Abstract
Background
Stenotrophomonas maltophilia is one of several opportunistic pathogens of growing significance. Several studies on the molecular epidemiology of S. maltophilia have shown clinical isolates to be genetically diverse.
Materials and Methods
A total of 121 clinical isolates tentatively identified as S. malophilia from seven tertiary-care hospitals in Korea from 2007 to 2011 were included. Species and groups were identified using partial gyrB gene sequences and antimicrobial susceptibility testing was performed using a broth microdilution method. Multi locus variable number of tandem repeat analysis (MLVA) surveys are used for subtyping.
Results
Based on partial gyrB gene sequences, 118 isolates were identified as belonging to the S. maltophilia complex. For all S. maltophilia isolates, the resistance rates to trimethoprime-sulfamethoxazole (TMP/SMX) and levofloxacin were the highest (both, 30.5%). Resistance rate to ceftazidime was 28.0%. 11.0% and 11.9% of 118 S. maltophilia isolates displayed resistance to piperacillin/tazobactam and tigecycline, respectively. Clade 1 and Clade 2 were definitely distinguished from the data of MLVA with amplification of loci. All 118 isolates were classified into several clusters as its identification.
Conclusion
Because of high resistance rates to TMP/SMX and levofloxacin, the clinical laboratory department should consider providing the data about other antimicrobial agents and treatment of S. maltophilia infections with a combination of antimicrobials can be considered in the current practice. The MLVA evaluated in this study provides a fast, portable, relatively low cost genotyping method that can be employed in genotypic linkage or transmission networks comparing to analysis of the gyrB gene.
Keywords: Sternotrophomonas maltophilia, Trimethoprime-sulfamethoxazole, Multi locus variable number of tandem repeat analysis
Introduction
Stenotrophomonas maltophilia is one of several opportunistic pathogens of growing significance and is one of the most common nonfermentative Gram-negative bacillus isolated from clinical specimens [1,2]. The British Society for Antimicrobial Chemotherapy (BSAC, version 10.2, 2011) recommends disk diffusion and dilution testing for trimethoprim-sulfamethoxazole (TMP/SMX) only, while the Clinical and Laboratory Standards Institute (CLSI) recommends dilution testing for TMP/SMX, ceftazidime, chloramphenicol, levofloxacin, minocycline, and ticarcillin-clavulanate, and disk diffusion testing for only TMP/SMX, levofloxacin, and minocycline [3].
Several studies on the molecular epidemiology of S. maltophilia have shown clinical isolates to be genetically diverse [3,4]. Genotypic profiles have been determined by various methods, including restriction fragment length polymorphism analysis of the gyrB gene or the intergenic region between smeD and smeT genes, amplified fragment length polymorphism fingerprinting, Enterobacterial Repetitive Intergenic Consensus-PCR (ERIC-PCR), Multilocus sequence typing (MLST), and pulsed-field gel electrophoresis (PFGE) analysis [5,6]. Changes in the number of repeats among isolates can be checked by multilocus variable number of tandem repeat analysis (MLVA) surveys are used for subtyping purposes [5,7]. MLVA assays provide results that parallel PFGE data [5]. The MLVA technique involves amplification and size analysis of polymorphic DNA regions containing variable numbers of tandem repeated sequences, and is an established method to classify isolates of microbial species [8].
S. maltophilia GTAG (SMAGs) make up approximately 0.5% of the K279a genome, and are spread throughout the chromosome either as single units, or in pairs, separated by 5–80 bp long spacers [5]. The size of the SMAG family is shown for repetitive extragenic palindromic sequences [5]. SMAGs are reiterated in tandem at multiple chromosomal loci, along with tracts of variable length of DNA. The occurrence of SMAG arrays to set up PCR-based typing protocols, and comparison of the electrophoresis sizing observations was by DNA sequencing of selected PCR products.
Studies on the antimicrobial susceptibility of S. maltophilia treated with various antimicrobial agents including TMP/SMX, ceftazidime, chloramphenicol, levofloxacin, minocycline, piperacillin-tazobactam, ticarcillin-clavulanate, and tigecycline have been performed [3,4,9,10,11,12]. Infections caused by S. maltophilia are particularly difficult to manage because they show resistance to many classes of antimicrobial agents [9,10,11,12,13]. However, only two studies have focused on the antimicrobial susceptibility of S. maltophilia in Korea [3,14].
The aim of this study was to determine the antimicrobial susceptibility of recent clinical S. maltophilia complex isolates from Korea and the clonality of the clinical S. maltophilia should be assessed in order to detect genotype relationships. This study was undertaken to determine whether the strains could be rapidly and accurately genotyped with MLVA.
Materials and Methods
1. Bacterial isolates
A total of 121 clinical isolates tentatively identified as S. malophilia were included in this study. They were collected from seven tertiary-care hospitals in Korea from 2007 to 2011 and were identified conventionally using VITEK2 systems (bioMérieux, Inc., Haselwood, MO, USA) in the hospitals’ clinical microbiology laboratories. Among them, 85 isolates were from blood, and the others were from sputum (9 isolates), urine (8 isolates), endotracheal aspirate (5 isolates), transtracheal aspirate (5 isolates), bile (3 isolates), pericardial fluid (2 isolates), pus (2 isolates), and discharge from ear (1 isolate). The source of one isolate was unknown. 98 isolates were from intensive care unit.
2. Species identification
To identify the Stenotrophomonas spp. isolates, we determined the partial gyrB gene sequence of all isolates using XgyrB1F/XgyrB1R (5’-ACGAGTACAACCCGGACAA-3’ / 5’-CCCATCARGGTGCTGAAGAT-3’), which amplifies one of the variable regions of the gyrB gene, region2 [15,16]. Ambiguous gyrB gene sequences from three isolates were excluded in further analyses. We published these results previously [15].
