Summary
Errors in chromosome segregation during mitosis have been recognized as a hallmark of tumor cells since the late 1800s, resulting in the long-standing hypothesis that mitotic abnormalities drive tumorigenesis. Recent work has shown that mitotic defects can promote tumors, suppress them, or do neither, depending on the rate of chromosome missegregation. Here we discuss the causes of chromosome missegregation, their effects on tumor initiation and progression, and the evidence that increasing the rate of chromosome missegregation may be an effective chemotherapeutic strategy.
Introduction
Aneuploidy, an abnormal chromosome number that deviates from a multiple of the haploid, results in global changes in the cellular transcriptome and proteome, as well as in specific changes that depend on the imbalance of individual genes. The state of aneuploidy often coexists with an ongoing rate of karyotypic change, known as Chromosomal INstability (CIN). Although aneuploidy commonly occurs in human cancers where it is a prognostic indicator in many tumor types, it often causes a proliferative disadvantage in cultured cells, which may be partially dependent on the tumor suppressor p53. This apparent contradiction is known as the aneuploidy paradox. Attempts to determine the effects of aneuploidy and CIN on tumorigenesis have been complicated by the non-mitotic functions of most proteins that prevent aneuploidy and CIN. Despite this, mounting evidence suggests that the effect of aneuploidy on tumors is determined by the rate of CIN, with low CIN being weakly tumor promoting and high CIN causing cell death and tumor suppression. Here we review the causes of aneuploidy and CIN, the evidence that aneuploidy due to low rates of CIN promotes tumors in the presence and absence of p53, and the data supporting the conclusion that there is a maximally tolerated threshold of CIN. We conclude by discussing the potential utility of increasing the rate of CIN as a chemotherapeutic strategy.
One of the first distinguishing features of tumor cells to be identified was abnormal configurations of chromosomes (von Hansemann, 1890). These chromosomal abnormalities can be produced by a variety of mitotic defects. Abnormally large nuclei are formed by mitotic exit without chromosome segregation or cytokinesis, which produces a single tetraploid cell that contains both copies of the replicated chromosomes and has a 4n content of DNA. Subsequent rounds of cytokinesis failure without chromosome segregation can produce even larger, polyploid nuclei. Cytokinesis failure after chromosome segregation results in binucleate cells with two nuclei that can be similar or distinct in size, depending on the accuracy of chromosome segregation. Successful cytokinesis following unequal segregation of chromosomes produces aneuploid progeny. Although aneuploid cells contain an aberrant number of chromosomes, they are distinct from tetraploid cells in that their chromosome number deviates from a multiple of the haploid, resulting in an incomplete set of chromosomes. Some aberrant divisions result in cells with one or more micronuclei, small nuclei that form in addition to the main nucleus. Though many aneuploid and tetraploid progeny survive, major defects in mitosis that result in substantial delay or missegregation of large numbers of chromosomes often result in cell death. The prevalence of mitotic errors in tumor cells led to the proposal by Theodor Boveri in the early 1900s that aneuploid cells are the originating cells of tumors (Boveri, 1902, 1914), although his own experiments showed that high rates of chromosome missegregation led to cell death (Boveri, 1914).
Two types of aneuploidy commonly occur in tumor cells. Numerical aneuploidy refers to gain and loss of whole chromosomes and is a typical consequence of chromosome missegregation during mitosis. Structural aneuploidy indicates gain or loss of large portions of chromosomes, often in the context of chromosomal rearrangements. Both numerical and structural aneuploidy can be stably maintained over multiple generations, or can evolve due to an ongoing rate of CIN.
Causes of aneuploidy and CIN
Chromosome segregation during mitosis is monitored by a major cell cycle regulatory pathway known as the mitotic checkpoint or spindle assembly checkpoint. This checkpoint delays mitotic progression until the chromosomes are attached to the mitotic spindle in a manner consistent with accurate segregation. Chromosomes enter mitosis as replicated pairs of sister chromatids (Fig. 1). The mitotic checkpoint delays their irreversible separation, which occurs at anaphase onset, until each sister chromatid pair has made stable attachments to both poles of the mitotic spindle through spindle microtubules. Sister chromatids attach to spindle microtubules through their kinetochores, protein complexes that assemble at the centromeric region of DNA. Unattached kinetochores generate the mitotic checkpoint signal, which is an inhibitory signal that prevents anaphase onset. Molecularly, mitotic checkpoint proteins including Mad1, Mad2, Bub1, BubR1, Bub3, CENP-E and Mps1 are recruited to unattached kinetochores to generate inhibitors of the Anaphase Promoting Complex/Cyclosome (APC/C) bound to its specificity factor Cdc20 [reviewed in (Cheeseman, 2014; Lischetti and Nilsson, 2015; Sacristan and Kops, 2015). Amphitelic attachments, in which the chromatids in a sister chromatid pair are attached to opposite spindle poles (Fig. 1A), silence the mitotic checkpoint and result in accurate segregation. Divisions in which every sister chromatid pair makes amphitelic attachments produce euploid daughter cells which both contain one and only one copy of each chromosome.
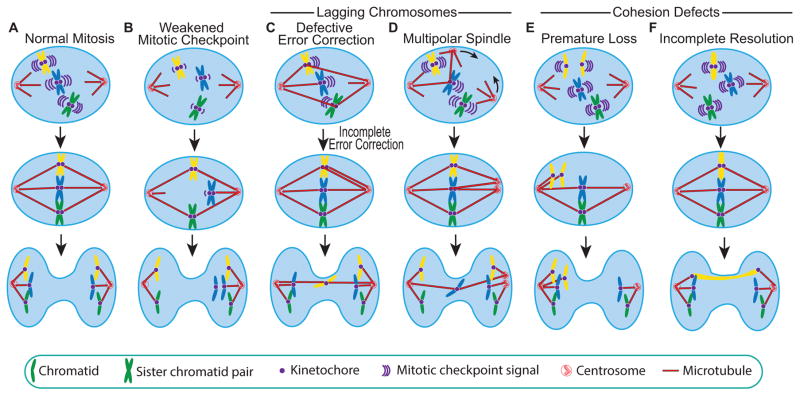
Figure 1. Causes of chromosome missegregation.
(A) A normal mitosis in which sister chromatid pairs make amphitelic attachments (in which one sister chromatid is attached to each spindle pole) produces two genetically identical daughter cells. (B) Weakened mitotic checkpoint signaling permits cells to enter anaphase before each sister chromatid pair has made stable attachments to both spindle poles, resulting in chromosome missegregation. (C–D) Chromatids that lag behind the segregating masses of DNA (lagging chromosomes) represent another source of mitotic errors. (C) Incorrect kinetochore-microtubule attachments, such as merotelic attachments in which a single kinetochore is attached to microtubules from both spindle poles, are generally corrected by the Aurora B error correction pathway. Defects in Aurora B signaling and/or hyperstable kinetochore-microtubules permit incorrect attachments to persist. Shown here is an example of uncorrected merotely, which results in a lagging chromosome. (D) Focusing of supernumerary centrosomes into a pseudo-bipolar spindle can produce merotelic attachments, resulting in lagging chromosomes. (E–F) Defects in establishing and releasing cohesion between sister chromatid pairs can produce chromosome missegregation. (E) Sister chromatid cohesion contributes to a geometric configuration that facilities amphitelic attachment. Premature sister chromatid separation results in random segregation of the sister chromatids. (F) After incomplete resolution of sister chromatids, despite poleward movement of kinetochores, the chromatid arms remain entangled, resulting in a chromosome bridge.
Defects in mitotic checkpoint signaling result in chromosome missegregation, aneuploidy, and CIN [Fig. 1B; reviewed in (Kops et al., 2005; Thompson et al., 2010)]. Complete inactivation of the mitotic checkpoint due to nullizygosity for mitotic checkpoint genes or ≥90% depletion of Mad2 or BubR1 protein levels leads to embryonic lethality in vivo and cell death within 6 divisions in cell culture (Kops et al., 2004; Michel et al., 2004). Interestingly, 20% of mice survived selective excision of conditional alleles of Mad2 in the epidermis after birth, suggesting that the mitotic checkpoint may be dispensable in certain somatic tissues (Foijer et al., 2013) and/or that the subset of cells that survived reduced their reliance on Mad2 for accurate chromosome missegregation (Buffin et al., 2007; Burds et al., 2005). Consistent with the latter option, cultured cells that exhibit a delay in mitosis due to reduced APC/C-Cdc20 activity are no longer reliant on Mad2 for survival or for accurate chromosome segregation (Wild et al., 2016). Although homozygous loss of mitotic checkpoint genes results in embryonic lethality, mice heterozygous for mitotic checkpoint genes are viable and fertile, despite exhibiting aneuploidy and CIN (Ricke et al., 2008; Zasadil et al., 2013).
Weakening mitotic checkpoint signaling through reduced expression of mitotic checkpoint genes is a common method of experimentally inducing aneuploidy and CIN. However, phenotypic identification of a weakened mitotic checkpoint in primary and metastatic human cancers is difficult, since detection requires approval for human subjects testing, the acquisition of unfixed tumor material, and successful culturing of tumor cells in the context of experimental perturbations. Indirect assessment of checkpoint activity based on expression levels of mitotic checkpoint components is complicated by the fact that most of these genes are cell cycle regulated, so proliferative index impacts population based analysis of mitotic checkpoint component levels. Weakened mitotic checkpoint signaling can be inferred, however, in fixed tumor sections based on altered expression of mitotic checkpoint protein levels in individual cells. This is particularly useful for components whose expression is not cell cycle regulated, such as Mad1, which is commonly over- or under-expressed in human breast cancer (Ryan et al., 2012). Evidence for mitotic checkpoint dysfunction in cancer cell lines is varied. Some CIN cancer cell lines do not exhibit mitotic checkpoint defects (Gascoigne and Taylor, 2008; Thompson and Compton, 2008), while others do (Ryan et al., 2012).
Lagging chromosomes, which lag behind the segregating masses of chromatin during anaphase and telophase, are another type of mitotic defect associated with aneuploidy and CIN (Fig. 1C–D). Lagging chromosomes can arise due to defects in the Aurora B-mediated error correction pathway and/or because of hyperstable kinetochore microtubule attachments (Fig. 1C). Aurora B activity destabilizes kinetochore microtubules that are not under tension, such as syntelic attachments, in which both kinetochores of a sister chromatid pair are connected to microtubules emanating from the same spindle pole. Aurora B also corrects merotelic attachments, in which a single kinetochore is connected to microtubules from both poles (Cimini et al., 2006). Merotelically attached kinetochores are not detected by the mitotic checkpoint since the kinetochore is attached and the mitotic checkpoint protein Mad2, which is considered a readout for mitotic checkpoint activation, is not recruited (Cimini et al., 2002; Cimini et al., 2001). Merotelically attached kinetochores also lack another marker of actively signaling kinetochores, the tension-sensing 3F3/2 phosphoepitope (Cimini et al., 2002; Cimini et al., 2001). Aurora B destabilizes improper kinetochore microtubule attachments by phosphorylating proteins critically involved in stabilizing these attachments, including Ndc80/Hec1 and Knl1 (Cheeseman et al., 2006; Welburn et al., 2010). Consistent with this, inhibition or depletion of Aurora B increases the frequency of lagging chromosomes (Cimini et al., 2006; Hauf et al., 2003).
