Abstract
Protein glycosylation and other post-translational modifications are involved in potentially all aspects of human growth and development. Defective glycosylation has adverse effects on human physiological conditions and accompanies many chronic and infectious diseases. Altered glycosylation can occur at the onset and/or during tumor progression. Identifying these changes at early disease stages may aid in making decisions regarding treatments as early intervention can greatly enhance survival. This review highlights some of the efforts being made to identify N- and O-glycosylation profile shifts in cancer using mass spectrometry. The analysis of single or panels of potential glycoprotein cancer markers are covered. Other emerging technologies like global glycan release and site-specific glycosylation analysis and quantitation are also discussed.
Keywords: Mass spectrometry, Cancer, Disease biomarker, Glycomics, Glycoproteomics, Site- specific glycosylation
Introduction
Cancer is a major cause of death in most parts of the world [1–4]. In the USA alone, more than 500 thousand deaths are projected to be associated with cancer and about 1.6 million new cancer cases are expected to be diagnosed in 2016 [5]. These deleterious effects of cancer continue despite the fact that when cancer is diagnosed early it can be contained or even cured. Efforts to identify biomarkers that can detect cancer early and discriminate a given type of cancer from other diseases face many challenges. Some of these challenges include the complexity of biological samples from which potential biomarkers are derived, extensive heterogeneity of potential analytes among different diseases and limitations of the current analytical methods [6].
A biomarker is a biologically important signature that unambiguously identifies a certain physiological condition. It could be a single measured entity or a panel of indicator substances [6]. A biomarker can be used to screen for a given disease condition, monitor patients undergoing therapy or even identify the re-occurrence of a disease condition. Markers are also useful in identifying people who are at a higher risk of developing certain diseases. The progress being made in cancer research raises hopes for identification of early detection markers.
Screening for biomarkers requires a thorough investigation of the potential indicator analytes to ensure that they meet specific requirements. For example, they should be able to identify when the condition exists (sensitivity) and when it does not exist (specificity). Most potential cancer biomarkers do not pass this test, resulting in false positives and false negatives, which are not favorable for the patients as well as those seeking prognosis [7,8]. The samples should be amenable to robust and reproducible instrumentation to minimize errors that may lead to incorrect diagnosis. Where possible, the samples should be easily obtainable in a noninvasive manner, and the assay should be affordable so that it can be accessible to a large population of people. It is also important that a biomarker is validated across a broad range of populations and at different sites (laboratories).
The current methods for discovering cancer biomarkers have been reviewed recently [9]. These methods include gene expression measurements using DNA, RNA and microRNA gene arrays [10,9] and protein microarray technologies [11,12]. For proteins, 2D-gel electrophoresis has been widely used because it is readily available. Although it remains a very useful technique, it requires large amounts of samples due to poor sensitivity, and it is a laborious and slow process. Mass spectrometry has essentially replaced gels because of its versatility in profiling biological compounds. The methods for proteomics, peptidomics, metabolomics, proteoglycomics, glycomics and MS imaging are all generally based on MS [12].
Several biological fluids have been used for cancer biomarker research including saliva [13], urine [14], nipple aspirate fluid [15], cerebrospinal fluids and tumor intestinal fluids [16], However, serum and plasma are the most commonly used human biological fluids for cancer biomarker research partly because obtaining blood is relatively noninvasive compared to other methods like biopsy [9,17], Additionally, blood can provide multiple molecular elements of cancer in the form of circulating cells, proteins, peptides, metabolites and cell-free DNA and RNA [18]. The proteins in serum and plasma have been the major target analytes for cancer diagnosis. Advancements in proteomics technologies have enabled more accurate profiling of proteins [19]. However, the dominance of a few proteins in serum/plasma and the presence of post-translational modifications make their complete characterization more challenging than analysis of the genes [20].
Protein modifications, which occur as translational, post-translational, regulatory and/or degradation products, increase the amount of useful cancer-related information that can be obtained from the proteins. The common post-translational modifications (PTMs) include but are not limited to glycosylation, phosphorylation, sulfation, and acetylation. These PTMs play important roles in biological processes including regulation of proper protein folding, host pathogen interactions, immune responses and also in inflammation. In this review, we highlight the recent efforts in identifying potential glycosylated proteins and released glycans as cancer biomarkers using mass spectrometry (MS). We will begin with a basic introduction to the mass spectrometry technology and its utility in identifying single glycoprotein markers, panels of glycoprotein markers and released glycan markers. We will then discuss advancements in site-specific glycosylation analysis for cancer diagnosis.
Mass Spectrometry
Mass spectrometry is a versatile technique that has increasingly been used in cancer biomarkers discovery due to its sensitivity, potential for high throughput, and the capability for tolerating diverse biological mixtures [21–27]. MS has been used in the analysis of various analytes in many fields including but not limited to proteomics, metabolomics, lipidomics, glycoproteomics, glycomics and proteoglycomics [28–31]. Samples can be introduced to the mass spectrometer via direct infusion, however, most of the potential cancer markers are found in biological mixtures requiring some pre-separation method. Separation techniques such as liquid chromatography (LC), ion mobility and capillary electrophoresis (CE) are typically used in conjunction with mass spectrometry.
Recent developments in microchip-based separation and ionization methods have enabled analysis of extremely small amounts of samples with high sensitivity and reproducibility [33,34]. This capability is particularly important because most of the analytes in biological samples are found in minute amounts that are not readily amenable to other analytical techniques. MS analysis generates a tremendous amount of data that requires the development of bioinformatics methods that can aid with data interpretation and expedite the analysis of large data sets [35,36].
Lone (Glyco)protein Markers for Cancer
Most of the Food and Drug Administration (FDA) approved cancer biomarkers are single proteins derived from serum [37], and the majority of these proteins are glycosylated. CA 125 antigen, for example, is a well-documented glycoprotein standard marker for ovarian cancer [38–40]. CA 19-9 has been used as a pancreatic cancer biomarker [41]. CA 15-3 is a useful breast cancer biomarker especially for monitoring patients who have already been diagnosed with the disease in evaluating the recurrence of the disease [42].
Other glycosylated protein markers include prostate-specific antigen (PSA), which is present in elevated amounts in seminal fluids and plasma, for screening and monitoring prostate cancer patients [43,44]. Carcinoembryonic antigen (CEA), which has been found to change in colorectal, bladder, breast, pancreatic and lung cancer, is highly N-glycosylated [45–47]. Multiple studies have also indicated that haptoglobin, which is also glycosylated, could be used as a cancer biomarker [48–50]. In one study using in-house ELISA, haptoglobin was found to be correlated with the clinical stage of epithelial ovarian cancer and patient survival [51]. Using statistical analysis, the study found that elevated serum haptoglobin levels were associated with poor outcome in overall patient survival. The isoforms of haptoglobin-1 precursor have been found to be significantly upregulated in all groups of ovarian cancer patients by other researchers [52].
Another study that identified single glycoprotein potential cancer markers utilized 2-D differential gel electrophoresis (2-D DIGE) and MALDI-TOF/TOF to identify depleted serum markers for breast cancer. Proapolipoprotein A1, transferrin and hemoglobin were found to be elevated in breast cancer compared to control samples while apolipoprotein A1, apolipoprotein C-III and alpha 2 haptoglobin were decreased. Immunochemical reactions were used to validate the selected candidate markers. In this work, it was demonstrated that 2-D DIGE can separate protein isoforms that are not amenable to immunochemical separation due to lack of isomer specific antibodies [53].
(Glyco)protein Panels as Markers for Cancer
A single protein biomarker may not provide the specificity and sensitivity necessary for disease identification. In this regard, identifying a number of individual potential markers that when combined would identify a certain disease condition is a common approach. Although it is a huge challenge to assemble and validate a panel of biomarker proteins [17], Table 1 shows representative studies that have identified panels of potential cancer protein markers that are all post-translationally modified. Ralhan et al [54] identified a panel of proteins by multidimensional LC-MS/MS that were differentially expressed when head and neck cancer patient samples were compared to the controls. For the analysis, the samples were labeled with isobaric mass tags for relative and absolute quantitation (iTRAQ) on a QSTAR Pulsar i MS. Using this approach, a panel of three cancer protein markers that are potentially modified on their serine residues were identified that achieved remarkable sensitivity of 0.92 and specificity of 0.91: 14-3-3 protein zeta/delta (phosphorylated), stratifin (phosphorylated), and S100-A7 (some serine residues are acetylated). These MS results were confirmed using immunoprecipitation. When evaluated individually, the sensitivity and specificity of each of the proteins were poor [54].
Table 1.
Contains a group of proteins that have been identified as potential biomarkers when combined. The columns represent the cancer type, the protein marker panel, the analytical technique that were used for the analysis and the reference number in the review.
| Cancer type | Panel of protein markers | Technique | Reference no. |
|---|---|---|---|
| Lung | carcinoembryonic antigen, retinol binding protein alpha 1-antitrypsin squamous cell carcinoma antigen |
2D-DIGE and MALDI-TOF | 55 |
| Ovarian | Corticosteroid-binding globulin (CBG) serum amyloid p component (SAP) complement factor B (CFAB) |
lectin array and quantitative LC-MS/MS |
59 |
| Head and neck | 14-3-3 protein zeta/delta (YWHAZ), stratifin S100-A7 |
Multidimensional LC-MS/MS | 54 |
| Pancreatic Cancer | α-1-antichymotrypsin (AACT) thrombospondin-1 (THBS1) haptoglobin (HPT) |
Label-free and TMT strategies LC-MS/MS |
67 |
In another study combining 2D-DIGE and MALDI-TOF analysis, serum samples from 50 lung cancer patients and 50 controls were screened for protein biomarkers. A panel of four proteins was able to discriminate healthy controls from patients who had lung cancer. This panel of four biomarkers included carcinoembryonic antigen, retinol binding protein, alpha-1-antitrypsin and squamous cell carcinoma antigen. The first three proteins are glycosylated while the latter is phosphorylated. The authors used an independent blinded validation set of sera from 49 lung cancer patients and 48 matched controls. The protein markers were able to classify most of the cancer patients in the independent validation set with sensitivity of 77.8% and specificity of 75.4% [55].
