Abstract
When there is no recombination (achiasmy) in one sex, it is in the heterogametic one. This observation is so consistent that it constitutes one of the few patterns in biology that may be regarded as a ‘rule’ and Haldane (Haldane 1922 J. Genet. 12, 101–109. (doi:10.1007/BF02983075)) proposed that it might be driven by selection against recombination in the sex chromosomes. Yet differences in recombination rates between the sexes (heterochiasmy) have also been reported in hermaphroditic species that lack sex chromosomes. In plants—the vast majority of which are hermaphroditic—selection at the haploid stage has been proposed to drive heterochiasmy. Yet few data are available for hermaphroditic animals, and barely any for hermaphroditic vertebrates. Here, we leverage reciprocal crosses between two black hamlets (Hypoplectrus nigricans, Serranidae), simultaneously hermaphroditic reef fishes from the wider Caribbean, to generate high-density egg- and sperm-specific linkage maps for each parent. We find globally higher recombination rates in the eggs, with dramatically pronounced heterochiasmy at the chromosome peripheries. We suggest that this pattern may be due to female meiotic drive, and that this process may be an important source of heterochiasmy in animals. We also identify a large non-recombining region that may play a role in speciation and local adaptation in Hypoplectrus.
Keywords: recombination, meiosis, heterochiasmy, linkage map, hermaphrodites, Hypoplectrus
1. Introduction
Reports of variable recombination rates between the sexes go back over a century [1]. Haldane [2] and Huxley [3] were quick to point out that when recombination is absent in one sex (achiasmy), it is in the heterogametic one. They proposed that this pattern may be due to a pleiotropic effect of selection against recombination in the sex chromosomes and Nei [4] established the plausibility of this scenario from a theoretical perspective. The observation that when one sex does not recombine it is the heterogametic one appears to hold and there are now close to 30 evolutionarily independent origins of this pattern reported in animals [5]. Nevertheless, when both sexes recombine the heterogametic sex does not systematically present lower recombination rates and differences in recombination rates between the sexes (heterochiasmy) are also observed in both hermaphroditic and gonochoric species that lack sex chromosomes [6–8]. Distinct processes may therefore underlie the evolution of achiasmy and heterochiasmy.
A variety of hypotheses including metabolic rate [9], dispersal [6,10], sexual selection [10,11], neutrality [6], haploid selection [7,12], and female meiotic drive (which may be viewed as a particular type of haploid selection) [8,13] have been put forward to explain the occurrence of heterochiasmy. Under the female meiotic drive hypothesis, alleles exploit asymmetric cell division during oogenesis to increase in frequency. Only one of the four products of female meiosis is passed on to the offspring, which sets the stage for competition between alleles for representation in the egg and for drive, i.e. the overrepresentation of specific alleles in the next generation. The number of recombination events and their position relative to the centromere and locus of interest are fundamental for the segregation of alleles and spread of meiotic drivers, generating the specific prediction that differences in recombination rates between the sexes should be heterogeneous along chromosomes and particularly pronounced around the centromeres [8]. Yet this prediction remains largely untested because the vast majority of studies in non-model organisms only consider heterochiasmy globally and do not provide information about the distribution of heterochiasmy along chromosomes.
Hermaphrodites constitute an interesting category in the context of heterochiasmy because they provide the opportunity to address female (ovule/egg) versus male (pollen/sperm) recombination in the absence of sex chromosomes, within individuals (i.e. with the same genetic background), at the same time in the case of simultaneous hermaphrodites and with various degrees of selfing. As most plants are hermaphroditic, this category has been well studied in the context of heterochiasmy [6–8]. Recombination rates tend to be lower in females in gymnosperms but higher in non-selfing angiosperms and the differential potential for haploid selection in males and females in these two groups led Lenormand & Dutheil [7] to propose that this may constitute a major source of heterochiasmy in plants. Yet there are important differences between plants and animals in this context. In particular, the small number of genes expressed in the sperm and the lack of a haploid phase in eggs, as meiosis is usually completed at or just before fertilization in animals, are expected to reduce the potential for haploid selection [14,15].
In sharp contrast with the situation in plants, only eight of the 164 species considered in the last review on heterochiasmy [8] are hermaphroditic animals, and we found just a few additional cases in the literature (table 1). In these studies, heterochiasmy estimates are derived from chiasma counts (i.e. histological observations of chromosomes during meiosis) or linkage maps. As mentioned above, one interesting aspect of hermaphrodites is that they provide the opportunity to address female versus male recombination in the same individuals and using the same markers in the case of linkage maps. This is relevant in the context of heterochiasmy because it allows disentangling the effect of sex versus individual variation on recombination. Yet all the linkage map studies on hermaphroditic animals listed in table 1 used different parents to generate male and female maps and we are not aware of other studies in animals where sperm- and egg-specific linkage maps have been generated from the same parents (but see e.g. [16] for chiasma counts).
Table 1.
