Abstract
Abstract: Background
Isotopic Ratio Outlier Analysis (IROA) is an untargeted metabolomics method that uses stable isotopic labeling and LC-HRMS for identification and relative quantification of metabolites in a biological sample under varying experimental conditions.
Objective
We demonstrate a method using high-sensitivity 13C NMR to identify an unknown metabolite isolated from fractionated material from an IROA LC-HRMS experiment.
Methods
IROA samples from the nematode Caenorhabditis elegans were fractionated using LC-HRMS using 5 repeated injections and collecting 30 sec fractions. These were concentrated and analyzed by 13C NMR.
Results
We isotopically labeled samples of C. elegans and collected 2 adjacent LC fractions. By HRMS, one contained at least 2 known metabolites, phenylalanine and inosine, and the other contained tryptophan and an unknown feature with a monoisotopic mass of m/z 380.0742 [M+H]+. With NMR, we were able to easily verify the known compounds, and we then identified the spin system networks responsible for the unknown resonances. After searching the BMRB database and comparing the molecular formula from LC-HRMS, we determined that the fragments were a modified anthranilate and a glucose modified by a phosphate. We then performed quantum chemical NMR chemical shift calculations to determine the most likely isomer, which was 3’-O-phospho-β-D-glucopyranosyl-anthranilate. This compound had previously been found in the same organism, validating our approach.
Conclusion
We were able to dereplicate previously known metabolites and identify a metabolite that was not in databases by matching resonances to NMR databases and using chemical shift calculations to determine the correct isomer. This approach is efficient and can be used to identify unknown compounds of interest using the same material used for IROA.
Keywords: NMR, IROA, Metabolomics, MS, Carbon-13, LC-MS, C.elegans
Introduction
Nuclear magnetic resonance (NMR) spectroscopy and liquid-chromatography high-resolution mass spectrometry (LC-HRMS) have complementary strengths in metabolomics. NMR is ideal for compound identification and quantification but suffers from low sensitivity. In contrast, LC-HRMS has extremely high sensitivity but has limitations in compound identification, especially for unknowns not in databases and isomers that have identical fragmentation patterns and are not separated chromatographically. In an attempt to improve the ability of MS to identify important features, isotopic LC-HRMS experiments are becoming increasingly popular [1-4]. We recently demonstrated a LC-HRMS experiment called isotopic ratio outlier analysis (IROA), which compares samples that have been isotopically labeled with 95% 13C or 5% 13C [5]. In our initial IROA experiment, we labeled the nematode Caenorhabditis elegans and subjected the 5% 13C “test” animals to a heat shock before mixing with 95% 13C “control” animals. The combined material was analyzed using LC-HRMS, yielding unique patterns of isotopic peaks for biosynthesized metabolites that can be easily discriminated from noise. Such isotopic patterns allow the calculation of the number of carbons in a metabolite as well as the relative abundance for the test vs. control samples [5, 6]. Although we used the standard IROA protocol with 5% and 95% 13C enrichment, a related technique called phenotypic IROA can be used with an unlabeled sample combined with another 95% 13C-labeled sample, which doesn’t need to be the same organism. In this case, the 95% 13C material serves as a labeled isotopic standard and the compounds in the labeled standard create a targeted assay. Thus, with phenotypic IROA, the study material does not need to be labeled as long as the metabolites of interest are in the 95% 13C standard [6]. The technique presented here could be applied without modification to phenotypic IROA, because the majority of the NMR signal comes from the 95% 13C material.
