Abstract
Background:
The prevalence of resistant Pseudomonas aeruginosa isolates is increasing and it is considered as one of the major public health concerns in the world. The association between integrons and drug resistance has been proven and evidences suggest that integrons are coding and responsible for dissemination of antibiotic resistance among P. aeruginosa isolates.
Objective:
This study is aimed to evaluate the relationship between class 1 integrons and drug resistance genes in clinical isolates of P. aeruginosa from burn patients.
Methods:
100 isolates of P. aeruginosa were collected from burn patients hospitalized in the skin ward of Shahid Motahari hospital and susceptibility testing was performed by disk diffusion method (Kirby-Bauer). Then DNA was extracted and PCR technique was performed for the detection of class 1 integrons and drug resistance genes. Then data was analyzed using SPSS software.
Results:
The most effective antibiotic was polymyxin B with sensitivity 100%, and the most resistance was observed to the ciprofloxacin (93%) and amikacin (67%), respectively. The maximum and lowest frequencies of drug resistance genes belonged to the aac (6 ') - 1, VEB-1 with prevalence rate 93% and 10%, respectively. The statistical Chi-square test did not find any significant correlation between class 1 integrons and drug resistance genes (p˃ 0.05).
Conclusion:
Although no significant correlation between class 1 integrons and drug resistance was observed, but the resistance rate to antibiotics tested among P. aeruginosa isolates was high. So, surveillance, optimization and strict consideration of antimicrobial use and control of infection are necessary.
Keywords: Burn ward, Integrons, Resistance, Pseudomonas aeruginosa
INTRODUCTION
Pseudomonas aeruginosa is the common cause of nosocomial infection, particularly in burn wounds [1]. One of the most important health concerns in many parts of the world is the treatment of burn wounds because contact with opportunistic bacteria such as Pseudomonas in patients with these wounds puts them at risk of nosocomial infections. Sometimes, for reasons such as colonization, in burn wounds with Pseudomonas or lack of proper neutrophilic response to tissue invasion, the microorganisms can even take the patient to the phase of bacteremia and sepsis that can result in death of the patient. The sepsis resulting from this bacterium is a serious complication after burn [2]. Sepsis risk is increased proportional to the degree of the burn wound [3]. Recently, the use of new surveillance methods has reduced infections in burn wounds. However, there is still a possibility of fatal infections in severe burns, especially in developing countries. Generally, they cause an increase in disease and mortality throughout the world [1]. Although, the antibiotics are the main way to control the infections but one of the most worrying features of P. aeruginosa is low biological sensitivity to antibiotics. Meanwhile, of which, the selective pressure of an inappropriate antibiotic user and increased dose are the greatest causes. The prevalence of P. aeruginosa resistant isolates is increasing and it is considered one of the major public health concerns in the world. The most serious problems resulting from increasing resistance are morbidity, mortality and restrictions of economic costs for therapeutic use. Regarding the importance of P. aeruginosa in burn wound infections, pneumonia, the urinary tract, meningitis and in high resistance to drugs and disinfectants as well as the potential threat for patients with impaired immune systems, detection of antibiotic resistance genes was performed in all parts of the world and, to determine the appropriate pattern, reporting these results is very important [4]. Mobile elements such as plasmids, transposons and integrons are the most effective genetic elements which have a role in acquiring and spreading resistance of gram-negative bacteria. In this regard, several studies show that multidrug resistance in these bacteria is significantly associated with integrons and gene cassettes [5]. In hospitals, multi-resistant strains of P. aeruginosa have been isolated from the environment and from personnel. Studies carried out in Iran have found that the incidence of multi-resistant P. aeruginosa isolated from nosocomial infections has reached alarming levels [6]. The association between integrons and drug resistance has been proven and evidence suggests that integrons are coding and responsible for dissemination of antibiotic resistance among P. aeruginosa isolates. For this reason, this study is aimed to evaluate the relationship between Class 1 integrons and drug resistance genes in clinical isolates of P. aeruginosa from burn patients.
MATERIALS AND METHODS
Source of Isolates and Susceptibility Testing of Isolates Using Disk Diffusion Method
Isolates were collected from burn patients hospitalized in the skin ward of Shahid Motahari Hospital. One hundred P. aeruginosa strains were isolated and confirmed by different biochemical and microbiologic differential tests. Standard strain PAO1 of P. aeruginosa and E. coli with ATCC 25922 were applied as controls.
As has already been mentioned, susceptibility testing was performed by disk diffusion method (Kirby-Bauer) [7], in accordance with the standards of CLSI 2014.8 The accuracy of antibiotic disks was approved using standard strains of E. coli with ATCC 25922. Antibiotic disks used in this study consisted of polymyxin B (300 units), amikacin (30 μg), tobramycin (10 μg), ciprofloxacin (5 μg), gentamicin (10 μg) and ceftazidime (30 μg) (Company MAST).
