Summary
The cellular proteome is a complex microcosm of structural and regulatory networks that requires continuous surveillance and modification to meet the dynamic needs of the cell. It is therefore crucial that the protein flux of the cell remains in balance to ensure proper cell function. Genetic alterations that range from chromosome imbalance to oncogene activation can affect the speed, fidelity and capacity of protein biogenesis and degradation systems, which often results in proteome imbalance. An improved understanding of the causes and consequences of proteome imbalance is helping to reveal how these systems can be targeted to treat diseases such as cancer.
Introduction
The cellular proteome is exceedingly complex (Box 1). Of the 20,000 or so protein-coding genes of the human genome, a typical cell transcribes about 10,000 genes, resulting in the production of at least as many proteins, which have a cumulative copy number of 109–1011 protein molecules per cell1–3. Although impressive, these numbers fail to demonstrate the true complexity of the cellular proteome due to three important reasons.
BOX 1. Current knowledge of proteome identity and abundance.
The human genome is predicted to encode 20,687 proteins, although its annotation continues to be refined131. Although RNA sequencing makes it possible to determine which genes in a particular cell type are transcribed, it is more challenging to determine the identity and relative abundance of proteins that are present in that cell type, partly because of the dynamic range of protein abundance and the difficulty in identifying certain types of proteins (such as membrane proteins)132. The translation products of 17,294 (ref. 15) and 18,097 (ref. 18) genes have been detected across a wide range of tissues and cell types, including putative translation products from what were previously considered to be non-coding RNAs. Studies in commonly used cell lines have attempted to identify all of the proteins in these cells and some have also tried to compare protein abundance with mRNA abundance1–3,8,9,133. Among the deepest proteomic content measured so far for a single cell line comes from studies of HEK293T cells134 and HeLa cells3. In the HeLa cell analysis, in which 10,596 proteins were detected, mRNA transcripts for up to an extra 20% of genes were identified3. Similarly, in the HEK293T cells, in which 10,326 proteins were detected, large-scale interaction proteomic experiments identified 10% more proteins than could be identified through total deep proteomic analysis134. Available descriptions of proteomes for such cell lines therefore probably underestimate the total number of proteins that are expressed by 10–20%. Proteomics also enables a means by which to estimate the copy number of individual proteins within a single cell. These estimates range from 1.7×1011 proteins to 3×109 proteins, depending on the cell type and depth of the analysis1–3. Several features of the HeLa cell proteome demonstrate its complexity1. For instance, of the 3 × 109 protein molecules per HeLa cell, the top 40 most abundant proteins constitute 25% of the entire proteome by mass, the top 600 most-abundant proteins constitute 75% of the proteome by mass and the least-abundant half of proteins accounts for less than 2% of the proteome by mass. These parameters, albeit incomplete, provide a glimpse into the intricacies of the proteome.
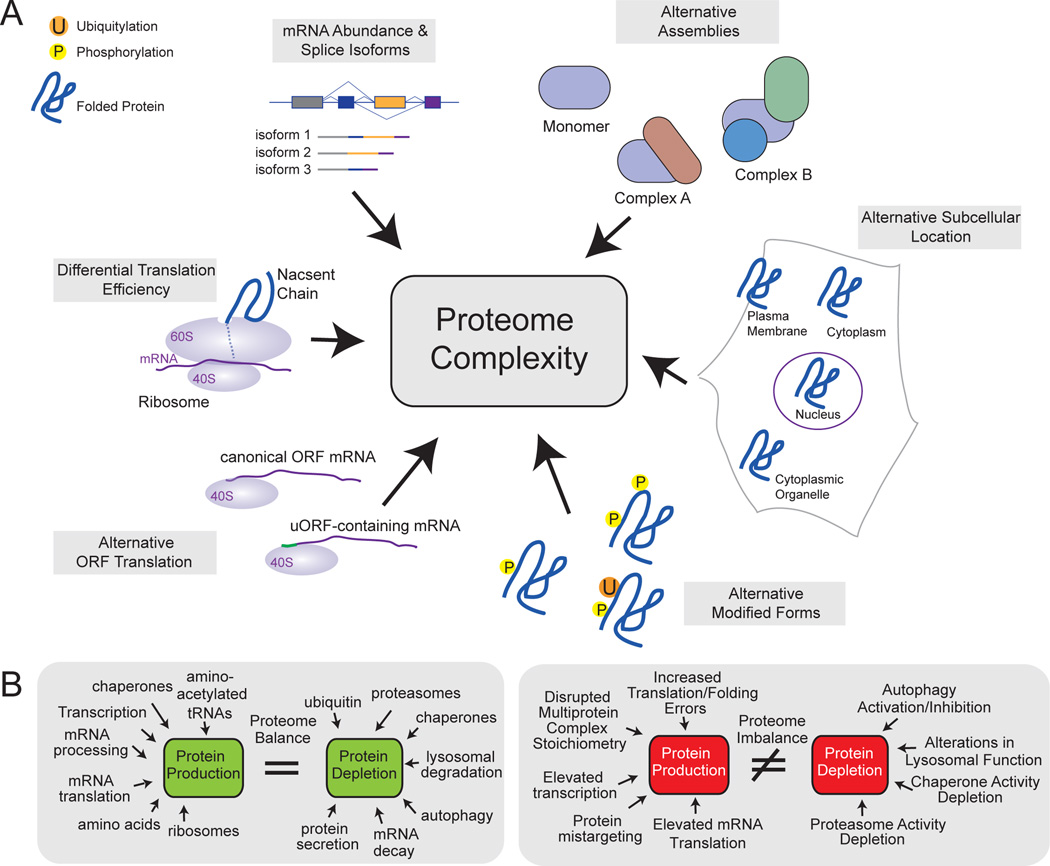
First, individual proteins often exist in several modified forms and they also often engage in numerous dynamically regulated protein complexes during their life cycle. For example, large-scale proteomic studies have identified thousands of sites of modification (including sites of phosphorylation, ubiquitylation, methylation and acetylation) in roughly 50% of proteins in humans, the combinatorial nature of which is mostly unknown4,5. It is also estimated that about 100,000 distinct protein isoforms can be generated through alternative splicing from the 20,000 protein-coding genes6. The mechanisms that underlie the dynamics, interactions, stoichiometry and turnover of most individual species of protein are poorly understood at the global level (Fig. 1).
Figure 1. An overview of proteome complexity.
a, Numerous factors contribute to the generation of complex proteomes. These include (clockwise from top left): alternative splicing; the assembly of protein complexes with varied compositions; the subcellular location of proteins; the attachment of various modifications to proteins; the use of alternative upstream open reading frames (uORFs, purple) in mRNA translation; and the efficiency of mRNA translation. b, In a balanced proteome (green), the level of protein production does not exceed the capacity of protein depletion systems. To achieve this equilibrium, many factors contribute to the generation or degradation of proteins. Cellular events and states such as chromosome imbalance, oncogenic activation and errors in translation alter the proteome in ways that promote proteotoxic stress and lead to imbalance in the proteome (red). This can be buffered by an enhanced capacity for protein degradation or be exacerbated by the depletion of factors that facilitate protein folding or degradation. P, phosphate; Ub, ubiquitin.
Second, cells of different lineages can express distinct sets of genes, including those that promote cellular identity. We are only beginning to understand how differential gene expression in individual cell types translates into differences in the organization and dynamics of the proteome7–9.
Third, variation in the human genome between individuals occurs at the level of around 106 differences, a subset of which might alter the abundance and the interactions of the resulting protein products10. This genomic variation is elevated further in cancer genomes, some of which contain thousands of single base-pair mutations; large-scale chromosomal abnormalities are so prevalent across a range of cancers that about 25% of a typical cancer-cell genome will have undergone a loss or gain in copy number11,12. Our knowledge of how genetic variation alters proteomes in the context of somatic mutations in specific cancers is still in its infancy13.
A complete description of the mechanisms that establish cellular proteomes requires not only an understanding of how proteins and their multimeric assemblies are built, but also of the rules that determine how proteins are selected for degradation when they are unable to assemble properly with components of cognate networks. In the past decade, methods have emerged that enable the quantitative analysis of rates of transcription and translation, as well as the determination of relative protein abundance2,8,9,14–18. Moreover, methods for quantifying the dynamics of protein turnover through the ubiquitin–proteasome and lysosomal systems, which include autophagy, are beginning to yield insights into mechanisms of substrate selection and how these pathways are integrated with stress-response and chaperone networks.
In this Review, we present our knowledge of the cellular systems that monitor and control the abundance and stoichiometry of proteins, as well as the mechanisms of quality control that underlie the cellular response to conditions in which the production and degradation of proteins deviates from the steady state — which we refer to as ‘proteome imbalance’. An emerging understanding of these systems is providing potential avenues for the development of therapeutics that are directed towards diseases of proteome imbalance.
Causes of proteome imbalance
In contrast to the complexity of the genome, we are unlikely to ever know the upper limits of proteome complexity with complete certainty. The protein biogenesis and degradation machineries determine the precise abundance of each protein within the proteome, and both biogenesis and degradation are highly regulated to execute the dynamic control of proteome complexity19–21. The interplay between protein anabolism and catabolism is evident in the consequences of the imbalance in protein homeostasis that can be observed with large-scale genetic variation or an increased demand for ribosomal output (Fig. 1b).
Cancer as a model for proteome imbalance
A number of diverse disorders in people, including many cancers, can be characterized by an imbalance in the proteome that results in chronic proteotoxic stress22,23. Two specific features of cancer cells are considered to contribute to proteome imbalance.
