Abstract
The suprachiasmatic nucleus (SCN) is the principal circadian clock of the brain, directing daily cycles of behavior and physiology. SCN neurons contain a cell-autonomous transcription-based clockwork but, in turn, circuit-level interactions synchronize the 20,000 or so SCN neurons into a robust and coherent daily timer. Synchronization requires neuropeptide signaling, regulated by a reciprocal interdependence between the molecular clockwork and rhythmic electrical activity, which in turn depends on a daytime Na+ drive and nighttime K+ drag. Recent studies exploiting intersectional genetics have started to identify the pacemaking roles of particular neuronal groups in the SCN. They support the idea that timekeeping involves nonlinear and hierarchical computations that create and incorporate timing information through the interactions between key groups of neurons within the SCN circuit. The field is now poised to elucidate these computations, their underlying cellular mechanisms, and how the SCN clock interacts with subordinate circadian clocks across the brain to determine the timing and efficiency of the sleep–wake cycle, and how perturbations of this coherence contribute to neurological and psychiatric illness.
The suprachiasmatic nucleus is a group of brain cells that regulates daily cycles of behavior and physiology. These cells produce coherent, robust oscillations in activity by combining both autonomous and circuit-based mechanisms.
We wake and sleep each day. Hormones reach peak plasma levels at specified times, for example cortisol peaks in the early morning. These, and many other physiological and behavioral, daily rhythms depend on an internal circadian clock, the suprachiasmatic nucleus (SCN) of the hypothalamus. Prior reviews have provided excellent summaries of research progress in the location and function of this body clock (Weaver 1998). This work focuses on recent advances in our understanding of the genetic basis for cell-autonomous generation of circadian time, and how cells within the SCN synchronize their daily rhythms across the circuit to produce a coherent oscillation in neuronal activity. It is these circuit-level emergent properties of the SCN that ultimately direct daily behaviors such as wake and sleep.
A BRIEF TIMELINE OF THE SCN CLOCK
The SCN is the principal circadian pacemaker in mammals, autonomously capable of defining temporal cycles with a period of ∼24 hours, and are necessary for the expression of coherent daily rhythms of physiology, behavior, and metabolism in the intact animal (Fig. 1). The principal discoveries regarding the clock function of the SCN are reviewed extensively elsewhere (Weaver 1998), but the key observations are as follows. Although ablation studies had indicated a hypothalamic site for the circadian clock, the SCN only came to attention once autoradiographic tracing methods revealed it as a site of retinal innervation, the principal termination site of the retinohypothalamic tract (RHT). Subsequent lesion studies showed that behavioral, endocrine, and seasonal rhythms were compromised when the SCN was damaged. Autoradiographic metabolic imaging and electrophysiological studies showed that activity in the SCN is rhythmic in vivo. In addition, slice electrophysiology showed that the electrical circadian rhythms were sustained in vitro, even when disconnected from the rest of the brain. The SCN, therefore, is a tissue-based clock. The potency of this clock function was shown by intracerebral grafting, in vivo, of fetal SCN into the brain of rodents carrying SCN lesions. These grafts restored circadian patterning to the arrhythmic activity/rest behaviors, with a period determined by the genotype of the grafted tissue. This showed, definitively, that the SCN was necessary and sufficient to sustain circadian behaviors. The cell-autonomous nature of timekeeping was shown in dispersed cultures of SCN, in which the spontaneous electrical activity of individual neurons was circadian but free-ran independent of other neurons in the same culture. Indeed, fully isolated SCN neurons can express daily rhythms in repetitive firing rates and gene expression (Webb et al. 2009). Circuit-level properties of the SCN are nevertheless important; the ventrolateral (core) and dorsomedial (shell) subdivisions have been defined on the basis of innervation and neuropeptidergic phenotype. Whereas all SCN neurons are GABAergic, the shell and the core subdivisions show, respectively, localized expression of arginine vasopressin (AVP) or vasoactive intestinal peptide (VIP), and gastrin-releasing peptide (GRP). Anatomical studies have shown that the SCN is densely innervated by retinal axonal projections (Hattar et al. 2006; McNeill et al. 2011), the core subdivision being the principal site of direct and indirect retinal innervation. The discovery that light-mediated resetting of the SCN clock was accompanied by the induced expression of immediate-early genes such as cfos in the retinorecipient core directed the analysis of circadian timekeeping in mammals toward signal transduction and transcriptional regulation. These studies involving the conversion of light-induced biochemical changes to behavioral phase shifts paved the way for subsequent interrogation of the molecular genetic basis of the clock.
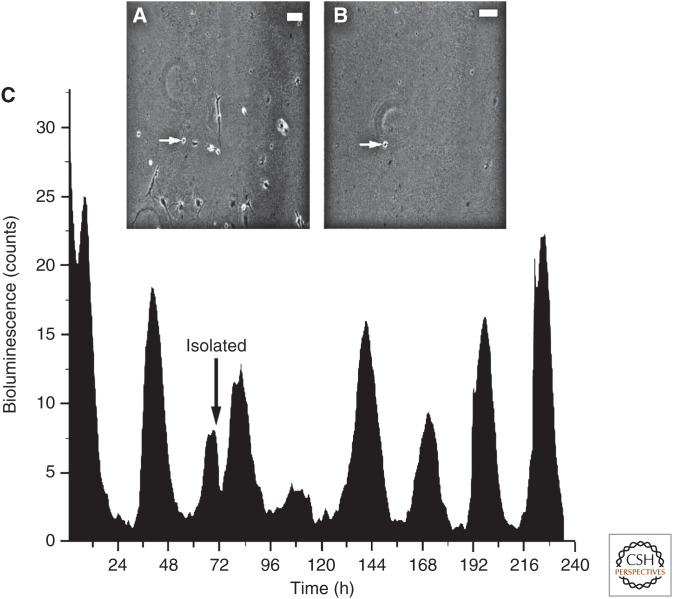
Figure 1.
Isolated neurons of the suprachiasmatic nucleus (SCN) are competent, cell-autonomous circadian pacemakers. (A,B) Micrographs of cultured SCN neurons before A, and after B, physically isolating a single neuron (arrow). Scale bars, 50 μm. (C) Recording of PER2 expression using a bioluminescent reporter of PER2 abundance reveals persistent daily rhythms before and after the neuron was isolated. (From Webb et al. 2009; reproduced, with permission, from the authors.)