Bioinformatics Study
Selection of one target gene and primers for detection of resistance genes were performed according to conducted scientific studies and bioinformatics software. After designing, they were evaluated by in silico PCR Amplification Software [8, 9]. Table 1 shows the characteristics of primers used for PCR performance. The conditions for PCR are abstracted in Table 2.
Table 1. The characteristics of used primers for PCR performance.
| Name of primers | Primers sequence(5′-3′) | Amplicon length(bp) | Target gene | s |
|---|---|---|---|---|
| PF-per1 | ATGAATGTCATTATAAAAGC | 933 | per1 | (10) |
| PR-per1 | AATTTGGGCTTAGGGCAGAA | |||
| PF_VIM1 | TTGCAATGCTGAATGGAGAG | 182 | VIM | This study |
| PR_VIM1 | CAACTCAACCAGTGCTCGAA | |||
| PF_ aac(6')-1 | CCCAGTGGACATAAGCCTGT | 229 | aac (6')-1 | This Study |
| PR_ aac(6')-1 | GTAACATCGTTGCTGCTCCA | |||
| PF-TEM1 | ATAAAATTCTTGAAGACGAAA | 1080 | TEM1 | (11) |
| PR_TEM1 | GACAGTTACCAATGCTTAATCA | |||
| PF_VEB1 | CGACTTCCATTTCCCGATGC | 634 | VEB1 | (12) |
| PR_VEB1 | GGACTCTGCAACAAATACGC | |||
| PF_SHV-1 | CTGCTCCACCAGGGCTAAAA | 576 | SHV | This Study |
| PR_SHV-1 | ATGAACGCGCAAATAGCGAC | |||
| PF_AMPC | CTCGACCTCGCGACCTATAC | 150 | AMPC | This Study |
| PR_AMPC | GATGCTCGGGTTGGAATAGA | |||
| PF_AAC(3) | CACTTCCAAGAACGCAGACA | 222 | َaac(3)-III | This study |
| PR_AAC(3) | GTACATGTCCATGCCGAGTG | |||
| PF -INT1 | ATGCCCGTTCCATACAGAAG | 184 | INT1 | This study |
| PR -INT1 | CGGCCTTGCTGTTCTTCTAC |
Table 2. The condition for PCR performance of different genes in this study.
| Target gene | Primary denaturation | Denaturation | Annealing | Extension | Final extension |
|---|---|---|---|---|---|
| blaSHV-1 | 72 ° C | 72 ° C | 57 ° C | 93° C | 95° C |
| 5 minutes | 60 seconds | 60 seconds | 45 seconds | 3 minutes | |
| blaTEM-1 | 72 ° C | 72 ° C | 48 °C | 93° C | 95° C |
| 5 minutes | 60 seconds | 60 seconds | 45 seconds | 3 minutes | |
| blaVEB-1 | 72 ° C | 72 ° C | 57 ° C | 93° C | 95° C |
| 5 minutes | 60 seconds | 60 seconds | 45 seconds | 3 minutes | |
| blaVIM-1 | 72 ° C | 72 ° C | 55°C | 93° C | 95° C |
| 5 minutes | 60 seconds | 60 seconds | 45 seconds | 3 minutes | |
| blaPER-1 | 72 ° C | 72 ° C | 46°C | 93° C | 95° C |
| 5 minutes | 60 seconds | 60 seconds | 45 seconds | 3 minutes | |
| blaAMPC | 72 ° C | 72 ° C | 57 ° C | 93° C | 95° C |
| 5 minutes | 60 seconds | 60 seconds | 45 seconds | 3 minutes | |
| INT-1 | 72 ° C | 72 ° C | 56°C | 93° C | 95° C |
| 5 minutes | 60 seconds | 60 seconds | 45 seconds | 3 minutes | |
| aac(3)-III | 72 ° C | 72 ° C | 56°C | 93° C | 95° C |
| 5 minutes | 60 seconds | 60 seconds | 45 seconds | 3 minutes | |
| aac(6')-1 | 72 ° C | 72 ° C | 55°C | 93° C | 95° C |
| 5 minutes | 60 seconds | 60 seconds | 45 seconds | 3 minutes |
DNA Extraction By Kit
In this study, for DNA extraction, the kit of Cinnagen Company was used [10-12]. After extraction, DNA was held in -20 °C until PCR performance.
PCR Technique Performance
Used kit for PCR was Amplicon Kit which was prepared from Pishgam Company. The required values for each PCR reaction were as follows; primers (F and R): 1 µl (0.4 μm); DNA Template: 1 µl (50 pg); Master Mix: 12.5 µl (1X); DDW: 9.5 µl, in final volume 25 µl.