First, the explosion of cancer-genomics data has led to a large catalogue of single base-pair alterations and structural alterations that affect gene copy numbers in numerous types of cancer12. A potential consequence of this is proteome imbalance caused by mutations that affect the ability of proteins to assemble with cognate complexes or that alter the proteins’ rate of turnover.
Second, sustained proliferation of cells in the absence of growth signals results in deregulated protein production24. In turn, this leads to an elevated demand on the translational apparatus in human cancers. Deviations in the rates of translation are sometimes evident only when translation rates are compared in normal and tumorigenic cells after the loss of permissive growth signals. This loss of growth control is often achieved through the mutation or genetic amplification of important regulatory proteins, including Myc, phosphoinositide 3-kinases (PI3Ks) or translation initiation factors such as eIF4E25–27.
The combination of enhanced genetic alteration and increased translational output suggests that tumorigenic cells must acquire an ability to survive despite the persistent generation of an increasingly unstable proteome.
Protein homeostasis and errors in translation
Cellular growth depends absolutely on protein synthesis. The proliferation of unicellular organisms under ideal growth conditions is limited by the speed of translation in such organisms. In Escherichia coli, cell division occurs as soon as enough ribosomes have been made to support the growth of another cell28,29. However, rapid protein biogenesis comes at the cost of fidelity. The rate of amino-acid misincorporation in E. coli is a single residue every 1,000 to 10,000 amino acids, with lower rates observed in eukaryotic cells30,31. Measurements of error rates for protein biogenesis that take into account mistakes at each step in the messenger RNA translation process are difficult to obtain on a global scale. A conservative estimate provides an overall protein synthesis error rate of 1 in 10,000: for a typical protein of 500 amino acids in length, the synthesis of 1 in every 20 such proteins will deviate from perfect synthesis, and a subset of these defective proteins will be intrinsically unstable. Although this proportion seems to be large at first, the impact of erroneous protein synthesis can be understood only in the context of the cell’s capacity for eliminating defective translation products through protein degradation20. Protein half-lives have been determined for a considerable percentage of the proteome using metabolic pulse labelling followed by mass spectrometry. A number of studies report a median protein half-life of 20–46 hours, which reflects an averaged turnover number of the probable most-stable protein isoforms2,32,33. It can be challenging or even impossible to extract quantitative information from these types of studies about the rates of and capacity for proteasomal degradation, or the relative contribution of distinct pools of proteins, including defective translation products, to the overall protein half-life.
An alternative way of estimating the scale of error in protein synthesis and its impact on protein homeostasis is to examine the fraction of total nascent chains that are marked with ubiquitin and targeted for degradation. About 2% of nascent chains in yeast and 12–15% of those in human cells are ubiquitylated34,35. There are between 1 million and 10 million ribosomes in a typical mammalian cell17; however, the percentage of actively translating ribosomes is unknown in such cells. If only 50% of 5 million cellular ribosomes translate an mRNA that encodes a protein of 500 amino acids at a speed of 5 amino acids per second, 1.5 million proteins will be synthesized every minute36. Under the assumption that 12% of all nascent chains are ubiquitylated and targeted for degradation as the result of an unresolvable error in translation, 180,000 proteins will require ubiquitin-dependent degradation every minute. And assuming a protein degradation rate of 10 amino acids per second (under idealized in vitro conditions37) and that there are 1 million proteasomes per cell (ref. 3), 50% of which are active, 600,000 proteins of 500 amino acids in length can be degraded every minute.
These estimates suggest that the defective products of translation are unlikely to burden the ubiquitin–proteasome system, at least under steady-state conditions, and that proteasomes are underloaded. However, careful quantitative studies are needed to more accurately define unknown parameters such as the fraction of active proteasomes in a cell and the in vivo rates of turnover for particular pools of individual proteins. The suggestion that there is spare proteasomal capacity in cells is in agreement with cryo-electron microscopy studies that estimate that about 20% of proteasomes in hippocampal neurons are in a substrate-engaged conformation38. And the observation that proteins that are targeted for proteasomal degradation accumulate only after more than 60% of cellular proteasomal activity is inhibited is consistent with the idea that the excess capacity for degradation exists to buffer cells from fluctuations in proteome stress39.
Such model systems do not take into account shifts in the genetic and environmental landscapes that occur over the lifetime of an organism. For example, in mouse models, genetic perturbations of translation-fidelity mechanisms that increase the output of defective translation products result in neurodegenerative phenotypes40,41. The observation that defects in systemic protein quality control often manifest as neurological phenotypes suggests that neurons might be particularly sensitive to conditions that promote proteome imbalance. Indeed, mice with reduced function of the E3 ubiquitin-protein ligase listerin (Ltn1), which is involved in the ubiquitylation of defective nascent chains, have numerous neurological anomalies42. These examples hint at the possibility that degradation capacities might be eclipsed, over considerable periods of time, by cellular environments that are permissive to lower-fidelity protein biogenesis.
The response of cells to low-fidelity translation
The balance between the speed and the fidelity of translation has been tuned over time to maximize fitness. Classic studies in E. coli have revealed mutations in ribosomal proteins that result in higher-fidelity translation31,43,44. However, this increase in fidelity comes at a considerable fitness cost, as demonstrated by slower rates of growth overall. Decreases in fidelity are also undesirable: in the yeast Saccharomyces cerevisiae, the introduction of a transfer RNA that miscodes a leucine codon as serine results in the chronic mistranslation of the entire transcriptome45. This reduction in fidelity produces a transcriptional response that resembles the environmental stress response, as well as a decrease in ribosomal protein mRNAs, a decrease in the overall rates of protein synthesis and an overall reduction in fitness45. To survive, cells that have unstable genomes, such as cancer cells, might require compensatory adaptation to such prolonged activation of stress responses. Laboratory evolution experiments performed in yeast that miscode a leucine codon to serine result in rapid adaptation to amino-acid miscoding and the recovery of growth rates46. Sequencing has revealed that the adapted strains often contain large-scale genomic deletions and amplifications. The enhanced proteome instability that these genetic changes generate does not seem to be deleterious to the cells, which already have elevated levels of proteome stress. It has been proposed that large-scale chromo- somal abnormalities and the sudden increase in proteome stress that they produce force cells to adapt to widespread proteome imbalance46. These observations could explain why a large proportion of tumour cells contain chromosomal abnormalities. Aneuploidy may not lead directly to tumorigenesis, but the resulting adaptation to proteome stress could be of benefit to tumour formation.
Ribosome-associated mechanisms of quality control
Errors in translation can occur at numerous points in the process, resulting in the production of defective nascent chains that are removed by ribosome quality control (RQC) pathways. RQC pathways engage stalled ribosomes and catalyse the degradation of the associated nascent chain. This process involves splitting of the 80S ribosome, recruitment of Ltn1, ubiquitylation of the nascent chain, extraction of the nascent chain by the ATPase Cdc48 (the yeast orthologue of p97) and proteasomal turnover of the nascent chain47–54. Considerable insight into this quality control system and a potential signaling arm of the pathway came from the discovery of Rqc2, a component of the complex that is responsible for RQC, and its role in adding C-terminal alanine and threonine extensions (known as CAT tails) to stalled nascent chains that cannot be ubiquitylated (for example, in Ltn1-mutant cells)55. Because Rqc2 was discovered in a screen for activators of heat shock factor protein 1 (HSF1)56, it is possible that the addition of CAT tails might directly or indirectly signal heat-shock activation, although further studies are needed to define the underlying mechanisms. A role for Rqc2, Ltn1 and CAT-tail formation in mediating the aggregation and toxicity of both stall-inducing nascent chains and mutated Huntingtin that contains expanded and pathogenic polyglutamine repeats has been demonstrated in S. cerevisiae57–59. Despite the elegant biochemical characterization of the RQC complex in mammalian cells, a physiological role for the complex or CAT-tail formation in mammals has yet to be demonstrated, and endogenous Ltn1 substrates are lacking. The abundance of ribosomes is about 300 times greater than that of Ltn1 (ref. 17), which suggests that the capacity of Ltn1 and the function of the RQC complex could be easily overcome by small perturbations in the proteome, or that alternative factors and pathways also function in regulating RQC. Ribosome stalling that is induced by translation elongation inhibitors has been shown to stimulate the site-specific regulatory ubiquitylation of particular 40S ribosomal proteins; it is therefore possible that post-translational mechanisms contribute to sensing or clearing stalled ribosomal complexes60. However, a direct role for these ubiquitylation events in mediating RQC has yet to be determined.
Another important consideration is the physiological conditions under which ribosome stalling occurs at high enough levels to elicit an adaptive response. Such conditions might include the limited availability of particular amino acids. Ribosome profiling studies in cells that were treated with l-asparaginase, which converts asparagine to aspartic acid, revealed the robust accumulation of ribosomes that were stalled on asparagine codons61. And in patient-derived clear cell renal cell carcinoma tumours, ribosomes were found to stall at proline codons. The same tumours showed an increase in a crucial proline biosynthetic enzyme as part of a possible compensatory feedback loop61. Together, these results suggest that altered amino-acid metabolism in some cancers might result in enhanced ribosome stalling. Whether these cancers display an increased need for RQC has yet to be examined.
Alterations in translational output
Reoccurring tumorigenic mutations in diverse genes stimulate or sustain mRNA translation under conditions that would otherwise dampen protein output24,26. One such mutation is the amplification of the oncogene Myc, which occurs in 50% of human cancers25. Myc functions as a transcription factor and it is estimated that the transcription of 15% of human genes is stimulated by the presence of activated Myc62. Myc has the ability to stimulate not only the production of ribosomal proteins but also ribosomal RNA synthesis by activating all three RNA polymerases25,62 (Fig. 2a), which directly leads to an increase in ribosome biogenesis and translational capacity. The global increases in ribosome biogenesis and the overall capacity for translation that are observed on Myc activation are thought to facilitate tumorigenesis. In support of this, the loss of translational capacity that results from the deletion of a single copy of the ribosomal protein gene RPL24 limits tumorigenesis in a widely used mouse model of B-cell lymphoma that is driven by Myc overproduction63. This result provides a clear example in which rebalancing the proteome stifles the development of cancer.