A SHORT HISTORY OF THE MAMMALIAN MOLECULAR CLOCKWORK
The Tau mutant hamster, in which behavioral and metabolic cycles free-run with a period of 20 hours in homozygotes, illustrated that the mammalian clock could be analyzed at a single gene level. Identification of the genetic components of the clock came, however, from de novo gene discovery in mice and by homology with known elements of the Drosophila clockwork (see Ode 2016). For example, Period1 (Per1), which encodes a negative transcriptional regulator within the clock mechanism, was cloned on the basis of conserved sequence identity with the PAS dimerization domains of dPer, but it was also discovered independently as a transcript encoded by human chromosome 17 and initially named Rigui. Per2 and Per3 were then identified by sequence homology with Per1. In contrast, the positive transcriptional regulator Clock was identified de novo in a mutagenesis screen and transgenic rescue studies in the mouse, independent of the discovery of the Drosophila paralog. Bmal1 (also called MOP3 or Arntl) was initially identified as encoding a binding partner to CLOCK proteins in a yeast two-hybrid screen, whereas the fly paralog, Cycle, was identified by mutagenesis. Finally, the genes encoding the cryptochromes (Cry1 and Cry2), the second set of negative regulators in the mammalian clock, were originally identified on the basis of their homology with photolyase DNA repair enzymes. Following the identification of dCry as a circadian photoreceptor in the fly, it was shown that CRY1 and CRY2 are essential negative elements of the mammalian feedback loop.
Notwithstanding the intriguing recent discoveries of circadian oscillations in peroxiredoxin superoxidation in transcriptionally incompetent anucleate erythrocytes (see Reddy and Rhee 2016) and the expression of such cycles in SCN slice culture (Edgar et al. 2012), the consensus model of how the mammalian clockwork operates at a molecular level involves an intracellular, autoregulatory, delayed negative feedback model, incorporating transcriptional and posttranslational feedback loops (TTFLs). In this scheme, early circadian day is marked in the SCN by the initiation of transcriptional activation of Per and Cry genes mediated by heterodimers of CLOCK and BMAL1 acting on “E-box” enhancer sequences (Fig. 2). Over the course of the day, transcript levels increase, accompanied by an increase in the levels of the relevant proteins. Circadian regulation of translational efficiency via mTOR and MAPK signaling cascades, converging on phosphorylation of the cap-binding protein eIF4E may facilitate this increase, such that by the end of circadian day nuclear complexes of PER and CRY suppress the activity of CLOCK and BMAL1 (Cao et al. 2013). Crystal structures recently revealed that CRY1 binds to a PAS domain within CLOCK and a TAD domain within the carboxyl terminus of BMAL1 to switch off transcriptional activation by CLOCK-BMAL1 (Xu et al. 2015). Consequently, over the course of circadian night, Per and Cry transcript levels decrease, followed by a progressive decline in PER and CRY proteins because of proteasomal degradation. By the end of circadian night, the negative feedback mediated by PER and CRY is dissipated and the cycle is renewed with a new “circadian dawn.” Beyond this inner loop, further feedback cycles confer stability and precision to the SCN clock, in particular one involving CLOCK/BMAL1/E-box-dependent expression of RORA and REV-ERBα and β proteins that in turn direct Bmal1 expression (Sato et al. 2004). More recently, a role for cytosolic BMAL1 in the circadian control of translation in peripheral tissues has been reported (Lipton et al. 2015), adding an additional layer of regulatory complexity beyond the canonical feedback loop. The molecular clock consists, therefore, of a temporally ordered, self-sustaining and self-initiating sequence of transcriptional and translational events.
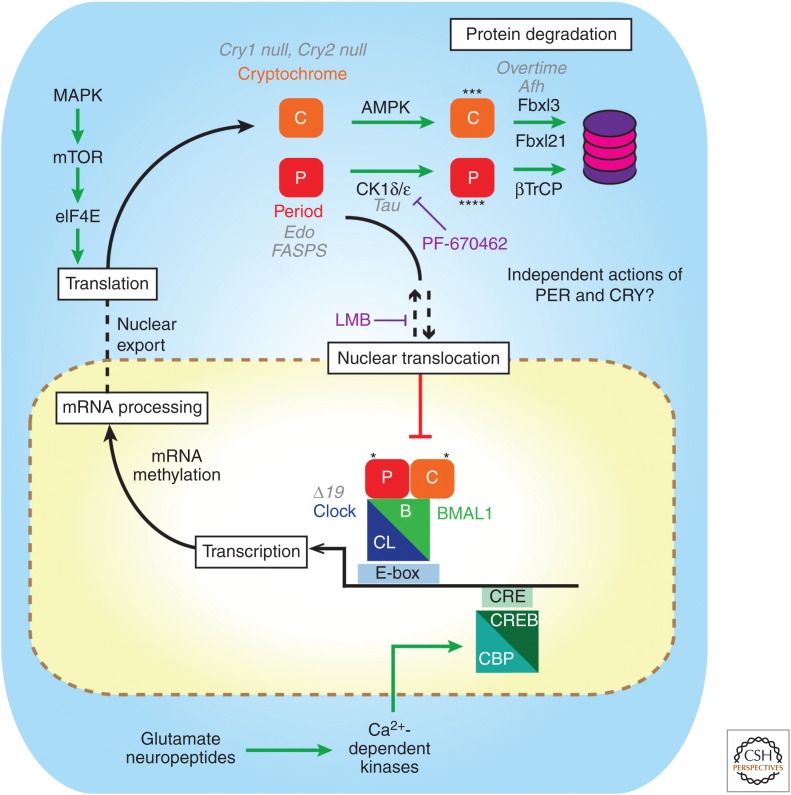
Figure 2.