RESULTS
The Results of the Susceptibility Test and the Occurrence Rate of Resistance Genes
For the detection of resistance isolates, susceptibility testing using the abovementioned antibiotics in the Materials and Methods section was conducted. The most effective antibiotic was polymyxin B with 100% susceptibility and the most resistance was observed to ciprofloxacin (93%) and amikacin (67%), respectively. The results are shown in Table 3. As shown in Table 4, the maximum and lowest frequencies of drug resistance genes belonged to the aac (6')-1 and VEB-1 with prevalence 93% and 10%, respectively Figs. (1-4). According to the statistical analysis, no significant association was observed between the presence of Class I integrons and the pattern of genes multidrug resistance blaVEB-1, blaTEM-1, blaPER-1, blaSHV-1, blaTEM-1, blaVIM-1, blaAMPC, aac (6')-1, aac (3) -III (p ˃ 0.05).
Table 3. The Results of the susceptibility testing using disk diffusion method.
| Antibiotics | ||||||
|---|---|---|---|---|---|---|
| Polymyxin B | Amikacin | Tobramycin | Ciprofloxacin | Gentamicin | Ceftazidime | |
| Resistance (%) | 0 | 87 | 83 | 93 | 83 | 85 |
| Scuseptibility(%) | 100 | 13 | 17 | 7 | 17 | 15 |
Table 4. PCR results of drug resistance genes in isolates of P. aeruginosa.
| Prevalence of resistance genes %) | Number | Genes |
|---|---|---|
| 89 | 89 | VIM-1 |
| 10 | 10 | VEB-1 |
| 17 | 17 | SHV-1 |
| 15 | 15 | PER-1 |
| 74 | 74 | AMPC |
| 13 | 13 | TEM-1 |
| 93 | 93 | aac(3)-III |
| 78 | 78 | aac(6')-1 |
| 90 | 90 | INT-1 |
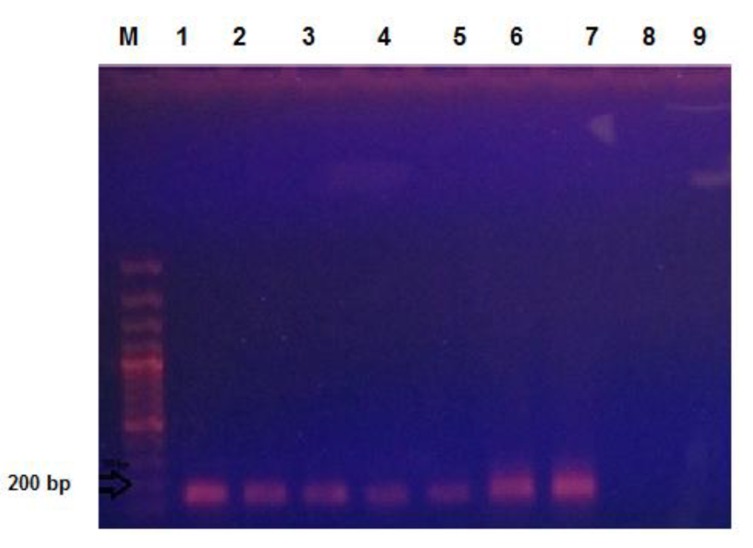
Fig. (1).
Gel electrophoresis image of VIM-1 gene with product size 182 bp. Wel ll M. 100 bp Marker, well 1-6. The positive samples for VIM-1 gene, well 7. Shows the positive control, well 8. It is corresponds with negative control and well 9 shows the negative sample for VIM-1 gene.
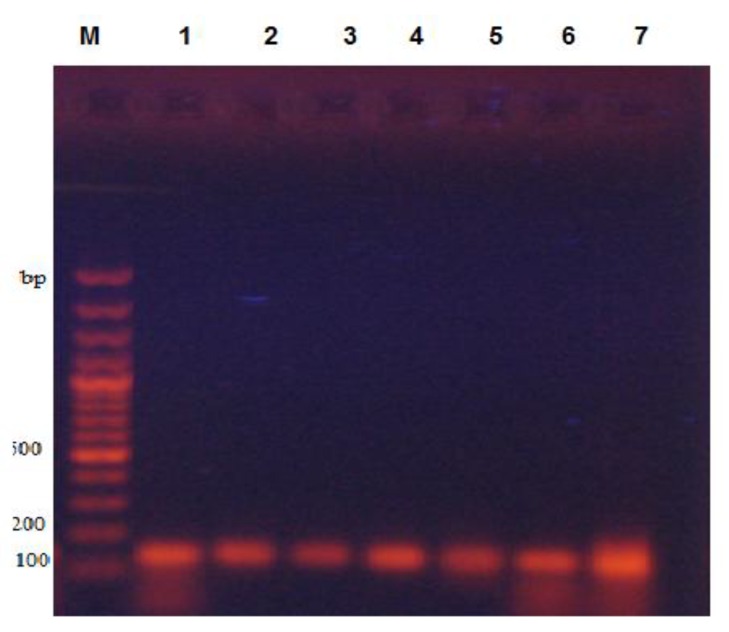
Fig. (4).