A major challenge with IROA, like other LC-HRMS techniques, is the identification of unknown metabolites that are not in existing databases. One solution to this general problem is to utilize NMR, either directly coupled in-line with the LC-HRMS (i.e. LC-NMR-HRMS) or in the off-line analysis of fractions isolated from an LC-HRMS experiment [7]. Joint use of NMR and LC-HRMS approaches has successfully resulted in metabolite identification in metabolomics datasets [8-11]. In-line LC-NMR-HRMS is limited by the NMR sensitivity and is generally not considered a practical method for most samples with many metabolites at low concentrations [7]. The most popular and useful approach to combine NMR with LC-HRMS is the use of preparative chromatography followed by solid phase extraction (SPE) of the fractions of interest. Using LC-SPE, multiple injections of a sample can be used to concentrate low abundance compounds for NMR analysis [12]. The method described here is similar to LC-SPE, but we used a standard fraction collector and concentrated fractions by evaporation using a Centrivap vacuum concentrator (Labconco). The Brüschweiler laboratory has developed a technique called SUMMIT MS/NMR (Structures of Unknown Metabolomic Mixture components by MS/NMR), which utilizes a combined MS and NMR approach for global metabolomics without isotopic labeling or sample fractionation [11]. The SUMMIT approach is very powerful and utilizes information from HRMS to obtain molecular formulas to determine all the possible structures consistent with the experimental molecular formula. Then, NMR chemical shifts are calculated for all possible structures and compared with experimental NMR data on the same mixture. SUMMIT could be applied to our method by using the molecular formula derived from IROA data and then comparing all possible structures to the 13C NMR data. In this study, we demonstrate the feasibility of using 13C NMR on isotopically labeled samples that have been fractionated by LC-HRMS. In addition, we utilize 13C chemical shift calculations, which are more accurate than 1H chemical shift calculations, to determine the most accurate chemical structure. While straightforward conceptually and completely compatible with both LC-SPE and SUMMIT MS/NMR, there are several experimental challenges that had to be considered, most importantly the overall sensitivity of the 13C NMR measurement using the small volumes and the small quantities of material available from LC-HRMS fractionation.
The samples used in this study were duplicates produced in our previous study in which we demonstrated IROA using heat shocked C. elegans [5]. We combined 3 samples, each containing 125,000 of 95% 13C and 5% 13C worms (total of 250,000 worms). This material was resuspended in 120 μL of water. Five 20 μL aliquots of this material were injected and analyzed using a Thermo Scientific Q-Exactive Orbitrap mass spectrometer with Doinex UHPLC, autosampler, and fraction collector. Fractions were collected every 30 sec, and the final samples were dried, resuspended in 50 μL D2O and added to a 1.5-mm NMR tube for analysis (See Supplementary Methods). We used a custom designed 13C-optimized NMR probe [13] with coils made from high temperature superconducting material, which along with the 13C and 1H preamplifiers are cryogenically cooled. The probe operates at 14.1 T (600 MHz 1H frequency), and the sample chamber accommodates 1.5-mm tubes and is regulated near room temperature. This probe has 2-3 x greater 13C mass sensitivity than cryogenic 13C optimized commercial probes available today [13]. The 13C S/N of our probe using the ASTM standard (40% dioxane in benzene-d6) is about 400:1 for a 40 μL sample volume. This is ideal for mass-limited samples. However, 13C-optimized 5-mm DCH cryogenic commercial probes are now available that offer 3000:1 for a 600 MHz system (Bruker Biospin). Such a probe would provide 7.5 x the sensitivity using 15 x the sample volume when compared to our probe. Thus, if one were to use the same 1.5-mm NMR tubes that we used in this study in a 5-mm commercial 13C cryoprobe, the S/N would be about a factor of two lower than we are reporting here. That would be satisfactory for many metabolites with the same level of concentration used here, but if sample volume is not limited, additional sample concentration could be used to get the same results.