Cases of heterochiasmy in hermaphroditic animals. CC, chiasma count; LM, linkage map; F/M, female/male recombination ratio; Pos. info, positional information on recombination frequencies along linkage groups (LGs); Same inds., male and female maps generated from the same parents. SNP: single nucleotide polymorphism; AFLP: amplified fragment length polymorphism; EST: expressed sequence tag; μsats: microsatellites.
| species | mating system | method (markers) | F/M | Pos. info. | Same inds. | reference |
|---|---|---|---|---|---|---|
| Dendrocoelum lacteum (flatworm) | simultaneous hermaphrodite | CC | 1.7 | no | no | [16] |
| Notoplana igiliensis (flatworm) | simultaneous hermaphrodite | CC | 1.5 | no | no | [17] |
| Paradistomoides orientalis (flatworm) | simultaneous hermaphrodite | CC | ∼1 | no | no | [18] |
| Gyratrix hermaphroditus (flatworm) | simultaneous hermaphrodite | CC | — | no | no | [19] |
| Schmidtea polychroa (flatworm) | simultaneous hermaphrodite | LM (4 μsats) | 3.3 | no | no | [20] |
| Acropora millepora (coral) | simultaneous hermaphrodite | LM (393 SNPs + 36 μsats) | 1.3 | no | no | [21] |
| Argopecten irradians (scallop) | simultaneous hermaphrodite | LM (161 μsats) | 1.1 | no | no | [22] |
| Pinctada maxima (oyster) | facultative protandrous hermaphrodite | LM (887 SNPs) | 1.2 | yes | no | [23] |
| Crassostrea virginica (oyster) | facultative protandrous hermaphrodite | LM (198 AFLPs + 5 μsats + 2 ESTs) | 1.5 | no | no | [24] |
| Crassostrea gigas/angulata (oyster) | facultative protandrous hermaphrodite | LM (3 367 SNPs) | 1.4 | yes | no | [25] |
| Sparus aurata (fish) | protandrous hermaphrodite | LM (204 μsats) | 1.2 | no | no | [26] |
| Hypoplectrus nigricans (fish) | simultaneous hermaphrodite | LM (2 697 SNPs) | 1.3 | yes | yes | this study |
The hamlets (Hypoplectrus spp., Serranidae) provide a rare opportunity to investigate patterns of recombination in the eggs and sperm within individuals in vertebrates. These simultaneously hermaphroditic reef fishes from the wider Caribbean are the poster child for egg trading, whereby the two partners of a mating pair divide their egg clutch into parcels and alternate sex roles to reciprocate the release and fertilization of eggs [27]. Spawning can be observed on a daily basis throughout the year during the hour preceding sunset; individuals engage in an elaborate courtship and spawn in pairs, alternating sex roles up to seven times within a single spawning period that typically lasts about 40 min. There does not appear to be selfing in the hamlets [27], fertilization is external, eggs and larvae are planktonic, and there is no parental care.
The hamlets are also known for the diversity of species within the genus, which differ in terms of colour pattern but are otherwise morphologically and ecologically very similar [28]. Several hypotheses have been put forward to explain speciation in the hamlets including sea-level fluctuations [29], aggressive mimicry [30], and sexual selection [31]. Yet few genomic resources are available for Hypoplectrus. Karyotypic information is lacking, but most species in the Perciformes in general and in the Serranidae in particular have a diploid number of 48 chromosomes that are often acrocentric [32]. Two population genomic studies have identified candidate loci for speciation and local adaptation [33,34], yet their location in the genome remains unknown. Here, we leverage reciprocal crosses between two black hamlets (Hypoplectrus nigricans) to generate egg- and sperm-specific linkage maps for both parents.
2. Material and methods
(a). Experimental crosses
Crosses were carried out in Bocas del Toro (Panama) as detailed in the electronic supplementary material, methods. F1 larvae were collected at 70 h post-fertilization, at which point most of the yolk was resorbed but active feeding had not started yet. This strategy maximized the number of offspring, reduced the potential for DNA contamination from consumed prey items and maternal yolk cells, and ensured that larvae had enough DNA for sequencing.
(b). DNA extraction and sequencing
DNA was extracted from two parents and 120 larvae with DNeasy Blood & Tissue columns using fin clip tissue for the parents and entire larvae for the progeny. Restriction-site associated DNA (RAD) libraries were prepared as detailed in the electronic supplementary material, methods, and sequenced on two lanes of a HiSeq 2000 Illumina sequencer. Because hamlets are simultaneously hermaphroditic and alternate sex roles during spawning [27], we expected to have larvae from two reciprocal crosses in our progeny sample: parent 1 as male × parent 2 as female and vice versa. In order to distinguish between these two situations, we sequenced a 655 bp cytochrome c oxidase subunit I (COI) gene region of mitochondrial DNA in all samples as detailed in the electronic supplementary material, methods.
(c). Filtering, assembly, and single-nucleotide polymorphism selection
Raw sequences were filtered as detailed in the electronic supplementary material, methods. This included the removal of low-quality reads, of reads with an ambiguous index or restrictions site, of reads including adapter sequences, and of pairs of paired-end reads that matched exactly (putative polymerase chain reaction (PCR) clones). In the absence of a reference genome for Hypoplectrus, reads were assembled de novo as detailed in the electronic supplementary material, methods. We applied stringent filtering for single-nucleotide polymorphism (SNP) selection, considering only data from stacks (putative loci) that were present in at least 90% of the larvae with at least 15× coverage. A single SNP was considered per stack (the first one).
(d). Linkage analysis and map construction
One map was generated for each parent using the markers that were heterozygous in that parent and homozygous in the other parent, as well as markers that were heterozygous in both parents. A χ2 test of Mendelian proportions was applied to test for segregation distortion. Linkage analysis and map construction were done with R/qtl v. 1.39-5 [35]. Recombination frequencies and logarithm of the odds (LOD) scores between all pairs of markers were estimated using the est.rf function with a maximum recombination frequency of 0.41 and a minimum LOD score of 4.0. Marker order was estimated with the order.markers function and optimized with ripple using a window size of eight markers. Genetic distances between markers were then estimated with the est.map function using the Kosambi map option. The maps were inspected visually and recombination events supported by less than two markers were removed. Linkage group (LG) sizes were estimated following [36] and [37] and the average between these two estimates was taken as our final LG size estimate. Genome coverage was estimated as the ratio between observed and estimated LG sizes.