Gel electrophoresis image of AMPC gene with product size 150 bp. Lane M. DNA marker, lane 1shows the positive control and lane 2-9 are associated with positive samples for this gene.
DISCUSSION
P. aeruginosa is an opportunistic pathogenic bacterium which is a major factor in the prevalence of death in patients with a weakened immune system. Owing to the presence of widespread antibiotic resistance genes and the high speed in dissemination of environmental resistance, control and eradication of this pathogen have been made difficult. Inherent resistance to antimicrobial agents in the treatment of infections caused by bacteria has formed a situation which is worsening. This is due to the increase in antibiotic-resistant organisms which, particularly in multidrug isolates, has created many difficulties in the treatment of infections. Studies show that there is a significant correlation between the antibiotics resistance of P. aeruginosa and antibiotic use [13]. ESBLs genes can cause multiple resistance to other antibiotics as incidence and dissemination of different genes of ESBLs due to the increased incidence of antibiotic resistance particularly as a multi-drug, have caused many problems in the treatment of infections which are caused by these genes [7, 14]. MDR strains of P. aeruginosa include isolates which are resistant at least to three different classes of antibiotics, including all penicillins and cephalosporins, even inhibitor compounds (fluoroquinolones and aminoglycosides).
Extensively drug-resistant (XDR) strains consist of isolates of bacteria which in addition are resistant to three classes of the abovementioned antibiotics (strains of MDR), and also are resistant to carbapenems [15]. More P. aeruginosa isolates are susceptible to aminoglycosides and broad-spectrum penicillins. Quinolones and carbapenems are two kinds of new antibiotics with activity against Pseudomonas. The main mechanisms of resistance to P. aeruginosa are mutation of purine proteins thus reducing the permeability of the drug into bacteria. Beta-lactamases also are involved in producing resistance [16]. Aminoglycosides are considered as a part of anti-Pseudomonas drugs that are used to treat a variety of infections. These anti-bacterial agents with less toxic activity play an important role in the treatment of serious infections which are caused by gram-negative bacilli [17]. In a study conducted by Ranjbar and et al. in Baqiyatallah Hospital in Tehran in 2011, the rate of resistance to antibiotics, including ceftazidime (57.5%), amikacin (90%), ciprofloxacin (65%), gentamicin (67.5%) and imipenem (97.5%), was reported [18]. In another study in 2015, resistance to amikacin, tobramycin, ciprofloxacin, gentamicin and ceftazidime antibiotics has been shown as 96%, 88%, 88%, 92% and 96%, respectively [19], in which the results of two mentioned studies are in line with our study results. In correlation with the study carried out by Fazel and et al., the resistance rate to amikacin, gentamicin and ciprofloxacin was reported at 50%, 60% and 65%, respectively [20]. The study of Adabi et al. also shows increasing resistance and, like our study, confirmed the fact that the resistance rate increased in 2015 compared to 2013 [21].
In the present study, all isolates were susceptible to the polymyxin B, as in the studies conducted by Rostampour and colleagues (2015) and in the study conducted by Hammami et al., where similar results were observed [22]. The results of this study showed that bacterial resistance to ceftazidime, ciprofloxacin, amikacin, gentamicin and tobramycin antibiotics was very high, where this necessitates vigilance and proper use of these antibiotics. On the other hand, the emergence of multidrug clinical isolates of P. aeruginosa is rising, which is considered a serious challenge for health system. It is to be noted for consideration that the resistance patterns are constantly changing. According to the antibiotics tested in this study, polymyxin B currently is the only antibiotic that can be used to treat P. aeruginosa infection in burn wounds. Regarding the importance of this issue today, ways of prevention of infection transmission and monitoring the rising antibiotic resistance of P. aeruginosa in hospitals should be emphasized. In this regard, application of an active monitoring system to determine the hospital-resistant strains, especially in burn treatment centers, seems necessary. Therefore, convenient and fast solutions to prevent the spread of Pseudomonas infection in the burn wards of hospitals should be implemented. In this respect, special attention to the following cases is necessary: According to statistical Chi-square test, no significant association was observed between the presence of Class I integrons and the pattern of genes of multidrug resistance blaVEB-1, blaTEM-1, blaPER-1, blaSHV-1, blaTEM-1, blaVIM-1, blaAMPC, aac (6')-1, aac (3)-III (p ˃ 0.05).
