Abstract
Background
Cytomegalovirus (CMV) and Epstein-Barr virus (EBV) are increasingly important in immunocompromised patients. Nucleic acid extraction methods could affect the results of viral nucleic acid amplification tests. We compared two automated nucleic acid extraction systems for detecting CMV and EBV using real-time PCR assays.
Methods
One hundred and fifty-three whole blood (WB) samples were tested for CMV detection, and 117 WB samples were tested for EBV detection. Viral nucleic acid was extracted in parallel by using QIAsymphony RGQ and QIAcube (Qiagen GmbH, Germany), and real-time PCR assays for CMV and EBV were performed with a Rotor-Gene Q real-time PCR cycler (Qiagen). Detection rates for CMV and EBV were compared, and agreements between the two systems were analyzed.
Results
The detection rate of CMV and EBV differed significantly between the QIAsymphony RGQ and QIAcube systems (CMV, 59.5% [91/153] vs 43.8% [67/153], P=0.0005; EBV, 59.0% [69/117] vs 42.7% [50/117], P=0.0008). The two systems showed moderate agreement for CMV and EBV detection (kappa=0.43 and 0.52, respectively). QIAsymphony RGQ showed a negligible correlation with QIAcube for quantitative EBV detection. QIAcube exhibited EBV PCR inhibition in 23.9% (28/117) of samples.
Conclusions
Automated nucleic acid extraction systems have different performances and significantly affect the detection of viral pathogens. The QIAsymphony RGQ system appears to be superior to the QIAcube system for detecting CMV and EBV. A suitable sample preparation system should be considered for optimized nucleic acid amplification in clinical laboratories.
Keywords: Cytomegalovirus, Epstein-Barr virus, QIAsymphony RGQ, QIAcube, Nucleic acid, Extraction, Performance
INTRODUCTION
Cytomegalovirus (CMV) and Epstein-Barr virus (EBV), members of the human herpesviridae (HHV) family, have double-stranded, linear DNA genomes of 230 kb and 172 kb encoding approximately 170 genes and 85 genes, respectively. CMV and EBV infections are common worldwide with some geographical variability. CMV seroprevalence ranges from 45% to 100% in women of reproductive age and tends to be highest in South America, Africa, and Asia [1]; EBV seroprevalence also ranges from 50% to 93% [2,3]. As with all HHV, CMV and EBV can establish a lifelong presence, exhibiting persistent and latent infection following primary infection that can be reactivated with shedding of infectious viruses.
Serological methods such as viral culture, antigen detection, and viral nucleic acid amplification tests (NAAT) can be used for the diagnosis and monitoring of active viral infections. Serology may be used to determine ongoing susceptibility to community-acquired disease in patients who are seronegative prior to transplantation and who do not develop infection or disease post transplantation [4,5]. The advantages of NAAT include high sensitivity, rapid results, and the ability to provide quantitative viral load measurements; PCR is one of the most widely applied tests for the diagnosis and monitoring of viral pathogens. The term DNAemia is used instead of viremia to reflect the detection of CMV or EBV DNA in blood.
Several automated sample preparation systems are currently available in clinical molecular laboratories. There is no single method for every application; many factors, including the target viral pathogen(s), sample volume, final volume needed for testing, yield, and upstream concentration, should be considered when a new sample preparation system is introduced into clinical laboratories [6,7]. In addition, various sample types have been used for detecting CMV and EBV DNA, with no consensus regarding the optimal sample type [3,8,9,10]. Extraction of viral DNA from whole blood (WB) is of great importance in clinical laboratories because host hemoglobin and DNA can interfere with the extraction and amplification of target viral DNA.
Several comparison studies examining CMV or EBV DNA detection have been published; however, it is unclear whether the same nucleic acid extraction system was used to detect CMV or EBV DNA, or, if different systems were used, which was optimal for the targeted viruses [11,12,13,14,15,16,17,18,19,20]. In this study, we evaluated two automated nucleic acid extraction systems used for detecting CMV and EBV DNA from WB: the QIAsymphony system and the QIAcube system (Qiagen GmbH, Hilden, Germany). The QIAcube system consists of the QIAcube and Rotor-Gene Q real-time PCR cycler (RGQ, Qiagen). The QIAsymphony RGQ system is a recently available fully automated sample preparation system, which consists of the QIAsymphony Sample Processing (SP)/Assay Set-up (AS) system (Qiagen) and RGQ. We compared the effect of these nucleic acid extraction methods on the analytical performance of quantitative PCR assays for detecting CMV and EBV from WB.