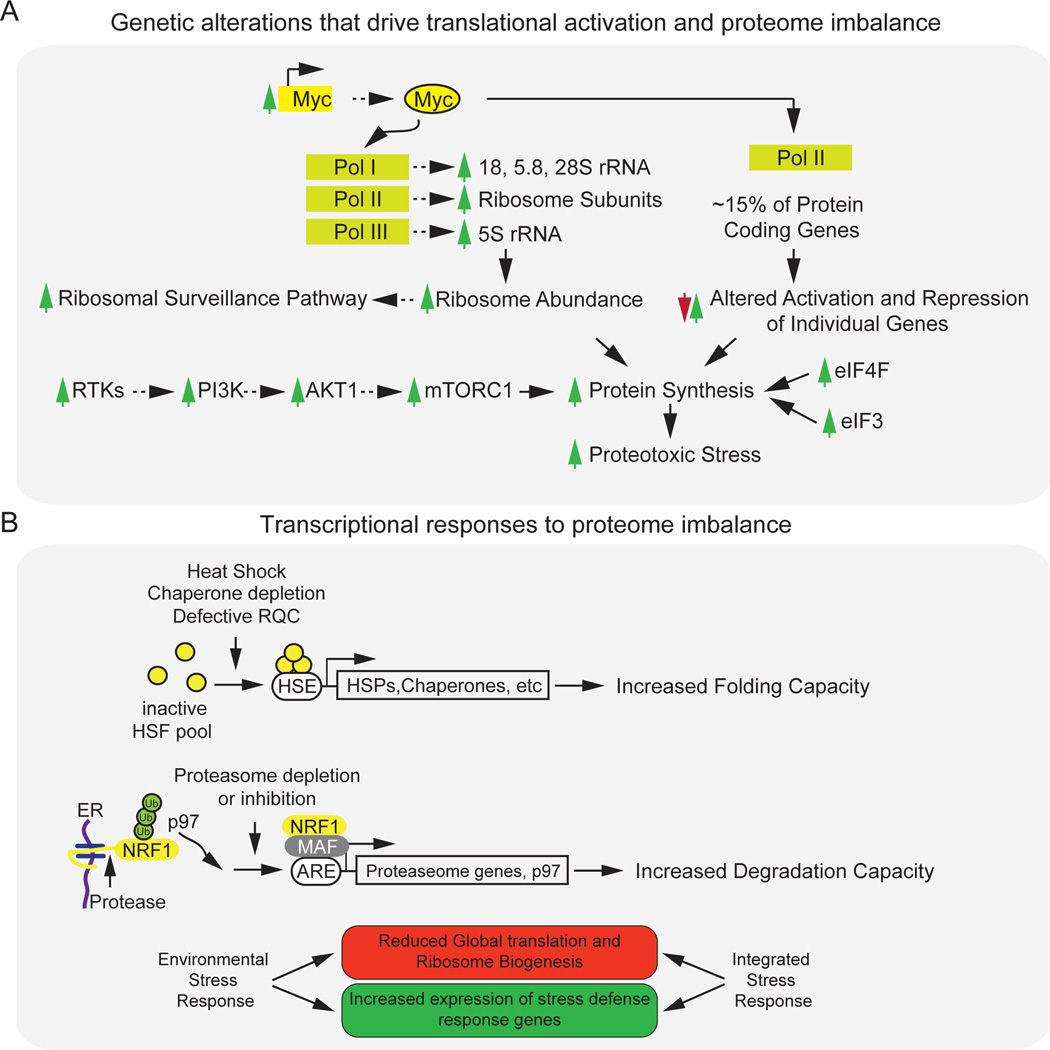
Figure 2. Mechanisms that contribute to proteome imbalance and transcriptional responses.
a, Genetic alterations in human cancers can generate proteome imbalance. Myc protein promotes the RNA polymerase (RNA pol) I- and RNA pol III-dependent transcription of rRNAs; it also promotes the RNA pol II-dependent transcription of ribosomal proteins to induce ribosome production. Elevated levels of some ribosomal proteins in excess of the assembled ribosome can lead to the activation of the ribosomal surveillance pathway, which stimulates cell death pathways. At the same time, Myc activates about 15% of protein-coding genes that are transcribed by RNA pol II, thereby increasing the protein-synthesis load of the cell. This is likely to increase the number of defective translation products, which lead to an increase in proteotoxic stress. Similarly, the activation of receptor tyrosine kinases (RTKs) and the PI3K signalling pathways that stimulate AKT1 and mTORC1 activity, or the overexpression of translation initiation factors eIF4F and eIF3 can increase protein synthesis, also leading to increased levels of proteotoxic stress. b, Two transcriptional response systems sense imbalance in the proteome and can elevate the cell’s capacity for protein folding or degradation. One mode of control involves the conversion of a pool of inactive HSF proteins in response to perturbations such as heat shock into an active nuclear transcription complex that binds a promoter known as a heat-shock element (HSE). This leads to the expression of heat-shock proteins (HSPs), including chaperones, and increases the cell’s capacity for protein folding. Another mode of control is the production of proteasome subunits to increase the cellular capacity for protein degradation. Under basal conditions, transcription factor NRF1 in the endoplasmic reticulum (ER) is targeted for p97-dependent proteasomal degradation through ER-associated degradation (ERAD) (not shown). However, when the activity of the proteasome is inhibited or depleted, retrotranslocated NRF1 is cleaved by an unknown protease, which facilitates the translocation of NRF1 into the nucleus where it activates the transcription of proteasome subunit genes. Global environmental and integrated stress-response pathways also attempt to rebalance the proteome through reduced translational output (red box) and enhanced cellular defence systems (green box) such as protein folding and degradation machineries. ARE, antioxidant response element; MAF, transcription factor MAF.
The activation of cellular signaling pathways that stimulate either translation initiation or elongation is common among tumorigenic cells. This activation is achieved often through mutations that stimulate PI3K signaling27. Such mutations can occur in upstream receptor tyrosine kinases, in PI3Ks themselves or in downstream effectors such as the kinase Akt1, with each resulting in the general, sustained activation of the kinase-containing signaling complex mTORC1 (refs 24 and 27). Although mTORC1 signaling impinges on a large array of cellular systems, it directly stimulates mRNA translation through activating phosphorylation of the ribosomal protein S6 kinase β-1 (RPS6KB1) and inhibitory phosphorylation of eIF4E binding protein 1 (EIF4EBP1) (ref. 24). The sustained hyperactivation of translation initiation and elongation that is observed on mTORC1 activation might lead to a further loss of fidelity in mRNA translation and to the increased production of defective products. Indeed, cells with chronically activated mTORC1 show a decrease in translational fidelity, which results in an enhanced sensitivity to proteotoxic stress64. This suggests that tumorigenic cells with mutations that activate translation must adapt to a heightened basal level of proteome imbalance. Accordingly, cells that are exposed to proteasome inhibitors respond by increasing the transcription of genes that encode proteasome components through activation of the transcription factor NRF1 (refs 65–67) (Fig. 2b). The disruption of NRF1 in cancer might therefore limit their ability to adapt to proteome imbalance.
Another mechanism by which cancer cells enhance translation is the overproduction of translation initiation factors22,26. Components of both the eIF3 and eIF4F initiation complexes are overexpressed in a wide range of cancers22,26. The idea that enhanced translation initiation can promote tumour formation is best described for eIF4E, the mRNA 5ʹ-cap-binding component of the eIF4F complex. Overexpression of eIF4E mediates both cellular transformation in cell-culture models and enhanced susceptibility to tumours in mouse models68–70. The ability of eIF4E to stimulate the initiation of translation is dependent on mTORC1 activity as the loss of inhibitory EIF4EBP1 phosphorylation sequesters eIF4E from the eIF4F complex71. Interestingly, the overexpression of a mutant EIF4EBP1 that evades inhibitory mTORC1 phosphorylation or a 50% reduction in the levels of eIF4E does not lead to global alterations in basal translation72,73. However, the expression of mutant EIF- 4EBP1 blocks eIF4-driven tumorigenesis in haematological cancers and prostate cancer in mice, and mice with reduced levels of eIF4E show suppressed development of GTPase KRAS-driven lung cancers71,73,74. Together, these results indicate that the suppression of hyperactivated translation can inhibit tumorigenesis. However, it remains unknown how the enhanced translation in these cases affects protein degradation systems, and specifically RQC, or whether adapting to the increased demand for protein degradation is a necessary step for tumorigenesis.
Changes in the stoichiometry of multisubunit complexes
Keeping the proteome in balance requires more than just quality control processes at the ribosome: it also involves mechanisms that control the stoichiometry of complexes. Imbalance in subunit stoichiometry can have detrimental effects by disturbing the assembly or dynamics of multisubunit complexes. This is demonstrated by the α–β-tubulin complex, which forms dynamic filaments that are crucial for cell division and organelle trafficking. In classic experiments, the 1.4-fold over-expression of β-tubulin leads to the disassembly of microtubules, the formation of alternative β-tubulin-positive structures and a loss of cell viability, which can be rescued by the expression of α-tubulin75,76. This phenotype reflects the sequestration of factors that are necessary for the formation of α–β-heterodimers by elevated β-tubulin. By contrast, the 30-fold overexpression of α-tubulin has no obvious phenotype75,76. This example reinforces the idea that cells require mechanisms for controlling the stoichiometry of multisubunit complexes. Proportional synthesis through tuned efficiencies of transcription and translation probably controls the stoichiometry of some complexes77 (Fig. 3a). However, turnover of excess or orphan proteins through the proteasome or the lysosome also plays a considerable part in coupling protein abundance with the assembly of complexes.
