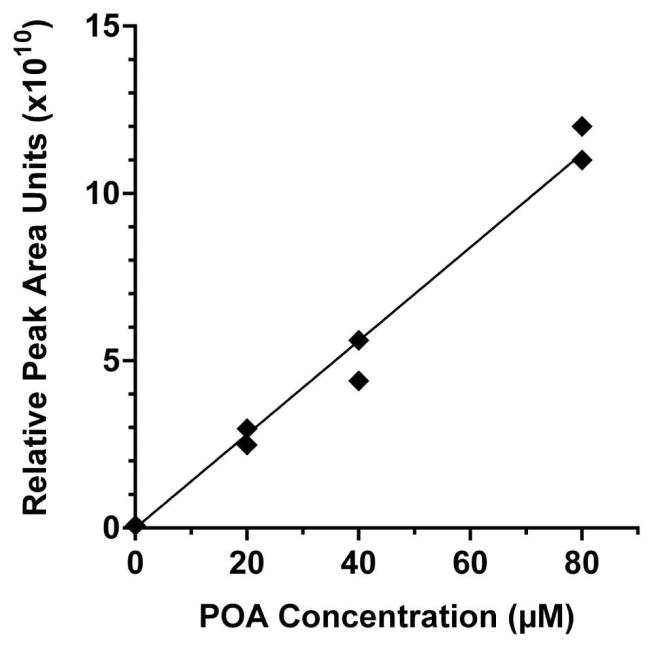
To determine the reproducibility of injections using the chip, a 50 μM standard of palmitoleic acid (16:1, [M−H]− = 253.2 Da) was perfused through the cell chamber, filled the loop, and then loaded onto the SPE bed and eluted to the MS repeatedly. It was found that the average peak area of 5 replicate injections was 5.8 ± 0.41 × 1010 (8% RSD). Similarly, to demonstrate the potential for quantification of NEFAs by chip-MS, a calibration curve of was created using palmitoleic acid, shown in Figure 4. With standards measured in duplicate, the calibration curve had a R2 of 0.98. The limit of detection for palmitoleic acid was 1.4 μM, as determined by the concentration of the signal that was 3 times the standard deviation of the two blank injections. We estimate the SPE system resulted in an approximately 12-fold sample concentration based on the observation that samples were loaded over a 10 min time and eluted peaks were ~50 s wide at the same flow rate.
Figure 4.
Calibration curve of palmitoleic acid (POA) standard, performed in duplicate. POA was dissolved in basal buffer and perfused through the chip at the inlet upstream of the cell chamber. Standard filled the loop, was loaded onto the SPE bed, washed and eluted to the MS in an on-line format.
We found that chips could be used for multiple experiments over a period of a few weeks. The re-sealable chamber made it feasible to load different batches of cells easily. The loading of cells on glass inserts allowed the cells to be removed and reloaded without need for extensive cleaning. The reversible sealing design allows daily loading of fresh adipocytes and reuse of the chip over 6–8 experiments.
NEFA Detection
Using the injection loop chip, select NEFAs secreted by adipocytes were analyzed by coupling on-chip cell perfusion to ESI-MS. Negative ion mode ESI was used for NEFA detection as the preferred ionization mechanism for NEFAs is the removal of a proton from the acidic carboxyl group [52]. All experiments were performed with adipocytes under basal conditions, followed by the perfusion of solutions containing isoproterenol and forskolin to stimulate lipolysis.
