Abstract
MicroRNAs are small non-coding RNAs that inhibit protein expression post-transcriptionally. They have been implicated in many different physiological processes, but little is known about their individual involvement in learning and memory. We recently identified several miRNAs that either increased or decreased intermediate-term memory when inhibited in the central nervous system, including miR-iab8-3p. We report here a new developmental role for this miRNA. Blocking the expression of miR-iab8-3p during the development of the organism leads to hypertrophy of individual mushroom body neuron soma, a reduction in the field size occupied by axonal projections, and adult intellectual disability. We further identified four potential mRNA targets of miR-iab8-3p whose inhibition modulates intermediate-term memory including ceramide phosphoethanolamine synthase, which may account for the behavioral effects produced by miR-iab8-3p inhibition. Our results offer important new information on a microRNA required for normal neurodevelopment and the capacity to learn and remember normally.
Keywords: MicroRNA, Drosophila, Learning, Memory, Neurodevelopment, Mushroom bodies
1. Introduction
MicroRNAs (miRNAs) are ~21 nucleotide (nt)-long RNAs and part of a family of small non-coding RNAs implicated in many different broad processes extending from normal development, tumorigenesis, to human disease (Bartel, 2004; Krützfeldt and Stofell, 2006; Bushati and Cohen, 2007; Chang and Mendell, 2007; Kloosterman and Plasterk, 2006; Adams et al., 2014). They have spurred much interest due to their unique biological function and as possible blood biomarkers for disease progression and therapeutic targets (Hunsberger et al., 2009; Im and Kenny, 2012; Rao et al., 2013).
MiRNAs repress protein expression post-transcriptionally by binding to complementary sequences in the 3′ untranslated region (3′ UTR) of target mRNAs (Bartel, 2009). About two-thirds of miRNAs found in mammalian species are transcribed from individual genes by RNA polymerase II, but others are processed from intronic sequences in primary transcripts and are called ‘miRtrons’ (Bartel, 2004; Rodriguez et al., 2004; Filipowicz et al., 2008; Krol et al., 2010). MiRNA gene expression is regulated in ways analogous to protein-coding genes (Krol et al., 2010; Aksoy-Aksel et al., 2014). The primary miRNA transcript (pri-miRNA) is processed in the nucleus by the microprocessor protein complex, Drosha/Pasha, to produce a ~85 nt precursor hairpin miRNA (pre-miRNA). The pre-miRNAs are shuttled by Exportin-5 to the cytoplasm where they are further processed by the Dicer/Argonaute proteins to yield to a mature miRNA duplex. The guide strand of the miRNA duplex is integrated into a ribonucleoprotein complex named the miRNA-induced silencing complex (miRISC) while the passenger strand is, in some cases, degraded (Bartel, 2009). The target sequence in the mRNA, termed the miRNA recognition element (MRE), is usually recognized by a ‘seed’ sequence located between nt 2–8 in the 5′-end of the miRNA (Bartel, 2009). Depending on the extent of nucleotide complementarity, miRNAs inhibit translation and/or induce mRNA degradation (Bartel, 2009; Jonas and Izaurralde, 2015).
Many studies have shown that miRNAs are involved in neural plasticity and memory formation (Bredy et al., 2011; Aksoy-Aksel et al., 2014) and that miRNA dysregulation is part of the pathophysiology of neurological diseases and neurodevelopmental disorders (Hunsberger et al., 2009; Xu et al., 2010; Wang et al., 2012; Maciotta et al., 2013; Tan et al., 2015). Neurodevelopmental disorders, which account for more than 10% of the burden of disease globally (Durkin et al., 2006; Hunsberger et al., 2009), are characterized by a complex set of phenotypic traits, including learning disabilities and comorbidity with other diseases. A unique feature of miRNAs is that they target multiple mRNAs that could account for such pleiotropy (Hunsberger et al., 2009; Xu et al., 2010; Maciotta et al., 2013). MiRNA dysregulation has been associated with Down’s syndrome, Fragile X syndrome, schizophrenia, bipolar disorder, DiGeorge syndrome, Tourette’s syndrome, autism spectrum disorders and Rett syndrome (Xu et al., 2010; Feng and Feng, 2011; Olde Loohuis et al., 2012; Nowak and Michlewski, 2013; Saab and Mansuy, 2014; Sun and Shi, 2015). Obtaining insights into how individual miRNAs mediate the development of the central nervous system in ways that impact learning and memory processes may help disentangle the mechanisms underlying these disorders and offer new ways for the management of such disorders.
Two hundred and fifty six miRNAs sequences have been described so far in Drosophila melanogaster including 150 with a high level of confidence (www.mirbase.org, Griffiths-Jones, 2004). MiRNA function in memory formation was first deduced from studies showing miRISC pathway involvement in long-term memory formation (Ashraf et al., 2006). Until recently, however, only one specific miRNA, miR-276a, was identified as important for the physiology underlying memory formation (Li et al., 2013).
In Drosophila, olfactory classical conditioning is a robust type of learning and memory assay (Busto et al., 2010; Kahsai and Zars, 2011; Davis, 2011). In this paradigm, flies learn to associate the noxious stimulus of electric shock, the unconditioned stimulus (US), with an odor, the conditioned stimulus (CS) (Tully and Quinn, 1985; Beck et al., 2000). Although a growing number of neurons and circuits are involved in olfactory memory (Guven-Ozkan and Davis, 2014), a large part of the association between the CS and the US occurs in the mushroom body neurons (MBn; Tomchik and Davis, 2009). To obtain a comprehensive view of the roles for individual miRNAs in memory formation, we recently completed a large screen of ~140 individual miRNAs to identify those that are functionally involved (Busto et al., 2015). We employed the miRNA sponge strategy (Loya et al., 2009; Ebert and Sharp, 2010; Fulga et al., 2015) to selectively inhibit individual miRNAs in a CNS-wide fashion. From these studies, we identified five miRNAs whose inhibition reproducibly modulated intermediate-term memory (Busto et al., 2015). A subsequent secondary screen (unpublished) using two genomic copies of the miRNA sponge to increase potency allowed us to identify one additional miRNA candidate, miR-iab8-3p.
Here, we provide data showing that miR-iab8-3p function in the developing MBn is required for the adult fly to learn about olfactory cues. This cognitive deficit is associated with a requirement during development for normal miR-iab8-3p function. The cellular consequence of inhibiting miR-iab8-3p function is that individual MBn exhibit an increase in cell soma size and a decrease in the neuropil volume occupied by their axons. We further identify four potential miR-iab8-3p mRNA-targets that are required for normal olfactory memory. We provide evidence and arguments supporting the model that ceramide phosphoethanolamine synthase is a likely effector target for both the normal structural development of the MBn as well as adult learning capacity.
2. Results
2.1. MiR-iab8-3p inhibition in MBn impairs memory formation and expression
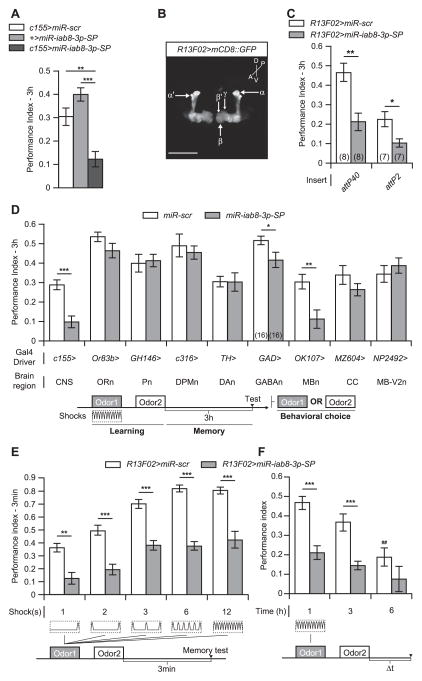
We used inducible complementary transcripts, called miRNA-sponges (uas-miR-SP) (Fulga et al., 2015), to decrease the expression of miR-iab8-3p in the CNS using the elavc155-gal4 driver (c155-gal4). Memory tested at 3 h after conditioning, which is regarded as intermediate-term memory, was impaired when compared to two control genotypes, one carrying the elavc155-gal4 driver crossed with miR-scr and the other carrying only the uas-miR-SP transgene (Fig. 1A). The miR-scr line carries a scrambled sequence instead of an authentic miR-SP inserted into the same vector and at the same defined genomic docking sites (attP40/attP2) as the miR-SP transgenes. The intermediate-term memory phenotype of the experimental flies (elavc155-gal4 > uas-miR-SP) was reproduced in multiple experiments, including that shown in Fig. 1D.
Fig. 1. MiR-iab8-3p inhibition in MBn impairs memory formation.