METHODS
1. Samples
A total of 270 WB samples were included in this study: 153 samples for CMV detection and 117 samples for EBV detection. All samples were obtained for routine diagnostic work-up or monitoring of CMV or EBV infection; remnant samples were used for this comparison study. The samples were tested retrospectively and kept frozen at -80℃ prior to being tested by the two systems. Viral nucleic acid was extracted in parallel by using the QIAsymphony RGQ system and QIAcube system and then amplified and detected by using RGQ. The tests were performed successively on both platforms by the same operator. The Institutional Review Board of Konkuk University Medical Center waived approval for this study (KUH1200042) because it used remnant samples for in vitro method comparison.
2. Test systems
QIAcube is a semi-automated nucleic acid extraction system, which uses a chemical enzymatic lysis and silica spin-column binding (centrifugation) isolation method with a batch capacity of 12 samples. The QIAsymphony RGQ system, an integrated fully automated nucleic acid extraction and sample preparation platform comprises three components: QIAsymphony SP, QIAsymphony AS, and RGQ. QIAsymphony uses a chemical lysis and paramagnetic bead binding and ethanol wash isolation method with a batch capacity of 24 samples.
3. Preparation
Nucleic acid was extracted according to the manufacturer's instructions. Using the QIAamp DSP DNA Mini Kit (Qiagen) with QIAcube, 100 µL eluate was obtained from 200 µL WB. Next, 30 µL of either the artus CMV RG Kit (Qiagen) or artus EBV RG Kit (Qiagen) master mix was added to 20 µL of the 100 µL eluate, for a final reaction volume of 50 µL.
Using the QIAsymphony DNA Mini Kit with QIAsymphony SP, 90 µL eluate was obtained from 200 µL (300 µL minus 100 µL dead volume) WB. A total of 60 µL internal control (IC) plasmid plus either CMV or EBV buffer was added to each sample prior to extraction. For the QIAsymphony AS component, 30 µL of either the artus CMV QS-RGQ kit (Qiagen) or artus EBV QS-RGQ kit (Qiagen) master mix was added to 20 µL (of the 60 µL) eluate, for a final reaction volume of 50 µL (Table 1).
Table 1. Nucleic acid extraction and amplification systems for the detection of cytomegalovirus and Epstein-Barr virus.
| System | QIAsymphony RGQ | QIAcube | ||
|---|---|---|---|---|
| Extraction kit | QIAsymphony DNA Mini Kit | QIAamp DSP DNA Mini Kit | ||
| Available sample type* | Whole blood, buffy coat, cultured cells, tissue, bacterial cultures | Whole blood, plasma, serum, buffy coat, lymphocytes, dried blood spot, body fluids, cultured cells, swabs, tissue | ||
| Lysis | Chemical | Chemical enzyme | ||
| Isolation method | Paramagnetic bead binding | Silica spin-column binding (centrifugation) | ||
| Batch capacity | 24 | 12 | ||
| Minimum sample requirement (µL) | 300 (dead volume, 100) | 200 | ||
| Elution volume (µL) | 90 | 100 | ||
| Final reaction volume (template + master mix, µL) | 20 + 30 | 20 + 30 | ||
*Technical specifications as indicated in the manufacturer's instructions; †Analytical sensitivity (limit of quantification) was defined as the concentration at which 95% of replicates were detected; ‡Corresponding to 1.0 copy/mL.
Abbreviations: CMV, cytomegalovirus; EBV, Epstein-Barr virus; CSF, cerebrospinal fluid; Ct, threshold cycle; IC, internal control; MIE, major immediate early.