Although lagging chromosomes are often merotelically attached, most merotelic attachments do not result in chromosome lagging. While merotelically attached kinetochores occur in ~30% of untreated prometaphase cells, only ~1% of these cells contain lagging chromosomes during anaphase (Cimini et al., 2003). Interestingly, high resolution timelapse microscopy revealed that 23 of 25 merotelically attached kinetochores in anaphase had more attachments to the correct spindle pole and were ultimately accurately segregated without lagging (Cimini et al., 2004).
Lagging chromosomes also occur as a consequence of hyperstable kinetochore microtubules. Microtubules are repeating polymers of α- and β-tubulin heterodimers that both polymerize and depolymerize throughout their lifetime, a characteristic known as dynamic instability. A ≥1.5 fold increase in the stability of kinetochore microtubules is sufficient to substantially increase the incidence of lagging chromosomes. Mechanistically, depletion of the microtubule destabilizing kinesin 13 family members MCAK and Kif2b, which are dependent upon Aurora B for localization, increases kinetochore microtubule stability and the incidence of lagging chromosomes (Bakhoum et al., 2009). Similarly, increases in kinetochore microtubule stability caused by overexpression of another Aurora kinase family member, Aurora A, or loss of the DNA damage checkpoint kinase Chk2, both of which also increase the rate of microtubule polymerization, result in an elevated frequency of lagging chromosomes (Ertych et al., 2014).
An additional cause of lagging chromosomes is transient spindle multipolarity (Fig. 1D). Daughter cells that arise from divisions of diploid or near-diploid cells into ≥3 daughters generally do not survive (Boveri, 1914; Ganem et al., 2009). However, multipolar mitotic spindles are often focused into a bipolar spindle prior to anaphase, resulting in a two-way DNA division that produces two daughter cells. A kinetochore that attaches to two nearby spindle poles prior to focusing can be pulled in opposite directions after spindle pole focusing, resulting in a merotelically attached lagging chromosome [(Ganem et al., 2009); Fig. 1D]. Similarly, incomplete centrosome separation at the time of nuclear envelope breakdown allows a single kinetochore to make attachments to both spindle poles before they have substantially separated, increasing the incidence of merotelic attachments and lagging chromosomes (Cimini et al., 2003; Silkworth et al., 2012).
Although lagging chromosomes occur in CIN cancer cell lines and multiple experimental perturbations increase both lagging chromosomes and CIN, lagging chromosomes are not necessarily missegregated (Gascoigne and Taylor, 2008; Thompson and Compton, 2008, 2011). The fate of lagging chromosomes was examined in chromosomally stable HCT116 colorectal cancer cells that had a single chromosome marked by incorporation of a LacO array and visualized by expression of LacI-GFP. These experiments revealed that lagging chromosomes that eventually form micronuclei, small nuclei that are separate from the main nucleus, segregated to the correct daughter cell ~2.3 fold more often than they segregated inappropriately. Interestingly, in experimentally manipulated mitoses that had an elevated incidence of lagging chromosomes, the marked chromosome missegregated without lagging almost as often as it missegregated by lagging (Thompson and Compton, 2011). Thus, chromosomes that missegregate are not necessarily discernable due to separation from the masses of segregating DNA, but lagging chromosomes are a visible sign of aberrant mitoses in which chromosome missegregation occurs. As such, lagging chromosomes are a useful indication of chromosome missegregation, even though the lagging chromosomes themselves are not necessarily missegregated (Thompson and Compton, 2011).
Lagging chromosomes that are segregated correctly but form micronuclei may still result in effective aneuploidy. In the human osteosarcoma cell line U2OS, the nuclear envelope surrounding approximately two-thirds of micronuclei collapsed, resulting in an inability to retain RFP tagged with a nuclear localization signal or exclude GFP fused to a nuclear export signal (Hatch et al., 2013). Nuclear envelope collapse, which correlated with the occurrence of a breach in the nuclear rim of lamin B1 localization, resulted in decreased transcription and replication. Thus, even if lagging chromosomes are segregated into the appropriate daughter cell, the transcriptional capacity of that cell may be 2n-1 if the lagging chromosome resides in a micronucleus (Hatch et al., 2013).
Defects in sister chromatid cohesion represent an additional mechanism for chromosome missegregation, aneuploidy and CIN (Fig. 1E–F). Premature sister chromatid separation results in random segregation since the connection between the sisters is necessary to ensure attachment of one sister to each spindle pole (Fig. 1E). Conversely, incomplete separation of sister chromatids due to a failure of cohesin removal or a failure to decatenate the sister chromatids leads to chromatin bridges, which stretch between the daughter nuclei forming a bridge of DNA [(Pampalona et al., 2016); Fig. 1F].
Connections between structural and numerical CIN
Traditionally, structural and numerical chromosomal aberrations were thought to occur independently. However, recent evidence has shown that lagging chromosomes can produce subsequent structural alterations and that DNA damage can result in whole chromosome gains and losses.
Lagging chromosomes were found to produce structural alterations by two mechanisms in nontransformed, telomerase immortalized human retinal pigment epithelial RPE cells. First, lagging chromosomes can be caught in the cytokinetic furrow, resulting in DNA damage (Janssen et al., 2011). This damage was present from telophase through at least 6 hours thereafter, as evidenced by foci of γH2AX, a marker of double strand DNA breaks, and 53BP1, a regulator of double strand break repair. This produced a bona fide DNA damage response, with activation of the DNA damage checkpoint proteins ATM and Chk2, as well as activation of p53. However, this damage was largely repaired by 8 hours post telophase, in part by nonhomologous end joining. Consistent with repair through the error prone nonhomologous end joining pathway, RPE cells with lagging chromosomes had an increased incidence of chromosomal translocations in a subsequent division (Janssen et al., 2011).
Lagging chromosomes in RPE cells were also shown to result in structural alterations through an independent mechanism. In this case, rather than being damaged during cytokinesis, lagging chromosomes that are incorporated into micronuclei become damaged because they enter the subsequent mitosis without being completely replicated. Micronuclei recruit lower levels of origin recognition complexes, which license DNA for replication during S phase (Crasta et al., 2012). Timed addition of the thymidine analog BrdU revealed that micronuclei replicate late as compared to primary nuclei, with ~30% of micronuclei incorporating BrdU at a timepoint expected to be in G2. A small portion (7.6%) of chromosome spreads made after induction of lagging chromosomes contained “pulverized” pieces of DNA. Most of these pulverized chromosomes (25 of 30) incorporated BrdU in the 2 hours preceding preparation, suggesting that they were incompletely replicated at the time the cells began condensing their chromatin. Spectral karyotyping, a method of identifying each chromosome as well as translocations between chromosomes in mitotic cells, revealed that in 72% of cases, the DNA with the pulverized appearance came from a single chromosome (Crasta et al., 2012). This was proposed as a mechanism for chromothripsis, a condition reported to occur in a small percentage of cancers in which extensive structural rearrangements occur on a single chromosome (Crasta et al., 2012; Zhang et al., 2015).
Chromothripsis was also reported to occur after segregation of dicentric chromosomes, which contained two centromeres due to chromosome fusion because of telomere erosion (Maciejowski et al., 2015). Dicentric chromosomes formed chromosome bridges in which the centromeres were segregated to opposite poles and the region of the chromosome between the centromeres was stretched between the two daughter nuclei. In this situation, the random rearrangements of chromothripsis were found to occur only in small portions of chromosomes, presumably the portions that formed the chromatin bridge between the two centromeres (Maciejowski et al., 2015).
Additional chromosomes can also cause structural chromosomal changes when they are not acquired via mitotic defects. Cells with additional chromosomes can be generated using microcell-mediated chromosome transfer, in which chromosomes of donor cells are isolated by inducing multinucleation followed by disruption of the actin cytoskeleton and centrifugation to permit enucleation. The resulting microcells containing isolated chromosomes are then fused with recipient cells through standard treatments such as polyethylene glycol. HCT116 and RPE subclones that contain an extra one or two chromosomes due to microcell-mediated chromosome transfer exhibited delayed replication timing and enhanced sensitivity to replication stress, resulting in chromosome bridges and structural CIN (Passerini et al., 2016). Subclones of chromosomally stable HCT116 cells containing one or two additional copies of chromosome 5 showed de novo structural rearrangements of the chromosomes consisting of duplications, deletion, or inversion, which are consistent with replication mediated rearrangements (Passerini et al., 2016). Thus, numerical chromosome changes can result in structural abnormalities through multiple mechanisms.
DNA damage has been shown to cause mitotic defects as well. DNA replication stress during S phase results in an increase in lagging acentric fragments and chromosome bridges, leading to variation in the number of chromosomes per cell (Burrell et al., 2013). Additionally, DNA damage during mitosis stabilizes kinetochore microtubules, leading to an increase in lagging chromosomes without causing an increase in acentric fragments or chromatin bridges (Bakhoum et al., 2014). The increases in kinetochore microtubule stability and in lagging chromosomes were dependent upon Chk2 activity as well as Plk1 and Aurora A, all of which have previously been implicated in regulating kinetochore microtubule stability (Ertych et al., 2014; Liu et al., 2012). Interestingly, activation of Chk2 without causing DNA damage by treatment with chloroquine was sufficient to increase lagging chromosomes, suggesting that DNA damage exerts its effects on chromosome missegregation indirectly through activation of Chk2 (Bakhoum et al., 2014).
The complex relationship between aneuploidy and Chromosomal INstability (CIN)
Aneuploidy is the state of having an imbalanced set of chromosomes, while CIN indicates an ongoing rate of chromosome gain and loss. In principle, aneuploid karyotypes can remain stable over many generations, or they can exhibit CIN and continue to evolve. It has long been proposed that aneuploidy causes CIN due to an imbalance of genes and protein complexes involved in chromosome segregation during mitosis (Duesberg et al., 1998), and aneuploidy has been shown to cause profound changes in the transcriptome and proteome, some of which are conserved among distinct aneuploid karyotypes (Pavelka et al., 2010; Stingele et al., 2012; Torres et al., 2007b; Upender et al., 2004). Meiosis in yeast strains with an uneven ploidy (3n or 5n) results in the production of both stably aneuploid and CIN yeast strains, with CIN yeast strains predominating (Pavelka et al., 2010; Sheltzer et al., 2011; Zhu et al., 2012). Similarly, in human cells the addition of one copy of chromosome 7 (in the chromosomally stable colorectal cancer cell line DLD1) or one copy of chromosome 13 (in DLD1 cells or in amniotic fibroblasts) leads to increased rates of chromosome missegregation, predominantly of the triploid chromosome (Nicholson et al., 2015). Gain of chromosome 13 but not 7 causes an increase in tetraploidy due to cytokinesis failure. Molecularly, cytokinesis failure in DLD1 cells containing an extra copy of chromosome 13 was attributed to overexpression of the gene encoding Spartin, a gene previously implicated in cytokinesis, which is located on chromosome 13. Consistent with this, overexpression of Spartin caused cytokinesis failure in control DLD1 cells while partial depletion of Spartin rescued cytokinesis failure in DLD1 cells and amniotic fibroblasts containing an extra copy of chromosome 13 (Nicholson et al., 2015). Thus, though there are common effects of aneuploidy, individual aneuploid karyotypes also have specific defects depending on the genes on the imbalanced chromosome(s).
