Abstract
Human brains are markedly asymmetric in structure and lateralized in function, which suggests a relationship between these two properties. The brains of other closely related primates, such as chimpanzees, show similar patterns of asymmetry, but to a lesser degree, indicating an increase in anatomical and functional asymmetry during hominin evolution. We analysed the heritability of cerebral asymmetry in chimpanzees and humans using classic morphometrics, geometric morphometrics, and quantitative genetic techniques. In our analyses, we separated directional asymmetry and fluctuating asymmetry (FA), which is indicative of environmental influences during development. We show that directional patterns of asymmetry, those that are consistently present in most individuals in a population, do not have significant heritability when measured through simple linear metrics, but they have marginally significant heritability in humans when assessed through three-dimensional configurations of landmarks that reflect variation in the size, position, and orientation of different cortical regions with respect to each other. Furthermore, genetic correlations between left and right hemispheres are substantially lower in humans than in chimpanzees, which points to a relatively stronger environmental influence on left–right differences in humans. We also show that the level of FA has significant heritability in both species in some regions of the cerebral cortex. This suggests that brain responsiveness to environmental influences, which may reflect neural plasticity, has genetic bases in both species. These results have implications for the evolvability of brain asymmetry and plasticity among humans and our close relatives.
Keywords: brain evolution, primates, environment, geometric morphometrics, fluctuating asymmetry, quantitative genetics
1. Introduction
For more than a century, anatomical observations and functional studies have demonstrated that human brains are markedly asymmetric. This asymmetry is especially notable in areas of the cerebral cortex that are involved in higher-order cognition and language, such as the inferior frontal, superior temporal, and inferior parietal regions [1–4]. For example, functional studies have shown a high density of unilateral activation peaks for language-related tasks in these frontal and parietal perisylvian areas, particularly in the left hemisphere [5]. These findings suggest that anatomical asymmetry is linked to functional lateralization [6,7], which is thought to optimize processing speed and synchronization through minimized wiring in large brains [8].
Subsequent studies have demonstrated that chimpanzees, one of the closest living relatives of humans, also show similar anatomical asymmetries, although to a lesser degree [9–12]. Other studies have further demonstrated that behavioural lateralization, especially handedness for different tasks, is common in chimpanzees and other great apes, although population-level handedness is not as pronounced as in humans [13–15]. Additionally, neuroimaging studies of chimpanzees have shown functional lateralization in Broca's area homologue related to communicative behaviour [16] and in the hand knob, the motor-hand region of the precentral gyrus, in relation to reach-and-grasping responses [17]. These observations, together with endocranial changes evident in the hominin fossil record [18–20], indicate that cerebral asymmetry was likely present in the last common ancestor of chimpanzees and humans by 6–8 Ma and in early hominins, but that it has increased during hominin evolution, probably in parallel with the evolution of greater functional lateralization [21].
Most previous studies have focused on directional patterns of cerebral asymmetry. Directional asymmetries are defined as those that are consistently identified in most individuals in a given population and are considered to have a genetic origin. We have recently shown, however, that the human brain is characterized not only by a strong degree of directional asymmetry (DA) when compared with chimpanzees, but also by a high degree of fluctuating asymmetry (FA) [12]. FA corresponds to random departures from the population-specific mean DA, and it is usually considered to result from the impact of environmental influences on developmental processes [22]. The most classic account for FA is that it is the outcome of developmental instability, that is the inability of individuals to buffer the effects of various perturbations during development [23]. We have proposed, however, that the high degree of FA observed in healthy human brains is more likely indicative of a high level of developmental plasticity, a hypothesis that is further supported by the low heritability for cortical anatomy observed in human brains compared to chimpanzees [24].
The available evidence, therefore, indicates that certain aspects of brain lateralization are genetically determined, whereas other features of anatomical asymmetry might be the result of environmental influences during development. In order to tease apart the causal factors underlying the phenotypic expression of brain asymmetries and their evolution, in the current study we evaluate the heritability of different forms of brain asymmetry and the genetic correlations between variables measured in the left and right sides in humans and chimpanzees. Based on the observation that human brains are structurally and functionally more asymmetric than chimpanzee brains, as well as more plastic, we have three major hypotheses. First, we hypothesize that heritability for directional cerebral asymmetry will be higher in human than in chimpanzee brains. Second, we hypothesize that environmental influences on brain asymmetry will be stronger in humans. Third, we hypothesize that FA will be genetically heritable, reflecting the capacity for plasticity to evolve.
2. Material and methods
(a). Samples and magnetic resonance imaging scans
A sample of 206 chimpanzee (79 males, 127 females, age range: 8–53 years) and 218 human (87 males, 131 females, age range: 22–30 years) magnetic resonance imaging (MRI) scans was used. Chimpanzees used in this study were housed at the Yerkes National Primate Research Center (YNPRC) in Atlanta, GA, USA, and at the National Center for Chimpanzee Care (NCCC) at The University of Texas MD Anderson Cancer Center (UTMDACC) in Bastrop, TX, USA. Chimpanzees were scanned using a 3T scanner (Siemens Trio, Siemens Medical Solutions, Malvern, PA, USA) or a 1.5T scanner (Phillips, Model 51, Philips Medical Systems, N.A., Bothell, WA, USA). Technical details regarding scanning procedures and processing can be found in [25]. No paternity tests were conducted for the purposes of this study, but a well-documented pedigree is available for these chimpanzees, which includes information on mother, father, and offspring identity for many individuals.
Human MRI scans were obtained from the Human Connectome Project (HCP) database [26]. Individuals were scanned with a Siemens Skyra 3T scanner. Technical details regarding scanning procedures and processing in human subjects can be found in [26,27]. Consent from human participants was obtained in the context of the HCP, and data-use terms for open and restricted data were accepted and observed as per HCP requirement [28]. The HCP database includes monozygotic twins, non-monozygotic twins, and non-twin siblings. To maximize sample size and minimize inter-population variability due to genetic ancestry, which might correlate with general brain anatomy [29], only individuals with the same ancestry (as self-reported) were selected.