(A) Three hour memory was impaired by miR-iab8-3p-SP expression in the CNS driven by the elavc155-gal4 driver. Memory expression was significantly different from flies carrying only the uas-miR-iab8-3p-SP transgene or the gal4 driver crossed with the miR-scr control. Results are presented as the mean±s.e.m. with n = 6. ANOVA was used to measure the effect of genotype on PIs followed by a Bonferroni’s post hoc test to compare performances between groups. ***: p<0.001,**: p<0.01. (B) The R13F02-gal4 element drives expression of a uas-mCD8::GFP reporter specifically in the MBn. MBn are classed as three major types based on their axonal projection pattern: αβ, α′β′ and γ MBn. The αβ and α′β′ MBn extend their axonal projections into the vertical and the horizontal lobes while the γ MBn extend axons into the horizontal lobe. Scale bar: 100 μm. (C) The uas-miR-iab8-3p-SP transgenes, either on the 2nd chromosome at the attP40 docking site or on the 3rd chromosome at the attP2 docking site, reduced 3 h memory when expressed in the MBn. For each case, the miR-scr control transgene was inserted at the same attP docking site. Results are presented as the mean±s.e.m. with n = 7–8. Two-sample, bilateral Student t-tests were used to compare PIs between the two genotypes. **: p<0.01,*: p<0.05. (D) MiR-iab8-3p-SP impaired 3 h memory when expressed in the MBn and weakly in GABAergic neurons. MiR-iab8-3p-SP was expressed in specific types of neurons and in different brain regions and parts of the olfactory nervous system using different gal4 driver lines. Expression domains for the gal4 drivers: CNS, central nervous system; ORn, olfactory receptor neurons; Pn, projection neurons; DPMn, dorsal paired medial neurons; DAn, dopaminergic neurons; GABAn, GABAergic neurons; MBn, mushroom body neurons; CC, central complex; MB-V2n, mushroom body extrinsic neurons V2. Three hour memory performance for flies carrying a gal4 transgene and uas-miR-iab8-3p-SP was compared to flies carrying miR-scr and the same gal4 element. Results are presented as the mean±s.e.m. with n = 12–16. Two-sample, bilateral Student t-tests were used to assess an effect of the genotype on PIs. ***: p<0.001,**: p<0.01,*: p<0.05. (E) MiR-iab8-3p-SP dramatically impaired memory acquisition when expressed in MBn. Memory acquisition was assessed by testing immediate performance after an increasing number of electric shocks (from 1 to 12, bottom) paired with odor during training. Although flies expressing miR-iab8-3p-SP learned the task, their performance plateaued at PI~0.4, while control flies expressing miR-scr plateaued at ~0.8. For all training protocols used, immediate performance was reduced with miR-iab8-3p-SP expression in the MBn. Results are presented as the mean±s.e. m. with n = 8. Two-sample, bilateral Student t-tests were used to assess the effect of genotype on PIs. ***: p<0.001,**: p<0.01. (F) MiR-iab8-3p-SP impaired memory retention when expressed in the MBn. Memory retention was measured at 1, 3 and 6 h following a 12 shock conditioning protocol. Memory expression was reduced to less than half of the control value at all time points tested. The PI at 6 h for the experimental group was not significantly different from zero. Results are presented as the mean±s.e.m. with n = 8. Two-sample, bilateral Student t-tests were used to compare PIs between the two genotypes while a one-sample, bilateral Student t-test was used to compare PIs with the null value. ***: p<0.001,**: p<0.01 for two-samples. ##: p<0.01 for one-sample.
We subsequently mapped the impairing effect of the miR-iab8-3p-SP in the CNS by expressing it with a panel of gal4 drivers with some focus on the MB. Expression of the disrupting sponge sequences with a very specific MB gal4 driver, R13F02, or a gal4 driver with preferential expression in the MB, OK107, reproduced the 3 h memory impairment whereas gal4 elements that drive expression in other neurons of the olfactory nervous system were ineffective (Fig. 1B–D). One exception to this was the GAD-gal4 driver, which also produced modest memory impairment (Fig. 1D). The memory impairment was observed with R13F02 using flies that contain two copies of the miR-iab8-3p-SP or when these were separated into single copies at either the attP40 or attP2 integration sites (Fig. 1C), despite the marked difference in performance between the control miR-scr inserted at the two different docking sites. We and others have recently shown that luciferase reporters and sponge transgenes can exhibit different expression levels or behavioral performance depending on their attP insertion sites (Markstein et al., 2008; Busto et al., 2015). In addition, there exists substantial variation in memory expression for the various gal4 > miR-scr controls, presumably due to genetic background effects (Fig. 1D). This restricts the possible comparisons to be between each gal4 > miR-scr control and its corresponding gal4 > miR-SP experimental genotype.
We tested for normal avoidance behavior to the olfactory cues and shock used during conditioning. Odor and shock avoidances were not significantly different between flies expressing miR-iab8-3p-SP or the miR-scr control in the MBn, indicating that the impairment in memory expression was not attributable to a defect in odor or shock perception (Table S1).
We next asked whether the 3 h memory impairment was due to a deficit in learning – the acquisition of the odor/shock association –or a deficit in the stability of memory of the association. We submitted flies to an increasing number of electric shocks along with the paired odor during training and measured the performance immediately (3 min) after such conditioning (Fig. 1E). Memory expression was drastically reduced for 1, 2, 3, 6, or 12 shock pairings. These data show that flies expressing miR-iab8-3p-SP were able to acquire the task but were limited in their learning abilities, with expression plateauing at a Performance Index (PI) of ~0.4 compared to ~0.8 for control flies. We also tested memory expression at different time points (1, 3 and 6 h) following 12 shock conditioning (Fig. 1F). Memory was reduced for flies expressing miR-iab8-3p-SP relative to the miR-scr control at 1 and 3 h after conditioning with no memory expression detectable in the experimental group at 6 h. Memory retention was also tested with another odorant combination (BEN/MCH instead of BEN/OCT) and in an alternative genetic background (w1118) with similar results (Fig. S1A,B). We measured the memory retention relative to the 3 min memory score following 12 shock training to discriminate between acquisition and memory stability impairment. Memory expression when transformed in this way was identical between the experimental and control genotypes (Fig. S1C), indicating that acquisition is the primary process impaired by miR-iab8-3p inhibition in MBn.
2.2. MiR-iab8-3p inhibition in all classes of MBn reduces three-hour memory
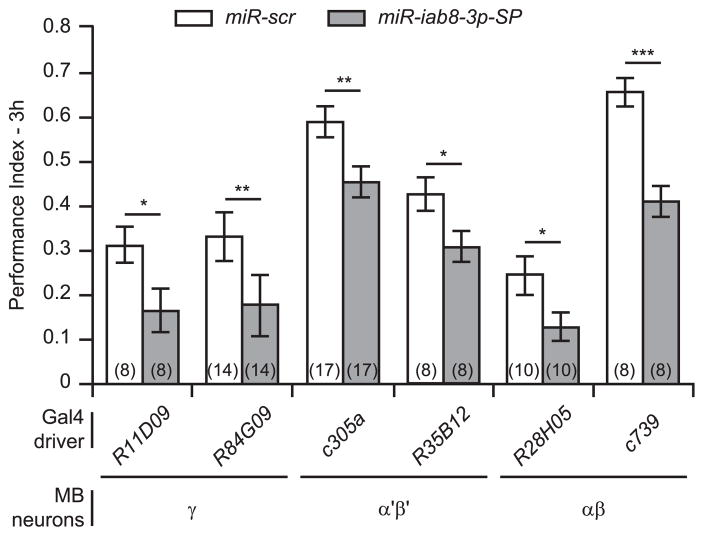
Three major classes of MBn have been identified from their birth during development and according to the axonal projection patterns in the MB lobes (Kunz et al., 2012). The γ neurons are born first in early larval stages followed by the α′β′ MBn in late larval stages and finally by the αβ MBn during the pupal stage. The axons of the γ, β′ and β MBn extend into the horizontal lobes of the MB while the α′ and α MBn extend collateral projections into the vertical lobes. In addition, evidence has accumulated that these MBn subtypes are differentially involved in the sequential phases of memory formation (Davis, 2011). The γ MBn are most strongly implicated in short-term memory while the α′β′ MBn are implicated in intermediate-term memory and consolidation. The αβ MBn are important for long-term memory and for the expression of all temporal phases of memory (Yu et al., 2006; Krashes et al., 2007; Akalal et al., 2011; Cervantes-Sandoval et al., 2013).
To determine whether miR-iab8-3p was required in a specific sub-population of MBn for normal adult memory, we expressed miR-iab8-3p-SP using a battery of Gal4 drivers expressing in the γ, α′β′ or αβ MBn, sampling at least two different Gal4 drivers for each class of MBn (Fig. 2). Although there exists substantial variation in memory expression across the controls for the various Gal4 drivers (see above), the miR-iab8-3p-SP expression in all three classes of MBn significantly decreased 3 h memory relative to their genotype-matched miR-scr control. These data indicate that normal levels of miR-iab8-3p expression are required in all classes of MBn for normal adult memory expression.
Fig. 2. MiR-iab8-3p inhibition in all three classes of MBn impairs 3 h memory expression.
MiR-iab8-3p inhibition specifically in the γ neurons using the R11D09-and R84G09-gal4 driver lines reduced 3 h memory. MiR-iab8-3p inhibition in α′β′ neurons using c305a- and R35B12-gal4 drivers also reduced 3 h memory. In addition, miR-iab8-3p inhibition in αβ MBn using the c739- and R28H05-gal4 drivers decreased 3 h memory. Results are presented as the mean±s.e.m. with n = 8–17. Two-sample, bilateral Student t-tests were used to assess significant PI differences between the two genotypes. ***: p<0.001,**: p<0.01,*: p<0.05.