3. Antimicrobial susceptibility testing
In vitro susceptibility testing was performed with 118 isolates identified as belonging to the S. maltophila complex in this study using the broth microdilution method according to the CLSI guidelines for five antimicrobial agents including ceftazidime, levofloxacin, piperacillin/tazobactam, TMP/SMX, and tigecycline [17]. The broth microdilution method always tends to give slightly higher minimum inhibitory concentrations (MICs), indicating its potency in the identification of isolates resistant to TMP/SMX because of trailing endpoints with TMP/SMX in the broth microdilution method [18]. TMP/SMX was tested by using the standard agar dilution method according to the CLSI guidelines [12]. The interpretive criteria used were those established in the CLSI standard M100-S21 [19]. Regarding tigecycline, interpretive criteria were defined based on the USA-FDA breakpoint criteria for Enterobacteriaceae (susceptible ≤2 mg/L, intermediate 4 mg/L, and resistant ≥8 mg/L). Escherichia coli ATCC25922 and Pseudomonas aeruginosa ATCC27853 were used as control strains. Multi-drug resistant (MDR) isolate was defined as one showing resistance to two or more antimicrobial agents [1].
4. Multilocus variable number of tandem repeat analysis (MLVA)
The DNA of single colonies derived from the final subcultures was analyzed by PCR amplification of DNA regions of interest. Genomic DNA was extracted and PCR reactions were carried out by incubating 20 ng of DNA with 160 ng of each primer in the presence of dXTPs (200 nanomoles), 1.5mM magnesium chloride and the Taq DNA polymerase Recombinant (Invitrogen, Midland, ON, Canada). Because of the high GC content of the S. maltophilia genome (> 66%), all PCR reactions were carried out in GC-rich buffer (Roche, Alameda, CA, USA) [5]. The oligomers used as primers, and the annealing temperatures, are from the paper by Emanuela Roscetto et al, 2008 [5]. Samples were incubated at 95°C for 5 minutes, and subsequently for 1 minute at 95°C, 1 minute at the annealing temperature and 1 minute at 72°C, for a total of 3 cycles. At the end of the cycle, samples were kept at 72°C for 7 minutes before harvesting. PCR products were electrophoresed on 1.5–2% agarose gels in 0.5×TBE buffer (45 mM Tris pH 8, 45 mM Borate, 0.5 mM EDTA) at 120 V (constant voltage). The 100 bp ladder (Fermentas, Carlsbad, CA, USA) was used as molecular weight marker [13]. PCR products were resolved by agarose electrophoresis in 1.5% multipurpose agarose gel (Fermentas, France), stained with ethidium bromide and photographed under UV light (BioRad, Berkeley, CA, USA).
PCR fragment length estimation of the SMAGs alleles was by reference to a 100-bp DNA size standard. Confirmation of the electrophoresis sizing observations was by DNA sequencing of selected PCR products.
5. Data analysis and statistics.
Cluster analysis of the MLVA typing data was performed in PHYLOVIZ softwear (downloading from http://www.phyloviz.net/) with the results of amplification and size analysis of polymorphic DNA regions containing variable numbers of tandemly repeated sequences of S. maltophilia complex isolates. Data were analyzed using SPSS 11.0 for Windows 2000 (SPSS, Chicago, IL, USA). Categorical data were tested using Chi-square analysis. Differences were considered statistically significant at a P-value of <0.05 for all tests.
Results
1. Antimicrobial susceptibility testing
Table 1 shows the MICs of antimicrobial agents and the resistance rates of the S. maltophilia complex isolates tested. For all S. maltophilia complex isolates, the resistance rates to TMP/SMX and levofloxacin were the highest (both were 30.5%) among five antimicrobial agents (Table 1). MIC50 and MIC90 of TMP/SMX were 2/38 and 32/608 mg/L, respectively. Resistance rate to ceftazidime was 28.0%. 11.0% and 11.9% of 118 S. maltophilia complex isolates displayed resistance to piperacillin/tazobactam and tigecycline, respectively. Thirty-eight MDR isolates (32.2%) were identified.
Table 1. Antimicrobial activity of antimicrobial agents against the 118 tested clinical Stenotrophomonas maltophilia complex isolates.
| Agent | MIC (µg/mL) | Number of isolates (%) | ||
|---|---|---|---|---|
| MIC50 | MIC90 | Susceptible | Resistant | |
| Ceftazidime | 8 | 64 | 70 (59.3) | 33 (28.0) |
| Levofloxacin | 2 | 16 | 61 (51.7) | 36 (30.5) |
| TMP/SMX | 2/38 | 32/608 | 82 (69.5)a | 36 (30.5)a |
| 65 (55.1) | 53 (44.9) | |||
| TZP | 16/4 | 128/4 | 84 (71.2) | 13 (11.0) |
| Tigecycline | 2 | 8 | 83 (70.3) | 14 (11.9) |
| MDR | 38 (32.2) | |||
aResults from standard agar dilution methods
MIC, minimum inhibitory concentration; TMP/SMX, trimethoprim-sulfamethoxazole; TZP, piperacillin/tazobactam; MDR, multidrug resistance
2. Multilocus variable number of tandem repeat analysis (MLVA)
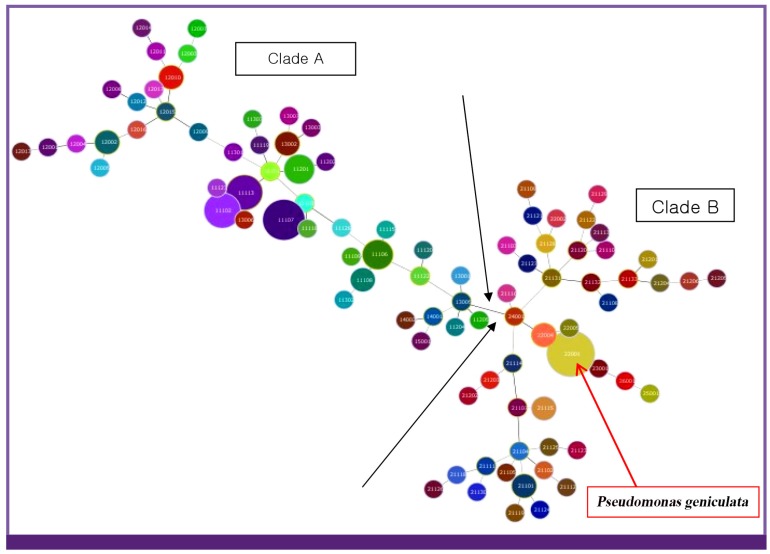
Considering previous identification of S. maltophilia complex, there were scattered clustering of S. maltophilia complex without relationship to identification in all data from amplification of 12 selected SMAGs loci, because the results showed that the majority of the isolates were not amplified successfully in loci I, IV, VI, and X. The number of alleles at each of the loci ranged from 0 to 21 numbers of allele repeat units on the 118 isolates analyzed (Table 2). The clonal profiles of the 118 S. maltophilia isolates were determined by random primer PCR fingerprinting and divided into two groups (clade A and B) as described previous study [15]. Antimicrobial resistance rates were varied by species or groups of S. maltophilia complex. Isolates of Clade A showed significantly lower antimicrobial resistance rates than those of Clade B as previous study [15]. Clade A and Clade B were also definitely distinguished from the data of MLVA with amplification of loci. All 118 isolates were classified into several clusters as its identification (Fig. 1).