(b). Three-dimensional reconstructions and landmarks
Three-dimensional models of the cerebral cortical surface were reconstructed from MRI scans using BrainVisa software [30] for chimpanzees and FreeSurfer software [31] for humans (three-dimensional models were directly obtained from the HCP database for the human sample). Thirty-two anatomically homologous landmarks (16 bilateral landmarks) were placed on the intersections and extreme points of the most constant sulci on the chimpanzee and human cortical surface [12,24] (electronic supplementary material, figure S1 and table S1). Because of the anatomical complexity of the human cortical surface, which makes it difficult to identify some sulci, landmark placement was aided by a comparison with automatically parcellated models. These parcellated models, obtained with FreeSurfer software v. 5.3.0 according to the Desikan surface atlas [32], are provided in the HCP database. These or similar configurations of landmarks have been previously used in our other studies of brain variation in chimpanzees and humans [12,24,33].
(c). Linear metrics and asymmetry quotients
Linear distances were calculated between several pairs of landmarks as a measure of the general proportions of the major lobes of the brain and of the length of the most prominent sulci (electronic supplementary material, table S2). These distances are linear approximations and they do not include variation along the course of a given sulcus. Linear distances were measured separately for the right and left sides in order to measure heritabilities for each side and genetic correlations between correspondent variables in each hemisphere (see below). Additionally, linear distances were used to measure asymmetry quotients (AQs) for all the variables, the heritability of which was estimated as well. AQs were calculated as the value of a variable in the right hemisphere minus the value of that variable in the left hemisphere, divided by the mean of that variable in both hemispheres ((R − L) × 100)/((R + L) × 0.5). Linear metrics were measured in Procrustes-superimposed configurations of landmarks (see below) because original distances are highly influenced by brain size, even if brain size has a quantitatively very small effect on sulcal anatomy [12]. However, some of the studied variables, such as AQs, are independent of total size, so this transformation does not have any effect in this case.
(d). Geometric morphometrics
Configurations of landmarks were also studied in a geometric morphometric context. Original configurations of landmarks were Procrustes-superimposed to remove information regarding the location, orientation, and size of the original specimens [34]. Each configuration was later mirror-imaged and relabelled following Klingenberg et al. [35]. The mean of the original and mirror-imaged configurations yielded a symmetric consensus configuration for each individual, whereas the difference between both configurations corresponded to the asymmetric component of shape variation [35]. The asymmetric component of variation was analysed through separate principal component analyses (PCAs) for each species. The first five principal components (PCs) for each species were explored in further detail.
PCs were tested for their association with the pattern of DA typical of each species, which was calculated by averaging the asymmetric components of shape variation of each individual for each species (in other words, DA in shape was calculated simply as the mean shape asymmetry for each species). These comparisons tested if variation associated with each PC is similar to the pattern of DA observed in the population or whether variation is not aligned with this population-typical pattern. The association between each PC and DA was measured by calculating the angle (α) between each eigenvector and the species-specific DA vector, which was calculated as the arccosine of the inner product of both vectors. An angle of α = 0° indicates a correlation of 1 between two vectors, whereas an angle of α = 90° indicates a correlation of 0. Significance was tested against a null distribution of 1 000 angles formed between randomly selected vectors. For vectors of the length included in our study, α = 78.42° is the significance threshold above which vectors are uncorrelated.
Additionally, FA scores were calculated for each individual as the difference between individual configurations of landmarks and the norm DA configuration typical of each species [36,37]. FA scores are calculated across all landmarks and represent the extent to which each individual departs from the norm DA pattern. An FA score of 0 indicates that a given individual shows exactly the same pattern of asymmetry that is defined as characteristic of the population, whereas a high FA score indicates that individuals depart from this population-specific pattern, regardless of the identity of the particular anatomical variation that is driving this departure.
(e). Quantitative genetics
Variance components and heritabilities were estimated using an animal model approach implemented in the R package MCMCglmm [38]. In evolutionary biology and quantitative genetics, an ‘animal model’ is a particular type of mixed-effects statistical model that can be used to decompose phenotypic variance into different genetic and environmental sources and to estimate key parameters such as the heritability and the genetic correlation between traits [39]. For humans, the classic implementation of MCMCglmm was changed as proposed in [40] to use the kinship matrix instead of a pedigree, which was necessary to include the degree of genetic similarity corresponding to monozygotic twins. All data were standardized prior to analysis by subtracting the mean from each individual value and dividing the difference by the standard deviation. Sex, age, and the interaction between sex and age were used as fixed effects in both species. Additionally, scanner type was included in chimpanzee analyses to account for the possible effect of using two different scanners. Phenotypic and genetic correlations between corresponding left and right variables were tested using bivariate animal models, which used the same fixed effects. Following other studies [41], we used slightly informative priors of the form (V = Vp/r, ν = 1), where Vp is the phenotypic variance and r the number of random factors, modified as (V = diag(n) × Vp/r, ν = n), where n is the number of traits, for bivariate analyses. Because all variables were standardized to a variance of 1 and all models included only one random factor, priors had the form (V = 1, ν = 1) for univariate models and (V = diag(2), ν = 2) for bivariate models. Parameter-expanded priors [42,43] yielded similar overall results, but they more often tended to result in null estimates. Models were run for 1 000 000 iterations, during which model parameters were updated. The first 500 000 iterations were discarded as a burn-in period and posterior distributions were sampled every 100th iteration to a final amount of 5 000 samples.
Significance of fixed effects was evaluated by assessing if 95% highest posterior density intervals include 0, which is indicative of non-significance. The significance of phenotypic and genetic correlations can be tested in the same way. Variance components from which heritability is estimated, however, are bound to be positive and posterior distributions will not overlap 0, even if their effect is not significant. We tested the significance of heritability estimates by comparing the deviance information criterion (DIC) in models including pedigree/kinship information and in models excluding it, which yielded a DIC differential value (ΔDIC). The significance of heritability was assessed using a simulation approach consisting of measuring the heritability of random variables using the same population structure and models [44]. By construction, these variables do not have significant heritability as values are randomly assigned to individuals. P-values were calculated as the proportion of 1 000 simulations yielding higher ΔDIC than each evaluated variable.