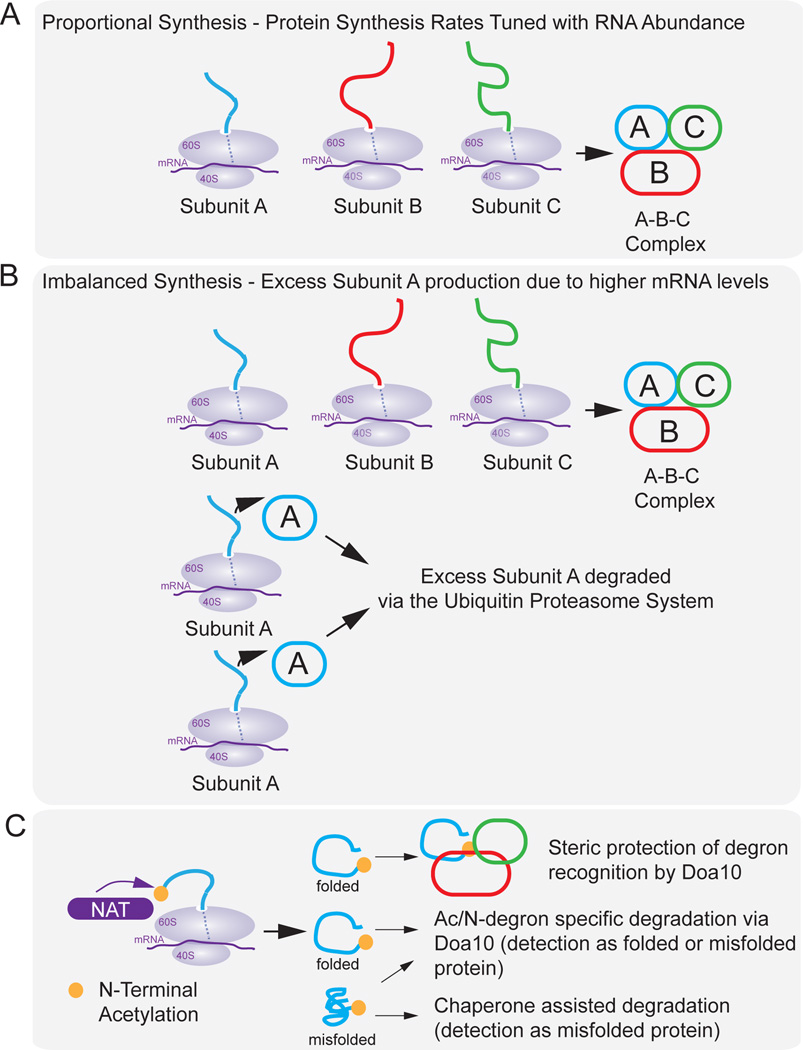
Figure 3. Regulating the stoichiometry of protein complexes.
a, In proportional synthesis, the abundance and translation rates of mRNA are tuned to produce the appropriate stoichiometry for the formation of multiprotein complexes with the subunits A, B and C. b, In imbalanced synthesis, subunit A is expressed at a higher rate than subunits B and C (top), either as a result of increased transcription and translation via gene dosage effects or oncogene activation, or through genetic programming. Excess subunit A is then degraded by the ubiquitin–proteasome system. N-terminal acetylation (blue circle) through the N-acetyltransferase (NAT) system (bottom) is also proposed as a mechanism by which supernumerary subunits are marked for degradation. After the N-acetylated protein is assembled into a complex, the N-terminal acetyl residue (Ac–N degron) can no longer be recognised by the E3 ligase Doa10 due to steric blockage, and the protein is not degraded. Folded N-acetylated proteins that have not been incorporated into complexes are detected by Doa10 and marked for degradation, and misfolded proteins that contain Ac–N degrons are degraded with assistance from chaperone proteins.
The response of cells to increased gene dosage
Our understanding of how cells respond to changes in protein abundance has been enhanced by examining the consequences of large-scale changes in chromosome copy number on the proteomes of S. cerevisiae, as well as in plants and mammalian cells, to a lesser extent. The analysis of haploid S. cerevisiae cells into which 13 of the 16 yeast chromosomes were individually introduced to produce a ‘disome’ has revealed several common principles by which cells respond to proteome imbalance78–82 (Fig. 1a, b). In a manner that is largely independent of the particular chromosomes that are duplicated, a two fold increase in transcription from the supernumerary chromosome triggers a chain of events that decrease the fitness of the cell and reduce the cell’s ability to cope with proteome imbalance83. Most evidence suggests that reduced fitness and the presence of proteotoxic stress do not reflect the expression of particular proteins from the disome84. Instead, they are a reflection of a greater reliance on protein chaperones and degradation machinery within the cell that attempts to manage the higher levels of protein production, the ensuing increase in translational errors that give rise to misfolded proteins, the imbalance in subunits of multiprotein complexes that accompanies enhanced expression from the disome and a reduction in the capacity of chaperones83,85 (Fig. 1b).
Although transcription from the disome produced a two fold increase in the abundance of mRNA, protein abundance diverged from a normal distribution, and the abundance of about 20% of disome-derived proteins increased by only 1.6-fold, compared to a two fold increase78,82 in mRNA. Interestingly, this group of disome-derived proteins was enriched in those that participate in multiprotein complexes82, which implies that protein degradation systems might participate in the maintenance of the stoichiometry of complexes. Although some proteins might be ‘immune’ to post-translational copy number control, others are therefore regulated by a form of dosage compensation that probably balances the abundance of subunits for multiprotein complexes. Interestingly, a transcriptional response that is related to the environmental stress response82,86 seen in cells that are exposed to external stress agents87 is layered on top of post-translational control (Fig. 2b). The environmental stress response is observed widely in cells from aneuploid fission yeast, mice, humans and plants, which implies that it is a conserved transcriptional response to proteome imbalance86.
Two lines of evidence suggest that a reduction in the dosage of disome-derived proteins can promote cellular fitness. First, deletion of the proteasome-associated deubiquitylating enzyme Ubp6 — a negative regulator of protein turnover — reverses the slow-growth phenotype of disomic cells82,88,89. Proteomic analysis hints that this effect is a reflection of the enhanced turnover of relatively abundant proteins, which reduces proteotoxic stress to promote fitness, although the removal of specific toxic proteins cannot be ruled out. Second, diploid cells that receive an extra chromosome are less sensitive to the perturbation than haploid cells, which suggests that excess protein derived from the supernumerary chromosome is more easily buffered under these conditions79,83. Genetic screens in S. cerevisiae for genes that increase the viability of disomic cells have led to the identification of mutant alleles in several genes that are related to protein homeostasis86. These include genes that encode the proteasome subunit Rpt1, two ubiquitin ligases (Rsp5 and Ubr1), Lad2 (the yeast orthologue of the cullin–RING E3 regulator CAND1), a protein involved in rRNA production called Utp1 and a protein involved in vacuolar targeting that is known as Vps64, indicating that there are other mechanisms by which cellular defects in protein imbalance can be ameliorated. Further mechanistic studies are needed to define the underlying pathways. However, the spectrum of genes that have been identified so far suggest that mechanisms that reduce flux through the ubiquitin–proteasome system probably contribute to the toxic effects of aneuploidy in disomic yeast cells.
Demands on the cellular machinery
Widespread proteome imbalance that arises from supernumerary chromosomes has two main effects: the loss of the buffering capacity of protein-folding chaperones and the degradation of proteins that are unable to join cognate complexes (Fig. 3b, top). Although a sizable proportion of the proteome is degraded as a result of either proteome imbalance or errors in translation or folding, we do not yet have a systematic understanding of the rules that dictate the turnover of excess subunits of complexes. This is partly because of a lack of methods that measure the ubiquitylation and turnover dynamics of proteins that are unable to assemble properly into complexes, or that measure rates of turnover for the same proteins in distinct complexes or subcellular compartments.
What are the mechanisms by which proteins that are unable to assemble with their partners are marked for degradation? N-terminal acetylation and a variation of the N-end rule90 have been implicated in one such mechanism (Fig. 3b, bottom). This pathway involves the E3 ligase Doa10, which recognizes acetylated N-terminal residues (or Ac–N-degrons) in target proteins91. Because N-terminal acetylation typically occurs at the same time as translation, the model predicts that newly synthesized proteins will be marked with this signal during their generation. If such proteins are incorporated successfully into cognate complexes, the Ac–N-degrons will be masked by other subunits in the complexes. But if these proteins remain unassembled, they are ubiquitylated by Doa10 and are then degraded by the proteasome. This mechanism has been demonstrated in S. cerevisiae for the proteins Cog1 (part of the conserved oligomeric Golgi complex) and Hcn1 (part of the anaphase-promoting complex)92. N-terminal proteomics studies suggest that about 85% of detected human proteins are at least partially N-acetylated93,94. Although such studies do not analyse the N terminus of every protein in a sample, the high degree of N-acetylation observed indicates that the Ac–N-degron recognition signal might not be universal because most soluble proteins that do not form tight stoichiometric complexes would be expected to be destroyed. However, a more careful analysis of the N termini of both monomeric proteins and proteins that participate in complexes is needed. N-acetyl groups can also mediate functional interactions with complexes in a dynamic setting without targeting the N-acetylated protein for degradation95. A bipartite signal would therefore be the minimum requirement for recognizing orphan proteins that have been stranded from their complexes. One possibility involves the use of hydrophobic surfaces as a second signal that could be recognized by protein chaperones. Indeed, orphan subunits of fatty acid synthase (FASN) as well as orphan von Hippel–Lindau disease tumour suppressor are recognized by the Hsp40–70–90 heat-shock protein system, and FASN is known to be directed to Ubr1, which is a principal quality control ligase96–98. FASN is also one of the most highly ubiquitin-modified proteins by abundance, as are other proteins that are obligate members of complexes, which indicates that this pathway is active under steady-state conditions and that these substrates would be the first to accumulate on proteasome inhibition99. Excess components of multisubunit complexes could offer an abundant source of degradation substrates that greatly exceed the amount of substrates that arise from errors in translation.