Table 2. MLVA data from amplification of 12 selected SMAGs loci of Stenotrophomonas maltophilia .
| No. | Classification No. | gyrB | Species | I | II | III | IV | V | VI | VII | VIII | IX | X | XI | XII |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 12001 | Spav | S. pavanii | 2 | 4 | 4 | 1 | 0 | 4 | 0 | 3 | ||||
| 2 | 21133 | Smal I | S. maltophilia I-2 | 6 | 1 | 1 | 1 | 3 | 12 | 4 | 1 | 5 | 4 | 5 | 3 |
| 3 | ambiguous gyrB gene sequences | ||||||||||||||
| 4 | 21201 | Smal I | S. maltophilia I-2 | 11 | 1 | 1 | 2 | 3 | 10 | 4 | 1 | 5 | 6 | 5 | 2 |
| 5 | 21101 | Smal I | S. maltophilia I-1 | 6 | 1 | 2 | 3 | 4 | 11 | 3 | 3 | 5 | 3 | 5 | 3 |
| 6 | 11101 | Smal II | S. maltophilia II-1 | 2 | 3 | 0 | 1 | 0 | 1 | 1 | 1 | ||||
| 7 | 13001 | Smal III | S. maltophilia III | 2 | 1 | 2 | 0 | 4 | 1 | 2 | 2 | 1 | |||
| 8 | 21102 | Smal I | S. maltophilia I-1 | 1 | 2 | 4 | 3 | 0 | 2 | 4 | 2 | ||||
| 9 | 21103 | Smal I | S. maltophilia I-1 | 11 | 1 | 2 | 3 | 4 | 12 | 4 | 1 | 6 | 18 | 4 | 3 |
| 10 | 21104 | Smal I | S. maltophilia I-1 | 1 | 2 | 4 | 3 | 0 | 2 | 4 | 3 | ||||
| 11 | 14001 | Pbet | P. beteli | 1 | 2 | 0 | 2 | 0 | 0 | 1 | 2 | 0 | 0 | ||
| 12 | 13002 | Smal III | S. maltophilia III | 2 | 1 | 0 | 15 | 0 | 1 | 1 | 1 | ||||
| 13 | 21105 | Smal I | S. maltophilia I-1 | 1 | 2 | 4 | 2 | 0 | 3 | 3 | 3 | ||||
| 14 | 22001 | Pgen | P. geniculata | 1 | 1 | 0 | 2 | 0 | 3 | 1 | 2 | 2 | 2 | 1 | |
| 15 | 21106 | Smal I | S. maltophilia I-1 | 8 | 1 | 2 | 4 | 4 | 3 | 3 | 3 | 5 | 5 | 5 | 3 |
| 16 | 21107 | Smal I | S. maltophilia I-1 | 1 | 2 | 2 | 3 | 0 | 2 | 4 | 3 | ||||
| 17 | 11301 | Smal II | S. maltophilia II-3 | 3 | 4 | 0 | 1 | 0 | 4 | 1 | 2 | ||||
| 18 | 11102 | Smal II | S. maltophilia II-1 | 2 | 3 | 1 | 1 | 0 | 1 | 1 | 2 | ||||
| 19 | 21202 | Smal I | S. maltophilia I-2 | 1 | 1 | 2 | 2 | 1 | 3 | 4 | 2 | ||||
| 20 | 11103 | Smal II | S. maltophilia II-1 | 2 | 3 | 0 | 1 | 0 | 2 | 1 | 4 | ||||
| 21 | 21108 | Smal I | S. maltophilia I-1 | 2 | 1 | 2 | 2 | 3 | 12 | 2 | 1 | 1 | 4 | 1 | 3 |
| 22 | 13003 | Smal III | S. maltophilia III | 2 | 1 | 0 | 21 | 0 | 1 | 1 | 1 | ||||
| 23 | 12002 | Spav | S. pavanii | 2 | 4 | 3 | 1 | 0 | 4 | 4 | 3 | ||||
| 24 | 12003 | Spav | S. pavanii | 2 | 4 | 3 | 3 | 0 | 3 | 5 | 2 | ||||
| 25 | 21109 | Smal I | S. maltophilia I-1 | 3 | 1 | 2 | 0 | 2 | 10 | 2 | 2 | 1 | 1 | 5 | 2 |
| 26 | 11201 | Smal II | S. maltophilia II-2 | 2 | 2 | 0 | 1 | 0 | 1 | 1 | 1 | ||||
| 27 | 12004 | Spav | S. pavanii | 2 | 4 | 3 | 1 | 0 | 4 | 0 | 3 | ||||
| 28 | 11104 | Smal II | S. maltophilia II-1 | 2 | 3 | 1 | 1 | 0 | 1 | 1 | 2 | ||||
| 29 | 11105 | Smal II | S. maltophilia II-1 | 2 | 3 | 1 | 1 | 0 | 1 | 1 | 2 | ||||
| 30 | 11302 | Smal II | S. maltophilia II-3 | 4 | 3 | 4 | 1 | 4 | 0 | 3 | 1 | 2 | 2 | 4 | |
| 31 | 11106 | Smal II | S. maltophilia II-1 | 2 | 3 | 2 | 2 | 4 | 1 | 1 | 1 | 2 | 4 | ||
| 32 | 23001 | Phib | P. hibisciola | 2 | 1 | 0 | 2 | 2 | 3 | 2 | 2 | 4 | 2 | 1 | |
| 33 | 11107 | Smal II | S. maltophilia II-1 | 2 | 3 | 1 | 1 | 0 | 2 | 1 | 4 | ||||
| 34 | 13004 | Smal III | S. maltophilia III | 2 | 1 | 0 | 15 | 0 | 1 | 1 | 1 | ||||
| 35 | 12005 | Spav | S. pavanii | 2 | 4 | 2 | 1 | 0 | 3 | 4 | 3 | ||||
| 36 | 11202 | Smal II | S. maltophilia II-2 | 2 | 2 | 0 | 1 | 0 | 1 | 1 | 3 | ||||
| 37 | 21110 | Smal I | S. maltophilia I-1 | 2 | 1 | 2 | 2 | 4 | 12 | 3 | 1 | 1 | 4 | 1 | 4 |
| 38 | 11108 | Smal II | S. maltophilia II-1 | 2 | 2 | 3 | 1 | 4 | 3 | 1 | 1 | 1 | 4 | 2 | 4 |
| 39 | 12006 | Spav | S. pavanii | 2 | 4 | 3 | 1 | 0 | 4 | 4 | 3 | ||||
| 40 | 21111 | Smal I | S. maltophilia I-1 | 1 | 2 | 3 | 3 | 0 | 1 | 2 | 3 | ||||
| 41 | 12007 | Spav | S. pavanii | 2 | 4 | 4 | 3 | 0 | 3 | 7 | 5 | ||||
| 42 | 11109 | Smal II | S. maltophilia II-1 | 5 | 2 | 3 | 2 | 3 | 4 | 1 | 1 | 1 | 4 | 2 | 4 |
| 43 | 12008 | Spav | S. pavanii | 2 | 4 | 1 | 2 | 0 | 4 | 6 | 2 | ||||
| 44 | 21112 | Smal I | S. maltophilia I-1 | 1 | 2 | 4 | 5 | 0 | 2 | 3 | 2 | ||||
| 45 | 21113 | Smal I | S. maltophilia I-1 | 1 | 2 | 4 | 5 | 1 | 4 | 3 | 3 | ||||