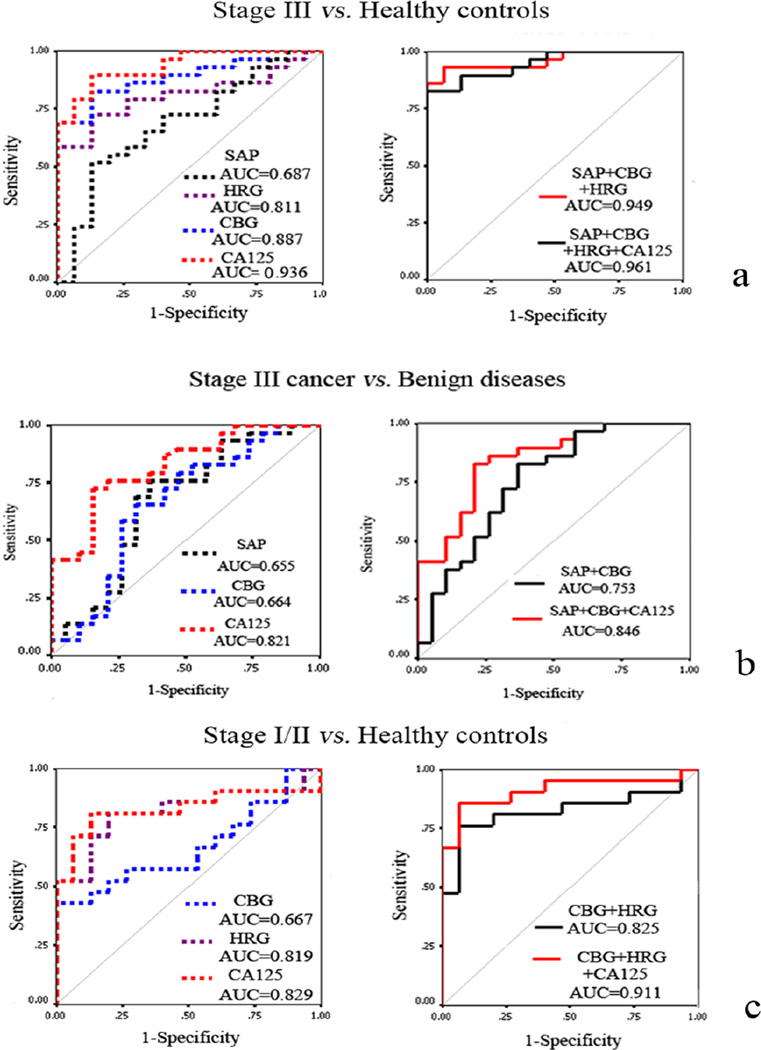
Selective capture of glycoproteins using lectins or other glycan-binding antibodies is another complementary method for the discovery of potential glycoprotein biomarkers. Lectins are proteins that recognize and bind specific glycan moieties and can be used to isolate target glycoproteins from a sample mixture. Thus, they provide the advantage of minimizing the number of compounds to be analyzed by MS [56–58]. Lectin-based methods have been used to detect differential glycosylation between cancer and healthy controls [59–62]. In a recent study, a glycoprotein binding lectin array was used to identify glycosylation changes in the sera of patients with benign diseases when compared to those with stage III ovarian cancer. Lectins whose binding response differed significantly between benign conditions and cancer were used to pull isotopically tagged glycoproteins for cancer biomarker screening. Quantitative LC-MS/MS on an LTQ-ion trap was used to distinguish between 15 healthy controls, 19 benign ovarian diseases, 21 stage I/II ovarian cancer and 30 stage III ovarian cancer cases. A panel of four glycoproteins including corticosteroid-binding globulin (CBG), serum amyloid p component (SAP), complement factor B (CFAB) and histidine-rich glycoprotein (HRG) were found to differentiate ovarian cancer patients from benign diseases and healthy controls. Combining CBG, SAP and CA 125 showed improved discrimination of stage III ovarian cancer from benign diseases when compared to CA 125 alone (Figure 1) [59].
Figure 1.
ROC curve analyses of significantly changed proteins detected by ELISA. (a) ROC curve for differentiating stage III ovarian cancer from normal healthy controls; (b) ROC curve for differentiating stage III ovarian cancer from benign diseases; (c) ROC curve for distinguishing stage I/II ovarian cancer from normal healthy controls. Reprinted from ref. 59. Copyright © 2012, American Chemical Society.
N-Acetylglucosaminyltransferase-V (GnT-V) catalyzes the formation of an unusual β-1,6-linked N-acetylglucosamine which has been found to be upregulated in cancer cells [63–65]. A breast cancer investigation used the lectin phytohemagglutinin-L4 (L-PHA) which binds specifically to β-(1,6)-branched N-linked structures on glycoproteins [66]. This lectin showed selective binding to breast epithelial cells as the disease progresses to invasive carcinoma, but not to normal healthy cells. Using this lectin, β-1,6-N-acetylglucosamine (β-1,6-GlcNAc) branched N-linked glycan-containing glycoproteins were enriched from the tissues of normal healthy controls and invasive carcinoma patients. In contrast to the healthy controls, 12 glycoproteins were enriched in the tissues of all four patients with invasive ductal breast carcinoma tissues [66].
Tandem mass tags (TMT) are chemical labels used to quantify multiple peptides or proteins in biological samples simultaneously [67]. Quantitative proteomics using TMT labeling was performed to identify a panel of three glycoproteins that could potentially distinguish pancreatic cancer and other pancreatic conditions like diabetes, cyst, chronic pancreatitis and obstructive jaundice from controls [68]. Depletion of the 14 most abundant proteins was first carried out and fucosylated glycoproteins were extracted using agarose-bound Aleuria aurantia lectin (AAL). A combination of α-1-antichymotrypsin (AACT), thrombospondin-1 (THBS1) and haptoglobin (HPT) showed good sensitivity in diagnosing pancreatic cancer when compared with the following conditions: normal controls (AUC = 0.95), diabetes (AUC = 0.89), cyst (AUC = 0.82), and chronic pancreatitis (AUC = 0.90). Adding CA 19-9 to the panel distinguished pancreatic cancer from patients with and without obstructive jaundice [68].
The studies highlighted in the above discussions are additional proofs that modified proteins have an influence in cancer and their modifications could potentially be used as cancer biomarkers. Due to this fact, substantial method development efforts are being made to identify the protein modifications and profile their levels in biological fluids in a high throughput manner. Some of these efforts have begun to bear fruits but a lot remain to be done. In the subsequent text we highlight some of the progress made to identify and profile protein modifications, especially O- and N-linked carbohydrates in the search for cancer biomarkers.
Post-Translational Modifications of Proteins in Cancer
Post-translational modifications (PTMs) of proteins include the covalent attachment of the molecules to amino acid(s) enzymatically during protein biosynthesis [69]. PTMs provide valuable additional details to the biological functions of proteins. The most common PTMs include the following: glycosylation (N- and O-linked), phosphorylation, acetylation, methylation and sulfation [69,70]. Differential expression of modifications between normal and cancerous cells occurs at the onset or during cancer progression [71,72]. These changes could be due to a single modification or multiple modifications.
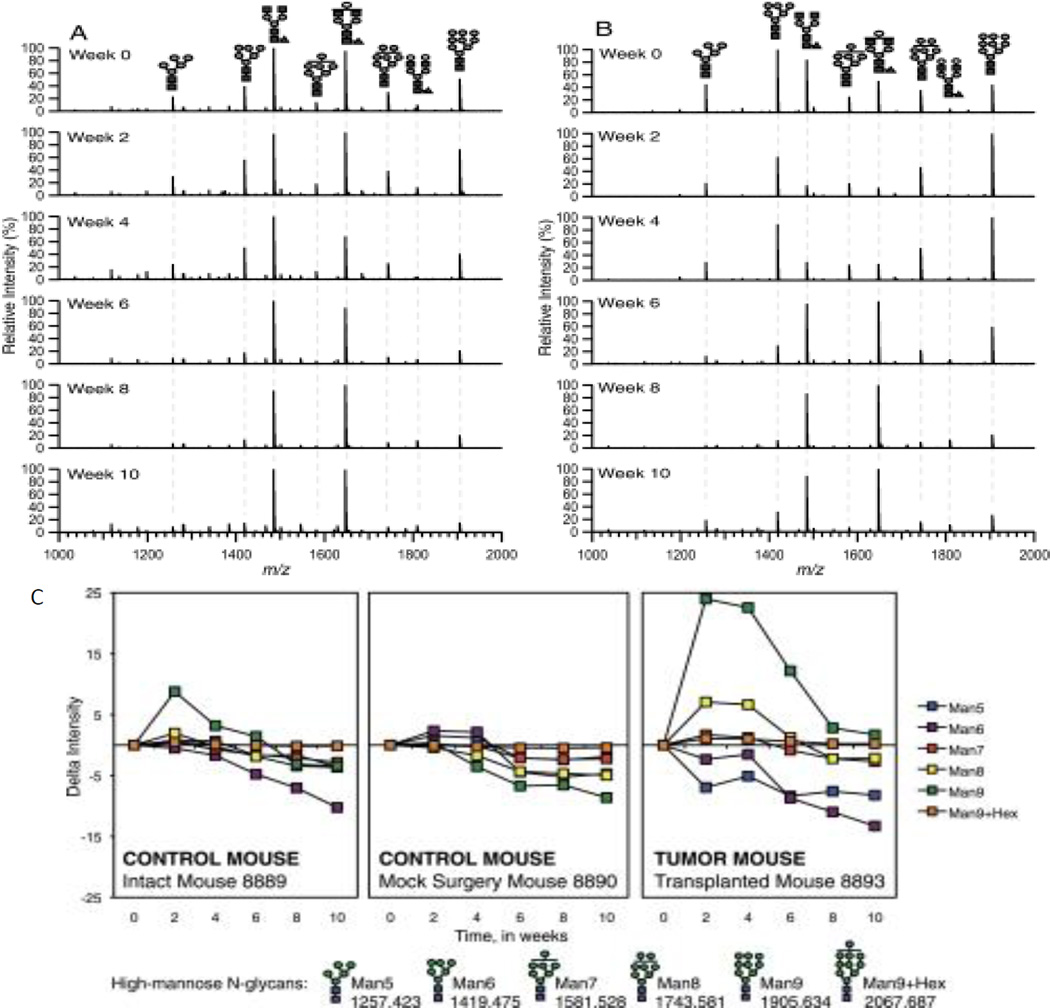
Glycosylation is a substantial PTM with respect to size and occurrence, where more than 50% of proteins are estimated to be glycosylated [73,25]. For humans, this value may be larger, and for some fluids such as human milk, glycosylated proteins may be as high as 80% [74]. Glycans are often found attached to proteins on the cell surface and in the extracellular matrix where they mediate many biological activities [75,76]. Major types of glycans include N-linked glycans attached to the nitrogen atom in the asparagine side chain within a consensus amino acid sequence Asn-X-Ser/Thr (X should not be proline), and O-linked glycans attached to the oxygen atom of several amino acid residues including serine and threonine. Other types of glycans include glycosaminoglycans usually found attached to the proteins (proteoglycans) and also lipid chains as in glycolipids [77]. There is now a considerable body of work showing that glycans released from cancer cells differ from those in healthy cells [76,78–80,71,81]. Tumors have been found to overexpress certain types of glycoproteins and glycolipids [80,82]. For example, there is an increase in the size and branching of N-glycans in tumors [76]. High mannose glycans have also been found to increase in other cancers (Figure 2) [83].
Figure 2.
Representative MALDI FT-ICR mass spectra of A, mock surgery control and B, tumor-transplanted mouse sera at different points. m/z 1000 –2000 region of 10% acetonitrile fraction taken in the positive ion mode is shown. C, Change in intensities from MALDI FT-ICR MS of high-mannose N-linked glycans in sera of an intact control mouse, mock surgery control mouse, and a tumor-transplanted mouse during breast cancer progression. All values relative to Week 0 intensity. Error bars are expressed as standard error of the mean (S.E.) from three spectral scans per mouse sample. Symbol representations of glycans: N-acetylglucosamine, blue square; mannose, green circle; glucose, blue circle. Reprinted from ref. 82. Copyright © 2011, by the American Society for Biochemistry and Molecular Biology.
Glycan analysis is more challenging compared to proteins. The non-template nature of their biosynthesis and involvement of a wide variety of essentially competing enzymes during their synthesis leads to more diverse glycan molecules having minor structural differences. Glycans are, however, easier to quantitate in relative amounts. Recent developments in liquid chromatography separation, ionization, and mass spectrometry methods have enabled extensive characterization of oligosaccharides and high throughput analysis of larger sample sizes [32]. These developments could potentially lead to the discovery of glycan specific cancer biomarkers.