In addition, we generated sex-specific maps for each parent. Using the mtDNA haplotypes to infer which parent was the mother of each offspring, this time we only considered the larvae for which the focal parent was the mother (egg map) and only the larvae for which the focal parent was the father (sperm map). The same methods described above were used for linkage analysis and map construction, resulting in four parent/sex-specific maps. For each parent, a G-test was used to test for differences in recombination rates between the sexes considering all pairs of adjacent markers.
(e). Chromosome type and synteny analysis
For each LG, chromosome type was inferred following [38]. Briefly, this approach consists of tracking the cumulative recombination frequency (RFm) along LGs starting from both ends of each LG. Assuming strong chromosome interference, RFm is expected to increase linearly from 0 at the terminal reference marker towards a value of 0.5 at the opposite end for acrocentric and telocentric chromosomes. By contrast, RFm is expected to plateau along the chromosome arm opposite to the terminal reference marker for metacentric chromosomes [38].
A high-resolution synteny analysis is beyond the scope of this study, but a broad-scale synteny analysis was performed between the linkage maps generated here and the genome of the three-spined stickleback (Gasterosteus aculeatus) as detailed in the electronic supplementary material, methods. This was done to validate our maps because relatively good synteny is expected among the Perciformes [39] and to name homologous LGs in a way that is consistent with the stickleback genome to facilitate comparative analyses.
3. Results
The outcomes of the experimental crosses, filtering, assembly, and SNP selection are detailed in the electronic supplementary material, results. The COI haplotypes confirmed that the two parents spawned as both male and female, with 55% and 45% of the larvae matching parent 1 and 2, respectively. A total of 184 182 283 paired-end reads were retained after filtering, corresponding to 54.9% of the raw reads. Preliminary analyses indicated that the 25 larvae with the lowest sequencing coverage provided a large proportion of missing data so these were excluded from downstream analyses, leaving a total of 95 F1 offspring. Mean coverage (number of reads per RAD site per sample) for parent 1 and 2 was 100× and 143×, respectively, and mean coverage for the 95 larvae was 19× (min = 12×, max = 28×).
(a). Linkage analysis and map construction
A total of 1 637 and 1 568 informative SNP markers were recovered for parent 1 and parent 2, respectively, representing 2 697 markers altogether (double heterozygote markers were used in both maps and hence the total number of markers is lower than the sum of the markers used for each map; table 2 and figure 1; electronic supplementary material, figures S2–S4). All SNPs were bi-allelic and no segregation distortion was found (χ2-test p-value > 0.05), suggesting that maternal yolk cells did not contaminate offspring genotypes. Both maps identified 24 LGs (table 2), which is consistent with karyotypic analyses in the Perciformes and Serranidae [32]. Average LG size (52.4 cM) and number of recombination events per meiosis and LG (0.56) were consistent with the occurrence of strong crossover interference and one crossover per bivalent (i.e. a chromosome size of 50 cM and 0.5 recombination events per meiosis and LG [15]).
