Abstract
The axoneme is the main extracellular part of cilia and flagella in eukaryotes. It consists of a microtubule cytoskeleton, which normally comprises nine doublets. In motile cilia, dynein ATPase motor proteins generate sliding motions between adjacent microtubules, which are integrated into a well-orchestrated beating or rotational motion. In primary cilia, there are a number of sensory proteins functioning on membranes surrounding the axoneme. In both cases, as the study of proteomics has elucidated, hundreds of proteins exist in this compartmentalized biomolecular system. In this article, we review the recent progress of structural studies of the axoneme and its components using electron microscopy and X-ray crystallography, mainly focusing on motile cilia. Structural biology presents snapshots (but not live imaging) of dynamic structural change and gives insights into the force generation mechanism of dynein, ciliary bending mechanism, ciliogenesis, and evolution of the axoneme.
Motile cilia are cell-surface structures that move (e.g., to sweep dirt from the lungs). Recent structural studies have provided insight into the components, dynamics, and evolution of the axoneme—the main internal structure.
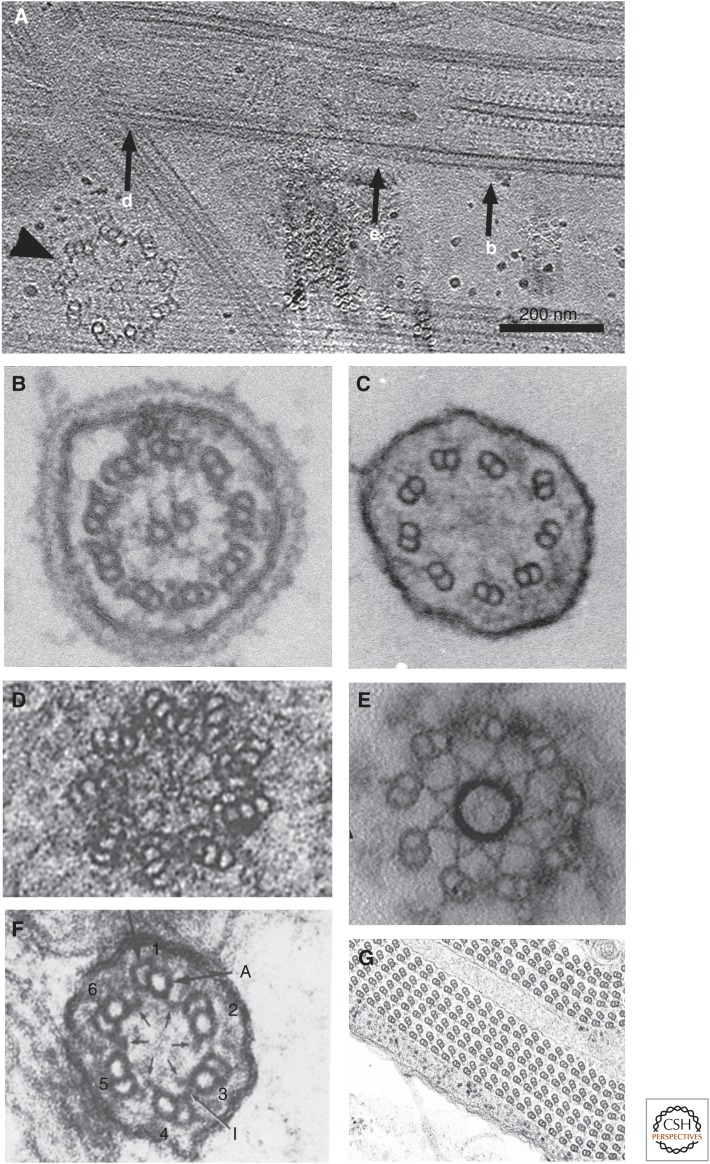
Flagella and cilia are appendage-like organelles in eukaryotic cells. Cilia and flagella (these two terms are used interchangeably) are categorized into two classes by function and structure. One is motile cilia, which have dynein, a family of ATPase motor proteins, and generate motion, whereas primary cilia are nonmotile and play roles in sensory function and transportation. The axoneme is the main part of flagella and cilia and is located outside of the cell body (Fig. 1A–C). The part inside the cell that anchors cilia is called a basal body (Fig. 1D). These two regions are connected by a transition zone (TZ) (Fig. 1E). These three regions are connected continuously with microtubule cytoskeleton—normally as triplet microtubules in the basal body (Fig. 1D) and doublets in the axoneme (Figs. 1B,C, and 2) as well as in the TZ (Fig. 1E). The axoneme consists of microtubule doublets (MTDs) and many other proteins encapsulated in the plasma membrane. The axoneme from most of the species has a 5- to 10-μm length and an ∼300-nm diameter.
Figure 1.
Axoneme structure from various species and locations of cilia. (A) Horizontal section from tomographic reconstruction of ice-embedded cilia including the basal body (centriole) and the transition zone (TZ) (M Hirono and T Ishikawa, unpubl.). A probasal body (a daughter centriole), which is positioned perpendicular to the basal body, is indicated by an arrowhead. (B–G) Electron micrographs of cross sections from plastic embedded cilia. (B) Typical 9+2 structure of the axoneme from Chlamydomonas. (Figure courtesy of Dr. Dennis Diener, Yale University.) (C) 9+0 structure from mouse nodal cilia. (Figure courtesy of Dr. Svetlana Markova, Dr. Dennis Diener, and Prof. Martina Brueckner, Yale University.) (D) Section of a centriole, showing microtubule triplets and the cartwheel (Berns et al. 1977). (E) The TZ from Chlamydomonas flagella. (Image modified from Awata et al. 2014.) (F) Unusual 6+0 structure (Schrevel and Besse 1975). (G) Abnormal axoneme with more than 100 microtubule doublets (MTDs) from the gall-midge fly Asphondylia ruebsaameeni Kertesz (Mencarelli et al. 2000). (Figure courtesy of Prof. Romano Dallai and Prof. Pietro Lupetti, University of Siena.) Corresponding position of cross sections in B, D, and E are indicated in A by b, d, and e, respectively.
Figure 2.
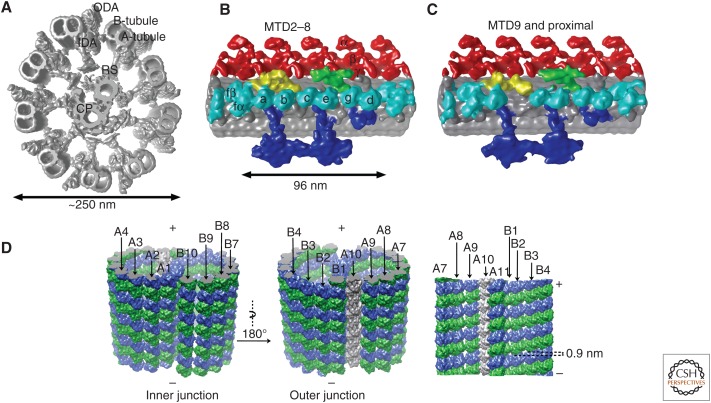
Overall structure of the 96-nm periodic unit from the axoneme. (A) Structure of the “9+2” axoneme based on cryoelectron tomography (cryo-ET). Outer and inner dynein arms (ODA and IDA, respectively), radial spokes (RS), A- and B-tubules from a microtubule doublet (MTD) as well as a central pair (CP) are indicated. (Figure modified from Bui and Ishikawa 2013.) (B,C) Enlarged views of one 96-nm periodic unit of regular MTDs. MTD2–8 in the distal region and MTD9 are shown (Bui et al. 2009, 2012). Dynein isoforms are indicated in B. (Image based on Bui et al. 2012.) MTD2–8 in the proximal region also lack dynein b. Red, ODA; cyan, IDA; purple, adjacent MTD; green, dynein regulatory complex (DRC); yellow, IC/LC (intermediate chain/light chain); blue, RS. Density maps are available as EMD2113-2130. (D) Molecular arrangement of tubulins in an MTD revealed by single particle cryoelectron microscopy (cryo-EM) (Maheshwari et al. 2015). Green, α-tubulin; blue, β-tubulin. Numbering of the protofilaments in A- and B-tubules is identical to that in Figure 3D.
The axoneme of motile cilia is a closed system—when it is isolated from the basal body and the cell body, it can still generate bending motion on addition of ATP. In this sense, eukaryotic flagella/cilia are different from bacterial flagella, which are driven by rotational motion of the base complex (Minamino et al. 2008). Structure and function of axonemes of motile cilia have been studied by genetics, physiology, and biochemistry for decades. Recently, progress in structural biology, genetic engineering, and proteomics has shed light on various new aspects of this organelle.
