Abstract
Anton van Leeuwenhoek’s startling microscopic observations in the 1600s first stimulated fascination with the way that cells use cilia to generate currents and to swim in a fluid environment. Research in recent decades has yielded deep knowledge about the mechanical and biochemical nature of these organelles but only opened a greater fascination about how such beautifully intricate and multifunctional structures arose during evolution. Answers to this evolutionary puzzle are not only sought to satisfy basic curiosity, but also, as stated so eloquently by Dobzhansky (Am Zool 4: 443 [1964]), because “nothing in biology makes sense except in the light of evolution.” Here I attempt to summarize current knowledge of what ciliary organelles of the last eukaryotic common ancestor (LECA) were like, explore the ways in which cilia have evolved since that time, and speculate on the selective processes that might have generated these organelles during early eukaryotic evolution.
The last eukaryotic common ancestor was remarkably complex, possessing motile cilia built on a typical 9+2 microtubule array and capable of gliding motility, beating motility, and display of sensory receptors.
Several excellent papers have appeared in the recent past that follow evolution of structures or complexes essential to ciliary function, including reviews on basal bodies and centrioles (Hodges et al. 2010; Ross and Normark 2015) and centrosomes (Azimzadeh 2014), trafficking and signaling modules (Johnson and Leroux 2010; Lim et al. 2011; Sung and Leroux 2013; Malicki and Avidor-Reiss 2014), tubulins (Findeisen et al. 2014), transition-zone complexes (Barker et al. 2014), and cilia themselves (Satir et al. 2008; Carvalho-Santos et al. 2011). Rather than attempting to cover all of the same ground in a single review, I will refer the reader to other sources where appropriate and instead try to look at the bigger pictures that are emerging from these details. Importantly, many model organisms have contributed to studies of cilia-related processes such as intraflagellar transport (IFT) trafficking, centriole assembly, and motility regulation, and these organisms include representatives of phylogenetically distant groups (green algae, ciliates, excavates, metazoa). Together with genomic data, these functional and structural studies provide a basis for understanding common features of cilia that would have been present in the last common ancestor as well as changes that may have occurred since that time. These aspects of ciliary evolution can, therefore, be presented with only a modest level of speculation, and speculative aspects are likely to be short-lived as additional data is collected.
Knowing how cilia evolved in the first place, during those dark ages of eukaryotic evolution after formation of true eukaryotes but before diversification into currently recognized supergroups, provides greater challenges. Evolutionary change includes continuous (as well as sudden and catastrophic) extinctions, which for the most part leave no trace in the genomic record of extant organisms. For larger, multicellular species and for those with hard shells or tests, fossils may provide some clues to morphologic features of extinct clades. For single-celled eukaryotes that existed before the last eukaryotic common ancestor (LECA), and especially for evidence of their motile mechanisms, fossils provide little information. We are, therefore, reduced to analysis of genomic changes between the closest living prokaryotic relatives of eukaryotes, as a proxy for the first eukaryotic ancestor, and the inferred common set of genes of the LECA, to learn how cilia evolved. Missing from this equation are data on other evolutionary paths leading to development of motile organelles (perhaps resembling cilia or perhaps not) that might have flourished for some time but that became extinct and left no trace in the current record.
THE CILIATED ORGANELLE OF THE LECA
When it comes to single-celled organisms, there is only so much to be learned from morphological features. Many species that were once thought to be closely related, based on outward appearance, have turned out to belong to quite distant branches of the tree of life. Such is certainly the case with prokaryotes, whose division into eubacteria and archaea was largely dependent on sequence comparisons, not shape, but similar discoveries have been made among eukaryotes such as oomycetes, which look like fungi but clearly belong in stramenopiles, and more recently with apusozoa, which resemble amoebae but are more closely related to us (Paps et al. 2013; Cavalier-Smith et al. 2014). The tree in Figure 1 summarizes many recent studies on the relationships among extant eukaryotes. Disagreement remains on some of the deep connections in this tree, in part because we still lack genomic sequences of some well-known eukaryotes and in part because some genomes appear to be evolving so fast that relationships are difficult to establish. Of course many other species remain to be discovered and some of these could reveal previously unappreciated relationships, but such trees remain useful because structures, mechanisms, and individual proteins that are found in multiple branches must have existed in the last common ancestor of those divergent species, regardless of their true phylogenetic relationships.
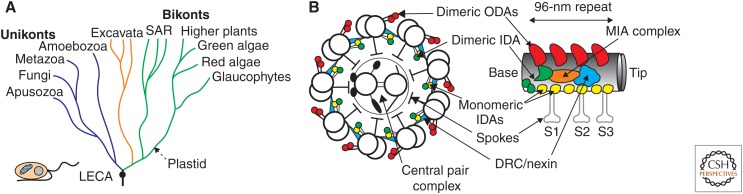
Figure 1.
(A) Diagram of major eukaryotic clades diverging from a last eukaryotic common ancestor (LECA), with the root placed between unikont and bikont superclades. Branches are color-coded according to differences in orientation of ciliary central-pair microtubules, which appear perpendicular to the bend plane and fixed in unikonts (blue), perpendicular to the bend plane but floating in excavates (orange), and twisted (helical) and rotating in other bikonts (green). The LECA is cartooned as a single cell with a nucleus, a mitochondrion, and two flagella: an anterior motile flagellum and a posterior gliding flagellum. The dashed arrow indicates the first acquisition of a plastid through endosymbiosis of a cyanobacterium. (B) Diagrams of ciliary axonemal structures that were present in the LECA. (Left) Cross-sectional view from inside the cell. (Right) Longitudinal view of one 96-nm repeat along an outer doublet. MIA, Modifier of inner arms; ODA, outer dynein arm; IDA, inner dynein arm; DRC, dynein regulatory complex.
Comparison of many recently generated trees of eukaryotic phylogeny leads to two striking conclusions. First, it is very difficult to sort out the branch order at the base of the tree, because all the major branches of eukaryotes seem to have diverged from a common ancestor within a short time (Fig. 1A). As with evolution anywhere, anytime, branching is a continuous process matched only by the frequency of extinction, so trees need to be interpreted by imagining what has been lost as well as what remains (Jablonski 1986). The effects of horizontal gene transfer can sometimes make trees look more like webs (Soucy et al. 2015), but most existing eukaryotes fit into one of several major clades related primarily by their decent from a common ancestor. These include the Stramenopiles, Alveolates, Rhizaria (SAR); the Plantae (probably a sister group to the SAR); the excavates (Heterolobosea, Euglenozoa, Diplomonads), which may be more closely related to the SAR than to the Plantae; the Amoebozoa; and the opisthokonts (Fungi, Choanozoa, Metazoa), which are more closely related to Amoebozoa than to the other clades. These branches appear to have originated in an evolutionary big bang, probably about one billion years ago (Chernikova et al. 2011; Parfrey et al. 2011). Second, the last common ancestor of all of these branches was highly complex (Koumandou et al. 2013). It had to have everything that is now found, in at least some representatives, in every branch. Based on features common to eukaryotes in all of these clades, the LECA must have divided by mitosis but also reproduced through a meiotic sexual cycle. It had a nucleus, endomembrane system, cytoskeleton, vesicular transport, mitochondria, and quite surely a cilium.