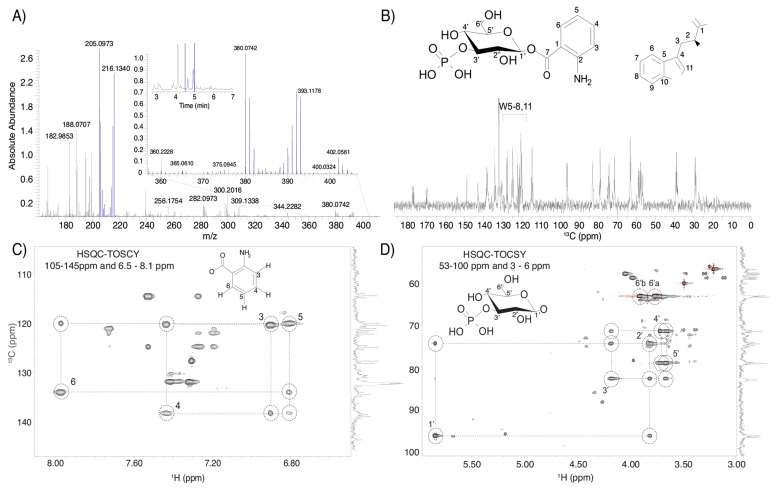
The purpose of this study was to demonstrate that 13C NMR can be used in conjunction with LC-HRMS for 13C-labeled samples, so we analyzed two adjacent 30 sec fractions from an IROA LC-HRMS study. Fraction 9 from 4 to 4.5 min (red in Fig. S1) contained two peaks—phenylalanine and inosine—that were easily identified from IROA LC-HRMS data alone. We were able to easily verify the LC-HRMS data following a query of the BMRB 13C database [14] using COLMAR (Complex Mixture Analysis by NMR) [15]. Fraction 10 from 4.5-5 min (blue in Fig. 1A) contained tryptophan and an unknown feature containing 13 carbons, with an observed monoisotopic mass of the [M+H]+ ion at m/z 380.0742, determined by the presence of the sodiated ion at the same retention time (4.65 min). The molecular formula was determined to be C13H18NO10P (1.40 ppm mass error, and only one possible formula with less than 2 ppm error had the correct number of carbons). Tandem MS of the totally 12C monoisotopic peak and the (1 12C)/(12 13C) monoisotopic peak with a mass window of ± 1 amu in both positive and negative ionization mode pointed to the compound containing a phosphorylated 6 carbon carbohydrate and an aromatic ring with molecular formula C7H7NO2 (Fig. S2). The ring was suspected to be anthranilate due to matches with the METLIN database [16] of the ions at m/z 120.0446 ([M-NH3+H]+) and m/z 138.0552 ([M+H]+), but structures such as para-aminobenzoic acid could not be ruled out.
The13C 1D NMR spectrum of fraction 10 was easily dereplicated by matching the BMRB 13C spectrum of tryptophan, labeled with “W” in Fig. 1B. The 13C-13C J-couplings in the NMR spectra from isotopically labeled material provided clear signatures of the carbon connectivity (Fig. S3). The remaining resonances were consistent with a carbohydrate, with 6 carbons from about 64 to 96 ppm and an aromatic system with resonances from about 120 to 140 ppm. We note that due to the large sample volumes injected in the fractionation, there was some bleed-through of phenylalanine and inosine from fraction 9 (typical injection volumes are 2-5 μL, but we injected 20 μL to reduce the number of total injections needed for NMR).
To establish the carbohydrate and aromatic spin systems, we collected 2D 13C-HSQC-TOCSY (Fig. 1C-D) and 13C-HSQC (Fig. S4) data. The 13C-HSQC-TOCSY was especially useful in defining correlated 13C and 1H resonances in both regions of interest. The carbohydrate region (Fig. 1D and Fig. S4B) had chemical shifts that matched closely to a substituted glucose moiety. We only observed one set of glucose resonances that are consistent with the β anomer, suggesting a bulky substituent attached to the 1’ carbon. The 13C-HSQC-TOCSY aromatic region (Fig. 1) provided 1H and 13C correlations that had several matches in the BMRB [14] and HMDB [17] databases. The HSQC assignments of anthranilate from the HMDB were close to our experimental data.
Fig. (1).
A) HRMS spectrum averaged across the chromatographic range 4.5 – 4.9 min. MS peaks from the known compound tryptophan are highlighted in blue from m/z 205.0973 to 216.1340. MS peaks from the unknown compound investigated in this study are shown in the expansion with [M+H]+ at m/z 380.0742. The total ion chromatogram is shown in the inset, with the fractionated region marked. B) 13C NMR spectra of this fraction contained tryptophan resonances and the unknown resonances (U) that were assigned to 3’-O-phospho-β-D-glucopyranosyl-anthranilate, as described in the text. C) HSQC-TOCSY of the unknown compound. The aromatic region provided evidence of a 1,2 bi-substituted benzene. Chemical shifts are consistent with anthranilate. D) The aliphatic region provided evidence of a β-glucose.