| 46 | 11110 | Smal II | S. maltophilia II-1 | 2 | 3 | 1 | 1 | 0 | 2 | 1 | 4 | ||||
| 47 | 12009 | Spav | S. pavanii | 2 | 4 | 1 | 3 | 0 | 4 | 6 | 2 | ||||
| 48 | 21203 | Smal I | S. maltophilia I-2 | 1 | 1 | 2 | 4 | 1 | 2 | 4 | 3 | ||||
| 49 | 11111 | Smal II | S. maltophilia II-1 | 2 | 3 | 1 | 1 | 0 | 2 | 1 | 4 | ||||
| 50 | 12010 | Spav | S. pavanii | 2 | 4 | 1 | 3 | 0 | 4 | 5 | 3 | ||||
| 51 | 11112 | Smal II | S. maltophilia II-1 | 2 | 3 | 1 | 1 | 0 | 2 | 1 | 4 | ||||
| 52 | 11113 | Smal II | S. maltophilia II-1 | 2 | 3 | 1 | 1 | 0 | 1 | 1 | 1 | ||||
| 53 | 21114 | Smal I | S. maltophilia I-1 | 1 | 2 | 2 | 4 | 0 | 2 | 4 | 3 | ||||
| 54 | 21115 | Smal I | S. maltophilia I-1 | 1 | 2 | 2 | 2 | 0 | 1 | 4 | 1 | ||||
| 55 | 11203 | Smal II | S. maltophilia II-2 | 2 | 2 | 0 | 1 | 0 | 1 | 1 | 1 | ||||
| 56 | 22002 | Pgen | P. geniculata | 1 | 0 | 2 | 0 | 3 | 1 | 1 | 1 | 2 | |||
| 57 | 21116 | Smal I | S. maltophilia I-1 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 3 | ||||
| 58 | 11303 | Smal II | S. maltophilia II-3 | 3 | 4 | 0 | 2 | 0 | 1 | 2 | 1 | ||||
| 59 | 21117 | Smal I | S. maltophilia I-1 | 1 | 2 | 2 | 2 | 0 | 1 | 4 | 1 | ||||
| 60 | 11114 | Smal II | S. maltophilia II-1 | 5 | 2 | 3 | 2 | 4 | 4 | 1 | 1 | 1 | 4 | 2 | 4 |
| 61 | 21118 | Smal I | S. maltophilia I-1 | 2 | 1 | 2 | 1 | 3 | 10 | 3 | 2 | 1 | 1 | 5 | 3 |
| 62 | 26001 | Smal IV | B0811-107 | 3 | 1 | 1 | 2 | 0 | 3 | 2 | 2 | 0 | 1 | ||
| 63 | 11115 | Smal II | S. maltophilia II-1 | 2 | 3 | 2 | 5 | 4 | 1 | 1 | 1 | 2 | 4 | ||
| 64 | 11116 | Smal II | S. maltophilia II-1 | 2 | 3 | 2 | 2 | 4 | 1 | 1 | 1 | 4 | 2 | 4 | |
| 65 | 21119 | Smal I | S. maltophilia I-1 | 11 | 1 | 2 | 2 | 4 | 3 | 4 | 3 | 5 | 5 | 5 | 4 |
| 66 | 11204 | Smal II | S. maltophilia II-2 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 1 | 2 | 4 | 2 | 1 |
| 67 | 21120 | Smal I | S. maltophilia I-1 | 1 | 1 | 2 | 2 | 4 | 12 | 4 | 1 | 1 | 4 | 1 | 3 |
| 68 | 22003 | Pgen | P. geniculata | 1 | 1 | 0 | 2 | 0 | 3 | 1 | 2 | 4 | 2 | 1 | |
| 69 | 21204 | Smal I | S. maltophilia I-2 | 4 | 1 | 1 | 1 | 3 | 10 | 5 | 1 | 5 | 5 | 5 | 3 |
| 70 | 22004 | Pgen | P. geniculata | 2 | 1 | 0 | 2 | 0 | 4 | 1 | 2 | 2 | 2 | 1 | |
| 71 | 12011 | Spav | S. pavanii | 2 | 4 | 3 | 2 | 0 | 4 | 5 | 3 | ||||
| 72 | 12012 | Spav | S. pavanii | 2 | 4 | 1 | 2 | 0 | 4 | 6 | 3 | ||||
| 73 | 11205 | Smal II | S. maltophilia II-2 | 2 | 2 | 2 | 2 | 2 | 0 | 3 | 1 | 2 | 2 | 1 | |
| 74 | 11117 | Smal II | S. maltophilia II-1 | 2 | 3 | 1 | 1 | 0 | 2 | 1 | 4 | ||||
| 75 | 21205 | Smal I | S. maltophilia I-2 | 5 | 1 | 1 | 1 | 2 | 10 | 6 | 2 | 5 | 4 | 5 | 4 |
| 76 | 12013 | Spav | S. pavanii | 2 | 2 | 4 | 4 | 6 | 4 | 0 | 7 | 9 | 3 | ||
| 77 | 13005 | Smal III | S. maltophilia III | 2 | 2 | 2 | 2 | 0 | 4 | 1 | 2 | 2 | 1 | ||
| 78 | 11206 | Smal II | S. maltophilia II-2 | 2 | 2 | 0 | 1 | 0 | 1 | 1 | 1 | ||||
| 79 | 12014 | Spav | S. pavanii | 2 | 4 | 0 | 2 | 0 | 4 | 5 | 3 | ||||
| 80 | 21121 | Smal I | S. maltophilia I-1 | 3 | 1 | 2 | 0 | 2 | 10 | 3 | 3 | 1 | 1 | 5 | 2 |
| 81 | 21122 | Smal I | S. maltophilia I-1 | 7 | 1 | 2 | 3 | 5 | 12 | 6 | 1 | 1 | 4 | 3 | |
| 82 | 13006 | Smal III | S. maltophilia III | 2 | 1 | 1 | 4 | 0 | 1 | 1 | 1 | ||||
| 83 | 13007 | Smal III | S. maltophilia III | 2 | 1 | 0 | 3 | 0 | 1 | 1 | |||||
| 84 | 25001 | 08-B-253 | 1 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | |||||
| 85 | 21123 | Smal I | S. maltophilia I-1 | 7 | 1 | 2 | 3 | 4 | 1 | 5 | 3 | 1 | 5 | 4 | 2 |
| 86 | 11118 | Smal II | S. maltophilia II-1 | 2 | 3 | 0 | 1 | 0 | 2 | 0 | 4 | ||||
| 87 | ambiguous gyrB gene sequences | 3 | 4 | ||||||||||||
| 88 | 12015 | Spav | S. pavanii | 2 | 4 | 1 | 3 | 0 | 4 | 6 | 3 | ||||
| 89 | 21124 | Smal I | S. maltophilia I-1 | 7 | 1 | 2 | 0 | 4 | 4 | 4 | 3 | 0 | 1 | 5 | 3 |
| 90 | 12016 | Spav | S. pavanii | 2 | 4 | 1 | 3 | 0 | 4 | 4 | 3 | ||||
| 91 | 12017 | Spav | S. pavanii | 2 | 4 | 3 | 3 | 0 | 4 | 1 | 3 | ||||
| 92 | 21125 | Smal I | S. maltophilia I-1 | 7 | 1 | 2 | 0 | 4 | 3 | 2 | 3 | 1 | 18 | 4 | 3 |
| 93 | 14002 | Pbet | P. beteli | 2 | 2 | 0 | 2 | 0 | 0 | 3 | 2 | 0 | 0 | ||