Table 2.
Summary of linkage maps for parents 1 and 2 considering (i) all offspring, (ii) only the larvae for which the focal parent is the mother (eggs map), and (iii) only the larvae for which the focal parent is the father (sperm map).
| length (cM) |
length (cM) |
length (cM) |
length (cM) |
length (cM) |
length (cM) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| parent 1 |
parent 2 |
|||||||||
| LG | N. markers | Av. spacing (cM) | all | eggs | sperm | N. markers | Av. spacing (cM) | all | eggs | sperm |
| I | 80 | 0.7 | 57.1 | 57.0 | 56.9 | 64 | 0.7 | 41.1 | 50.3 | 34.4 |
| II | 72 | 0.9 | 64.4 | 68.0 | 54.9 | 77 | 0.7 | 52.7 | 67.9 | 41.7 |
| III | 80 | 0.7 | 51.7 | 49.3 | 47.0 | 75 | 0.7 | 50.6 | 61.2 | 42.8 |
| IV | 69 | 0.8 | 51.6 | 55.3 | 39.9 | 69 | 0.8 | 54.4 | 56.5 | 51.5 |
| V | 79 | 0.6 | 48.7 | 57.3 | 36.7 | 65 | 0.8 | 48.1 | 80.9 | 28.3 |
| VI | 70 | 0.8 | 52.2 | 51.3 | 47.5 | 57 | 0.9 | 52.1 | 66.9 | 36.8 |
| VII | 89 | 0.6 | 52.4 | 57.1 | 41.6 | 85 | 0.5 | 45.4 | 65.3 | 45.7 |
| VIII | 59 | 0.9 | 50.5 | 52.6 | 42.2 | 43 | 1.0 | 40.9 | 60.5 | 34.1 |
| IX | 92 | 0.6 | 57.5 | 51.4 | 60.5 | 72 | 0.7 | 50.5 | 49.9 | 34.6 |
| X | 77 | 0.8 | 57.1 | 50.7 | 51.5 | 85 | 0.6 | 48.6 | 70.7 | 31.9 |
| XI | 72 | 0.6 | 45.8 | 41.6 | 50.0 | 70 | 0.6 | 42.4 | 50.0 | 36.1 |
| XII | 73 | 0.8 | 54.9 | 51.3 | 53.0 | 85 | 0.4 | 36.2 | 45.8 | 28.4 |
| XIII | 44 | 1.6 | 67.4 | 66.4 | 41.9 | 56 | 0.9 | 48.5 | 55.5 | 43.9 |
| XIV | 52 | 1.0 | 48.7 | 41.5 | 48.3 | 38 | 1.4 | 51.0 | 65.6 | 40.9 |
| XV | 59 | 1.0 | 59.3 | 51.4 | 52.5 | 27 | 1.7 | 44.0 | 86.7 | 43.7 |
| XVI | 68 | 1.0 | 67.8 | 41.8 | 58.5 | 86 | 0.6 | 47.5 | 66.2 | 28.7 |
| XVII | 86 | 0.6 | 48.0 | 60.8 | 32.3 | 89 | 0.6 | 56.0 | 59.9 | 37.9 |
| XVIII | 68 | 0.8 | 56.7 | 61.0 | 45.7 | 49 | 1.1 | 55.1 | 74.7 | 41.1 |
| XIX | 81 | 0.7 | 55.3 | 45.7 | 57.3 | 87 | 0.7 | 57.5 | 53.2 | 55.5 |
| XX | 66 | 0.9 | 61.1 | 57.1 | 54.2 | 57 | 0.7 | 40.2 | 70.0 | 17.3 |
| XXI | 38 | 1.5 | 54.5 | 52.9 | 47.3 | 59 | 0.6 | 37.2 | 42.8 | 35.1 |
| XXII | 44 | 1.2 | 50.0 | 47.3 | 52.5 | 45 | 1.3 | 55.9 | 68.6 | 47.2 |
| XXIII | 54 | 1.1 | 59.6 | 63.6 | 50.6 | 53 | 1.0 | 50.8 | 58.4 | 46.4 |
| XXIV | 65 | 1.0 | 64.9 | 63.1 | 55.0 | 75 | 0.9 | 70.1 | 71.6 | 71.0 |
| overall | 1 637 | 0.8 | 1 337.2 | 1 295.8 | 1 177.8 | 1 568 | 0.8 | 1 177.0 | 1 499.2 | 954.8 |
| estimated | — | 0.9 | 1 370.6 | 1 328.5 | 1 208.6 | — | 0.8 | 1 201.8 | 1 536.9 | 983.3 |
| coverage (%) | — | — | 97.6 | 97.5 | 97.4 | 97.9 | 97.5 | 97.1 | ||
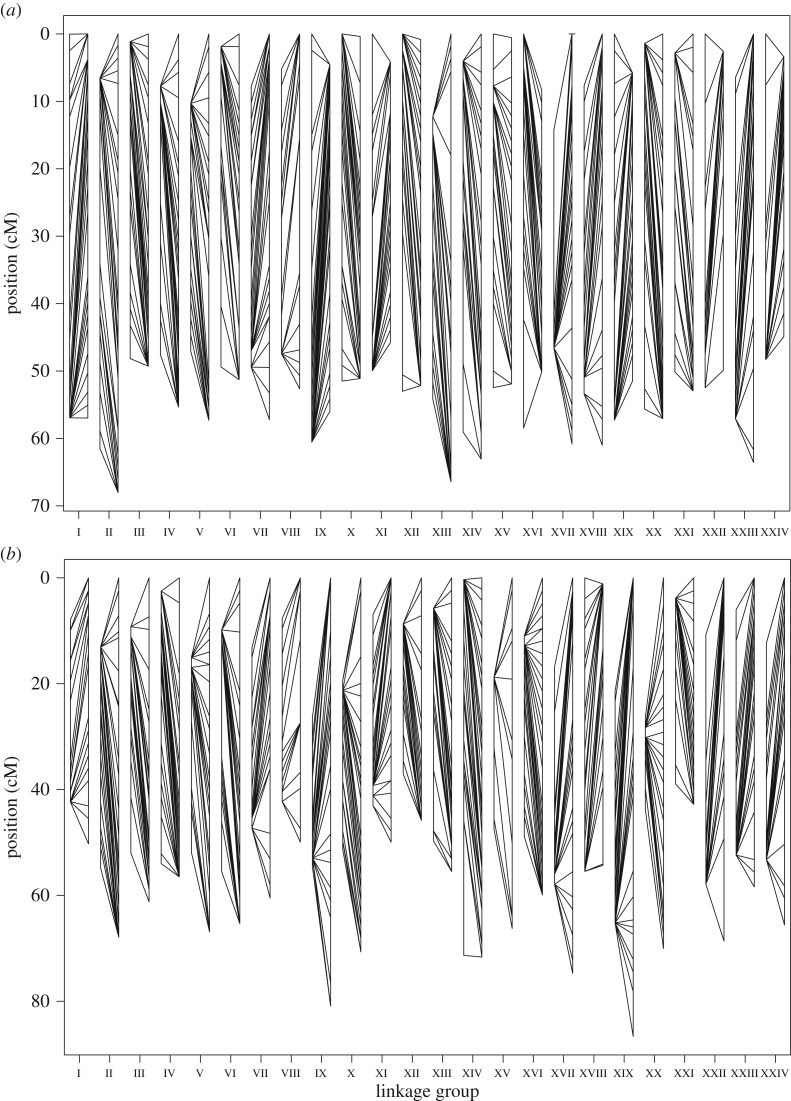
Figure 1.
Linkage maps for parent 1 (a) and parent 2 (b), including both eggs and sperm for each parent. (c,d) Estimated recombination fractions (above diagonal) and LOD scores (below diagonal). Red and blue indicate strong and weak linkage, respectively. Numbers of markers are shown in Latin numbers and LGs in Roman numerals. The occurrence of a large non-recombining region is revealed by a bold red square that covers a large proportion of LG VIII in both parents (highlighted).