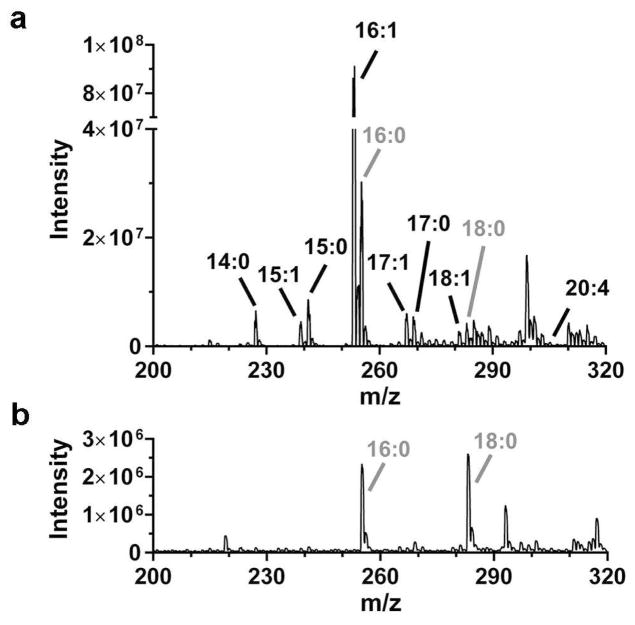
Initial experiments were aimed at evaluating what compounds were detected. Figure 5 shows a mass spectrum collected from the cells on chip compared to background. For this data, the mass spectrometer was scanned from m/z 200 to 320, which covers the range expected for most common NEFAs. To determine the source of the signals, we matched the signal to m/z values for NEFAs known to be present in 3T3-L1 adipocytes. Previous work using GC-MS has shown that NEFAs from C8 to C20, both saturated and monounsaturated, are present in adipocytes with C14-C18 being the most prevalent [53]. In agreement with those findings, we found peaks matching the nominal mass of C14:0, C15:1, C15:0, C16:1, C16:0, C17:1, C17:0, C18:1, C18:0, and C20:4. Small peaks for C16:0 and C18:0 were also found in background signals (e.g. when no adipocytes were in the chip) and therefore not further investigated. Previous studies have also reported background signal from 16:0 and 18:0 in fresh LC solvents, suggesting they may represent a ubiquitous background contaminant [54].
Figure 5.
Full scan analysis of m/z ranging from 200–320 was performed for all experiments on a QQQ-MS. (a) Mass spectrum of on-line elution profile from 3T3-L1 adipocytes. Adipocytes were loaded in the cell chamber and stimulated with isoproterenol/forskolin in this example spectrum. Perfusate was loaded onto the on-line SPE bed. The SPE bed was subsequently washed and sample was eluted to the MS. Peaks are labeled with the NEFAs that associate with the appropriate m/z. (b) Mass spectrum of direct injection of elution buffer, showing background signal of 16:0 and 18:0.
To better verify the identity of the signals, we also analyzed cell conditioned media by HPLC-TOF-MS (see ESM). We found matching peaks for all of the NEFAs detected from the chip system but at higher mass accuracy (as low as 1 ppm) using the TOF-MS. These results match what was found in the prior GC-MS studies and provide better confidence in the identifications shown in Figure 5. Collision-induced dissociation (CID), a fragmentation method used to more accurately identify and quantify compounds, was attempted but did not prove successful with the native NEFAs of interest in our study, as has also been observed in other reports [55].