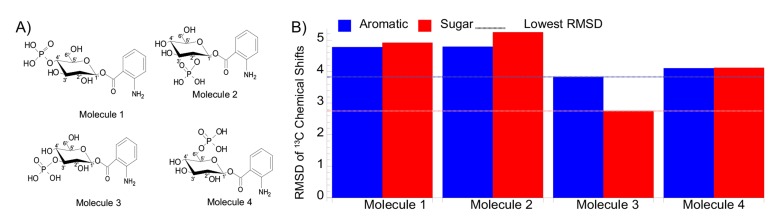
Chemical shift calculations described previously [18] were performed for four different phospo- substituted glucose moieties to confirm the position of the phosphate on glucose (Fig. 2). These calculations are very robust, especially for 13C. In our earlier work we were able to distinguish between several diastereomers of a monoterpene solely with chemical shifts [18]. The root mean squared standard deviation (RMSD) between experimental and calculated chemical shifts was computed for both 13C and 1H for each possible molecule (Fig. 2). Molecule 3 with the 3’ phosphate substitution gave a lower RMSD for both the aromatic and sugar carbon chemical shifts (Fig. 2). Molecule 3 also gave the lowest RMSD for the glucose protons, though molecule 2 gave lower RMSD for the aromatic protons. Given these results as well as evidence from the 2D NMR and MS, we hypothesized that the 3’-O-phospho-β-D-glucopyranosyl-anthranilate is the unknown compound in fraction 10. 13C chemical shifts were much better than 1H for computational matches with experimental data (Fig. 2), further demonstrating the utility of 13C NMR for unknown compound identification. A spike with the synthetic compound would be required for conclusive proof of an unknown compound identification, but after we finished our analysis, we discovered that the same molecule had previously been reported in the same organism. Schroeder and coworkers discovered that anthranilate glucoside and 3’-phospho-glucoside from C. elegans were the chemical components responsible for blue fluorescence upon the death of an animal [19]. We were unaware of this finding when we conducted our study, but use it as a more definitive confirmation of our proposed structure and a validation of this approach.
Fig. (2).
A) Four variants of a phospho-glucose moiety. B) Bar plot of all aromatic (blue) and all sugar (red) 13 C RMSDs. Molecule 3 (3’ phosphate substitution) has the lowest RMSD, indicated by the dashed line.
With the Schroeder laboratory, we previously identified several worm-produced metabolites of the bacterial small-molecule toxins 1-hydroxyphenazine and indole that led to reduced toxicity of these compounds [20]. One of the modifications was a 3’-phospho glucoside, a relatively uncommon modification that we now have also observed in an anthranilate glucoside. Additionally, we found nonphosphorylated mono-, di-, and tri-saccharide modifications of 1-hydroxyphenazine, and so attempted to verify additional such modifications of anthranilate. Similar to the previous report [19], we also found evidence of an anthranilate glucoside (4.91 min, [M+H]+: 300.1078) but found no evidence of di- or tri-saccharides. Both detoxification [20] and death fluorescence indicators [19] utilize an unusual 3’-phospho glucoside modification, but more work needs to be done to determine if these are biologically related or not.
By combining high-sensitivity, small volume 13C NMR measurements with isotopically-enriched samples [5], we have demonstrated efficient identification of compounds not found in existing databases. This approach can easily be adapted to LC-SPE methods and also could utilize the SUMMIT MS/NMR method for compound identification. The advantages of 13C NMR include greater chemical shift dispersion, easily interpreted J-coupling patterns with enriched samples, and the ability to detect quaternary carbons. In our situation, we were able to obtain database matches to two separate fragments and then propose a structure that we subsequently found in the literature [19] using high-level ab initio NMR chemical shift calculations of related isomers. This method has a few limitations including the need for a sensitive 13C NMR probe, computing power for ab initio chemical shift calculations, and previously described limitations with IROA [5]. As described in the introduction, similar results could be obtained using a commercial 5-mm DCH 13C-optimized Bruker cryoprobe. Depending on the complexity and quantity of the unknown compound, the entire process of NMR ID with chemical shift calculations as validation can be done in a few days, making this a useful approach to annotate important features from an IROA LC-HRMS experiment.