Schematic diagram of the intracellular suprachiasmatic nucleus (SCN) clockwork with points of regulation by mutations and drugs. The canonical clockwork involves a transcription–translational negative feedback loop in which PER–CRY dimers inhibit their own transcription by repressing the actions of CLOCK–BMAL1 dimers on E-box elements in clock genes. Beyond this, mRNA maturation and posttranslational regulation of clock gene products including phosphorylation by AMPK and CK1, ubiquitinylation by FBXL3, FBXL21, and βTrCP, and translational regulation by eIF4E and mRNA methylation contribute to oscillation and determine circadian period. Deletions of Cry1 or Cry2, circadian mutations (gray) in PER genes (Edo and FASP), CK1ɛ (Tau), Fbxl3 (Afterhours, Overtime), and CLOCK (Δ19) as well as drug manipulations (purple, Leptomycin B and PF-670462) highlight key points of regulatory control of the molecular clock.
A DISTRIBUTED CLOCK NETWORK
In addition to elucidating the timing mechanism of the SCN clock, identification of mammalian clock genes had additional important consequences. To build on similar technical approaches used in Arabidopsis and Drosophila, it now became possible to develop fluorescence- and bioluminescence-based reporters of circadian gene and protein expression to monitor cell- and tissue-based (and, more recently, behaving mice) (Saini et al. 2013) circadian oscillations with exquisite spatial and temporal resolution. This allowed for the characterization of circadian clock gene expression in any tissue- and cell-type, and led to the paradigm-shifting discovery that most, if not all, major organ systems have local tissue-based clocks that use the same core genetic timing mechanism as the SCN (Nagoshi et al. 2004; Yoo et al. 2004). The SCN is not, therefore, a driver of peripheral rhythms; rather, it is a coordinator and synchroniser at the head of a distributed network of cellular clocks. Its defining properties are, first, that it is the point of entrainment of the entire system by the retinally sensed light–dark cycle, conveyed directly by the RHT and indirectly via afferents from other retinorecipient, areas such as the ventral thalamus (Delogu et al. 2012; LeGates et al. 2014). The second defining property of the SCN is that, in contrast to the oscillations of peripheral clocks, ex vivo, it will continue to show high amplitude molecular and electrical circadian oscillations effectively indefinitely.
This resilience and robustness of the SCN arises from its circuitry (Liu et al. 2007). Although individual SCN neurons have intrinsic cell-autonomous clocks, as do fibroblasts, what makes the SCN an effective oscillator is the circuit-based mechanisms whereby the individual cellular clocks are mutually sustained and synchronized. Put another way, SCN cellular clocks operate more effectively when functioning as a circuit. This review seeks to explore what is known of the roles and mechanisms of intercellular communication in coordinated daily rhythms. In doing so, it will emphasize how new approaches based on real-time imaging, intersectional genetics, and opto- and chemo-genetic manipulations have started to provide new opportunities to unravel the underlying genetic and neuronal mechanisms of SCN circadian timekeeping.
SETTING THE SPEED OF THE SCN MOLECULAR CLOCK
Implicit in the TTFL model of the clockwork is the expectation that events that alter transcription rates and protein trafficking and stability of clock elements like PER and CRY will influence the period of the molecular oscillation (Fig. 2). Indeed, computer simulations have been developed to test the assumptions of the TTFL model. A variety of models have predicted, for example, that rhythms occur within a narrow range of rates of transcription and translation (Forger and Peskin 2003; Mirsky et al. 2009; Meeker et al. 2011; Korencic et al. 2014). They have inspired experiments revealing that different phosphorylation patterns of PER2 can either stabilize or degrade PER2 and, thereby, accelerate or slow down circadian cycling (Virshup et al. 2007; Zhou et al. 2015). Often, however, we lack the necessary details (e.g., rate constants) or data (e.g., temporal resolution) to validate or revise the existing dogma. For example, models based on existing observations have not resolved whether isolated SCN cells should be described as noisy, sustained, or slowly damping circadian oscillators (Westermark et al. 2009).
From an experimental perspective, the period-defining role within the TTFL of the rate of transcription was provided initially by the ClockΔ19 mutant mouse, in which the compromised transcriptional activation of E-boxes by CLOCK was associated with its characteristic longer circadian period (Partch et al. 2014). More recently, the rate of RNA processing as determined by RNA-methylation has also been shown to control the speed of the circadian clock (Fustin et al. 2013). The bulk of experimental evidence for setting SCN clock speed has come, however, from studies of PER and CRY protein stability.
Accumulation, degradation, and localization of PER proteins play a role in setting the period of circadian rhythms. For example, the rate of PER2 accumulation in a cell on one day predicts, with 83% accuracy, whether that cell will show a circadian rhythm on the next day (Webb et al. 2009). Proteasomal degradation of PER proteins is triggered by casein kinase 1 (CK1)-mediated phosphorylation of various serine residues (including S477–S479 in mPER2) that, in turn, directs ubiquitinylation by the ubiquitin ligase β-TRCP (Zhou et al. 2015), and mutations or pharmacological inhibition of β-TRCP compromise the clock (Reischl et al. 2007). In addition, PER2 is also phosphorylated by an unknown priming kinase at a more carboxy-terminal serine (FASP site), and this triggers serial phosphorylations by other kinases, including CK1. Phosphorylation of this FASP site is thought to negatively regulate phosphorylation at the β-TRCP site, possibly by inducing a conformational change of the PER protein, and thereby stabilizing it. Importantly, phosphorylation at the β-TRCP and FASP sites is differentially sensitive to temperature, with the result that the rate of degradation of PER2 is constant over a range of temperatures. Temperature compensation of the circadian period is a canonical property of circadian pacemakers, including the SCN (Herzog and Huckfeldt 2003; Buhr et al. 2010), and these recent findings suggest that the opposing phosphorylation events on PER2 (and possibly PER1) are at least one molecular contribution to this (Zhou et al. 2015). Importantly, mutations in human PER2 or CK1δ that affect the phosphorylation at the carboxy-terminal site are associated with familial sleep disorders consistent with an accelerated SCN clockwork and associated with faster PER2 degradation (Jones et al. 2013). A similar phenotype of an accelerated circadian period in behavior and the SCN clock is also observed in the Tau mutation of CK1ɛ first identified in hamsters (Ralph et al. 1990) and reengineered in mouse (Meng et al. 2008). It has been argued that this mutation is a gain-of-function at the β-TRCP site arising from reduced phosphorylation at the competing FASP site (Zhou et al. 2015). The accelerated degradation of PER2 can be attenuated by selective inhibition of CK1ɛ (Meng et al. 2010) and this reverses the shortened period of the Tau mutant SCN clock. Selective inhibition of CK1ɛ does not, however, affect the period of wild-type (WT) SCN, whereas selective inhibition of CK1δ potently lengthens the period in WT, CK1ɛTau mutant and CK1ɛnull SCN. Equally, inhibition of CK1δ, but not CK1ɛ, stabilizes PER2 levels in the SCN and lengthens the period of activity/ rest behavioral rhythms in WT, CK1ɛTau mutant, and CK1ɛnull mice. Taken together with evidence of period lengthening in the neonatal SCN lacking CK1δ but not CK1ɛ (Etchegaray et al. 2010), it appears that, under natural conditions, CK1δ is the predominant regulator of PER stability, but gain or loss of CK1ɛ function can nevertheless alter PER dynamics and thereby the intrinsic period of the SCN and/or its response to resetting cues (Pilorz et al. 2014). Given the importance of phosphorylation in setting the circadian period, it is not surprising that in cell cultures the balance between protein phosphatase 1 and CK1 kinase activity directs circadian period (Lee et al. 2011). The importance of PER2 stability in setting the circadian period has recently been reinforced by analysis of the ENU-induced Early doors mutant of Per2 (Militi et al. 2016), in which a conserved isoleucine is replaced by asparagine (I324N). This destabilizes the PAS A/PAS B dimerization domain of PER2, accelerating CK1-mediated degradation of PER2Edo. As a result, double homozygous mutant Edo and Tau mice have unprecedentedly short periods (<18 h) of behavior and SCN molecular pacemaking. The precision and stability of the clockwork are nevertheless unaffected, highlighting again its remarkable robustness (Militi et al. 2016).