2.3. MiR-iab8-3p-SP expression limits acquisition and subsequent memory retention
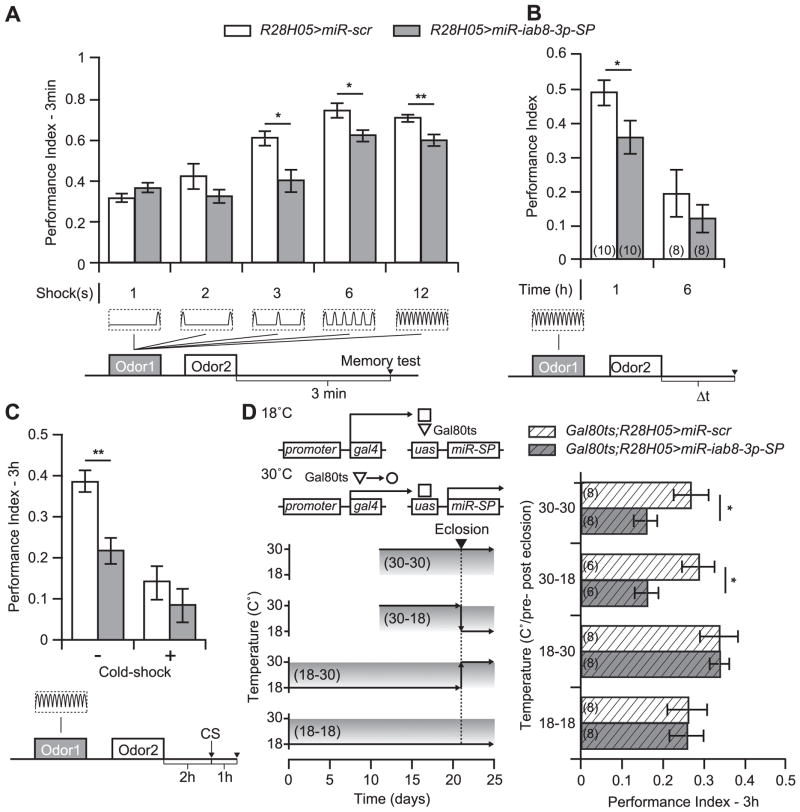
We focused our subsequent studies on the roles for miR-iab8-3p in memory processes such as acquisition and memory retention. We initially tested memory acquisition after expressing miR-iab8-3p-SP in the αβ MBn. Memory expression in the experimental flies was tested immediately (3 min) after conditioning with 1, 2, 3, 6, or 12 electric shock pulses, with the sponge-expressing flies showing poor performance after 3, 6 and 12 shocks conditioning (Fig. 3A). The ceiling level of performance for flies expressing miR-iab8-3p-SP was ~0.6 while the control group reached a ceiling of ~0.75. These data strongly suggest a deficit in acquisition processes in the experimental group. Similar behavioral results were obtained upon expressing miR-iab8-3p-SP in the α′β′ or γ MBn (Fig. S1A, S3A). This limitation was not associated with a deficit in odor or shock perception as reflected by avoidance behavior (Table S2). Note that the degree of memory impairment with disruption in individual classes of MBn (Fig. 3A, S2A, S3A) was milder than that observed with pan-MB inhibition (Fig. 1E), consistent with a requirement in all classes of MBn. Therefore, these data argue that normal expression of miR-iab8-3p in all three classes of MBn is required for normal acquisition.
Fig. 3. Inhibition of miR-iab8-3p in αβ MBn impairs memory in adults.
(A) Memory acquisition was constrained by miR-iab8-3p inhibition in αβ MBn compared to the miR-scr control group. Three-minute memory was assessed after conditioning flies with an increasing number of electric shocks (1-12, bottom panel). After 1 and 2 shocks, both genotypes exhibited similar performance. For increasing shock numbers, flies expressing the miR-iab8-3p-SP exhibited reduced memory performance. Results are presented as the mean±s.e.m. with n = 6. Two-sample, bilateral Student t-tests were used to test the effect of the genotype on PIs. **: p<0.01;*: p<0.05. (B) Memory retention was reduced by miR-iab8-3p-SP expression in αβ MBn. Memory retention was measured at 1 and 6 h following a 12 shocks conditioning protocol. One hour after training flies expressing miR-iab8-3p-SP exhibited a significant decrease in memory performance relative to flies expressing miR-scr. After 6 h, memory performance was nearly zero and indistinguishable between genotypes. Results are presented as the mean±s.e.m. with n = 8–10. Two-sample, bilateral Student t-tests were used to test the effect of the genotype on PIs. **: p<0.01;*: p<0.05. (C) Anesthesia-Resistant Memory (ARM) was not affected by miR-iab8-3p-SP expression. A cold-shock (CS, bottom panel) was delivered 2 h after training to remove ASM. The residual memory (ARM) was tested 1 h later. MiR-iab8-3p-SP expression had no effect on ARM but significantly reduced ASM. Results are presented as the mean±s.e.m. with n = 6. Two-way ANOVA was used to assess the effects of the genotype and cold-shock on PIs. Multiple comparisons between groups were performed using Tukey’s post hoc tests. **: p<0.01. (D) MiR-iab8-3p is required in αβ MBn during development. The Gal80ts protein (TARGET system) allows temporal control of miR-iab8-3p-SP expression through control of environmental temperature across the life cycle starting at mating (Time 0). At 18 °C, Gal80ts and Gal4 proteins interact together preventing Gal4 binding to the uas sequences and consequently preventing miR-SP expression. When the organism is cultured at 30 °C, Gal80ts undergoes a conformational change releasing Gal4 from inhibition. Gal4 is thus free to bind to the uas sequences and promote miR-SP expression (top of the left panel). Temperature shifts between 18 °C and 30 °C were performed at eclosion (dotted vertical line). For flies grown at 18 °C across the life cycle (18-18, bottom), miR-iab8-3p-SP expression is inhibited and memory performance is similar to the miR-scr control. When flies are cultured at 30 °C across the life cycle (30-30), miR-iab8-3p-SP is expressed causing a reduction in memory performance relative to miR-scr. To determine whether miR-iab8-3p is necessary before or after eclosion, two groups were generated and cultured at 30 °C prior to eclosion (30-18), or after eclosion (18–30). MiR-iab8-3p inhibition restricted to developmental periods prior to eclosion reduced memory and phenocopied the flies kept at 30 °C across the life cycle. MiR-iab8-3p-SP expression restricted to periods after eclosion had no effect on 3 h memory. Results are presented as the mean±s.e.m. with n = 6–8. Two-sample, bilateral Student t-tests were used to test the effect of the genotype on PIs. *: p<0.05.
In addition to our tests of memory retention at 3 h after conditioning (Fig. 2), we tested memory retention at 1 and 6 h after conditioning. The performance of flies expressing miR-iab8-3p-SP in the αβ MBn was reduced at the 1 h time point, probably reflecting the deficit in acquisition (Fig. 3B). This was also true for flies that express miR-iab8-3p-SP in the α′β′ or γ MBn (Fig. S2B, S3B).
Memory expression within a few hours after conditioning has been dissociated in two major components including anesthesia-resistant memory (ARM) and anesthesia-sensitive memory (ASM), named for their sensitivity to cold anesthesia (Isabel et al., 2004). We exposed control and experimental groups of flies to a cold-shock after conditioning in order to remove the ASM component and measure the remaining ARM (Fig. 3C). We observed a similar level of performance following cold-shock in flies expressing miR-iab8-3p-SP or miR-scr, indicating that ARM was preserved. We conclude from this result that a specific deficit in ASM was produced by miR-iab8-3p inhibition (Fig. 3C). Similar conclusions were obtained with miR-iab8-3p inhibition in α′β′ and γ MBn (Fig. S2C, S3C).
We next determined the temporal requirement for miR-iab8-3p expression using the TARGET system (Fig. 3D, left panel, McGuire et al., 2003). Raising experimental genotype Drosophila throughout development and adulthood at 18 °C (negative control) until conditioning failed to produce a significant memory phenotype (Fig. 3D, right panel). In contrast, culturing the organism throughout development and adulthood at 30 °C induced miR-iab8-3p-SP expression (positive control) and impaired memory as previously observed. Culturing the organism at 18 °C across embryonic, larval and pupal stages and imposing a temperature shift to 30 °C upon eclosion to inhibit miR-iab8-3p in adult flies failed to produce a phenotype. However, culturing the organism at 30 °C until eclosion, with a transfer to 18 °C for adulthood, reduced memory expression in a quantitative way similar to the positive control. These results indicate that developmental knockdown in the R28H05-gal4 expressing neurons results in an adult memory deficit. A similar experiment was performed with disruption of expression in the α′β′ MBn with nearly identical results (Fig. S2D).
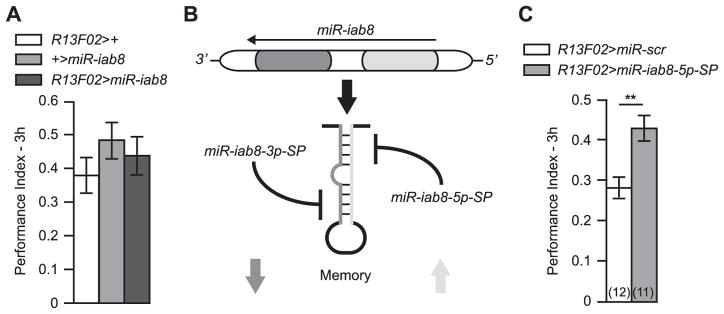
2.4. MiR-iab8 has a bi-functional role on memory
The results above showing that diminishing miR-iab8-3p expression in the MBn impairs memory beg the question of what occurs when miR-iab8 is overexpressed. We used the MBn driver R13F02 to overexpress this miRNA and found no change in memory expression when tested at 3 h (Fig. 4A). For this experiment, in which the uas-miR-iab8 transgene was inserted in the attP-86Fb landing site, we compared the performance of the bi-genic progeny to that of progeny carrying only the gal4 driver or only the uas-miR-iab8 transgene. We obtained the same conclusion of no phenotype using the elavc155-gal4 driver (data not shown). However, the miR-iab8 gene produces two different mature miRNAs, including miR-iab8-3p and miR-iab8-5p (Fig. 4C; http://www.mirbase.org/). To determine whether this alternative miR-iab8-5p is involved in memory expression, we blocked its expression in the MBn using miR-iab8-5p-SP at the same characterized attP docking sites used for miR-iab8-3p-SP and compared memory expression relative to the miR-scr control. Surprisingly, 3 h memory expression was increased by the inhibition of miR-iab8-5p in the MBn (Fig. 4B). This result indicates that each strand of miR-iab8 participates in memory processes with opposite effects, such that over-expression of the complete transgene nullifies the effect of the two encoded miRNAs (Fig. 5C).