4. Amplification
Real-time PCR for quantitation of CMV and EBV DNA from both extraction systems was performed in conjunction with RGQ according to the manufacturer's instructions (hold, 95℃ for 10 minutes; 45 cycles of 95℃ for 15 sec, 65℃ for 30 sec, and 72℃ for 20 sec; auto-gain optimization setup, 65℃). The green signal indicates the viral target, and the yellow signal indicates the IC.
In both systems, the viral DNA load was calculated by applying a calibration curve generated with four calibrators, including the heterologous IC. The QIAcube system reports the viral load results as copies/µL and copies/mL, while the QIAsymphony RGQ system reports them as IU/mL, copies/µL, and copies/mL. In addition to the manufacturer-supplied QIAsymphony RGQ system conversion factor, we measured the conversion factors in both systems using the WHO international standard [21]. Briefly, the WHO international standard was diluted 1:10 (500,000 IU/mL), 1:100 (50,000 IU/mL), 1:1,000 (5,000 IU/mL), and 1:10,000 (500 IU/mL) in WB samples that previously tested negative for CMV and EBV DNA. These dilutions were aliquoted and tested in duplicate in three separate experiments. The quantitative PCR results were multiplied by their initial dilution to obtain experimental WHO values. To obtain the results in IU/mL, the results obtained in copies/mL were multiplied by the conversion factor. The major characteristics of the CMV and EBV DNA amplification systems are summarized in Table 1.
5. Study design
The linearity of the QIAsymphony RGQ system was determined by using the WHO international standard. Briefly, the standard was serially diluted (1:10, 1:100, 1:1,000, 1:10,000) in WB samples that did not contain detectable CMV and EBV DNA. These dilutions were aliquoted and tested in duplicate in three separate experiments. Prior to determining linearity, the manufacturer-specified limit of detection (LOD) and limit of quantification (LOQ) were verified: CMV, LOD 2.22 copies/mL and LOQ 164.55 copies/mL; EBV, LOD 2.46 copies/mL and LOQ 288.42 copies/mL [22].
We compared the detection rates for CMV and EBV DNA using the QIAsymphony RGQ and QIAcube systems. The agreement between the qualitative results of both systems was analyzed. The correlation between the CMV and EBV DNA concentrations detected by the two systems was evaluated. In addition, we compared the quantitative results of CMV and EBV detection of the two systems.
6. Statistical analysis
Linearity was assessed by using regression analysis. The Chi-squared test was performed to compare the detection rate of CMV and EBV DNA. For qualitative comparison of CMV and EBV detection, the inter-rater agreement statistic (kappa value) was used. The Wilcoxon test was used to analyze discrepant CMV and EBV detection results. To compare the quantitative CMV and EBV detection results of the QIAsymphony RGQ and QIAcube systems, Passing-Bablok regression was performed, and Bland-Altman plots were used to identify mean differences of averaged logs for the positive results of both systems. The correlation coefficients (r) were defined as: ≤0.30, negligible correlation; 0.30–0.50, low correlation; 0.50–0.70, moderate correlation; 0.70–0.90, high correlation; and ≥0.90, very high correlation [23]. Statistical analysis was performed by using Analyse-it Software (version 3.90.5 Analyse-it Software, Ltd., Leeds, UK) and MedCalc Software (version 14.12.0, MedCalc Software, Mariakerke, Belgium). P values less than 0.05 were considered statistically significant.
RESULTS
1. Linearity
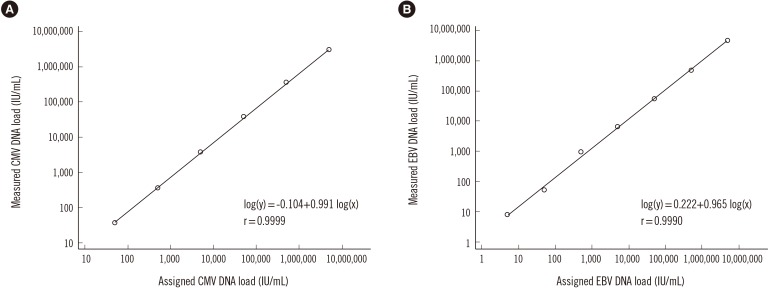
The QIAsymphony RGQ system was linear for approximately seven orders of magnitude and exhibited first-order log transformation. The linear range was 50–5,000,000 IU/mL for CMV detection and 5–5,000,000 IU/mL for EBV detection (Fig. 1). There was no significant deviation from linearity for CMV and EBV detections (P=0.99 and P=0.97, respectively).