Because cilia function in various tissues at respiratory tracts, lungs, kidneys, oviducts, sperm, brains, and embryos, defects of cilia cause diverse symptoms such as lung and kidney diseases, male and female infertility, mental disorder, and developmental abnormality. These diseases are called ciliopathy (Fliegauf et al. 2007). Ciliopathy takes place in some cases because of lack of the entire cilia in specific tissues and in other cases because of malfunction of ciliary motion. Therefore, research on both cilia formation (ciliogenesis) and cilia motility can have medical relevance.
More than 600 proteins were detected from the Chlamydomonas axoneme by mass spectroscopy–based proteomics (Pazour et al. 2005). Chlamydomonas is widely studied as a model organism for cilia research, because its protein composition and physiological function and structure have much in common with other species, including humans. Major components of the axoneme in both Chlamydomonas and Tetrahymena are motor proteins, signal transduction proteins, and membrane proteins (Pazour et al. 2005; Smith et al. 2005), whereas components in primary cilia vary, depending on their functions (Liu et al. 2007; Mayer et al. 2009). It has not been established yet which among the more than 600 proteins of motile cilia (Pazour et al. 2005) are indispensable for bending motion. Microtubules and dynein motor proteins play central roles for force generation, driven by ATP-induced conformational change of dyneins (Figs. 3B and 4A). Dyneins are associated with a variety of intermediate chains (ICs), light chains (LCs), and other regulators (see review by King 2016).
Figure 3.
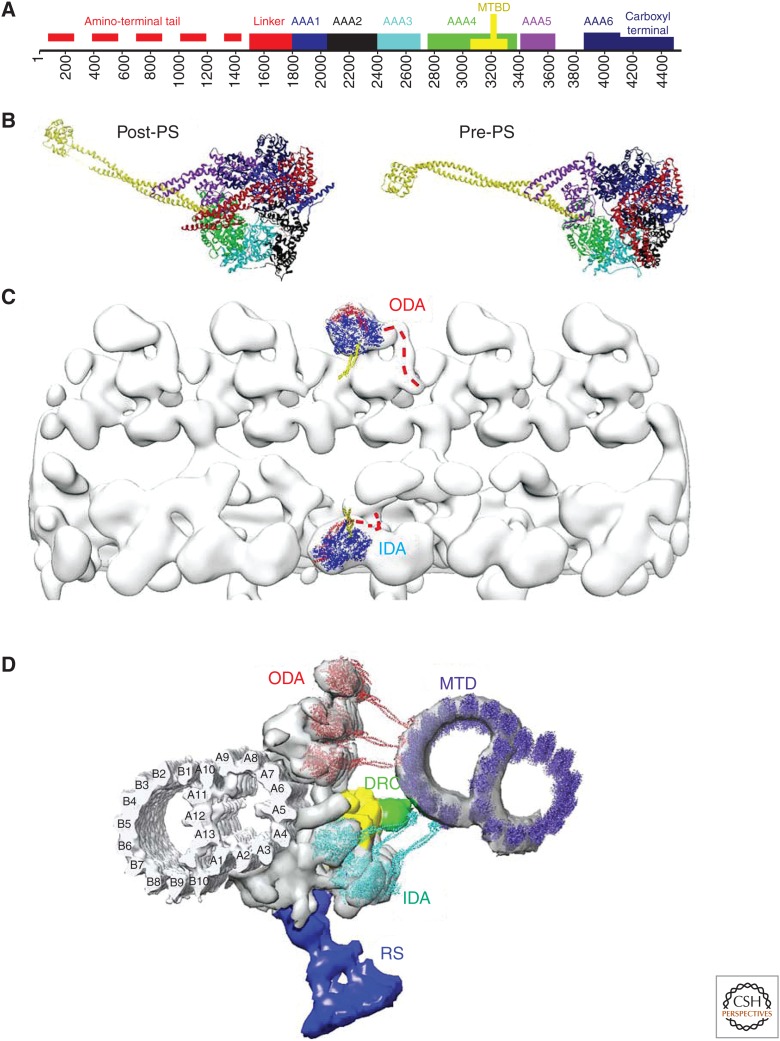
Dynein structure and arrangement in the axoneme. (A) Sequence motif of dynein. (B) Atomic structure of dynein in the post–power stroke (post-PS) conformation (PDB ID: 3VKH) (Kon et al. 2012) and in the pre–power stroke (pre-PS) conformation (4RH7) (Schmidt et al. 2014). In A and B, the color code is as follows: red, amino terminus and linker; blue, AAA1; black, AAA2; cyan, AAA3; green, AAA4; yellow, coiled-coil stalk (which is a part of AAA4); purple, AAA5; dark blue, AAA6 and carboxyl terminus. (C) Atomic models (3VKH) fitted to cryoelectron tomography (cryo-ET) structure (EMDB ID: 2117) (Bui et al. 2012) to show the front and back of dyneins in outer dynein arms (ODAs) and inner dynein arms (IDAs), respectively. The linker (red) is above the AAA ring (blue) in ODAs, whereas it is below the ring in IDAs. Yellow, stalks. (D) Atomic models in which density maps from cryo-ET (Bui et al. 2012) and single-particle analysis (ciliary microtubule doublet [MTD)] (Maheshwari et al. 2015) are fitted. Red, ODA; cyan, IDA; purple, adjacent MTD; green, dynein regulatory complex (DRC); yellow, intermediate and light chains (IC/LC); blue, radial spoke (RS).
Figure 4.
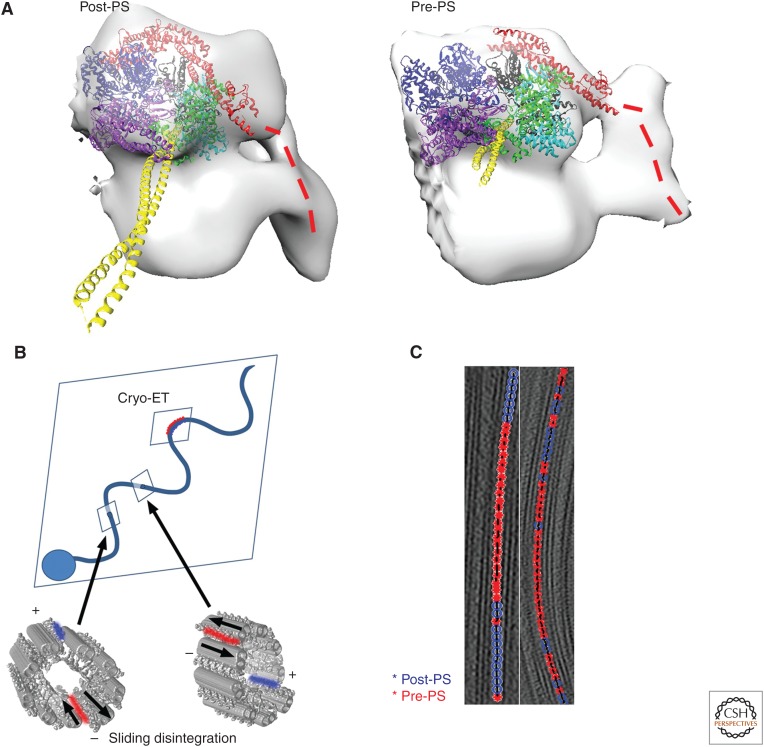
Dynein structural change induced by nucleotides. (A) Atomic model fitting to cryoelectron tomography (cryo-ET) (Ueno et al. 2014). The overall orientation of the linker at pre–power stroke (pre-PS) and post–power stroke (post-PS) condition (red) is the same as crystallography (Fig. 3B). The color code is the same as in Figure 3A,B. (B) Switching models based on sliding disintegration (Hayashi and Shingyoji 2008) and cryo-ET (Lin et al. 2014) of sea urchin sperm. Sliding disintegration occurs by sliding between two adjacent microtubule doublets (MTDs) (red in the surface-rendered models of two axonemes), whereas dyneins at the opposite side of the axoneme are relaxed (blue). In intact cilia, straight areas between principle and reverse bending (indicated by arrows) should be where sliding takes place. If sliding moves toward the opposite directions at these two sites, bending can happen. The sliding plane is perpendicular to the bending plane—switching occurs between the upper and lower sides of the bending plane. However, switching was found at the area of curvature by cryo-ET (Lin et al. 2014). Image classification of outer arm dynein structure showed localization of two dynein conformations at external and internal sides of curved cilia, suggesting switch within the bending plane. (C) Distribution of structure of outer dyneins along MTDs in the axoneme in the presence of the nonhydrolyzable ATP analog, ADP.Vi, show clustering, in which 10 to 20 dyneins in the post-PS conformation make a row (blue), followed by a row of dyneins in the pre-PS conformation (red). Under the same ADP.Vi concentration, almost all the dyneins turn to the pre-PS conformation without coexistence of multiple forms. This suggests cooperative conformational change of neighboring dyneins in the axoneme (Movassagh et al. 2010).