Fortunately model organisms from distantly related branches have been used extensively in studies of ciliary structure, function, and biochemistry, so that features common to the LECA and the extent of evolutionary change since that time can be reconstructed. Particularly useful has been the intense focus on Trypanosoma (excavata), Tetrahymena (SAR), Chlamydomonas (plantae), and several metazoa (opisthokonts). Based on features common to cilia in these distantly related organisms, the LECA had (most likely two) cilia with a typical 9+2 architecture of outer doublet and central singlet microtubules, assembled on basal bodies built from nine triplet microtubules. Motility generated by both outer and inner rows of dynein arms was regulated by interactions between radial spokes and a central-pair complex, and by nexin/dynein regulatory complex (DRC) interdoublet links (Fig. 1B). Ciliary assembly required trafficking on IFT complexes, the resulting organelles possessed sensory capabilities maintained in part by Bardet–Beidl syndrome (BBS) complex-dependent receptor trafficking, and movement of transmembrane proteins could be used for transport of material along the ciliary surface or gliding motility of the cell. In short, these cilia would have been structurally and functionally indistinguishable from typical cilia seen today.
TUBULINS, DOUBLETS, AND TRIPLETS
Phylogenomics shows that this ciliated LECA expressed α-, β-, γ-, δ-, ɛ-, and ζ-tubulin (Dutcher 2003; Findeisen et al. 2014). α-, β-, and γ-tubulin are essentially universal in eukaryotes, whereas δ-, ɛ-, and ζ-tubulins are only found in species with cilia. Although not all organisms with cilia retain all three of these more divergent tubulin isoforms, they occur in diverse taxa, supporting their presence in the LECA (see Azimzadeh 2014 and Turk et al. 2015 for other recent overviews of centrioles and tubulin diversity). δ- and ɛ-tubulin have been implicated in formation of basal bodies (Dutcher et al. 2002), and recent evidence suggests that ζ-tubulin is important for formation of basal feet, structures equivalent to subdistal appendages, which organize interactions of basal bodies with other cell structures and maintain basal body orientation in multiciliated cells (Turk et al. 2015).
Perhaps one of the most elusive features of basal bodies is the way in which triplet microtubules form. High-resolution electron microscopy of ciliary doublet (Nicastro et al. 2011; Maheshwari et al. 2015) and basal body triplet microtubules (Li et al. 2012; Guichard and Gonczy 2016) has revealed not only the underlying tubulin lattice structure but also many nontubulin components, including structures important for closing the walls of these doublet and triplet microtubules. The protein that forms one of these nontubulin “protofilaments,” the elusive protofilament 11 that closes the wall of the B-tubule, has recently been identified through a combined genetic and structural study in Chlamydomonas (Yanagisawa et al. 2014). Equally elusive until relatively recently was the basis for ninefold symmetry of basal bodies, which depends on self-association of SAS-6 into a cartwheel (Kitagawa et al. 2011; van Breugel et al. 2011) and on its attachment to intertriplet linkers (Hiraki et al. 2007; for more on centriole assembly, see reviews by Azimzadeh and Marshall 2010; Hirono 2014; Winey and O’Toole 2014).
More than a dozen phylogenetically widespread nontubulin proteins have also been identified as common components of centrioles and basal bodies, and many of these play specific roles in centriolar assembly or function (Hodges et al. 2010; Carvalho-Santos et al. 2011). Others have a more limited phylogenetic distribution and likely evolved to fill organism-specific functions. Despite a number of proteomic and transcriptomic studies (Keller et al. 2005; Kilburn et al. 2007; Fritz-Laylin and Cande 2010; Firat-Karalar et al. 2014) and genetic screens (Dobbelaere et al. 2008; Lin and Dutcher 2015), additional proteins involved in evolutionarily conserved basal body functions likely remain to be discovered.
MOTILITY
Motility in cilia of the LECA would have been generated by a two-headed outer row of dynein arms (ODAs) and an inner row that had both a two-headed inner dynein arm (similar to the I1 dynein of Chlamydomonas) and single-headed inner dynein arms (IDAs) generated from three different heavy chain isoforms (Fig. 1B) (Wickstead and Gull 2007; Wilkes et al. 2008). A pair of WD-repeat-containing intermediate chains was likely associated with each of the two-headed axonemal dyneins (Wickstead and Gull 2007; Patel-King et al. 2013). These intermediate chains (ICs) were similar to, and likely evolved from, the intermediate chain homodimers that are associated with all cytoplasmic dyneins. Both ODA and I1 axonemal dyneins have heterodimeric intermediate chains. Sequence comparisons suggest the formation first of an axonemal IC heterodimer, followed by a round of duplication to create separate heterodimers for outer dynein arms and I1 inner dynein arms. Interestingly, IFT dynein (unlike cytoplasmic dynein) appears to have heterodimeric intermediate chains that most closely resemble those of axonemal dyneins in the ODA-IC2 family (Patel-King et al. 2013). A potential evolutionary pathway would start with a two-headed IFT dynein that had an IC2-like homodimer, followed by evolution of a two-headed axonemal dynein from association of IFT dynein heads with a new heterodimeric IC base. This new base with its new IC1/IC2 heterodimer would allow IFT dynein to associate with A-tubules as cargoes and thus become the first axonemal dynein. Through a later duplication of the single IC2-like IFT dynein intermediate chain gene, IFT dynein itself evolved to its present form with a base made from heterodimeric intermediate chains.