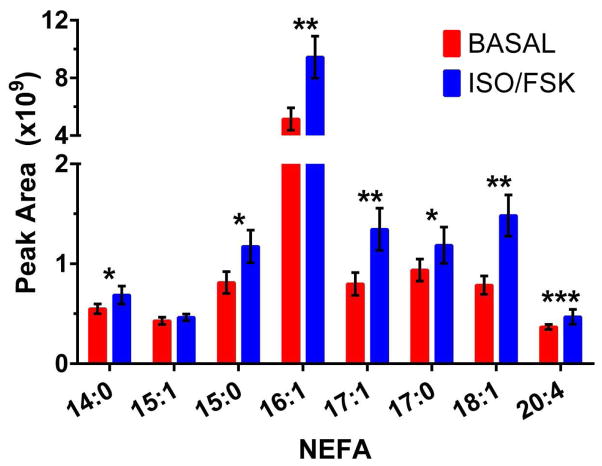
We then used the chip to monitor the effect of treatment with forskolin and isoproterenol on NEFA secretion (Figure 6). Both agents are known to stimulate lipolysis and were used in conjunction to evoke strong release of NEFAs from adipocytes, as previous MS reports of measuring NEFA release from 3T3-L1 cells have not involved the use of lipolysis-stimulating agents. To measure the various NEFAs, the peak areas of the extracted ion chromatograms (EICs) for the NEFA m/z values were quantitated. As shown in Figure 6, a statistically significant increase in 7 NEFAs was found with the application of lipolysis stimulating agents. The increase measured with the MS detection method was 1.2- to 1.9-fold over basal levels. This increase is less than what has been detected using enzyme assays for all NEFAs [28]. Stimulation of lipolysis with forskolin or isoproterenol generally results in a 2- to 10-fold increase in NEFA secretion over basal conditions as measured by enzyme assay in static culture or on-chip perfusion [26, 28, 56–58]. The smaller magnitude of stimulation detected with this technique could be accounted for by the fact that the enzyme assays measure every NEFA released from the adipocytes, whereas with the current method, two of the most abundant NEFAs were not able to be accurately quantitated. Furthermore, the relative abundance of secreted NEFAs might vary depending on culture conditions (i.e. various NEFAs are present in FBS, which is the source of lipid accumulation in cultured 3T3-L1 adipocytes). Despite these deviations from other reports of adipocyte lipid content, these results demonstrate the first on-line MS measurement of secreted NEFAs from living, perfused adipocytes.
Figure 6.
Averaged peak area of various NEFAs detected by MS from on-chip adipocyte secretion under basal and isoproterenol/forskolin (iso/fsk) stimulation. Data is average of 3 cell experimental replicates ± standard deviation, statistical significance determined by paired, 2-tailed Student’s t-test, * P < 0.1, ** P < 0.05, *** P < 0.01.
Conclusions
An integrated microfluidic chip has been developed that is capable of perfusing cultured cells and monitoring secreted metabolites with MS detection. This continuous flow device allows cells to be cultured under physiologically-relevant conditions while coupling to MS enables potential detection of a wide variety of secreted metabolites. PDMS as the microfluidic substrate permits multilayer valve construction, similar to conventional 4- or 6-port valves, increasing the automation of analysis. The injection loop valve system allowed discrete volumes of sample to be loaded onto an SPE bed, while isolating the cells from the backpressure generated by downstream clean-up or separation techniques. Using adipocytes as a model cell system, a variety of NEFAs were detected with this on-line perfusion format. NEFA secretion was measured under basal and stimulated conditions, demonstrating the capability of performing a range of biologically significant experiments on cultured cell lines on one miniaturized platform.
Although NEFA secretion was measured from adipocytes, this basic design could, in principle, be used for measuring release of many other secretions from a variety other cell types. The advantage of the MS system, compared to enzyme and immunoassay previously used for monitoring cell secretion on chips, is the versatility to detect a wider variety of compounds. Further, the ability to detect multiple compounds with a rapid scan enables multi-analyte monitoring. In the current system, a perfusate was collected in a loop and then extracted by SPE. The integration of SPE enables removal of ion suppressing components and concentrates the analytes 12-fold for improved sensitivity. Indeed, the NEFAs would not be detectable at the flow rates used without SPE. A weakness of the system as described here is the low temporal resolution of monitoring (30 min to fill loop and elute one sample). Chips with other materials may allow higher pressures to be used to increase flow rate over the SPE bed. Alternatively, other on-line extraction methods might prove useful.
Supplementary Material
Fig. S1 Extracted ion chromatogram of 8 NEFAs in adipocyte conditioned buffer, measured with HPLC-TOF-MS (a) along with the associated mass spectra of the most abundant NEFAs (b)
Table S1 Comparison of measured NEFA mass accuracy on TOF and QQQ
Acknowledgments
This research was funded by NIH R37 DK046960 to RTK. JPG was supported by NIH F32 EB019800. Spray tips and PicoClear unions were generously provided by New Objective Inc. TOF-MS experiments utilized Core Services of the Michigan Regional Comprehensive Metabolomics Resource Core (MRC2) supported by grant DK097153 of NIH to the University of Michigan.