Although one species of cytoplasmic dynein recognizes a variety of cargos with the help of adaptors, in cilia there are a number of axonemal dynein isoforms, similar to the case of kinesin. Axonemal dyneins form two complexes, inner and outer dynein arms (IDAs and ODAs, respectively), on MTDs. ODAs form either a heterodimer or heterotrimer. Although the functional difference between an ODA with dimer and ODA with trimer is not known, species with dimers are mostly unikont with calaxin as an associated calcium sensor and species with trimers are mostly bikont with LC4 as a sensor (Inaba 2015), suggesting their diversification during evolution. In Chlamydomonas, all the axonemal dyneins regularly functioning in cilia are identified and named as α, β, γ (ODAs) or a, b, c, d, e, fα, fβ, and g (IDAs) (Fig. 2B) (Yagi et al. 2009). They have different characteristics revealed by in vitro motility assay, such as velocity, ATPase, and duty ratio (Kagami and Kamiya 1992; Kamiya 2002). As described in detail later, all the species of dynein have fixed loci in the 96-nm periodic unit (Bui et al. 2008, 2012). It is not known whether they need to be in the proper positions to orchestrate bending motion in a designed manner or whether they only need to be on MTDs at high density to generate intense force. Judging from the beating of a deletion mutant, which lacks an entire ODA, with smaller amplitude but normal waveform (Kagami and Kamiya 1992), an ODA is an accelerator and not essential for waveform determination. Another important protein complex is the radial spoke (RS), which protrudes from an MTD toward the central pair (CP) singlet microtubules. In motile cilia, mutants lacking the RS (Ebersold et al. 1962; Huang et al. 1981; Yang et al. 2004) or CP (Starling and Randall 1971) are paralyzed under physiological beating conditions, and many IDA mutants have abnormal beating (summarized in Kamiya 2002), suggesting a signal transduction pathway from the CP and RS to the IDAs for bending motion. However, RS/CP mutants can be activated for motion in the presence of high ADP concentration, indicating an alternative pathway, independent of RS/CP (Kamiya and Yagi 2014). Two pathways, one by CP/RS/IDA and the other by ODA, could either make the system robust by redundancy or enable finer regulation.
“9+2”
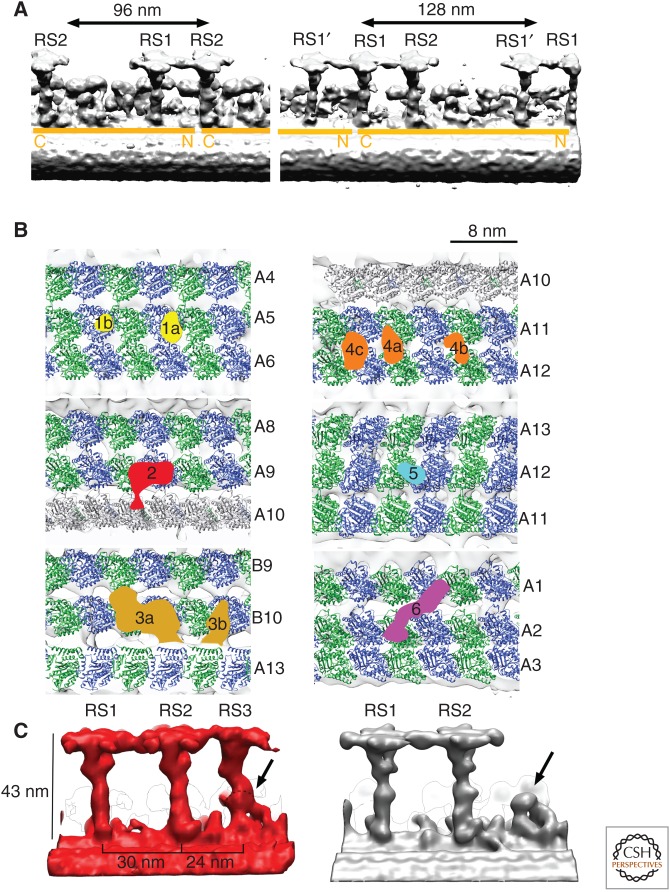
Most species—from the green algae Chlamydomonas to humans—share a similar ultrastructure of axoneme in motile cilia. Nine MTDs surround two singlet microtubules (the CP). This common architecture is called the “9+2” axonemal structure (Figs. 1B and 2A). An MTD consists of one complete cylindrical microtubule (A-tubule) and one incomplete tubule (B-tubule) attached on the A-tubule (Figs. 2A and 3D) and extends from a triplet microtubule in the basal body (centriole). Adjacent MTDs are linked by dyneins and dynein regulatory complex (DRC) (nexin), whereas RSs are between the MTD and the CP (Figs. 2A and 3D). Dyneins, RSs, and the DRC form a regular 96-nm periodicity along MTDs. This periodic length is determined by two coiled-coil proteins, FAP59 and FAP172, proved by extension of the periodicity induced by elongation of these proteins (Fig. 5A) (Oda et al. 2014a). Nine MTDs show pseudo ninefold symmetry, but not exact symmetry, as mentioned above (see the section Uneven Distribution of Axonemal Dynein Isoforms). Differently from cytoplasmic dynein, there are a number of axonemal dynein isoforms. These dyneins are composed of ODAs and IDAs (Fig. 2B). Within one 96-nm periodic unit, there are four ODAs with 24-nm spacing (Fig. 2B). One IDA consists of eight axonemal dynein molecules, whereas one ODA is either heterodimer or heterotrimer. Each proximal end of the axoneme (1 µm or less) has no dynein or RS.
Figure 5.
Rulers, microtubule inner proteins (MIPs), and radial spokes (RSs). (A) Location of FAP59 and FAP172, two coiled-coil proteins extending along the microtubule doublet (MTD) as proved by genetically tagged labels, which determine the periodic length, 96 nm. When amino-terminal subdomains of these proteins are duplicated by genetic engineering, the MTD has a 128-nm periodicity, allowing the third RS. Surface-rendered from EMD6108 and EMD6115 (Oda et al. 2014a). (B) Unidentified proteins binding to the inside of MTDs. Most of these MIPs bind two or more protofilaments. (From Maheshwari et al. 2015; reproduced, with permission, from the authors.) (C) Comparison of structure of RSs from Tetrahymena (left) and Chlamydomonas (right). The common structural motif at the base of RS3 is shown by arrows. (Modified from Pigino et al. 2011.)
MOTILE CILIA WITH EXCEPTIONAL STRUCTURE
There are also exceptional motile cilia. Nodal cilia and Anguilla (eel) sperm (Woolley 1997) are both motile, but lack the RS and CP (“9+0” with dynein) (Fig. 1C). Their conical or screw-like motion (Woolley 1998), instead of planar beating, likely originates from this “9+0” structure. Monogenean Pseudodactylogyrus sp. has “9+1” with nine MTDs, an RS, and, instead of a CP, one central singlet microtubule (Mollaret and Justine 1997). Although they are rare, there are even axonemes with different numbers of MTDs. Motile flagella from gametes of the gregarine Lecdina tuzetae and Deplauxis hatti have 6+0 and 3+0 structure, respectively (Fig. 1F) (Schrevel and Besse 1975; Prensier et al. 1980). Flagella from some insect sperms have large numbers of MTDs, such as Cecidomyiinae Monarthoropalpus flavus, in which more than 100 MTDs form laminae, instead of cylinders (Fig. 1G). They have only ODAs, which are structurally similar to those ODAs in “9+2” axonemes (Lupetti et al. 2005). Details of such insect sperm axonemes are reviewed in Dallai (2014). Except for these unusual flagella, all axonemes have nine MTDs. Nonmotile primary cilia have the “9+0” structure, without a CP, an RS, and dyneins. 3D reconstruction of primary cilia was done by cryoelectron tomography (cryo-ET) (Gilliam et al. 2012; Doroquez et al. 2014), but periodicity and other structural detail are still to be investigated. In this review, we will focus on the 3D structure of the “9+2” axoneme from motile cilia and flagella.