Fig. 1. Linearity of quantification using 1:10 serial dilutions of the first WHO international standards for (A) cytomegalovirus (CMV) and (B) Epstein-Barr virus (EBV). Viral load results were obtained using the artus CMV QS-RGQ Kit and the artus EBV QS-RGQ kit in combination with the Rotor Gene Q thermal cycler. The “0” results (N=2 for CMV and N=1 for EBV) are not demonstrated in this figure because of the log transformation. Solid lines indicate the linear regression fit.
2. Detection rate and agreement
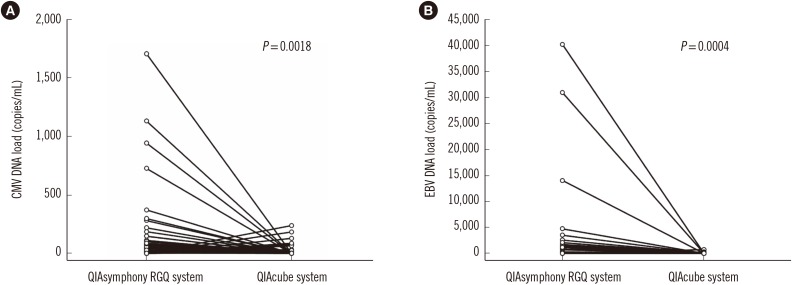
As shown in Table 2 and Fig. 2, CMV was detected in 91 (59.5%) and 67 (43.8%) of 153 samples by using the QIAsymphony RGQ system or QIAcube system, respectively. The detection rate for CMV differed significantly between the two systems (P=0.0005) with a concordance rate of 71.2% (109/153), indicating a moderate agreement (kappa=0.43, 95% confidence interval [CI]=0.30–0.57). The 44 discrepant results between the two systems were statistically significant (P=0.0018).
Table 2. Detection of cytomegalovirus and Epstein-Barr virus using the QIAsymphony RGQ and QIA cube systems.
| QIAcube system | ||||||||
|---|---|---|---|---|---|---|---|---|
| CMV | Positive | Negative | Total | EBV | Positive | Negative | Total | |
| QIAsymphony RGQ system | Positive | 57 | 34 | 91 | Positive | 45 | 24 | 69 |
| Negative | 10 | 52 | 62 | Negative | 5 | 43 | 48 | |
| Total | 67 | 86 | 153 | Total | 50 | 67 | 117 | |
| Kappa (95% CI) | 0.43 (0.30–0.57) | Kappa (95% CI) | 0.52 (0.37–0.66) | |||||
| Detection rate | 59.5% (91/153) vs 43.8% (67/153) P = 0.0005 |
Detection rate | 59.0% (69/117) vs 42.7% (50/117) P = 0.0008 |
|||||
Abbreviations: CMV, cytomegalovirus; EBV, Epstein-Barr virus; CI, confidence interval.
Fig. 2. Discrepant cytomegalovirus (CMV, N=44; left panel) and Epstein-Barr virus (EBV, N=29; right panel) results using the QIAsymphony RGQ and QIAcube systems.
As shown in Table 2, EBV was detected in 69 (59.0%) and 50 (42.7%) of 117 samples by using the QIAsymphony RGQ system and QIAcube system, respectively, demonstrating a significant difference (P=0.0008). The concordance rate for EBV between the two systems was 75.2% (88/117), signifying moderate agreement (kappa=0.52, 95% CI=0.37–0.66). The 29 discrepant results between the two systems were statistically significant (P=0.0004).