Fig. 4. MiR-iab8 has a bi-functional effect on memory.
(A) Overexpression of miR-iab8 in the MBn had no effect on 3 h memory expression relative to control genotypes carrying only the R13F02-gal4 driver or the miR-iab8 transgene. Results are presented as the mean±s.e.m. with n = 6. ANOVA was used to test the effect of genotype on PI. (B) Each miRNA hairpin is composed of two miRNAs, one called the guide strand and the other called the passenger strand. In most cases, the guide strand is integrated in the miRISC while the passenger strand is degraded. In some cases, both strands can be integrated into the miRISC to regulate protein expression. (C) MiR-iab8-5p inhibition in MBn increases 3 h memory. MiR-iab8-5p-SP expression in MBn using the R13F02-gal4 driver increased 3 h memory relative to the expression of the miR-scr control. Results are presented as the mean±s.e.m. with n = 11–12. Two-sample, bilateral Student t-test was used to compare the two genotypes. **: p<0.01.
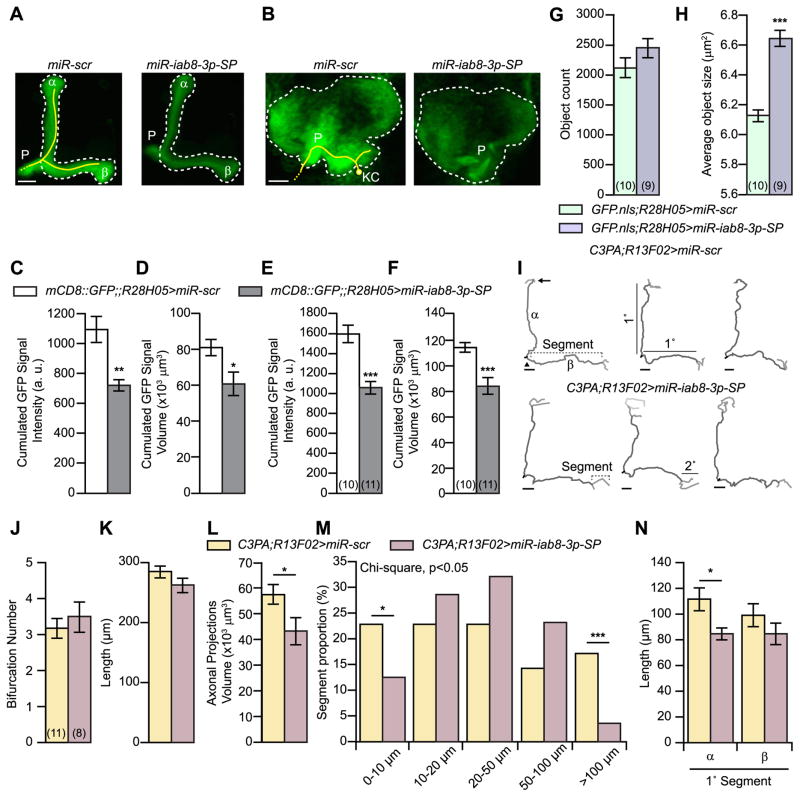
Fig. 5. MiR-iab8-3p inhibition alters the structure of αβ MBn.
(A) Representative maximal projection image of a confocal stack of the right αβ MB lobes of flies co-expressing mCD8::GFP with miR-scr (left panel) or miR-iab8-3p-SP (right panel) and driven by the R28H05-gal4 driver. The white dashed line was added to help visualize the contour of the lobes and a yellow line to illustrate the projections from a single MBn neuron. α: alpha lobe, β: beta lobe, P: peduncle. Scale bar: 10 μm. (B) Representative maximal projection image of a confocal stack of the right αβ MB calyx from flies co-expressing mCD8::GFP with miR-scr (left panel) or miR-iab8-3p-SP (right panel) and driven by R28H05-gal4 driver. A yellow line was added to help visualize the projection in the MB calyx from a single neuron and a white dashed line to identify the boundaries for the calyx. KC: Kenyon Cell, P: peduncle. Scale bar: 10 μm. (C) MiR-iab8-3p-SP expression decreases mCD8::GFP intensity in the αβ MB lobes. The mCD8::GFP average intensity was calculated for each slice of a stack and then summed across all slices. Results are presented as the mean±s.e.m. with n = 11. A two-sample, bilateral Student t-test was used to compare the two genotypes with Welch’s correction to account for the inequality of the variances. **: p<0.01. (D) MiR-iab8-3p-SP expression decreased the signal area identifying the αβ MB lobes. The area covered by the mCD8::GFP signal was computed for each slice of the stack and summed across slices. The signal was thresholded using the same cut-off intensity value. MiR-iab8-3p inhibition reduced the volume occupied by the mCD8::GFP signal in the αβ MB lobes. Results are presented as the mean±s.e.m. with n = 11. A two-sample, bilateral Student t-test was used to compare the two genotypes. *: p<0.05. (E) MiR-iab8-3p-SP expression decreases mCD8::GFP intensity in the calyx. The mCD8::GFP average intensity was calculated for each slice of a stack and then summed across all slices. Results are presented as the mean±s.e.m. with n = 10–11. A two-sample, bilateral Student t-test was used to compare the two genotypes. ***: p<0.001. (F) MiR-iab8-3p-SP expression decreased the signal area identifying the ccalyx of αβ MBn. The area covered by the mCD8::GFP signal was computed for each slice of the stack and summed across slices. Results are presented as the mean±s.e.m. with n = 10–11. A two-sample, bilateral Student t-test was used to compare the two genotypes. ***: p<0.001. (G) MiR-iab8-3p-SP expression had no significant effect on the total number of αβ MBn. The number of neurons estimated in flies expressing miR-iab8-3p-SP was not significantly different from flies expressing the miR-scr control. Results are presented as the mean±s.e.m. with n = 9–10. (H) MiR-iab8-3p-SP expression increased the size of αβ MBn nuclei as estimated from the GFP.nls signal. The area occupied by the nuclei of the αβ MBn was increased in flies expressing miR-iab8-3p-SP relative to flies expressing the miR-scr control. Results are presented as the mean±s.e.m. with n = 9–10. A two-sample, bilateral Student t-test was used to compare the two genotypes. ***: p<0.001. (I) Representative traces of axonal projections of the αβ neurons obtained after photo-conversion of C3PA-GFP in the cell body. The top panel shows three traces from flies expressing miR-scr together with uas-C3PA-GFP in MBn. The lower panels show ones obtained from flies with miR-iab8-3p-SP expressed in the MBn. The arrowhead identifies the initial bifurcation of the axon entering the lobes from the peduncle. The arrow identifies a bifurcation of the axon in the lobe. A segment is defined as the three dimensional distance of the axon located between two successive bifurcations or between a bifurcation and the end of a projection. Segments lengths were located between 1.4 and 161.8 μm and were ranked accordingly by class.α: alpha lobe, β: beta lobe. 1°: primary segments emerging from the initial bifurcation. Scale bars: 10 μm. (J) MiR-iab8-3p-SP expression had no effect on the number of bifurcations of αβ MBn in the αβ lobes. This number includes the initial bifurcation of the peduncular axon into the vertical and horizontal projections. The number of bifurcations was assessed using the ‘Branched Structure Analysis’ of the Neurolucida explorer software after tracing the projections. Results are presented as the mean±s.e. m. with n = 8–11. (K) MiR-iab8-3p-SP expression has no effect on the total length of the αβ MBn axonal projections in the αβ lobes. The length was assessed using the ‘Branched Structure Analysis’ of the Neurolucida explorer software after tracing the projections. Results are presented as the mean±s.e.m. with n = 8–11. (L) MiR-iab8-3p-SP expression decreased the neuropil volume occupied by the axonal projections in the αβ lobes. The volume was computed using the ‘Complex Hull Analysis’ of the Neurolucida explorer software after tracing the projections. Results are presented as the mean±s.e.m. with n = 8–11. A two-sample, bilateral Student t-test was used to compare the two genotypes. *: p<0.05. (M) MiR-iab8-3p-SP expression altered the length distribution of axonal segments. The definition of segment is provided in panel I. All segments were ranked according to their length and separated by genotype. MiR-iab8-3p-SP expression significantly reduced the fraction of small (<10 μm) and long axonal segments (> 100 μm). Results are presented as the proportion of segments assigned to a distribution of size classes and relative to the total number of branches with n = 56–70. Chi-square tests were used to compare the distributions and proportions. *: p<0.05,***: p<0.001. (N) MiR-iab8-3p-SP expression reduces the length of the primary segment of the αβ MBn in the α lobe but not in the β lobe. The average length of the primary segment was computed for the α and β lobes independently and according to the genotype. Results are presented as the mean±s.e.m. with n = 8–11. A bilateral Mann-Whitney test was used to compare genotypes. *: p<0.05.
2.5. MiR-iab8-3p inhibition alters αβ MBn projections, distribution and cell body size
The precise development of neural circuitry that mediates cognitive abilities is critical for normal cognition during adulthood (Xu et al., 2010; Feng and Feng, 2011; Olde Loohuis et al., 2012; Nowak and Michlewski, 2013; Sun and Shi, 2015). Given that normal miR-iab8-3p expression was necessary in αβ MBn during development for normal adult memory, we asked whether the structure of αβ MBn in adults was perturbed by miR-iab8-3p inhibition during development (Fig. 5).