It is unclear why most species share the axonemal structure with nine doublets. Because nine MTDs stem from nine triplets in the basal body, ninefold symmetry is likely determined by centriolar structure, rather than ciliary structure. The mechanism to maintain the ninefold symmetry of centriole is still to be investigated, but it is surely linked to the SAS-6 protein, which builds a ninefold cartwheel in the centriole and also can form a 9-mer in vitro (Kitagawa et al. 2011; van Breugel et al. 2011). In Chlamydomonas bld12 mutants, which are composed of eight to 12 MTDs and have abnormal motility, the CP is often missing in axomenes with eight MTDs, whereas two pairs of CPs are found in axonemes with more than nine MTDs, indicating that the CP is formed properly in an optimum space (Nakazawa et al. 2014). Although the diameter of the axoneme increases proportionally to the number of MTDs, the distance between MTDs is regular regardless of the number of MTDs (Nakazawa et al. 2014). This suggests that the proteins that contribute to the maintenance of axoneme structure, such as dyneins, RSs, and DRC proteins, are tuned to be accommodated in the “9+2” axoneme during evolution.
METHODS OF STUDY
Similar to studies of other cellular ultrastructures, the axoneme has been investigated by transmission electron microscopy (TEM), resin embedding and staining, freeze substitution of high-pressure frozen specimen, or freeze-fracture deep etch. These classical specimen preparations provide micrographs with high contrast, with which single-dynein molecules can be observed directly and many fine details of the axoneme can be revealed, such as arrangement of dyneins (Avolio et al. 1984; Goodenough and Heuser 1985a), RSs (Warner and Satir 1974; Goodenough and Heuser 1985b), and the DRC (Gardner et al. 1994). By these methods, periodicity of the axoneme, the T shape of the RS (Goodenough and Heuser 1985b), and dynein structure with a head and a tail domain (Goodenough and Heuser 1984) were visualized. By combining these classical techniques with image analysis, arrangement of dynein heavy chains in ODAs (Lupetti et al. 2005) and IDAs (Mastronarde et al. 1992) was modeled, which is consistent with our knowledge today. However, recent progress of cryo-ET brought a new scope to cilia studies. It enables 3D reconstruction, imaging conformation of individual molecules, and intermolecular network. By comparing deletion mutant structures, labeling by genetic tagging to add extra density to target molecules, their position can be located. Details of this method for the axoneme are described and findings revealed by this method are reviewed elsewhere (Bui and Ishikawa 2013; Ishikawa 2015).
One major reason for the enormous success of cryo-ET of the axoneme is its periodicity. To overcome the disadvantage of cryo-ET (i.e., poor signal-to-noise ratio [S/N]) and visualize molecules, many volumes, including the identical molecule with the same conformation (called subtomograms), must be computationally extracted, aligned three-dimensionally, and averaged. Structure along MTDs can be extracted and aligned, using 96-nm periodicity and ninefold symmetry, and is thus suitable for subtomogram averaging from cryotomography. Although another part of the cilia with less symmetry could also be a target to extract and average, until now successful 3D structural analysis is limited to exceptionally long stacks (therefore with periodicity) of centriolar SAS-6 cartwheels in Trichonympha (Guichard et al. 2012).
A few component molecules, such as dynein and tubulin, are solved at atomic resolution by X-ray and electron crystallography, respectively (Nogales et al. 1998; Kon et al. 2012; Schmidt et al. 2012). Dyneins and MTDs were analyzed in vitro at intermediate resolution by single-particle cryoelectron microscopy (cryo-EM) analysis (Roberts et al. 2012, 2009; Maheshwari et al. 2015). Although the spatial resolution of in vivo cryo-ET even after subtomogram averaging is moderate (∼25 Å), the axoneme could be studied in greater molecular detail by fitting high-resolution in vitro structure to the in vivo structure by tomography (Roberts et al. 2012; Maheshwari et al. 2015).
OVERALL STRUCTURE OF THE “9+2” AXONEME
The triplet of the basal body consists of one complete microtubule (A-tubule) and two incomplete microtubules (B- and C-tubules) (Fig. 1D). The axonemal MTDs are extensions of A- and B-tubules of the basal body. Loss of the C-tubule happens at 300–500 nm from the proximal end of the basal body. However, the orientations of A- and B-tubules are slightly different between the doublet in the axoneme (Fig. 1B,C) and the triplet in the basal body (Fig. 1D). The factor to cause this twist has not been identified yet. The TZ, in which a characteristic star-shaped object (Diener et al. 2015) connects inside of MTDs, is located on an MTD (Fig. 1E) at the area close to the interface to the triplet. The point on the MTD where dyneins, RSs, and the CP start is located distal to the TZ. The structure of the area in between has not been analyzed. Axonemal dyneins, RSs, and DRCs are arranged with 96-nm periodicity (Mastronarde et al. 1992). Within this repeating unit, one set of IDAs, which contains eight inner arm dynein heavy chains, and four ODAs with 24-nm spacing (in which each ODA consists of two or three heavy chains), as well as ICs and LCs associated with the heavy chains, are found.
MTD STRUCTURE
The A-tubule of an MTD consists of 13 protofilaments, which are parallel to the axis of the tubule without twist, similar to the reconstituted 13-protofilament microtubule (Chrétien and Wade 1991). The arrangement of α- and β-tubulins in the A-tubule is also the same as the reconstituted microtubule—tubulin dimers form a left-handed stagger with one helical discontinuity between two adjacent protofilaments, called a B-lattice. Helical discontinuity occurs at the external side of the axoneme (Fig. 2D) (Maheshwari et al. 2015). The B-tubule is slightly larger in diameter than the A-tubule, corresponding to a 15-protofilament microtubule. However, although a 15-protofilament microtubule reconstituted from tubulin has helical twist (Chrétien and Wade 1991; Ray et al. 1993), protofilaments of the B-tubule are positioned straight—parallel to the axis of the MTD. This suggests a different mechanism of microtubule formation or stabilization of the MTD from reconstituted microtubules. Because this special structure is maintained after the removal of all the major proteins, such as dynein, RSs, and the DRC, proteins located inside an MTD may play an essential role for the tubulin arrangement in MTDs.
A number of proteins are found inside MTDs (Sui and Downing 2006). Many of them are common among nine MTDs from various species (Pigino et al. 2012), although some of them are specific. MTDs 1, 5, and 9 have a large complex named a beak, which occupies the inside of the B-tubule. Although its protein composition is not known, interestingly the beaks in MTD5 and MTD6 are missing in mbo-1, a backward swimming mutant (Segal et al. 1984). More inside proteins were found by cryo-EM tomography (Nicastro et al. 2011) and single-particle analysis (Fig. 4B) (Maheshwari et al. 2015). Some proteins inside MTDs have been identified and functionally characterized. Rib72, an EF-hand protein, was located in the region between the A- and B-tubules, called the ribbon region (Ikeda et al. 2003). The ribbon binds tektin, which forms a filament, as well as other EF-hand proteins, Rib74 and Rib85.5 (Linck et al. 2014). Other structures are not biochemically characterized yet. They connect adjacent protofilaments of A- and B-tubules (Fig. 5B), which could enable them to bundle protofilaments during the bending motion and keep their lattice arrangements different from those of reconstituted microtubules (Maheshwari et al. 2015).
AXONEMAL DYNEIN STRUCTURE
Dynein is a gigantic ATP-driven motor protein family with ∼4500 amino acids (Fig. 3A). The catalytic domain of dynein consists of ∼3000 amino acids, forming six AAA domains (Neuwald et al. 1999). A tail region with ∼1500 amino acids is at the amino terminus and responsible for cargo binding in cytoplasmic dynein and permanent (independent of ATP hydrolysis) binding to the A-tubule. Sequences of amino-terminal tail domains of dynein isoforms are diverse. Amino-terminal tails of axonemal dynein could be essential to determine the loci of dyneins, conformation of dynein arms, and connection between dyneins, RSs, and the DRC. There are two textbooks on dynein, edited by King (2011) and by Amos and Hirose (2012).