Outer dynein arms of the LECA were probably anchored onto doublets with the aid of a docking complex made from a heterodimer of coiled-coil proteins, based on localization studies and similarities between the phenotypes of vertebrate (Jerber et al. 2013; Knowles et al. 2013; Onoufriadis et al. 2013; Hjeij et al. 2014) and algal (Koutoulis et al. 1997; Takada et al. 2002) docking complex mutants. However, homologs of one of the two proteins thought to function as a docking complex in vertebrates (CCDC151) is more closely related at the primary sequence level to a protein (ODA10) that functions in outer dynein assembly rather than docking in the alga (Dean and Mitchell 2013, 2015). Thus some caution is needed before all proteins identified on the basis of sequence similarity are assumed to be functionally equivalent in different organisms, including the LECA. Many additional proteins associated with axonemal dyneins in Chlamydomonas that presumably act as axonemal dynein cargo adaptors (Yamamoto et al. 2006, 2008) or regulatory complexes (Yamamoto et al. 2013) have easily identified homologs in the genomes of other ciliated organisms (Hom et al. 2011), but direct tests of their functional equivalency are lacking. As a further example of more recently acquired diversity underlying apparent homology, regulation of ciliary motility by calcium-dependent modulation of outer dynein arm activity is widespread, but the actual calcium sensors likely evolved independently in different clades (Inaba 2015).
An interesting point of speculation is how the three single-headed IDAs expressed in the LECA (Wickstead and Gull 2007) were arranged along the axoneme. Conservation of 96-nm periodicity (Oda et al. 2014) and of multiple structures associated with IDAs in such diverse organisms as Chlamydomonas and sea urchins (Nicastro et al. 2006; O’Toole et al. 2012; Pigino et al. 2012) argues for the presence in the LECA of not three, but six single-headed dyneins in each repeating unit (Fig. 1B). Perhaps future studies of organisms with simplified genomes that have reduced numbers of single-headed dynein genes will provide clues to how three single-headed inner arm dyneins may have been anchored in these six sites in cilia of the LECA.
Ancestral axonemal dyneins would have been regulated by at least three complexes (Fig. 1B): the modifier of inner arms (MIA) complex associated with I1 dynein (Yamamoto et al. 2013), the DRC/nexin link that joins adjacent doublets and likely transmits information about the extent or rate of interdoublet sliding (Heuser et al. 2009; Bower et al. 2013), and the radial spoke–central-pair complex that may transmit information across the axoneme and provide a more global level of control (Oda et al. 2014). The MIA complex and interdoublet DRC/nexin links occur once every 96 nm along each doublet. They form interactions with multiple dyneins as well as with radial spokes (Heuser et al. 2009; Oda et al. 2015) and play important, if poorly understood, roles in dynein regulation. Similar links have been seen by electron microscopy in motile cilia from many diverse organisms, and mutations in DRC subunits have been shown to alter the activity of axonemal dyneins in Chlamydomonas (Brokaw et al. 1982; Awata et al. 2015) and vertebrates (Wirschell et al. 2013; Olbrich et al. 2015). Most of the known Chlamydomonas DRC subunits have clear homologs in vertebrate genomes, confirming the overall high level of conservation of this complex (Bower et al. 2013).
The regulatory network supplied by interactions between the central-pair complex and radial spokes must also have evolved before the LECA, based on overall similarities between central-pair complexes (Pigino et al. 2012; Carbajal-Gonzalez et al. 2013; Oda et al. 2014) and radial spokes (Lin et al. 2012; Pigino et al. 2012) of sea urchin, Tetrahymena and Chlamydomonas, as well as conservation of many central-pair and radial spoke proteins at the primary sequence level. We lack biochemical and genetic data on the functions of many of these central-pair proteins, but continued exploration of mutations that generate ciliary motility defects in humans and in model organisms such as mice, zebrafish, Trypanosoma, and Chlamydomonas promises further tests of functional as well as sequence-level evolutionary conservation.
TRAFFICKING, ASSEMBLY, AND SIGNALING
The way in which cilia are assembled and maintained as segregated cytoplasmic and membrane compartments requires specialized docking of basal bodies to the plasma membrane through interactions that involve the centriolar proximal and distal appendages, and functions related to endosomal trafficking. Once an axoneme begins to assemble, transition zone components are added that provide a selective barrier, and specialized transport systems are needed to supply and retain material targeted to cilia. Many known components of each of these systems have been conserved in multiple eukaryotic clades and were therefore already present in the LECA. These include proteins associated with distal appendages needed for membrane anchoring and structures such as basal feet and rootlets involved in anchoring motile cilia in the cytoplasm (Carvalho-Santos et al. 2011).
IFT complexes and their associated dynein and kinesin motors are needed both for assembling the organelle and for maintaining its normal length and composition in many diverse species (Briggs et al. 2004; van Dam et al. 2013). The gliding motility associated with flagella in many single-celled eukaryotes likely depends on IFT motors as well (Collingridge et al. 2013; Shih et al. 2013), although as yet direct analysis has unfortunately been limited to one organism. The BBS complex, which plays a conserved role in maintaining normal concentrations of ciliary membrane-associated proteins, likely evolved from gene duplication and repurposing of IFT components, which in turn were derived from vesicle coat proteins (Satir et al. 2008; van Dam et al. 2013), before evolution of the LECA.
Signaling pathways associated with ciliary membranes have been modified extensively to fit the needs of each organism or cell type, with relatively few signaling modules retained since the LECA. One that appears to have associated with cilia comparatively early is the transient receptor potential (TRP) channel-linked mechanical gating, including PKD1/2-like channels. In vertebrates, polycystic kidney disease (PKD) channels have been linked to signaling in embryonic nodal cilia (Field et al. 2011; Kamura et al. 2011; Yoshiba et al. 2012) as well as in purely sensory cilia such as those in kidney tubules that give these proteins the PKD label (Nauli et al. 2003). In Chlamydomonas, a PKD2-related TRP channel has been shown to play a role in the mating reaction, which involves adhesion-dependent ciliary signaling (Huang et al. 2007), and similar channels are localized to ciliary membranes in Paramecium (Valentine et al. 2012). Chlamydomonas CAV2 is a voltage-dependent calcium channel important for control of waveform (Fujiu et al. 2009), and another Chlamydomonas TRPV-related protein, which shows homology with the mechanosensitive Drosophila TRPV ciliary channels, is involved in ciliary mechanosensation (Fujiu et al. 2011).
The regulation of trafficking into this compartment by small G proteins of the Arl/Rab/Arf family has been well documented in diverse organisms, and must therefore have also been present in the LECA (Sung and Leroux 2013). Some other modules associated with trafficking and signaling likely evolved in association with cilia before or soon after the LECA, such as the RJL family of small G proteins (Elias and Archibald 2009) and some cyclic nucleotide-based sensory systems (Johnson and Leroux 2010; Sung and Leroux 2013), whereas prominent cilia-associated developmental signaling pathways such as Hedgehog are widespread among metazoa, but not universally associated with cilia (Roy 2012).