3. Results
(a). Description of asymmetry
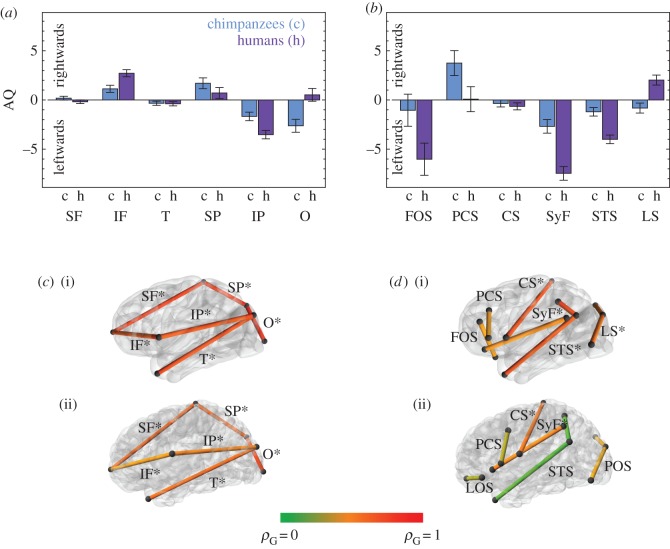
AQs based on interlandmark distances are roughly consistent with previous studies of AQs based on detailed sulcal anatomy [25]. In general, chimpanzees and humans show the same direction of AQ patterns, although values are greater in humans (figure 1). Distances related to the perisylvian region, such as the inferior parietal length and the lengths of the Sylvian fissure and of the superior temporal sulcus show a clear leftward bias in both species, although it is stronger in humans than in chimpanzees. Variables related to other regions, such as the frontal and occipital lobes, do not show as consistent asymmetry patterns, either between species or across different variables within each region.
Figure 1.
Analysis of asymmetry based on interlandmark linear distances. (a) Asymmetry quotients (AQs) for lobe proportions (mean AQs and standard errors). (b) AQs for sulcal lengths. (c) Genetic correlations between left and right lobe proportions in chimpanzees (i) and humans (ii). (d) Genetic correlations between left and right sulcal lengths in chimpanzees (i) and humans (ii). Asterisks mark significant genetic correlations in (c) and (d). No AQ shows significant heritability in (a) and (b). Numerical values for heritabilities and colour-coded genetic correlations are provided in the electronic supplementary material, tables S5, S8–S10. SF, superior frontal length; IF, inferior frontal length; T, temporal length; SP, superior parietal length; IP, inferior parietal length; O, occipital length; FOS, fronto-orbital sulcus (latero-orbital sulcus—LOS—in humans); PCS, precentral sulcus; CS, central sulcus; SyF, Sylvian fissure; STS, superior temporal sulcus; LS, lunate sulcus (parieto-occipital sulcus—POS—in humans); ρG, genetic correlation.
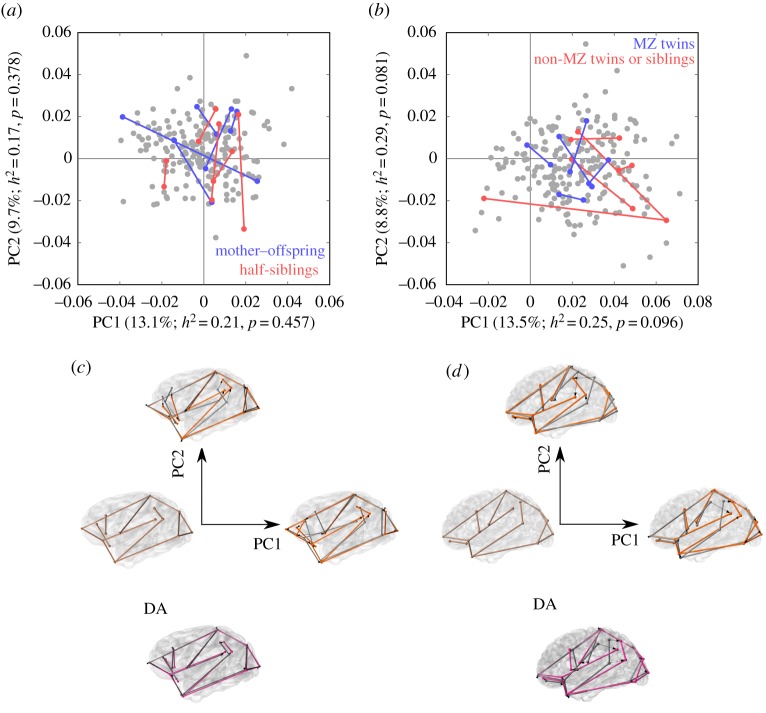
Geometric morphometric analyses show that directional asymmetric variation is concentrated in the inferior parietal area in both species, although those changes are much more marked in humans, where they also involve a strong reorientation of the Sylvian fissure that is not observed in chimpanzees (figure 2). The general pattern of DA in humans also includes some changes in the inferior frontal and in the occipital regions. The distribution of individuals in PCA plots shows additional evidence of the stronger degree of DA in humans, as demonstrated by the off-centred position of more symmetric individuals with respect to the range of variation of the population in humans, but not in chimpanzees (figure 2).
Figure 2.
Geometric morphometric analysis of asymmetry. (a) Principal component analysis (PCA) of asymmetric shape variation in chimpanzees showing five randomly selected mother–offspring and half-sibling pairs (50 versus 25% genetic similarity). (b) PCA of asymmetric shape variation showing five randomly selected pairs of monozygotic twins and of non-monozygotic twins or non-twin siblings (100 versus 50% genetic similarity). PCA plots in (a,b) are centred on a hypothetical perfectly symmetric individual. The percentage of variance explained by each PC and their heritabilities and p-values are provided (see the electronic supplementary material, tables S11 and S12 for extended information). (c,d) Major patterns of shape variation in (c) chimpanzees and (d) humans, showing the symmetric consensus for each species and major patterns of variation corresponding to the positive extremes of PC1 and PC2 (grey for the right hemisphere and orange for the left). The DA pattern for each species is shown on the bottom panels. For DA, grey corresponds to the right hemisphere and magenta to the left hemisphere. PC1, PC2, and DA shape variation has been exaggerated beyond the range observed in actual data to facilitate visualization.