The stability of CRY proteins is also a factor in setting the pace of the SCN clock and behavioral rhythms. Most obviously, loss of CRY1 accelerates the clock, whereas loss of CRY2 lengthens period. Under normal circumstances, therefore, the WT period of ca. 24 h arises from functional interactions between the two CRY isoforms. Proteasomal degradation of CRY proteins is directed by the combined, antagonistic actions of two Skp1-Cul1-Fbox (SCF) ubiquitin ligases. FBXL3 (Godinho et al. 2007; Siepka et al. 2007) promotes degradation of nuclear-localized CRY, and loss-of-function mutations prolong the period of behavioral and SCN molecular rhythms. Conversely, FBXL21 promotes degradation of cytoplasmic-localized CRY but opposes the action of FBXL3 in the nucleus (Hirano et al. 2013; Yoo et al. 2013) and loss-of-function mutations of FBXL21 present as a shortened circadian period. Under normal circumstances, circadian period is therefore set by the balance and cellular localization of CRY proteins and these competing ligases. The circadian period is also sensitive to the relative abundance of CRY isoforms, CRY1 and CRY2. Analysis of the compound Cry and Fbxl3 mutants has revealed that, although CRY2 is a negative regulator within the SCN clockwork, it is less potent than CRY1 and indeed antagonizes CRY1 inhibition (Anand et al. 2013). Under normal circumstances, circadian period is therefore set by the abundance and cellular localization of CRY1 and CRY2 and these competing ligases.
Upstream of E3 ligases, AMP-dependent kinase (AMPK) plays a role in phosphorylating CRY1 and thereby targeting it for ubiquitinylation (Lamia et al. 2009). Although AMPK1 or AMPK2 single-null mice show modest effects on circadian behavior, the double mutant has not been tested (Um et al. 2011). Importantly, AMPK activity is regulated by cellular ATP levels (Oakhill et al. 2011) and so may provide a conduit for the action of metabolic cues on the clock, especially in peripheral tissues for which metabolic stimuli may be critical entraining factors (Dibner et al. 2010). An additional avenue for metabolic signaling to the clock is provided by SIRT1, the NAD+-dependent deacetylase, which can stimulate Clock and Bmal1 expression (Chang and Guarente 2013). These observations have led to the speculation that the progressive fragmentation of circadian behavior with age may reflect compromised CLOCK/BMAL1 function because of declining SIRT1 levels.
Thus, a general model has been proposed that CKI primarily regulates the accumulating phase of the PER–CRY repressive complex by controlling the nuclear import rate, whereas FBXL3/FBXL21 separately regulate the duration of transcriptional repression in the nucleus (St John et al. 2014). Given the dramatic individual effects of PER and CRY stability on SCN and behavioral periodicity, the question arises of how do they interact? Intercrosses between the short period CK1ɛTau and the long period Fbxl3Afh mice revealed that these mutations act independently and additively: there was no evidence for epistasis (Maywood et al. 2011a). This suggests that, contrary to the accepted view that PER and CRY function as heteromeric complexes, they more likely act separately in time and/or cellular space. Evidence for the former is provided by chromatin immunoprecipitation studies of E-box occupancy in mouse liver across circadian time (Koike et al. 2012). Whereas E-box-mediated transcriptional activation during the circadian day (CT6-10) is associated with peak occupancy of CLOCK and BMAL1 on the E-boxes, during the early repressive phase (CT15–18) PER1, PER2, and CRY2 proteins are in position, consistent with their negative feedback role. In contrast, CRY1 peak occupancy on E-boxes occurs later in the repressive phase, as PER1/2 and CRY2 decrease. Assuming that a similar pattern occurs in the SCN, this would explain why reduced PER stability and enhanced CRY stability have additive effects on overall SCN period-truncation of one element of the molecular sequence (PER-dependent feedback) would be compensated for by prolongation of a later phase (CRY1-dependent feedback). Moreover, the temporal segregation of E-box occupation by CRY2 and CRY1 suggests that CRY2 may antagonize CRY1 function by excluding it from E-boxes early in circadian night.