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
References
- 1.Nge PN, Rogers CI, Woolley AT. Advances in microfluidic materials, functions, integration, and applications. Chem Rev. 2013;113:2550–83. doi: 10.1021/cr300337x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roman GT, Kennedy RT. Fully integrated microfluidic separations systems for biochemical analysis. J Chromatogr A. 2007;1168:170–88. doi: 10.1016/j.chroma.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 3.Hung PJ, Lee PJ, Sabounchi P, Aghdam N, Lin R, Lee LP. A novel high aspect ratio microfluidic design to provide a stable and uniform microenvironment for cell growth in a high throughput mammalian cell culture array. Lab Chip. 2005;5:44–8. doi: 10.1039/b410743h. [DOI] [PubMed] [Google Scholar]
- 4.Kim L, Toh YC, Voldman J, Yu H. A practical guide to microfluidic perfusion culture of adherent mammalian cells. Lab Chip. 2007;7:681–94. doi: 10.1039/b704602b. [DOI] [PubMed] [Google Scholar]
- 5.El-Ali J, Sorger PK, Jensen KF. Cells on chips. Nature. 2006;442:403–11. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 6.Vyawahare S, Griffiths AD, Merten CA. Miniaturization and parallelization of biological and chemical assays in microfluidic devices. Chem Biol. 2010;17:1052–65. doi: 10.1016/j.chembiol.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Salieb-Beugelaar GB, Simone G, Arora A, Philippi A, Manz A. Latest developments in microfluidic cell biology and analysis systems. Anal Chem. 2010;82:4848–64. doi: 10.1021/ac1009707. [DOI] [PubMed] [Google Scholar]
- 8.Primiceri E, Chiriaco MS, Rinaldi R, Maruccio G. Cell chips as new tools for cell biology - results, perspectives and opportunities. Lab Chip. 2013;13:3789–802. doi: 10.1039/c3lc50550b. [DOI] [PubMed] [Google Scholar]
- 9.Culbertson CT, Mickleburgh TG, Stewart-James SA, Sellens KA, Pressnall M. Micro total analysis systems: Fundamental advances and biological applications. Anal Chem. 2014;86:95–118. doi: 10.1021/ac403688g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker GM, Zeringue HC, Beebe DJ. Microenvironment design considerations for cellular scale studies. Lab Chip. 2004;4:91–7. doi: 10.1039/b311214d. [DOI] [PubMed] [Google Scholar]
- 11.Dhumpa R, Roper MG. Temporal gradients in microfluidic systems to probe cellular dynamics: A review. Anal Chim Acta. 2012;743:9–18. doi: 10.1016/j.aca.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kovarik ML, Gach PC, Ornoff DM, Wang Y, Balowski J, Farrag L, et al. Micro total analysis systems for cell biology and biochemical assays. Anal Chem. 2012;84:516–40. doi: 10.1021/ac202611x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sankar KS, Green BJ, Crocker AR, Verity JE, Altamentova SM, Rocheleau JV. Culturing pancreatic islets in microfluidic flow enhances morphology of the associated endothelial cells. PLoS ONE. 2011:6. doi: 10.1371/journal.pone.0024904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–U10. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung PJ, Lee PJ, Sabounchi P, Lin R, Lee LP. Continuous perfusion microfluidic cell culture array for high-throughput cell-based assays. Biotechnol Bioeng. 2005;89:1–8. doi: 10.1002/bit.20289. [DOI] [PubMed] [Google Scholar]