In contrast to the finding that mitotic errors are more common in trisomic cells, experiments examining interphase cells from human tissue with single chromosome gains support the conclusion that aneuploidy is not sufficient to drive CIN. In this analysis, it was expected that CIN would result in whole chromosome gains and/or losses that would be detectable using Fluorescence In Situ Hybridization (FISH) analysis of chromosome copy numbers. Interphase fibroblasts from human tissue with constitutional gain of one copy of chromosome 8 (Warkany syndrome 2), 13 (Patau syndrome), 18 (Edwards syndrome), or 21 (Down syndrome) had no increase in copy number alterations of chromosomes 2 or 17 (Valind et al., 2013), suggesting that aneuploidy did not result in CIN in this context. During mitosis, the primary chromosome missegregated was found to be the trisomic chromosome itself (Nicholson et al., 2015), which was not examined in the interphase study (Valind et al., 2013), suggesting that trisomic cells without additional mitotic defects, such as cells trisomic for chromosome 7, revert to a diploid state. Consistent with this, cells trisomic for chromosome 3 were found to have late or incomplete replication of the pericentromeric region of the third copy of chromosome 3, which after division can produce one trisomic and one diploid daughter cell. Together, these data suggest that whether aneuploidy is sufficient to initiate CIN depends on the specific aneuploid karyotype.
Although CIN results in the production of aneuploid progeny, this does not necessarily result in a predominantly aneuploid population. Populations of HCT116 colorectal cancer cells maintain a stable consensus karyotype despite the finding that subclones derived from single cells exhibit low rates of structural and numerical CIN (Roschke et al., 2002). This suggests that the consensus karyotype of HCT116 cells is optimized for growth under standard cell culture conditions, and that altering this karyotype results in a proliferative disadvantage. Consistent with this, induced chromosome missegregation in HCT116 cells or nontransformed human RPE cells produces only transient increases in aneuploidy (Thompson and Compton, 2008), suggesting that the aneuploid cells were outcompeted by cells with the consensus karyotype. Supporting this hypothesis, HCT116 cells that missegregate a marked chromosome survive for 5 days but show elevated levels of stabilized nuclear p53 and its downstream effector p21, consistent with cell cycle arrest (Thompson and Compton, 2010). Thus, for aneuploidy and CIN, the presence of one does not necessitate the widespread presence of the other.
The aneuploidy paradox
Aneuploidy is common in tumors but is generally associated with a proliferative delay. This incongruous observation is known as the aneuploidy paradox. Whole chromosome aneuploidy is prevalent in a wide variety of cancers, with approximately 86% of solid tumors and 72% of hematopoietic cancers having gained or lost chromosomes to become aneuploid. Analysis of whole chromosome karyotypic variability indicates that 44% of solid and 14% of hematopoietic cancers exhibit CIN (Zasadil et al., 2013). These features are associated with a worse prognosis and increased disease progression. In diffuse large B-cell lymphoma, a tumor type with an unusually high mitotic index, histological evidence of lagging chromosomes portends a worse prognosis and a 24% decrease in overall survival from 8.76 to 6.62 years (Bakhoum et al., 2011). In a panel of tumor types assessed for structural CIN by gene expression data, CIN correlated with decreased survival across the board (Carter et al., 2006). Cancers exhibiting CIN are also more likely to relapse. After treatment, diffuse large B-cell lymphoma patients exhibiting CIN had a 48% decrease in relapse-free survival (Bakhoum et al., 2011), and breast cancers exhibiting whole chromosome CIN had reduced breast cancer-related overall survival times (Denu et al., 2016). Consistent with this, benign lesions exhibited lower levels of whole chromosomal aneuploidy and CIN than those seen in cancers (Zasadil et al., 2013), and metastatic sites had a higher CIN gene expression signature than primary tumors (Carter et al., 2006). Thus, aneuploidy and CIN are common features that confer poor prognosis in a variety of cancers.
Although aneuploidy commonly occurs in proliferative tumors where it serves as a marker of poor prognosis, aneuploidy due to chromosome gains causes an overall growth defect in several systems. In budding yeast, aneuploid strains containing a single additional chromosome display a growth disadvantage under conditions optimal for euploid yeast growth (Torres et al., 2007a), a delay which is attributable to an extended duration of G1 (Thorburn et al., 2013). In mice, trisomy for a single chromosome, generated by crossing animals with chromosome fusions with wild type animals, causes embryonic lethality. However, mouse embryonic fibroblasts (MEFs) generated from these embryos are viable in culture. As was seen in yeast, MEFs with an additional chromosome showed a proliferative defect as compared to euploid controls (Williams et al., 2008). Similarly, fibroblasts from human trisomy 21 patients proliferated more slowly than diploid controls (Segal and McCoy, 1974). Consistent with this, trisomy 21 patients have reduced stature and head circumference (Van Gameren-Oosterom et al., 2012). HCT116 cells with extra copies of chromosome 3 or 5 also showed marked growth delay under standard conditions (Stingele et al., 2012). Therefore in yeast, mice, and human, chromosome gains confer a growth disadvantage under conditions optimized for euploid cell growth.
Chromosome losses may also cause a proliferative disadvantage. Expression of one hypomorphic and one null allele of the mitotic checkpoint component Bub1 (Bub1−/H) decreases Bub1 protein levels to 20% of normal. This hypomorphic expression of Bub1 causes chromosome missegregation (Jeganathan et al., 2007) and accelerates the formation of lymphomas in animals heterozygous for the p53 tumor suppressor (Baker et al., 2009). Lymphomas are relatively rare in p53+/− animals but predominate in p53−/− animals. Concordantly, lymphomas in Bub1−/H;p53+/− animals had lost the wild type copy of p53. Interestingly, however, all of the Bub1−/H;p53+/− lymphomas examined had 2 copies of chromosome 11, which contains the p53 gene, suggesting that gain of a mutated chromosome 11 occurred prior to or contemporaneous with loss of the wild type chromosome 11. Thus, simple loss of the copy of chromosome 11 containing wild type p53 was apparently insufficient to permit these tumors to form (Baker et al., 2009), suggesting that monosomy of murine chromosome 11 induces a fitness penalty that is sufficient to prevent tumor growth even in the absence of p53. A subsequent study from the same authors came to a similar conclusion for murine chromosome 18, which contains the tumor suppressor Adenomatous Polyposis Coli (Apc) (Baker and van Deursen, 2010). Mice expressing the Multiple Intestinal Neoplasia (Min) allele of Apc develop intestinal tumors after loss of the wild type copy of Apc. Reduction of Bub1 in ApcMin/+ animals resulted in an increase in colon tumor formation (Baker et al., 2009). 7 of 7 colon tumors examined had lost the wild type copy of Apc but still contained two copies of chromosome 18 (Baker and van Deursen, 2010). These two examples in mice suggest that loss of a single copy of an autosome in an otherwise diploid cell confers a fitness penalty even in the absence of a tumor suppressor, although direct experiments rigorously testing the consequences of single chromosome loss in diploid cells have not yet been reported.
Chromosome gains generally lead to concomitant increases in transcription that mirror chromosome copy numbers in yeast and human cells (Pavelka et al., 2010; Stingele et al., 2012; Torres et al., 2007a). Proteomic analysis of HCT116 and RPE cells with one or two additional chromosomes revealed that, while transcription reflects gene copy number, protein levels for ~25% of genes on the extra chromosomes are lower than what would be predicted based on gene copy number and are instead similar to the levels in diploid cells (Stingele et al., 2012). Proteins whose expression was downregulated post-transcriptionally included those that function in DNA replication and repair, which is consistent with the observed growth defect during interphase, as well as with a delay in DNA synthesis (Passerini et al., 2016). One mechanism contributing to the detrimental effects of chromosome gains is the induction of a stress response, in part due to additional genetic copies of individual subunits of multiprotein complexes (Dephoure et al., 2014; Stingele et al., 2012).
Despite a growth delay under standard conditions, aneuploid yeast strains with specific chromosome gains grow better under suboptimal conditions, including treatment with concentrations of chemotherapeutic and antifungal drugs that inhibit euploid cell growth (Pavelka et al., 2010). Acquisition of an aneuploid karyotype accompanies resistance to antifungal drugs in pathogenic yeast (Selmecki et al., 2006; Selmecki et al., 2009). Similarly, one study found aneuploidy and CIN in murine liver promote regeneration after hepatic injury (Duncan et al., 2012), although two subsequent studies using single cell sequencing did not identify aneuploidy in wild type hepatocytes (Choi et al., 2016; Knouse et al., 2014). Human cells with an extra chromosome 7 or 13 have a proliferative advantage over diploid cells under multiple stresses (Rutledge et al., 2016). Together, these data suggest that aneuploidy and CIN generate phenotypic variation, which allows for adaptation to environmental stresses and could explain their prevalence in aggressive human cancers.
Testing Boveri’s hypothesis: does aneuploidy drive tumorigenesis?
The tumor phenotype of numerous mouse models exhibiting genetically induced aneuploidy and/or CIN has been determined [reviewed in (Holland and Cleveland, 2009; Ricke et al., 2008; Zasadil et al., 2013)] and a wide range of tumor phenotypes has been observed. Some models exhibiting aneuploidy and CIN do not have elevated levels of spontaneous, chemically-induced, or genetically driven tumors (Malureanu et al., 2010; Ricke et al., 2012). In contrast, other models exhibiting similar levels of aneuploidy and CIN do display an elevated tumor incidence. Some of these animals develop tumors in a small number of tissues with low penetrance near the end of their normal lifespan (Iwanaga et al., 2007; Michel et al., 2001; Weaver et al., 2007). Others develop early onset tumors with higher penetrance in a wider range of tissues (Li et al., 2009; Ricke et al., 2011; Sotillo et al., 2007). Perhaps most intriguingly, in some cases aneuploidy and/or CIN are tumor suppressive (Chesnokova et al., 2005; Rowald et al., 2016; Silk et al., 2013; Sussan et al., 2008; Weaver et al., 2007; Yang and Reeves, 2011; Zasadil et al., 2016). Despite the complexity of the tumor phenotypes, several themes emerge. First, most aneuploid karyotypes are not sufficient for tumor initiation and progression. Second, the rate of chromosome missegregation predicts whether CIN will promote or suppress tumors. Low rates of chromosome missegregation (1 or a few chromosomes missegregated every few divisions) are weakly tumor promoting, while high rates of chromosome missegregation (≥5 chromosomes per division) lead to cell death and suppression of tumor growth due to loss of both copies of one or more essential chromosomes (Silk et al., 2013; Zasadil et al., 2016). Third, non-mitotic functions of the experimentally altered genes have a substantial impact on tumor phenotypes.