(b). Heritabilities and genetic correlations
Our results show that both chimpanzees and humans have significant heritability in most lobe proportions, with the exception of frontal dimensions in the left hemisphere in humans (electronic supplementary material, tables S3 and S4). Although some studies have evaluated the evolution of lateralization through differential heritability in the left and right sides [45,46], as well as through different evolutionary trends of variables measured in the left and right hemispheres [47], our study does not show consistently higher heritabilities for one hemisphere or the other, barring the two non-significant values in humans, which correspond to the left hemisphere. Genetic correlations between corresponding left and right lobe proportions are high in chimpanzees (figure 1; electronic supplementary material, table S5). Genetic correlations are also high in humans, although they are slightly lower than in chimpanzees (figure 1; electronic supplementary material, table S5).
Heritability for sulcal lengths is substantially higher in chimpanzees than in humans, as has been demonstrated previously [24]. As with lobe proportions, no consistent pattern of higher heritabilities in the left or right hemisphere is observed in either species (electronic supplementary material, tables S6 and S7). Genetic correlations between matching left and right sulcal lengths are in general significant and relatively high for chimpanzees, although there are some exceptions (figure 1; electronic supplementary material, table S8). In humans, most genetic correlations between sulcal lengths in the left and right hemispheres are not significant, with the exception of the correlation between the left and right central sulci, and the left and right Sylvian fissures (figure 1; electronic supplementary material, table S8). These results indicate that covariation between the left and the right hemispheres is more strongly genetically determined in chimpanzees, whereas it is exposed to higher environmental influence in humans.
(c). Heritability of asymmetry
The analysis of the heritability of AQs for lobe proportions and sulcal lengths results in generally non-significant values and in marginally significant values only for a few AQs (electronic supplementary material, tables S9 and S10). This result is initially surprising, because some of these patterns of asymmetry are known to represent very consistent DA patterns, which are expected to be genetically determined. However, it is possible that AQs based on linear metrics do not have sufficient resolution to detect the genetic origin of brain asymmetries. We further explored this point by measuring the heritability of particular aspects of asymmetric shape variation summarized by PC1–PC5 (electronic supplementary material, tables S11 and S12). These principal components of shape are based on three-dimensional configurations of landmarks, and include all aspects of shape variation, such as the size, position, and orientation of the cortical regions included in those configurations. In humans, PC1 and PC2 are the only principal components of asymmetric shape variation that have marginally significant heritability as inferred from our simulation-based significance threshold (PC1: h2 = 0.25, ΔDIC = 16.15, p = 0.096; PC2: h2 = 0.29, ΔDIC = 17.86, p = 0.081; figure 2; electronic supplementary material, table S12). Interestingly, PC1 is the principal component of shape variation that shows the closest correspondence with DA in humans (α = 36.4°; p < 0.001; electronic supplementary material, table S12). In chimpanzees, no single PC is strongly associated with the DA vector, although PC2 shows a slight correlation with DA (α = 64.7°; p < 0.001; electronic supplementary material, table S11). Principal components of asymmetric shape variation in chimpanzees do not to show significant or marginally significant heritability.
Individual FA scores are substantially higher in humans than in chimpanzees (figure 3), which is consistent with our previous report based on Procrustes ANOVAs [12]. When calculating FA scores for total cortical anatomy and for the three major lobes of the brain (frontal, temporo-parietal, and occipital), we observed that one of these values has significant heritability for each species (table 1): occipital FA for humans (h2 = 0.43, ΔDIC = 42.6, p = 0.005) and total FA for chimpanzees (h2 = 0.41, ΔDIC = 33.3, p = 0.028), with chimpanzees also showing marginally significant heritability for the frontal lobe (h2 = 0.32, ΔDIC = 23.7, p = 0.074). This result shows that the general level of FA, which is indicative of the propensity to have a brain that departs from species' typical configurations regardless of the particular changes driving this departure, is in part genetically heritable in both species.
Figure 3.
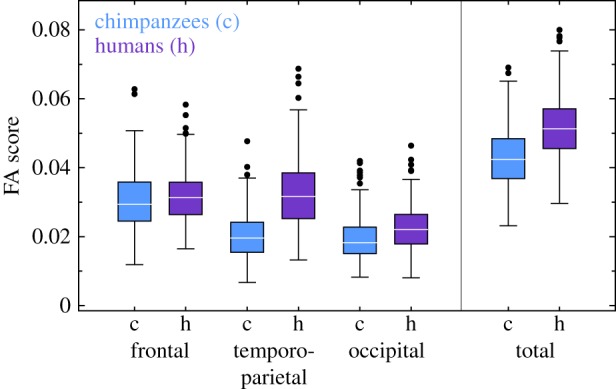
FA scores for chimpanzees and humans. FA scores have been calculated as the residual variation in each individual after removing the DA pattern typical of each species. Heritabilities of FA scores are provided in table 1. (Online version in colour.)
Table 1.