Circadian period is, therefore, an emergent property of a constellation of molecular functions involving posttranslational modifications, suggesting that a number of control points may be subject to pharmacological manipulation. This raises the possibility of developing therapeutic interventions in clock-relevant diseases, such as metabolic syndrome (Green et al. 2008). To identify suitable compounds, targeted drug development, for example, CK1δ/ɛ inhibitors (Walton et al. 2009), has been complemented by unbiased screening assays, taking advantage of circadian function in convenient cell lines. Several CK1 inhibitors were identified by their prolongation of circadian period (Chen et al. 2012). In addition, a broad-acting CK1 inhibitor, Longdaysin, highlighted unanticipated CK1a participation in PER degradation and determination of SCN period (Hirota et al. 2010). Similarly, a small molecule that activated CRY function by preventing ubiquitinylation by FBXL3 prolonged circadian period in peripheral cells (Hirota et al. 2012). In another example, inhibitors of GSK-3β, which phosphorylates REV-ERB and BMAL1, were shown to accelerate the clock in cells (Hirota et al. 2008) and SCN neurons (Besing et al. 2015). Although in its infancy, the intimate relationship between the clock and basic physiology associated with systemic diseases means that a chemical biological approach offers considerable promise for therapeutic development. This potential is highlighted by the observation that “the majority of best-selling drugs and World Health Organization essential medicines directly target the products of rhythmic genes” (Zhang et al. 2014).
DAILY REGULATION OF SCN NEURONAL EXCITABILITY
Although the molecular clockwork defines circadian time, if the SCN is to direct behavior, the molecular clockwork needs to regulate the electrophysiological activity of SCN neurons and thus its downstream signaling to targets in the local hypothalamus and brainstem. The remarkable vitality of the SCN clock when maintained in an organotypic slice culture has facilitated the real-time monitoring of molecular and electrophysiological events of SCN neurons over many days (Fig. 3A,B). For example, fluorescent and bioluminescent reporters have been used to map circadian activation of PER1 (Kuhlman et al. 2000; Yamaguchi et al. 2003), PER2 (Yoo et al. 2004), Cry1 (Maywood et al. 2013), and Bmal1 (Noguchi et al. 2010) in the SCN taken from genetically modified mice. Furthermore, combined electrophysiological and bioluminescent recordings in SCN slices have shown that spontaneous firing rate peaks at about 4–10 Hz in the middle of the circadian day, as Per expression is rising, and falls to a nadir in the middle of circadian night, coincident with transcriptional repression of PER2 (Atkinson et al. 2011). Importantly, even though the molecular clocks of SCN cells are synchronized, one to another, across the circuit their activity is not simultaneous. Different subpopulations express stereotypical phases of gene expression that generate a circadian wave of Per and Cry1 activation that tracks across the SCN. This trajectory can be expressed as a center-of-luminescence plot, in which the center of gravity of the bioluminescent signal on CCD recordings is plotted in x-y coordinates through time (Fig. 3C). In CRY-deficient SCN, the trajectory is absent but can be activated within two or three circadian cycles by virally mediated (AAV) expression of CRY1 or CRY2, driven by a minimal Cry1 promoter (Edwards et al. 2016). The spatial configuration that directs the wave is therefore a latent property of the SCN circuit (Pauls et al. 2014) that requires CRY-dependent temporal function for its expression. This network-dependent and CRY-dependent wave of daily gene expression is also age-dependent, maturing over the first 2 weeks after birth (Ono et al. 2013).
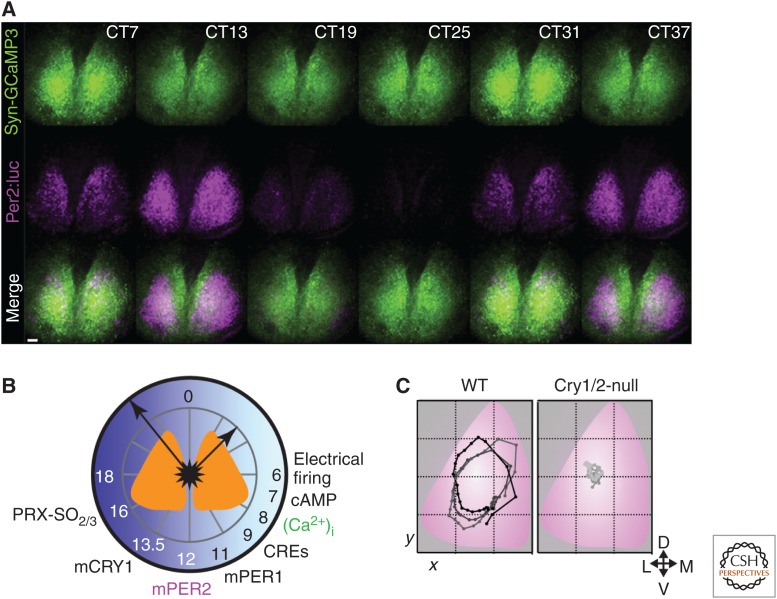
Figure 3.
Temporal and spatial circadian programs in the suprachiasmatic nucleus (SCN): calcium and gene expression. (A) Representative images from combined bioluminescent and fluorescent recordings of circadian PER2 (cyan) and [Ca2+]i (green) in organotypic SCN slice culture. (B) Schematic plot of “A day in the life of the SCN” assembled from a series of combined recordings as in A, registered via PER2 and [Ca2+]i rhythms. This reveals phase-specific circadian cycles of electrical activity, calcium-dependent gene expression (CRE), and transcriptional and posttranslational feedback loop (TTFL) functions, clustered around circadian day, and electrical inactivity, and peroxiredoxin overoxidation in circadian night. The electrical activity peak and molecular cycle are intimately interdependent. (C) Circuit-level SCN timekeeping is embodied in a spatiotemporal wave of circadian gene expression, plotted as the center of mass of PER2 bioluminescent signal over three sequential circadian cycles of wild-type (WT) SCN (left) (overlaid with a standardized SCN schematic). This structure is lost in CRY-deficient SCN (right). (Redrawn from data in Edgar et al. 2012, Brancaccio et al. 2013, and Edwards et al. 2016.)
The assumption is that the wave of molecular activity reads out to a coordinated wave of electrical activity, although this is currently difficult to monitor with the spatial and temporal precision of bioluminescence imaging. What is clear is that perturbations in the TTFL that disturb circadian rhythms in gene expression have similar effects on firing rate rhythms (Herzog et al. 1998; Albus et al. 2002; Nakamura et al. 2002). If the gene expression wave is indeed accompanied by a corresponding electrical wave, it means, first, that different subpopulations of SCN neurons hold contrasting activity phases. This differential phasing may be evidence of reciprocally inhibitory interactions between clock cells, as recently shown for the Drosophila clock circuit (Liang et al. 2016). Second, the various outputs of the SCN that are innervated by particular subpopulations will be regulated differentially by temporally specific SCN programs. These programs may cut across the conventional divisions of the SCN defined by neuropeptide expression, such that, for example, different subpopulations of SCN neurons will activate and suppress their particular targets with unique temporal patterns. The relationship between the temporal structure of overt behavior and SCN cell populations is considered further below.