Most proteins that function in mitosis and the mitotic checkpoint have known additional roles outside of chromosome segregation (Fig. 2). These non-mitotic functions often occur in pathways that are likely to influence tumor phenotypes, such as cell death, DNA damage, and cell migration. The BUB family of mitotic checkpoint proteins (Bub1, BubR1, Bub3) has been implicated in cell death pathways. Bub1 facilitates cell death following chromosome missegregation events (Jeganathan et al., 2007) and is cleaved in a caspase-dependent manner in response to multiple pro-apoptotic conditions to produce cleavage fragments that increase cell death (Perera and Freire, 2005). BubR1 is also cleaved by caspase-dependent mechanisms following mitotic arrest, and expression of a caspase-resistant BubR1 mutant in HeLa cells increases apoptosis after mitotic arrest with the microtubule poison nocodazole (Baek et al., 2005). Depletion of either Bub3 or its binding partner BuGZ results in apoptosis due to disruption of their interphase function in pre-mRNA splicing. Bub3 or BuGZ depletion causes splicing defects, predominantly exon skipping, and increases the formation of RNA-DNA hybrids leading to DNA damage, p53 activation (as assessed by phosphorylation on Serine 15), and apoptosis (Wan et al., 2015). Thus, there is evidence that all three Bub family members have non-mitotic effects on cell death pathways, which are likely to impact tumor phenotypes in animals which express altered levels or mutated versions of these genes.
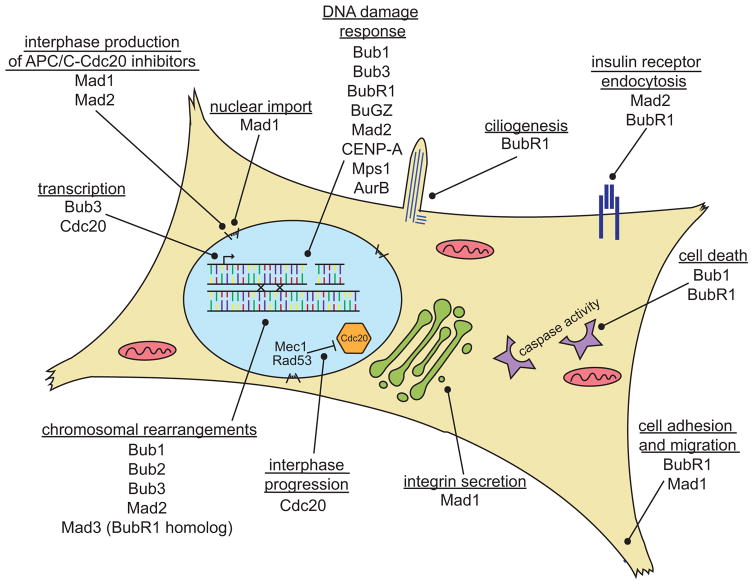
Figure 2. Interphase roles of ‘mitotic’ proteins.
Many proteins with known mitotic functions that have been classically considered to function primarily or exclusively during mitosis function during interphase as well. Reported interphase roles of these proteins are depicted here. See text for references.
Alterations in mitotic checkpoint components often also result in heightened DNA damage through increased formation of DNA breaks and/or impaired DNA repair. In a panel of yeast strains with high rates of gross chromosomal rearrangements, defects in the mitotic checkpoint genes BUB1, BUB3, MAD2 and the BubR1 homolog MAD3 suppressed the incidence of these rearrangements, suggesting that overexpression of these genes promotes structural rearrangements of chromosomes (Myung et al., 2004). Consistent with this, overexpression of Mad2 resulted in structural as well as numerical aneuploidy in mice (Sotillo et al., 2007). Mad2 overexpression has been shown to impair DNA damage repair (Fung et al., 2008). Mad2 loss also affects the DNA damage response, and resulted in a decreased ability to maintain arrest and delay entry into mitosis in the presence of double strand breaks or incompletely replicated DNA (Dotiwala et al., 2010; Sugimoto et al., 2004). Bub proteins also impact DNA damage and repair pathways. Bub1 is phosphorylated by ATM in response to DNA damage and facilitates DNA damage repair. Knockdown of Bub1 prolongs the time required to repair DNA, as evidenced by a delay in resolution of γH2AX foci, and increased sensitivity to ionizing radiation (Yang et al., 2012). BubR1 facilitates cell cycle arrest following DNA damage, and reduction of BubR1 decreases the accumulation of γH2AX, p53, and p21 under these conditions (Fang et al., 2006). Depletion of Bub3 induces DNA damage due to splicing defects (Wan et al., 2015). The Mps1 mitotic checkpoint kinase also participates in the DNA damage response (Wei et al., 2005; Yu et al., 2016). Thus, components of the mitotic checkpoint are required to prevent and repair DNA damage through multiple mechanisms.
Recently, the mitotic checkpoint components Mad1 and BubR1 have been shown to promote cell migration. During interphase Mad1 functions at the Golgi to facilitate α5 integrin secretion (Wan et al., 2014). α5 integrin bound to β1 integrin serves as a major cellular receptor for the extracellular matrix component fibronectin. Concordantly, Mad1 levels are directly related to cell attachment, adhesion, and motility on fibronectin (Wan et al., 2014). Similarly, cytosolic BubR1 expression correlates with motility in oral squamous cell carcinoma cell lines and knockdown of BubR1 decreases migration as well as the activity of matrix metalloproteases (Chou et al., 2015). Thus, these proteins traditionally considered to function specifically during mitosis also have interphase activities with implications for metastasis.
Mitotic proteins have also been implicated in ciliogenesis (Miyamoto et al., 2011), nuclear import (Cairo et al., 2013; Iouk et al., 2002), transcriptional repression (Yoon et al., 2004), and cell cycle progression during interphase (Clarke et al., 2003), which may impact tumor phenotypes in animals which express altered levels or mutants of these genes (Fig. 2). Recently, both Mad2 and BubR1 have been shown to regulate insulin signaling (Choi et al., 2016). Additionally, Mad1 and Mad2 localize to nuclear pores during interphase (Campbell et al., 2001; Iouk et al., 2002), where Mad1 functions in nuclear import, at least in yeast (Cairo et al., 2013; Iouk et al., 2002). Nuclear pore localization of Mad1 and Mad2 also facilitates interphase production of so-called “mitotic checkpoint complexes” (Rodriguez-Bravo et al., 2014), which consist of Mad2, BubR1, Bub3 and Cdc20 (Sudakin et al., 2001) and inhibit APC/C-Cdc20.
Mouse models with alterations in most mitotic checkpoint genes develop aneuploidy and CIN in the context of additional defects that can have a substantial impact on their tumor phenotype. Considering the large number of animal studies available and taking their known and potential interphase defects into account, the weight of the evidence suggests that, at least in part, Boveri was correct: aneuploidy due to low CIN is weakly tumor promoting. However, high rates of chromosome missegregation cause cell death, as originally shown by Boveri himself after inducing tripolar or tetrapolar mitosis (Boveri, 1914).
The role of p53 in monitoring aneuploidy and CIN
The finding that CIN does not necessarily produce an aneuploid population of cells suggested that certain genes prevent proliferation after chromosome missegregation, and that these genes must be mutated or otherwise inactivated to permit the propagation of aneuploid cells. The leading candidate for this role is the transcription factor and tumor suppressor p53. Evidence from this comes from studies with HCT116 cells induced to form lagging chromosomes. While control HCT116 cells showed only a transient increase in aneuploidy at the population level after induced chromosome missegregation, p53−/− HCT116 cells continued to exhibit aneuploidy over the course of the 6 day experiment. p53 levels are normally kept low due to continuous ubiquitination and degradation. However, in response to a number of stress stimuli, including DNA damage, oncogene activation, hypoxia, and nucleotide imbalance, p53 is stabilized and accumulates in the nucleus where it binds DNA, leading to changes in gene transcription. 76% of HCT116 cells that missegregated a marked chromosome showed nuclear localization of p53 and its downstream effector p21 (Thompson and Compton, 2010). Similarly, depletion of the centromeric protein CENP-A in diploid human fibroblasts, which causes chromosome segregation defects, resulted in a senescence associated growth defect that was partially dependent on p53 (Maehara et al., 2010). Thus, p53 appears to be activated in at least a subset of aneuploid cells.
However, p53 is not uniformly activated in aneuploid cells. In another study, although CIN caused by a variety of genetic insults correlated with an increase in activated (phosphorylated) p53, p53 was only activated in a fraction of the aneuploid cells, and many aneuploid cells continued to divide (Li et al., 2010). In the numerous mouse models of aneuploidy and CIN in which p53 was not intentionally mutated, aneuploid cells continued to survive and divide, even in non-tumorous tissues.
It therefore remains unclear whether p53 recognizes chromosome missegregation per se, rather than other stresses that can occur as a later consequence of an imbalanced genome. DNA damage is detected in some, but not all, studies after chromosome missegregation. RPE cells that exhibited stabilization of p53 and p21 after missegregation of lagging chromosomes were not found to have DNA damage, as measured by γH2AX, 24 hours after missegregation (Thompson and Compton, 2010). However, another study that did identify DNA damage after chromosome lagging found that foci of 53BP1, a regulator of double stranded DNA break repair, were largely absent by 8 hours after chromosome missegregation (Janssen et al., 2011). Thus, DNA damage that stabilized p53 may have been resolved 24 hours after chromosome missegregation. Alternatively, another study demonstrated that CIN caused an increase in oxidative DNA damage that was not sufficient to cause an increase in γH2AX reactivity, but could have been sufficient to stabilize p53 (Li et al., 2010). Similarly, tetraploidy was reported to stabilize p53 through oxidative DNA damage that did not correlate with γH2AX staining, but was dependent on the DNA damage checkpoint kinase ATM (Kuffer et al., 2013). A recent study reported that Serine 31 on histone H3 is phosphorylated on missegregated chromosomes that are separated from the masses of segregating DNA during anaphase, and that this phosphorylation was sufficient to stabilize p53 in the subsequent G1 in a manner independent of the DNA damage checkpoint kinases ATM, ATR and Chk1 (Hinchcliffe et al., 2016).
As a test of whether p53 restrains the tumorigenic potential of aneuploidy, numerous CIN mouse models have been mated with animals deficient for p53. Interestingly, loss of p53 or its effector p21 results in overexpression of Mad2 (Schvartzman et al., 2011). Mad2 overexpression in the presence of wild type p53 causes aneuploidy, tetraploidy and CIN (Sotillo et al., 2007) and promotes lymphomagenesis and lung tumour relapse (Sotillo et al., 2007; Sotillo et al., 2010), but suppresses mammary tumor formation in models with a preexisting rate of CIN (Rowald et al., 2016). Mad2 expression has not been examined in most CIN models crossed into p53 deficient backgrounds, and the potential contributions of Mad2 overexpression to aneuploidy, tetraploidy, CIN, DNA damage, chromosomal rearrangements and insulin signaling remain undefined. Similar to models containing wild type p53, a variety of phenotypes have been observed in CIN models with reduced p53 function. Most models of CIN either decrease tumor latency in p53+/− (Table 1) and p53−/− (Table 2) animals or have no substantial effect. However, reduction of Mad1 in Mad2+/−;p53+/− animals suppresses their incidence of T-cell anaplastic lymphoblastic lymphoma, which is associated with splenic enlargement, (Table 1) and decreases splenic weight by >60% (Chi et al., 2009).