3. Quantitative comparison between the QIAsymphony RGQ and QIAcube systems
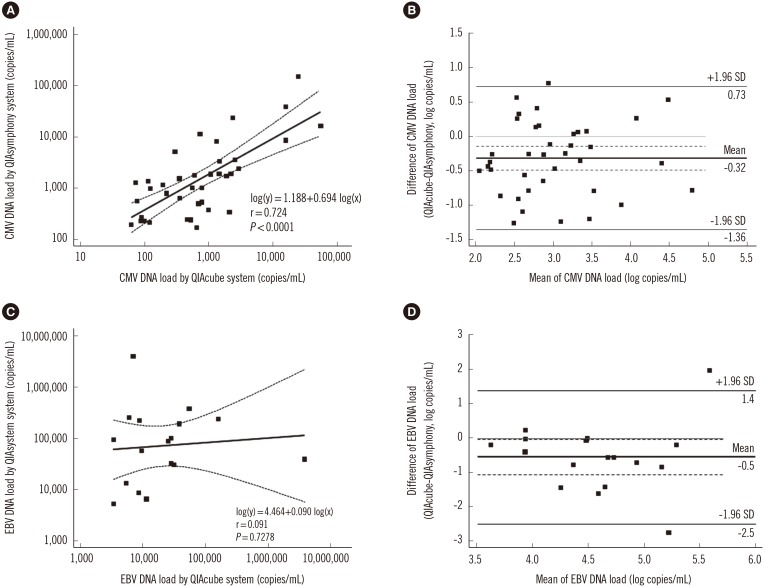
The viral load results obtained by using the QIAsymphony RGQ and QIAcube systems were compared. For CMV detection, 38 of 153 (24.8%) samples showed higher values than the LOQ of both systems. Passing-Bablok regression of correlation demonstrated high correlation (r=0.724). The mean difference was -0.32 log copies/mL and the QIAsymphony RGQ system values were higher than the QIAcube system values (Fig. 3A and B).
Fig. 3. Comparison of cytomegalovirus (CMV, N=38) and Epstein-Barr virus (EBV, N=17) results using the QIAsymphony RGQ and QIA-cube systems. (A) Passing-Bablok regression for CMV detection. (B) Bland-Altman plot for CMV detection. (C) Passing-Bablok regression for EBV detection. (D) Bland-Altman plot for EBV detection. In the regression plot, the solid line indicates the regression line and dashed lines indicate 95% confidence interval. In the Bland-Altman plot, the bold line indicates the mean difference between values, the dashed lines indicate 95% confidence interval, and the solid lines indicate mean difference ±1.96 standard deviation.
For EBV detection, 17 of 117 (14.5%) samples showed higher values than the LOQ of both systems. Passing-Bablok regression of correlation showed a negligible correlation (r=0.091). The mean difference was -0.5 log copies/mL, and the QIAsymphony RGQ system values were mostly higher than the QIAcube system values (Fig. 3C and D).
4. Inhibition
For CMV detection, none of the 153 samples showed PCR inhibition for either the QIAsymphony RGQ or QIAcube systems. For EBV detection, although no samples exhibited PCR inhibition for the QIAsymphony RGQ system, 28 of 117 (23.9%) samples demonstrated PCR inhibition for the QIAcube system.
DISCUSSION
The purpose of this study was to evaluate two automated nucleic acid extraction systems for detecting CMV and EBV DNA from WB samples. The QIAsymphony RGQ and QIAcube systems were compared to determine the effect of nucleic acid extraction method on the analytical characteristics of quantitative PCR assays. We evaluated the detection rate, agreement, and quantitative comparison between the QIAsymphony RGQ system (using the artus CMV QS-RGQ Kit and the artus EBV QS-RGQ Kit) and the QIAcube system (using the artus CMV RG PCR Kit and the EBV RG PCR Kit) in conjunction with RGQ.
There were considerable differences between the QIAsymphony RGQ and QIAcube systems. The detection rate of the QIAsymphony RGQ system was significantly higher than that of the QIAcube system, and the discrepant results between the two systems were statistically significant (Table 2 and Fig. 2). The two PCR systems showed a concordance rate of 71.2% and 75.2% for CMV and EBV, respectively, with moderate agreement. Previous studies have reported that the overall agreement between QIAsymphony and other molecular assays based on automated sample preparation and real-time PCR was 86% (kappa=0.67) for CMV detection and 82.7% (kappa=0.64) for EBV detection, with good agreement, respectively [14,20]. Moreover, the EBV viral load detected by using the QIAsymphony RGQ system showed a negligible correlation with that detected by using the QIAcube system (Fig. 3).