Among the six AAA domains of cytoplasmic dynein, one AAA domain (AAA1) is capable of ATP hydrolysis and three (AAA2, AAA3, and AAA4) bind nucleotides, whereas AAA5 and AAA6 do not have p-loops and therefore do not bind nucleotides. AAA3 hydrolyzes ATP (but less than AAA1) and is thought to be regulatory (Takahashi et al. 2004). AAA4 has a long coiled-coil protrusion called a stalk. The microtubule-binding domain (MTBD) at the tip of the stalk is responsible for ATP-dependent interaction to the microtubule (in the case of axonemal dynein, the B-tubule of the adjacent MTD). Atomic structures of the catalytic domain of cytoplasmic dynein in the absence of nucleotides (Schmidt et al. 2012), in the presence of ADP (Kon et al. 2012), and in the presence of ADP.Vi, which mimics the ADP.Pi state (Schmidt et al. 2014), are available. The first two are in the post-PS structure, whereas the ADP.Vi structure is in the pre-PS structure (Shimizu and Johnson 1983; Johnson 1985). Between the amino-terminal tail and AAA1 is an α-helix-rich linker domain (Carter et al. 2011; Kon et al. 2011). In the post-PS structure, the linker domain stems from AAA1 and extends across the ring to dock on AAA4 in the ADP state (Kon et al. 2012) and on AAA5 in the apo state (Schmidt et al. 2012). In the pre-PS state, the linker kinks and bridges AAA1 and AAA2 (Schmidt et al. 2014).
Although no atomic structure of axonemal dynein has been solved, based on high homology with the sequence of catalytic domains of cytoplasmic and axonemal dyneins (47.7% homology, 26.4% identity) (Paschal et al. 1992), we assume that axonemal dynein has a structure similar to that of cytoplasmic dynein. Indeed, the cryo-EM structure of the inner arm dynein c at ∼20 Å resolution and its nucleotide-induced change are consistent with the high-resolution structure of cytoplasmic dynein (Roberts et al. 2009, 2012, 2013). On that basis, we can model axonemal dynein heads by fitting atomic models of cytoplasmic dynein to the density map of the axoneme from tomography (Lin et al. 2014; Ueno et al. 2014).
Orientation of dyneins in both ODAs and IDAs is similar along the longitudinal direction. In the 3D structure reconstructed from cryo-ET, the amino-terminal tail extends from the AAA ring toward the direction of the distal end of the axoneme, which corresponds to the plus end of the microtubule (Fig. 2B) (Bui et al. 2008). The stalks from two or three outer arm dyneins also show nearly the same orientation—downward when seen from the adjacent MTD with the proximal end left, but tilted ∼35° toward the proximal direction (yellow in Fig. 3C) (Bui et al. 2009, 2008), both with and without nucleotides. By fitting the atomic model of cytoplasmic dynein to the ODAs in the tomogram, the linker domain turns out to be above the AAA ring—the linker is located at the opposite side of the ring from the A-tubule, and the amino-terminal tail must bend and wrap one AAA domain to extend toward the A-tubule (red dotted line in Fig. 3C). Conformational change of axonemal dyneins induced by nucleotides also shows high coincidence with crystallography of cytoplasmic dynein. The linker extends from AAA1 toward AAA2 in the presence of ADP.Vi (Ueno et al. 2014). The orientation of the inner dynein heads is different from that of the outer dynein heads. Although inner dyneins have the amino termini extending toward the tip of cilia, similar to outer dyneins, the orientation of the stalk should be upward in Figure 3C to interact with the adjacent MTD (yellow in Fig. 3C). This means that the orientation of the inner dynein head is “flipped” with the linker down toward the A-tubule. Thus, the amino terminus should extend toward the A-tubule without wrapping the AAA domain (red dotted line in Fig. 3C).
DYNEIN STRUCTURAL CHANGE IN THE AXONEME AND BENDING MECHANISM
How are the conformational changes of the axonemal dyneins integrated into ciliary bending motion? This ultimate question will be decomposed into a few questions. How is the conformational change of each dynein in the axoneme? Is dynein activity in the axoneme correlated to the geometrical bending pattern? Do the dimeric axonemal dyneins (OADs and IADs) behave similarly to dimeric cytoplasmic dynein?
The switching model (switch model hypothesis)—in which one side of curved cilia has dyneins in one state of the force generation cycle and dyneins in the other side of the curved cilia are in another state to cause or maintain asymmetry and the two sides switch when the curvature flips—was proposed based on negative stain of cilia (Sale and Satir 1977; reviewed in Satir et al. 2014). The switching model was formulated by simulation (Sugino and Naitoh 1982) and is consistent with sliding disintegration studies, in which MTDs from enzymatically treated (i.e., the linkage between MTDs is disconnected) axonemes slide to split the axoneme into two parts (each consists of three to six MTDs) at the splitting plane perpendicular to the bending plane (arrows in Fig. 4B) (Hayashi and Shingyoji 2008). The direction of sliding disintegration indicates that dyneins on one MTD slide past each other (in Fig. 4B, MTDs at the red area in the surface model of the axonemes), whereas dyneins on the other side of the axoneme do not (in Fig. 4B, MTDs at the blue area). These MTDs are located on the same plane as the CP, suggesting the role of RS/CP for switching. On the contrary, cryo-ET showed outer arm dyneins along MTDs at the external side of a curved part of native sea urchin sperm flagella are in one conformation, whereas dyneins at the internal side of curvature are in the other conformation (Fig. 4B, indicated by “cryo-ET”) (Lin et al. 2014). Switching of sliding should take place in the straight area of bending cilia (indicated by arrows in Fig. 4B) in the model based on sliding disintegration, whereas in cryo-EM analysis conformational heterogeneity appeared at the area with curvature (Fig. 4C), suggesting these two experiments may observe different phenomena.
In the switching model, dynein activity must be suppressed locally. Indeed, ATPase activity of dynein in the axoneme is approximately five times lower than expected from microtubule-activated dynein ATPase (Maheshwari and Ishikawa 2012). Localization of suppression is supposed to occur either by a well-organized program (including 3D positioning of RS/CP) or by a spontaneous self-organization of dyneins; it could be the combination of both. Cryo-ET showed cooperative behavior of outer arm dyneins on MTDs—dynein conformations have a tendency to cluster, with 10 to 20 dyneins showing the same form in a row (Fig. 4C) (Movassagh et al. 2010). Dynein may sense mechanical strain from the conformational change of adjacent dyneins. This hypothesis is also consistent with force-triggered ciliary oscillation by a microneedle (Ishikawa and Shingyoji 2007).
Although the switching hypothesis presents a straightforward model to correlate molecular function of dynein and physiological bending, it must be examined from mechanical and kinetical aspects. Can outer and inner dyneins make as frequent ATPase as ciliary beating? ATPase turnover of microtubule-activated axonemal dyneins is two to five ATP hydrolyses per one dynein head (e.g., Shimizu et al. 1992, 2014). This means one dynein molecule needs ∼300 msec for turn over on average. Because frequency of ciliary beating is ∼50 Hz in wild-type and ∼25 Hz in ODA depleted oda mutants (Kamiya and Okamoto 1985), the ATPase cycle of axonemal dynein is too slow to make the transition from one curvature to the flipped curvature—it is unlikely that dyneins can turn from the pre-PS form to the post-PS form and come back to the pre-PS form in 20 msec, unless unknown factors drastically reduce the energy barrier to accelerate the ATPase cycle (however, it was shown that ATPase activity in the axoneme is not higher than in vitro) (Maheshwari and Ishikawa 2012). This suggests that, even if switching of MTD sliding is the mechanism of bending, it is not as simple as was thought. The hypothesis attributing the bending mechanism to asymmetrical distribution of conformations of dyneins is still attractive. However, dynein states in the real functional cilia will be more stochastic than the simplified switching of an entire row of dyneins.
UNEVEN DISTRIBUTION OF AXONEMAL DYNEIN ISOFORMS
It has been known that one of the nine MTDs lacks an outer arm in Chlamydomonas. The MTD without the ODA is numbered as MTD1 and is apposed (i.e., MTD1s of two cilia in one Chlamydomonas cell face each other) (Hoops and Witman 1983). Detailed analysis of Chlamydomonas flagella by cryo-ET proved that some inner dynein species are missing in parts of the axoneme (Bui et al. 2009). MTD1 and MTD9 lack dynein b. In MTD1, dynein c and e are also missing or fold abnormally (Bui et al. 2009). This indicates weaker sliding force at the inner side of the axoneme, suggesting that the mechanism generates an asymmetrical waveform. Dynein b is also missing in all of the MTDs in the proximal region (Bui et al. 2012), showing not only circumvention but also longitudinal asymmetry (Fig. 2B,C).