Table 1. Tumor incidence, survival, and spectrum in p53+/− mice with secondary CIN-inducing mutations.
Tumor incidence is shown as percentage of cohort with tumor in double mutants versus p53+/− mice, respectively. Median tumor free survival is shown as the median survival of the double mutant cohort versus the p53+/− cohort, respectively. Studies in which effect on tumor phenotype is listed as unchanged are those in which incidence and/or survival between p53+/− and doubly mutant mice did not reach statistical significance.
| Secondary Mutation | Tumor Incidence | Median Tumor Free Survival (days) | Primary Tumor Type$ | Effect on Tumor Phenotype | Reference |
|---|---|---|---|---|---|
| Bub1+/H ^ | 57% vs 43% | 495 vs 471* | sarcoma | Unchanged | (Baker et al., 2009) |
| Bub1+/− | 57% vs 43% | 469 vs 471* | sarcoma | Unchanged | |
| Bub1H/H | 78% vs 43% | 346 vs 471* | lymphoma | Exacerbated | |
| Bub1−/H | 76% vs 43% | 202 vs 471* | lymphoma | Exacerbated | |
| Bub3+/− | 47% vs 62% | ~560 vs ~525 | osteosarcoma | Unchanged | (Kalitsis et al., 2005) |
| Mad1+/− | 76% vs 67% | osteosarcoma | Unchanged | (Chi et al., 2009) | |
| Mad2+/− | 88% vs 67% | lymphoma | Exacerbated | ||
| Mad1+/−;Mad2+/− | 95% vs 67% | osteosarcoma | Exacerbated | ||
| Mad1+/−;Mad2+/− | 25% vs 53% in Mad2+/−;p53+/− # | osteosarcoma | Suppressed | ||
| Separase OE MMTV-LTR promoter (mammary) | 100% | 439 vs ~580 | mammary carcinoma | Exacerbated | (Mukherjee et al., 2014) |
| Separase+/H | 100% vs ~50% | 460 vs ~530 | Exacerbated | (Mukherjee et al., 2011) | |
| Mps1f/f p53f/+; Lck-Cre (T cells)& | 100% vs ~0% in p53f/+;Lck-Cre | ~105 vs >360 | T-cell acute lymphoblastic lymphoma | Exacerbated | (Foijer et al., 2014) |
| Plk4 OE;p53f/+; ERT-CRE (β-actin promoter) | ~565 vs ~575 | lymphoma | Unchanged | (Vitre et al., 2015) |
the primary tumor type in p53+/− animals is sarcoma
H indicates a hypomorphic allele
average tumor latency
lymphoma incidence
f indicates a floxed allele
Table 2. Tumor-free survival and tumor spectrum in p53−/− mice with secondary CIN-inducing mutations.
Median tumor-free survival is shown as the median survival of the double mutant cohort versus the p53−/− cohort, respectively. Studies in which effect on tumor phenotype is listed as unchanged are those in which the difference in survival between p53−/− and doubly mutant mice was not statistically significant.
| Secondary Mutation | Median Tumor Free Survival (days) | Primary Tumor Type | Effect on Tumor Phenotype | Reference |
|---|---|---|---|---|
| Bub1+/H ^ | ~175 vs ~175 | thymic lymphoma | Unchanged | (Li et al., 2010) |
| Bub1+/− | ~175 vs ~175 | thymic lymphoma | Unchanged | |
| Bub1H/H | ~90 vs ~175 | thymic lymphoma | Exacerbated | |
| Bub1−/H | ~85 vs ~175 | thymic lymphoma | Exacerbated | |
| Cdc20+/AAA | ~120 vs ~210 | thymic lymphoma | Exacerbated | |
| Separase+/H | 118 vs 151 | lymphoma | Exacerbated | (Mukherjee et al., 2011) |
| Mps1f/+;p53f/f; Lck-Cre (T cells)& | ~140 vs 150 in p53f/f;Lck-Cre | T-cell acute lymphoblastic lymphoma | Unchanged | (Foijer et al., 2014) |
| Mps1f/f p53f/f; Lck-Cre (T cells) | ~105 vs ~150 in p53f/f;Lck-Cre | T-cell acute lymphoblastic lymphoma | Exacerbated | |
| Mps1f/f p53f/f; MMTV-Cre (mammary, T cells) | ~100 vs ~200 in p53f/f:MMTV-Cre | T-cell acute lymphoblastic lymphoma (no mammary) | Exacerbated | |
| Plk4OE; p53f/f; ERT-Cre (β-actin promoter) | ~ 180 vs ~180 in p53f/f;ERT-Cre | thymic lymphoma | Unchanged | (Vitre et al., 2015) |
| Plk4OE p53f/f K14-Cre (epidermis) | 175 vs 266 in p53f/f ;K14-Cre* | skin carcinoma | Exacerbated | (Sercin et al., 2016) |
| Plk4OE/OEp53−/− (ROSA26 locus) | 105 vs 140 | sarcoma | Exacerbated | (Coelho et al., 2015) |
H indicates hypomorphic allele
f indicates a floxed allele
average time to tumor onset
Thus, p53 is an important, but incompletely understood, regulator of cellular proliferation in aneuploid cells. Since homozygous loss of p53 is not sufficient to prevent the embryonic lethality caused by homozygous mutation or loss of mitotic checkpoint genes (Burds et al., 2005; Li et al., 2010), it is likely that CIN cells can be removed from the cycling population by p53 independent mechanisms, at least during development.
Induction of high CIN as a chemotherapeutic strategy
CIN can be subdivided into low CIN and high CIN. Several mouse models that missegregate 1 or a few chromosomes every few divisions – a rate consistent with low CIN – such as those heterozygous for Mad1, Mad2 or CENP-E, developed late onset low penetrance tumors (Iwanaga et al., 2007; Michel et al., 2001; Silk et al., 2013; Weaver et al., 2007). However, increasing the rate of CIN by combining two insults that each cause low CIN resulted in cell death and suppressed tumors in multiple contexts (Silk et al., 2013; Weaver et al., 2007; Zasadil et al., 2016; Rowald et al., 2016). Similarly, in cell culture experiments, combining a genetic cause of low CIN with chemically induced low CIN leads to high CIN and cell death (Janssen et al., 2009). Research over the past 15 years has shown that partial depletion of mitotic checkpoint components leads to CIN, while ≥90% depletion of these components leads to cell death both in cell culture and in animal models (Babu et al., 2003; Baker et al., 2004; Dobles et al., 2000; Jeganathan et al., 2007; Kops et al., 2004; Meraldi et al., 2004; Michel et al., 2004; Putkey et al., 2002; Weaver et al., 2007). Together, these data suggest that, while low CIN is weakly tumor promoting, high rates of CIN cause cell death and tumor suppression. Consistent with this, mathematical modeling shows that a low rate of CIN optimizes tumor heterogeneity and survival and that increasing this rate decreases tumor fitness (Bakhoum et al., 2015; Komarova and Wodarz, 2004).
These data suggest that increasing the rate of CIN over a critical threshold could be an effective tumor therapy. For this to be clinically useful, high CIN must be capable of inhibiting tumor growth and progression, not just tumor initiation. Consistent with this, increasing the rate of CIN in intestinal tumors in ApcMin/+ animals inhibits tumor progression without suppressing tumor initiation (Zasadil et al., 2016). Concordantly, in a mouse model of triple-negative breast cancer, a combination of pharmacological inhibition of the mitotic kinase Mps1/TTK and treatment with the microtubule stabilizing drug docetaxel, each of which cause low CIN, showed increased efficacy as compared to either treatment as a single agent (Maia et al., 2015), supporting the hypothesis that increasing chromosome missegregation is a viable therapeutic strategy.
In human cancers, tumors with the highest level of CIN often have better outcomes. In ovarian, gastric, non-small cell lung, and ER-negative breast cancers, patients with a high gene expression measure of CIN have improved outcomes relative to patients with lower levels of this CIN gene signature (Birkbak et al., 2011; Jamal-Hanjani et al., 2015; Roylance et al., 2011). Similarly, rectal tumors with high CIN, as assessed by lagging and bridge chromosomes, have improved pathological responses to radiation and DNA damaging chemotherapy than tumors with low CIN (Zaki et al., 2014), perhaps due to an increased rate of chromosome missegregation caused by hyperstable kinetochore microtubules induced by DNA damage (Bakhoum et al., 2014).
Paclitaxel (Taxol™) is the best selling chemotherapy drug in history. Although it is considered highly effective, only approximately half of breast cancer patients benefit from paclitaxel therapy. Currently, there is no reliable method to identify these patients prior to treatment. For the more than three decades paclitaxel has been used in patients, it has been expected to exert its anti-tumor effects by arresting cells in mitosis, as it does at typically used concentrations in cell culture. However, in human breast cancers, mitotic arrest is not necessary for paclitaxel to exert its anti-cancer effects (Symmans et al., 2000; Zasadil et al., 2014). Moreover, measured levels of paclitaxel in primary breast cancers are lower than those necessary to effect mitotic arrest (Zasadil et al., 2014). Concentrations of paclitaxel measured in brain, ovarian, uterine and cervical cancers are similar or lower than those measured in breast cancer, suggesting that paclitaxel concentrations are insufficient to cause mitotic arrest in these contexts as well (Fernández-Peralbo et al., 2014; Fine et al., 2006; Koshiba et al., 2009; Mori et al., 2006).
One alternative hypothesis that is consistent with the finding that mitotic arrest is not necessary for paclitaxel to exert its anti-cancer effects is that paclitaxel causes cytotoxicity during interphase. Interphase microtubules provide structural support for cilia and flagella, and serve as tracks for the transport and secretion of a wide variety of cargoes. It has therefore been proposed that paclitaxel, and other microtubule poisons, exert their cytotoxic effects by interfering with these interphase functions of microtubules (Komlodi-Pasztor et al., 2011; Komlodi-Pasztor et al., 2012; Mitchison, 2012). This hypothesis awaits direct experimental validation and is largely based on the slow doubling rates of human tumors. Calculations of cell cycle times based on these rates predict that too few cells pass through mitosis in the presence of drug for effective tumor clearance. However, it has long been realized that these calculations do not account for the levels of cell death that occur in untreated tumors (Kerr et al., 1972; Lowe and Lin, 2000; Searle et al., 1973), and that measured proliferation rates are substantially higher than those calculated based on tumor doubling times (Frindel et al., 1968; Kerr and Searle, 1972; Weinstein and Frost, 1970). Since paclitaxel is retained in gynecological tumors for at least 5 to 6 days (Koshiba et al., 2009; Mori et al., 2006), it has an extended window of time to exert its effects on slowly proliferating cells as they progress through mitosis.