Moreover, in the study conducted by Iraj Nikokar et al., resistance rates to various antibiotics were as follows: cloxacillin (91.8%), cotrimoxazole (86%), cephazolin (83.7%), carbenicillin (74.4%), piperacillin (69.9%), ceftazidime (68.8%), ciprofloxacin (66.3%), tobramycin (58.2%), amikacin (48.8%) and gentamicin (37.2%). The most effective antibiotic was imipenem with a resistance rate of 23.3. Their results were similar with our findings but in our study, unlike in this mentioned study, the best antibiotic was polymixin B, for which no resistant isolate was found. Also, their PCR results showed that 37 (43%) P. aeruginosa isolates and 27 (69.2%) multidrug resistant strains contained Class 1 integrons. A significant correlation was obtained between the presence of integrons and resistance against imipenem, ceftazidime, piperacillin and ciprofloxacin (p < 0.0). But, in present study, regarding the 90% prevalence of Class 1 integrons as compared to their results, the prevalence rate was higher and no significant relationship was discovered between the presence of Class I integrons and the pattern of multidrug resistance genes (p ˃ 0.05) [23]. Based on our results, the prevalence of Class I integrons in the present study was 90%, but in the study carried out by Rajabnia et al. in Babol in 2013, the prevalence of Class I integrons was reported at 39.4% [24]. This is lower than our study, but this rate is high too. This difference probably is a result of the geographical location and the bacteria isolates. But unlike our study, a correlation between Class I integrons and bacterial antibiotics was found. Consistent with our study, in other previous studies conducted in China and Brazil [25], the prevalence of Class I integrons was 40.8% and 41.5%, respectively. Studies conducted in China and Spain revealed that there was a significant association between resistance genes and some antibiotics such as aminoglycosides, beta-lactams, and quinolons [26, 27], but our study did not show such a relationship between Class I integrons and the mentioned antibiotic classes. In our study, the most prevalent gene belonged to the aac(6')-1, with occurrence of 93%. In accordance with the present study, a study in southern Taiwan showed that among 735 P. aeruginosa isolates, 305 (41.4%) were positive for existence of Class 1 integrons, and aac(6')-II-catB2-aadA2 (23.5%) was the most dominant gene. The same study has shown the presence of high variability of gene cassettes through Class 1 integrons among multidrug-resistant strains of P. aeruginosa [28]. Similarly, our results in a study done in Guilan in 2012 for detection of integrons found that 37 (43%) of P. aeruginosa isolates and 27 (69.2%) multidrug resistant strains contained Class 1 integrons, and a significant correlation, unlike in our study, was found between the presence of integrons and resistance to antibiotics. The most effective antibiotic in our study was polymyxin B, but in the stated investigation the most effective antibiotic was imipenem with a resistance frequency of 23.3% [29]. Again, in confirmation of our results, Nahaei et al. in Tabriz, in 2010, presented that 90 cases (56.3%) of P. aeruginosa isolates contained Class 1 integrons, but, against our results, they proved association between Class 1 integrons and resistance to imipenem and also reported that the best antibiotics were imipenem, meropenem and piperacillin/tazobactam [30]. These results are contrary to our findings, because we had polymixin B as the most effective antibiotic against P. aeruginosa. Based on previous studies, this high drug resistance is perhaps the reason for the extensive range of antibiotics utilized by hospitalized patients. In addition, it appears that former exposure to antibiotics is one of the key factors in drug resistance [31]. Therefore, the enhancement in clinically-resistant isolates may possibly be explained by antibiotic selective pressure and the prevalent presence of integrons [32]. According to our results, no significant correlation between Class 1 integrons and drug resistance was found. This is possibly due to many isolates having other antibiotic resistances which are not encoded by integrons-related resistance gene cassettes. These resistances may be owing to the plasmid acquisition, chromosomal mutation, or even the presence of other integrons in addition to Class 1 types [33].
CONCLUSION
Although no significant correlation between class 1 integrons and drug resistance was observed, but the resistance rate to antibiotics tested among P. aeruginosa isolates was high. So, surveillance, optimization and strict consideration of antimicrobial use and control of infection are necessary.