Global N- and O-Glycan Analysis
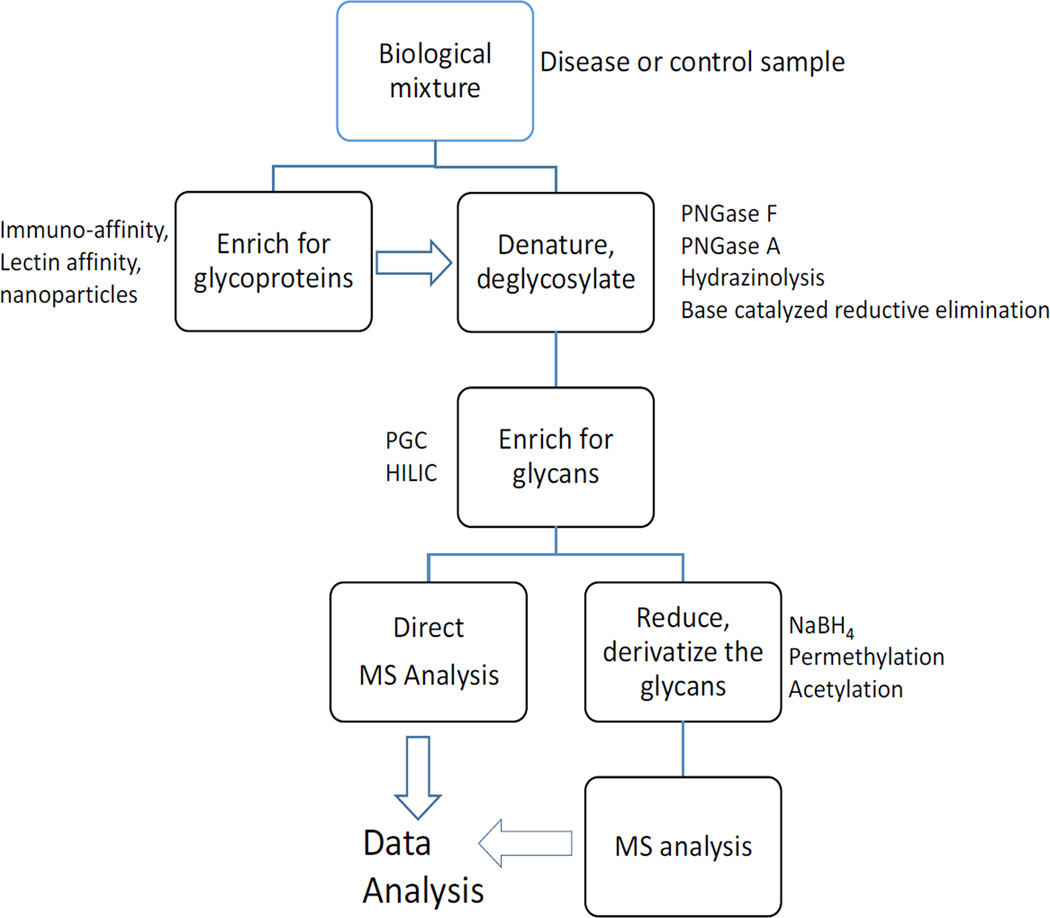
Efforts to identify cancer biomarkers from released glycans have been reviewed recently [79,41,84,85]. The common approach is a global analysis of glycosylation involving cleaving the glycans from the proteins using enzymatic or chemical means. Usually, peptide-N-glycosidase F (PNGase F) enzyme is used to cleave the N-glycans from purified proteins or from biological mixtures. O-Glycans can be cleaved from the proteins using alkaline sodium borohydride (NaBH4) in a β-elimination reaction, while both N- and O-glycans can be cleaved simultaneously using hydrazinolysis (Figure 3) [79,86]. This step is usually followed by glycan enrichment using solid phase extraction (SPE) on a porous graphitized carbon (PGC) or hydrophilic interaction chromatography (HILIC) column. The samples are then analyzed using methods such as MALDI-TOF, MALDI-Fourier transform ion cyclotron resonance (FTICR), Chip-TOF and Chip-Q-TOF mass spectrometry for compositional or structural analysis. The glycans can be analyzed in their native state or chemically reduced to minimize the multiplicity of MS peaks due to different glycan anomeric configurations especially when a high resolving column (e.g., PGC) is used for separation. Glycans may also be derivatized through permethylation or acetylation to achieve uniform ionization in the mass spectrometer [34,86,87]. There are numerous important studies involving glycan analysis in the search for cancer biomarkers [34,41,79,85,86,88–91].
Figure 3.
The general procedure for the released glycan analysis.
MALDI-MS has been widely used to profile glycans as potential cancer biomarkers due to its sensitivity and high ionization efficiency even at higher mass range [32,88,92–98]. In a study to identify distinguishing features between ovarian cancer patients and healthy persons, O-glycans were cleaved from conditioned media and from serum and profiled using high resolution MALDI-FTICR-MS. Tandem mass spectrometry was used for further structural identification of glycans using infrared multiphoton dissociation (IRMPD). Several unique glycans observed in ovarian cancer patient serum samples, mostly neutral structures composed only of hexose and N-acetylhexosamine residues, were absent in the controls [88]. This work also investigated four ovarian cancer cell lines, OVCAR-3, Caov-3, SK-OV-3 and ES-2, and obtained similar glycan profiles. Multiple oligosaccharides observed in the supernatants of the cancer cell lines were also observed in the patient sera. This was one of the first studies to show that glycans harvested directly from cells or serum can be used as potential cancer markers without requiring isolation of proteins using antibodies [88]. Recent work has identified stage-specific N-glycans released from case-control plasma samples that could potentially be used as ovarian cancer markers [99].
In a related study, O-glycans were chemically cleaved from breast cancer tumor cell lines, sera from mice transplanted with mammary tumors, sera from breast cancer patients and those collected from healthy individuals. This was accomplished using beta elimination with sodium borohydride dissolved in sodium hydroxide solution [100]. High resolution MALDI-FTICR-MS and IRMPD was used to characterize the glycan structures. Differences in intensities were found in the O-glycan profiles that distinguished breast cancer patients from the controls. This study also identified changes in specific structures during tumor growth which may be used as potential indicators of disease onset [100].
Global release methods have also been conducted for the analysis of N-glycans. Using MALDI-MS, permethylated N-glycans from 10 healthy males and 24 prostate cancer patients were compared using statistical methods, principle component analysis (PCA), analysis of variance (ANOVA) and receiver operating characteristic (ROC). Out of the 50 N-glycans identified, 12 glycan structures, half of which were fucosylated, were found to vary significantly between disease and control specimens [95]. The relative abundances of high mannose and complex biantennary structures decreased in cancer while the fucosylated complex biantennary and complex tetraantennary N-glycans were elevated in cancer in comparison to the healthy controls. A separate study by the same group noted increased fucosylation of serum N-glycans in breast cancer patients [96].
A recent study investigated the possibility of using permethylated N-glycans from serum and isolated immunoglobulin G (IgG) as ovarian cancer markers. The samples were obtained from healthy controls and from patients with late-stage recurrent ovarian cancer (OC) before they started an experimental drug treatment (baseline) and prior to the second treatment cycle. Compared to the controls, the level of bisecting glycans decreased in late stage recurrent ovarian cancer patients and this trend was accentuated in patients who underwent the first cycle of the trial treatment. Triantennary and tetraantennary glycans that were sialylated and fucosylated seemed to increase in the baseline OC samples and also showed further increases in the patients’ serum after the experimental drug treatment [101]. Given that patients exhibited increased tumor burden post-experimental treatment, these glycan changes were further assessed as potential markers for disease progression. Contrastingly, when IgG-specific glycans were investigated, fucosylation seemed not to be a strong determinant. Galactosylated glycan levels decreased while the agalactosylated glycans increased in patients with OC when compared to the healthy controls [101]. Other studies have reported increased agalactosylated IgG glycans in ovarian cancer [102], lung cancer [103] and non-small cell lung cancer (NSCLC) patients [104].
As researchers seek more specific and sensitive markers for cancer, there is a greater need to increase the amount of information obtained from the samples. In this regard, efforts to identify the structures of the glycan biomarkers are gaining interest. Porous graphitized carbon (PGC), which separates glycans based on their size, polarity and three dimensional structure, is a good platform for released glycan separation. PGC has high sensitivity, low sample consumption, minimal ion suppression and large instrumental dynamic range, which are all important for clinical glycomics [105–107].
A novel method that uses rapid throughput nano-flow liquid chromatography on a PGC column to separate isomeric glycans from serum was recently developed. Using this method, more than 100 unique N-glycan compositions having more than 300 glycan species were identified and their abundances analyzed in two classes of prostate cancer patients. It was found that fucosylated, non-sialylated complex/hybrid glycans were significantly higher in the good prognosis (G) group (4 subjects) compared to the four subjects in the poor prognosis group (P). In addition, sialylated, non-fucosylated complex/hybrid glycans were more abundant in the P compared to the G group [105]. This strategy increases the level of information obtained for biomarker analysis and may lead to structural specific markers.
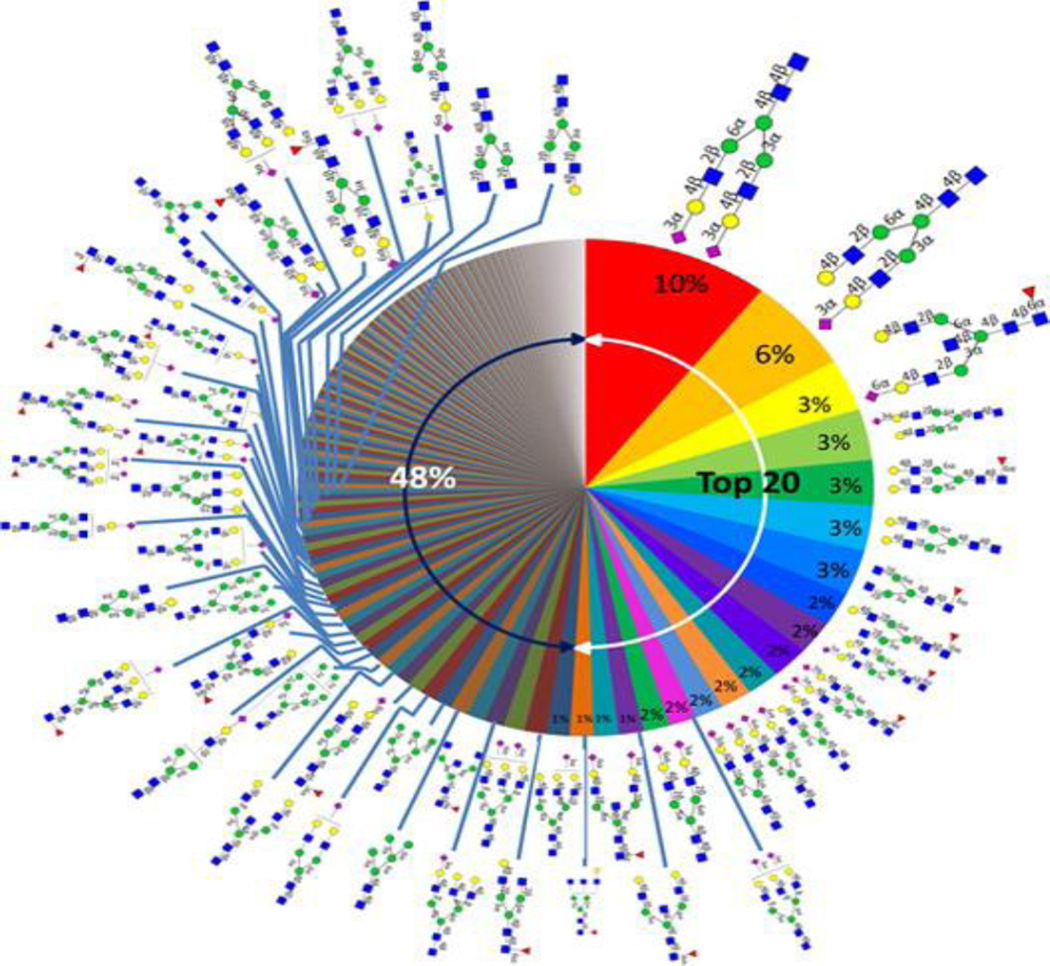
Advanced chip-based LC separations have also enabled the analysis of N-linked glycans from serum with very high reproducibility [108]. A library composed of serum N-glycans including fine details like the residue linkages was recently created [109]. The library was created by first utilizing PNGase F to release the glycans from whole serum. The glycans were then reduced using NaBH4 and enriched using graphitized carbon cartridges. The samples were analyzed using nano-LC-chip-TOF MS. The retention times and accurate mass derived from the library enable rapid sequencing of serum N-glycans. This library provides additional structural specific information that can be very useful in cancer biomarker research. The library contains diverse N-glycan groups including high mannose, sialylated, fucosylated, biantenary and triantennary complex and hybrid structures (Figure 4). The library included approximately 50 complete structures and 170 compositions with nearly complete were elucidated using this method. This technology provides a deeper level of analysis of the glycan structural species that change with disease, which aids in understanding the mechanisms underlying these biological changes [109].