PCR efficiency can be affected by several parameters, including sample type, primer and probe design, inaccurate sample and reagent pipetting, and inappropriate standard curves. PCR inhibitors present in samples include heparin, proteins such as hemoglobin, polysaccharides, and others [24]; a higher probability of PCR inhibition exists in WB samples than in the other sample types. Interestingly, the WB samples did not exhibit PCR inhibition using the QIAsymphony RGQ system; however, when the QIAcube system was applied, 23.9% of the WB samples showed PCR inhibition for EBV detection. QIAcube relies on silica-DNA binding for the removal of non-nucleic acid components. QIAsymphony uses magnetic-particle chemistry and built-in UV lamps that provide effective decontamination (Table 1). This may have caused the difference in PCR inhibition between the two systems. We were unable to determine the exact reason why PCR inhibition occurred only in EBV PCR and not in CMV PCR using the QIAcube system; further studies are required to elucidate the reasons for these differences.
Although viral load measurement using quantitative PCR is important in the prevention, diagnosis, and monitoring of EBV and CMV, multicenter evaluations of CMV and EBV NAAT have demonstrated significant inter-laboratory variability in viral quantitative reporting. Thus, international CMV and EBV DNA reference standard materials and units are needed to enable laboratories to achieve comparable numeric results [5,25,26,27]. In 2010 and 2012, the WHO produced the first international reference standard (NIBSC 09/162, 09/260, Hertfordshire, UK) for CMV and EBV viral load NAAT; the U.S. National Institute of Standards and Technology also provided standard reference material (SRM 2366) for CMV DNA that served as the basis for establishing metrological traceability of assay calibrants [28,29]. Although the availability of the international reference materials may standardize viral load reporting, several considerations remain that could independently or collectively account for assay-specific variability: differences in nucleic acid extraction methods, type and volume of clinical samples, selection of primers and probes, target-specific amplification efficiencies, detection chemistries and reagents, instrumentation, and operator-dependent variability [28,30,31,32].
The strength of this study lies in the comparison of both CMV and EBV DNA quantification PCR assays using the QIAsymphony RGQ and QIAcube systems under identical laboratory settings. The two systems exhibited significant differences for the detection of CMV and EBV DNA in clinical WB samples. The limitation of this study is that we used two different extraction methods; therefore, it is difficult to determine whether the difference was the result of the nucleic acid extraction process (including isolation methods), detection chemistries and reagents, instrumentation, or operator dependent variability. In addition, we were unable to determine the weakest step (extraction or amplification) of the QIAcube system; considering the manufacturer's instruction regarding intended use, it was impossible to test discrepant samples by combining the QIAcube extraction and the QIAsymphony platform quantitative PCR assay and vice-versa. Nevertheless, the present study demonstrates that the fully automated QIAsymphony RGQ nucleic acid extraction system is more effective for CMV and EBV DNA detection than a semi-automated QIAcube system. The QIAsymphony RGQ system has the potential to improve the agreement and clinical utility of CMV and EBV DNA measurements and to enable standardization and uniformity in clinical laboratories. A suitable sample preparation system should be considered for optimized nucleic acid amplification in clinical laboratories.
Acknowledgments
This work was supported by Konkuk University Medical Center Research Grant 2014.
Footnotes
Authors' Disclosures of Potential Conflicts of Interest: No potential conflicts of interest relevant to this article were reported.