An important example that has emerged from the 13 yeast disome strains is the dosage compensation of essentially all detected ribosomal proteins82. Considering the cellular resources that are used in ribosome production, it is unsurprising that cells actively control the stoichiometry of ribosomes. Early experiments in mammalian cells demonstrated that a disruption in the synthesis of rRNA or in the overproduction of an individual ribosome subunit had little impact on total ribosomal-protein synthesis but that it did result in the turnover of ribosomal proteins though unknown mechanisms100,101. In S. cerevisiae, several ribosomal proteins do not accumulate when overexpressed, and these excess subunits are degraded by the proteasome, seemingly without the contribution of autophagy102. Unlike ribosomal proteins that have been assembled into subunits, which are mainly found in the cytoplasm under steady-state conditions, excess ribosomal protein Rpl26a accumulates in the nucleus. Interestingly, when the proteasome is inhibited, endogenous newly synthesized ribosomal proteins aggregate in the nucleus, suggesting that an important quality control mechanism is present to ensure that the proper stoichiometry of ribosomal proteins is maintained102. Understanding the machinery that is involved in this quality control process will be a crucial next step. Intriguingly, a subset of extra-ribosomal proteins participates in the ribosome surveillance pathway in which excess ribosomal proteins RPL11 and RPL5 physically inhibit the p53 ubiquitin ligase Mdm2 (refs 103 and 104), thereby promoting p53-dependent cell death. However, it is still unknown how quality control pathways can distinguish this subset of ribosomal proteins from others that are to be degraded rapidly.
Mislocalized proteins
The stoichiometry of protein complexes is only one of several types of alterations that can promote imbalance in the proteome. For example, newly synthesized membrane proteins occasionally fail to translocate properly into the endoplasmic reticulum, which results in their aggregation in the cytoplasm through hydrophobic transmembrane domains. Proteins that contain transmembrane domains can be recognized by a surveillance complex containing the proteins BAG6, TRC35 (also known as GET4), UBL4A and SGTA105–107, which then facilitates the ubiquitin-dependent degradation of the transmembrane client protein through the ubiquitin ligase RNF126 (ref. 108). A distinct system that involves members of the ubiquilin protein family has been identified for proteins containing a C-terminal transmembrane sequence that fails to insert into the mitochondrial outer membrane109. Ubiquilins (UBQLN1, UBQLN2, UBQLN3 and UBQLN4 in humans) contain an N-terminal ubiquitin-like domain, a central domain that is related to a chaperone-binding domain from heat-shock protein STI1 that associates with hydrophobic sequences, and a C-terminal ubiquitin-associated domain, which binds ubiquitin. The association of client proteins with ubiquilins promotes the ubiquitylation of client proteins by an unknown E3 ligase as well as rapid proteasomal degradation that is mediated by the association of ubiquilin’s ubiquitin-like domain with the protea- some109. The impairment of mitochondrial import or the overproduction of mislocalized mitochondrial proteins initiates a cytoplasmic stress response that includes activation of the proteasome and a reduction in translation110. Similarly to when cells are exposed to stress through chronic mistranslation, cells that experience high levels of mislocalized proteins adapt through enhancements to protein-degradation systems46. Further studies are needed to understand the global role of such membrane-protein quality control pathways and to elucidate the contributions of membrane proteins to proteome imbalance.
Autophagy and the control of proteome imbalance
Autophagy is a catabolic process in which proteins and cellular organelles are targeted to the autophagosome for delivery to the lysosome, a site of degradation. This process involves the lipidation of ubiquitin-like ATG8 proteins (MAP1LC3A, MAP1LC3B and MAP1LC3C and GABARAP, GABARAPL1 and GABARAPL2 in mammalian cells) to promote autophagosome formation, maturation and cargo recruitment111. Evidence to support an increase in the number of autophagosomes and in the expression of autophagic machinery was initially observed in human colon cancer cells (HCT116) that contain supernumerary chromosomes112. However, ubiquitin-binding protein p62 (also known as SQSTM1) — a cargo adaptor that associates with ubiquitylated protein aggregates in HCT116 cells — increases in abundance, which is counter-intuitive if the autophagy flux has increased112. Lipidation of MAP1LC3s and the levels of p62 were also increased in retinal pigment epithelial cells that contain supernumerary chromosomes113; however, although these MAP1LC3-positive structures were delivered to the lysosome, they were not efficiently degraded. The inability of lysosomes to degrade MAP1LC3s did not reflect the inhibition of lysosomal proteases, but it was accompanied by the activation of a lysosome stress response that involved the transcription factor TFEB113. It is unclear whether this is a response to the broad ubiquitylation of misfolded proteins. Further studies should help to explain the role that autophagy has in the catabolism of excess proteins that emerge from supernumerary chromosomes as well as the relationship between autophagy and the ubiquitin system in this context.
Therapeutic targeting of proteome imbalance
At present, there is great interest in determining whether interference in stress-response pathways in cells with elevated proteome imbalance could be used therapeutically, especially given that the threshold for cell viability could be altered (Fig. 4a). Perhaps the most advanced example of using proteome imbalance to treat diseases in people is the application of proteasome inhibitors (such as bortezomib, carfilzomib and ixazomib) in malignant bone-marrow cells114,115. These cells are particularly sensitive to proteasome inhibition because they show a high rate of protein synthesis through the secretory pathway116. This concept has been extended to the p97 (or VCP) AAA-ATPase, which functions as a segregase for the extraction of ubiquitylated proteins from complexes and membranes, especially in the context of endoplasmic-reticulum-associated degradation (ERAD), and from chromatin117. After retrotranslocation, misfolded and ubiquitylated proteins in the endoplasmic reticulum are extracted and delivered to the proteasome through p97 (ref. 118) (Fig. 4b). ATP-competitive inhibitors of p97 such as CB-5083, which block the delivery of ERAD substrates and other ubiquitylated proteins to the proteasome, have been shown to inhibit the growth of myeloma cells both in vitro and in mouse xenograft experiments119,120. But unlike bortezomib, CB-5083 also shows activity towards solid tumours in mice, which generally have more complex karyotypes than do multiple myeloma cells, suggesting that the inhibition of p97 might have an enhanced effect on cells with greater imbalance in the proteome. Interestingly, p97 has roles in the extraction of ubiquitylated proteins from stalled ribosomes54 (Fig. 4b). It is possible that some of the effects of p97 inhibition on cell proliferation are a reflection of increased proteotoxic stress that results from a decreased ability to free the ribosome of defective products of translation.
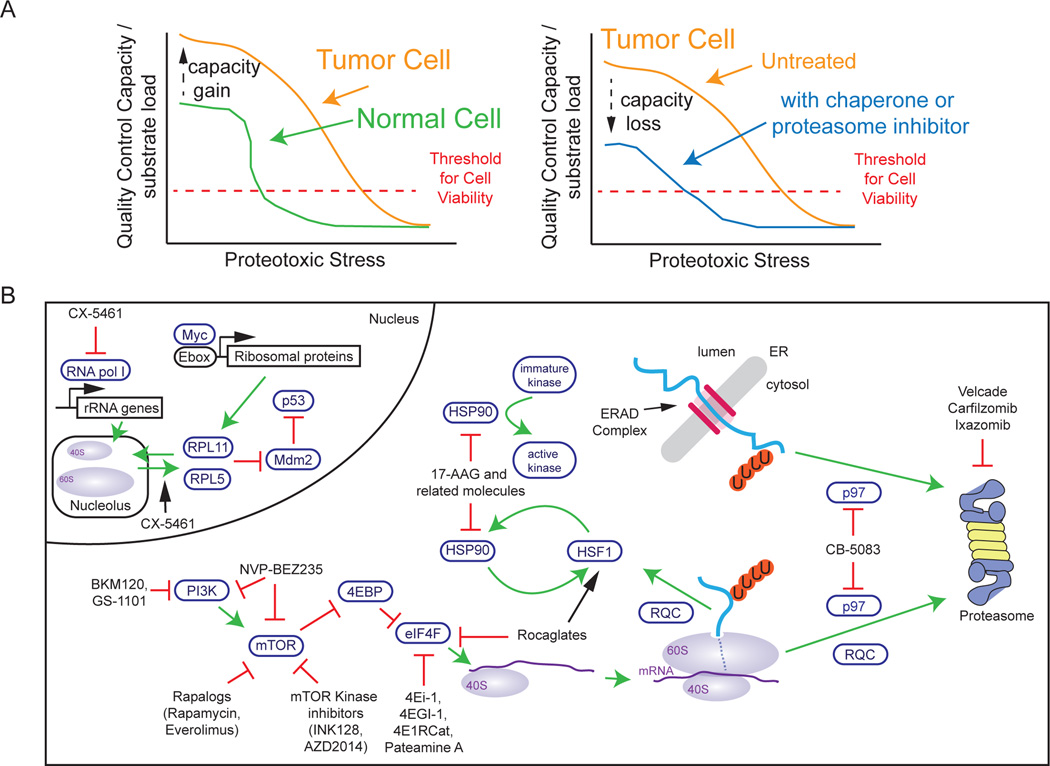
Figure 4. Therapeutic strategies that target proteome maintenance.