Figure 4.
Human serum “glycan wheel” based on the relative abundances averaged from nine individual sera. Reprinted with permission from Ting Song, Danielle Aldredge, Carlito B. Lebrilla. Ref. 108. Copyright © 2015, American Chemical Society
Protein-Specific Glycan Markers
Another way to analyze glycosylation is to first isolate the possible glycoprotein targets from biological mixtures, cleave off the glycans and analyze them using mass spectrometry [90,89,110]. The glycans obtained from this method are assumed to be from the glycoproteins isolated. This procedure does not provide site-specific information about the glycoforms. Nonetheless, the information obtained is useful in protein-specific biomarker studies. This method was used to investigate possible glycan stomach cancer markers by first isolating the most abundant serum proteins IgG, haptoglobulin, transferrin and alpha-1-acid glycoprotein using affinity purification or 2D gel electrophoresis. The study employed LC-MS/MS to identify the proteins and then ultra-performance liquid chromatography (UPLC) with UV detection to separate and identify the glycans on a HILIC stationary phase. Core fucosylated biantennary glycans were found to be elevated in IgG with increased disease progression while sialylated and galactosylated core fucosylated biantennary glycans decreased with disease progression. Contrastingly, sialylated glycans in haptoglobin, transferrin and alpha-1-acid glycoprotein increased with disease progression [90].
Another glycan study was conducted to distinguish between non-atrophic gastritis (NAG), duodenal ulcer (DU) and gastric cancer (GC) cases. IgG was isolated from the serum using protein G. The N-glycans were released from IgG using PNGase F, purified by PGC and analyzed using nHPLC-chip-TOF-MS. A total of 48 IgG glycans were observed in this research. Statistical data analysis showed that among these, eight glycans were significantly altered between GC and NAG. These changes include increased levels of truncated glycans and decreased levels of biantennary galactosylated glycans in the GC group compared to the NAG group. In addition, lower sialylation was observed in GC compared to the DU and NAG [110].
In a pancreatic cancer study, a western blot using Aleuria aurantica lectin (AAL) which preferentially binds to fucosylated structures was used to identify fucosylated proteins in the serum of cancer patients and healthy controls. Among these proteins, fucosylated haptoglobin was found to be elevated in pancreatic cancer patients compared to the controls. Subsequently, haptoglobin was purified from serum and analyzed using LC-ESI-MS and MALDI-TOF, which showed that alpha-(1–3)-, alpha-(1–4)- and alpha-(1–6)-fucosyl linkages increased in pancreatic cancer patients compared to the controls [49]. Although fucosylation has been identified to change in other diseases, the elevation of these analytes was more pronounced in pancreatic cancer patients. It has also been observed that haptoglobin alpha subunit levels change with the small cell lung cancer stage and could also be a potential biomarker [89].
From the above discussion, it is clear that global glycan analysis provides useful information about potential cancer biomarkers. Isolation of specific glycoproteins from biological sample mixtures is also gaining interest. From the glycosylation analysis of IgG isolated from serum, similarities in glycan patterns are observed in a protein-specific manner in different cancer types. For instance, increased levels of truncated glycans without galactose have been reported in gastric cancer, ovarian cancer and lung cancer. Similar studies from other serum glycoproteins may uncover protein-specific glycosylation changes that may serve as potential cancer indicators.
Site-Specific Glycan Markers
Although the analyses of global glycosylation changes provide useful information about cancer and other diseases, one drawback of this method is that the glycan cannot be traced back to the protein site of origin. In order to identify subtle glycosylation differences between disease conditions and gain more knowledge about cancer biology, it is important to not only identify the glycan but also its site of attachment within the protein. This information is obtained by thoroughly investigating the glycosylated peptides (glycopeptides) identified from tandem mass spectrometry of the glycoproteins.
Glycopeptide analysis is more challenging than protein or global glyco-analysis due to several reasons. A single glycan composition in a peptide may contain a large number of isomeric structures due to differences in glycosidic linkages between monosaccharides, branching, and the existence of many monosaccharides bearing the same mass. The presence of multiple unique glycans that share the same peptide backbone causes the MS signal to be split into various glycoforms thus lowering their individual abundances compared to the peptides that are not glycosylated. It is also challenging to obtain comprehensive glycopeptide fragmentation for both the glycan and the peptide moieties as they have different fragmentation efficiencies [111]. Nonetheless, recent advancements in mass spectrometry technologies have enabled characterization of the glycans with their peptide backbone [112–114].
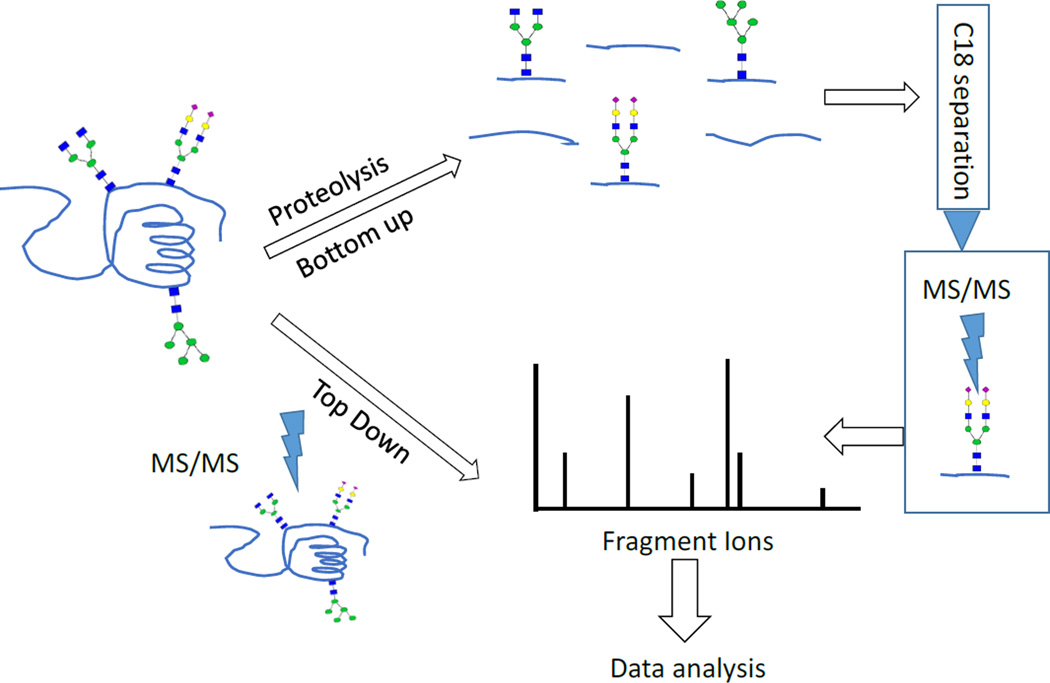
Due to the above-mentioned problems, it has been challenging to develop algorithms that can automatically identify glycans and their peptide moieties from the tandem MS data. Despite these limitations, progress is being made in our laboratory and elsewhere to develop softwares that can identify glycopeptides spectra [115,111,116–122]. Site-specific glycosylation information can be obtained directly from tandem mass spectrometry of intact proteins (top down) [123]. Molecular ions from intact glycoproteins are selected and subjected to multiple stage MS (MSn) analysis (Figure 5). This method has been limited to few types of MS instruments that can detect ions at high mass range and the ones equipped with electron based fragmentation techniques. These techniques include electron capture dissociation (ECD), electron transfer dissociation (ETD), and negative electron transfer dissociation (NETD) that retain labile protein modifications in the peptide backbone [124].
Figure 5.
The glycoprotein and glycopeptide analysis procedure
Alternatively, another common strategy for site-specific glycosylation analysis involves digesting the proteins using specific or non-specific proteases to produce glycosylated and nonglycosylated peptides (bottom up). The samples can then be analyzed by MS directly to identify the glycopeptides. However, since the glycopeptides ionize relatively less efficiently compared to the nonglycosylated peptides, a fractionation step can be added to enrich for the glycopeptides [125]. The fractionation step, although a useful step, may affect the levels of anaytes in different proportions which may lead to inaccurate quantitative information. Tandem mass spectrometry (MS/MS or MSn) is used to fragment glycopeptides to help in identifying both the glycan and its peptide moiety. Diagnostic glycan and the peptide backbone fragments are necessary for unambiguous identification of the site of glycosylation [112,126]. Another aspect of glycosylation analysis is the extent to which the site of glycosylation is occupied with the glycans (site occupancy). The common method to measure the glycosylation site occupancy involves releasing the N-glycans using PNGase F, which deaminates the glycosylated asparagine residues (N) to aspartic acid (D). The mass difference between the N and D (0.984 Da) allows them to be distinguished using MS [127].
The assignment of the glycopeptides is usually carried out using computer algorithms designed to identify the unique fragmentation patterns of the glycopeptides [128,112]. Glycopeptides produce abundant glycan diagnostic protonated fragments such as HexNAc (204.08 Da), Hex (163.06 Da), Hex+HexNAc (366.14 Da), Neu5Ac-2H2O (274.09 Da) and Neu5Ac-H2O (292.10 Da). We provide a few but not exhaustive examples where site-specific analysis of the glycans provided useful information about potential glycopeptide biomarkers.
In a recent study, the protein clusterin was pulled from the plasma of clear cell renal cell carcinoma (ccRCC) patients before and after curative nephrectomy for localized disease using immunoaffinity purification. Site-specific N-glycan heterogeneity analysis was carried out using LTQ-orbitrap elite tandem mass spectrometry. Biantennary digalactosylated disialylated oligosaccharides with and without fucose found in glycosite N374 were significantly decreased in disease RCC (+) samples [129].
As mentioned earlier, global analysis of the glycosylation of haptoglobin show elevation of fucosylation in pancreatic cancer patients’ serum [49]. Another study was carried out to identify the sites where fucosylation changed. Among the four sites of glycosylation, fucosylation of the bi-antennary glycans in two sites (N207 and N241) increased while that of the tri-antennary glycans increased in all the sites [50].
A method for screening and quantifying core fucosylated (CF) peptides in the depleted serum was recently developed [130]. The method uses lens culinaris agglutinin (LCA) to enrich for the core fucosylated peptides. The presence of metal ions Na+, Ca2+, and Mn2+ increased the lectin binding. The same metal ions were added during the elution step to increase the elution of CF. This work identified 630 sites containing core fucosylated structures in 322 proteins using the Orbitrap Elite MS. Additionally, eight potential CF glycopeptide cancer markers that differed between pancreatic cancer and healthy controls or chronic pancreatitis were identified.
Another method to measure the level of core fucosylation in alpha-2-macroglobulin (A2MG) extracted from serum using immunoprecipitation was recently developed. This method utilizes endoglycosidase F to release a portion of the glycan, leaving the inner core GlcNAc residue attached to the peptide. The heterogeneity due to multiple glycan structures per peptide was reduced, the ionization efficiency was increased, and the analysis of the data was simplified [131]. Nano-LC-linear trap quadrupole (LTQ)-MS was used to analyze the samples. The study found lower levels of core-fucosylation at sites N396 and N1424 in both chronic pancreatitis and pancreatic cancer compared to normal controls [131]. The method used in this work can be used for researchers targeting core fucosylation, which has been found to change in different types of cancer [132,133,49].
Multiple Reaction Monitoring for Glycopeptide Analysis
Multiple reaction monitoring (MRM) is a targeted analytical method used for quantitation of biomolecules. MRM is usually performed on a triple quadrupole mass spectrometer (QqQ). The precursor of interest is isolated in the first quadrupole, fragmented in the second, and only specific fragment ions are transmitted through the third quadrupole for detection. This method can quantify analytes from biological samples with high accuracy and precision. MRM, which has been widely applied in quantification of small molecules and also proteins, is gaining interest in the glycoproteomics field [134–139].