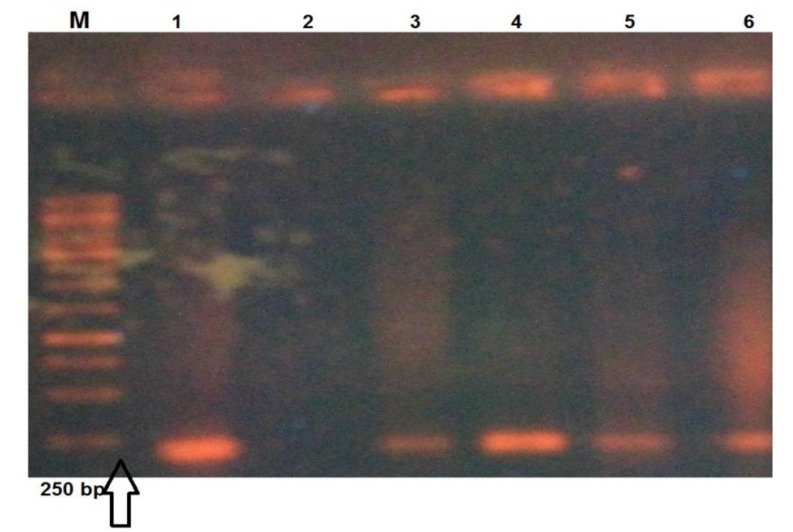
Fig. (2).
The image of INT-1 gene with amplicon 184 bp. Lane M. Marker 1000 bp, lane 1 and 2 show the positive and negative controls and lane 3-6 present the positive samples with product size 184 bp for INT-1 gene.
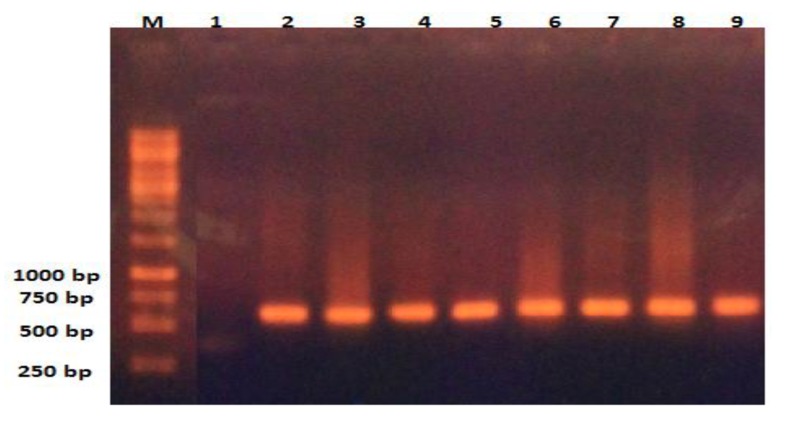
Fig. (3).
Analysis by agarose gel (2%) electrophoresis of the PCR products for blaVEB-1gene. Lane M. Marker, lane 2 and 3 show the negative and positive controls, respectively and lane 3-9 are related to the positive samples for blaVEB-1 with product size 634 bp.
ACKNOWLEDGEMENTS
We would like to thank medical laboratory personnel of Shahid Motahari hospital for their cooperation.
FUNDING SOURCE
This study did not receive any funding support.
AUTHORS’ CONTRIBUTION
All the authors contributed to the study concept and design; conduct, draft of the manuscript or revising it critically for important intellectual content. All the authors have read and approved every part of the work. All the authors accept the responsibility of the content of the paper.
CONFLICT OF INTREST
The author confirms that this article content has no conflict of interest.
REFERENCES
- 1.Erol S., Altoparlak U., Akcay M.N., Celebi F., Parlak M. Changes of microbial flora and wound colonization in burned patients. Burns. 2004;30(4):357–361. doi: 10.1016/j.burns.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 2.Thompson J.T., Meredith J.W., Molnar J.A. The effect of burn nursing units on burn wound infections. J. Burn Care Rehabil. 2002;23:281–286. doi: 10.1097/00004630-200207000-00011. [DOI] [PubMed] [Google Scholar]
- 3.De Vos D., Lim A., Jr, Pirnay J-P., Struelens M., Vandenvelde C., Duinslaeger L., Vanderkelen A., Cornelis P. Direct detection and identification of Pseudomonas aeruginosa in clinical samples such as skin biopsy specimens and expectorations by multiplex PCR based on two outer membrane lipoprotein genes, oprI and oprL. J. Clin. Microbiol. 1997;35(6):1295–1299. doi: 10.1128/jcm.35.6.1295-1299.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basiri S., Esmaily H., Vosough-Ghanbari S., Mohammadirad A., Yasa N., Abdollahi M. Improvement by Satureja khuzestanica essential oil of malathion-induced red blood cells acetylcholinesterase inhibition and altered hepatic mitochondrial glycogen phosphorylase and phosphoenolpyruvate carboxykinase activities. Pestic. Biochem. Physiol. 2007;89(2):124–129. doi: 10.1016/j.pestbp.2007.04.006. [DOI] [Google Scholar]
- 5.Zarei-Yazdeli M, Eslami G, Zandi H, Mousavi SM, Kosha H, Akhavan F. Relationship between antimicrobial resistance and class I integronsin Pseudomonas aeruginosa isolated from clinical specimens in Yazd during 2012-2013. . Feyz. J. Kashan Univ. Med. Sci. 2014;18(1):60–7. [Google Scholar]
- 6.Rastegar Lari A., Bahrami Honar H., Alaghehbandan R. Pseudomonas infections in Tohid Burn Center, Iran. Burns. 1998;24(7):637–641. doi: 10.1016/S0305-4179(98)00090-4. [DOI] [PubMed] [Google Scholar]
- 7.Khaledi A, Khademi F, Esmaeili D, Esmaeili S-A, Rostami H. The role of HPaA protein as candidate vaccine against Helicobacter pylori. Der Pharma Chemica. 2016;(3):235–7. [Google Scholar]
- 8.Díez-Aguilar M., Morosini M-I., López-Cerero L., Pascual Á., Calvo J., Martínez-Martínez L., Marco F., Vila J., Ortega A., Oteo J., Cantón R. Performance of EUCAST and CLSI approaches for co-amoxiclav susceptibility testing conditions for clinical categorization of a collection of Escherichia coli isolates with characterized resistance phenotypes. J. Antimicrob. Chemother. 2015;70(8):2306–2310. doi: 10.1093/jac/dkv088. [DOI] [PubMed] [Google Scholar]