Heritability of FA scores. h2, heritability; HPDI, 95% highest posterior density interval (credible intervals indicating that the heritability of each trait has 95% of probability to lie between the lower and the upper bounds); ΔDIC (p), difference in the DIC between the model with and without pedigree/kinship information (p-value); fixed, significant fixed effects.
| chimpanzees |
humans |
|||||||
|---|---|---|---|---|---|---|---|---|
| h2 | HPDI | ΔDIC (p) | fixed | h2 | HPDI | ΔDIC (p) | fixed | |
| frontal | 0.32 | 0.12–0.58 | 23.68 (0.074) | — | 0.17 | 0.07–0.39 | 5.13 (0.415) | — |
| temporo-parietal | 0.21 | 0.08–0.47 | 9.30 (0.358) | — | 0.17 | 0.08–0.36 | 2.58 (0.546) | — |
| occipital | 0.23 | 0.10–0.45 | 9.00 (0.373) | — | 0.43 | 0.17–0.68 | 42.65 (0.005) | — |
| total | 0.41 | 0.14–0.63 | 33.28 (0.028) | — | 0.19 | 0.08–0.40 | 5.56 (0.338) | — |
4. Discussion
Comparisons of heritability values across different populations or species are unavoidably influenced by the different environmental conditions in which different groups live. Indeed, heritability estimates are specific to the groups and conditions in which they were obtained, and they cannot be generalized to other circumstances. This point is particularly important because of the very different environmental conditions corresponding to our chimpanzee and human samples, with chimpanzees living in the more homogeneous conditions typical of captive habitats. These differences, however, are much more likely to be reflected in behavioural phenotypes than in neuroanatomical phenotypes. However, differences in the relatedness structure of the chimpanzee and human samples are likely to have a stronger effect on our results. Analyses of brain size have shown that heritability estimates based on twins (as in our human sample) tend to be higher than those based on extended pedigrees (as in our chimpanzee sample) [48]. An implication of this observation is that human heritabilities yielded by our analyses are likely to be overestimated in comparison with chimpanzee heritabilities. With this in mind, we focus our discussion on the comparison of the heritability of different variables within each species.
Our results shed light on the heritability of directional and fluctuating brain asymmetry in humans and chimpanzees. These two types of asymmetry have different bases in genetics and development, each with distinct implications for the evolutionary origin of neural structure and function. Classic studies of human brain anatomy have focused on directional asymmetries [1,4,45], as they are more consistent and, therefore, easier to identify, and because they have well known functional correlates. Our study, however, highlights the importance of FA, which, according to various lines of evidence [12,24], may be interpreted to reflect variation due to plasticity in normal brain development.
(a). Directional asymmetry and functional lateralization
Because DA of the brain is usually assumed to be genetically determined, our finding that most AQs do not show significant heritability in either species does not fit our hypotheses and is initially surprising. Studies of heritability in human neuroanatomy have reported differential heritability for some variables (lobar volume and grey matter distribution) in both hemispheres [45,46]. However, direct evaluations of the heritability of brain asymmetry in humans are not common in the literature [49], which may reflect a publication bias resulting from negative results. In chimpanzees, however, it has been reported that the AQ of grey matter volume shows low but significant heritability in the posterior region of the superior temporal gyrus, but not in the inferior frontal gyrus [49]. Because our previous studies have demonstrated that FA is preferentially located in the inferior frontal region in chimpanzees [12], we hypothesize that significant heritability for DA may be harder to identify in brain regions with strong FA. However, our study does not identify significant heritability for the AQ of the superior temporal sulcus, even though this region does not show particularly high FA in chimpanzees. This difference may result from the lack of separation between the anterior and posterior segments of the superior temporal sulcus in our study, or it may indicate that DA in grey matter volume is more heritable than landmark-based sulcal lengths.
When exploring more complex patterns of asymmetric shape variation as described by the three-dimensional configurations of landmarks, chimpanzees and humans show some similarities in their major patterns of DA, namely the difference in size and orientation between the left and right superior temporal sulci. In humans, the major pattern of DA is associated with the first principal component of shape variation, which is one of the PCs that show marginally significant heritability as determined by our simulation-based significance threshold. These results indicate that complex patterns of asymmetry, which include all parameters of shape variation (size, position, and orientation of the different cortical regions with respect to each other), may show moderate but significant heritability in larger samples and, therefore, some level of genetic control.
Our results are consistent with studies showing low to moderate heritability for neuroanatomical asymmetries in primates [49–51], which contrasts with other studies yielding substantially higher heritability for behavioural lateralization in chimpanzees and humans, usually measured as handedness [52,53]. This apparent paradox highlights the difficulty in drawing direct associations between structural and functional asymmetry. Studies of heritability based on functional neuroimaging in humans, which might serve as an interface between neuroanatomical and behavioural studies, are particularly uncommon [54], which makes it challenging to bridge both types of observations.
(b). Fluctuating asymmetry and plasticity
FA was indirectly measured in our study through the analysis of genetic correlations between the left and right hemispheres. These results show that inter-hemispheric genetic correlations are high for all variables in chimpanzees. In humans, however, general lobe proportions and evolutionary and developmentally primary sulci (such as the central sulcus and the Sylvian fissure) show high genetic correlations between the left and right sides, whereas other sulci show low and not significant correlations. This difference points to a greater environmental influence on left–right differences in humans. Some authors have suggested that ‘in the absence of differential developmental effects, the correlation between the two sides of the same organ should be 1’ ([55], p. 708). This expectation is true for perfectly symmetric organs and for those showing genetically determined DA. Lower inter-hemispheric genetic correlations observed in human brains indicate greater non-genetic developmental effects than in chimpanzees. We interpret this result to reflect a high level of developmental plasticity in human brains, which is consistent with other lines of evidence (see also [12,24]). Our results have been obtained from a healthy human population for which a high level of developmental instability due to stress or illness, which may be another cause of FA, would not be expected. In addition, microstructural and gene expression studies show that human evolution has been characterized by increases in the level of cerebral plasticity, as evident by an extended period of environment-dependent myelination [56] and upregulation of genes associated with synaptogenesis [57]. The results of the current study provide further support from an analysis of brain anatomy that elevated plasticity characterizes the human cerebral cortex compared to other primate species. In addition, developmental changes are known to have occurred during hominin evolution that have extended the period of time during which brain maturation is exposed to a complex extra-uterine environment [58]. Studies based on endocranial anatomy, furthermore, also show that the level of FA observed in modern human endocasts is higher than that observed in great apes, including chimpanzees, bonobos, and gorillas [59].