Stable isotope standards and capture by anti-peptide antibodies (SISCAPA) is a useful technique to enrich specific peptides for quantitation [140]. This method was used in combination with the lectin L-PHA (described above) to investigate aberrantly glycosylated metallopeptidase inhibitor 1 (TIMP1) as a marker for colon cancer [141]. First, L-PHA was used to enrich for serum glycoproteins containing β1,6-GlcNAc N-linked glycans. A monoclonal anti-peptide TIMP1 antibody coupled to magnetic beads was used to enrich for a specific TIMP1 peptide that was subsequently analyzed using MRM with isotopically labelled standards. Using this method, an aberrantly glycosylated TIMP1 isoform was quantified at a level of 0.8 ng/mL in the serum of colorectal cancer patients.
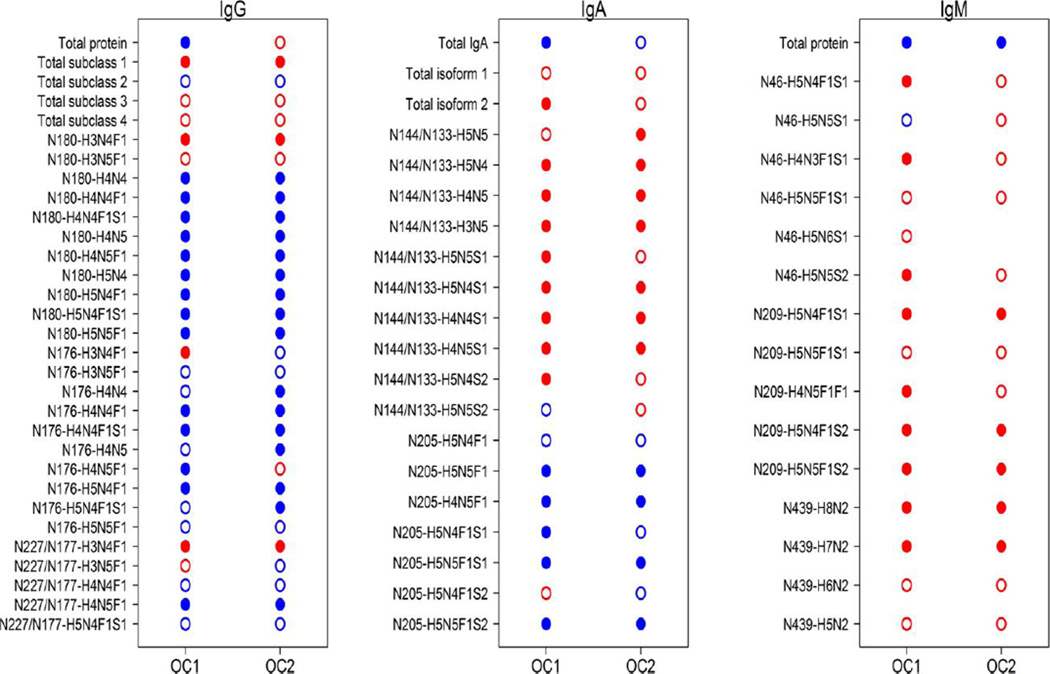
An MRM method to quantify IgG and its subclasses directly from un-depleted serum was recently developed in our laboratory [137]. In this method, trypsin digested IgG glycoprotein standard was used to optimize the QqQ-MS response for glycopeptide analysis. The LC solvent gradient, the MS front end ionization parameters and the fragmentation energy for the glycopeptides were optimized. The method is able to measure the amount of the protein and how its site-specific glycosylation changes between samples. Robust nonglycosylated peptides found in all the IgG subclasses were used to obtain the total amount of IgG in the serum. The glycopeptides from the four IgG subclasses (IgG1, IgG2, IgG3 and IgG4) were normalized to the response of their respective protein subclasses. Normalizing the glycopeptide to the level of the protein helps to identify the actual glycosylation changes irrespective of the changes in the level of protein expression between samples [137]. This method has been extended to include other glycoproteins in serum[142] and it is currently applied in our laboratory to screen for disease biomarkers for cancer [143] autoimmune conditions and other diseases. A recent study investigated the immunoglobulins in ovarian cancer patients compared to the controls. Several glycopeptides were found to vary significantly between the ovarian cancer cases and controls (Figure 6).
Figure 6.
Differential analysis of peptide and glycopeptide variables from immunoglobulins in EOC. Closed dots indicate significantly different abundance levels between EOC cases and healthy controls, while open dots indicate no significance was achieved at the false discovery rate (FDR) < 0.05. Red dots indicate increased levels in EOC cases compared with controls, while blue dots indicate decreased levels in EOC compared with controls. Reprinted with permission from ref. 137. L. Renee Ruhaak, Kyoungmi Kim, Carol Stroble, Sandra L. Taylor, Qiuting Hong, Suzanne Miyamoto, Carlito B. Lebrilla, and Gary Leiserowitz J. Proteome Res., 2016, 15, 1002–1010. Copyright © 2016, American Chemical Society.
Conclusions and Overview
Glycosylation is a post-translational modification of proteins that affects the biochemical activities of the glycoproteins, and they play a substantial role in cancer biology. Glycomics and glycoproteomics methods are less developed than proteomics partly due to the inherent challenges associated with their analysis. However, as the awareness of the roles of glycosylation and other PTMs in cancer biology continues to increase, multidisciplinary efforts are likely to expand the field. Advanced analytical instruments will enable scientists to develop new and robust methods that will uncover potential cancer biomarkers. The complex and laborious glycosylation data analysis process will be improved by applying new or improved bioinformatics methods. These methods are likely to unravel hidden information that would otherwise be lost and also enable fast data analysis to increase the MS obtainable glycan and glycopeptide structural details. Global glycan analysis and protein-specific analysis from human fluids would ultimately add more specific potential cancer markers. A relatively more challenging but useful site-specific glycosylation analysis is likely to provide crucial and detailed information about protein glycosylation patterns. Targeted methods like MRM are gaining interest in the glycoproteomics field. These methods are expected to provide precise quantitative information about the glycosylation site heterogeneity in cancerous cells, tissues or bio-fluids compared to non-cancerous ones.
Despite the advances in glycan analysis, there remain challenges in developing glycan markers. Further progress in analysis will be made to yield exact structures with extensive site-specific heterogeneity in a quantitative manner. The major limitation is that these methods require considerable expertise as well as expensive instrumentation. Their implementation may be limited to selective groups for some time. To obtain truly relevant clinical markers, extensive multidisciplinary and collaborative efforts between those with analytical tools and clinicians are necessary today. Furthermore, considerable efforts should also be placed in making these methods more widely available and easier to use.
Footnotes
Compliance with Ethical Standards
The authors have declared no conflict of interest.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, Boyle P. Estimates of the cancer incidence and mortality in Europe in 2006. Ann Oncol. 2007;18(3):581–592. doi: 10.1093/annonc/mdl498. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 4.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 6.Sawyers CL. The cancer biomarker problem. Nature. 2008;452(7187):548–552. doi: 10.1038/nature06913. [DOI] [PubMed] [Google Scholar]
- 7.Kohn EC, Azad N, Annunziata C, Dhamoon AS, Whiteley G. Proteomics as a tool for biomarker discovery. Dis Markers. 2007;23(5–6):411–417. doi: 10.1155/2007/967023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamandis EP, van der Merwe D-E. Plasma protein profiling by mass spectrometry for cancer diagnosis: opportunities and limitations. Clin Cancer Res. 2005;11(3):963–965. [PubMed] [Google Scholar]
- 9.Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Prac Oncol. 2008;5(10):588–599. doi: 10.1038/ncponc1187. [DOI] [PubMed] [Google Scholar]
- 10.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. doi: http://www.nature.com/nature/journal/v415/n6871/suppinfo/415530a_S1.html. [DOI] [PubMed] [Google Scholar]
- 11.Espina V, Woodhouse EC, Wulfkuhle J, Asmussen HD, Petricoin Iii EF, Liotta LA. Protein microarray detection strategies: focus on direct detection technologies. J Immunol Methods. 2004;290(1–2):121–133. doi: 10.1016/j.jim.2004.04.013. doi: http://dx.doi.org/10.1016/j.jim.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 12.Wulfkuhle JD, Liotta LA, Petricoin EF. Proteomic applications for the early detection of cancer. Nat Rev Cancer. 2003;3(4):267–275. doi: 10.1038/nrc1043. [DOI] [PubMed] [Google Scholar]
- 13.Nagler RM. Saliva as a tool for oral cancer diagnosis and prognosis. Oral Oncol. 2009;45(12):1006–1010. doi: 10.1016/j.oraloncology.2009.07.005. doi: http://dx.doi.org/10.1016/j.oraloncology.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R, Lonigro RJ, Tsodikov A, Wei JT, Tomlins SA, Chinnaiyan AM. A First-Generation Multiplex Biomarker Analysis of Urine for the Early Detection of Prostate Cancer. Cancer Res. 2008;68(3):645–649. doi: 10.1158/0008-5472.CAN-07-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexander H, Stegner AL, Wagner-Mann C, Du Bois GC, Alexander S, Sauter ER. Proteomic Analysis to Identify Breast Cancer Biomarkers in Nipple Aspirate Fluid. Clin Cancer Res. 2004;10(22):7500–7510. doi: 10.1158/1078-0432.CCR-04-1002. [DOI] [PubMed] [Google Scholar]
- 16.Teng P-n, Bateman NW, Hood BL, Conrads TP. Advances in Proximal Fluid Proteomics for Disease Biomarker Discovery. J Proteome Res. 2010;9(12):6091–6100. doi: 10.1021/pr100904q. [DOI] [PubMed] [Google Scholar]
- 17.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452(7187):571–579. doi: 10.1038/nature06916. [DOI] [PubMed] [Google Scholar]
- 18.Hassanein M, Callison JC, Callaway-Lane C, Aldrich MC, Grogan EL, Massion PP. The State of Molecular Biomarkers for the Early Detection of Lung Cancer. Cancer Prevention Research. 2012;5(8):992–1006. doi: 10.1158/1940-6207.CAPR-11-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polanski M, Anderson NL. A list of candidate cancer biomarkers for targeted proteomics. Biomarker insights. 2006;1:1. [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Shan Q, Jiang L, Zhu B, Xi X. Quantitative proteomic analysis by iTRAQ for identification of candidate biomarkers in plasma from acute respiratory distress syndrome patients. Biochem Biophys Res Commun. 2013;441(1):1–6. doi: 10.1016/j.bbrc.2013.09.027. [DOI] [PubMed] [Google Scholar]
- 21.Lam YW, Mobley JA, Evans JE, Carmody JF, Ho SM. Mass profiling-directed isolation and identification of a stage-specific serologic protein biomarker of advanced prostate cancer. Proteomics. 2005;5(11):2927–2938. doi: 10.1002/pmic.200401165. [DOI] [PubMed] [Google Scholar]
- 22.Palmblad M, Tiss A, Cramer R. Mass spectrometry in clinical proteomics - from the present to the future. Proteomics Clinical Applications. 2009;3(1):6–17. doi: 10.1002/prca.200800090. [DOI] [PubMed] [Google Scholar]
- 23.Miller RA, Spellman DS. Mass Spectrometry-Based Biomarkers in Drug Development. Advancements of Mass Spectrometry in Biomedical Research, vol 806. In: Woods AG, Darie CC, editors. Advances in Experimental Medicine and Biology. 2014. pp. 341–359. [DOI] [PubMed] [Google Scholar]
- 24.Pusch W, Flocco MT, Leung SM, Thiele H, Kostrzewa M. Mass spectrometry-based clinical proteomics. Pharmacogenomics. 2003;4(4):463–476. doi: 10.1517/phgs.4.4.463.22753. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Jiao J, Yang P, Lu H. Mass spectrometry-based N-glycoproteomics for cancer biomarker discovery. Clin Proteomics. 2014;11(1):18–18. doi: 10.1186/1559-0275-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jimenez CR, Verheul HMW. Mass spectrometry-based proteomics: from cancer biology to protein biomarkers, drug targets, and clinical applications. American Society of Clinical Oncology educational book / ASCO American Society of Clinical Oncology Meeting. 2014:e504–e510. doi: 10.14694/EdBook_AM.2014.34.e504. [DOI] [PubMed] [Google Scholar]