We first used mCD8::GFP expression in the αβ MBn to gain insight into the global structure of these neurons in adults (Fig. 5A–F). We imaged the MB lobes and calyx (Fig. 5A,B) using confocal microscopy and quantified the GFP signal intensity in sub-stacks for miR-iab8-3p-SP or the miR-scr expressing flies. We uncovered a significant decrease in GFP intensity in the lobes and calyces of flies expressing miR-iab8-3p-SP (Fig. 5C,E). In addition, the volume occupied by the GFP signal was reduced in flies expressing miR-iab8-3p-SP compared to the control (Fig. 5D,F). We also examined the GFP signal intensity across the dorsal/ventral, medial/lateral, and anterior/posterior axes of the MB lobes using a sliding window and found that the distribution intensity was altered slightly in the miR-iab8-3p-SP expressing flies across the medial/lateral axis of the vertical lobes (Fig. S4A,B) and across the dorsal/ventral axis of the horizontal lobes (Fig. S4C,D). These results together suggest that miR-iab8-3p-SP expression decreases the number and spatial distribution of neurites extending from αβ MBn.
A decreased number of axonal or dendritic fibers might be due to fewer MB cells. We estimated the number of αβ MBn cell bodies from image stacks of adult brains. To achieve this goal, we co-expressed miR-iab8-3p-SP or miR-scr together with a nuclear-localized GFP (uas-GFP.nls). We processed images of confocal stacks to obtain an estimate of the number of αβ MBn nuclei. Each brain (both hemispheres) contained ~2300 αβ MBn counted in this way, but there was no significant difference between genotypes (Fig. 5G). Surprisingly, we identified a significant increase in the size of the nuclear signal in the miR-iab8-3p-SP expressing flies (Fig. 5H). This conclusion was confirmed by measuring the size of αβ MBn nuclei in dissociated neurons (Fig. S4E,F). We conclude from these structural analyses that MBn cell body size is increased in the miR-iab8-3p-SP expressing flies, using the proxy of nuclear area, and that there is a decrease in the neuropil volume occupied by the MBn axonal and dendritic fibers.
To gain a more precise understanding of the effect of miR-iab8-3p-SP on the structure of individual αβ MBn, we activated C3PA-GFP (Patterson and Lippincott-Schwartz, 2002; Ruta et al., 2010; Caron et al., 2013) in the cell body of MBn to visualize the neuronal projections in the αβ MB lobes (Fig. 5I). The C3PA-GFP was expressed together with miR-iab8-3p-SP or miR-scr in the MBn. After photoactivation and diffusion of the C3PA-GFP, the projections were manually traced using Neurolucida and analyzed with Neurolucida explorer. The total length (cumulative length of all segments) and the number of bifurcations were not affected (Fig. 5J, K). But the global volume occupied by the axonal projections was significantly decreased (Fig. 5L) in miR-iab8-3p-SP expressing flies. This result confirmed the decreased cumulative area of the mCD8:: GFP signal observed in flies expressing miR-iab8-3p-SP (Fig. 5D). We also compared the distribution of axonal segment sizes and found a significant change, with a decrease in the small and large size axonal segments in flies expressing miR-iab8-3p-SP (Fig. 5M). We asked whether this difference was due to α or β primary (1°) segments (Fig. 5N) and found that the difference was restricted to the α neurite. Together, these results show that the α lobe neurite is shortened with the expression of miR-iab8-3p-SP.
2.6. Potential targets of MiR-iab8-3p
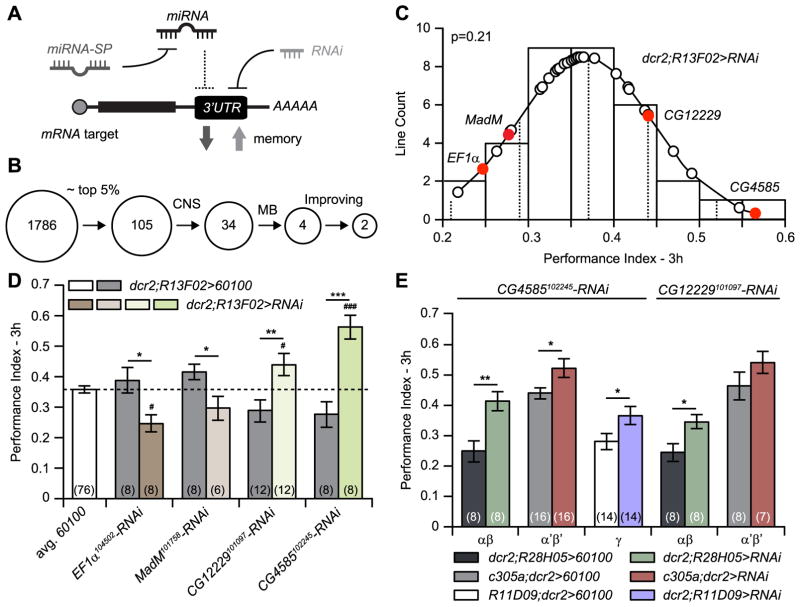
We next sought to obtain preliminary information about the mRNA targets for miR-iab-3p that might influence memory formation (Fig. 6A). We used the bio-informatic tool microRNA.org to predict potential mRNA targets based on multiple parameters: sequence complementarity at the seed region and 3′ end of the mRNA, A/U composition near the target site, secondary structure accessibility, length of the UTR, relative position of the target site in the UTR and conservation score (Betel et al., 2010). The micro-RNA.org algorithm predicted 1786 potential mRNA targets (Fig. 6). We selected the top ~5% (105) of these based on mirSVR scores (≤ – 1.2) and tested for the involvement of these candidates in memory formation using RNAi interference (Walkinshaw et al., 2015), expressing RNAi transgene for each in the CNS using the pan-CNS, n-syb-gal4 driver, and subsequently in the MBn (Table S3). Thirty-four genes modulated 3 h memory when inhibited in the CNS, and 4 of these reproducibly influenced memory scores when subsequently tested in the MBn and relative to the 60100-control line, host genotype for producing the RNAi transgenics (Fig. 6C,D and Table S3).
Fig. 6. CG12229 and CG4585 are potential mRNA targets for miR-iab8-3p.
(A) Rational for the miR-iab8-3p mRNA-target screen. MiR-iab8-3p-SP binds to miR-iab8-3p, preventing its interaction with the 3′UTR of mRNA targets and consequently increasing protein expression. Since miR-iab8-3p-SP expression impairs memory formation, we adopted the working hypothesis that inhibiting its targets using an RNAi approach would decrease protein expression and increase memory formation. (B) Schematic representation of the workflow used to identify the potential mRNA-targets. A computational approach (microRNA.org) identified 1786 potential targets. We selected the most likely targets picking lines with a mirSVR score ≤ – 1.2 and obtained a primary list of 105 candidate genes. Three-hour memory was tested for flies expressing RNAi (uas-RNAi) to each of these candidates using the CNS-wide, n-syb-gal4 driver. This primary screen reduced the list to 34 RNAi candidates that influenced memory formation. These RNAi’s were subsequently expressed in the MBn using R13F02-gal4. We identified 4 genes in this manner that reproducibly modulated 3 h memory. Two of these increased memory formation. (C) PI distribution of miR-iab8-3p potential targets when inhibited in MBn. Three-hour memory was measured in RNAi-expressing flies using the R13F02-gal4 driver. The average PI for 3 h memory was 0.37±0.01 (mean±s.e.m.); the PIs followed a Normal distribution (p = 0.21). PIs obtained for genes ultimately identified are highlighted in red. PIs for genes represented with open circles were discarded since not confirmed by the following experimental validation. (D) Validation for the four potential targets. The memory performance for flies carrying each individual uas-RNAi and the R13F02-gal4 driver was compared to R13F02-gal4 > 60100 flies, host genotype used to produce the uas-RNAi lines. For each of the four RNAis, the PIs were significantly different from the paired 60100 control flies, and to the average PI for R13F02-gal4 > 60100 flies across the screen. Results are presented as the mean±s.e.m. with n = 6–12. Two-sample, bilateral Student t-tests were used to compare the two genotypes. ***: p<0.001,**: p<0.01,*: p<0.05. For comparisons to the average 60100 control (n = 76): ###: p<0.001, #: p<0.05. (E) CG12229 and CG4585 inhibition in subclasses of MBn increased memory expression. Each uas-RNAi transgene was combined with either R11D09, c305a or R28H05-gal4 drivers to obtain expression in the, α′β′ or αβ neurons, respectively. CG12229 inhibition in αβ neurons increased memory expression relative to the 60100 control. CG4585 inhibition in, α′β′ and αβ neurons increased memory expression. Results are presented as the mean±s.e.m. with n = 8–16. Two-sample, bilateral Student t-tests were used to compare the two genotypes. **: p<0.01, *: p<0.05.
We focused our interest on CG12229 and CG4585 because the RNAi knockdowns increased the memory performance, the prediction as an authentic target based on the opposite effect produced by the miR-iab8-3p-SP sponge (Fig. 6A). CG12229 is predicted to code for a pyruvate kinase and CG4585 an ethanolamine phosphotransferase. Since expression of miR-iab8-3p-SP decreases memory when expressed in any of the three major classes of MBn, we tested RNAi knockdowns for CG12229 and CG4585 in these same three classes of neurons. CG12229 increased memory only when knocked down in the αβ MBn, while CG4585 increased memory when inhibited in all three classes of MBn (Fig. 6E). These results identify ethanolamine phosphotransferase as an attractive target of miR-iab8-3p for mediating perhaps some of the structural and behavioral effects of miR-iab8-3p inhibition.
3. Discussion
In the present report, we offer compelling results indicating that miR-iab8-3p exerts an important and specific role during development of MBn, a cell population that is critical for memory formation in adult flies (Busto et al., 2010; Davis, 2011). We found that developmental inhibition of miR-iab8-3p perturbs the structure of adult αβ neurons, including an increased cell body size and a decreased volume of neuropil occupancy by MBn axons. The latter cellular phenotype seems primarily attributable to a decrease, specifically in the α lobe, in the length of the very long, and short axonal segments, as we found no difference in branching. It seems probable that the limitation in territory occupied by the MBn axons affects their connectivity with either extrinsic input and/or output neurons, leading to poor learning of olfactory cues with this alteration in circuit arrangement (Perisse et al., 2013).