In breast cancer cell lines in culture, clinically relevant doses of paclitaxel cause chromosome missegregation on multipolar spindles, resulting in CIN (Zasadil et al., 2014). At least in cultured cells, these clinically relevant doses do not cause cell death in interphase cells that have not previously traversed mitosis in the presence of drug (Zasadil et al., 2014). Evidence from animal models and cell culture indicates that low CIN + low CIN = high CIN, cell death, and tumor suppression. Both genetic and pharmaceutical methods of inducing low CIN are sufficient (Janssen et al., 2009; Maia et al., 2015; Silk et al., 2013; Zasadil et al., 2016). Together, this suggests that the ~50% of breast cancers that exhibit CIN prior to therapy are the same ~50% that respond to paclitaxel therapy.
Conclusions and future directions
Aneuploidy and CIN are common characteristics of human tumors that are associated with poor prognosis. Despite this correlation, the mere presence of aneuploidy and/or CIN is not sufficient to uniformly sensitize animals to tumor formation. The rate of CIN, rather than the overall level of accumulated aneuploidy, determines the effect on tumor phenotype with low levels of CIN being weakly tumor promoting and high levels of CIN leading to cell death and tumor suppression. Two insults that both cause a low rate of CIN can be combined to cause high CIN and tumor suppression. Both genetic and pharmacologic causes of low CIN can be used, suggesting that increasing the rate of CIN may be a successful therapeutic strategy. Consistent with this, the widely used chemotherapy drug paclitaxel was recently shown to cause CIN – rather than mitotic arrest as had long been presumed – at clinically relevant doses. Together these data suggest that paclitaxel is most effective in patient tumors that exhibit low CIN prior to treatment, a hypothesis that must now be rigorously tested.
Other outstanding questions of critical importance remain. These include determining whether chromosome losses cause similar phenotypes as chromosome gains and identifying the stimulus that stabilizes p53 in a subset of aneuploid cells. It remains unclear whether p53 mediated apoptosis is necessary for cell death due to high rates of CIN or whether other genes and mechanisms of cell death contribute or can compensate when p53 function is compromised. Though consequences of aneuploidy and CIN on tumor initiation have been well studied, their consequences on tumor progression remain relatively uncharacterized. Further delineation of the interphase roles of mitotic genes will assist in interpreting tumor phenotypes in models with altered expression of these genes.
From a clinical perspective, although the relative contribution of various mechanisms of CIN has been described in cultured cancer cell lines, it remains to be determined whether these models faithfully recapitulate the types of CIN found in primary and metastatic tumors. Additionally, the genetic and epigenetic causes and consequences of CIN in human tumors remain to be elucidated. With respect to treatment, a thorough understanding of the mechanism of effective anti-mitotic drugs is likely to provide dual benefits. First, it will facilitate identification of a biomarker to predict which tumors are likely to respond to these drugs. Second, it will inform the development of novel anti-mitotic drugs. Vinca alkaloids and paclitaxel were both expected to exert their anti-cancer effects by causing mitotic arrest. Though paclitaxel has now been shown to cause multipolar divisions rather than mitotic arrest in patient tumors, it remains to be determined whether vinca alkaloids cause mitotic arrest or abnormal mitotic divisions. This is highly relevant, since biomarkers that can predict response to these drugs will likely depend on their clinically relevant mechanism of action. If all microtubule poisons with proven clinical utility exert their anti-cancer effects by causing multipolar divisions rather than eliciting mitotic arrest, this would suggest that novel anti-mitotic therapies should be developed based on their ability to cause CIN rather than accumulation in mitosis. Although many questions remain unanswered, it is clear that an improved fundamental understanding of mitosis and the cellular responses to mitotic defects can be leveraged to improve patient outcomes after treatment with anti-mitotic chemotherapy.
Acknowledgments
We apologize to those whose work could not be described due to space limitations. Beth A. Weaver, PhD was supported by a Research Scholar Grant, RSG-15-006-01-CCG from the American Cancer Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;160:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek KH, Shin HJ, Jeong SJ, Park JW, McKeon F, Lee CW, Kim CM. Caspases-dependent cleavage of mitotic checkpoint proteins in response to microtubule inhibitor. Oncol Res. 2005;15:161–168. doi: 10.3727/096504005776367906. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36:744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, van Deursen JM. Chromosome missegregation causes colon cancer by APC loss of heterozygosity. Cell Cycle. 2010;9:1711–1716. doi: 10.4161/cc.9.9.11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Danilova OV, Kaur P, Levy NB, Compton DA. Chromosomal instability substantiates poor prognosis in patients with diffuse large B-cell lymphoma. Clin Cancer Res. 2011;17:7704–7711. doi: 10.1158/1078-0432.CCR-11-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Kabeche L, Murnane JP, Zaki BI, Compton DA. DNA-damage response during mitosis induces whole-chromosome missegregation. Cancer Discov. 2014;4:1281–1289. doi: 10.1158/2159-8290.CD-14-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Kabeche L, Wood MD, Laucius CD, Qu D, Laughney AM, Reynolds GE, Louie RJ, Phillips J, Chan DA, et al. Numerical chromosomal instability mediates susceptibility to radiation treatment. Nat Commun. 2015;6:5990. doi: 10.1038/ncomms6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkbak NJ, Eklund AC, Li Q, McClelland SE, Endesfelder D, Tan P, Tan IB, Richardson AL, Szallasi Z, Swanton C. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011;71:3447–3452. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T. Ueber mehrpolige Mitosen als Mittel zur Analyse des Zellkerns. Vehr d phys med Ges zu Wurzburg, NF. 1902 (available in English translation at: http://8edevbiocom/articlephp?ch=4&id=24) Bd. 35.
- Boveri T. The Origin of Malignant Tumors by Theodor Boveri; Translated by Marcella Boveri. Vol. 1929 Williams and Wilkins; Baltimore: 1914. [Google Scholar]
- Buffin E, Emre D, Karess RE. Flies without a spindle checkpoint. Nat Cell Biol. 2007;9:565–572. doi: 10.1038/ncb1570. [DOI] [PubMed] [Google Scholar]
- Burds AA, Lutum AS, Sorger PK. Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proc Natl Acad Sci U S A. 2005;102:11296–11301. doi: 10.1073/pnas.0505053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N, Domingo E, Kanu N, Dewhurst SM, Gronroos E, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairo LV, Ptak C, Wozniak RW. Mitosis-specific regulation of nuclear transport by the spindle assembly checkpoint protein Mad1p. Mol Cell. 2013;49:109–120. doi: 10.1016/j.molcel.2012.10.017. [DOI] [PubMed] [Google Scholar]
- Campbell MS, Chan GK, Yen TJ. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J Cell Sci. 2001;114:953–963. doi: 10.1242/jcs.114.5.953. [DOI] [PubMed] [Google Scholar]
- Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM. The kinetochore. Cold Spring Harb Perspect Biol. 2014;6:a015826. doi: 10.1101/cshperspect.a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Chesnokova V, Kovacs K, Castro AV, Zonis S, Melmed S. Pituitary hypoplasia in Pttg−/− mice is protective for Rb+/− pituitary tumorigenesis. Mol Endocrinol. 2005;19:2371–2379. doi: 10.1210/me.2005-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi YH, Ward JM, Cheng LI, Yasunaga J, Jeang KT. Spindle assembly checkpoint and p53 deficiencies cooperate for tumorigenesis in mice. Int J Cancer. 2009;124:1483–1489. doi: 10.1002/ijc.24094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E, Zhang X, Xing C, Yu H. Mitotic Checkpoint Regulators Control Insulin Signaling and Metabolic Homeostasis. Cell. 2016 doi: 10.1016/j.cell.2016.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CK, Wu CY, Chen JY, Ng MC, Wang HM, Chen JH, Yuan SS, Tsai EM, Chang JG, Chiu CC. BubR1 Acts as a Promoter in Cellular Motility of Human Oral Squamous Cancer Cells through Regulating MMP-2 and MMP-9. Int J Mol Sci. 2015;16:15104–15117. doi: 10.3390/ijms160715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Cameron LA, Salmon ED. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr Biol. 2004;14:2149–2155. doi: 10.1016/j.cub.2004.11.029. [DOI] [PubMed] [Google Scholar]
- Cimini D, Fioravanti D, Salmon ED, Degrassi F. Merotelic kinetochore orientation versus chromosome mono-orientation in the origin of lagging chromosomes in human primary cells. J Cell Sci. 2002;115:507–515. doi: 10.1242/jcs.115.3.507. [DOI] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Moree B, Canman JC, Salmon ED. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci. 2003;116:4213–4225. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- Cimini D, Wan X, Hirel CB, Salmon ED. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol. 2006;16:1711–1718. doi: 10.1016/j.cub.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Clarke DJ, Segal M, Andrews CA, Rudyak SG, Jensen S, Smith K, Reed SI. S-phase checkpoint controls mitosis via an APC-independent Cdc20p function. Nat Cell Biol. 2003;5:928–935. doi: 10.1038/ncb1046. [DOI] [PubMed] [Google Scholar]
- Coelho PA, Bury L, Shahbazi MN, Liakath-Ali K, Tate PH, Wormald S, Hindley CJ, Huch M, Archer J, Skarnes WC, et al. Over-expression of Plk4 induces centrosome amplification, loss of primary cilia and associated tissue hyperplasia in the mouse. Open Biol. 2015;5:150209. doi: 10.1098/rsob.150209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denu RA, Zasadil LM, Kanugh C, Laffin J, Weaver BA, Burkard ME. Centrosome amplification induces high grade features and is prognostic of worse outcomes in breast cancer. BMC Cancer. 2016;16:47. doi: 10.1186/s12885-016-2083-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Hwang S, O’Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. Elife. 2014;3:e03023. doi: 10.7554/eLife.03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- Dotiwala F, Harrison JC, Jain S, Sugawara N, Haber JE. Mad2 prolongs DNA damage checkpoint arrest caused by a double-strand break via a centromere-dependent mechanism. Curr Biol. 2010;20:328–332. doi: 10.1016/j.cub.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P, Rausch C, Rasnick D, Hehlmann R. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc Natl Acad Sci U S A. 1998;95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Hanlon Newell AE, Bi W, Finegold MJ, Olson SB, Beaudet AL, Grompe M. Aneuploidy as a mechanism for stress-induced liver adaptation. J Clin Invest. 2012;122:3307–3315. doi: 10.1172/JCI64026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertych N, Stolz A, Stenzinger A, Weichert W, Kaulfuß S, Burfeind P, Aigner A, Wordeman L, Bastians H. Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nat Cell Biol. 2014;16:779–791. doi: 10.1038/ncb2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Liu T, Wang X, Yang YM, Deng H, Kunicki J, Traganos F, Darzynkiewicz Z, Lu L, Dai W. BubR1 is involved in regulation of DNA damage responses. Oncogene. 2006 doi: 10.1038/sj.onc.1209392. [DOI] [PubMed] [Google Scholar]