| 94 | 15001 | K01-43 | 2 | 2 | 0 | 2 | 0 | 1 | 2 | 2 | 0 | 3 | |||
| 95 | 11119 | Smal II | S. maltophilia II-1 | 2 | 3 | 0 | 1 | 0 | 1 | 2 | 1 | ||||
| 96 | 21126 | Smal I | S. maltophilia I-1 | 1 | 2 | 3 | 3 | 2 | 2 | 1 | 3 | ||||
| 97 | 11120 | Smal II | S. maltophilia II-1 | 3 | 2 | 3 | 2 | 2 | 3 | 1 | 1 | 2 | 2 | 5 | |
| 98 | 21206 | Smal I | S. maltophilia I-2 | 7 | 1 | 1 | 4 | 2 | 7 | 5 | 1 | 1 | 5 | 4 | |
| 99 | 22005 | Pgen | P. geniculata | 10 | 1 | 0 | 0 | 9 | 0 | 4 | 1 | 2 | 4 | 2 | |
| 100 | 11121 | Smal II | S. maltophilia II-1 | 2 | 3 | 1 | 1 | 0 | 1 | 1 | 1 | ||||
| 101 | 11122 | Smal II | S. maltophilia II-1 | 2 | 3 | 2 | 2 | 4 | 1 | 1 | 2 | 2 | 1 | ||
| 102 | 11123 | Smal II | S. maltophilia II-1 | 2 | 3 | 1 | 1 | 0 | 1 | 0 | 1 | ||||
| 103 | 22006 | Pgen | P. geniculata | 2 | 1 | 0 | 2 | 0 | 4 | 1 | 2 | 2 | 1 | ||
| 104 | ambiguous gyrB gene sequences | -5 | |||||||||||||
| 105 | 24001 | Sarf | S. africana | 2 | 1 | 2 | 2 | 0 | 4 | 1 | 2 | 2 | 1 | ||
| 106 | 21127 | Smal I | S. maltophilia I-1 | 2 | 1 | 2 | 2 | 2 | 7 | 4 | 1 | 6 | 5 | 1 | 3 |
| 107 | 11124 | Smal II | S. maltophilia II-1 | 2 | 3 | 1 | 1 | 0 | 1 | 1 | 1 | ||||
| 108 | 21128 | Smal I | S. maltophilia I-1 | 1 | 2 | 2 | 3 | 1 | 1 | 1 | 3 | ||||
| 109 | 21129 | Smal I | S. maltophilia I-1 | 8 | 1 | 2 | 3 | 5 | 6 | 6 | 1 | 3 | 5 | 4 | 2 |
| 110 | 22007 | Pgen | P. geniculata | 2 | 1 | 0 | 2 | 0 | 3 | 1 | 2 | 4 | 2 | 1 | |
| 111 | 11125 | Smal II | S. maltophilia II-1 | 2 | 3 | 1 | 2 | 4 | 1 | 1 | 1 | 4 | 2 | 4 | |
| 112 | 22008 | Pgen | P. geniculata | 1 | 1 | 0 | 1 | 2 | 0 | 3 | 1 | 2 | 4 | 2 | 1 |
| 113 | 11126 | Smal II | S. maltophilia II-1 | 2 | 3 | 1 | 1 | 0 | 1 | 1 | 1 | ||||
| 114 | 11127 | Smal II | S. maltophilia II-1 | 2 | 3 | 1 | 1 | 0 | 1 | 1 | 2 | ||||
| 115 | 12018 | Spav | S. pavanii | 2 | 4 | 1 | 3 | 0 | 4 | 5 | 3 | ||||
| 116 | 22009 | Pgen | P. geniculata | 1 | 1 | 0 | 2 | 0 | 3 | 1 | 2 | 3 | 2 | 1 | |
| 117 | 21130 | Smal I | S. maltophilia I-1 | 1 | 2 | 3 | 5 | 0 | 2 | 2 | 4 | ||||
| 118 | 21131 | Smal I | S. maltophilia I-1 | 2 | 1 | 2 | 1 | 2 | 7 | 4 | 1 | 3 | 2 | 1 | 3 |
| 119 | 22010 | Pgen | P. geniculata | 1 | 1 | 0 | 2 | 0 | 3 | 1 | 2 | 2 | 1 | ||
| 120 | 21132 | Smal I | S. maltophilia I-1 | 2 | 1 | 2 | 1 | 3 | 7 | 4 | 1 | 3 | 2 | 1 | 3 |
| 121 | 11128 | Smal II | S. maltophilia II-1 | 2 | 3 | 2 | 1 | 0 | 2 | 1 | 4 | ||||
The PCR products derived from the amplification of loci I to XII in the listed strains are labeled with the number of SMAG repeats present.
MLVA, multi locus variable number of tandem repeat analysis; SMAG, Sternotrophomonas maltophilia GTAG; PCR, Polymerase Chain Reaction.
Figure 1.
Dendrogram demonstrating genetic disimilarity of 118 Stenotrophomonas maltophilia complex isolates based on MLVA genotyping method in amplifications of all eight SMAGs loci.
Considering previous identification of S. maltophilia complex, there were also scatterred clustering of S. maltophilia complex with relationship to identification in all data from eight fully amplified loci II, III, V, VII, VIII, IX, XI and XII. All Pseudomonas geniculata were from one single hospital from previous identification study (Red arrow in Fig. 1). With hierarchical clustering, the MLVA profiles of the members of the 118 S. maltophilia complex were classified in all data from twelve SMAGs loci.
Discussion
TMP/SMX alone, or in combination with other agents is still considered the treatment of choice for suspected or culture-proven S. maltophilia infections. Resistance rates to TMP/SMX have been reported to vary geographically [1,2,3,14,19], but were generally less than 20%. However, isolates from cystic fibrosis patients and from patients in some Asian countries, such as Taiwan, as well as Turkey, showed high resistance rates (31.3-100%) [19,20]. The most notable finding in this study is the high resistance rate to TMP/SMX. Worldwide surveillance studies have shown relatively low resistance rates, varying from 4% to 20% [1,2,3,12,14,20]. A recent