The miR-iab8 hairpin (including miR-iab8-5p and −3p), which is also called miR-iab4AS, was first described as a Hox miRNA gene, and a member of the bithorax complex (BX-C) necessary for fertility (Bender, 2008; Stark et al., 2008; Tyler et al., 2008). It resides between the abd-A and Abd-B genes as part of a noncoding RNA (ncRNA) on the antisense strand of the miR-iab4 locus (Bender, 2008; Stark et al., 2008; Tyler et al., 2008). The miR-iab8 hairpin decreases Ubx and abd-A protein expression and induces a haltere-to-wing homeotic transformation when ectopically expressed (Stark et al., 2008; Tyler et al., 2008). The primary transcript for miR-iab8 hairpin is detected in embryos, larvae, pupae and adult male and females (Stark et al., 2008). Deep sequencing results indicate that the miR-iab8-3p strand is expressed at all developmental phases at low levels with some preference in embryos (Ruby et al., 2007; Chung et al., 2008).
Our approach and results highlight the advantages of using miR-SP for dissecting miRNA function. Although they probably offer a hypomorphic insult, they have the great advantage of targeting individual mature miRNAs, while clean genomic knockouts may be difficult when multiple miRNA genes are clustered (Ebert and Sharp, 2010). In our specific case, two different mature miRNAs, miR-iab8-3p and miR-iab8-5p, are encoded from the same RNA hairpin embedded in the larger iab8 noncoding RNA (Bender, 2008; Stark et al., 2008; Tyler et al., 2008). Effects of a genomic deletion would consequently be difficult to interpret due to the loss of multiple miRNAs. The deletion of this locus (called Δmir) affects fertility, segment identity (Bender, 2008; Stark et al., 2008; Tyler et al., 2008) and self-righting behavior through the inhibition of miR-iab4, the miRNA hairpin encoded by the complementary DNA strand of miR-iab8 (Picao-Osorio et al., 2015). Remarkably, we found that each strand of the miR-iab8 hairpin, when inhibited, produced an opposite effect on memory performance. This indicates a dual role for this miRNA on memory processes and stresses the unique advantage of miR-SP to properly dissect the individual role of each strand of the miRNA hairpin. Both strands of the bifunctional miR-9 have been shown to regulate two molecular components (REST and CoREST) of the same repressor complex (Packer et al., 2008). This precedent offers the attractive hypothesis that miR-iab8 is similarly bifunctional and with both strands acting in opposite ways during the development of the MBn. Although intriguing, we currently have no data speaking to the mechanism whereby miR-iab8-5p disruption produces memory enhancement.
The effect of miR-iab8-3p inhibition on memory was primarily restricted to the MBn, although we observed a slight effect with inhibition in GABAergic neurons as well. Thus, the effects are largely localized to the MBn population. This is important as it shows that miR-iab8-3p has cell-type specific roles during development rather than exerting an effect on target genes in all types of brain neurons. However, we did find a role for this miRNA in all three major classes of MBn, suggesting that miR-iab8-3p function is engaged when neurons are differentiated into MBn and before they become further differentiated into one of the three individual classes of MBn.
Individual miRNAs serve to orchestrate the expression level of many target genes in tissue and developmental time-specific ways. This makes the task of identifying the specific effectors of any miRNA-associated phenotype a long and difficult task, especially given the possibility that a given phenotype may emerge from changing the expression level of multiple target effectors. But as an initial step in this direction, we chose to provide a preliminary identification of miR-iab8-3p targets using a behavioral approach rather than through mRNA quantification since some miRNAs decrease protein expression without changing mRNA expression (Griggs et al., 2013). In addition, the approach interrogates the important endpoint of function, rather than simple changes in abundance that may or may not relate to function.
We identified four genes whose inhibition in MBn modulated memory formation. These include EFα1, MadM, CG12229 and CG4585. We focused on the two genes whose inhibition increased memory as predicted by our model (Fig. 6A). We provide three arguments for favoring CG4585 as a major effector of the miR-iab8-3p role in learning. First, CG4585 improved memory when inhibited. Second, the effect was obtained with inhibition in all three classes of MBn. Third, the function of CG4585 aligns with the phenotypic observations.
Drosophila sphingolipids are critical for normal development with metabolic intermediates being involved in signal transduction cascades and progressive neurodegenerative disorders (Kraut, 2011). Drosophila lacks sphingomyelin (SM) but instead synthetizes the SM analog ceramide phosphoethanolamine (CPE) from ceramide (Vacaru et al., 2013). In general, dysfunction of sphingolipid biosynthesis leads to the accumulation of lipid substrates leading to cellular enlargement, dementia, and intellectual disability (Mencarelli and Martinez-Martinez, 2013). Interestingly, CG4585 has recently been shown to possess a CPE synthase activity from expression experiments in cultured S2 insect cells (Vacaru et al., 2013). MiR-iab8-3p inhibition should increase CG4585 expression leading to an excess of CPE production, analogous to the accumulation of SM observed in Niemann Pick’s disease (Mencarelli and Martinez-Martinez, 2013). These observations are generally consistent with the phenotypes of neuronal enlargement and behavioral alterations observed with miR-iab8-3p inhibition. An immediate goal from these results will be to further characterize the link between miR-iab8-3p and CG4585.
Our results begin to fill an important gap in knowledge about Drosophila learning and memory – the identification and characterization of genetic factors that influence the development of the nervous system important for adult cognitive functions. Past research on memory formation in Drosophila has emphasized biological functions that influence the adult physiology of MBn critical for memory formation (Davis, 2005, 2011). The critical experimental distinction in showing developmental vs adult physiological requirement has been temporal knockdown or rescue experiments, typified by adult-specific rescue of dunce and rutabaga functions (Dauwalder and Davis, 1995; McGuire et al., 2003; Mao et al., 2004). Understanding developmental genetic functions is critically important to elucidate the molecular logic underlying the development of adult brain structures important for cognition, such as MBn here for olfactory learning and help gain potential insights into the myriad of human neurodevelopmental disorders such as intellectual disability and autism.
4. Materials and methods
4.1. Fly lines
Drosophila was raised using standard food at room temperature. Fly crosses were performed at 25 °C with ~70% relative humidity across a 12 h light/12 h dark cycle. The uas-miR-iab8-3p-SP, uas-miR-iab8-5p-SP and uas-miR-scr fly lines were previously described (Busto et al., 2015; Fulga et al., 2015). These first two uas lines were crossed to various Gal4 lines to establish the experimental genotype. The control genotype, in general, was made by crossing the same Gal4 line to uas-miR-scr, a transgenic line containing scrambled sequences in the same vector and inserted at the same docking site (attP40/attP2) in the genome as the uas-miR-SP lines. The uas-miR-iab8 over-expression line was backcrossed in the wCS10 background for six generations. The other lines used in this study include: elavc155-gal4 (Lin and Goodman, 1994), R13F02-gal4 (Jenett et al., 2012), uas-mCD8::GFP (Lee and Luo, 1999), Or83b(orco)-gal4 (Wang et al., 2003), GH146-gal4 (Stocker et al., 1997), C316-gal4 (Waddell et al., 2000), TH-gal4 (Friggi-Grelin et al., 2003), Gad-gal4 (Ng et al., 2002), OK107-gal4 (Connolly et al., 1996), MZ604-gal4 (Suster et al., 2003), NP2492-gal4 (Tanaka et al., 2008), R11D09-gal4 (Jenett et al., 2012), R84G09-gal4 (Jenett et al., 2012), c305a-gal4 (Armstrong et al., 2006), R35B12-gal4 (Jenett et al., 2012), R28H05-gal4 (Jenett et al., 2012), c739-gal4 (O’Dell et al., 1995), tub-Gal80ts (McGuire et al., 2003), uas-GFP.nls (Shiga et al., 1996), n-syb-gal4 (Pauli et al., 2008), uas-dicer2 (Dietzl et al., 2007) and uas-PA-GFP (Patterson and Lippincott-Schwartz, 2002; Ruta et al., 2010; Caron et al., 2013). RNA interference (uas-RNAi) lines, and their control line (60100, host genotype used to construct the uas-RNAi transgenics), were obtained from the KK library of the Vienna Drosophila RNAi Center (VDRC) (Dietzl et al., 2007).
4.2. Behavioral analyses
One-to-four day old flies were collected for behavioral experiments ~16-to-24 h prior to experiments. Thirty minutes before conditioning, flies were transferred in fresh food vials and put in the behavioral test room to adapt to the conditions (dim red light, 25 °C, ~80% relative humidity). Groups of ~65 flies were trained using the standard aversive olfactory conditioning test already described (Beck et al., 2000). Flies were subjected to a succession of two odor stimuli lasting 1 min each and separated by 30 s. The first odor stimulus (CS+) was associated with 12 repeats of a 90-V electric shock (US). The second odor stimulus was not paired with electric shocks and thus constituted a non-conditioned stimulus (CS-). The odorants used were benzaldehyde (BEN), 3-octanol (OCT) or 4-methycyclohexanol (MCH) when specified. They were diluted in mineral oil at concentrations of ~0.05, ~0.2% and ~0.1%, respectively. Memory retention was tested by allowing flies to choose for 2 min between the sides of a T-maze containing the CS+ odor in one arm and CS− odor in the alternative arm.
Memory acquisition was tested giving an increasing number (from 1 to 12) of electric shocks during the CS+ stimulation (Fig. 1E, bottom). Memory was tested immediately following training. Anesthesia-sensitive (ASM) and anesthesia-resistant (ARM) memories were tested 3 h following conditioning (Fig. 3C, bottom). These two constituents of memory were distinguished by giving a brief cold-shock induced anesthesia (2 min, 4 °C) 2 h following conditioning to erase ASM. After that period, flies were transferred back in their food vial and kept unperturbed for an additional hour before the memory test. For the TARGET experiments (Fig. 3D, left panel), three-to-five day old flies were used; conditioning and memory tests were performed at 25 °C (McGuire et al., 2003).