- 16.Huh D, Fujioka H, Tung Y-C, Futai N, Paine R, Grotberg JB, et al. Acoustically detectable cellular-level lung injury induced by fluid mechanical stresses in microfluidic airway systems. P Natl Acad Sci USA. 2007;104:18886–91. doi: 10.1073/pnas.0610868104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Godwin LA, Pilkerton ME, Deal KS, Wanders D, Judd RL, Easley CJ. Passively operated microfluidic device for stimulation and secretion sampling of single pancreatic islets. Anal Chem. 2011;83:7166–72. doi: 10.1021/ac201598b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Croushore CA, Supharoek S-a, Lee CY, Jakmunee J, Sweedler JV. Microfluidic device for the selective chemical stimulation of neurons and characterization of peptide release with mass spectrometry. Anal Chem. 2012;84:9446–52. doi: 10.1021/ac302283u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kane BJ, Zinner MJ, Yarmush ML, Toner M. Liver-specific functional studies in a microfluidic array of primary mammalian hepatocytes. Anal Chem. 2006;78:4291–8. doi: 10.1021/ac051856v. [DOI] [PubMed] [Google Scholar]
- 20.Godwin LA, Brooks JC, Hoepfner LD, Wanders D, Judd RL, Easley CJ. A microfluidic interface for the culture and sampling of adiponectin from primary adipocytes. Analyst. 2015;140:1019–25. doi: 10.1039/c4an01725k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ong TH, Tillmaand EG, Makurath M, Rubakhin SS, Sweedler JV. Mass spectrometry-based characterization of endogenous peptides and metabolites in small volume samples. BBA-Proteins Proteomics. 2015;1854:732–40. doi: 10.1016/j.bbapap.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goto M, Sato K, Murakami A, Tokeshi M, Kitamori T. Development of a microchip-based bioassay system using cultured cells. Anal Chem. 2005;77:2125–31. doi: 10.1021/ac040165g. [DOI] [PubMed] [Google Scholar]
- 23.Shackman JG, Dahlgren GM, Peters JL, Kennedy RT. Perfusion and chemical monitoring of living cells on a microfluidic chip. Lab Chip. 2005;5:56–63. doi: 10.1039/b404974h. [DOI] [PubMed] [Google Scholar]
- 24.Shackman JG, Reid KR, Dugan CE, Kennedy RT. Dynamic monitoring of glucagon secretion from living cells on a microfluidic chip. Anal Bioanal Chem. 2012;402:2797–803. doi: 10.1007/s00216-012-5755-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dishinger JF, Reid KR, Kennedy RT. Quantitative monitoring of insulin secretion from single islets of langerhans in parallel on a microfluidic chip. Anal Chem. 2009;81:3119–27. doi: 10.1021/ac900109t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark AM, Sousa KM, Chisolm CN, MacDougald OA, Kennedy RT. Reversibly sealed multilayer microfluidic device for integrated cell perfusion and on-line chemical analysis of cultured adipocyte secretions. Anal Bioanal Chem. 2010;397:2939–47. doi: 10.1007/s00216-010-3897-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark AM, Sousa KM, Jennings C, MacDougald OA, Kennedy RT. Continuous-flow enzyme assay on a microfluidic chip for monitoring glycerol secretion from cultured adipocytes. Anal Chem. 2009;81:2350–6. doi: 10.1021/ac8026965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dugan CE, Cawthorn WP, MacDougald OA, Kennedy RT. Multiplexed microfluidic enzyme assays for simultaneous detection of lipolysis products from adipocytes. Anal Bioanal Chem. 2014;406:4851–9. doi: 10.1007/s00216-014-7894-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowen AL, Martin RS. Integration of on-chip peristaltic pumps and injection valves with microchip electrophoresis and electrochemical detection. Electrophoresis. 2010;31:2534–40. doi: 10.1002/elps.201000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price AK, Fischer DJ, Martin RS, Spence DM. Deformation-induced release of ATP from erythrocytes in a poly(dimethylsiloxane)-based microchip with channels that mimic resistance vessels. Anal Chem. 2004;76:4849–55. doi: 10.1021/ac0495992. [DOI] [PubMed] [Google Scholar]