report from a Korean hospital also documented a low resistance rate of 6% to TMP/SMX using the agar dilution method [3]. For all clinical S. maltophilia complex isolates, the resistance rate to TMP/SMX was 45.5% with the broth microdilution test and 29.8% with the agar dilution test, which was an unexpected result. We re-confirmed that the broth microdilution method always tends to give slightly higher MICs, indicating its potency in identifying isolates resistant to TMP/SMX because of trailing endpoints with TMP/SMX in the broth microdilution method [18]. So, the broth microdilution method can be somewhat useful in clinical practice to detect potential resistant isolate. In the agar dilution test, the resistance rate to TMP/SMX was 30.5%. This rate was much higher than the rate in previous studies, although the patient group in this study did not include patients with cystic fibrosis who usually show high resistance rates to TMP/SMX according to previous studies. The resistance rate to levofloxacine was 30.5%, which was higher than the rates of 3 to 13% seen in a previous studies [3,12,13,14,20,21,22]. The tetracycline derivatives minocycline and tigecycline have shown good in vitro activity against clinical isolates of S. maltophilia [3]. Previous studies in Taiwan, Brazil, Spain, and the USA showed that there was no or low resistance to tigecycline [3,20,21,22]. However, this study showed that 11.9% of clinical 118 S. maltophilia complex isolates displayed resistance to tigecycline. The high resistance rates to TMP/SMX, levefloxacin, and tigecycline in this study should be noted.
The isolates showed low rates of susceptibilities (23 to 43%) to ceftazidime, ticarcillin-clavulanic acid, and piperacillin/tazobactam [23]. However, susceptibility rates to piperacillin/tazobactam and ceftazidime in this study were 71.2% and 59.3%, respectively, which were higher than the susceptibility rates in other studies [2,3].
As a whole, the most notable finding in this study is the high resistance rate to TMP/SMX. The differences in the TMP/SMX resistance rates between studies in Korea may be due to differences in tertiary hospitals, isolation periods, and numbers of isolates. In addition, most of our isolates (98 isolate) were from intensive care units. The high TMP/SMX resistance rate found in this study is of concern because the preferred treatment option of S. maltophilia infections is TMP/SMX [1]. Thus, continuous surveillance of antimicrobial resistance of S. maltophilia is recommended. In contrast to the high resistance to TMP/SMX and levofloxacin, piperacillin/tazobactam and tigecycline showed potent activities against S. maltophilia complex isolates. The resistance rate to the tigecycline was 11.9%, which was the highest resistance rate in the world. This relatively high resistance rate may lead to tigecycline being considered an alternative therapeutic option as a component of combination therapy [1,2]. Treatment strategies have included the use of select antibiotics in synergy [1]. Piperacillin/tazobactam can be a candidate for combination therapy in treating S. maltophilia because isolates show a high susceptibility rate to this combination. However, piperacillin/tazobactam cannot be used as monotherapeutic drugs to treat S. maltophilia because this microorganism has a high intrinsic resistance to most penicillins and cephalosporins, as well as to all carbapenems. Treatment of S. maltophilia infections with a combination of two or three antimicrobials can be considered in the current practice because S. maltophilia has a high resistance rate to TMP/SMX and levofloxacin. In clinical practice, the clinical laboratory department usually provides information about susceptibility to TMP/SMX and levofloxacin. Physicians usually choose one of these agents or both agents. Therefore, the clinical laboratory department should consider providing the data about other antimicrobial agents such as ceftazidime, piperacillin/tazobactam, and tigecycline to physicians.