ACKNOWLEDGEMENTS
Funding for this study was from the NIH (1U24DK097209-01A1 and R01EB009772 to A.S.E.) and the National High Magnetic Field Laboratory’s AMRIS Facility, which is supported by National Science Foundation Cooperative Agreement No. DMR-1157490 and the State of Florida. A.S.E. receives support from the Georgia Research Alliance. We are grateful to Bill Brey, Jerris Hooker, and Vijay Ramaswamy for the HTS NMR probe and continued support during this project. Jim Rocca provided support in the NMR data collection. Ramadan Ajredini provided help with sample preparation. Chris Beecher provided valuable support for IROA experiments. We thank Rick Yost and Chris Beecher for helpful comments to this manuscript. Data will be available at the NIH Common Fund's Data Repository and Coordinating Center (supported by NIH grant, U01-DK097430: http://www.metabolomicsworkbench.org)
SUPPLEMENTARY MATERIAL
Supporting material is available on the publisher’s web site along with the published article.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Huang X., Chen Y.J., Cho K., Nikolskiy I., Crawford P.A., Patti G.J. X13CMS: global tracking of isotopic labels in untargeted metabolomics. Anal. Chem. 2014;86(3):1632–1639. doi: 10.1021/ac403384n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bueschl C., Kluger B., Lemmens M., Adam G., Wiesenberger G., Maschietto V., Marocco A., Strauss J., Bodi S., Thallinger G.G., Krska R., Schuhmacher R. A novel stable isotope labelling assisted workflow for improved untargeted LC-HRMS based metabolomics research. Metabolomics. 2014;10(4):754–769. doi: 10.1007/s11306-013-0611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bueschl C., Krska R., Kluger B., Schuhmacher R. Isotopic labeling-assisted metabolomics using LC-MS. Anal. Bioanal. Chem. 2013;405(1):27–33. doi: 10.1007/s00216-012-6375-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giavalisco P., Kohl K., Hummel J., Seiwert B., Willmitzer L. 13C isotope-labeled metabolomes allowing for improved compound annotation and relative quantification in liquid chromatography-mass spectrometry-based metabolomic research. Anal. Chem. 2009;81(15):6546–6551. doi: 10.1021/ac900979e. [DOI] [PubMed] [Google Scholar]
- 5.Stupp G.S., Clendinen C.S., Ajredini R., Szewc M.A., Garrett T., Menger R.F., Yost R.A., Beecher C., Edison A.S. Isotopic ratio outlier analysis global metabolomics of Caenorhabditis elegans. Anal. Chem. 2013;85(24):11858–11865. doi: 10.1021/ac4025413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Jong F.A., Beecher C. Addressing the current bottlenecks of metabolomics: Isotopic ratio outlier analysis™, an isotopic-labeling technique for accurate biochemical profiling. Bioanalysis. 2012;4(18):2303–2314. doi: 10.4155/bio.12.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfender J.L., Bohni N., Ndjoko-Ioset K., Edison A.S. In: Advanced spectroscopic detectors for identification and quantification: nuclear magnetic resonance; Fanali, S.; Haddad, P.R.; Poole, C.F.; Schoenmakers, P. Lloyd D., editor. Amsterdam: Elsevier; 2013. pp. 349–384. [Google Scholar]
- 8.Lambert M., Wolfender J.L., Stærk D., Christensen S.B., Hostettmann K., Jaroszewski J.W. Identification of natural products using HPLC-SPE combined with CapNMR. Anal. Chem. 2007;79(2):727–735. doi: 10.1021/ac0616963. [DOI] [PubMed] [Google Scholar]