References
- 1.Cannon MJ, Schmid DS, Hyde TB. Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol. 2010;20:202–213. doi: 10.1002/rmv.655. [DOI] [PubMed] [Google Scholar]
- 2.Chen CY, Huang KY, Shen JH, Tsao KC, Huang YC. A large-scale seroprevalence of Epstein-Barr virus in Taiwan. PLoS One. 2015;10:e0115836. doi: 10.1371/journal.pone.0115836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanakry JA, Hegde AM, Durand CM, Massie AB, Greer AE, Ambinder RF, et al. The clinical significance of EBV DNA in the plasma and peripheral blood mononuclear cells of patients with or without EBV diseases. Blood. 2016;127:2007–2017. doi: 10.1182/blood-2015-09-672030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotton CN, Kumar D, Caliendo AM, Asberg A, Chou S, Danziger-Isakov L, et al. Updated international consensus guidelines on the management of cytomegalovirus in solid-organ transplantation. Transplantation. 2013;96:333–360. doi: 10.1097/TP.0b013e31829df29d. [DOI] [PubMed] [Google Scholar]
- 5.Gulley ML, Tang W. Using Epstein-Barr viral load assays to diagnose, monitor, and prevent posttransplant lymphoproliferative disorder. Clin Microbiol Rev. 2010;23:350–366. doi: 10.1128/CMR.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thatcher SA. DNA/RNA preparation for molecular detection. Clin Chem. 2015;61:89–99. doi: 10.1373/clinchem.2014.221374. [DOI] [PubMed] [Google Scholar]
- 7.Rahman MM, Elaissari A. Nucleic acid sample preparation for in vitro molecular diagnosis: from conventional techniques to biotechnology. Drug Discov Today. 2012;17:1199–1207. doi: 10.1016/j.drudis.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Koidl C, Bozic M, Marth E, Kessler HH. Detection of CMV DNA: is EDTA whole blood superior to EDTA plasma? J Virol Methods. 2008;154:210–212. doi: 10.1016/j.jviromet.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Jones S, Webb EM, Barry CP, Choi WS, Abravaya KB, Schneider GJ, et al. Commutability of cytomegalovirus WHO international standard in different matrices. J Clin Microbiol. 2016;54:1512–1519. doi: 10.1128/JCM.03292-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hakim H, Gibson C, Pan J, Srivastava K, Gu Z, Bankowski MJ, et al. Comparison of various blood compartments and reporting units for the detection and quantification of Epstein-Barr virus in peripheral blood. J Clin Microbiol. 2007;45:2151–2155. doi: 10.1128/JCM.02308-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verheyen J, Kaiser R, Bozic M, Timmen-Wego M, Maier BK, Kessler HH. Extraction of viral nucleic acids: comparison of five automated nucleic acid extraction platforms. J Clin Virol. 2012;54:255–259. doi: 10.1016/j.jcv.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Costa C, Mantovani S, Balloco C, Sidoti F, Fop F, Cavallo R. Comparison of two nucleic acid extraction and testing systems for HCMV-DNA detection and quantitation on whole blood specimens from transplant patients. J Virol Methods. 2013;193:579–582. doi: 10.1016/j.jviromet.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 13.Miller S, Seet H, Khan Y, Wright C, Nadarajah R. Comparison of QIAGEN automated nucleic acid extraction methods for CMV quantitative PCR testing. Am J Clin Pathol. 2010;133:558–563. doi: 10.1309/AJCPE5VZL1ONZHFJ. [DOI] [PubMed] [Google Scholar]
- 14.Pillet S, Bourlet T, Pozzetto B. Comparative evaluation of the QIAsymphony RGQ system with the easyMAG/R-gene combination for the quantitation of cytomegalovirus DNA load in whole blood. Virol J. 2012;9:231. doi: 10.1186/1743-422X-9-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forman M, Wilson A, Valsamakis A. Cytomegalovirus DNA quantification using an automated platform for nucleic acid extraction and real-time PCR assay setup. J Clin Microbiol. 2011;49:2703–2705. doi: 10.1128/JCM.00721-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raggam RB, Bozic M, Salzer HJ, Hammerschmidt S, Homberg C, Ruzicka K, et al. Rapid quantitation of cytomegalovirus DNA in whole blood by a new molecular assay based on automated sample preparation and real-time PCR. Med Microbiol Immunol. 2010;199:311–316. doi: 10.1007/s00430-010-0164-z. [DOI] [PubMed] [Google Scholar]