We evaluated an MLVA to assess the molecular epidemiology of S.maltophilia complex. Studies based on MLST demonstrated that S. maltophilia isolates were heterogeneous, because several previously proposed species are recognized to be closely related with S. maltophilia (It might be referred to as ‘S. maltophilia complex’ including Stenotrophomonas pavanii, Stenotrophomonas africana, Pseudomonas geniculata, P. hibisciola, and P. beteli) [16,24,25]. Loci I, IV, VI, and X were not amplified successfully comparing to reference article [5]. We might postulate that unsuccessful amplification might related to genetically diversity of S. maltophilia clinical isolates [3,4,15,16,23,24]. In this study, MLVA of S. maltophilia isolates was somewhat heterogeneous, but large groups were distinguishable as clade A and B. Although the clustering based on MLVA was not found to link to any characteristics of isolates yet, it is important that the selected SMAGs loci used for typing S. maltophilia complex. The results of this study suggested that MLVA exhibited higher resolution in using SMAGs loci amplification. Amplification locus and the number of amplified loci are essential to investigate to obtain sufficiently large clusters without obscuring genotypic links. Epidemiologically, S. maltophilia isolates are genetically diverse, and thus clonal dissemination may be rare [16]. These results are in accordance with earlier studies demonstrating an elevated genetic diversity in S. maltophilia isolates even when recovered from the same hospital [6,23]. Although the isolates examined here exhibited high heterogeneity, homologous isolates (e.g., all P. geniculata from one hospital in Seoul) could be recovered from different patients on different ward and at different time points, which may suggest the settle-down of this species in particular hospital and the possibility horizontal transmission among patients (Table 3). These findings further reinforce the postulation that while S. maltophilia could be acquired by diverse routes, cross-transmission is also possible. Hence, management of S. maltophilia infections would be problematic as the agents may not behave uniformly and transmission may include multiple drug-resistant isolates. These results support efforts directed towards continuous surveillance for antimicrobial drug resistance and epidemiological monitoring, which may act as early warning systems for predicting resistance and preventing outbreaks. The MLVA evaluated in this study provides a fast, portable, relatively low cost genotyping method that can be employed in genotypic linkage or transmission networks comparing to gyrB analysis.
Table 3. Sources and antimicrobial activity of Pseudomonas geniculata from single hospital.
| No. | Collection year | Ward | Source | MIC (ug/mL) | ||||
|---|---|---|---|---|---|---|---|---|
| Tigecycline | Ceftazidime | TMP/SMX | Levofloxacin | TZP | ||||
| 14 | 2006 | ICU | Blood | 0.25 | 16 | 4 : 76 | 1 | 16 : 4 |
| 58 | 2008 | ICU | Blood | 4 | 16 | 64 : 1216 | 16 | 16 : 4 |
| 71 | 2009 | ICU | Blood | 4 | 32 | 16 : 304 | 1 | 16 : 4 |
| 73 | 2009 | GW | Blood | 0.5 | 8 | 0.25 : 4.75 | 1 | 4 : 4 |
| 102 | 2008 | ICU | Sputum | 8 | >64 | 64 : 1216 | 16 | 64 : 4 |
| 106 | 2010 | GW | Pus | 2 | >64 | 4 : 76 | 2 | > 256 : 4 |
| 113 | 2011 | GW | Blood | 1 | 4 | 32 : 608 | 2 | 8 : 4 |
| 115 | 2011 | GW | Sputum | 4 | >64 | 16 : 304 | 4 | > 256 : 4 |
| 119 | 2011 | ICU | Blood | 16 | >64 | 16 : 304 | 32 | > 256 : 4 |
| 122 | 2011 | ICU | Blood | 16 | >64 | 32 : 608 | 64 | 256 : 4 |
MIC, minimum inhibitory concentration; TMP/SMX, trimethoprim-sulfamethoxazole; TZP, piperacillin/tazobactam; ICU, intensive care unit; GW, general ward.
Footnotes
Conflict of Interest: No conflicts of interest.