In addition to the day–night differences in firing rate observed in SCN neurons, passive membrane properties also change across circadian time. For example, the mean resting membrane potential of SCN neurons is significantly more depolarized during the day and more hyperpolarized at night. Furthermore, the mean input resistance is higher during the day than at night (Colwell 2011). These observations have led to the hypothesis that daily oscillations in electrical activity in SCN neurons are driven by specific ionic conductances. Indeed, there are day–night differences in the current magnitude required to hold SCN neurons at –60 mV in which the mean holding current peaks during the subjective day and decreases during the subjective night. Additionally, K+ channel blockers have larger effects during the subjective night on resting membrane potential and input resistance in PER1-expressing SCN neurons (Kuhlman and McMahon 2004). These observations suggest that the day–night changes in the functional expression of K+ channels are primarily driving the daily oscillations in electrical activity observed in SCN neurons. For example, circadian changes in the activation of BK channels regulate excitability of SCN neurons (Whitt et al. 2016). In addition, there is a diurnal variation in a leak Na+ current encoded by the NALCN channels in mouse SCN neurons (as well as in Drosophila clock neurons) (Flourakis et al. 2015). This subthreshold, voltage-independent channel plays a critical role in regulating repetitive firing rates, resting membrane potential, and input resistance, particularly during the day. Pharmacological and genetic manipulations of NALCN channels disrupted daily oscillations in clock neuronal activity and locomotor behavior in the fly, and altered firing and membrane properties in SCN neurons. These observations suggest a conserved “bicycle model” in which antiphasic cycling of Na+ and K+ conductances regulates the daily oscillations of electrical activity in neurons (Fig. 4). Although NALCN channels are critical in regulating daytime firing properties, the hyperpolarized resting membrane potential and decreased input resistance observed at night in mouse SCN neurons suggests that the overall change in K+ conductance is primarily driving the daily oscillations of electrical activity. Studies focused on identifying specific ionic conductances that are regulated by the molecular clock to dictate daytime and/or nighttime firing properties in clock neurons are considered further in Allen et al. (2016). Thus, daily oscillations in K+ and Na+ conductances enable clock neurons to convert their TTFL into circadian rhythms in output.
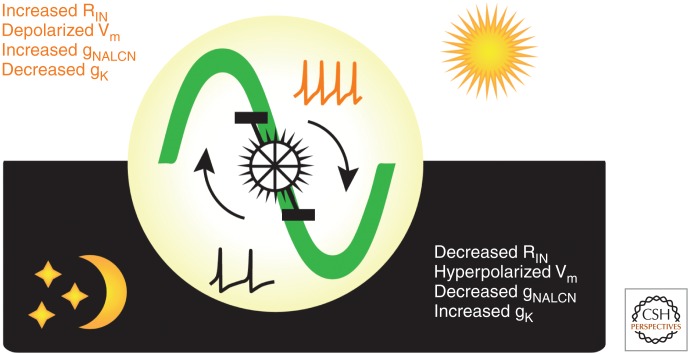
Figure 4.
Circadian regulation of suprachiasmatic nucleus (SCN) neuronal excitability. A schematic that summarizes the daily changes in excitability of SCN neurons as a “bicycle model.” Each neuron has the intrinsic capacity to generate daily oscillations in electrical activity. During the day or “up-state,” a Na+ leak conductance (gNALCN) increases while the overall K+ conductance (gK) decreases, thereby resulting in an increase in input resistance (RIN) and a more depolarized membrane potential (Vm) and higher firing rates. At night, the neurons enter a “down-state,” with lower RIN, hyperpolarized Vm, elevated gK, and lower gNALCN and firing rates. In this way, the excitatory drive of Na-currents during the day is opposed by elevated K-currents at night. (Redrawn from data in Flourakis et al. 2015.)
The changes in firing and membrane properties observed in SCN neurons, however, are not merely the output of the molecular clockwork, they can also affect the clockwork. Treatment with TTX, which blocks action potential firing, causes circadian gene expression cycles to lose amplitude and precision (Yamaguchi et al. 2003), confirming that the molecular clock is sensitive to electrical activity. Furthermore, toxin-induced blockade of synaptic vesicle recycling, an alternative method to curtail interneuronal signaling, also compromises the molecular clock (Deery et al. 2009). These results have been interpreted as evidence that cell–cell communication amplifies the intracellular circadian oscillator. Going one step further, there is some evidence that cell-autonomous membrane potential oscillations are important for daily cycles in the molecular clock. For example, chronic hyperpolarization of SCN neurons can blunt circadian rhythms in Per1 expression (Lundkvist et al. 2005). In addition, some ionic conductances, when altered, affect circadian clock gene expression. For example, loss of α subunits Kv1.4 and Kv4.2, which encode for the A-type K+ current, increases firing rate in SCN neurons during the day and night and significantly shortens the circadian period of PER2 expression (Granados-Fuentes et al. 2012, 2015). Finally, direct depolarization or hyperpolarization of SCN neurons with light-activated channelrhodopsin or halorhodopsin, respectively, phase-shifted the rhythms of PER2 expression in vitro and locomotor activity in vivo (Jones et al. 2015). More specifically, activation in early circadian night, when firing rate was declining, resulted in a phase delay, whereas direct activation in late circadian night, before the daily increase in firing, caused a phase advance of the PER2 rhythm. Phase shifts were blocked by simultaneous treatment with TTX, showing that action potential firing was necessary to reset the molecular clockwork. Taken together, this is strong evidence that changes in repetitive firing rates, an output of the clock, can in turn reset the molecular clockwork of circadian neurons. Future work should test whether daily rhythms in gene expression depend on specific ionic conductances in the SCN, and which ionic conductances are critical to initiate firing during the day and reduce firing at night.