Twenty-four LGs were again identified in the four sex-specific maps, with high homology and no conflicts between the sperm and eggs maps for each parent (table 2 and figure 2). For parent 1, the total length of the eggs and sperm map was 1 295.8 cM and 1 177.8 cM, respectively, resulting in an eggs/sperm ratio of 1.1. This ratio was 1.6 for parent 2 and 1.3 across both parents. The vast majority of pairs of adjacent markers presented recombination rates that were either eggs- or sperm-biased (electronic supplementary material, figure S5), and this difference was highly significant between the sexes for both parents (G-test p-values < 0.0001, d.f. = 70 and 104 for parent 1 and parent 2, respectively). Individual LGs tended to be larger in the eggs than in the sperm, but this trend was not entirely consistent (table 2 and figure 2). Yet our data provided the opportunity to characterize heterochiasmy not only at the LG level but also along LGs and a highly consistent pattern was revealed there. All LGs presented a marked difference in recombination patterns between the eggs and sperm, with relatively higher recombination in the eggs at one LG extremity and the opposite at the other extremity (figures 2 and 3; electronic supplementary material, figures S3 and S6). As detailed below, LG VIII stood out as the only exception.
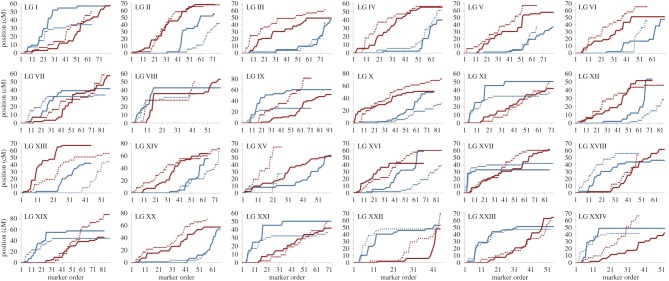
Figure 2.
Comparison between sperm (left) and egg (right) linkage maps for parent 1 (a) and parent 2 (b). The eggs and sperm maps are anchored at the LG midpoint with each line connecting the same marker in the two maps. Differences in recombination between the sexes are particularly pronounced at the LG peripheries (white triangles).
Figure 3.
Position of all markers in the sperm (blue) and eggs (red) in parent 1 (solid lines) and parent 2 (dashed lines) for each LG. All LGs present a marked difference between the eggs and sperm, with relatively higher recombination in the eggs and relatively lower recombination in the sperm at one LG extremity and the opposite at the other extremity. LG VIII stands out as the only exception, with a large non-recombining region in both sexes. The direction in which each map is generated is random.
Both linkage maps revealed a large non-recombining region on LG VIII in both sexes. This is indicated by the two bold red squares in figure 1 (also present in the four sex-specific maps, data not shown), the four horizontal lines in figure 3 for LG VIII, and the large number of markers that mapped to a single location for this LG (electronic supplementary material, figures S2 and S3). A closer look at LG VIII indicated that 37 out of 59 (62.7%) markers in parent 1 and 27 out of 43 (62.8%) markers in parent 2 are located within only 2.1 cM of each other, indicating very low recombination rates across about two-thirds of this LG.
(b). Chromosome type and synteny analysis
All LGs presented a pattern characteristic of acrocentric or telocentric chromosomes (electronic supplementary material, figure S7). Of the 2 697 RAD markers used to build the two maps, 105 (3.9%) mapped to the stickleback genome, representing an average of four to five markers per LG. As expected, high synteny was observed between the hamlet and stickleback LGs (electronic supplementary material, figure S8). For 96 markers out of the 105, markers from a single hamlet LG mapped to a single stickleback LG, allowing the identification of a hamlet homologous LG for the 21 stickleback LGs. As the hamlets have 24 LGs, this leaves three additional hamlet LGs that were named arbitrarily (XXII–XXIV).
4. Discussion
The generation of high-density egg- and sperm-specific linkage maps revealed that heterochiasmy is highly heterogeneous along chromosomes and dramatically pronounced at the chromosome peripheries in H. nigricans, that this pattern is highly consistent across chromosomes and individuals, and that it is not due to genetic background because the same individuals were used to generate egg- and sperm-specific maps. In addition, our analyses indicate that chromosomes are acrocentric or telocentric, i.e. that the centromeres are located close to the chromosome periphery. Among the different hypotheses put forward to explain heterochiasmy [6–14], meiotic drive stands out by precisely predicting heterogeneous patterns of heterochiasmy along chromosomes and stronger heterochiasmy close to the centromeres [8].
(a). Recombination in the eggs and sperm
To the best of our knowledge, this is the first animal study that compares linkage maps in the eggs versus sperm within individuals. The advantage of this approach is clear when contrasting the comparative plots between the two parents (electronic supplementary material, figure S4) versus between the sexes within parents (figure 2). In the former situation, a different set of markers is used because informative markers are the ones that are heterozygous in the focal parent and homozygous in the other parent. Only the double heterozygote markers are shared, and these are relatively few and more difficult to map because they are less informative (which explains why we observe few conflicts between the two parents for these markers). By contrast, the egg and sperm maps for each parent are based on the same markers, providing the opportunity to anchor them at a large number of positions and explore heterochiasmy along LGs. Furthermore, differences in recombination rates due to sex versus individual variation may be confounded in gonochoric species. Altogether, the use of different markers and individuals to compare recombination between the sexes can constitute a potentially important confounding factor, as the difference in recombination rate that we observed between the two parents (parent 1/parent 2 = 1.1) is not too far from the difference observed between the sexes in the hamlets (eggs/sperm = 1.3) and in other species (table 1). This observation also stresses the need to replicate the experiment presented here to draw general conclusions. Finally, and as discussed below, the comparison of recombination rates in the eggs and sperm along LGs provides the opportunity to better interpret heterochiasmy and the hypotheses put forward to explain this phenomenon.
Globally, we observed higher recombination rates in the eggs than in the sperm in the two parents (average female/male ratio = 1.3). In addition, our data show that heterochiasmy is heterogeneous along LGs and particularly pronounced at the LG peripheries. The Limborg et al. [38] method that we applied to our dataset indicates that the 24 hamlet LGs correspond to acrocentric or telocentric chromosomes, but it does not provide the opportunity to identify the centromere location. Thus, karyotypic or cytogenetic data are needed to determine whether higher recombination rates occur close to the centromeres or to the telomeres for each sex.