References
- 1.Brooke JS. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev. 2012;25:2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Looney WJ, Narita M, Mühlemann K. Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect Dis. 2009;9:312–323. doi: 10.1016/S1473-3099(09)70083-0. [DOI] [PubMed] [Google Scholar]
- 3.Chung HS, Hong SG, Lee Y, Kim M, Yong D, Jeong SH, Lee K, Chong Y. Antimicrobial susceptibility of Stenotrophomonas maltophilia isolates from a Korean tertiary care hospital. Yonsei Med J. 2012;53:439–441. doi: 10.3349/ymj.2012.53.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nicodemo AC, Araujo MR, Ruiz AS, Gales AC. In vitro susceptibility of Stenotrophomonas maltophilia isolates: comparison of disc diffusion, Etest and agar dilution methods. J Antimicrob Chemother. 2004;53:604–608. doi: 10.1093/jac/dkh128. [DOI] [PubMed] [Google Scholar]
- 5.Roscetto E, Rocco F, Carlomagno MS, Casalino M, Colonna B, Zarrilli R, Di Nocera PP. PCR-based rapid genotyping of Stenotrophomonas maltophilia isolates. BMC Microbiol. 2008;8:202. doi: 10.1186/1471-2180-8-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valdezate S, Vindel A, Martín-Dávila P, Del Saz BS, Baquero F, Cantón R. High genetic diversity among Stenotrophomonas maltophilia strains despite their originating at a single hospital. J Clin Microbiol. 2004;42:693–699. doi: 10.1128/JCM.42.2.693-699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho S, Boxrud DJ, Bartkus JM, Whittam TS, Saeed M. Multiple-locus variable-number tandem repeat analysis of Salmonella Enteritidis isolates from human and non-human sources using a single multiplex PCR. FEMS Microbiol Lett. 2007;275:16–23. doi: 10.1111/j.1574-6968.2007.00875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Belkum A. Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA) FEMS Immunol Med Microbiol. 2007;49:22–27. doi: 10.1111/j.1574-695X.2006.00173.x. [DOI] [PubMed] [Google Scholar]
- 9.Tan CK, Liaw SJ, Yu CJ, Teng LJ, Hsueh PR. Extensively drug-resistant Stenotrophomonas maltophilia in a tertiary care hospital in Taiwan: microbiologic characteristics, clinical features, and outcomes. Diagn Microbiol Infect Dis. 2008;60:205–210. doi: 10.1016/j.diagmicrobio.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Toleman MA, Bennett PM, Bennett DM, Jones RN, Walsh TR. Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul gene. Emerg Infect Dis. 2007;13:559–565. doi: 10.3201/eid1304.061378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fihman V, Le Monnier A, Corvec S, Jaureguy F, Tankovic J, Jacquier H, Carbonnelle E, Bille E, Illiaquer M, Cattoir V, Zahar JR. Stenotrophomonas maltophiliae-the most worrisome threat among unusual non-fermentative gram-negative bacilli from hospitalized patients: a prospective multicenter study. J Infect. 2012;64:391–398. doi: 10.1016/j.jinf.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Samonis G, Karageorgopoulos DE, Maraki S, Levis P, Dimopoulou D, Spernovasilis NA, Kofteridis DP, Falagas ME. Stenotrophomonas maltophilia infections in a general hospital: patient characteristics, antimicrobial susceptibility, and treatment outcome. PLoS One. 2012;7:e37375. doi: 10.1371/journal.pone.0037375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantó R, Valdezate S, Vindel A, Sáchez Del Saz B, Maíz L, Baquero F. Antimicrobial susceptibility profile of molecular typed cystic fibrosis Stenotrophomonas maltophilia isolates and differences with noncystic fibrosis isolates. Pediatr Pulmonol. 2003;35:99–107. doi: 10.1002/ppul.10216. [DOI] [PubMed] [Google Scholar]
- 14.Song JH, Sung JY, Kwon KC, Park JW, Cho HH, Shin SY, Ko YH, Kim JM, Shin KS, Koo SH. Analysis of acquired resistance genes in Stenotrophomonas maltophilia . Korean J Lab Med. 2010;30:295–300. doi: 10.3343/kjlm.2010.30.3.295. [DOI] [PubMed] [Google Scholar]
- 15.Rhee JY, Choi JY, Choi MJ, Song JH, Peck KR, Ko KS. Distinct groups and antimicrobial resistance of clinical Stenotrophomonas maltophilia complex isolates from Korea. J Med Microbiol. 2013;62:748–753. doi: 10.1099/jmm.0.053355-0. [DOI] [PubMed] [Google Scholar]
- 16.Svensson-Stadler LA, Mihaylova SA, Moore ER. Stenotrophomonas interspecies differentiation and identification by gyrB sequence analysis. FEMS Microbiol Lett. 2012;327:15–24. doi: 10.1111/j.1574-6968.2011.02452.x. [DOI] [PubMed] [Google Scholar]
- 17.Valenza G, Tappe D, Turnwald D, Frosch M, König C, Hebestreit H, Abele-Horn M. Prevalence and antimicrobial susceptibility of microorganisms isolated from sputa of patients with cystic fibrosis. J Cyst Fibros. 2008;7:123–127. doi: 10.1016/j.jcf.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Luber P, Bartelt E, Genschow E, Wagner J, Hahn H. Comparison of Broth Microdilution, E Test, and agar dilution methods for antibiotic susceptibility testing of Campylobacter jejuni and Campylobacter coli . J Clin Microbiol. 2003;41:1062–1068. doi: 10.1128/JCM.41.3.1062-1068.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing, 21st informational supplement. Wayne, PA: CLSI; 2011. p. M100-S21. [Google Scholar]
- 20.Wang WS, Liu CP, Lee CM, Huang FY. Stenotrophomonas maltophilia bacteremia in adults: four years’ experience in a medical center in northern Taiwan. J Microbiol Immunol Infect. 2004;37:359–365. [PubMed] [Google Scholar]
- 21.Galles AC, Jones RN, Sader HS. Antimicrobial susceptibility profile of contemporary clinical strains of Stenotrophomonas maltophilia isolates: can moxifloxacin activity be predicted by levofloxacin MIC results? J Chemother. 2008;20:38–42. doi: 10.1179/joc.2008.20.1.38. [DOI] [PubMed] [Google Scholar]
- 22.Vartivarian S, Anaissie E, Bodey G, Sprigg H, Rolston K. A changing pattern of susceptibility of Xanthomonas maltophilia to antimicrobial agents: implications for therapy. Antimicrob Agents Chemother. 1994;38:624–627. doi: 10.1128/aac.38.3.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gülmez D, Hascelik G. Stenotrophomonas maltophilia: antimicrobial resistance and molecular typing of an emerging pathogen in a Turkish university hospital. Clin Microbiol Infect. 2005;11:880–886. doi: 10.1111/j.1469-0691.2005.01257.x. [DOI] [PubMed] [Google Scholar]
- 24.Cho HH, Sung JY, Kwon KC, Koo SH. Expression of Sme efflux pumps and multilocus sequence typing in clinical isolates of Stenotrophomonas maltophilia . Ann Lab Med. 2012;32:38–43. doi: 10.3343/alm.2012.32.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaiser S, Biehler K, Jonas D. Stenotrophomonas maltophilia multilocus sequence typing scheme for inferring population structure. J Bacteriol. 2009;91:2934–2943. doi: 10.1128/JB.00892-08. [DOI] [PMC free article] [PubMed] [Google Scholar]