- 27.Yin H, Lin Z, Nie S, Wu J, Tan Z, Zhu J, Dai J, Feng Z, Marrero J, Lubman DM. Mass-Selected Site-Specific Core-Fucosylation of Ceruloplasmin in Alcohol-Related Hepatocellular Carcinoma. J Proteome Res. 2014;13(6):2887–2896. doi: 10.1021/pr500043k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griffiths WJ, Wang Y. Mass spectrometry: from proteomics to metabolomics and lipidomics. Chem Soc Rev. 2009;38(7):1882–1896. doi: 10.1039/b618553n. [DOI] [PubMed] [Google Scholar]
- 29.Ly M, Laremore TN, Linhardt RJ. Proteoglycomics: recent progress and future challenges. OMICS: J Integrative Biol. 2010;14(4):389–399. doi: 10.1089/omi.2009.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wuhrer M, Catalina MI, Deelder AM, Hokke CH. Glycoproteomics based on tandem mass spectrometry of glycopeptides. J Chromatogr B. 2007;849(1):115–128. doi: 10.1016/j.jchromb.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 31.Zaia J. Mass spectrometry and the emerging field of glycomics. Chem Biol. 2008;15(9):881–892. doi: 10.1016/j.chembiol.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kailemia MJ, Ruhaak LR, Lebrilla CB, Amster IJ. Oligosaccharide analysis by mass spectrometry: a review of recent developments. Anal Chem. 2013;86(1):196–212. doi: 10.1021/ac403969n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hua S, Lebrilla C, An HJ. Application of nano-LC-based glycomics towards biomarker discovery. Bioanalysis. 2011;3(22):2573–2585. doi: 10.4155/bio.11.263. [DOI] [PubMed] [Google Scholar]
- 34.Alley WR, Madera M, Mechref Y, Novotny MV. Chip-based Reversed-phase Liquid Chromatography–Mass Spectrometry of Permethylated N-Linked Glycans: A Potential Methodology for Cancer-biomarker Discovery. Anal Chem. 2010;82(12):5095–5106. doi: 10.1021/ac100131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 36.Kronewitter SR, De Leoz MLA, Strum JS, An HJ, Dimapasoc LM, Guerrero A, Miyamoto S, Lebrilla CB, Leiserowitz GS. The glycolyzer: automated glycan annotation software for high performance mass spectrometry and its application to ovarian cancer glycan biomarker discovery. Proteomics. 2012;12(15–16):2523–2538. doi: 10.1002/pmic.201100273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ludwig JA, Weinstein JN. Biomarkers in Cancer Staging, Prognosis and Treatment Selection. Nat Rev Cancer. 2005;5(11):845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 38.Yin BWT, Lloyd KO. Molecular Cloning of the CA125 Ovarian Cancer Antigen: IDENTIFICATION AS A NEW MUCIN, MUC16. J Biol Chem. 2001;276(29):27371–27375. doi: 10.1074/jbc.M103554200. [DOI] [PubMed] [Google Scholar]
- 39.Jacobs I, Bast RC. The CA 125 tumour-associated antigen: a review of the literature. Hum Reprod. 1989;4(1):1–12. doi: 10.1093/oxfordjournals.humrep.a136832. [DOI] [PubMed] [Google Scholar]
- 40.Miralles C, Orea M, España P, Provencio M, Sánchez A, Cantos B, Cubedo R, Carcereny E, Bonilla F, Gea T. Cancer Antigen 125 Associated With Multiple Benign and Malignant Pathologies. Ann Surg Oncol. 2003;10(2):150–154. doi: 10.1245/aso.2003.05.015. [DOI] [PubMed] [Google Scholar]
- 41.Adamczyk B, Tharmalingam T, Rudd PM. Glycans as cancer biomarkers. Biochimica et Biophysica Acta (BBA) - General Subjects. 2012;1820(9):1347–1353. doi: 10.1016/j.bbagen.2011.12.001. doi: http://dx.doi.org/10.1016/j.bbagen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Duffy M, Shering S, Sherry F, McDermott E, O'higgins N. CA 15-3: a prognostic marker in breast cancer. The International journal of biological markers. 1999;15(4):330–333. doi: 10.1177/172460080001500410. [DOI] [PubMed] [Google Scholar]
- 43.Drake RR, Jones EE, Powers TW, Nyalwidhe JO. Chapter Ten-Altered Glycosylation in Prostate Cancer. Adv Cancer Res. 2015;126:345–382. doi: 10.1016/bs.acr.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 44.Catalona W, Richie J, Ahmann F, Hudson M, Scardino P, Flanigan R, Dekernion J, Ratliff T, Kavoussi L, Dalkin B. Comparison of digital rectal examination and serum prostate specific antigen in the early detection of prostate cancer: results of a multicenter clinical trial of 6,630 men. The Journal of urology. 1994;151(5):1283–1290. doi: 10.1016/s0022-5347(17)35233-3. [DOI] [PubMed] [Google Scholar]
- 45.Hammarström S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol. 1999;9(2):67–81. doi: 10.1006/scbi.1998.0119. doi: http://dx.doi.org/10.1006/scbi.1998.0119. [DOI] [PubMed] [Google Scholar]
- 46.Benchimol S, Fuks A, Jothy S, Beauchemin N, Shirota K, Stanners CP. Carcinoembryonic antigen, a human tumor marker, functions as an intercellular adhesion molecule. Cell. 1989;57(2):327–334. doi: 10.1016/0092-8674(89)90970-7. doi: http://dx.doi.org/10.1016/0092-8674(89)90970-7. [DOI] [PubMed] [Google Scholar]
- 47.Moertel CG, Fleming TR, Macdonald JS, Haller DG, Laurie JA, Tangen C. An evaluation of the carcinoembryonic antigen (CEA) test for monitoring patients with resected colon cancer. JAMA. 1993;270(8):943–947. [PubMed] [Google Scholar]
- 48.Thompson S, Turner G. Elevated levels of abnormally-fucosylated haptoglobins in cancer sera. Br J Cancer. 1987;56(5):605. doi: 10.1038/bjc.1987.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okuyama N, Ide Y, Nakano M, Nakagawa T, Yamanaka K, Moriwaki K, Murata K, Ohigashi H, Yokoyama S, Eguchi H. Fucosylated haptoglobin is a novel marker for pancreatic cancer: a detailed analysis of the oligosaccharide structure and a possible mechanism for fucosylation. Int J Cancer. 2006;118(11):2803–2808. doi: 10.1002/ijc.21728. [DOI] [PubMed] [Google Scholar]
- 50.Miyoshi E, Nakano M. Fucosylated haptoglobin is a novel marker for pancreatic cancer: detailed analyses of oligosaccharide structures. Proteomics. 2008;8(16):3257–3262. doi: 10.1002/pmic.200800046. [DOI] [PubMed] [Google Scholar]
- 51.Zhao C, Annamalai L, Guo C, Kothandaraman N, Koh SCL, Zhang H, Biswas A, Choolani M. Circulating Haptoglobin Is an Independent Prognostic Factor in the Sera of Patients with Epithelial Ovarian Cancer. Neoplasia (New York, NY) 2007;9(1):1–7. doi: 10.1593/neo.06619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahmed N, Barker G, Oliva KT, Hoffmann P, Riley C, Reeve S, Smith AI, Kemp BE, Quinn MA, Rice GE. Proteomic-based identification of haptoglobin-1 precursor as a novel circulating biomarker of ovarian cancer. Br J Cancer. 2004;91(1):129–140. doi: 10.1038/sj.bjc.6601882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang HL, Stasyk T, Morandell S, Dieplinger H, Falkensammer G, Griesmacher A, Mogg M, Schreiber M, Feuerstein I, Huck CW, Stecher G, Bonn GK, Huber LA. Biomarker discovery in breast cancer serum using 2-D differential gel electrophoresis/MALDI-TOF/TOF and data validation by routine clinical assays. Electrophoresis. 2006;27(8):1641–1650. doi: 10.1002/elps.200500857. [DOI] [PubMed] [Google Scholar]
- 54.Ralhan R, DeSouza LV, Matta A, Tripathi SC, Ghanny S, Gupta SD, Bahadur S, Siu KM. Discovery and verification of head-and-neck cancer biomarkers by differential protein expression analysis using iTRAQ labeling, multidimensional liquid chromatography, and tandem mass spectrometry. Mol Cell Proteomics. 2008;7(6):1162–1173. doi: 10.1074/mcp.M700500-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patz EF, Campa MJ, Gottlin EB, Kusmartseva I, Guan XR, Herndon JE. Panel of serum biomarkers for the diagnosis of lung cancer. J Clin Oncol. 2007;25(35):5578–5583. doi: 10.1200/JCO.2007.13.5392. [DOI] [PubMed] [Google Scholar]
- 56.Drake RR, Schwegler EE, Malik G, Diaz J, Block T, Mehta A, Semmes OJ. Lectin capture strategies combined with mass spectrometry for the discovery of serum glycoprotein biomarkers. Mol Cell Proteomics. 2006;5(10):1957–1967. doi: 10.1074/mcp.M600176-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Zhao J, Simeone DM, Heidt D, Anderson MA, Lubman DM. Comparative serum glycoproteomics using lectin selected sialic acid glycoproteins with mass spectrometric analysis: application to pancreatic cancer serum. J Proteome Res. 2006;5(7):1792–1802. doi: 10.1021/pr060034r. [DOI] [PubMed] [Google Scholar]
- 58.Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K-i, Takahashi N, Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat Biotech. 2003;21(6):667–672. doi: 10.1038/nbt829. doi: http://www.nature.com/nbt/journal/v21/n6/suppinfo/nbt829_S1.html. [DOI] [PubMed] [Google Scholar]
- 59.Wu J, Xie X, Liu Y, He J, Benitez R, Buckanovich RJ, Lubman DM. Identification and Confirmation of Differentially Expressed Fucosylated Glycoproteins in the Serum of Ovarian Cancer Patients Using a Lectin Array and LC-MS/MS. J Proteome Res. 2012;11(9):4541–4552. doi: 10.1021/pr300330z. [DOI] [PubMed] [Google Scholar]
- 60.Alvarez-Manilla G, Warren NL, Atwood J, Orlando R, III, Dalton S, Pierce M. Glycoproteomic Analysis of Embryonic Stem Cells: Identification of Potential Glycobiomarkers Using Lectin Affinity Chromatography of Glycopeptides. J Proteome Res. 2010;9(5):2062–2075. doi: 10.1021/pr8007489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuno A, Kato Y, Matsuda A, Kaneko MK, Ito H, Amano K, Chiba Y, Narimatsu H, Hirabayashi J. Focused Differential Glycan Analysis with the Platform Antibody-assisted Lectin Profiling for Glycan-related Biomarker Verification. Mol Cell Proteomics. 2009;8(1):99–108. doi: 10.1074/mcp.M800308-MCP200. [DOI] [PubMed] [Google Scholar]
- 62.Li Y, Wen T, Zhu M, Li L, Wei J, Wu X, Guo M, Liu S, Zhao H, Xia S, Huang W, Wang P, Wu Z, Zhao L, Shui W, Li Z, Yin Z. Glycoproteomic analysis of tissues from patients with colon cancer using lectin microarrays and nanoLC-MS/MS. Mol Biosyst. 2013;9(7):1877–1887. doi: 10.1039/c3mb00013c. [DOI] [PubMed] [Google Scholar]
- 63.Dennis JW, Laferte S, Waghorne C, Breitman ML, Kerbel RS. Beta 1–6 branching of Asn-linked oligosaccharides is directly associated with metastasis. Science. 1987;236(4801):582–585. doi: 10.1126/science.2953071. [DOI] [PubMed] [Google Scholar]
- 64.Ihara S, Miyoshi E, Ko JH, Murata K, Nakahara S, Honke K, Dickson RB, Lin C-Y, Taniguchi N. Prometastatic Effect ofN-Acetylglucosaminyltransferase V Is Due to Modification and Stabilization of Active Matriptase by Adding β1–6 GlcNAc Branching. J Biol Chem. 2002;277(19):16960–16967. doi: 10.1074/jbc.M200673200. [DOI] [PubMed] [Google Scholar]