As immunolabeling and electron tomography showed recently, some dyneins are localized either in a proximal or a distal part of the axoneme. In Chlamydomonas, three dynein heavy chains, DHC3, DHC4, and DHC11, are localized at the proximal region (<∼2 μm from the basal body) (Yagi et al. 2009). They are called minor dyneins and replace major dyneins in the 96-nm unit—for example, DHC11 replaces dynein d (Bui et al. 2012).
In human cilia, replacement occurs in ODAs. DNAH5 is a homolog of Chlamydomonas dynein β and one cause of programmed cell death (PCD). It exists along the entire length of the cilia, whereas DNAH9 replaces DNAH5 partially but only at the distal region (Fliegauf et al. 2005); another outer arm dynein DNAH11 is localized at the proximal region (Dougherty et al. 2016). The mechanism and the meaning of the asymmetric distribution of dynein isoforms must be investigated further.
DYNEIN IC/LC
Although all the species of IADs and OADs are associated by ICs and LCs, structurally the most prominent and best analyzed are those of inner dynein f. ICs and LCs associated with the dynein f dimer form a large complex (40 nm in length along the MTD, 10 nm in width; yellow in Fig. 2B) and bridge the ODAs (by IC140 and IC2), dynein f, and other IDAs (by IC138) (Heuser et al. 2012; Oda and Kikkawa 2013), and the connection extends further to the DRC (by the modifier of inner arms [MIA] complex) (Yamamoto et al. 2013) and RSs (Fig. 2B).
RSs AND THE CP
The repeating unit contains two or three RSs and one DRC. Each RS has a T shape and is of ∼43 nm in height with a stalk and a head facing the CP. The first (RS1) and the second (RS2) are structurally similar, although not identical, suggesting similar components (Fig. 5C). RS1 and RS2 have branches between the stalk and the head (called a neck) (Pigino et al. 2011). At least 23 component proteins are identified from Chlamydomonas flagella (Yang et al. 2006). Comparison of the structures of deletion mutants enabled us to locate RSP1, 4, 6, 9, and 10 in the head, RSP2, 16, and 23 in the neck, and the rest in the stalk (Pigino et al. 2011). The head and the neck domains show pseudo twofold symmetry. It was an unexpected observation that symmetrical RS heads interact with the CP, which has clear polarity. Pseudo twofold symmetry can be explained as the sign of two preassembled subcomplexes proposed as two 12S complexes to be assembled into one 20S complex based on the L-shaped 12S complex from cytoplasm, whereas the entire RS is 20S (Diener et al. 2011). However, the assignment of RS head proteins by genetic tagging does not support the idea of twofold symmetrical arrangement of RS head proteins—RSP4 is located opposite from RSP6 (Oda et al. 2014c). When there are three RSs, the third RS (RS3) has a different structure than RS1 and RS2. Chlamydomonas, which has two RSs, has a short protrusion, which is at the same locus as RS3 and similar in shape to RS3. This suggests an evolutional origin of this RS3-like protrusion—either growth stopped or, once completed, RS3 was degenerated. Furthermore, mutants, which cause loss of RS1 or RS2, do not lose RS3. This indicates distinct protein components of RS1/2 and RS3 (Pigino et al. 2011; Pigino and Ishikawa 2012). The bases of RSs are connected to the tails of dyneins (Fig. 5C).
The CP consists of two singlet microtubules, composed of two sets of 13 protofilaments and various binding proteins with 32-nm periodicity. These decorating proteins form protrusions from the microtubules and linkers between two tubules (reviewed in Mitchell 2016). The CP seems straight in sperm flagella. This leads us to hypothesize that the planar bending motion of the “9+2” axoneme is defined by the CP. However, in Chlamydomonas and Tetrahymena, the CP twists shallowly (Mitchell and Nakatsugawa 2004; Pigino et al. 2012), although Chlamydomonas and Tetrahymena have planar waveforms. This hypothesis is still supported by the fact that the “9+0” axoneme from nodal cilia and eel sperm flagella makes a screw-like conical motion, instead of planar beating. The “6+0” axoneme from Gregarine (Schrevel and Besse 1975) makes a beating, but not planar, motion. Maybe the CP affects the waveform but with other influential factors. This mechanism must be further explored.
Interaction between RS heads and the CP will be central for ciliary regulation. Geometrically, there is a gap between them and it is difficult to build a model of direct protein interaction between RSs and the CP, taking twisting CP and RSs on the straight MTD into account. In addition, there is no predictable signal transduction subdomain in five RS head proteins. Lack of chemical interaction and ellipsoidal sectional view of the CP leads us to build a hypothesis that mechanical pressure from the CP to the RS head induces regulation on the RS. A paralyzed mutant lacking one protrusion on the CP toward the RS head was rescued by recovering the volume with a genetically tagged (but completely different) protein from the CP protrusion on RS head proteins, strongly supporting this “mechanical interaction” hypothesis (Oda et al. 2014c).
LINKERS BETWEEN MTDs
There are linkers connecting adjacent MTDs in Chlamydomonas flagella. The DRC connects all nine adjacent pairs of MTDs. The DRC consists of at least 11 proteins (Bower et al. 2013) building bilobed structure (Heuser et al. 2009). Among them, DRC1, DRC2, and DRC4 are likely bundled coiled-coil proteins with the amino termini toward the adjacent B-tubule and the carboxyl termini anchored on the A-tubule (Oda et al. 2014b). Two more linkers between limited pairs of MTDs were found (Bui et al. 2009). IDL2 extends from near IC/LC of dynein f of MTDs 4, 5, and 9 to MTDs 5, 6, and 1. IDL3 exists only between MTD1 and MTD2 and is located close to the DRC.
Functions of these linkers are not known yet. Currently, it is unclear whether these linkers detach from MTDs during ciliary bending, which causes sliding of adjacent MTDs, or whether they keep binding and stretch. In the case when they keep bundling the adjacent MTDs, they might determine waveforms. IDL2 and IDL3 could limit sliding of MTDs 4–6 and 9–2. It could be the mechanism to limit ciliary motion planar (Bui et al. 2009).
CONCLUDING REMARKS
In this article, we reviewed what we have learned about ciliary axonemes through recent progress in structural biology, X-ray crystallography, cryo-EM single-particle analysis, cryo-ET, and 3D-image analysis. The in vitro and in vivo conformation of dyneins and the arrangement of tubulins in MTDs and the RS have recently been revealed. By combining structural biology with genetic engineering techniques, a number of features of dyneins and RSs were visualized. Structural analysis provides us insight not only into detailed 3D structure, molecular arrangement, and conformation of individual molecules but also into dynamics, cooperativity, and evolution.
ACKNOWLEDGMENTS
Our work is supported by grants from the Swiss National Science Foundation (NF3100A0-107540; NF31003A-125131/1; NF31003A-144035/1).