(b). Possible causes of heterochiasmy
What processes may underlie heterochiasmy in the hamlets? The nature of our reciprocal cross provides the opportunity to identify several unlikely explanations. The effect of sex chromosomes can be ruled out because as simultaneous hermaphrodites the hamlets are not expected to have sex chromosomes, which is confirmed by our sex-specific maps. Differences in metabolic rates between males and females [9] appear unlikely because the reciprocal crosses were conducted with the same two parents and at the same time. Sex-specific differences in dispersal [6,10] can also be ruled out because the hamlets are simultaneously hermaphroditic and gametes do not disperse. In the same line, the particular mating system of the hamlets suggests that sexual selection is not driving heterochiasmy either [10,11]. While there is intense sexual selection in terms of competition among individuals for matings [31], this is not expected to translate into large differences between the sexes because mating success as a male depends on the previous release of eggs [27]. This leaves us with neutrality [6], haploid selection [7,12], and female meiotic drive [8,13] as potential drivers of heterochiasmy. Among these, female meiotic drive stands out by predicting heterogeneous recombination rates along LGs, with pronounced heterochiasmy close to the centromeres. As pointed out by Brandvain and Coop [8], this pattern is not expected otherwise because recombination events close to the centromere do not contribute to the cohesion of homologous chromosomes and are unlikely to be neutral because they can cause segregation problems during meiosis. Importantly, no particular direction of heterochiasmy (i.e. higher versus lower recombination rates in females close to the centromeres) is predicted by this hypothesis and as mentioned above we do not have this information for the hamlets because we do not know on what end of each LG the centromere is located. The formation of quadrivalents during meiosis has been proposed to account for the extreme heterochiasmy observed close to the centromere in rainbow trout [40], yet this phenomenon is due to the fourth round of genome duplication in salmonids [41] and is therefore not directly relevant for serranids. Regarding haploid selection, the relatively small number of genes expressed in sperm and the lack of a haploid phase in females are expected to reduce the potential for selection at the haploid stage, yet the possibility that selection may be operating in the hamlets at the haploid stage or on imprinted genes cannot be ruled out and certainly deserves further investigation through, e.g. gene expression or genomic analyses. Overall, the recombination patterns reported here are consistent with the female meiotic drive hypothesis. This, of course, does not constitute evidence that meiotic drive is indeed driving heterochiasmy in the hamlets, but until additional data are collected we believe that this is the most parsimonious explanation.
(c). Broader context
The observation of higher recombination rates in the eggs than in the sperm in the hamlets is consistent with the trend observed so far in animals, although there are exceptions [8]. No such trend is observed in plants globally, but as pointed out in the Introduction, female recombination rates tend to be lower in gymnosperms and higher in non-selfing angiosperms [7]. Another striking difference between plants and animals is that the vast majority of plants considered in the context of heterochiasmy are hermaphroditic while most animals are gonochoric, which may confound comparisons between the two groups due to the potential for self-fertilization in hermaphrodites and the occurrence of sex chromosomes in many gonochoric species. Few data are available for hermaphroditic animals, but females tend to present higher recombination rates in the cases reported so far (table 1). Yet as pointed out in the Introduction, the vast majority of studies in non-model organisms do not provide information about heterochiasmy along LGs, and hermaphroditic animals are no exception. The ones that do generally indicate higher recombination rates in females close to the centromeres in a variety of animal groups as molluscs [23,25], fishes [42–44], and mammals (including humans [8]). Here again there are exceptions, but the broad picture suggests that it might be quite common among animals and possibly driven by female meiotic drive.
(d). Large non-recombining region
One unexpected finding of this study is the identification of a large non-recombining region on LG VIII. Other low-recombining regions may also be present, but the one on LG VIII stands out in terms of number of markers involved, strength of linkage, and consistency between the four parent/sex-specific maps.
Chromosomal inversions are the usual suspects when it comes to large non-recombining regions, and this may well be the case in Hypoplectrus. Under this scenario, parent 1 would be homozygous for one orientation of the inversion and parent 2 for the opposite orientation, implying that this inversion is polymorphic in H. nigricans and suggesting that it does not have a strong impact on larval development because survival was high for this cross. Chromosomal inversions can facilitate local adaptation and speciation through the reduction of recombination and accumulation of genomic divergence if they encompass genes that are involved in these processes [45,46]. In this context, we note that Tpm4, the single candidate gene for local adaptation identified in the hamlets [34], maps to the stickleback LG VIII (e-value 5 × 10−25, location 14.35 Mb). Assuming synteny between the black hamlet and three-spined stickleback in this region, it would fall inside the putative inversion identified here, suggesting that it could facilitate local adaptation in Hypoplectrus.
(e). Perspectives
This study illustrates the potential of the hamlets to address heterochiasmy in vertebrates. The relatively high spawning success obtained in the experimental crosses, the large number of larvae recovered, and the approximately equal number of larvae produced as a male and female for each parent indicate that the experimental design used here may be replicated, with higher resolution if needed, in a variety of hamlet populations and species. In addition, the possibility to cross individuals from different species [29] provides the potential to explore the role played by recombination and heterochiasmy in hybridization and speciation. Finally, the linkage maps presented here will allow the assembly of a chromosome-level genome for the hamlets, providing the opportunity to characterize the genomic architecture of speciation and adaptation in this group and test our hypothesis that the non-recombining region evidenced here may play a role in these processes.
With the democratization of next-generation sequencing, we now have the ability to routinely genotype thousands of markers in non-model species. A new generation of high-density linkage maps will provide the opportunity to better describe the patterns associated with heterochiasmy in a variety of taxa and, in combination with reference genomes, take a more functional genomics approach to address this process.