- 65.Buckhaults P, Chen L, Fregien N, Pierce M. Transcriptional Regulation of N-Acetylglucosaminyltransferase V by the srcOncogene. J Biol Chem. 1997;272(31):19575–19581. doi: 10.1074/jbc.272.31.19575. [DOI] [PubMed] [Google Scholar]
- 66.Abbott KL, Aoki K, Lim J-M, Porterfield M, Johnson R, O'Regan RM, Wells L, Tiemeyer M, Pierce M. Targeted glycoproteomic identification of biomarkers for human breast carcinoma. J Proteome Res. 2008;7(4):1470–1480. doi: 10.1021/pr700792g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 68.Nie S, Lo A, Wu J, Zhu J, Tan Z, Simeone DM, Anderson MA, Shedden KA, Ruffin MT, Lubman DM. Glycoprotein Biomarker Panel for Pancreatic Cancer Discovered by Quantitative Proteomics Analysis. J Proteome Res. 2014;13(4):1873–1884. doi: 10.1021/pr400967x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotech. 2003;21(3):255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 70.Pagel O, Loroch S, Sickmann A, Zahedi RP. Current strategies and findings in clinically relevant post-translational modification-specific proteomics. Expert Review of Proteomics. 2015;12(3):235–253. doi: 10.1586/14789450.2015.1042867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Christiansen MN, Chik J, Lee L, Anugraham M, Abrahams JL, Packer NH. Cell surface protein glycosylation in cancer. Proteomics. 2014;14(4–5):525–546. doi: 10.1002/pmic.201300387. [DOI] [PubMed] [Google Scholar]
- 72.Holst S, Wuhrer M, Rombouts Y. Chapter Six - Glycosylation Characteristics of Colorectal Cancer. In: Richard RD, Lauren EB, editors. Advances in Cancer Research. Vol. 126. Academic Press; 2015. pp. 203–256. doi: http://dx.doi.org/10.1016/bs.acr.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 73.Walsh G, Jefferis R. Post-translational modifications in the context of therapeutic proteins. Nat Biotech. 2006;24(10):1241–1252. doi: 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- 74.An HJ, Froehlich JW, Lebrilla CB. Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Curr Opin Chem Biol. 2009;13(4):421–426. doi: 10.1016/j.cbpa.2009.07.022. doi: http://dx.doi.org/10.1016/j.cbpa.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shriver Z, Raguram S, Sasisekharan R. Glycomics: a pathway to a class of new and improved therapeutics. Nat Rev Drug Discov. 2004;3(10):863–873. doi: 10.1038/nrd1521. [DOI] [PubMed] [Google Scholar]
- 76.Dube DH, Bertozzi CR. Glycans in cancer and inflammation [mdash] potential for therapeutics and diagnostics. Nat Rev Drug Discov. 2005;4(6):477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 77.Reis CA, Osorio H, Silva L, Gomes C, David L. Alterations in glycosylation as biomarkers for cancer detection. J Clin Pathol. 2010;63(4):322–329. doi: 10.1136/jcp.2009.071035. [DOI] [PubMed] [Google Scholar]
- 78.Varki A, Kannagi R, Toole BP. Glycosylation changes in cancer. 2009 [PubMed] [Google Scholar]
- 79.An HJ, Kronewitter SR, de Leoz MLA, Lebrilla CB. Glycomics and disease markers. Curr Opin Chem Biol. 2009;13(5–6):601–607. doi: 10.1016/j.cbpa.2009.08.015. doi: http://dx.doi.org/10.1016/j.cbpa.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guo H, Abbott KL. Chapter Eight - Functional Impact of Tumor-Specific N-Linked Glycan Changes in Breast and Ovarian Cancers. In: Richard RD, Lauren EB, editors. Advances in Cancer Research. Vol. 126. Academic Press; 2015. pp. 281–303. doi: http://dx.doi.org/10.1016/bs.acr.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 81.Drake PM, Cho W, Li B, Prakobphol A, Johansen E, Anderson NL, Regnier FE, Gibson BW, Fisher SJ. Sweetening the Pot: Adding Glycosylation to the Biomarker Discovery Equation. Clin Chem. 2010;56(2):223–236. doi: 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nature Reviews Cancer. 2015 doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 83.de Leoz MLA, Young LJT, An HJ, Kronewitter SR, Kim J, Miyamoto S, Borowsky AD, Chew HK, Lebrilla CB. High-Mannose Glycans are Elevated during Breast Cancer Progression. Mol Cell Proteomics. 2011;10(1) doi: 10.1074/mcp.M110.002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruhaak LR, Miyamoto S, Lebrilla CB. Developments in the Identification of Glycan Biomarkers for the Detection of Cancer. Mol Cell Proteomics. 2013;12(4):846–855. doi: 10.1074/mcp.R112.026799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Budnik BA, Lee RS, Steen JAJ. Global methods for protein glycosylation analysis by mass spectrometry. Biochim Biophys Acta. 2006;1764(12):1870–1880. doi: 10.1016/j.bbapap.2006.10.005. doi: http://dx.doi.org/10.1016/j.bbapap.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 86.An HJ, Lebrilla CB. Functional Glycomics. Springer; 2010. A glycomics approach to the discovery of potential cancer biomarkers; pp. 199–213. [DOI] [PubMed] [Google Scholar]
- 87.JooáAn H. The prospects of glycan biomarkers for the diagnosis of diseases. Mol Biosyst. 2009;5(1):17–20. doi: 10.1039/b811781k. [DOI] [PubMed] [Google Scholar]
- 88.An HJ, Miyamoto S, Lancaster KS, Kirmiz C, Li B, Lam KS, Leiserowitz GS, Lebrilla CB. Profiling of Glycans in Serum for the Discovery of Potential Biomarkers for Ovarian Cancer. J Proteome Res. 2006;5(7):1626–1635. doi: 10.1021/pr060010k. [DOI] [PubMed] [Google Scholar]
- 89.Bharti A, Ma PC, Maulik G, Singh R, Khan E, Skarin AT, Salgia R. Haptoglobin α-Subunit and Hepatocyte Growth Factor can Potentially Serve as Serum Tumor Biomarkers in Small Cell Lung Cancer. Anticancer Res. 2004;24(2C):1031–1038. [PubMed] [Google Scholar]
- 90.Bones J, Mittermayr S, O'Donoghue N, Guttman A, Rudd PM. Ultra Performance Liquid Chromatographic Profiling of Serum N-Glycans for Fast and Efficient Identification of Cancer Associated Alterations in Glycosylation. Anal Chem. 2010;82(24):10208–10215. doi: 10.1021/ac102860w. [DOI] [PubMed] [Google Scholar]
- 91.Wada Y, Azadi P, Costello CE, Dell A, Dwek RA, Geyer H, Geyer R, Kakehi K, Karlsson NG, Kato K, Kawasaki N, Khoo K-H, Kim S, Kondo A, Lattova E, Mechref Y, Miyoshi E, Nakamura K, Narimatsu H, Novotny MV, Packer NH, Perreault H, Peter-Katalinić J, Pohlentz G, Reinhold VN, Rudd PM, Suzuki A, Taniguchi N. Comparison of the methods for profiling glycoprotein glycans—HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycobiology. 2007;17(4):411–422. doi: 10.1093/glycob/cwl086. [DOI] [PubMed] [Google Scholar]
- 92.Ressom HW, Varghese RS, Goldman L, An Y, Loffredo CA, Abdel-Hamid M, Kyselova Z, Mechref Y, Novotny M, Drake SK, Goldman R. Analysis of MALDI-TOF mass spectrometry data for discovery of peptide and glycan biomarkers of hepatocellular carcinoma. J Proteome Res. 2008;7(2):603–610. doi: 10.1021/pr0705237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rodrigo MAM, Zitka O, Krizkova S, Moulick A, Adam V, Kizek R. MALDI-TOF MS as evolving cancer diagnostic tool: A review. J Pharm Biomed Anal. 2014;95:245–255. doi: 10.1016/j.jpba.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 94.Powers TW, Neely BA, Shao Y, Tang H, Troyer DA, Mehta AS, Haab BB, Drake RR. MALDI Imaging Mass Spectrometry Profiling of N-Glycans in Formalin-Fixed Paraffin Embedded Clinical Tissue Blocks and Tissue Microarrays. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0106255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kyselova Z, Mechref Y, Al Bataineh MM, Dobrolecki LE, Hickey RJ, Vinson J, Sweeney CJ, Novotny MV. Alterations in the serum glycome due to metastatic prostate cancer. J Proteome Res. 2007;6(5):1822–1832. doi: 10.1021/pr060664t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kyselova Z, Mechref Y, Kang P, Goetz JA, Dobrolecki LE, Sledge GW, Schnaper L, Hickey RJ, Malkas LH, Novotny MV. Breast Cancer Diagnosis and Prognosis through Quantitative Measurements of Serum Glycan Profiles. Clin Chem. 2008;54(7):1166–1175. doi: 10.1373/clinchem.2007.087148. [DOI] [PubMed] [Google Scholar]
- 97.Balog CIA, Stavenhagen K, Fung WLJ, Koeleman CA, McDonnell LA, Verhoeven A, Mesker WE, Tollenaar RAEM, Deelder AM, Wuhrer M. N-glycosylation of Colorectal Cancer Tissues. Mol Cell Proteomics. 2012;11(9):571–585. doi: 10.1074/mcp.M111.011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.de Leoz MLA, An HJ, Kronewitter S, Kim J, Beecroft S, Vinall R, Miyamoto S, White RdV, Lam KS, Lebrilla C. Glycomic approach for potential biomarkers on prostate cancer: Profiling of N-linked glycans in human sera and pRNS cell lines. Dis Markers. 2008;25(4–5):243–258. doi: 10.1155/2008/515318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hecht ES, Scholl EH, Walker SH, Taylor AD, Cliby WA, Motsinger-Reif AA, Muddiman DC. Relative Quantification and Higher-Order Modeling of the Plasma Glycan Cancer Burden Ratio in Ovarian Cancer Case-Control Samples. J Proteome Res. 2015;14(10):4394–4401. doi: 10.1021/acs.jproteome.5b00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kirmiz C, Li B, An HJ, Clowers BH, Chew HK, Lam KS, Ferrige A, Alecio R, Borowsky AD, Sulaimon S. A serum glycomics approach to breast cancer biomarkers. Mol Cell Proteomics. 2007;6(1):43–55. doi: 10.1074/mcp.M600171-MCP200. [DOI] [PubMed] [Google Scholar]
- 101.Alley WR, Vasseur JA, Goetz JA, Svoboda M, Mann BF, Matei DE, Menning N, Hussein A, Mechref Y, Novotny MV. N-linked Glycan Structures and Their Expressions Change in the Blood Sera of Ovarian Cancer Patients. J Proteome Res. 2012;11(4):2282–2300. doi: 10.1021/pr201070k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saldova R, Royle L, Radcliffe CM, Abd Hamid UM, Evans R, Arnold JN, Banks RE, Hutson R, Harvey DJ, Antrobus R, Petrescu SM, Dwek RA, Rudd PM. Ovarian Cancer is Associated with Changes in Glycosylation in Both Acute-Phase Proteins and IgG. Glycobiology. 2007;17(12):1344–1356. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 103.Kanoh Y, Mashiko T, Danbara M, Takayama Y, Ohtani S, Imasaki T, Abe T, Akahoshi T. Analysis of the Oligosaccharide Chain of Human Serum Immunoglobulin G in Patients with Localized or Metastatic Cancer. Oncology. 2004;66(5):365–370. doi: 10.1159/000079484. [DOI] [PubMed] [Google Scholar]