- 9.Kalendar R., Lee D., Schulman A.H. Java web tools for PCR, in silico PCR, and oligonucleotide assembly and analysis. Genomics. 2011;98(2):137–144. doi: 10.1016/j.ygeno.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Fazeli H., Bafghi M.F., Faghri J., Akbari R. Molecular study of PER and VEB genes is multidrug resistant Pseudomonas aeroginosa isolated. J. Kerman Univ. Med. Sci. 2012;19(4):345–353. [Google Scholar]
- 11.Teo J., Ngan G., Balm M., Jureen R., Krishnan P., Lin R. Molecular characterization of NDM-1 producing Enterobacteriaceae isolates in Singapore hospitals. Western Pac. Surveill. Response J. 2012;3(1):19–24. doi: 10.5365/wpsar.2011.2.4.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang X., Zhang Z., Li M., Zhou D., Ruan F., Lu Y. Detection of extended-spectrum β-lactamases in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2006;50(9):2990–2995. doi: 10.1128/AAC.01511-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mansury D., Motamedifar M., Sarvari J., Shirazi B., Khaledi A. Antibiotic susceptibility pattern and identification of extended spectrum β-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae from Shiraz, Iran. Iran. J. Microbiol. 2016;8(1):55–61. [PMC free article] [PubMed] [Google Scholar]
- 14.Douglas M.W., Mulholland K., Denyer V., Gottlieb T. Multi-drug resistant Pseudomonas aeruginosa outbreak in a burns unitan infection control study. Burns. 2001;27(2):131–135. doi: 10.1016/S0305-4179(00)00084-X. [DOI] [PubMed] [Google Scholar]
- 15.Manchanda V., Sanchaita S., Singh N. Multidrug resistant acinetobacter. J. Glob. Infect. Dis. 2010;2(3):291–304. doi: 10.4103/0974-777X.68538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonomo R.A., Szabo D. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 2006;43(Suppl. 2):S49–S56. doi: 10.1086/504477. [DOI] [PubMed] [Google Scholar]
- 17.Katzung B.G., Masters S.B., Trevor A.J. Basic Clin Pharmacol 12/E Inkling (ENHANCED EBOOK). McGraw Hill Professional; 2012. [Google Scholar]
- 18.Ranjbar R., Owlia P., Saderi H., Mansouri S., Jonaidi-Jafari N., Izadi M., Farshad S., Arjomandzadegan M. Characterization of Pseudomonas aeruginosa strains isolated from burned patients hospitalized in a major burn center in Tehran, Iran. Acta Med. Iran. 2011;49(10):675–679. [PubMed] [Google Scholar]
- 19.Rajaie S., Mohammadi Sichani M., Mohammad Yousefi H. Study on the inhibitory activity of zinc oxide nanoparticles against Pseudomonas aeruginosa isolated from burn wounds. Qom. Uni. Med. Sci. J. 2015;9(1):30–37. [Google Scholar]
- 20.Fazeli H., Havaei S.A., Solgi H., Shokri D., Motallebirad T. Pattern of antibiotic resistance in Pesudomonas aeruginosa isolated from intensive care unit, isfahan, Iran. J. Isfahan Med. Sch. 2013;31(232) [Google Scholar]
- 21.Adabi M., Talebi T.M., Arbabi L., Afshar M., Fathizadeh S., Minaeian S. Determination of antibiotic resistance pattern of Pseudomonas aeruginosa strains isolated from patients with burn wounds. J. Ardabil. Univ. Med. Sci. 2015;15(1):66–74. [Google Scholar]
- 22.RostamPour S., Gorzin A., Motamedi Gh. Frequency of blaKHM-1, blaIMP-1, 2 and blaSPM-1 genes in clinical isolates of metallo β-lactamase producing Pseudomonas aeruginosa in hospitalized burned patients in Ghotbeddin Shirazi Hospital. J. Qazvin. Univ. Med. Sci. 2015;19(2):21–29. [Google Scholar]