- Fernández-Peralbo MA, Priego-Capote F, Luque de Castro MD, Casado-Adam A, Arjona-Sánchez A, Muñoz-Casares FC. LC-MS/MS quantitative analysis of paclitaxel and its major metabolites in serum, plasma and tissue from women with ovarian cancer after intraperitoneal chemotherapy. J Pharm Biomed Anal. 2014;91:131–137. doi: 10.1016/j.jpba.2013.12.028. [DOI] [PubMed] [Google Scholar]
- Fine RL, Chen J, Balmaceda C, Bruce JN, Huang M, Desai M, Sisti MB, McKhann GM, Goodman RR, Bertino JS, et al. Randomized study of paclitaxel and tamoxifen deposition into human brain tumors: implications for the treatment of metastatic brain tumors. Clin Cancer Res. 2006;12:5770–5776. doi: 10.1158/1078-0432.CCR-05-2356. [DOI] [PubMed] [Google Scholar]
- Foijer F, DiTommaso T, Donati G, Hautaviita K, Xie SZ, Heath E, Smyth I, Watt FM, Sorger PK, Bradley A. Spindle checkpoint deficiency is tolerated by murine epidermal cells but not hair follicle stem cells. Proc Natl Acad Sci U S A. 2013;110:2928–2933. doi: 10.1073/pnas.1217388110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foijer F, Xie SZ, Simon JE, Bakker PL, Conte N, Davis SH, Kregel E, Jonkers J, Bradley A, Sorger PK. Chromosome instability induced by Mps1 and p53 mutation generates aggressive lymphomas exhibiting aneuploidy-induced stress. Proc Natl Acad Sci U S A. 2014;111:13427–13432. doi: 10.1073/pnas.1400892111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frindel E, Malaise E, Tubiana M. Cell proliferation kinetics in five human solid tumors. Cancer. 1968;22:611–620. doi: 10.1002/1097-0142(196809)22:3<611::aid-cncr2820220317>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Fung MK, Han HY, Leung SC, Cheung HW, Cheung AL, Wong YC, Ling MT, Wang X. MAD2 interacts with DNA repair proteins and negatively regulates DNA damage repair. J Mol Biol. 2008;381:24–34. doi: 10.1016/j.jmb.2008.05.080. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe EH, Day CA, Karanjeet KB, Fadness S, Langfald A, Vaughan KT, Dong Z. Chromosome missegregation during anaphase triggers p53 cell cycle arrest through histone H3.3 Ser31 phosphorylation. Nat Cell Biol. 2016;18:668–675. doi: 10.1038/ncb3348. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol. 2002;159:807–819. doi: 10.1083/jcb.200205068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaga Y, Chi YH, Miyazato A, Sheleg S, Haller K, Peloponese JM, Jr, Li Y, Ward JM, Benezra R, Jeang KT. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–166. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- Jamal-Hanjani M, A’Hern R, Birkbak NJ, Gorman P, Grönroos E, Ngang S, Nicola P, Rahman L, Thanopoulou E, Kelly G, et al. Extreme chromosomal instability forecasts improved outcome in ER-negative breast cancer: a prospective validation cohort study from the TACT trial. Ann Oncol. 2015;26:1340–1346. doi: 10.1093/annonc/mdv178. [DOI] [PubMed] [Google Scholar]
- Janssen A, Kops GJ, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci U S A. 2009;106:19108–19113. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol. 2007;179:255–267. doi: 10.1083/jcb.200706015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P, Fowler KJ, Griffiths B, Earle E, Chow CW, Jamsen K, Choo KH. Increased chromosome instability but not cancer predisposition in haploinsufficient Bub3 mice. Genes Chromosomes Cancer. 2005;44:29–36. doi: 10.1002/gcc.20215. [DOI] [PubMed] [Google Scholar]
- Kerr JF, Searle J. A suggested explanation for the paradoxically slow growth rate of basal-cell carcinomas that contain numerous mitotic figures. J Pathol. 1972;107:41–44. doi: 10.1002/path.1711070107. [DOI] [PubMed] [Google Scholar]
- Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouse KA, Wu J, Whittaker CA, Amon A. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc Natl Acad Sci U S A. 2014;111:13409–13414. doi: 10.1073/pnas.1415287111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova NL, Wodarz D. The optimal rate of chromosome loss for the inactivation of tumor suppressor genes in cancer. Proc Natl Acad Sci U S A. 2004;101:7017–7021. doi: 10.1073/pnas.0401943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komlodi-Pasztor E, Sackett D, Wilkerson J, Fojo T. Mitosis is not a key target of microtubule agents in patient tumors. Nat Rev Clin Oncol. 2011;8:244–250. doi: 10.1038/nrclinonc.2010.228. [DOI] [PubMed] [Google Scholar]
- Komlodi-Pasztor E, Sackett DL, Fojo AT. Inhibitors targeting mitosis: tales of how great drugs against a promising target were brought down by a flawed rationale. Clin Cancer Res. 2012;18:51–63. doi: 10.1158/1078-0432.CCR-11-0999. [DOI] [PubMed] [Google Scholar]
- Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci U S A. 2004;101:8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, Weaver BA, Cleveland DW. On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer. 2005;5:773–785. doi: 10.1038/nrc1714. [DOI] [PubMed] [Google Scholar]
- Koshiba H, Hosokawa K, Mori T, Kubo A, Watanabe A, Honjo H. Intravenous paclitaxel is specifically retained in human gynecologic carcinoma tissues in vivo. Int J Gynecol Cancer. 2009;19:484–488. doi: 10.1111/IGC.0b013e3181a130db. [DOI] [PubMed] [Google Scholar]
- Kuffer C, Kuznetsova AY, Storchová Z. Abnormal mitosis triggers p53-dependent cell cycle arrest in human tetraploid cells. Chromosoma. 2013;122:305–318. doi: 10.1007/s00412-013-0414-0. [DOI] [PubMed] [Google Scholar]
- Li M, Fang X, Baker DJ, Guo L, Gao X, Wei Z, Han S, van Deursen JM, Zhang P. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:14188–14193. doi: 10.1073/pnas.1005960107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Fang X, Wei Z, York JP, Zhang P. Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. J Cell Biol. 2009;185:983–994. doi: 10.1083/jcb.200904020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischetti T, Nilsson J. Regulation of mitotic progression by the spindle assembly checkpoint. Mol Cell Oncol. 2015;2:e970484. doi: 10.4161/23723548.2014.970484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Davydenko O, Lampson MA. Polo-like kinase-1 regulates kinetochore-microtubule dynamics and spindle checkpoint silencing. J Cell Biol. 2012;198:491–499. doi: 10.1083/jcb.201205090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000;21:485–495. doi: 10.1093/carcin/21.3.485. [DOI] [PubMed] [Google Scholar]
- Maciejowski J, Li Y, Bosco N, Campbell PJ, de Lange T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell. 2015;163:1641–1654. doi: 10.1016/j.cell.2015.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehara K, Takahashi K, Saitoh S. CENP-A reduction induces a p53-dependent cellular senescence response to protect cells from executing defective mitoses. Mol Cell Biol. 2010;30:2090–2104. doi: 10.1128/MCB.01318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia AR, de Man J, Boon U, Janssen A, Song JY, Omerzu M, Sterrenburg JG, Prinsen MB, Willemsen-Seegers N, de Roos JA, et al. Inhibition of the spindle assembly checkpoint kinase TTK enhances the efficacy of docetaxel in a triple-negative breast cancer model. Ann Oncol. 2015;26:2180–2192. doi: 10.1093/annonc/mdv293. [DOI] [PubMed] [Google Scholar]
- Malureanu L, Jeganathan KB, Jin F, Baker DJ, van Ree JH, Gullon O, Chen Z, Henley JR, van Deursen JM. Cdc20 hypomorphic mice fail to counteract de novo synthesis of cyclin B1 in mitosis. J Cell Biol. 2010;191:313–329. doi: 10.1083/jcb.201003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Michel L, Diaz-Rodriguez E, Narayan G, Hernando E, Murty VV, Benezra R. Complete loss of the tumor suppressor MAD2 causes premature cyclin B degradation and mitotic failure in human somatic cells. Proc Natl Acad Sci U S A. 2004;101:4459–4464. doi: 10.1073/pnas.0306069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VV, Benezra R. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ. The proliferation rate paradox in antimitotic chemotherapy. Mol Biol Cell. 2012;23:1–6. doi: 10.1091/mbc.E10-04-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto T, Porazinski S, Wang H, Borovina A, Ciruna B, Shimizu A, Kajii T, Kikuchi A, Furutani-Seiki M, Matsuura S. Insufficiency of BUBR1, a mitotic spindle checkpoint regulator, causes impaired ciliogenesis in vertebrates. Hum Mol Genet. 2011;20:2058–2070. doi: 10.1093/hmg/ddr090. [DOI] [PubMed] [Google Scholar]
- Mori T, Kinoshita Y, Watanabe A, Yamaguchi T, Hosokawa K, Honjo H. Retention of paclitaxel in cancer cells for 1 week in vivo and in vitro. Cancer Chemother Pharmacol. 2006;58:665–672. doi: 10.1007/s00280-006-0209-6. [DOI] [PubMed] [Google Scholar]
- Mukherjee M, Ge G, Zhang N, Edwards DG, Sumazin P, Sharan SK, Rao PH, Medina D, Pati D. MMTV-Espl1 transgenic mice develop aneuploid, estrogen receptor alpha (ERalpha)-positive mammary adenocarcinomas. Oncogene. 2014;33:5511–5522. doi: 10.1038/onc.2013.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee M, Ge G, Zhang N, Huang E, Nakamura LV, Minor M, Fofanov V, Rao PH, Herron A, Pati D. Separase loss of function cooperates with the loss of p53 in the initiation and progression of T- and B-cell lymphoma, leukemia and aneuploidy in mice. PLoS One. 2011;6:e22167. doi: 10.1371/journal.pone.0022167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Smith S, Kolodner RD. Mitotic checkpoint function in the formation of gross chromosomal rearrangements in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101:15980–15985. doi: 10.1073/pnas.0407010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JM, Macedo JC, Mattingly AJ, Wangsa D, Camps J, Lima V, Gomes AM, Dória S, Ried T, Logarinho E, et al. Chromosome mis-segregation and cytokinesis failure in trisomic human cells. Elife. 2015:4. doi: 10.7554/eLife.05068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampalona J, Roscioli E, Silkworth WT, Bowden B, Genescà A, Tusell L, Cimini D. Chromosome Bridges Maintain Kinetochore-Microtubule Attachment throughout Mitosis and Rarely Break during Anaphase. PLoS One. 2016;11:e0147420. doi: 10.1371/journal.pone.0147420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passerini V, Ozeri-Galai E, de Pagter MS, Donnelly N, Schmalbrock S, Kloosterman WP, Kerem B, Storchová Z. The presence of extra chromosomes leads to genomic instability. Nat Commun. 2016;7:10754. doi: 10.1038/ncomms10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera D, Freire R. Human spindle checkpoint kinase Bub1 is cleaved during apoptosis. Cell Death Differ. 2005;12:827–830. doi: 10.1038/sj.cdd.4401620. [DOI] [PubMed] [Google Scholar]