Even if particular plastic changes are not genetically heritable, the general propensity to have a more plastic brain that will be more responsive to environmental influences can be coded by genes. This is what our results show, at least partially, by revealing significant heritability for FA scores in some brain areas in chimpanzees and humans. Indeed, our analyses yield unexpected results because the heritability of some aspects of FA is substantially higher than the heritability of AQs and principal components of asymmetric shape variation, which are more reflective of DA. Although this result should be confirmed in other samples and using additional methods to characterize cortical organization, it seems to indicate that the responsiveness of brain anatomy to environmental influences is more strongly genetically controlled than structural asymmetry itself. The finding of non-significant heritability for FA in some areas of the brain may reflect more complex patterns of inheritance, or the inability of our relatively small samples to detect heritability levels that are expected to be moderate [60]. In fact, several studies have demonstrated that human-specific variants of certain genes are associated with increases in the level of plasticity in the formation of cortico-basal neural circuits [61] and in the maturation of synaptic spines [62]. The evolution of neural plasticity can also be mediated in part by epigenetic mechanisms that allow for context-dependent changes of synapses and circuits [63]. Taken together with the findings from the current analysis, these observations indicate that the level of brain plasticity in the chimpanzee–human clade has a genetic basis and, therefore, is heritable and evolvable.
Supplementary Material
Ethics
Scanning procedures in chimpanzees were approved by the Institutional Animal Care and Use Committees at YNPRC and UTMDACC, and also followed the guidelines of the Institute of Medicine on the use of chimpanzees in research.
Data accessibility
The datasets supporting this article are available in Dryad database at http://dx.doi.org/10.5061/dryad.n04r6 [64].
Authors' contributions
A.G.-R. and C.C.S. conceived of the study; W.D.H. and S.J.S. collected chimpanzee scan data; A.G.-R. collected morphometric data, designed and performed analyses; A.G.-R. and C.C.S. wrote the manuscript, with contributions from W.D.H. and S.J.S.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was supported by a National Institutes of Health grant no. NS042867; and James S. McDonnell Foundation grant no. 220020293. Chimpanzees at UTMDACC were supported by NIH Cooperative Agreement U42 OD-011197. Chimpanzee brain data were provided by the National Chimpanzee Brain Resource, supported by NIH grant no. NS092988. Human data were provided by the Human Connectome Project, WU-Minn Consortium (principal investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University.
References
- 1.Geschwind N, Levitsky W. 1968. Human brain: left-right asymmetries in temporal speech region. Science 161, 186–187. ( 10.1126/science.161.3837.186) [DOI] [PubMed] [Google Scholar]
- 2.Amunts K, Schleicher A, Bürgel U, Mohlberg H, Uylings HB, Zilles K. 1999. Broca's region revisited: cytoarchitecture and intersubject variability. J. Comp. Neurol. 412, 319–341. ( 10.1002/(SICI)1096-9861(19990920)412:2%3C319::AID-CNE10%3E3.0.CO;2-7) [DOI] [PubMed] [Google Scholar]
- 3.Sowell ER, Thompson PM, Rex D, Kornsand D, Tessner KD, Jernigan TL, Toga AW. 2002. Mapping sulcal pattern asymmetry and local cortical surface gray matter distribution in vivo: maturation in perisylvian cortices. Cereb. Cortex 12, 17–26. ( 10.1093/cercor/12.1.17) [DOI] [PubMed] [Google Scholar]
- 4.Toga AW, Thompson PM. 2003. Mapping brain asymmetry. Nat. Rev. Neurosci. 4, 37–48. ( 10.1038/nrn1009) [DOI] [PubMed] [Google Scholar]
- 5.Vigneau M, et al. 2011. What is right-hemisphere contribution to phonological, lexico-semantic, and sentence processing? Insights from a meta-analysis. Neuroimage 54, 577–593. ( 10.1016/j.neuroimage.2010.07.036) [DOI] [PubMed] [Google Scholar]
- 6.Tzourio N, Nkanga-Ngila B, Mazoyer B. 1998. Left planum temporale surface correlates with functional dominance during story listening. Neuroreport 9, 829–833. ( 10.1097/00001756-199803300-00012) [DOI] [PubMed] [Google Scholar]
- 7.Powell HWR, et al. 2006. Hemispheric asymmetries in language-related pathways: a combined functional MRI and tractography study. Neuroimage 32, 388–399. ( 10.1016/j.neuroimage.2006.03.011) [DOI] [PubMed] [Google Scholar]
- 8.Ringo JL, Doty RW, Demeter S, Simard PY. 1994. Time is of the essence: a conjecture that hemispheric specialization arises from interhemispheric conduction delay. Cereb. Cortex 4, 331–343. ( 10.1093/cercor/4.4.331) [DOI] [PubMed] [Google Scholar]
- 9.Gannon PJ, Holloway RL, Broadfield DC, Braun AR. 1998. Asymmetry of chimpanzee planum temporale: humanlike pattern of Wernicke's brain language area homolog. Science 279, 220–222. ( 10.1126/science.279.5348.220) [DOI] [PubMed] [Google Scholar]