Supplementary Material
Acknowledgements
We thank Erin Datlof for her help in the field and in the laboratory, Kosmas Hench for his help with the synteny analysis, Morten T. Limborg for his help for the implementation of his method, and John Davey and Michael A. Quail for providing the RAD adapter sequences.
Ethics
All work was carried out under the Smithsonian Tropical Research Institute IACUC protocol no. 2013-0301-2016 and the Autoridad de los Recursos Acuáticos de Panamá research, collecting and export permit nos. 13 and 41.
Data accessibility
Theodosiou L, McMillan WO, Puebla O (2016). Data from: Recombination in the eggs and sperm in a simultaneously hermaphroditic vertebrate. Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.p61vk.
Authors' contributions
O.P. did the field and laboratory work, L.T. the data analysis, and all authors contributed to the writing.
Competing interests
We have no competing interests.
Funding
This study was funded by a grant from the Smithsonian Competitive Grants Program for Science to O.P., an individual DFG grant to O.P., a Smithsonian Tropical Research Institute short-term fellowship to Erin Datlof and the IMPRS program for Evolutionary Biology.
References
- 1.Morgan TH. 1912. Complete linkage in the second chromosome of the male of Drosophila. Science 36, 719–720. ( 10.1126/science.36.934.719) [DOI] [Google Scholar]
- 2.Haldane JBS. 1922. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 12, 101–109. ( 10.1007/BF02983075) [DOI] [Google Scholar]
- 3.Huxley JS. 1928. Sexual difference of linkage in Gammarus chevreuxi. J. Genet. 20, 145–156. ( 10.1007/BF02983136) [DOI] [Google Scholar]
- 4.Nei M. 1969. Linkage modification and sex difference in recombination. Genetics 63, 681–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell G. 1982. The masterpiece of nature: the evolution and genetics of sexuality, p. 685. Berkeley, CA: University of California Press. [Google Scholar]
- 6.Burt A, Bell G, Harvey PH. 1991. Sex differences in recombination. J. Evol. Biol. 4, 259–277. ( 10.1046/j.1420-9101.1991.4020259.x) [DOI] [Google Scholar]
- 7.Lenormand T, Dutheil J. 2005. Recombination difference between sexes: a role for haploid selection. PLoS Biol. 3, e63 ( 10.1371/journal.pbio.0030063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandvain Y, Coop G. 2012. Scrambling eggs: meiotic drive and the evolution of female recombination rates. Genetics 190, 709–723. ( 10.1534/genetics.111.136721) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein H, Hopf FA, Michod RE. 1988. Is meiotic recombination an adaptation for repairing DNA, producing genetic variation, or both? In The evolution of sex (eds Michod R, Levin BR), pp. 139–160. Sunderland, MA: Sinauer Press. [Google Scholar]
- 10.Mank JE. 2009. The evolution of heterochiasmy: the role of sexual selection and sperm competition in determining sex-specific recombination rates in eutherian mammals. Genet. Res. 91, 355–363. ( 10.1017/S0016672309990255) [DOI] [PubMed] [Google Scholar]
- 11.Trivers R. 1988. Sex differences in rates of recombination and sexual selection. In The evolution of sex (eds Michod R, Levin BR), pp. 270–286. Sunderland, MA: Sinauer Press. [Google Scholar]
- 12.Lenormand T. 2003. The evolution of sex dimorphism in recombination. Genetics 163, 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haig D. 2010. Games in tetrads: segregation, recombination, and meiotic drive. Am. Nat. 176, 404–413. ( 10.1086/656265) [DOI] [PubMed] [Google Scholar]
- 14.Joseph S, Kirkpatrick M. 2004. Haploid selection in animals. Trends Ecol. Evol. 19, 592–597. ( 10.1016/j.tree.2004.08.004) [DOI] [Google Scholar]
- 15.Lenormand T, Engelstädter J, Johnston SE, Wijnker E, Haag CR. 2016. Evolutionary mysteries in meiosis. Phil. Trans. R. Soc. B 371, 1–14, 20160001 ( 10.1098/rstb.2016.0001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pastor JB, Callan HG. 1952. Chiasma formation in spermatocytes and oocytes of the turbellarian Dendrocoelum lacteum. J. Genet. 50, 449–454. ( 10.1007/BF02986840) [DOI] [Google Scholar]
- 17.Galleni L, Puccinelli I.. 1975. Karyology of Notoplana igiliensis Galleni (Polycladida: Acotylea). Caryologia 28, 375–387. ( 10.1080/00087114.1975.10796626) [DOI] [Google Scholar]
- 18.Dhar VN, Sharma GP. 1984. Behaviour of chromosomes during gametogenesis and fertilization in Paradistomoides orientalis (Digenea: Trematoda). Caryologia 37, 207–218. ( 10.1080/00087114.1984.10797699) [DOI] [Google Scholar]
- 19.L'Hardy J-P. 1986. Karyology of a marine population of Gyratrix hermaphroditus (Turbellaria, Rhabdocoela) and chromosomal evolution in this species complex. Hydrobiologia 132, 233–238. ( 10.1007/BF00046254) [DOI] [Google Scholar]
- 20.Pongratz N, Gerace L, Alganza AM, Beukeboom LW, Michiels NK. 2001. Microsatellite development and inheritance in the planarian flatworm Schmidtea polychroa. Belg. J. Zool. 131, 71–75. [Google Scholar]