EVOLUTIONARY TRENDS SINCE THE DIVERGENCE OF EUKARYOTES
Evolutionary change at the level of individual proteins or protein complexes, such as those providing the functional elements of cilia, can be conceptually divided into simple categories. Gene or whole genome duplication can result in multiple copies of slightly divergent proteins that provide greater functional diversity, either within one organelle or as a way to differentiate organelles that provide unique abilities to a cell. Examples within an organelle include the frequent expansion of the number of genes encoding one or more of the three types of single-headed IDAs. Different eukaryotic clades have independently expanded the number of these single-headed axonemal dyneins, and at least in Chlamydomonas we know that this expansion results in a single cilium that uses multiple related dyneins within a single organelle, presumably to generate subtle levels of motility control (Kamiya and Yagi 2014). Even greater expansion of the axonemal dynein family in some organisms may allow assembly of cilia with unique traits, such as the different cilia types present in the oral zone, body wall, and caudal region of ciliates (Rajagopalan and Wilkes 2016), or the different cilia found on epithelia of mammalian airways, brain ventricles, fallopian tubes, and the embryonic node.
Alternatively, gene duplication can lead to diversification into proteins used in very different ways. Many proteins in cilia appear to be more or less closely related to cytoplasmic proteins, but in some cases these proteins have clearly acquired new functions quite independently in different eukaryotic clades, and were not likely present in cilia of the LECA. An example is the presence of metabolic enzymes involved in adenosine triphosphate (ATP) generation. Mammalian sperm have glycolytic enzymes and can use directly imported glucose for ATP generation (Mukai and Okuno 2004), whereas ATP concentrations in other cilia are maintained through phosphate shuttles (phosphocreatine in sea urchin sperm [Tombes et al. 1987], phosphoarginine in Paramecium cilia [Noguchi et al. 2001] and Trypanosoma flagella [Ooi et al. 2015]), or through independently acquired steps of the glycolytic pathway (Chlamydomonas flagella [Mitchell et al. 2005]).
Equally commonly seen is the loss of proteins or structures, including cilia themselves, when the energy inherent in maintaining such complexity is no longer needed. Examples include the loss of IFT complexes in organisms that only assemble cilia in the cytoplasm and then extrude them preassembled (e.g., apicomplexans such as Plasmodium); loss of outer row (e.g., mosses and ferns) or inner row (diatoms) dyneins; and loss of the central-pair/radial spoke regulatory network (e.g., eel sperm, mammalian nodal cilia). Comparative genomics backed by electron microscopy has also shown that transition zone structures, essential for maintaining correct abundance of signaling modules on ciliary membranes (Li et al. 2016), have been lost in some organisms (Barker et al. 2014).
Evolutionary changes in motility regulation are more difficult to follow phylogenetically, but one interesting case may add clarity to weakly supported deep relationships among major eukaryotic clades. Orientation of an asymmetric central-pair complex has been linked to planar beating of cilia. In many unrelated instances, loss of the central-pair has been correlated with a switch to a circular or helical bending pattern, as seen in mammalian nodal cilia, eel sperm, and the simplified cilia of some parasitic protozoa (Satir et al. 2008). Although detailed structural comparisons support a remarkable conservation of the central-pair complex, other evidence suggests that central-pair function changed in a dramatic way during divergence from the LECA (Fig. 1A). In all unikonts examined thus far, orientation of the central pair is fixed along the length of the cilium, with a plane through the two central singlets perpendicular to the bend plane. In all members of the SAR or Plantae superclades, including ciliates, stramenopiles, and green algae, the central pair rotates during bend propagation so that, within each bend, a plane through the two central singlets is parallel to the bend plane (Omoto et al. 1999; Mitchell 2003; Mitchell and Nakatsugawa 2004). In organisms with a rotating central pair this complex is inherently helical, twisting in interbend regions. Finally, in members of the Excavata superclade such as Euglena and Trypanosoma, central-pair orientation resembles that in unikonts, but is not as tightly fixed (Melkonian et al. 1982) and can “float” through as much as 180° when released by mutations that affect flagellar motility (Branche et al. 2006; Gadelha et al. 2006). Thus, rotation of a helical central pair is a synapomorphy of the SAR and Plantae that supports the early branching of excavates before separation of these two superclades, whereas endosymbiosis of a plastid precursor occurred later, after their separation.
EVOLUTION OF CILIA BEFORE EUKARYOTIC DIVERGENCE
Evolution of ciliary proteins in the pre-LECA era must have involved transformation of microtubule-based trafficking systems (single microtubules, cytoplasmic dynein and kinesin, and vesicle coat proteins) into cilia-specific forms (doublet microtubules, axonemal dyneins, IFT proteins). One view of this process starts with formation of doublet microtubules that act as primitive centrioles, followed by evolution of mechanisms to dock a protocentriole to the cell membrane and to extend the centriolar doublet into a cilium-like structure (Marshall 2009). Alternatively, projections of the cell membrane generated by assembly of multiple singlet microtubules may have come first, and doublet formation may have evolved afterward (Satir et al. 2008). In this latter scenario, microtubule-based membrane extensions function as both sensory antennae and as gliding organelles, based on properties common to cytoplasmic trafficking by dyneins and kinesins. Doublets would evolve later, with the selective advantage deriving from their differentiation into two sides that can uniquely act as dynein cargos (A-tubules) and tracks (B-tubules) so that unidirectional bends could form between doublet pairs.
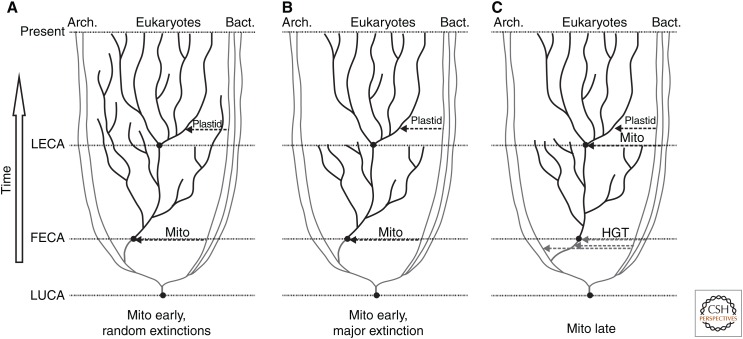
Many steps must have been needed to generate the 600 or so proteins and 9+2 architecture traceable to cilia of the LECA. Presumed intermediate steps included trials of many alternative mechanisms of microtubule-based cell motility, as well as more primitive versions of cilia, before a 9+2 organelle appeared. The puzzle therefore is why no eukaryotes exist today that descended from cells with any of these alternative proto-cilia. Why have no clades survived from earlier branches during the long evolutionary period needed to get from the first eukaryote common ancestor (FECA) to the last common ancestor of all eukaryotes? Was there an extreme bottleneck in eukaryotic evolution at the time of the LECA, which eliminated any traces of the previous evolutionary divergence of eukaryotes? Such an event could have opened multiple ecological niches for rapid expansion of the LECA into the major branches represented by existing species. If so, did this bottleneck result from a normal level of fluctuation in a relatively small community of eukaryotic organisms (Fig. 2A), a cataclysmic extinction event (Fig. 2B), or a sudden advantage that appeared in only one lineage (Fig. 2C)?