- 23.Nikokar I., Tishayar A., Flakiyan Z., Alijani K., Rehana-Banisaeed S., Hossinpour M., Amir-Alvaei S., Araghian A. Antibiotic resistance and frequency of class 1 integrons among Pseudomonas aeruginosa, isolated from burn patients in Guilan, Iran. Iran. J. Microbiol. 2013;5(1):36–41. [PMC free article] [PubMed] [Google Scholar]
- 24.Rajabnia R., Asgharpour F., Shahandashti E.F., Khalilian M., Norkhomami S., Moulana Z. Class 1 integron in Pseudomonas aeruginosa isolates from different places and devices of ICU in Babol, Iran. Jundishapur J. Microbiol. 2013;6(2) doi: 10.5812/jjm.4850. [DOI] [Google Scholar]
- 25.Gu B. Prevalence and characterization of class I integrons among Pseudomonas aeruginosa and Acinetobacter baumannii isolates from patients in Nanjing, China. J. Clin. Microbiol. . 2007;45(1):241–3. doi: 10.1128/JCM.01318-06. Jan; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu B., Tong M., Zhao W., Liu G., Ning M., Pan S., Zhao W. Prevalence and characterization of class I integrons among Pseudomonas aeruginosa and Acinetobacter baumannii isolates from patients in Nanjing, China. J. Clin. Microbiol. 2007;45(1):241–243. doi: 10.1128/JCM.01318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Juan C., Beceiro A., Gutiérrez O., Albertí S., Garau M., Pérez J.L., Bou G., Oliver A. Characterization of the new metallo-β-lactamase VIM-13 and its integron-borne gene from a Pseudomonas aeruginosa clinical isolate in Spain. Antimicrob. Agents Chemother. 2008;52(10):3589–3596. doi: 10.1128/AAC.00465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsiao K-Y., Lee M-F., Peng C-F. Detection and characterization of class 1 integron-associated gene cassettes from Pseudomonas aeruginosa isolates in southern Taiwan. Biomarkers Genomic Med. 2014;6(2):74–78. doi: 10.1016/j.bgm.2014.02.004. [DOI] [Google Scholar]
- 29.Nikokar I., Tishayar A., Flakiyan Z., Alijani K., Rehana-Banisaeed S., Hossinpour M., Amir-Alvaei S., Araghian A. Antibiotic resistance and frequency of class 1 integrons among Pseudomonas aeruginosa, isolated from burn patients in Guilan, Iran. Iran. J. Microbiol. 2013;5(1):36–41. [PMC free article] [PubMed] [Google Scholar]
- 30.Yousefi S., Nahaei M., Farajnia S., Ghojazadeh M., Akhi M., Sharifi Y., Milani M., Ghotaslou R. Class 1 integron and imipenem resistance in clinical isolates of Pseudomonas aeruginosa: prevalence and antibiotic susceptibility. Iran. J. Microbiol. 2010;2(3):115–121. [PMC free article] [PubMed] [Google Scholar]
- 31.Onguru P., Erbay A., Bodur H., Baran G., Akinci E., Balaban N., Cevik M.A. Imipenem-resistant Pseudomonas aeruginosa: risk factors for nosocomial infections. J. Korean Med. Sci. 2008;23(6):982–987. doi: 10.3346/jkms.2008.23.6.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tenover F.C. Mechanisms of antimicrobial resistance in bacteria. Am. J. Med. 2006;119(6) Suppl. 1:S3–10. doi: 10.1016/j.amjmed.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 33.Chen J., Su Z., Liu Y., Wang S., Dai X., Li Y., Peng S., Shao Q., Zhang H., Wen P., Yu J., Huang X., Xu H. Identification and characterization of class 1 integrons among Pseudomonas aeruginosa isolates from patients in Zhenjiang, China. Int. J. Infect. Dis. 2009;13(6):717–721. doi: 10.1016/j.ijid.2008.11.014. [DOI] [PubMed] [Google Scholar]