To test odor avoidance, naive flies were allowed to distribute for 2 min between the two arms of the T-maze with an odor stream on one side and a non-odorized air stream on the other. To test shock avoidance, naive flies were allowed to distribute for 2 min between the two arms of a T-maze containing copper grids with only one side being electrified. An avoidance index was computed with the following formula: [(number of flies in neutral arm) – (number of flies in odor/shocked arm)/(total number of flies in both arms)].
4.3. Immunohistochemistry
We followed the protocol described by the Fly Light Project (Jenett et al., 2012) using 1–6 day old female fly brains. The primary antibodies used were rabbit anti-GFP (1:1000, Invitrogen) and mouse anti-nc82 (1:50; DSHB). The secondary antibodies included Alexa 633 anti-mouse (1:400) and Alexa 488 anti-rabbit (1:800). Images were obtained using Leica TCS SP5 and SP8 confocal microscopes using the exact same set-up for both experimental groups.
4.4. Image analysis
Analyses of GFP immunohistochemistry on confocal images were performed using ImageJ (Abramoff et al., 2004). For each brain, the cumulative GFP signal intensity was established by calculating the average intensity of the GFP signal for each confocal plane using the ‘mean gray value’ function. The average intensity values for each confocal plane were then summed for all the brains to obtain a quantitative value of GFP intensity representing the GFP signal throughout the brain. To assess the area spanned by the GFP signal, we used the same intensity threshold for all brains using the ‘threshold’ function of ImageJ. The area covered by the GFP signal was calculated for each confocal plan and then summed for the all brain. To gain access to the distribution of the GFP signal in the three dimensions of the MB lobes, we made a maximal projection of the confocal stack of the lobes. We used the ‘plot profile’ function to obtain an average of the GFP signal every 0.36 μm (0.49 μm for the antero-posterior axis) along the axis of our choice. Samples were aligned using the section of the lobe with the highest GFP intensity value. Only regions represented with n ≥ 7 were considered for statistical analysis. Average intensity was plotted relative to the highest intensity region.
To estimate the cell body number, each original image was first processed using the ‘unsharp mask’ function of ImageJ to highlight the nuclei. A ‘threshold’ was then imposed to isolate the nuclei from the background. The ‘despeckle’ function was then used to remove the isolated pixels. The ‘watershed’ function was used to isolate individual objects. Eventually, we used the ‘analyze particles’ tools to count objects. Values were summed for the all stacks.
4.5. Brain dissociation
One-to-six day old individual female fly brains were dissected in insect’s Ringer solution, the optic lobes removed, and brains washed once with dissecting solution. Brains were then incubated for 30 min in activated papain solution then replaced with insect’s Ringer solution and triturated in a microcentrifuge tube first with a 200 μl and then a 20 μl pipet tip. Cells were ultimately transferred in 384 well plate and allowed to settle at the bottom of a well for 3 h and then centrifuged at 12.000 RPM for 5 min. Neurons were imaged for GFP with the GE Incell 6000 high-content screening microscope at 10 ×. Cells were counted and measured using custom segmentation algorithms in GE Developer.
4.6. Photo-activation experiments
One-to-six day old individual female fly brains were dissected in ice-cold saline (in mM NaCl 124, KCl 3, MOPS 20, CaCl2 1.5, MgCl2(H2O)6 4, NaHCO3 5, NaH2PO4(H2O) 1, trehalose 10, sucrose 7, glucose 10). Brains were subsequently transferred and stuck at the bottom of a petri dish filled with saline. Individual MBn cell body was identified using the 488 nm laser of a confocal microscope (Leica). A circular 1-by-1 μm region of interest was defined at the center of a MBn and C3PA-GFP activated using a multi-photon laser (Coherent Inc.) set at 710 nm. The photoactivation stimuli consisted of three stimuli (0.325 ms each) separated by 2 min and for 1 h (90 stimuli total) with a 40x objective. The photoactivation laser power was typically between 4 and 40 mW at the objective. Brains were later flipped and axonal MBn projections imaged using the 63 × objective with the confocal set at 488 nm.
Neuronal projections were manually traced using the Neurolucida software (MBF Bioscience). Total projections length, bifurcations, neuronal projection volume and length of the segments were computed using the ‘Convex Hull Analysis’ and ‘Branched Structure Analysis’ functions of the Neurolucida explorer software (MBF bioscience). α′β′ and γ neurons were discarded as the neurons with no C3PA-GFP diffusion in the calyx or in the lobes or with major gaps or unresolvable structures.
4.7. Statistics
Statistical analyses were performed using XLSTAT®, Prism® and R (R Core Team, 2008). Memory Performance Indices are known to follow a normal distribution (Tully et al., 1994). Normality of samples was additionally assessed using the Kolmogorov-Smirnov test or the D’Agostino & Pearson test when possible. When average PIs of two groups were compared we used two-sample, bilateral Student t-tests if samples followed a normal distribution. In cases where they did not, we used Mann-Whitney tests for non-parametric data. In case where the variances were not the same between samples we used Welch’s correction for inequality of variances. To compare the average performance of a group with a theoretical value, we used one-sample bilateral Student t-tests. For multiple group comparisons, we used ANOVA followed by Bonferroni’s or two-way ANOVA followed by Tukey’s post hoc tests corrected for multiple comparisons. To compare distribution and proportions we used χ-square tests. For confocal image analysis, one outlier was removed using the Grubb’s test.
Supplementary Material
Summary statement.
MiR-iab8-3p is necessary during development for normal olfactory learning. Inhibition of this miRNA alters the size of mushroom body neurons and their axonal projections, which impacts learning abilities in the adult.
Acknowledgments
Funding
Research was funded by NIH grants P01NS090994 and 2R37N519904 to RLD.
We want to thank all members of the laboratory for their help and comments and more specifically Erica Walkinshaw, Daniel Richter and Caitlin Farkas for their help with behavioral tests of potential miR-iab8-3p-SP targets and fly husbandry, and Kyle Vick for the analysis of GFP signals of dissociated brains. We are grateful to David Van Vactor and Tudor Fulga for the sponge lines. For supplying Drosophila lines, we thank the Bloomington Stock Center, Janelia Farms, the VDRC, FlyORF and Richard Axel.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2016.09.010.
Footnotes
Competing interests
No competing interests declared.
Author contributions
GUB, TGO and RLD designed the project, analyzed the data and wrote the manuscript. GUB performed the experiments. MC performed immunohistochemistry and brain dissociations.
References
- Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with Image. J Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762–R776. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akalal DB, Yu D, Davis RL. The long-term memory trace formed in the Drosophila α/β mushroom body neurons is abolished in long-term memory mutants. J Neurosci. 2011;31:5643–5647. doi: 10.1523/JNEUROSCI.3190-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksoy-Aksel A, Zampa F, Schratt G. MicroRNAs and synaptic plasticity–a mutual relationship. Philos Trans R Soc Lond B Biol Sci. 2014:369. doi: 10.1098/rstb.2013.0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong JD, Texada MJ, Munjaal R, Baker DA, Beckingham KM. Gravitaxis in Drosophila melanogaster: a forward genetic screen. Genes Brain Behav. 2006;5:222–239. doi: 10.1111/j.1601-183X.2005.00154.x. [DOI] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CD, Schroeder B, Davis RL. Learning performance of normal and mutant Drosophila after repeated conditioning trials with discrete stimuli. J Neurosci. 2000;20:2944–2953. doi: 10.1523/JNEUROSCI.20-08-02944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W. MicroRNAs in the Drosophila bithorax complex. Genes Dev. 2008;22:14–19. doi: 10.1101/gad.1614208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredy TW, Lin Q, Wei W, Baker-Andresen D, Mattick JS. MicroRNA regulation of neural plasticity and memory. Neurobiol Learn Mem. 2011;96:89–94. doi: 10.1016/j.nlm.2011.04.004. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Busto GU, Cervantes-Sandoval I, Davis RL. Olfactory learning in Drosophila. Physiol (Bethesda) 2010;25:338–346. doi: 10.1152/physiol.00026.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busto GU, Guven-Ozkan T, Fulga TA, Van Vactor D, Davis RL. microRNAs that promote or inhibit memory formation in Drosophila melanogaster. Genetics. 2015;200:569–580. doi: 10.1534/genetics.114.169623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron SJ, Ruta V, Abbott LF, Axel R. Random convergence of olfactory inputs in the Drosophila mushroom body. Nature. 2013;497:113–117. doi: 10.1038/nature12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Sandoval I, Martin-Peña A, Berry JA, Davis RL. System-like consolidation of olfactory memories in Drosophila. J Neurosci. 2013;33:9846–9854. doi: 10.1523/JNEUROSCI.0451-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genom Hum Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- Chung WJ, Okamura K, Martin R, Lai EC. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr Biol. 2008;18:795–802. doi: 10.1016/j.cub.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly JB, Roberts IJ, Armstrong JD, Kaiser K, Forte M, Tully T, O’Kane CJ. Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science. 1996;274:2104–2107. doi: 10.1126/science.274.5295.2104. [DOI] [PubMed] [Google Scholar]