- 31.Nunemaker CS, Dishinger JF, Dula SB, Wu R, Merrins MJ, Reid KR, et al. Glucose metabolism, islet architecture, and genetic homogeneity in imprinting of [Ca2+]i and insulin rhythms in mouse islets. PLoS ONE. 2009;4:e8428. doi: 10.1371/journal.pone.0008428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Yi L, Mukhitov N, Schrell AM, Dhumpa R, Roper MG. Microfluidics-to-mass spectrometry: A review of coupling methods and applications. J Chromatogr A. 2015;1382:98–116. doi: 10.1016/j.chroma.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nge PN, Pagaduan JV, Yu M, Woolley AT. Microfluidic chips with reversed-phase monoliths for solid phase extraction and on-chip labeling. J Chromatogr A. 2012;1261:129–35. doi: 10.1016/j.chroma.2012.08.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mellors JS, Gorbounov V, Ramsey RS, Ramsey JM. Fully integrated glass microfluidic device for performing high-efficiency capillary electrophoresis and electrospray ionization mass spectrometry. Anal Chem. 2008;80:6881–7. doi: 10.1021/ac800428w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazar IM, Trisiripisal P, Sarvaiya HA. Microfluidic liquid chromatography system for proteomic applications and biomarker screening. Anal Chem. 2006;78:5513–24. doi: 10.1021/ac060434y. [DOI] [PubMed] [Google Scholar]
- 36.Liuni P, Rob T, Wilson DJ. A microfluidic reactor for rapid, low-pressure proteolysis with on-chip electrospray ionization. Rapid Commun Mass Spec. 2010;24:315–20. doi: 10.1002/rcm.4391. [DOI] [PubMed] [Google Scholar]
- 37.Ohla S, Belder D. Chip-based separation devices coupled to mass spectrometry. Curr Opin Chem Biol. 2012;16:453–9. doi: 10.1016/j.cbpa.2012.05.180. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Hu H, Zhao S, Liu Y-M. Microfluidic Platform with In-Chip Electrophoresis Coupled to Mass Spectrometry for Monitoring Neurochemical Release from Nerve Cells. Anal Chem. 2016;88:5338–44. doi: 10.1021/acs.analchem.6b00638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Enders JR, Marasco CC, Wikswo JP, McLean JA. A dual-column solid phase extraction strategy for online collection and preparation of continuously flowing effluent streams for mass spectrometry. Anal Chem. 2012;84:8467–74. doi: 10.1021/ac3021032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gasilova N, Qiao L, Momotenko D, Pourhaghighi MR, Girault HH. Microchip emitter for solid-phase extraction–gradient elution–mass spectrometry. Anal Chem. 2013;85:6254–63. doi: 10.1021/ac400171e. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S, Sahore V, Rogers CI, Woolley AT. Development of an integrated microfluidic solid-phase extraction and electrophoresis device. Analyst. 2016;141:1660–8. doi: 10.1039/c5an02352a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao D, Wei H, Guo G-S, Lin J-M. Microfluidic cell culture and metabolism detection with electrospray ionization quadrupole time-of-flight mass spectrometer. Anal Chem. 2010;82:5679–85. doi: 10.1021/ac101370p. [DOI] [PubMed] [Google Scholar]
- 43.Wei H, Li H, Gao D, Lin J-M. Multi-channel microfluidic devices combined with electrospray ionization quadrupole time-of-flight mass spectrometry applied to the monitoring of glutamate release from neuronal cells. Analyst. 2010;135:2043–50. doi: 10.1039/c0an00162g. [DOI] [PubMed] [Google Scholar]