- 10.Cantalupo C, Hopkins WD. 2001. Asymmetric Broca's area in great apes. Nature 414, 505 ( 10.1038/35107134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilissen EP. 2001. Structural symmetries and asymmetries in human and chimpanzee brains. In Evolutionary anatomy of the primate cerebral cortex (eds Falk D, Gibson KR), pp. 187–215. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 12.Gómez-Robles A, Hopkins WD, Sherwood CC. 2013. Increased morphological asymmetry, evolvability and plasticity in human brain evolution. Proc. R. Soc. B 280, 20130575 ( 10.1098/rspb.2013.0575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopkins WD. 2006. Comparative and familial analysis of handedness in great apes. Psychol. Bull. 132, 538–559. ( 10.1037/0033-2909.132.4.538) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopkins WD, Russell JL, Lambeth S, Schapiro SJ. 2007. Handedness and neuroanatomical asymmetries in captive chimpanzees: a summary of 15 years of research. In The evolution of hemispheric specialization in primates (ed. Hopkins WD.), pp. 146–181. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 15.Hopkins WD. 2013. Comparing human and nonhuman primate handedness: challenges and a modest proposal for consensus. Dev. Psychobiol. 55, 621–636. ( 10.1002/dev.21139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taglialatela JP, Russell JL, Schaeffer JA, Hopkins WD. 2008. Communicative signaling activates ‘Broca's’ homolog in chimpanzees. Curr. Biol. 18, 343–348. ( 10.1016/j.cub.2008.01.049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hopkins WD, Taglialatela JP, Russell JL, Nir TM, Schaeffer J. 2010. Cortical representation of lateralized grasping in chimpanzees (Pan troglodytes): a combined MRI and PET study. PLoS ONE 5, e13383 ( 10.1371/journal.pone.0013383) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeMay M. 1976. Morphological cerebral asymmetries of modern man, fossil man, and nonhuman primate. Ann. N. Y. Acad. Sci. 280, 349–366. ( 10.1111/j.1749-6632.1976.tb25499.x) [DOI] [PubMed] [Google Scholar]
- 19.Holloway RL, Broadfield DC, Yuan MS, Schwartz JH, Tattersall I. 2004. The human fossil record. Brain endocasts—the paleoneurological evidence, vol. 3 New York, NY: Wiley-Liss. [Google Scholar]
- 20.Balzeau A, Holloway RL, Grimaud-Hervé D. 2012. Variations and asymmetries in regional brain surface in the genus Homo. J. Hum. Evol. 62, 696–706. ( 10.1016/j.jhevol.2012.03.007) [DOI] [PubMed] [Google Scholar]
- 21.Falk D. 1987. Brain lateralization in primates and its evolution in hominids. Am. J. Phys. Anthropol. 30, 107–125. ( 10.1002/ajpa.1330300508) [DOI] [Google Scholar]
- 22.Palmer AR, Strobeck C. 2003. Fluctuating asymmetry analyses revisited. In Developmental instability: causes and consequences (ed. Polak M.), pp. 279–319. Oxford, UK: Oxford University Press. [Google Scholar]
- 23.Dongen SV. 2006. Fluctuating asymmetry and developmental instability in evolutionary biology: past, present and future. J. Evol. Biol. 19, 1727–1743. ( 10.1111/j.1420-9101.2006.01175.x) [DOI] [PubMed] [Google Scholar]
- 24.Gómez-Robles A, Hopkins WD, Schapiro SJ, Sherwood CC. 2015. Relaxed genetic control of cortical organization in human brains compared with chimpanzees. Proc. Natl Acad. Sci. USA 112, 14 799–14 804. ( 10.1073/pnas.1512646112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogart SL, Mangin JF, Schapiro SJ, Reamer L, Bennett AJ, Pierre PJ, Hopkins WD. 2012. Cortical sulci asymmetries in chimpanzees and macaques: a new look at an old idea. Neuroimage 61, 533–541. ( 10.1016/j.neuroimage.2012.03.082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Essen DC, et al. 2012. The Human Connectome Project: a data acquisition perspective. Neuroimage 62, 2222–2231. ( 10.1016/j.neuroimage.2012.02.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glasser MF, et al. 2013. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124. ( 10.1016/j.neuroimage.2013.04.127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K. 2013. The WU-Minn Human Connectome Project: an overview. Neuroimage 80, 62–79. ( 10.1016/j.neuroimage.2013.05.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan CC, et al. 2015. Modeling the 3D geometry of the cortical surface with genetic ancestry. Curr. Biol. 25, 1988–1992. ( 10.1016/j.cub.2015.06.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cointepas Y, Mangin J-F, Garnero L, Poline J-B, Benali H. 2001. BrainVISA: Software platform for visualization and analysis of multi-modality brain data. Neuroimage 13, 98 ( 10.1016/S1053-8119(01)91441-7) [DOI] [Google Scholar]
- 31.Fischl B. 2012. FreeSurfer. Neuroimage 62, 774–781. ( 10.1016/j.neuroimage.2012.01.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desikan RS, et al. 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980. ( 10.1016/j.neuroimage.2006.01.021) [DOI] [PubMed] [Google Scholar]
- 33.Gómez-Robles A, Hopkins WD, Sherwood CC. 2014. Modular structure facilitates mosaic evolution of the brain in chimpanzees and humans. Nat. Commun. 5, 4469 ( 10.1038/ncomms5469) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohlf FJ, Slice D. 1990. Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst. Zool. 39, 40–59. ( 10.2307/2992207) [DOI] [Google Scholar]
- 35.Klingenberg CP, Barluenga M, Meyer A. 2002. Shape analysis of symmetric structures: quantifying variation among individuals and asymmetry. Evolution 56, 1909–1920. ( 10.1554/0014-3820(2002)056%5B1909:SAOSSQ%5D2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 36.Klingenberg CP, Monteiro LR. 2005. Distances and directions in multidimensional shape spaces: implications for morphometric applications. Syst. Biol. 54, 678–688. ( 10.1080/10635150590947258) [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez PN, Lotto FP, Hallgrímsson B. 2014. Canalization and developmental instability of the fetal skull in a mouse model of maternal nutritional stress. Am. J. Phys. Anthropol. 154, 544–553. ( 10.1002/ajpa.22545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hadfield JD. 2010. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Softw. 33, 1–22. ( 10.18637/jss.v033.i02)20808728 [DOI] [Google Scholar]
- 39.Wilson AJ, Réale D, Clements MN, Morrissey MM, Postma E, Walling CA, Kruuk LEB, Nussey DH. 2010. An ecologist's guide to the animal model. J. Anim. Ecol. 79, 13–26. ( 10.1111/j.1365-2656.2009.01639.x) [DOI] [PubMed] [Google Scholar]
- 40.Zhao JH. 2015. gap: Genetic Analysis Package. R package. See http://cran.r-project.org/package=gap.