CONTROLLING THE MOLECULAR CLOCKWORK
How might the electrical firing of neurons engage with the molecular clockwork? One likely avenue is via increases in intracellular calcium levels ([Ca2+]i) because of the opening of voltage-gated calcium channels and release from intracellular stores (see Allen et al. 2016). Consistent with this, fluorescent imaging using a genetically encoded calcium reporter has revealed a pronounced circadian cycle of [Ca2+]i in SCN neurons that peaks at CT06, coincident with high firing rates and increasing Per expression, as revealed by simultaneous bioluminescent imaging (Brancaccio et al. 2013). Moreover, although this rhythm was initially thought to be independent of action potential firing and reach its maximum about 4 h in advance of peak electrical activity (Ikeda et al. 2003), more recent analysis using genetically encoded reporters indicates simultaneous peaks of cytosolic [Ca2+]i and electrical activity. Furthermore, the cytosolic [Ca2+]i rhythm is abrogated when action potentials are blocked by TTX (Enoki et al. 2012; Brancaccio et al. 2013), as observed in Per gene expression. The regulatory elements of Per1 and Per2 contain calcium-response elements (CREs) that may mediate the effect of the [Ca2+]i rhythm on Per transcription (Travnickova-Bendova et al. 2002), and indeed CRE-dependent gene expression (as reported by a CRE-luciferase viral construct) shows a pronounced circadian cycle that peaks after the daily surge in [Ca2+]i and before peak Per1 expression (Brancaccio et al. 2013). Furthermore, direct pharmacogenetic activation of [Ca2+]i in SCN neurons, using DREADD technology to stimulate Gq signaling, not only increases CRE-dependent transcription, but also acutely induces Per expression and chronically disorganizes the ongoing rhythm (Brancaccio et al. 2013). These observations show that [Ca2+]i signaling plays a critical role in maintaining the molecular clockwork.
The axis of [Ca2+]i and CRE-dependent gene expression is also the means by which activity in the RHT is able to entrain the SCN clock to solar time. The RHT terminals are glutamatergic, and a series of in vivo and ex vivo studies have shown that the RHT acts via NMDA- and AMPA-type ionotropic glutamate receptors and [Ca2+]i-dependent signaling cascades (MAPK/ERK) to shift rhythms of electrical activity, gene expression, and behavior. Importantly, resetting is accompanied by increased firing rate of SCN neurons and transcriptional activation, especially of immediate-early genes such as c-fos that contain CRE sequences. In this respect, Per1 and Per2 can also be thought of as immediate-early genes being acutely and directly activated in the SCN by retinally derived glutamatergic signaling (Kuhlman et al. 2003). Glutamate-induced phosphorylation of the CRE-binding protein (CREB) by calcium-dependent kinases is a critical stage in this transduction process. By delivering an additional bolus of PER protein, a nocturnal light pulse will either delay or advance the TTFL, as a function of whether the bolus is delivered, respectively, on the falling or the rising phase of the ongoing oscillation of PER levels. Conversely, light pulses delivered during the circadian day when spontaneous firing rate (SFR), [Ca2+]i levels and PER expression are already high will have little effect on the clock and behavior. Thus, a common transduction mechanism mediates the impact of spontaneous electrical activity on the molecular clockwork and the glutamatergic induction of phase shifts of the TTFL in retinorecipient SCN neurons.
The precise molecular details of the signaling cascade activated in retinorecipient neurons have recently been further elaborated. DNA microarray studies have shown that several hundred genes are down-regulated and ∼100 genes are acutely up-regulated by a delaying light pulse (Jagannath et al. 2013). The latter group included the coactivator of CREB, CRTC1 (Sakamoto et al. 2013), and salt inducible kinase 1 (Sik1). The significance of this dual-activation is that, whereas CRTC1 facilitates the transcriptional actions of pCREB, Sik1 deactivates CRTC1 by phosphorylating it and thereby curtails pCREB activity. The transcriptional actions of a light-pulse in the SCN are therefore tightly defined, and circumscribed in time. Transcription is not, however, the only control point, and it is interesting, therefore, that nocturnal light pulses increase the abundance of inducible PER proteins by phosphorylating eIF4E to control their translation (Cao et al. 2015). This is triggered by the MAPK pathway, a known target of glutamatergic cues in the retinorecipient SCN, and the upstream regulator of the kinases (MNK) that phosphorylate eIF4E. Importantly, this regulatory step is also under direct circadian control, such that the spontaneous surge of PER expression in circadian daytime is also enhanced at the translational level. This observation further reinforces the view that the molecular mechanisms of resetting are also components of the spontaneous cycle within the TTFL: inputs to the clock can function within the oscillatory mechanism itself.
Beyond these qualitative models on the intracellular responses, a quantitative understanding of how the SCN clock is entrained by light pulses is not yet available. Moreover, even though events in the retinorecipient VIP and GRP neurons of the SCN core likely initiate photic phase shifts in the SCN, resetting ultimately requires circuit-wide readjustments of the core and shell in combination, which is dependent on interneuronal signaling and shows complex spatiotemporal patterns (Nagano et al. 2003). In this regard, neuropeptidergic signaling via G-coupled receptors may be an important mechanism for engagement of [Ca2+]i and CRE-dependent control of the TTFL in nonretinorecipient SCN neurons. To address this issue, it is necessary to move from cell-autonomous mechanisms and to consider the circuit-level properties of the SCN.
INTRINSIC MECHANISMS OF SYNCHRONIZATION
How do SCN cells synchronize to each other to produce a coherent daily rhythm in the neuronal ensemble and its associated spatiotemporal wave? This mechanistic question has inspired many computational modelers because of its profound implications for understanding how emergent properties arise from a network of coupled oscillators (Gonze et al. 2005; Bernard et al. 2007; Locke et al. 2008; Vasalou et al. 2011; Hafner et al. 2012; DeWoskin et al. 2015). For example, Webb and colleagues (2012) predicted that SCN cells would likely synchronize to each other faster if the most highly connected cells are intrinsically weaker (e.g., damped) circadian oscillators. Additional studies have also predicted that increasing connectivity from the SCN core to the shell during winter, for example, could explain the seasonally tighter distribution of phases among SCN cells, whereas the loss of connections from the core to the shell could explain the fragmentation of daily rhythms with aging (Vasalou et al. 2011; Bodenstein et al. 2012). Others have predicted that the topology of connections within the SCN opposes large shifts and thus underlies the robust nature of SCN rhythmicity and its modern consequence, jet lag (Hafner et al. 2012).