a, The ability of cells to respond to fluctuations in proteome balance is dictated by the cellular ratio of quality control capacity to substrate abundance. When the capacity to load ratio (y axis) is high, levels of proteotoxic stress (x axis) are low. Tumour cells may adapt to increase their quality control capacity, which enables them to become more resistant to the effects of proteotoxic stress (left). Pharmacological targeting of the pathways that regulate the balance of the proteome with drugs that inhibit chaperones or the proteasome can alter the cell’s capacity to load ratio in favour of enhanced cell death at lower levels of proteotoxic stress (right). b, A number of pathways that regulate proteome balance have been targeted using small molecules. In the nucleus, inhibitors of RNA pol I, such as CX-5461, block the production of rRNA. This isolated reduction of rRNA leads to an excess of ribosomal proteins RPL11 and RPL5, which induces p53 stabilization through inhibition of the ubiquitin ligase Mdm2. In the cytoplasm, translational control mechanisms, protein chaperones such as HSP90 and the AAA-ATPase p97 control a network of interactions that regulate the production and turnover of the defective products of translation at the ribosome. The excess of ribosomal proteins such as RPL11 and RPL5 is also increased in the context of Myc overexpression, which promotes the production of ribosomal proteins. Several small-molecule inhibitors, including the HSP90 inhibitor tanespimycin (17-AAG), the PI3K and mTOR inhibitor NVP-BEZ235 and various elongation factor 4E (EIF4E) inhibitors (4EI-1 related molecules and the natural product pateamine A), have been developed to target distinct steps in the pathway. p97 also controls the proteasomal turnover of proteins through the ERAD pathway, which might be important for determining the clinical activity of p97 inhibitor CB-5083. Ub, ubiquitin.
A further target for therapeutics that exploit proteome imbalance has emerged from two independent lines of investigation that point to inhibitors of the heat-shock protein HSP90 as a possible treatment for aneuploid tumours. Two studies in aneuploid model systems121,122 led to the finding that HSP90 inhibitors, including tanespimycin (also known as 17-AAG), enhance cell death in cells with aneuploidy compared to non-aneuploid cells. This is consistent with a role for the Hsp90–Cdc37 system in folding and activating protein kinases that are important for proliferation; for example, the toxicity of human SRC kinase expression in yeast is relaxed in disomic cells85. This effect is interpreted as there being insufficient levels of the Hsp90–Cdc37 system to support the maturation of kinases in aneuploid cells85. The selective inhibition of cells with supernumerary chromosomes by Hsp90 inhibitors has implications for the many clinical trials in progress that are targeting Hsp90 (ref. 123). More broadly, HSF1 might represent an important target for therapeutics. HSF1 promotes homeostasis through the transcription of molecular chaperones, but it is also linked to energy metabolism124 (Fig. 2b). The inhibition of translation with small-molecule rocaglates leads to the inactivation of HSF1, a reduction in cellular energy, the loss of chaperone capacity and a selective reduction in the proliferation of cancer cells124. Further studies will improve understanding of the mechanistic regulation of HSF1 by alterations in protein synthesis.
Considerable focus has been placed on targeting pathways that promote translation22,26 (Fig. 4b), including efforts that have yielded inhibitors of the binding of eIF4E to eIF4F, some of which show efficacy in xenograft models of cancer125. Because of the prevalence of Myc activation in cancer, numerous strategies to target Myc function have been implemented62, including the inhibition of ribosome biogenesis through RNA polymerase I (ref. 126). The idea that underlies this approach is to inhibit rRNA synthesis without affecting the synthesis of ribosomal proteins, which leads to the continuous production of ribosomal proteins that require degradation. This strategy simultaneously activates the ribosomal surveillance pathway and reduces the capacity for degradation (owing to an excess of orphan ribosomal proteins), and it has proven effective and synergistic with mTOR inhibition in mouse models of Myc-driven B-cell lymphomas127. Although the small-molecule-mediated inhibition of RNA polymerase I has not been tested specifically in cells with supernumerary chromosomes, it is intriguing to postulate that a sudden increase in unassembled ribosomal proteins that require quality-control-dependent degradation might have large effects on cells that are already burdened by excess ribosomal protein synthesis. These approaches underscore the general hypothesis that therapeutics aimed at increasing proteotoxic stress in cells with an already overburdened protein-degradation system might represent anti-cancer strategies with broad benefits (Fig. 4a).
Future directions
We now have an extensive picture of how imbalance in the proteome, whether it is generated through the activation of oncogenes or by alterations in gene dosage, limits the capacity of systems that chaperone or degrade proteins. Studies of gene dosage, in particular, have revealed the existence of widespread dosage compensation that is mediated through the ubiquitin–proteasome system, and this finding is beginning to shed light on how the stoichiometry of proteins is established within the cell. Numerous questions have emerged (Box 2), such as how and from where do cells orchestrate the degradation of excess subunits from protein complexes, and whether there are differences in the capacity of distinct types of cells to handle proteome imbalance. In this regard, haematopoietic stem cells have lower rates of protein synthesis in vivo in comparison to those of differentiated lineages, and they are sensitive to genetic perturbations that lead to either increases or decreases in protein synthesis128,129. Attempts to quantify proteomes have focused on identification of the relative abundances of proteins within ensembles of cells130. However, such studies do not address features of the proteome that are crucial for understanding how balance is achieved. In particular, studies that are limited to measuring the total abundance of proteins do not address the complexity that individual proteins exhibit through various free and complexed forms that might have broadly different stabilities. Understanding turnover rates for both orphan subunits and their assemblies on a global scale will require innovative methods that rely on the integration of quantitative proteomics and imaging. Moreover, a present limitation is that most studies that analyse protein abundance do not reach the depth that is required to fully address the status of low- abundance proteins (Box 1), and it is unclear whether the behaviour of abundant proteins accurately models that of proteins that are near the limits of detection. The reliance on using bulk measurements to understand most assembly and turnover pathways limits our ability to appreciate fully the underlying regulatory systems. In the future, methods based on analyses of single cells will help to unravel the complex interplay between protein quality control and proteome imbalance.
BOX 2. Proteome imbalance at a glance.
Which E3 ubiquitin-ligase systems and mechanisms control the turnover of excess subunits from complexes? Our understanding of the roles and mechanisms of E3 ligases that are involved in quality control is limited, and the rules for selecting orphan subunits of multimeric complexes for degradation are unclear.
What are the mechanisms that underlie spatially distinct quality- control pathways? Misfolded proteins in yeast are often channelled into three locations: the juxtanuclear quality-control compartment in which active re-folding is promoted; the cytoplasmic insoluble protein deposits compartment for irreconcilably misfiled proteins; or the Q-bodies, which are processing centres for soluble misfolded proteins that are controlled by the Hsp104 disaggregase135,136. Comparatively little is known in mammalian cells about the spatial organization of the synthesis and degradation machinery and whether they are coupled.
Which mechanisms underlie the capacity of the cell for degradation? The occupancy of the proteasome is estimated to be about 20%, based on tomography in neurons38, but it is unclear whether this reflects the full capacity of the cell’s proteolytic machinery. The finding that deletion of the deubiquitinase Ubp6 in yeast (or of USP14 in mammalian cells) can increase flux through the proteasome suggests that it is possible to increase proteasomal activity, although perhaps at the expense of specificity.
What is the role of autophagy in controlling proteome imbalance? Links between autophagy and proteotoxic stress in the context of chromosome imbalance are limited and the circumstances under which autophagy is activated or inhibited are unclear. The extent to which proteotoxic stress produces protein aggregates that require autophagy for degradation is unknown. The basis for the apparent inhibition of protein degradation within lysosomes in the context of chromosome imbalance113 is also unknown.
What are the molecular determinants that specify the ubiquitylation and turnover of defective products of translation? Ribosomes that lack the ubiquitin ligase Ltn1 are unable to degrade misfolded nascent chains efficiently; this leads to the accumulation of nascent proteins with C-terminal alanine and threonine extensions, which points to a central role for Ltn1 in the removal of defective nascent chains. However, Ltn1 is present at levels that are much lower than those of the ribosome. It will be important to understand the dynamics of Ltn1 and whether there are mechanisms that can compensate for its loss or operate in parallel in the context of particular types of translational errors.
What contributions do membrane proteins make to proteome imbalance? The answer to this question will probably require the development of improved methods to quantify the turnover of the pools of individual proteins that are contained within vesicular structures as well as the turnover of membrane proteins themselves.
Acknowledgments
J.W.H. is supported by grants from the US National Institutes of Health (AG011085, R37NS083524 and GM095567) and by grants from Biogen, Inc. E.J.B. is supported by grants from the National Institutes of Health (1DP2GM119132 and 2P50GM085764) and by a New Scholar in Aging award from The Ellison Medical Foundation (AG-NS-0902-12). We thank A. Amon at the Massachusetts Institute of Technology for providing comments and insight during the preparation of this manuscript.
Footnotes
Note added in proof: Recent work has demonstrated a conserved pathway in which mTORC1 negatively regulates the production of assembly factors and chaperones called ribosome-associated complexes (RACs) for the proteasome (A. Rousseau and A. Bertolotti. An evolutionarily conserved pathway controls proteasome homeostasis Nature 536, 184–189; 2016). mTORC1 inhibition leads to the phosphorylation of MAP kinase family members, which promotes RAC expression. This suggests that proteasome activity is controlled by a highly integrated network and provides a therapeutic target.