Figure 2.
Three alternative branching and extinction pathways leading from the last universal common ancestor (LUCA) to the first eukaryotic common ancestor (FECA), the last eukaryotic common ancestor (LECA), and present eukaryotic clades are diagrammed to explore mechanisms that selected for a LECA with 9+2 cilia. (A) Mitochondrial (Mito) endosymbiosis early, together with formation of a nucleus, transforms an archaeum into the FECA. Present diversity results from random branching and extinction, and alternate ciliary architectures are lost by chance. (B) Mitochondria are acquired early. Present clades result from rapid divergence from the LECA following a major extinction event, which chanced to preserve an organism with 9+2 cilia. (C) Horizontal gene transfer (HGT) between eubacteria and archaea generates an amitochondriate FECA. Much later, endosymbiosis of an α-proteobacterium creates the LECA, which rapidly diverges into present clades, whereas clades that lack mitochondria become extinct. The host for endosymbiosis, by chance, has 9+2 cilia. Arch., Archaea; Bact., bacteria.
The spectacular finding of Lokiarchaea and their kin in deep sea vents has opened a new vista on the origins of eukaryotes (Spang et al. 2015). These archaeal genomes reveal present-day organisms that have all the components needed for vesicular trafficking, including coat proteins and small G proteins. However, these trafficking components have not differentiated into the specific families of coat proteins (IFT and BBS complex subunits) and G proteins (Arls, Rabs, IFT22, IFT27) that are known to function in ciliary trafficking. Likewise, Lokiarchaeotes have actin-related sequences, but lack canonical eukaryotic actin and Arps. The available genomic sequences also lack homologs of the GTPase-activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs) that regulate trafficking in modern eukaryotes, and, most important, have no obvious homologs of tubulin. Based on recent analysis of orthologous groups, proto-eukaryotes likely formed through acquisition of a large set of bacterial genes by an archaeote closely related to existing Lokiarchaeota, perhaps through multiple HGTs (Spang et al. 2015; Pittis and Gabaldon 2016) as indicated in Figure 2C. Then, at a much later time, endosymbiosis of an α proteobacterium in one descendant branch led to mitochondrial formation (Pittis and Gabaldon 2016). The Big Bang of eukaryotic diversification, which probably occurred about one billion years ago, may thus have been stimulated by the energetic and metabolic benefits conferred by mitochondria (Lane 2014).
Because eukaryotic tubulins are not specifically more closely related to α-proteobacterial tubulin homologs than to tubulin homologs from other prokaryotes (Findeisen et al. 2014), an early HGT (perhaps one leading specifically to eukaryote formation, as in Fig. 2C) likely included a bacterial FtsZ homolog that became the ancestor of all tubulins. Duplication and diversification resulted in the full repertoire of tubulins of the LECA. Multiple tubulin-based motility mechanisms may have coexisted in divergent pre-LECA branches, with dominance of 9+2 cilia only occurring post-LECA. One apparent difficulty with this view is that if an ATP-rich intracellular environment is needed to drive ciliary motility (Chen et al. 2015), then mitochondria would need to already be present to support evolutionary selection of an energy-demanding motility mechanism. However, although abundant ATP might have been necessary to support initial selection of the many steps needed during cilia evolution, flagellated anaerobic (and virtually amitochondriate) organisms are known among both parasitic (e.g., Giardia) and free-living (e.g., Hexamita and Mastigamoeba) genera, so mitochondrial-based ATP formation is not essential for a cell to maintain flagellar function. Thus, an organism that had already evolved a primitive cilium might have had a selective advantage over competing organisms when its motility was further enhanced by acquisition of an endosymbiotic α-proteobacterial power source.
CONCLUDING REMARKS
The last eukaryotic common ancestor was a remarkably complex cell with motile cilia built on a typical 9+2 microtubule array, capable of gliding motility, beating motility, and display of sensory receptors. Genomic, metagenomic, and transcriptomic sequence analyses have revealed that most of the proteins present in cilia today were also present in this common ancestor, pushing the initial evolution of cilia into the interval between divergence of eukaryotes from archaeal prokaryotes and divergence of extant eukaryotic clades beginning about one billion years ago. Diversification into modern organisms may be traced by relatively subtle changes in ciliary architecture but by more radical changes in regulation of bending patterns and by many instances of reduction or loss of cilia. Ciliary based signaling through sensory receptors, although present ancestrally, has shown the greatest clade- and cell-type-specific expansion in the repertoire of these versatile organelles.