- 41.Teplitsky C, Mouawad NG, Balbontin J, De Lope F, Møller AP. 2011. Quantitative genetics of migration syndromes: a study of two barn swallow populations. J. Evol. Biol. 24, 2025–2039. ( 10.1111/j.1420-9101.2011.02342.x) [DOI] [PubMed] [Google Scholar]
- 42.Gelman A. 2006. Prior distributions for variance parameters in hierarchical models (comment on article by Browne and Draper). Bayesian Anal. 1, 515–534. ( 10.1214/06-BA117A) [DOI] [Google Scholar]
- 43.Hadfield J. 2015. MCMCglmm course notes. See http://cran.us.r-project.org/web/packages/MCMCglmm/vignettes/CourseNotes.pdf.
- 44.Leder EH, McCairns RJS, Leinonen T, Cano JM, Viitaniemi HM, Nikinmaa M, Primmer CR, Merilä J. 2015. The evolution and adaptive potential of transcriptional variation in sticklebacks—signatures of selection and widespread heritability. Mol. Biol. Evol. 32, 674–689. ( 10.1093/molbev/msu328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson PM, et al. 2001. Genetic influences on brain structure. Nat. Neurosci. 4, 1253–1258. ( 10.1038/nn758) [DOI] [PubMed] [Google Scholar]
- 46.Geschwind DH, Miller BL, DeCarli C, Carmelli D. 2002. Heritability of lobar brain volumes in twins supports genetic models of cerebral laterality and handedness. Proc. Natl Acad. Sci. USA 99, 3176–3181. ( 10.1073/pnas.052494999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smaers JB, Steele J, Case CR, Cowper A, Amunts K, Zilles K. 2011. Primate prefrontal cortex evolution: human brains are the extreme of a lateralized ape trend. Brain Behav. Evol. 77, 67–78. ( 10.1159/000323671) [DOI] [PubMed] [Google Scholar]
- 48.Peper JS, Brouwer RM, Boomsma DI, Kahn RS, Hulshoff Pol HE. 2007. Genetic influences on human brain structure: a review of brain imaging studies in twins. Hum. Brain Mapp. 28, 464–473. ( 10.1002/hbm.20398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hopkins WD, Misiura M, Pope SM, Latash EM. 2015. Behavioral and brain asymmetries in primates: a preliminary evaluation of two evolutionary hypotheses. Ann. N. Y. Acad. Sci. 1359, 65–83. ( 10.1111/nyas.12936) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fears SC, et al. 2011. Anatomic brain asymmetry in vervet monkeys. PLoS ONE 6, e28243 ( 10.1371/journal.pone.0028243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheverud JM, Falk D, Hildebolt C, Moore A, Helmkamp RC, Vannier M. 1990. Heritability and association of cortical petalias in rhesus macaques (Macaca mulatta). Brain. Behav. Evol. 35, 368–372. ( 10.1159/000115881) [DOI] [PubMed] [Google Scholar]
- 52.Hopkins WD, Adams MJ, Weiss A. 2013. Genetic and environmental contributions to the expression of handedness in chimpanzees (Pan troglodytes). Genes Brain Behav. 12, 446–452. ( 10.1111/gbb.12044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lien Y-J, Chen WJ, Hsiao P-C, Tsuang H-C. 2015. Estimation of heritability for varied indexes of handedness. Laterality 20, 469–482. ( 10.1080/1357650X.2014.1000920) [DOI] [PubMed] [Google Scholar]
- 54.Jansen AG, Mous SE, White T, Posthuma D, Polderman TJC. 2015. What twin studies tell us about the heritability of brain development, morphology, and function: a review. Neuropsychol. Rev. 25, 27–46. ( 10.1007/s11065-015-9278-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Atkinson EG, Rogers J, Cheverud JM. 2016. Evolutionary and developmental implications of asymmetric brain folding in a large primate pedigree. Evolution 70, 707–715. ( 10.1111/evo.12867) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller DJ, et al. 2012. Prolonged myelination in human neocortical evolution. Proc. Natl Acad. Sci. USA 109, 16 480–16 485. ( 10.1073/pnas.1117943109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cáceres M, Suwyn C, Maddox M, Thomas JW, Preuss TM. 2007. Increased cortical expression of two synaptogenic thrombospondins in human brain evolution. Cereb. Cortex 17, 2312–2321. ( 10.1093/cercor/bhl140) [DOI] [PubMed] [Google Scholar]
- 58.Hublin J-J, Neubauer S, Gunz P. 2015. Brain ontogeny and life history in Pleistocene hominins. Phil. Trans. R. Soc. B 370, 20140062 ( 10.1098/rstb.2014.0062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balzeau A, Gilissen E, Grimaud-Hervé D. 2012. Shared pattern of endocranial shape asymmetries among great apes, anatomically modern humans, and fossil hominins. PLoS ONE 7, e29581 ( 10.1371/journal.pone.0029581) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leamy LJ, Klingenberg CP. 2005. The genetics and evolution of fluctuating asymmetry. Annu. Rev. Ecol. Evol. Syst. 36, 1–21. ( 10.1146/annurev.ecolsys.36.102003.152640) [DOI] [Google Scholar]
- 61.Enard W, et al. 2009. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell 137, 961–971. ( 10.1016/j.cell.2009.03.041) [DOI] [PubMed] [Google Scholar]
- 62.Charrier C, et al. 2012. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell 149, 923–935. ( 10.1016/j.cell.2012.03.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krubitzer L, Stolzenberg DS. 2014. The evolutionary masquerade: genetic and epigenetic contributions to the neocortex. Curr. Opin. Neurobiol. 24, 157–165. ( 10.1016/j.conb.2013.11.010) [DOI] [PubMed] [Google Scholar]
- 64.Gómez-Robles A, Hopkins WD, Schapiro SJ, Sherwood CC. 2016. Data from: The heritability of chimpanzee and human brain asymmetry. Dryad Digital Repository. ( 10.5061/dryad.n04r6) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article are available in Dryad database at http://dx.doi.org/10.5061/dryad.n04r6 [64].