- 44.Luni C, Giulitti S, Serena E, Ferrari L, Zambon A, Gagliano O, et al. High-efficiency cellular reprogramming with microfluidics. Nat Meth. 2016;13:446–52. doi: 10.1038/nmeth.3832. [DOI] [PubMed] [Google Scholar]
- 45.Patabadige DEW, Mickleburgh T, Ferris L, Brummer G, Culbertson AH, Culbertson CT. High-throughput microfluidic device for single cell analysis using multiple integrated soft lithographic pumps. Electrophoresis. 2016;37:1337–44. doi: 10.1002/elps.201500557. [DOI] [PubMed] [Google Scholar]
- 46.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–6. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 47.Arner P. Human fat cell lipolysis: Biochemistry, regulation and clinical role. Best Pract Res Clin Endoc Metab. 2005;19:471–82. doi: 10.1016/j.beem.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 48.Lafontan M, Langin D. Lipolysis and lipid mobilization in human adipose tissue. Prog Lipid Res. 2009;48:275–97. doi: 10.1016/j.plipres.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 49.Cellar NA, Burns ST, Meiners J-C, Chen H, Kennedy RT. Microfluidic chip for low-flow push-pull perfusion sampling in vivo with on-line analysis of amino acids. Anal Chem. 2005;77:7067–73. doi: 10.1021/ac0510033. [DOI] [PubMed] [Google Scholar]
- 50.Ceriotti L, de Rooij NF, Verpoorte E. An integrated fritless column for on-chip capillary electrochromatography with conventional stationary phases. Anal Chem. 2002;74:639–47. doi: 10.1021/ac0109467. [DOI] [PubMed] [Google Scholar]
- 51.Maiolica A, Borsotti D, Rappsilber J. Self-made frits for nanoscale columns in proteomics. Proteomics. 2005;5:3847–50. doi: 10.1002/pmic.200402010. [DOI] [PubMed] [Google Scholar]
- 52.Banerjee S, Mazumdar S. Electrospray ionization mass spectrometry: A technique to access the information beyond the molecular weight of the analyte. Int J Anal Chem. 2012:40. doi: 10.1155/2012/282574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberts LD, Virtue S, Vidal-Puig A, Nicholls AW, Griffin JL. Metabolic phenotyping of a model of adipocyte differentiation. Physiol Genomics. 2009;39:109–19. doi: 10.1152/physiolgenomics.90365.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bollinger JG, Rohan G, Sadilek M, Gelb MH. LC/ESI-MS/MS detection of FAs by charge reversal derivatization with more than four orders of magnitude improvement in sensitivity. J Lipid Res. 2013;54:3523–30. doi: 10.1194/jlr.D040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murphy RC. Tandem mass spectrometry of lipids: Molecular analysis of complex lipids. The Royal Society of Chemistry; 2015. Fatty Acids; pp. 1–39. [Google Scholar]
- 56.Rosenstock M, Greenberg AS, Rudich A. Distinct long-term regulation of glycerol and non-esterified fatty acid release by insulin and TNF-α in 3T3-L1 adipocytes. Diabetologia. 2001;44:55–62. doi: 10.1007/s001250051580. [DOI] [PubMed] [Google Scholar]
- 57.Zhou D, Samovski D, Okunade AL, Stahl PD, Abumrad NA, Su X. CD36 level and trafficking are determinants of lipolysis in adipocytes. FASEB J. 2012;26:4733–42. doi: 10.1096/fj.12-206862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang ZG, Pini M, Yao T, Zhou ZX, Sun CH, Fantuzzi G, et al. Homocysteine suppresses lipolysis in adipocytes by activating the AMPK pathway. Am J Physiol Endocrinol Metab. 2011;301:E703–E12. doi: 10.1152/ajpendo.00050.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Extracted ion chromatogram of 8 NEFAs in adipocyte conditioned buffer, measured with HPLC-TOF-MS (a) along with the associated mass spectra of the most abundant NEFAs (b)
Table S1 Comparison of measured NEFA mass accuracy on TOF and QQQ