- 17.Lee AV, Atkinson C, Manuel RJ, Clark DA. Comparative evaluation of the QIAGEN QIAsymphony® SP system and bioMérieux NucliSens easyMAG automated extraction platforms in a clinical virology laboratory. J Clin Virol. 2011;52:339–343. doi: 10.1016/j.jcv.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 18.Buelow D, Sun Y, Tang L, Gu Z, Pounds S, Hayden R. Comparative evaluation of four real-time PCR methods for the quantitative detection of Epstein-Barr virus from whole blood specimens. J Mol Diagn. 2016;18:527–534. doi: 10.1016/j.jmoldx.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laus S, Kingsley LA, Green M, Wadowsky RM. Comparison of QIAsymphony automated and QIAamp manual DNA extraction systems for measuring Epstein-Barr virus DNA load in whole blood using real-time PCR. J Mol Diagn. 2011;13:695–700. doi: 10.1016/j.jmoldx.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raggam RB, Wagner J, Bozic M, Michelin BD, Hammerschmidt S, Homberg C, et al. Detection and quantitation of Epstein-Barr virus (EBV) DNA in EDTA whole blood samples using automated sample preparation and real time PCR. Clin Chem Lab Med. 2010;48:413–418. doi: 10.1515/CCLM.2010.064. [DOI] [PubMed] [Google Scholar]
- 21.Semenova T, Lupo J, Alain S, Perrin-Confort G, Grossi L, Dimier J, et al. Multicenter evaluation of whole-blood Epstein-Barr viral load standardization using the WHO international standard. J Clin Microbiol. 2016;54:1746–1750. doi: 10.1128/JCM.03336-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.CLSI. Evaluation of detection capability for clinical laboratory measurement procedures; approved guideline-Second ed. CLSI document EP17-A2. Wayne, PA: Clinical and Laboratory Standards Institute; 2012. [Google Scholar]
- 23.Mukaka MM. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med J. 2012;24:69–71. [PMC free article] [PubMed] [Google Scholar]
- 24.Schrader C, Schielke A, Ellerbroek L, Johne R. PCR inhibitors - occurrence, properties and removal. J Appl Microbiol. 2012;113:1014–1026. doi: 10.1111/j.1365-2672.2012.05384.x. [DOI] [PubMed] [Google Scholar]
- 25.Pang XL, Fox JD, Fenton JM, Miller GG, Caliendo AM, Preiksaitis JK, et al. Interlaboratory comparison of cytomegalovirus viral load assays. Am J Transplant. 2009;9:258–268. doi: 10.1111/j.1600-6143.2008.02513.x. [DOI] [PubMed] [Google Scholar]
- 26.Wolff DJ, Heaney DL, Neuwald PD, Stellrecht KA, Press RD. Multi-Site PCR-based CMV viral load assessment-assays demonstrate linearity and precision, but lack numeric standardization: a report of the association for molecular pathology. J Mol Diagn. 2009;11:87–92. doi: 10.2353/jmoldx.2009.080097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gärtner B, Preiksaitis JK. EBV viral load detection in clinical virology. J Clin Virol. 2010;48:82–90. doi: 10.1016/j.jcv.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Razonable RR, Hayden RT. Clinical utility of viral load in management of cytomegalovirus infection after solid organ transplantation. Clin Microbiol Rev. 2013;26:703–727. doi: 10.1128/CMR.00015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.First WHO International Standard for Epstein–Barr Virus for Nucleic Acid Amplification Techniques. [Updated on Sep 2014]. https://www.nibsc.org/documents/ifu/09-260.pdf.
- 30.Hirsch HH, Lautenschlager I, Pinsky BA, Cardeñoso L, Aslam S, Cobb B, et al. An international multicenter performance analysis of cytomegalovirus load tests. Clin Infect Dis. 2013;56:367–373. doi: 10.1093/cid/cis900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayden RT, Yan X, Wick MT, Rodriguez AB, Xiong X, Ginocchio CC, et al. Factors contributing to variability of quantitative viral PCR results in proficiency testing samples: a multivariate analysis. J Clin Microbiol. 2012;50:337–345. doi: 10.1128/JCM.01287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park JE, Kim JY, Yun SA, Lee MK, Huh HJ, Kim JW, Ki CS. Performance evaluation of the Real-Q cytomegalovirus (CMV) Quantification Kit using two real-time PCR systems for quantifying CMV DNA in whole blood. Ann Lab Med. 2016;36:603–606. doi: 10.3343/alm.2016.36.6.603. [DOI] [PMC free article] [PubMed] [Google Scholar]