- 21.Wang S, Zhang L, Meyer E, Matz MV. 2009. Construction of a high-resolution genetic linkage map and comparative genome analysis for the reef-building coral Acropora millepora. Genome Biol. 10, R126 ( 10.1186/gb-2009-10-11-r126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Liu X, Zhang G. 2012. A consensus microsatellite-based linkage map for the hermaphroditic bay scallop (Argopecten irradians) and its application in size-related QTL analysis. PLoS ONE 7, e46926 ( 10.1371/journal.pone.0046926) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones DB, Jerry DR, Khatkar MS, Raadsma HW, Zenger KR. 2013. A high-density SNP genetic linkage map for the silver-lipped pearl oyster, Pinctada maxima: a valuable resource for gene localisation and marker-assisted selection. BMC Genomics 14, 810 ( 10.1186/1471-2164-14-810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu Z, Guo X.. 2003. Genetic linkage map of the eastern oyster Crassostrea virginica gmelin. Biol. Bull. 204, 327–338. ( 10.2307/1543603) [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Li L, Zhang G. 2016. A high-density SNP genetic linkage map and QTL analysis of growth-related traits in a hybrid family of oysters (Crassostrea gigas × Crassostrea angulata) using genotyping-by-sequencing. G3 6, 1417–1426. ( 10.1534/g3.116.026971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franch R, et al. 2006. A genetic linkage map of the hermaphrodite teleost fish Sparus aurata L. Genetics 174, 851–861. ( 10.1534/genetics.106.059014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer EA. 1980. The relationship between mating system and simultaneous hermaphroditism in the coral reef fish, Hypoplectrus nigricans (Serranidae). Anim. Behav. 28, 620–633. ( 10.1016/S0003-3472(80)80070-4) [DOI] [Google Scholar]
- 28.Lobel PS. 2011. A review of the Caribbean hamlets (Serranidae, Hypoplectrus) with description of two new species. Zootaxa 3096, 1–17. [Google Scholar]
- 29.Domeier ML. 1994. Speciation in the serranid fish Hypoplectrus. Bull. Mar. Sci. 54, 103–141. [Google Scholar]
- 30.Puebla O, Bermingham E, Guichard F, Whiteman E. 2007. Colour pattern as a single trait driving speciation in Hypoplectrus coral reef fishes? Proc. R. Soc. B 274, 1265–1271. ( 10.1098/rspb.2006.0435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puebla O, Bermingham E, Guichard F. 2012. Pairing dynamics and the origin of species. Proc. R. Soc. B 279, 1085–1092. ( 10.1098/rspb.2011.1549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arai R. 2011. Fish karyotypes. Tokyo, Japan: Springer. [Google Scholar]
- 33.Puebla O, Bermingham E, McMillan WO. 2014. Genomic atolls of differentiation in coral reef fishes (Hypoplectrus spp., Serranidae). Mol. Ecol. 23, 5291–5303. ( 10.1111/mec.12926) [DOI] [PubMed] [Google Scholar]
- 34.Picq S, McMillan WO, Puebla O. 2016. Population genomics of local adaptation versus speciation in coral reef fishes (Hypoplectrus spp, Serranidae). Ecol. Evol. 6, 2109–2124. ( 10.1002/ece3.2028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broman KW, Wu H, Sen Ś, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890. ( 10.1093/bioinformatics/btg112) [DOI] [PubMed] [Google Scholar]
- 36.Chakravarti A, Lasher LK, Reefer JE. 1991. A maximum likelihood method for estimating genome length using genetic linkage data. Genetics 128, 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fishman L, Kelly AJ, Morgan E, Willis JH. 2001. A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159, 1701–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Limborg MT, McKinney GJ, Seeb LW, Seeb JE. 2016. Recombination patterns reveal information about centromere location on linkage maps. Mol. Ecol. Resour. 16, 655–661. ( 10.1111/1755-0998.12484) [DOI] [PubMed] [Google Scholar]
- 39.Tine M, et al. 2014. European sea bass genome and its variation provide insights into adaptation to euryhalinity and speciation. Nat. Commun. 5, 5770 ( 10.1038/ncomms6770) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakamoto T, et al. 2000. A microsatellite linkage map of rainbow trout (Oncorhynchus mykiss) characterized by large sex-specific differences in recombination rates. Genetics 155, 1331–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allendorf FW, Thorgaard GH. 1984. Tetraploidy and the evolution of salmonid fishes. In Evolutionary genetics of fishes (ed. Turner BJ.), pp. 1–53. New York, NY: Plenum Press. [Google Scholar]
- 42.Singer A, Perlman H, Yan Y, Walker C, Corley-Smith G, Brandhorst B, Postlethwait J. 2002. Sex-specific recombination rates in zebrafish (Danio rerio). Genetics 160, 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reid DP, Smith C-A, Rommens M, Blanchard B, Martin-Robichaud D, Reith M. 2007. A genetic linkage map of Atlantic halibut (Hippoglossus hippoglossus L.). Genetics 177, 1193–1205. ( 10.1534/genetics.107.075374) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lien S, Gidskehaug L, Moen T, Hayes BJ, Berg PR, Davidson WS, Omholt SW, Kent MP et al. 2011. A dense SNP-based linkage map of Atlantic salmon (Salmo salar) reveals extended chromosome homeologies and striking differences in sex-specific recombination patterns. BMC Genomics 12, 615 ( 10.1186/1471-2164-12-615) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones FC, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484, 55–61. ( 10.1038/nature10944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berg PR, Star B, Pampoulie C, Sodeland M, Barth JMI, Knutsen H, Jakobsen KS, Jentoft S et al. 2016. Three chromosomal rearrangements promote genomic divergence between migratory and stationary ecotypes of Atlantic cod. Sci. Rep. 6, 23246 ( 10.1038/srep23246) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Theodosiou L, McMillan WO, Puebla O (2016). Data from: Recombination in the eggs and sperm in a simultaneously hermaphroditic vertebrate. Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.p61vk.