References
- 1.Nagaraj N, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 3.Wiśniewski JR, Hein MY, Cox J, Mann M. A “proteomic ruler” for protein copy number and concentration estimation without spike-in standards. Mol. Cell. Proteomics. 2014;13:3497–3506. doi: 10.1074/mcp.M113.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jünger MA, Aebersold R. Mass spectrometry-driven phosphoproteomics: patterning the systems biology mosaic. Wiley Interdiscip. Rev. Dev. Biol. 2014;3:83–112. doi: 10.1002/wdev.121. [DOI] [PubMed] [Google Scholar]
- 5.Hornbeck PV, et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 7.Buszczak M, Signer RA, Morrison SJ. Cellular differences in protein synthesis regulate tissue homeostasis. Cell. 2014;159:242–251. doi: 10.1016/j.cell.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jovanovic M, et al. Dynamic profiling of the protein life cycle in response to pathogens. Science. 2015;347:1259038. doi: 10.1126/science.1259038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 10.The 1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Zhang B, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingolia NT. Ribosome profiling: new views of translation, from single codons to genome scale. Nature Rev. Genet. 2014;15:205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- 15.Kim MS, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nature Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, Herrmann CJ, Simonovic M, Szklarczyk D, von Mering C. Version 4.0 of PaxDb: protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics. 2015;15:3163–3168. doi: 10.1002/pmic.201400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilhelm M, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 19.Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodrigo-Brenni MC, Hegde RS. Design principles of protein biosynthesis-coupled quality control. Dev. Cell. 2012;23:896–907. doi: 10.1016/j.devcel.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhat M, et al. Targeting the translation machinery in cancer. Nature Rev. Drug Discov. 2015;14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 23.Deshaies RJ. Proteotoxic crisis, the ubiquitin-proteasomesystem, and cancer therapy. BMC Biol. 2014;12:94. doi: 10.1186/s12915-014-0094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsieh AL, Walton ZE, Altman BJ, Stine ZE, Dang CV. MYC and metabolism on the path to cancer. Semin. Cell Dev. Biol. 2015;43:11–21. doi: 10.1016/j.semcdb.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruggero D. Translational control in cancer etiology. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong KK, Engelman JA, Cantley LC. Targeting the PI3K signaling pathway in cancer. Curr. Opin. Genet. Dev. 2010;20:87–90. doi: 10.1016/j.gde.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basan M, et al. Inflating bacterial cells by increased protein synthesis. Mol. Syst. Biol. 2015;11:836. doi: 10.15252/msb.20156178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scott M, Klumpp S, Mateescu EM, Hwa T. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol. Syst. Biol. 2014;10:747. doi: 10.15252/msb.20145379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nature Rev. Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doherty MK, Hammond DE, Clague MJ, Gaskell SJ, Beynon RJ. Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J. Proteome Res. 2009;8:104–112. doi: 10.1021/pr800641v. [DOI] [PubMed] [Google Scholar]
- 33.Boisvert F-M, et al. A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol. Cell. Proteomics. 2012;11:M111.011429. doi: 10.1074/mcp.M111.011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duttler S, Pechmann S, Frydman J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol. Cell. 2013;50:379–393. doi: 10.1016/j.molcel.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang F, Durfee LA, Huibregtse JM. A cotranslational ubiquitination pathway for quality control of misfolded proteins. Mol. Cell. 2013;50:368–378. doi: 10.1016/j.molcel.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peth A, Nathan JA, Goldberg AL. The ATP costs and time required to degrade ubiquitinated proteins by the 26 S proteasome. J. Biol. Chem. 2013;288:29215–29222. doi: 10.1074/jbc.M113.482570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asano S, et al. A molecular census of 26S proteasomes in intact neurons. Science. 2015;347:439–442. doi: 10.1126/science.1261197. [DOI] [PubMed] [Google Scholar]
- 39.Gendron JM, et al. Using the ubiquitin-modified proteome to monitor distinct and spatially restricted protein homeostasis dysfunction. Mol. Cell. Proteomics. 2016;15:2576–2593. doi: 10.1074/mcp.M116.058420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ishimura R, et al. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science. 2014;345:455–459. doi: 10.1126/science.1249749. This paper and ref. 41 identify mutations in mice that increase miscoding during translation and result in neurodegenerative phenotypes.
- 41.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 42.Chu J, et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Natl Acad. Sci. USA. 2009;106:2097–2103. doi: 10.1073/pnas.0812819106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Triman KL. Mutational analysis of the ribosome. Adv. Genet. 2007;58:89–119. doi: 10.1016/S0065-2660(06)58004-6. [DOI] [PubMed] [Google Scholar]
- 44.Zaher HS, Green R. Hyperaccurate and error-prone ribosomes exploit distinct mechanisms during tRNA selection. Mol. Cell. 2010;39:110–120. doi: 10.1016/j.molcel.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paredes JA, et al. Low level genome mistranslations deregulate the transcriptome and translatome and generate proteotoxic stress in yeast. BMC Biol. 2012;10:55. doi: 10.1186/1741-7007-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kalapis D, et al. Evolution of robustness to protein mistranslation by accelerated protein turnover. PLoS Biol. 2015;13:e1002291. doi: 10.1371/journal.pbio.1002291. This paper provides evidence of adaptation to proteotoxic stress through large chromosomal alterations and the genetic modification of protein- degradation systems.
- 47.Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010;467:470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Defenouillère Q, et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Natl Acad. Sci. USA. 2013;110:5046–5051. doi: 10.1073/pnas.1221724110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shao S, Hegde RS. Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol. Cell. 2014;55:880–890. doi: 10.1016/j.molcel.2014.07.006. This paper and ref. 50 reconstitute Ltn1-mediated RQC and establish the order of operations.
- 51.Shao S, von der Malsburg K, Hegde RS. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol. Cell. 2013;50:637–648. doi: 10.1016/j.molcel.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verma R, Oania RS, Kolawa NJ, Deshaies RJ. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. eLife. 2013;2:e00308. doi: 10.7554/eLife.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brandman O, Hegde RS. Ribosome-associated protein quality control. Nature Struct. Mol. Biol. 2016;23:7–15. doi: 10.1038/nsmb.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shen PS, et al. Rqc2p and 60S ribosomal subunits mediate mRNA- independent elongation of nascent chains. Science. 2015;347:75–78. doi: 10.1126/science.1259724. Refs 55 57 and 59 identify the mechanism of RQC-mediated CAT-tail formation and demonstrate that defective tail formation suppresses the protein aggregation that results from RQC events.
- 56.Brandman O, et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choe YJ, et al. Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature. 2016;531:191–195. doi: 10.1038/nature16973. [DOI] [PubMed] [Google Scholar]
- 58.Yang J, Hao X, Cao X, Liu B, Nyström T. Spatial sequestration and detoxification of Huntingtin by the ribosome quality control complex. eLife. 2016;5:e11792. doi: 10.7554/eLife.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yonashiro R, et al. The Rqc2/Tae2 subunit of the ribosome-associated quality control (RQC) complex marks ribosome-stalled nascent polypeptide chains for aggregation. eLife. 2016;5:e11794. doi: 10.7554/eLife.11794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Higgins R, et al. The unfolded protein response triggers site-specific regulatory ubiquitylation of 40S ribosomal proteins. Mol. Cell. 2015;59:35–49. doi: 10.1016/j.molcel.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loayza-Puch F, et al. Tumour-specific proline vulnerability uncovered by differential ribosome codon reading. Nature. 2016;530:490–494. doi: 10.1038/nature16982. [DOI] [PubMed] [Google Scholar]
- 62.Poortinga G, Quinn LM, Hannan RD. Targeting RNA polymerase I to treat MYC-driven cancer. Oncogene. 2015;34:403–412. doi: 10.1038/onc.2014.13. [DOI] [PubMed] [Google Scholar]
- 63. Barna M, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. This paper demonstrates that a Myc-driven model of lymphoma can be suppressed by the haploinsufficiency of specific ribosomal genes.
- 64.Conn CS, Qian SB. Nutrient signaling in protein homeostasis: an increase in quantity at the expense of quality. Sci. Signal. 2013;6:ra24. doi: 10.1126/scisignal.2003520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mitsiades N, et al. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc. Natl Acad. Sci. USA. 2002;99:14374–14379. doi: 10.1073/pnas.202445099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meiners S, et al. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of mammalian proteasomes. J. Biol. Chem. 2003;278:21517–21525. doi: 10.1074/jbc.M301032200. [DOI] [PubMed] [Google Scholar]
- 67.Radhakrishnan SK, et al. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell. 2010;38:17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ruggero D, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nature Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 69.Avdulov S, et al. Activation of translation complex eIF4F is essential for the genesis and maintenance of the malignant phenotype in human mammary epithelial cells. Cancer Cell. 2004;5:553–563. doi: 10.1016/j.ccr.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 70.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5' cap. Nature. 1990;345:544–547. doi: 10.1038/345544a0. [DOI] [PubMed] [Google Scholar]
- 71.Hsieh AC, et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP–eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Truitt ML, et al. Differential requirements for eIF4E dose in normal development and cancer. Cell. 2015;162:59–71. doi: 10.1016/j.cell.2015.05.049. Refs 73 and 74 establish that tumorigenesis can be suppressed in vivo by genetically limiting eIF4E.
- 74.Hsieh AC, et al. Cell type-specific abundance of 4EBP1 primes prostate cancer sensitivity or resistance to PI3K pathway inhibitors. Sci. Signal. 2015;8:ra116. doi: 10.1126/scisignal.aad5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Katz W, Weinstein B, Solomon F. Regulation of tubulin levels and microtubule assembly in Saccharomyces cerevisiae: consequences of altered tubulin gene copy number. Mol. Cell. Biol. 1990;10:5286–5294. doi: 10.1128/mcb.10.10.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weinstein B, Solomon F. Phenotypic consequences of tubulin overproduction in Saccharomyces cerevisiae: differences between alpha-tubulin and beta-tubulin. Mol. Cell. Biol. 1990;10:5295–5304. doi: 10.1128/mcb.10.10.5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. This study provided the first systematic analysis of both the effects of aneuploidy of intact chromosomes in yeast and the effects of an increased chromosome copy number on cell fitness.