- Dauwalder B, Davis RL. Conditional rescue of the dunce learning/memory and female fertility defects with Drosophila or rat transgenes. J Neurosci. 1995;15:3490–3499. doi: 10.1523/JNEUROSCI.15-05-03490.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- Davis RL. Traces of Drosophila memory. Neuron. 2011;70:8–19. doi: 10.1016/j.neuron.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Durkin MS, Schneider H, Pathania VS, Nelson KB, Solarsh GC, Bellows N, Scheffler RM, Hofman KJ. Learning and developmental disabilities. In: Jamison DT, Breman JG, Measham AR, Alleyne G, Claeson M, Evans DB, Jha P, Mills A, Musgrove P, editors. Disease Control Priorities in Developing Countries. 2. Chapter 49. World Bank; Washington (DC): 2006. [Google Scholar]
- Ebert MS, Sharp PA. MicroRNA Sponges: progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Feng Y. MicroRNAs in neural cell development and brain diseases. Sci China Life Sci. 2011;54:1103–1112. doi: 10.1007/s11427-011-4249-8. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 2003;54:618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- Fulga TA, McNeill EM, Binari R, Yelick J, Blanche A, Booker M, Steinkraus BR, Schnall-Levin M, Zhao Y, DeLuca T, et al. A transgenic resource for conditional competitive inhibition of conserved Drosophila microRNAs. Nat Commun. 2015;6:7279. doi: 10.1038/ncomms8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs EM, Young EJ, Rumbaugh G, Miller CA. MicroRNA-182 regulates amygdala-dependent memory formation. J Neurosci. 2013;33:1734–1740. doi: 10.1523/JNEUROSCI.2873-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guven-Ozkan T, Davis RL. Functional neuroanatomy of Drosophila olfactory memory formation. Learn Mem. 2014;21:519–526. doi: 10.1101/lm.034363.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunsberger JG, Austin DR, Chen G, Manji HK. MicroRNAs in mental health: from biological underpinnings to potential therapies. Neuromol Med. 2009;11:173–182. doi: 10.1007/s12017-009-8070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabel G, Pascual A, Preat T. Exclusive consolidated memory phases in Drosophila. Science. 2004;304:1024–1027. doi: 10.1126/science.1094932. [DOI] [PubMed] [Google Scholar]
- Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- Kahsai L, Zars T. Learning and memory in Drosophila: behavior, genetics, and neural systems. Int Rev Neurobiol. 2011;99:139–167. doi: 10.1016/B978-0-12-387003-2.00006-9. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S. Sequential use of mushroom body neuron subsets during drosophila odor memory processing. Neuron. 2007;53:103–115. doi: 10.1016/j.neuron.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut R. Roles of sphingolipids in Drosophila development and disease. J Neurochem. 2011;116:764–778. doi: 10.1111/j.1471-4159.2010.07022.x. [DOI] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Krützfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 2006;4:9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Kunz T, Kraft KF, Technau GM, Urbach R. Origin of Drosophila mushroom body neuroblasts and generation of divergent embryonic lineages. Development. 2012;139:2510–2522. doi: 10.1242/dev.077883. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Li W, Cressy M, Qin H, Fulga T, Van Vactor D, Dubnau J. MicroRNA-276a functions in ellipsoid body and mushroom body neurons for naive and conditioned olfactory avoidance in Drosophila. J Neurosci. 2013;33:5821–5833. doi: 10.1523/JNEUROSCI.4004-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DM, Goodman CS. Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron. 1994;13:507–523. doi: 10.1016/0896-6273(94)90022-1. [DOI] [PubMed] [Google Scholar]
- Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciotta S, Meregalli M, Torrente Y. The involvement of microRNAs in neurodegenerative diseases. Front Cell Neurosci. 2013;7:265. doi: 10.3389/fncel.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Roman G, Zong L, Davis RL. Pharmacogenetic rescue in time and space of the rutabaga memory impairment by using Gene-Switch. Proc Natl Acad Sci USA. 2004;101:198–203. doi: 10.1073/pnas.0306128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Mencarelli C, Martinez-Martinez P. Ceramide function in the brain: when a slight tilt is enough. Cell Mol Life Sci. 2013;70:181–203. doi: 10.1007/s00018-012-1038-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–483. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Roorda RD, Lima SQ, Zemelman BV, Morcillo P, Miesenbock G. Transmission of olfactory information between three populations of neurons in the antennal lobe of the fly. Neuron. 2002;36:463–474. doi: 10.1016/s0896-6273(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Nowak JS, Michlewski G. miRNAs in development and pathogenesis of the nervous system. Biochem Soc Trans. 2013;41:815–820. doi: 10.1042/BST20130044. [DOI] [PubMed] [Google Scholar]
- O’Dell KMC, Armstrong JD, Yang MY, Kaiser K. Functional dissection of the Drosophila mushroom bodies by selective feminization of genetically defined subcompartments. Neuron. 1995;15:55–61. doi: 10.1016/0896-6273(95)90064-0. [DOI] [PubMed] [Google Scholar]
- Olde Loohuis NF, Kos A, Martens GJ, Van Bokhoven H, Nadif Kasri N, Aschrafi A. MicroRNA networks direct neuronal development and plasticity. Cell Mol Life Sci. 2012;69:89–102. doi: 10.1007/s00018-011-0788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell. 2008;14:239–251. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perisse E, Yin Y, Lin AC, Lin S, Huetteroth W, Waddell S. Different kenyon cell populations drive learned approach and avoidance in Drosophila. Neuron. 2013;79:945–956. doi: 10.1016/j.neuron.2013.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picao-Osorio J, Johnston J, Landgraf M, Berni J, Alonso CR. MicroRNA-encoded behavior in Drosophila. Science. 2015;350:815–820. doi: 10.1126/science.aad0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. [Google Scholar]
- Rao P, Benito E, Fischer A. MicroRNAs as biomarkers for CNS disease. Front Mol Neurosci. 2013;6:39. doi: 10.3389/fnmol.2013.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruta V, Datta SR, Vasconcelos ML, Freeland J, Looger LL, Axel R. A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature. 2010;468:686–690. doi: 10.1038/nature09554. [DOI] [PubMed] [Google Scholar]
- Saab BJ, Mansuy IM. Neuroepigenetics of memory formation and impairment: the role of microRNAs. Neuropharmacology. 2014;80:61–69. doi: 10.1016/j.neuropharm.2014.01.026. [DOI] [PubMed] [Google Scholar]
- Shiga Y, Tanaka-Matakatsu M, Hayashi S. A nuclear GFP/beta-galactosidase fusion protein as a marker for morphogenesis in living Drosophila. Dev Growth Differ. 1996;38:99–106. [Google Scholar]
- Stark A, Bushati N, Jan CH, Kheradpour P, Hodges E, Brennecke J, Bartel DP, Cohen SM, Kellis M. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 2008;22:8–13. doi: 10.1101/gad.1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker RF, Heimbeck G, Gendre N, de Belle JS. Neuroblast ablation in Drosophila P[GAL4] lines reveals origins of olfactory interneurons. J Neurobiol. 1997;32:443–456. doi: 10.1002/(sici)1097-4695(199705)32:5<443::aid-neu1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Sun E, Shi Y. MicroRNAs: Small molecules with big roles in neurodevelopment and diseases. Exp Neurol. 2015;268:46–53. doi: 10.1016/j.expneurol.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Suster ML, Martin JR, Sung C, Robinow S. Targeted expression of tetanus toxin reveals sets of neurons involved in larval locomotion in Drosophila. J Neurobiol. 2003;55:233–246. doi: 10.1002/neu.10202. [DOI] [PubMed] [Google Scholar]
- Tan L, Yu JT, Tan L. Causes and consequences of MicroRNA dysregulation in neurodegenerative diseases. Mol Neurobiol. 2015;51:1249–1262. doi: 10.1007/s12035-014-8803-9. [DOI] [PubMed] [Google Scholar]
- Tanaka NK, Tanimoto H, Ito K. Neuronal assemblies of the Drosophila mushroom body. J Comp Neurol. 2008;508:711–755. doi: 10.1002/cne.21692. [DOI] [PubMed] [Google Scholar]
- Tomchik SM, Davis RL. Dynamics of learning-related cAMP signaling and stimulus integration in the Drosophila olfactory pathway. Neuron. 2009;64:510–521. doi: 10.1016/j.neuron.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A. 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- Tully T, Preat T, Boynton SC, Del Vecchio M. Genetic dissection of consolidated memory in Drosophila. Cell. 1994;79:35–47. doi: 10.1016/0092-8674(94)90398-0. [DOI] [PubMed] [Google Scholar]
- Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, Hannon GJ, Lai EC. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacaru AM, van den Dikkenberg J, Ternes P, Holthuis JC. Ceramide phosphoethanolamine biosynthesis in Drosophila is mediated by a unique ethanolamine phosphotransferase in the Golgi lumen. J Biol Chem. 2013;288:11520–11530. doi: 10.1074/jbc.M113.460972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell. 2000;103:805–813. doi: 10.1016/s0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- Walkinshaw E, Gai Y, Farkas C, Richter D, Nicholas E, Keleman K, Davis RL. Identification of genes that promote or inhibit olfactory memory formation in Drosophila. Genetics. 2015;199:1173–1182. doi: 10.1534/genetics.114.173575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R, Shigeo JF. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 2003;112:271–282. doi: 10.1016/s0092-8674(03)00004-7. [DOI] [PubMed] [Google Scholar]
- Wang W, Kwon EJ, Tsai LH. MicroRNAs in learning, memory, and neurological diseases. Learn Mem. 2012;19:359–368. doi: 10.1101/lm.026492.112. [DOI] [PubMed] [Google Scholar]
- Xu B, Karayiorgou M, Gogos JA. MicroRNAs in psychiatric and neurodevelopmental disorders. Brain Res. 2010;1338:78–88. doi: 10.1016/j.brainres.2010.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Akalal DB, Davis RL. Drosophila alpha/beta mushroom body neurons form a branch-specific, long-term cellular memory trace after spaced olfactory conditioning. Neuron. 2006;52:845–855. doi: 10.1016/j.neuron.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.