Footnotes
Editors: Wallace Marshall and Renata Basto
Additional Perspectives on Cilia available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Avolio J, Lebduska S, Sati P. 1984. Dynein arm substructure and the orientation of arm-microtubule attachments. J Mol Biol 173: 389–401. [DOI] [PubMed] [Google Scholar]
- Awata J, Takada S, Standley C, Lechtreck KF, Bellvé KD, Pazour GJ, Fogarty KE, Witman GB. 2014. NPHP4 controls ciliary trafficking of membrane proteins and large soluble proteins at the transition zone. J Cell Sci 127: 4714–4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berns MW, Rattner JB, Brenner S, Meredith S. 1977. The role of the centriolar region in animal cell mitosis. A laser microbeam study. J Cell Biol 72: 351–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R, Tritschler D, Vanderwaal K, Perrone CA, Mueller J, Fox L, Sale WS, Porter ME. 2013. The N-DRC forms a conserved biochemical complex that maintains outer doublet alignment and limits microtubule sliding in motile axonemes. Mol Biol Cell 24: 1134–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Ishikawa T. 2013. 3D structural analysis of flagella/cilia by cryo-electron tomography. Methods Enzymol 524: 305–323. [DOI] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. 2008. Molecular architecture of inner dynein arms in situ in Chlamydomonas reinhardtii flagella. J Cell Biol 183: 923–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. 2009. Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J Cell Biol 186: 437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Yagi T, Yamamoto R, Kamiya R, Ishikawa T. 2012. Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J Cell Biol 198: 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Cho C, Jin L, Vale RD. 2011. Crystal structure of the dynein motor domain. Science 331: 1159–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétien D, Wade RH. 1991. New data on the microtubule surface lattice. Biol Cell 71: 161–174. [DOI] [PubMed] [Google Scholar]
- Dallai R. 2014. Overview on spermatogenesis and sperm structure of Hexapoda. Arthropod Struct Dev 43: 257–290. [DOI] [PubMed] [Google Scholar]
- Diener DR, Yang P, Geimer S, Cole DG, Sale WS, Rosenbaum JL. 2011. Sequential assembly of flagellar radial spokes. Cytoskeleton (Hoboken) 68: 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener DR, Lupetti P, Rosenbaum JL. 2015. Proteomic analysis of isolated ciliary transition zones reveals the presence of ESCRT proteins. Curr Biol 25: 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroquez DB, Berciu C, Anderson JR, Sengupta P, Nicastro D. 2014. A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans. eLife 3: e01948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty GW, Loges NT, Klinkenbusch JA, Olbrich H, Pennekamp P, Menchen T, Raidt J, Wallmeier J, Werner C, Westermann C, et al. 2016. DNAH11 localization in the proximal region of respiratory cilia defines distinct outer dynein arm complexes. Am J Respir Cell Mol Biol 10.1165/rcmb.2015-0353OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersold WT, Levine RP, Levine EE, Olmsted MA. 1962. Linkage maps in Chlamydomonas reinhardi. Genetics 47: 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M, Olbrich H, Horvath J, Wildhaber JH, Zariwala MA, Kennedy M, Knowles MR, Omran H. 2005. Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am J Respir Crit Care Med 171: 1343–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, Omran H. 2007. When cilia go bad: Cilia defects and ciliopathies. Nat Rev Mol Cell Biol 8: 880–893. [DOI] [PubMed] [Google Scholar]
- Gardner LC, O’Toole E, Perrone CA, Giddings T, Porter ME. 1994. Components of a “dynein regulatory complex” are located at the junction between the radial spokes and the dynein arms in Chlamydomonas flagella. J Cell Biol 127: 1311–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliam JC, Chang JT, Sandoval IM, Zhang Y, Li T, Pittler SJ, Chiu W, Wensel TG. 2012. Three-dimensional architecture of the rod sensory cilium and its disruption in retinal neurodegeneration. Cell 151: 1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Heuser J. 1984. Structural comparison of purified dynein proteins with in situ dynein arms. J Mol Biol 180: 1083–1118. [DOI] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE. 1985a. Outer and inner dynein arms of cilia and flagella. Cell 41: 341–342. [DOI] [PubMed] [Google Scholar]
- Goodenough UW, Heuser JE. 1985b. Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J Cell Biol 100: 2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P, Desfosses A, Maheshwari A, Hachet V, Dietrich C, Brune A, Ishikawa T, Sachse C, Gönczy P. 2012. Cartwheel architecture of Trichonympha basal body. Science 337: 553. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Shingyoji C. 2008. Mechanism of flagellar oscillation-bending-induced switching of dynein activity in elastase-treated axonemes of sea urchin sperm. J Cell Sci 121: 2833–2843. [DOI] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D. 2009. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol 187: 921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Barber CF, Lin J, Krell J, Rebesco M, Porter ME, Nicastro D. 2012. Cryoelectron tomography reveals doublet-specific structures and unique interactions in the I1 dynein. Proc Natl Acad Sci 109: E2067–E2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops HJ, Witman GB. 1983. Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J Cell Biol 97: 902–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Piperno G, Ramanis Z, Luck DJ. 1981. Radial spokes of Chlamydomonas flagella: Genetic analysis of assembly and function. J Cell Biol 88: 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Brown JA, Yagi T, Norrander JM, Hirono M, Eccleston E, Kamiya R, Linck RW. 2003. Rib72, a conserved protein associated with the ribbon compartment of flagellar A-microtubules and potentially involved in the linkage between outer doublet microtubules. J Biol Chem 278: 7725–7734. [DOI] [PubMed] [Google Scholar]
- Inaba K. 2015. Calcium sensors of ciliary outer arm dynein: Functions and phylogenetic considerations for eukaryotic evolution. Cilia 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T. 2015. Cryo-electron tomography of motile cilia and flagella. Cilia 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, Shingyoji C. 2007. Induction of beating by imposed bending or mechanical pulse in demembranated, motionless sea urchin sperm flagella at very low ATP concentrations. Cell Struct Funct 32: 17–27. [DOI] [PubMed] [Google Scholar]
- Johnson KA. 1985. Pathway of the microtubule-dynein ATPase and the structure of dynein: A comparison with actomyosin. Annu Rev Biophys Biophys Chem 14: 161–188. [DOI] [PubMed] [Google Scholar]
- Kagami O, Kamiya R. 1992. Translocation and rotation of microtubules caused by multiple species of Chlamydomonas inner-arm dynein. J Cell Sci 103: 653–664. [Google Scholar]
- Kamiya R. 2002. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int Rev Cytol 219: 115–155. [DOI] [PubMed] [Google Scholar]
- Kamiya R, Okamoto M. 1985. A mutant of Chlamydomonas reinhardtii that lacks the flagellar outer dynein arm but can swim. J Cell Sci 74: 181–191. [DOI] [PubMed] [Google Scholar]
- Kamiya R, Yagi T. 2014. Functional diversity of axonemal dyneins as assessed by in vitro and in vivo motility assays of Chlamydomonas mutants. Zoolog Sci 31: 633–644. [DOI] [PubMed] [Google Scholar]
- *.King SM. 2016. Axonemal dynein arms. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a028100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Flückiger I, Gönczy P, et al. 2011. Structural basis of the 9-fold symmetry of centrioles. Cell 144: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon T, Sutoh K, Kurisu G. 2011. X-ray structure of a functional full-length dynein motor domain. Nat Struct Mol Biol 18: 638–642. [DOI] [PubMed] [Google Scholar]
- Kon T, Oyama T, Shimo-Kon R, Imamula K, Shima T, Sutoh K, Kurisu G. 2012. The 2.8 Å crystal structure of the dynein motor domain. Nature 484: 345–350. [DOI] [PubMed] [Google Scholar]
- Lin J, Okada K, Raytchev M, Smith MC, Nicastro D. 2014. Structural mechanism of the dynein power stroke. Nat Cell Biol 16: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linck R, Fu X, Lin J, Ouch C, Schefter A, Steffen W, Warren P, Nicastro D. 2014. Insights into the structure and function of ciliary and flagellar doublet microtubules: Tektins, Ca2+-binding proteins and stable protofilaments. J Biol Chem 89: 17427–17444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Rux JJ, Speicher DW, Pierce EA. 2007. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics 6: 1299–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupetti P, Lanzavecchia S, Mercati D, Cantele F, Dallai R, Mencarelli C. 2005. Three-dimensional reconstruction of axonemal outer dynein arms in situ by electron tomography. Cell Motil Cytoskeleton 62: 69–83. [DOI] [PubMed] [Google Scholar]
- Maheshwari A, Ishikawa T. 2012. Heterogeneity of dynein structure implies coordinated suppression of dynein motor activity in the axoneme. J Struct Biol 179: 235–241. [DOI] [PubMed] [Google Scholar]