- Putkey FR, Cramer T, Morphew MK, Silk AD, Johnson RS, McIntosh JR, Cleveland DW. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev Cell. 2002;3:351–365. doi: 10.1016/s1534-5807(02)00255-1. [DOI] [PubMed] [Google Scholar]
- Ricke RM, Jeganathan KB, Malureanu L, Harrison AM, van Deursen JM. Bub1 kinase activity drives error correction and mitotic checkpoint control but not tumor suppression. J Cell Biol. 2012;199:931–949. doi: 10.1083/jcb.201205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke RM, Jeganathan KB, van Deursen JM. Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J Cell Biol. 2011;193:1049–1064. doi: 10.1083/jcb.201012035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke RM, van Ree JH, van Deursen JM. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 2008;24:457–466. doi: 10.1016/j.tig.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Bravo V, Maciejowski J, Corona J, Buch HK, Collin P, Kanemaki MT, Shah JV, Jallepalli PV. Nuclear pores protect genome integrity by assembling a premitotic and mad1-dependent anaphase inhibitor. Cell. 2014;156:1017–1031. doi: 10.1016/j.cell.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschke AV, Stover K, Tonon G, Schäffer AA, Kirsch IR. Stable karyotypes in epithelial cancer cell lines despite high rates of ongoing structural and numerical chromosomal instability. Neoplasia. 2002;4:19–31. doi: 10.1038/sj.neo.7900197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowald K, Mantovan M, Passos J, Buccitelli C, Mardin BR, Korbel JO, Jechlinger M, Sotillo R. Negative Selection and Chromosome Instability Induced by Mad2 Overexpression Delay Breast Cancer but Facilitate Oncogene-Independent Outgrowth. Cell Rep. 2016;15:2679–2691. doi: 10.1016/j.celrep.2016.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roylance R, Endesfelder D, Gorman P, Burrell RA, Sander J, Tomlinson I, Hanby AM, Speirs V, Richardson AL, Birkbak NJ, et al. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol Biomarkers Prev. 2011;20:2183–2194. doi: 10.1158/1055-9965.EPI-11-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge SD, Douglas TA, Nicholson JM, Vila-Casadesús M, Kantzler CL, Wangsa D, Barroso-Vilares M, Kale SD, Logarinho E, Cimini D. Selective advantage of trisomic human cells cultured in non-standard conditions. Sci Rep. 2016;6:22828. doi: 10.1038/srep22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan SD, Britigan EM, Zasadil LM, Witte K, Audhya A, Roopra A, Weaver BA. Up-regulation of the mitotic checkpoint component Mad1 causes chromosomal instability and resistance to microtubule poisons. Proc Natl Acad Sci U S A. 2012;109:E2205–2214. doi: 10.1073/pnas.1201911109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacristan C, Kops GJ. Joined at the hip: kinetochores, microtubules, and spindle assembly checkpoint signaling. Trends Cell Biol. 2015;25:21–28. doi: 10.1016/j.tcb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- Schvartzman JM, Duijf PH, Sotillo R, Coker C, Benezra R. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell. 2011;19:701–714. doi: 10.1016/j.ccr.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle J, Collins DJ, Harmon B, Kerr JF. The spontaneous occurrence of apoptosis in squamous carcinomas of the uterine cervix. Pathology. 1973;5:163–169. doi: 10.3109/00313027309060831. [DOI] [PubMed] [Google Scholar]
- Segal DJ, McCoy EE. Studies on Down’s syndrome in tissue culture. I. Growth rates and protein contents of fibroblast cultures. J Cell Physiol. 1974;83:85–90. doi: 10.1002/jcp.1040830112. [DOI] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 2009;5:e1000705. doi: 10.1371/journal.pgen.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sercin O, Larsimont JC, Karambelas AE, Marthiens V, Moers V, Boeckx B, Le Mercier M, Lambrechts D, Basto R, Blanpain C. Transient PLK4 overexpression accelerates tumorigenesis in p53-deficient epidermis. Nat Cell Biol. 2016;18:100–110. doi: 10.1038/ncb3270. [DOI] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk AD, Zasadil LM, Holland AJ, Vitre B, Cleveland DW, Weaver BA. Chromosome missegregation rate predicts whether aneuploidy will promote or suppress tumors. Proc Natl Acad Sci U S A. 2013;110:E4134–4141. doi: 10.1073/pnas.1317042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Paul R, Mogilner A, Cimini D. Timing of centrosome separation is important for accurate chromosome segregation. Mol Biol Cell. 2012;23:401–411. doi: 10.1091/mbc.E11-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, Benezra R. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–440. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol. 2012;8:608. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto I, Murakami H, Tonami Y, Moriyama A, Nakanishi M. DNA replication checkpoint control mediated by the spindle checkpoint protein Mad2p in fission yeast. J Biol Chem. 2004;279:47372–47378. doi: 10.1074/jbc.M403231200. [DOI] [PubMed] [Google Scholar]
- Sussan TE, Yang A, Li F, Ostrowski MC, Reeves RH. Trisomy represses Apc(Min)-mediated tumours in mouse models of Down’s syndrome. Nature. 2008;451:73–75. doi: 10.1038/nature06446. [DOI] [PubMed] [Google Scholar]
- Symmans WF, Volm MD, Shapiro RL, Perkins AB, Kim AY, Demaria S, Yee HT, McMullen H, Oratz R, Klein P, et al. Paclitaxel-induced apoptosis and mitotic arrest assessed by serial fine-needle aspiration: implications for early prediction of breast cancer response to neoadjuvant treatment. Clin Cancer Res. 2000;6:4610–4617. [PubMed] [Google Scholar]
- Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc Natl Acad Sci U S A. 2011;108:17974–17978. doi: 10.1073/pnas.1109720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn RR, Gonzalez C, Brar GA, Christen S, Carlile TM, Ingolia NT, Sauer U, Weissman JS, Amon A. Aneuploid yeast strains exhibit defects in cell growth and passage through START. Mol Biol Cell. 2013;24:1274–1289. doi: 10.1091/mbc.E12-07-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007a;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007b;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, Ried T. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res. 2004;64:6941–6949. doi: 10.1158/0008-5472.CAN-04-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valind A, Jin Y, Baldetorp B, Gisselsson D. Whole chromosome gain does not in itself confer cancer-like chromosomal instability. Proc Natl Acad Sci U S A. 2013;110:21119–21123. doi: 10.1073/pnas.1311163110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gameren-Oosterom HB, Van Dommelen P, Oudesluys-Murphy AM, Buitendijk SE, Van Buuren S, Van Wouwe JP. Healthy growth in children with Down syndrome. PLoS One. 2012;7:e31079. doi: 10.1371/journal.pone.0031079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitre B, Holland AJ, Kulukian A, Shoshani O, Hirai M, Wang Y, Maldonado M, Cho T, Boubaker J, Swing DA, et al. Chronic centrosome amplification without tumorigenesis. Proc Natl Acad Sci U S A. 2015;112:E6321–6330. doi: 10.1073/pnas.1519388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hansemann D. Ueber asymmetrische Zelltheilung in Epithelkrebsen und deren biologische Bedeutung. Virchow’s Arch Path Anat. 1890;119:299–326. [Google Scholar]
- Wan J, Zhu F, Zasadil LM, Yu J, Wang L, Johnson A, Berthier E, Beebe DJ, Audhya A, Weaver BA. A Golgi-localized pool of the mitotic checkpoint component Mad1 controls integrin secretion and cell migration. Curr Biol. 2014;24:2687–2692. doi: 10.1016/j.cub.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Zheng X, Chen H, Guo Y, Jiang H, He X, Zhu X, Zheng Y. Splicing function of mitotic regulators links R-loop-mediated DNA damage to tumor cell killing. J Cell Biol. 2015;209:235–246. doi: 10.1083/jcb.201409073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Wei JH, Chou YF, Ou YH, Yeh YH, Tyan SW, Sun TP, Shen CY, Shieh SY. TTK/hMps1 participates in the regulation of DNA damage checkpoint response by phosphorylating CHK2 on threonine 68. J Biol Chem. 2005;280:7748–7757. doi: 10.1074/jbc.M410152200. [DOI] [PubMed] [Google Scholar]
- Weinstein GD, Frost P. Cell proliferation in human basal cell carcinoma. Cancer Res. 1970;30:724–728. [PubMed] [Google Scholar]
- Welburn JP, Vleugel M, Liu D, Yates JR, Lampson MA, Fukagawa T, Cheeseman IM. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild T, Larsen MS, Narita T, Schou J, Nilsson J, Choudhary C. The Spindle Assembly Checkpoint Is Not Essential for Viability of Human Cells with Genetically Lowered APC/C Activity. Cell Rep. 2016;14:1829–1840. doi: 10.1016/j.celrep.2016.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Reeves RH. Increased survival following tumorigenesis in Ts65Dn mice that model Down syndrome. Cancer Res. 2011;71:3573–3581. doi: 10.1158/0008-5472.CAN-10-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Wang H, Xu Y, Brinkman KL, Ishiyama H, Wong ST, Xu B. The kinetochore protein Bub1 participates in the DNA damage response. DNA Repair (Amst) 2012;11:185–191. doi: 10.1016/j.dnarep.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YM, Baek KH, Jeong SJ, Shin HJ, Ha GH, Jeon AH, Hwang SG, Chun JS, Lee CW. WD repeat-containing mitotic checkpoint proteins act as transcriptional repressors during interphase. FEBS Lett. 2004;575:23–29. doi: 10.1016/j.febslet.2004.07.089. [DOI] [PubMed] [Google Scholar]
- Yu ZC, Huang YF, Shieh SY. Requirement for human Mps1/TTK in oxidative DNA damage repair and cell survival through MDM2 phosphorylation. Nucleic Acids Res. 2016;44:1133–1150. doi: 10.1093/nar/gkv1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki BI, Suriawinata AA, Eastman AR, Garner KM, Bakhoum SF. Chromosomal instability portends superior response of rectal adenocarcinoma to chemoradiation therapy. Cancer. 2014;120:1733–1742. doi: 10.1002/cncr.28656. [DOI] [PubMed] [Google Scholar]
- Zasadil LM, Andersen KA, Yeum D, Rocque GB, Wilke LG, Tevaarwerk AJ, Raines RT, Burkard ME, Weaver BA. Cytotoxicity of Paclitaxel in Breast Cancer Is due to Chromosome Missegregation on Multipolar Spindles. Sci Transl Med. 2014;6:229ra243. doi: 10.1126/scitranslmed.3007965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasadil LM, Britigan EM, Ryan SD, Kaur C, Guckenberger DJ, Beebe DJ, Moser AR, Weaver BA. High rates of chromosome missegregation suppress tumor progression, but do not inhibit tumor initiation. Mol Biol Cell. 2016 doi: 10.1091/mbc.E15-10-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasadil LM, Britigan EM, Weaver BA. 2n or not 2n: Aneuploidy, polyploidy and chromosomal instability in primary and tumor cells. Semin Cell Dev Biol. 2013;24:370–379. doi: 10.1016/j.semcdb.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Pavelka N, Bradford WD, Rancati G, Li R. Karyotypic determinants of chromosome instability in aneuploid budding yeast. PLoS Genet. 2012;8:e1002719. doi: 10.1371/journal.pgen.1002719. [DOI] [PMC free article] [PubMed] [Google Scholar]