- 104.Ruhaak LR, Stroble C, Dai J, Barnett MJ, Taguchi A, Goodman GE, Miyamoto S, Gandara DR, Feng Z, Lebrilla C, Hanash SM. Serum glycans as risk markers for non-small cell lung cancer. Cancer Prevention Research. 2016 doi: 10.1158/1940-6207.CAPR-15-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hua S, An HJ, Ozcan S, Ro GS, Soares S, DeVere-White R, Lebrilla CB. Comprehensive native glycan profiling with isomer separation and quantitation for the discovery of cancer biomarkers. Analyst. 2011;136(18):3663–3671. doi: 10.1039/c1an15093f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ruhaak LR, Deelder A, Wuhrer M. Oligosaccharide analysis by graphitized carbon liquid chromatography–mass spectrometry. Anal Bioanal Chem. 2009;394(1):163–174. doi: 10.1007/s00216-009-2664-5. [DOI] [PubMed] [Google Scholar]
- 107.Stavenhagen K, Kolarich D, Wuhrer M. Clinical Glycomics Employing Graphitized Carbon Liquid Chromatography–Mass Spectrometry. Chromatographia. 2014;78(5):307–320. doi: 10.1007/s10337-014-2813-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chu CS, Niñonuevo MR, Clowers BH, Perkins PD, An HJ, Yin H, Killeen K, Miyamoto S, Grimm R, Lebrilla CB. Profile of native N-linked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Proteomics. 2009;9(7):1939–1951. doi: 10.1002/pmic.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Song T, Aldredge D, Lebrilla CB. A Method for In-Depth Structural Annotation of Human Serum Glycans That Yields Biological Variations. Anal Chem. 2015;87(15):7754–7762. doi: 10.1021/acs.analchem.5b01340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ruhaak LR, Barkauskas DA, Torres J, Cooke CL, Wu LD, Stroble C, Ozcan S, Williams CC, Camorlinga M, Rocke DM, Lebrilla CB, Solnick JV. The serum immunoglobulin G glycosylation signature of gastric cancer. EuPA Open Proteomics. 2015;6:1–9. doi: 10.1016/j.euprot.2014.11.002. doi: http://dx.doi.org/10.1016/j.euprot.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Strum JS, Nwosu CC, Hua S, Kronewitter SR, Seipert RR, Bachelor RJ, An HJ, Lebrilla CB. Automated Assignments of N- and O-Site Specific Glycosylation with Extensive Glycan Heterogeneity of Glycoprotein Mixtures. Anal Chem. 2013;85(12):5666–5675. doi: 10.1021/ac4006556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nwosu CC, Seipert RR, Strum JS, Hua SS, An HJ, Zivkovic AM, German BJ, Lebrilla CB. Simultaneous and Extensive Site-specific N- and O-Glycosylation Analysis in Protein Mixtures. J Proteome Res. 2011;10(5):2612–2624. doi: 10.1021/pr2001429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ueda K, Takami S, Saichi N, Daigo Y, Ishikawa N, Kohno N, Katsumata M, Yamane A, Ota M, Sato T-A, Nakamura Y, Nakagawa H. Development of Serum Glycoproteomic Profiling Technique; Simultaneous Identification of Glycosylation Sites and Site-Specific Quantification of Glycan Structure Changes. Mol Cell Proteomics. 2010;9(9):1819–1828. doi: 10.1074/mcp.2010/000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ueda K. Glycoproteomic strategies: From discovery to clinical application of cancer carbohydrate biomarkers. Proteomics Clinical Applications. 2013;7(9–10):607–617. doi: 10.1002/prca.201200123. [DOI] [PubMed] [Google Scholar]
- 115.Bern M, Kil YJ, Becker C. Byonic: advanced peptide and protein identification software. Current Protocols in Bioinformatics. 2012 doi: 10.1002/0471250953.bi1320s40. 13.20. 11-13.20. 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ozohanics O, Krenyacz J, Ludányi K, Pollreisz F, Vékey K, Drahos L. GlycoMiner: a new software tool to elucidate glycopeptide composition. Rapid Commun Mass Spectrom. 2008;22(20):3245–3254. doi: 10.1002/rcm.3731. [DOI] [PubMed] [Google Scholar]
- 117.He L, Xin L, Shan B, Lajoie GA, Ma B. GlycoMaster DB: Software To Assist the Automated Identification of N-Linked Glycopeptides by Tandem Mass Spectrometry. J Proteome Res. 2014;13(9):3881–3895. doi: 10.1021/pr401115y. [DOI] [PubMed] [Google Scholar]
- 118.Mayampurath A, Yu C-Y, Song E, Balan J, Mechref Y, Tang H. Computational Framework for Identification of Intact Glycopeptides in Complex Samples. Anal Chem. 2014;86(1):453–463. doi: 10.1021/ac402338u. [DOI] [PubMed] [Google Scholar]
- 119.Mayampurath AM, Wu Y, Segu ZM, Mechref Y, Tang H. Improving confidence in detection and characterization of protein N-glycosylation sites and microheterogeneity. Rapid Commun Mass Spectrom. 2011;25(14):2007–2019. doi: 10.1002/rcm.5059. [DOI] [PubMed] [Google Scholar]
- 120.Khatri K, Staples GO, Leymarie N, Leon DR, Turiák L, Huang Y, Yip S, Hu H, Heckendorf CF, Zaia J. Confident Assignment of Site-Specific Glycosylation in Complex Glycoproteins in a Single Step. J Proteome Res. 2014;13(10):4347–4355. doi: 10.1021/pr500506z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Go EP, Rebecchi KR, Dalpathado DS, Bandu ML, Zhang Y, Desaire H. GlycoPep DB; A Tool for Glycopeptide Analysis Using a “Smart Search”. Anal Chem. 2007;79(4):1708–1713. doi: 10.1021/ac061548c. [DOI] [PubMed] [Google Scholar]
- 122.Woodin CL, Maxon M, Desaire H. Software for automated interpretation of mass spectrometry data from glycans and glycopeptides. Analyst. 2013;138(10):2793–2803. doi: 10.1039/c2an36042j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Reid GE, Stephenson JL, McLuckey SA. Tandem Mass Spectrometry of Ribonuclease A and B; N-Linked Glycosylation Site Analysis of Whole Protein Ions. Anal Chem. 2002;74(3):577–583. doi: 10.1021/ac015618l. [DOI] [PubMed] [Google Scholar]
- 124.Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nat Methods. 2007;4(10):817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ito S, Hayama K, Hirabayashi J. Enrichment Strategies for Glycopeptides. Glycomics, vol 534. In: Packer N, Karlsson N, editors. Methods in Molecular Biology™. Humana Press; 2009. pp. 194–203. [DOI] [PubMed] [Google Scholar]
- 126.Wuhrer M, Deelder AM, Hokke CH. Protein glycosylation analysis by liquid chromatography–mass spectrometry. J Chromatogr B. 2005;825(2):124–133. doi: 10.1016/j.jchromb.2005.01.030. doi: http://dx.doi.org/10.1016/j.jchromb.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 127.Zhu Z, Go EP, Desaire H. Absolute quantitation of glycosylation site occupancy using isotopically labeled standards and LC-MS. J Am Soc Mass Spectrom. 2014;25(6):1012–1017. doi: 10.1007/s13361-014-0859-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dalpathado DS, Desaire H. Glycopeptide analysis by mass spectrometry. Analyst. 2008;133(6):731–738. doi: 10.1039/b713816d. [DOI] [PubMed] [Google Scholar]
- 129.Gbormittah FO, Bones J, Hincapie M, Tousi F, Hancock WS, Iliopoulos O. Clusterin Glycopeptide Variant Characterization Reveals Significant Site-Specific Glycan Changes in the Plasma of Clear Cell Renal Cell Carcinoma. J Proteome Res. 2015;14(6):2425–2436. doi: 10.1021/pr501104j. [DOI] [PubMed] [Google Scholar]
- 130.Tan Z, Yin H, Nie S, Lin Z, Zhu J, Ruffin MT, Anderson MA, Simeone DM, Lubman DM. Large-Scale Identification of Core-Fucosylated Glycopeptide Sites in Pancreatic Cancer Serum Using Mass Spectrometry. J Proteome Res. 2015;14(4):1968–1978. doi: 10.1021/acs.jproteome.5b00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lin Z, Yin H, Lo A, Ruffin MT, Anderson MA, Simeone DM, Lubman DM. Label-free relative quantification of alpha-2-macroglobulin site-specific core-fucosylation in pancreatic cancer by LC-MS/MS. Electrophoresis. 2014;35(15):2108–2115. doi: 10.1002/elps.201300376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Saldova R, Fan Y, Fitzpatrick JM, Watson RWG, Rudd PM. Core fucosylation and α2–3 sialylation in serum N-glycome is significantly increased in prostate cancer comparing to benign prostate hyperplasia. Glycobiology. 2011;21(2):195–205. doi: 10.1093/glycob/cwq147. [DOI] [PubMed] [Google Scholar]
- 133.Miyoshi E, Moriwaki K, Nakagawa T. Biological Function of Fucosylation in Cancer Biology. J Biochem. 2008;143(6):725–729. doi: 10.1093/jb/mvn011. [DOI] [PubMed] [Google Scholar]
- 134.Ruhaak LR, Lebrilla C. Applications of Multiple Reaction Monitoring to Clinical Glycomics. Chromatographia. 2015;78(5–6):335–342. doi: 10.1007/s10337-014-2783-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Meth. 2012;9(6):555–566. doi: 10.1038/nmeth.2015. doi: http://www.nature.com/nmeth/journal/v9/n6/abs/nmeth.2015.html#supplementary-information. [DOI] [PubMed] [Google Scholar]
- 136.Song E, Pyreddy S, Mechref Y. Quantification of glycopeptides by multiple reaction monitoring liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2012;26(17):1941–1954. doi: 10.1002/rcm.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hong Q, Lebrilla CB, Miyamoto S, Ruhaak LR. Absolute Quantitation of Immunoglobulin G and Its Glycoforms Using Multiple Reaction Monitoring. Anal Chem. 2013;85(18):8585–8593. doi: 10.1021/ac4009995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sanda M, Pompach P, Brnakova Z, Wu J, Makambi K, Goldman R. Quantitative Liquid Chromatography-Mass Spectrometry-Multiple Reaction Monitoring (LC-MS-MRM) Analysis of Site-specific Glycoforms of Haptoglobin in Liver Disease. Mol Cell Proteomics. 2013;12(5):1294–1305. doi: 10.1074/mcp.M112.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yuan W, Sanda M, Wu J, Koomen J, Goldman R. Quantitative analysis of immunoglobulin subclasses and subclass specific glycosylation by LC–MS–MRM in liver disease. J Proteomics. 2015;116:24–33. doi: 10.1016/j.jprot.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3(2):235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 141.Ahn YH, Lee JY, Lee JY, Kim Y-S, Ko JH, Yoo JS. Quantitative Analysis of an Aberrant Glycoform of TIMP1 from Colon Cancer Serum by L-PHA-Enrichment and SISCAPA with MRM Mass Spectrometry. J Proteome Res. 2009;8(9):4216–4224. doi: 10.1021/pr900269s. [DOI] [PubMed] [Google Scholar]
- 142.Hong Q, Ruhaak LR, Stroble C, Parker E, Huang J, Maverakis E, Lebrilla CB. A Method for Comprehensive Glycosite-Mapping and Direct Quantitation of Serum Glycoproteins. J Proteome Res. 2015;14(12):5179–5192. doi: 10.1021/acs.jproteome.5b00756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ruhaak LR, Kim K, Stroble C, Taylor SL, Hong Q, Miyamoto S, Lebrilla CB, Leiserowitz G. Protein-Specific Differential Glycosylation of Immunoglobulins in Serum of Ovarian Cancer Patients. J Proteome Res. 2016;15(3):1002–1010. doi: 10.1021/acs.jproteome.5b01071. [DOI] [PMC free article] [PubMed] [Google Scholar]