- Maheshwari A, Obbineni JM, Bui KH, Shibata K, Toyoshima YY, Ishikawa T. 2015. α- and β-tubulin lattice of the axonemal microtubule doublet and binding proteins revealed by single particle cryo-electron microscopy and tomography. Structure 23: 1584–1595. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN, O’Toole ET, McDonald KL, McIntosh JR, Porter ME. 1992. Arrangement of inner dynein arms in wild-type and mutant flagella of Chlamydomonas. J Cell Biol 118: 1145–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Küller A, Daiber PC, Neudorf I, Warnken U, Schnölzer M, Frings S, Möhrlen F. 2009. The proteome of rat olfactory sensory cilia. Proteomics 9: 322–334. [DOI] [PubMed] [Google Scholar]
- Mencarelli C, Lupetti P, Rosetto M, Dallai R. 2000. Morphogenesis of the giant sperm axoneme in Asphondylia ruebsaameni Kertesz (Diptera, Cecidomyiidae). Tissue Cell 32: 188–197. [DOI] [PubMed] [Google Scholar]
- Minamino T, Imada K, Namba K. 2008. Molecular motors of the bacterial flagella. Curr Opin Struct Biol 18: 693–701. [DOI] [PubMed] [Google Scholar]
- *.Mitchell DR. 2016. Evolution of cilia. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a028290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR, Nakatsugawa M. 2004. Bend propagation drives central pair rotation in Chlamydomonas reinhardtii flagella. J Cell Biol 166: 709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollaret I, Justine JL. 1997. Immunocytochemical study of tubulin in the 9+“1” sperm axoneme of a monogenean (Platyhelminthes), Pseudodactylogyrus sp. Tissue Cell 29: 699–706. [DOI] [PubMed] [Google Scholar]
- Movassagh T, Bui KH, Sakakibara H, Oiwa K, Ishikawa T. 2010. Nucleotide-induced global conformational changes of flagellar dynein arms revealed by in situ analysis. Nat Struct Mol Biol 17: 761–767. [DOI] [PubMed] [Google Scholar]
- Nakazawa Y, Ariyoshi T, Noga A, Kamiya R, Hirono M. 2014. Space-dependent formation of central pair microtubules and their interactions with radial spokes. PLoS ONE 9: e110513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. 1999. Assembly , operation , and disassembly of protein complexes AAA: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res 9: 27–43. [PubMed] [Google Scholar]
- Nicastro D, Fu X, Heuser T, Tso A, Porter ME, Linck RW. 2011. Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc Natl Acad Sci 108: E845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH. 1998. Structure of the αβ tubulin dimer by electron crystallography. Nature 391: 199–203. [DOI] [PubMed] [Google Scholar]
- Oda T, Kikkawa M. 2013. Novel structural labeling method using cryo-electron tomography and biotin-streptavidin system. J Struct Biol 183: 305–311. [DOI] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Kamiya R, Kikkawa M. 2014a. Cilia and flagella. A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science 346: 857–860. [DOI] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Kikkawa M. 2014b. Detailed structural and biochemical characterization of the nexin–dynein regulatory complex. Mol Biol Cell 26: 294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Yagi T, Kikkawa M. 2014c. Mechanosignaling between central apparatus and radial spokes controls axonemal dynein activity. J Cell Biol 204: 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschal BM, Mikami A, Pfister KK, Vallee RB. 1992. Homology of the 74-kD cytoplasmic dynein subunit with a flagellar dynein polypeptide suggests an intracellular targeting function. J Cell Biol 118: 1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Agrin N, Leszyk J, Witman GB. 2005. Proteomic analysis of a eukaryotic cilium. J Cell Biol 170: 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G, Ishikawa T. 2012. Axonemal radial spokes: 3D structure, function and assembly. Bioarchitecture 2: 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G, Bui KH, Maheshwari A, Lupetti P, Diener D, Ishikawa T. 2011. Cryoelectron tomography of radial spokes in cilia and flagella. J Cell Biol 195: 673–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G, Maheshwari A, Bui KH, Shingyoji C, Kamimura S, Ishikawa T. 2012. Comparative structural analysis of eukaryotic flagella and cilia from Chlamydomonas, Tetrahymena, and sea urchins. J Struct Biol 178: 199–206. [DOI] [PubMed] [Google Scholar]
- Prensier G, Vivier E, Goldstein S, Schrével J. 1980. Motile flagellum with a “3 + 0” ultrastructure. Science 207: 1493–1494. [DOI] [PubMed] [Google Scholar]
- Ray S, Meyhöfer E, Milligan RA, Howard J. 1993. Kinesin follows the microtubule’s protofilament axis. J Cell Biol 121: 1083–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Numata N, Walker ML, Kato YS, Malkova B, Kon T, Ohkura R, Arisaka F, Knight PJ, Sutoh K, et al. 2009. AAA+ ring and linker swing mechanism in the dynein motor. Cell 136: 485–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Malkova B, Walker ML, Sakakibara H, Numata N, Kon T, Ohkura R, Edwards TA, Knight PJ, Sutoh K, et al. 2012. ATP-driven remodeling of the linker domain in the dynein motor. Struct Lond Engl 20: 1670–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AJ, Kon T, Knight PJ, Sutoh K, Burgess SA. 2013. Functions and mechanics of dynein motor proteins. Nat Rev Mol Cell Biol 14: 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale WS, Satir P. 1977. Direction of active sliding of microtubules in Tetrahymena cilia. Proc Natl Acad Sci 74: 2045–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P, Heuser T, Sale WS. 2014. A structural basis for how motile cilia beat. Bioscience 64: 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Gleave ES, Carter AP. 2012. Insights into dynein motor domain function from a 3.3-Å crystal structure. Nat Struct Mol Biol 19: 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Zalyte R, Urnavicius L, Carter AP. 2014. Structure of human cytoplasmic dynein-2 primed for its power stroke. Nature 518: 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrevel J, Besse C. 1975. A functional flagella with a 6 + 0 pattern. J Cell Biol 66: 492–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal RA, Huang B, Ramanis Z, Luck DJ. 1984. Mutant strains of Chlamydomonas reinhardtii that move backwards only. J Cell Biol 98: 2026–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Johnson KA. 1983. Presteady state kinetic analysis of vanadate-induced inhibition of the dynein ATPase. J Biol Chem 258: 13833–13840. [PubMed] [Google Scholar]
- Shimizu T, Hosoya N, Hisanaga S, Marchese-Ragona SP, Pratt MM. 1992. Activation of ATPase activity of 14S dynein from Tetrahymena cilia by microtubules. Eur J Biochem 206: 911–917. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Sakakibara H, Kojima H, Oiwa K. 2014. Slow axonemal dynein e facilitates the motility of faster dynein c. Biophys J 106: 2157–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JC, Northey JGB, Garg J, Pearlman RE, Siu KWM. 2005. Robust method for proteome analysis by MS/MS using an entire translated genome: Demonstration on the ciliome of Tetrahymena thermophila. J Proteome Res 4: 909–919. [DOI] [PubMed] [Google Scholar]
- Starling D, Randall J. 1971. Flagella of temporary dikaryons of Chlamydomonas reinhardii. Genet Res 18: 107. [Google Scholar]
- Sugino K, Naitoh Y. 1982. Simulated cross-bridge patterns corresponding to ciliary beating in paramecium. Nature 295: 609–611. [Google Scholar]
- Sui H, Downing KH. 2006. Molecular architecture of axonemal microtubule doublets revealed by cryo-electron tomography. Nature 442: 475–478. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Edamatsu M, Toyoshima YY. 2004. Multiple ATP-hydrolyzing sites that potentially function in cytoplasmic dynein. Proc Natl Acad Sci 101: 12865–12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno H, Bui KH, Ishikawa T, Imai Y, Yamaguchi T, Ishikawa T. 2014. Structure of dimeric axonemal dynein in cilia suggests an alternative mechanism of force generation. Cytoskeleton (Hoboken) 71: 412–422. [DOI] [PubMed] [Google Scholar]
- van Breugel M, Hirono M, Andreeva A, Yanagisawa H, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong IO, Robinson CV, et al. 2011. Structures of SAS-6 suggest its organization in centrioles. Science 331: 1196–1199. [DOI] [PubMed] [Google Scholar]
- Warner FD, Satir P. 1974. The structural basis of ciliary bend formation. Radial spoke positional changes accompanying microtubule sliding. J Cell Biol 63: 35–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley DM. 1997. Studies on the eel sperm flagellum. I: The structure of the inner dynein arm complex. J Cell Sci 110: 85–94. [DOI] [PubMed] [Google Scholar]
- Woolley DM. 1998. Studies on the eel sperm flagellum. 2: The kinematics of normal motility. Cell Motil Cytoskeleton 39: 233–245. [DOI] [PubMed] [Google Scholar]
- Yagi T, Uematsu K, Liu Z, Kamiya R. 2009. Identification of dyneins that localize exclusively to the proximal portion of Chlamydomonas flagella. J Cell Sci 122: 1306–1314. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Song K, Yanagisawa HA, Fox L, Yagi T, Wirschell M, Hirono M, Kamiya R, Nicastro D, Sale WS. 2013. The MIA complex is a conserved and novel dynein regulator essential for normal ciliary motility. J Cell Biol 201: 263–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Yang C, Sale WS. 2004. Flagellar radial spoke protein 2 is a calmodulin binding protein required for motility in Chlamydomonas reinhardtii. Eukaryot Cell 3: 72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Diener DR, Yang C, Kohno T, Pazour GJ, Dienes JM, Agrin NS, King SM, Sale WS, Kamiya R, et al. 2006. Radial spoke proteins of Chlamydomonas flagella. J Cell Sci 119: 1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]