Footnotes
Editors: Wallace Marshall and Renata Basto
Additional Perspectives on Cilia available at www.cshperspectives.org
REFERENCES
- Awata J, Song K, Lin J, King SM, Sanderson MJ, Nicastro D, Witman GB. 2015. DRC3 connects the N-DRC to dynein g to regulate flagellar waveform. Mol Biol Cell 26: 2788–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J. 2014. Exploring the evolutionary history of centrosomes. Philos Trans R Soc Lond B Biol Sci 369: 20130453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J, Marshall WF. 2010. Building the centriole. Curr Biol 20: R816–R825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker AR, Renzaglia KS, Fry K, Dawe HR. 2014. Bioinformatic analysis of ciliary transition zone proteins reveals insights into the evolution of ciliopathy networks. BMC Genomics 15: 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R, Tritschler D, Vanderwaal K, Perrone CA, Mueller J, Fox L, Sale WS, Porter ME. 2013. The N-DRC forms a conserved biochemical complex that maintains outer doublet alignment and limits microtubule sliding in motile axonemes. Mol Biol Cell 24: 1134–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branche C, Kohl L, Toutirais G, Buisson J, Cosson J, Bastin P. 2006. Conserved and specific functions of axoneme components in trypanosome motility. J Cell Sci 119: 3443–3455. [DOI] [PubMed] [Google Scholar]
- Briggs LJ, Davidge JA, Wickstead B, Ginger ML, Gull K. 2004. More than one way to build a flagellum: Comparative genomics of parasitic protozoa. Curr. Biol. 14: R611–R612. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ, Luck DJL, Huang B. 1982. Analysis of the movement of Chlamydomonas flagella: The function of the radial-spoke system is revealed by comparison of wild-type and mutant flagella. J Cell Biol 92: 722–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbajal-Gonzalez BI, Heuser T, Fu X, Lin J, Smith BW, Mitchell DR, Nicastro D. 2013. Conserved structural motifs in the central pair complex of eukaryotic flagella. Cytoskeleton (Hoboken) 70: 101–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. 2011. Evolution: Tracing the origins of centrioles, cilia, and flagella. J Cell Biol 194: 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE, Snell EA, Berney C, Fiore-Donno AM, Lewis R. 2014. Multigene eukaryote phylogeny reveals the likely protozoan ancestors of opisthokonts (animals, fungi, choanozoans) and Amoebozoa. Mol Phylogenet Evol 81: 71–85. [DOI] [PubMed] [Google Scholar]
- Chen DT, Heymann M, Fraden S, Nicastro D, Dogic Z. 2015. ATP consumption of eukaryotic flagella measured at a single-cell level. Biophys J 109: 2562–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernikova D, Motamedi S, Csuros M, Koonin EV, Rogozin IB. 2011. A late origin of the extant eukaryotic diversity: Divergence time estimates using rare genomic changes. Biol Direct 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge P, Brownlee C, Wheeler GL. 2013. Compartmentalized calcium signaling in cilia regulates intraflagellar transport. Curr Biol 23: 2311–2318. [DOI] [PubMed] [Google Scholar]
- Dean AB, Mitchell DR. 2013. Chlamydomonas ODA10 is a conserved axonemal protein that plays a unique role in outer dynein arm assembly. Mol Biol Cell 24: 3689–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean AB, Mitchell DR. 2015. Late steps in cytoplasmic maturation of assembly-competent axonemal outer arm dynein in Chlamydomonas require interaction of ODA5 and ODA10 in a complex. Mol Biol Cell 26: 3596–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J, Josue F, Suijkerbuijk S, Baum B, Tapon N, Raff J. 2008. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol 6: e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK. 2003. Long-lost relatives reappear: Identification of new members of the tubulin superfamily. Curr Opin Microbiol 6: 634–640. [DOI] [PubMed] [Google Scholar]
- Dutcher SK, Morrissette NS, Preble AM, Rackley C, Stanga J. 2002. ɛ-Tubulin is an essential component of the centriole. Mol Biol Cell 13: 3859–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias M, Archibald JM. 2009. The RJL family of small GTPases is an ancient eukaryotic invention probably functionally associated with the flagellar apparatus. Gene 442: 63–72. [DOI] [PubMed] [Google Scholar]
- Field S, Riley KL, Grimes DT, Hilton H, Simon M, Powles-Glover N, Siggers P, Bogani D, Greenfield A, Norris DP. 2011. Pkd1l1 establishes left–right asymmetry and physically interacts with Pkd2. Development 138: 1131–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findeisen P, Muhlhausen S, Dempewolf S, Hertzog J, Zietlow A, Carlomagno T, Kollmar M. 2014. Six subgroups and extensive recent duplications characterize the evolution of the eukaryotic tubulin protein family. Genome Biol Evol 6: 2274–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Rauniyar N, Yates JR III, Stearns T. 2014. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr Biol 24: 664–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Cande WZ. 2010. Ancestral centriole and flagella proteins identified by analysis of Naegleria differentiation. J Cell Sci 123: 4024–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiu K, Nakayama Y, Yanagisawa A, Sokabe M, Yoshimura K. 2009. Chlamydomonas CAV2 encodes a voltage-dependent calcium channel required for the flagellar waveform conversion. Curr Biol 19: 133–139. [DOI] [PubMed] [Google Scholar]
- Fujiu K, Nakayama Y, Iida H, Sokabe M, Yoshimura K. 2011. Mechanoreception in motile flagella of Chlamydomonas. Nat Cell Biol 13: 630–632. [DOI] [PubMed] [Google Scholar]
- Gadelha C, Wickstead B, McKean PG, Gull K. 2006. Basal body and flagellum mutants reveal a rotational constraint of the central pair microtubules in the axonemes of trypanosomes. J Cell Sci 119: 2405–2413. [DOI] [PubMed] [Google Scholar]
- Guichard P, Gonczy P. 2016. Basal body structure in Trichonympha. Cilia 5: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser T, Raytchev M, Krell J, Porter ME, Nicastro D. 2009. The dynein regulatory complex is the nexin link and a major regulatory node in cilia and flagella. J Cell Biol 187: 921–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraki M, Nakazawa Y, Kamiya R, Hirono M. 2007. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr Biol 17: 1778–1783. [DOI] [PubMed] [Google Scholar]