- 79.Sheltzer JM, et al. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Thorburn RR, et al. Aneuploid yeast strains exhibit defects in cell growth and passage through START. Mol. Biol. Cell. 2013;24:1274–1289. doi: 10.1091/mbc.E12-07-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blank HM, Sheltzer JM, Meehl CM, Amon A. Mitotic entry in the presence of DNA damage is a widespread property of aneuploidy in yeast. Mol. Biol. Cell. 2015;26:1440–1451. doi: 10.1091/mbc.E14-10-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dephoure N, et al. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. eLife. 2014;3:e03023. doi: 10.7554/eLife.03023. This paper provides a comprehensive proteomic and transcriptional analysis of yeast disomic strains, demonstrating that a subset of genes on disomes undergo dosage compensation through active mechanisms of turnover.
- 83.Tang YC, Amon A. Gene copy-number alterations: a cost-benefit analysis. Cell. 2013;152:394–405. doi: 10.1016/j.cell.2012.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bonney ME, Moriya H, Amon A. Aneuploid proliferation defects in yeast are not driven by copy number changes of a few dosage-sensitive genes. Genes Dev. 2015;29:898–903. doi: 10.1101/gad.261743.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes Dev. 2012;26:2696–2708. doi: 10.1101/gad.207407.112. This paper reveals that aneuploidy in yeast generates proteotoxic stress, which affects the ability of yeast to properly fold proteins that are already prone to misfolding.
- 86.Sheltzer JM, Torres EM, Dunham MJ, Amon A. Transcriptional consequences of aneuploidy. Proc. Natl Acad. Sci. USA. 2012;109:12644–12649. doi: 10.1073/pnas.1209227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gasch AP, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Crosas B, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 89.Torres EM, et al. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu. Rev. Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Shemorry A, Hwang CS, Varshavsky A. Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol. Cell. 2013;50:540–551. doi: 10.1016/j.molcel.2013.03.018. This paper provides evidence for N- acetylation in the turnover of conserved oligomeric Golgi and anaphase-promoting complex proteins that are not properly assembled into their associated complexes.
- 93.Arnesen T, et al. Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl Acad. Sci. USA. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Van Damme P, et al. NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation. PLoS Genet. 2011;7:e1002169. doi: 10.1371/journal.pgen.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Scott DC, Monda JK, Bennett EJ, Harper JW, Schulman BA. N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science. 2011;334:674–678. doi: 10.1126/science.1209307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 97.Scazzari M, Amm I, Wolf DH. Quality control of a cytoplasmic protein complex: chaperone motors and the ubiquitin-proteasome system govern the fate of orphan fatty acid synthase subunit Fas2 of yeast. J. Biol. Chem. 2015;290:4677–4687. doi: 10.1074/jbc.M114.596064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schüller HJ, Förtsch B, Rautenstrauss B, Wolf DH, Schweizer E. Differential proteolytic sensitivity of yeast fatty acid synthetase subunits α and β contributing to a balanced ratio of both fatty acid synthetase components. Eur. J. Biochem. 1992;203:607–614. doi: 10.1111/j.1432-1033.1992.tb16590.x. [DOI] [PubMed] [Google Scholar]
- 99. Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. This study provided the first large-scale analysis of the ubiquitin-modified proteome.
- 100.Warner JR. In the absence of ribosomal RNA synthesis, the ribosomal proteins of HeLa cells are synthesized normally and degraded rapidly. J. Mol. Biol. 1977;115:315–333. doi: 10.1016/0022-2836(77)90157-7. [DOI] [PubMed] [Google Scholar]
- 101.Abovich N, Gritz L, Tung L, Rosbash M. Effect of RP51 gene dosage alterations on ribosome synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1985;5:3429–3435. doi: 10.1128/mcb.5.12.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sung MK, Reitsma JM, Sweredoski MJ, Hess S, Deshaies RJ. Ribosomal proteins produced in excess are degraded by the ubiquitin– proteasome system. Mol Biol Cell. 2016 doi: 10.1091/mbc.E16-05-0290. http://dx.doi.org/10.10.91/mbc.E16-05-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bursać S, et al. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc. Natl Acad. Sci. USA. 2012;109:20467–20472. doi: 10.1073/pnas.1218535109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein–Mdm2–p53 pathway. Oncogene. 2010;29:4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 105.Hessa T, et al. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature. 2011;475:394–397. doi: 10.1038/nature10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shao S, Hegde RS. Target selection during protein quality control. Trends Biochem. Sci. 2016;41:124–137. doi: 10.1016/j.tibs.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 107.Wang Q, et al. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol. Cell. 2011;42:758–770. doi: 10.1016/j.molcel.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rodrigo-Brenni MC, Gutierrez E, Hegde RS. Cytosolic quality control of mislocalized proteins requires RNF126 recruitment to Bag6. Mol. Cell. 2014;55:227–237. doi: 10.1016/j.molcel.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Itakura E, et al. Ubiquilins chaperone and triage mitochondrial membrane proteins for degradation. Mol. Cell. 2016;63:21–33. doi: 10.1016/j.molcel.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wrobel L, et al. Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature. 2015;524:485–488. doi: 10.1038/nature14951. [DOI] [PubMed] [Google Scholar]
- 111.Mancias JD, Kimmelman AC. Mechanisms of selective autophagy in normal physiology and cancer. J. Mol. Biol. 2016;428:1659–1680. doi: 10.1016/j.jmb.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stingele S, et al. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol. Syst. Biol. 2012;8:608. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Santaguida S, Vasile E, White E, Amon A. Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev. 2015;29:2010–2021. doi: 10.1101/gad.269118.115. This paper demonstrates that autophagy is induced rapidly after chromosome mis-segregation in mammalian cells and that undigested proteins accumulate in lysosomes after delivery by autophagosomes.
- 114.Dou QP, Zonder JA. Overview of proteasome inhibitor-based anti-cancer therapies: perspective on bortezomib and second generation proteasome inhibitors versus future generation inhibitors of ubiquitin- proteasome system. Curr. Cancer Drug Targets. 2014;14:517–536. doi: 10.2174/1568009614666140804154511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Moreau P, et al. Oral ixazomib, lenalidomide, and dexamethasone for multiple myeloma. N. Engl. J. Med. 2016;374:1621–1634. doi: 10.1056/NEJMoa1516282. [DOI] [PubMed] [Google Scholar]
- 116.San Miguel J. Multiple myeloma: a model for scientific and clinical progress. Hematology Am. Soc. Hematol. Educ. Program. 2014;2014:1–7. doi: 10.1182/asheducation-2014.1.1. [DOI] [PubMed] [Google Scholar]
- 117.Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nature Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 118.Christianson JC, Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nature Struct. Mol. Biol. 2014;21:325–335. doi: 10.1038/nsmb.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou HJ, et al. Discovery of a first-in-class, potent, selective, and orally bioavailable inhibitor of the p97 AAA ATPase (CB-5083) J. Med. Chem. 2015;58:9480–9497. doi: 10.1021/acs.jmedchem.5b01346. [DOI] [PubMed] [Google Scholar]
- 120. Anderson DJ, et al. Targeting the AAA ATPase p97 as an approach to treat cancer through disruption of protein homeostasis. Cancer Cell. 2015;28:653–665. doi: 10.1016/j.ccell.2015.10.002. This paper identified inhibitors of the AAA-ATPase p97 and demonstrated their activity against cancer cells both in vitro and in xenograft experiments.
- 121.Donnelly N, Passerini V, Durrbaum M, Stingele S, Storchova Z. HSF1 deficiency and impaired HSP90-dependent protein folding are hallmarks of aneuploid human cells. EMBO J. 2014;33:2374–2387. doi: 10.15252/embj.201488648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tang YC, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy- selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jhaveri K, Taldone T, Modi S, Chiosis G. Advances in the clinical development of heat shock protein 90 (Hsp90) inhibitors in cancers. Biochim. Biophys. Acta. 2012;1823:742–755. doi: 10.1016/j.bbamcr.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Santagata S, et al. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science. 2013;341:1238303. doi: 10.1126/science.1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res. 2015;75:250–263. doi: 10.1158/0008-5472.CAN-14-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Drygin D, et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011;71:1418–1430. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]
- 127.Devlin JR, et al. Combination therapy targeting ribosome biogenesis and mRNA translation synergistically extends survival in MYC-driven lymphoma. Cancer Discov. 2016;6:59–70. doi: 10.1158/2159-8290.CD-14-0673. [DOI] [PubMed] [Google Scholar]
- 128. Vilchez D, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–308. doi: 10.1038/nature11468. Refs 128 and 129 establish that some populations of stem cells require lower rates of protein synthesis and show elevated capacities for protein degradation.
- 129.Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics. 2014;13:2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vuckovic D, Dagley LF, Purcell AW, Emili A. Membrane proteomics by high performance liquid chromatography-tandem mass spectrometry: analytical approaches and challenges. Proteomics. 2013;13:404–423. doi: 10.1002/pmic.201200340. [DOI] [PubMed] [Google Scholar]
- 133.Marguerat S, et al. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Huttlin EL, et al. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Escusa-Toret S, Vonk WI, Frydman J. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nature Cell Biol. 2013;15:1231–1243. doi: 10.1038/ncb2838. [DOI] [PMC free article] [PubMed] [Google Scholar]