- 9.Tayyari F., Gowda G.A., Gu H., Raftery D. 15N-cholamine--a smart isotope tag for combining NMR- and MS-based metabolite profiling. Anal. Chem. 2013;85(18):8715–8721. doi: 10.1021/ac401712a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan T.W., Lane A.N., Higashi R.M., Farag M.A., Gao H., Bousamra M., Miller D.M. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM). Mol. Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bingol K., Bruschweiler-Li L., Yu C., Somogyi A., Zhang F., Bruschweiler R. Metabolomics beyond spectroscopic databases: A combined MS/NMR strategy for the rapid identification of new metabolites in complex mixtures. Anal. Chem. 2015;87(7):3864–3870. doi: 10.1021/ac504633z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sumner L.W., Lei Z., Nikolau B.J., Saito K. Modern plant metabolomics: advanced natural product gene discoveries, improved technologies, and future prospects. Nat. Prod. Rep. 2015;32(2):212–229. doi: 10.1039/c4np00072b. [DOI] [PubMed] [Google Scholar]
- 13.Ramaswamy V., Hooker J.W., Withers R.S., Nast R.E., Brey W.W., Edison A.S. Development of a (13)C-optimized 1.5-mm high temperature superconducting NMR probe. J. Magn. Reson. 2013;235C:58–65. doi: 10.1016/j.jmr.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ulrich E.L., Akutsu H., Doreleijers J.F., Harano Y., Ioannidis Y.E., Lin J., Livny M., Mading S., Maziuk D., Miller Z., Nakatani E., Schulte C.F., Tolmie D.E., Kent Wenger R., Yao H., Markley J.L. BioMagResBank. Nucleic Acids Res. 2008;36(Database issue):D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinette S.L., Zhang F., Bruschweiler-Li L., Bruschweiler R. Web server based complex mixture analysis by NMR. Anal. Chem. 2008;80(10):3606–3611. doi: 10.1021/ac702530t. [DOI] [PubMed] [Google Scholar]
- 16.Smith C.A., O' Maille, Want G., Qin E.J., Trauger C., Brandon S.A., Custodio T.R., Abagyan D.E., Siuzdak R., METLIN G. a metabolite mass spectral database. Ther. Drug Monit. 2005;27(6):747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 17.Wishart D.S., Tzur D., Knox C., Eisner R., Guo A.C., Young N., Cheng D., Jewell K., Arndt D., Sawhney S., Fung C., Nikolai L., Lewis M., Coutouly M.A., Forsythe I., Tang P., Shrivastava S., Jeroncic K., Stothard P., Amegbey G., Block D., Hau D.D., Wagner J., Miniaci J., Clements M., Gebremedhin M., Guo N., Zhang Y., Duggan G.E., MacInnis G.D., Weljie A.M., Dowlatabadi R., Bamforth F., Clive D., Greiner R., Li L., Marrie T., Sykes B.D., Vogel H.J., Querengesser L. HMDB: the human metabolome database. Nucleic Acids Res. 2007;35:D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang B., Dossey A.T., Walse S.S., Edison A.S., Merz K.M. Relative configuration of natural products using NMR chemical shifts. J. Nat. Prod. 2009;72(4):709–713. doi: 10.1021/np8005056. [DOI] [PubMed] [Google Scholar]
- 19.Coburn C., Allman E., Mahanti P., Benedetto A., Cabreiro F., Pincus Z., Matthijssens F., Araiz C., Mandel A., Vlachos M., Edwards S.A., Fischer G., Davidson A., Pryor R.E., Stevens A., Slack F.J., Tavernarakis N., Braeckman B.P., Schroeder F.C., Nehrke K., Gems D. Anthranilate fluorescence marks a calcium-propagated necrotic wave that promotes organismal death in C. elegans. PLoS Biol. 2013;11(7):e1001613. doi: 10.1371/journal.pbio.1001613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stupp G.S., von Reuss S.H., Izrayelit Y., Ajredini R., Schroeder F.C., Edison A.S. Chemical detoxification of small molecules by Caenorhabditis elegans. ACS Chem. Biol. 2013;8(2):309–313. doi: 10.1021/cb300520u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting material is available on the publisher’s web site along with the published article.