- Hirono M. 2014. Cartwheel assembly. Philos Trans R Soc Lond B Biol Sci 369: 20130458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjeij R, Onoufriadis A, Watson CM, Slagle CE, Klena NT, Dougherty GW, Kurkowiak M, Loges NT, Diggle CP, Morante NF, Gabriel GC, et al. 2014. CCDC151 mutations cause primary ciliary dyskinesia by disruption of the outer dynein arm docking complex formation. Am J Hum Genet 95: 257–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K. 2010. Reconstructing the evolutionary history of the centriole from protein components. J Cell Sci 123: 1407–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom EF, Witman GB, Harris EH, Dutcher SK, Kamiya R, Mitchell DR, Pazour GJ, Porter ME, Sale WS, Wirschell M, et al. 2011. A unified taxonomy for ciliary dyneins. Cytoskeleton 68: 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K, Diener DR, Mitchell A, Pazour GJ, Witman GB, Rosenbaum JL. 2007. Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella. J Cell Biol 179: 501–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K. 2015. Calcium sensors of ciliary outer arm dynein: Functions and phylogenetic considerations for eukaryotic evolution. Cilia 4: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski D. 1986. Background and mass extinctions: The alternation of macroevolutionary regimes. Science. 231: 129–133. [DOI] [PubMed] [Google Scholar]
- Jerber J, Baas D, Soulavie F, Chhin B, Cortier E, Vesque C, Thomas J, Durand B. 2013. The coiled-coil domain containing protein CCDC151 is required for the function of IFT-dependent motile cilia in animals. Hum Mol Genet 23: 563–577. [DOI] [PubMed] [Google Scholar]
- Johnson JL, Leroux MR. 2010. cAMP and cGMP signaling: Sensory systems with prokaryotic roots adopted by eukaryotic cilia. Trends Cell Biol 20: 435–444. [DOI] [PubMed] [Google Scholar]
- Kamiya R, Yagi T. 2014. Functional diversity of axonemal dyneins as assessed by in vitro and in vivo motility assays of Chlamydomonas mutants. Zoolog Sci 31: 633–644. [DOI] [PubMed] [Google Scholar]
- Kamura K, Kobayashi D, Uehara Y, Koshida S, Iijima N, Kudo A, Yokoyama T, Takeda H. 2011. Pkd1l1 complexes with Pkd2 on motile cilia and functions to establish the left–right axis. Development 138: 1121–1129. [DOI] [PubMed] [Google Scholar]
- Keller LC, Romijn EP, Zamora I, Yates JR III, Marshall WF. 2005. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol 15: 1090–1098. [DOI] [PubMed] [Google Scholar]
- Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH Jr, Culver BP, Yates JR III, Winey M. 2007. New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol 178: 905–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Fluckiger I, Gonczy P, et al. 2011. Structural basis of the 9-fold symmetry of centrioles. Cell 144: 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles MR, Leigh MW, Ostrowski LE, Huang L, Carson JL, Hazucha MJ, Yin W, Berg JS, Davis SD, Dell SD, et al. 2013. Exome sequencing identifies mutations in CCDC114 as a cause of primary ciliary dyskinesia. Am J Hum Genet 92: 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumandou VL, Wickstead B, Ginger ML, van der GM, Dacks JB, Field MC. 2013. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit Rev Biochem Mol Biol 48: 373–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutoulis A, Pazour GJ, Wilkerson CG, Inaba K, Sheng H, Takada S, Witman GB. 1997. The Chlamydomonas reinhardtii ODA3 gene encodes a protein of the outer dynein arm docking complex. J Cell Biol 137: 1069–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N. 2014. Bioenergetic constraints on the evolution of complex life. Cold Spring Harb Perspect Biol 6: a015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Fernandez JJ, Marshall WF, Agard DA. 2012. Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J 31: 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Jensen VL, Park K, Kennedy J, Garcia-Gonzalo FR, Romani M, De MR, Bruel AL, Gaillard D, Doray B, et al. 2016. MKS5 and CEP290 dependent assembly pathway of the ciliary transition zone. PLoS Biol 14: e1002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YS, Chua CE, Tang BL. 2011. Rabs and other small GTPases in ciliary transport. Biol Cell 103: 209–221. [DOI] [PubMed] [Google Scholar]
- Lin J, Heuser T, Carbajal-Gonzalez BI, Song K, Nicastro D. 2012. The structural heterogeneity of radial spokes in cilia and flagella is conserved. Cytoskeleton (Hoboken) 69: 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Dutcher SK. 2015. Genetic and genomic approaches to identify genes involved in flagellar assembly in Chlamydomonas reinhardtii. Methods Cell Biol 127: 349–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheshwari A, Obbineni JM, Bui KH, Shibata K, Toyoshima YY, Ishikawa T. 2015. α- and β-tubulin lattice of the axonemal microtubule doublet and binding proteins revealed by single particle cryo-electron microscopy and tomography. Structure 23: 1584–1595. [DOI] [PubMed] [Google Scholar]
- Malicki J, Avidor-Reiss T. 2014. From the cytoplasm into the cilium: Bon voyage. Organogenesis 10: 138–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF. 2009. Centriole evolution. Curr Opin Cell Biol 21: 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkonian M, Robenek H, Rassat J. 1982. Flagellar membrane specializations and their relationship to mastigonemes and microtubules in Euglena gracilis. J Cell Sci 55: 115–135. [DOI] [PubMed] [Google Scholar]
- Mitchell DR. 2003. Orientation of the central pair complex during flagellar bend formation in Chlamydomonas. Cell Motil Cytoskeleton 56: 120–129. [DOI] [PubMed] [Google Scholar]
- Mitchell DR, Nakatsugawa M. 2004. Bend propagation drives central pair rotation in Chlamydomonas reinhardtii flagella. J Cell Biol 166: 709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BF, Pedersen LB, Feely M, Rosenbaum JL, Mitchell DR. 2005. ATP production in Chlamydomonas reinhardtii flagella by glycotytic enzymes. Mol Biol Cell 16: 4509–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai C, Okuno M. 2004. Glycolysis plays a major role for adenosine triphosphate supplementation in mouse sperm flagellar movement. Biol Reprod 71: 540–547. [DOI] [PubMed] [Google Scholar]
- Nauli SM, Alenghat FJ, Luo Y, Williams E, Vassilev P, Li X, Elia AE, Lu W, Brown EM, Quinn SJ, et al. 2003. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat Genet 33: 129–137. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. 2006. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313: 944–948. [DOI] [PubMed] [Google Scholar]
- Nicastro D, Fu X, Heuser T, Tso A, Porter ME, Linck RW. 2011. Cryo-electron tomography reveals conserved features of doublet microtubules in flagella. Proc Natl Acad Sci 108: E845–E853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi M, Sawadas T, Akazawa T. 2001. ATP-regenerating system in the cilia of Paramecium caudatum. J Exp Biol 204: 1063–1071. [DOI] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Yagi T, Kikkawa M. 2014. Mechanosignaling between central apparatus and radial spokes controls axonemal dynein activity. J Cell Bio. 204: 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Yanagisawa H, Kikkawa M. 2015. Detailed structural and biochemical characterization of the nexin-dynein regulatory complex. Mol Biol Cell 26: 294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olbrich H, Cremers C, Loges NT, Werner C, Nielsen KG, Marthin JK, Philipsen M, Wallmeier J, Pennekamp P, Menchen T, et al. 2015. Loss-of-function GAS8 mutations cause primary ciliary dyskinesia and disrupt the nexin-dynein regulatory complex. Am J Hum Genet 97: 546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omoto CK, Gibbons IR, Kamiya R, Shingyoji C, Takahashi K, Witman GB. 1999. Rotation of the central pair microtubules in eukaryotic flagella. Mol Biol Cell 10: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoufriadis A, Paff T, Antony D, Shoemark A, Micha D, Kuyt B, Schmidts M, Petridi S, Dankert-Roelse JE, Haarman EG, et al. 2013. Splice-site mutations in the axonemal outer dynein arm docking complex gene CCDC114 cause primary ciliary dyskinesia. Am J Hum Genet 92: 88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi CP, Rotureau B, Gribaldo S, Georgikou C, Julkowska D, Blisnick T, Perrot S, Subota I, Bastin P. 2015. The flagellar arginine kinase in Trypanosoma brucei is important for infection in tsetse flies. PLoS ONE 10: e0133676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole ET, Giddings TH Jr, Porter ME, Ostrowski LE. 2012. Computer-assisted image analysis of human cilia and Chlamydomonas flagella reveals both similarities and differences in axoneme structure. Cytoskeleton (Hoboken) 69: 577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paps J, Medina-Chacon LA, Marshall W, Suga H, Ruiz-Trillo I. 2013. Molecular phylogeny of unikonts: New insights into the position of apusomonads and ancyromonads and the internal relationships of opisthokonts. Protist 164: 2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJ, Knoll AH, Katz LA. 2011. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci 108: 13624–13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel-King RS, Gilberti RM, Hom EF, King SM. 2013. WD60/FAP163 is a dynein intermediate chain required for retrograde intraflagellar transport in cilia. Mol Biol Cell 24: 2668–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G, Maheshwari A, Bui KH, Shingyoji C, Kamimura S, Ishikawa T. 2012. Comparative structural analysis of eukaryotic flagella and cilia from Chlamydomonas, Tetrahymena, and sea urchins. J Struct Biol 178: 199–206. [DOI] [PubMed] [Google Scholar]
- Pittis AA, Gabaldon T. 2016. Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature 531: 101–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan V, Wilkes DE. 2016. Evolution of the dynein heavy chain family in ciliates. J Eukaryot Microbiol 63: 138–141. [DOI] [PubMed] [Google Scholar]
- Ross L, Normark BB. 2015. Evolutionary problems in centrosome and centriole biology. J Evol Biol 28: 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S. 2012. Cilia and Hedgehog: When and how was their marriage solemnized? Differentiation 83: S43–S48. [DOI] [PubMed] [Google Scholar]
- Satir P, Mitchell DR, Jekely G. 2008. How did the cilium evolve? Curr Top Dev Biol 85: 63–82. [DOI] [PubMed] [Google Scholar]
- Shih SM, Engel BD, Kocabas F, Bilyard T, Gennerich A, Marshall WF, Yildiz A. 2013. Intraflagellar transport drives flagellar surface motility. eLife 2: e00744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy SM, Huang J, Gogarten JP. 2015. Horizontal gene transfer: Building the web of life. Nat Rev Genet 16: 472–482. [DOI] [PubMed] [Google Scholar]
- Spang A, Saw JH, Jorgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van ER, Schleper C, Guy L, Ettema TJ. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521: 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CH, Leroux MR. 2013. The roles of evolutionarily conserved functional modules in cilia-related trafficking. Nat Cell Biol 15: 1387–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Wilkerson CG, Wakabayashi K, Kamiya R, Witman GB. 2002. The outer dynein arm-docking complex: Composition and characterization of a subunit (Oda1) necessary for outer arm assembly. Mol Biol Cell 13: 1015–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombes RM, Brokaw CJ, Shapiro BM. 1987. Creatine kinase-dependent energy transport in sea urchin spermatozoa. Flagellar wave attenuation and theoretical analysis of high energy phosphate diffusion. Biophys J 52: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk E, Wills AA, Kwon T, Sedzinski J, Wallingford JB, Stearns T. 2015. ζ-Tubulin is a member of a conserved tubulin module and is a component of the centriolar basal foot in multiciliated cells. Curr Biol 25: 2177–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine MS, Rajendran A, Yano J, Weeraratne SD, Beisson J, Cohen J, Koll F, Van HJ. 2012. Paramecium BBS genes are key to presence of channels in cilia. Cilia 1: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel M, Hirono M, Andreeva A, Yanagisawa HA, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong IO, Robinson CV, et al. 2011. Structures of SAS-6 suggest its organization in centrioles. Science 331: 1196–1199. [DOI] [PubMed] [Google Scholar]
- van Dam TJ, Townsend MJ, Turk M, Schlessinger A, Sali A, Field MC, Huynen MA. 2013. Evolution of modular intraflagellar transport from a coatomer-like progenitor. Proc Natl Acad Sci 110: 6943–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstead B, Gull K. 2007. Dyneins across eukaryotes: A comparative genomic analysis. Traffic 8: 1708–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkes DE, Watson HE, Mitchell DR, Asai DJ. 2008. Twenty-five dyneins in Tetrahymena: A re-examination of the multidynein hypothesis. Cell Motil Cytoskeleton 65: 342–351. [DOI] [PubMed] [Google Scholar]
- Winey M, O’Toole E. 2014. Centriole structure. Philos Trans R Soc Lond B Biol Sci 369: 20130457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M, Olbrich H, Werner C, Tritschler D, Bower R, Sale WS, Loges NT, Pennekamp P, Lindberg S, Stenram U, et al. 2013. The nexin-dynein regulatory complex subunit DRC1 is essential for motile cilia function in algae and humans. Nat Genet 45: 262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Yanagisawa HA, Yagi T, Kamiya R. 2006. A novel subunit of axonemal dynein conserved among lower and higher eukaryotes. FEBS Lett 580: 6357–6360. [DOI] [PubMed] [Google Scholar]
- Yamamoto R, Yanagisawa HA, Yagi T, Kamiya R. 2008. Novel 44-kilodalton subunit of axonemal dynein conserved from Chlamydomonas to mammals. Eukaryot Cell 7: 154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Song K, Yanagisawa HA, Fox L, Yagi T, Wirschell M, Hirono M, Kamiya R, Nicastro D, Sale WS. 2013. The MIA complex is a conserved and novel dynein regulator essential for normal ciliary motility. J Cell Biol 201: 263–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa HA, Mathis G, Oda T, Hirono M, Richey EA, Ishikawa H, Marshall WF, Kikkawa M, Qin H. 2014. FAP20 is an inner junction protein of doublet microtubules essential for both the planar asymmetrical waveform and stability of flagella in Chlamydomonas. Mol Biol Cell 25: 1472–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba S, Shiratori H, Kuo IY, Kawasumi A, Shinohara K, Nonaka S, Asai Y, Sasaki G, Belo JA, Sasaki H, et al. 2012. Cilia at the node of mouse embryos sense fluid flow for left-right determination via Pkd2. Science 338: 226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]