To address directly the mechanisms and consequences of circadian synchrony, researchers have exploited the ability to record rhythms from many cells in the isolated SCN simultaneously. The most obvious manifestation of circuit-level organization of the SCN is the spatio-temporal wave of Per and Cry gene expression that starts from the dorsomedial lip and sweeps ventrally across the nucleus. First observed in ex vivo brain sections (Hastings et al. 1999; Koinuma et al. 2013), the wave can also be observed with exquisite precision in organotypic slices (Yamaguchi et al. 2003; Maywood et al. 2013). The transcriptional rhythm is also associated with, and likely synchronized by, a circadian wave of [Ca2+]i (Enoki et al. 2012; Brancaccio et al. 2013). As with CRE-dependent transcriptional rhythms, the [Ca2+]i wave is suppressed by TTX and coherence between dorsal and ventral compartments is progressively lost. The waves of [Ca2+]i and gene expression rely, therefore, on neuronal firing for their definition. As for the orientation, it has been suggested that neurons in the dorsomedial lip of the SCN, characterized by enriched expression of RGS16 (an inhibitor of Gi signaling), provide the point for initiation of the wave perhaps because they have an early disinhibition of cAMP signaling (Doi et al. 2011). Certainly, in mice lacking RGS16, the phase spread of the wave is curtailed and the periods of SCN and behavioral rhythms are lengthened. The expression of RGS16 may create a shorter cell-autonomous period in these neurons, causing them to phase-lead the circuit, but when seeking to identify the “phase leaders” there is a caveat. The waves of [Ca2+]i and gene expression may reflect a release from inhibition, rather than activation, and so the source of the wave may arise in inhibitory inputs to the dorsomedial lip projecting from elsewhere in the SCN circuit. The second caveat is that recent studies with temporally chimeric mice, in which the SCN contains a spatially heterogeneous mixture of neurons with either 24 h or 20 h genetically specified cell-autonomous periods, nevertheless show spatiotemporal waves of PER2::LUC expression no different from WT and CK1ɛTau mutants (Smyllie et al. 2016). This suggests that the generation of the daily wave remains intact despite a bimodal distribution of intracellular periods. This observation does not question the relevance of the RGS16-positive cells as potential “phase leaders,” but it does suggest other mechanisms that specify the spatiotemporal wave of gene expression. Given its prolonged time-base, acute signaling events such as action potential firing per se may not be relevant, although the demonstrated paracrine properties of VIP and other peptidergic signals (see below) provide one possible means of sculpting and coordinating the wave.
Role of VIP: A Master Synchronizer with the Power to Desynchronize
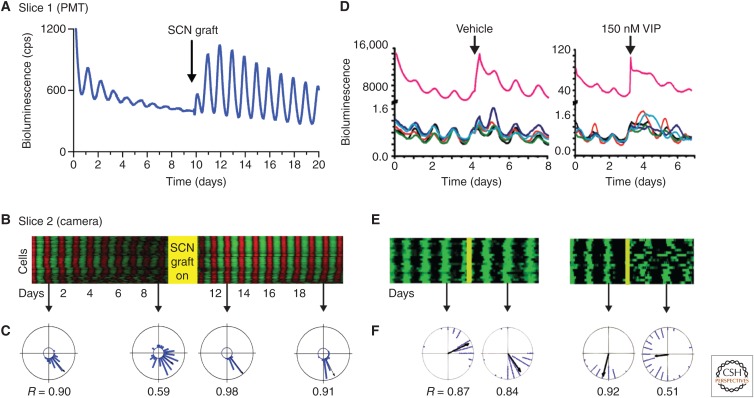
As recipients of RHT input, VIP cells are of primary importance in entraining the SCN, but they exercise a far more pervasive role in synchronizing the entire circuit. The loss of the genes encoding the VIP precursor peptide or the VIP receptor, VPAC2, compromises circadian control of behavioral and endocrine rhythms (Harmar et al. 2002; Colwell et al. 2003). At a neuronal level, individual cellular oscillations of electrical firing or gene expression are desynchronized (Fig. 5A–C) (Aton et al. 2005; Maywood et al. 2006). Moreover, the molecular oscillations lose amplitude, showing that VIP-mediated signaling is necessary both to sustain and to synchronize the cell-autonomous clockwork. Underlying the disorganized molecular and electrophysiological rhythms is a comparable desynchrony of individual neuronal rhythms of [Ca2+]i and CRE-dependent transcription (Brancaccio et al. 2013). VIP, therefore, is responsible for linking the SCN circuit together, likely through its parallel actions on adenylyl cyclase/cAMP and phospholipase C/[Ca2+]i signaling in cells expressing the VPAC2 receptor (An et al. 2011). In the absence of VPAC2 expression, ERK/MAPK and pCREB signaling in the SCN is responsive to retinal activation at all circadian phases. This loss of a “circadian gate” is likely the result of a proportion of desynchronized SCN neurons being in the responsive phase at any time point (Maywood et al. 2007). Under a light–dark cycle, this acute responsiveness is sufficient to synchronize the SCN and sustain a daily rhythm of behavior, but free-running rhythms break down on release to continuous darkness as the cellular rhythms dissociate in the absence of VIP signaling. Intriguingly, coherence can be restored in vivo by scheduled wheel-running activity (Power et al. 2010). This is likely mediated by arousal-relevant neurochemical inputs to the SCN, such as NPY and 5-HT from the ventral thalamus and brainstem, respectively, and highlights significant plasticity in the VIP-deficient circuit. In organotypic cultures, circuit level coordination can be restored in a VIP-deficient SCN slice by coculturing with a VIP-proficient SCN (Fig. 5A–C) (Maywood et al. 2011b). Importantly, the period of the restored molecular oscillation of the VIP-null SCN is determined by the genotype of the graft, confirming that VIP signaling transmits temporal information between SCN neurons and across circuits, and is not simply a permissive signal for the molecular clock of the VIP-null SCN to run at its own pace. Moreover, VIP-mediated restoration can occur via a paracrine mechanism, providing a different spatio-temporal perspective on the role of neuropeptides in SCN circuitry. In the absence of effective VIP signaling, not only is the spatiotemporal wave of circadian gene expression lost, but also there is no accompanying spatial structure to circadian gene expression, even where some temporal coherence could be identified in the SCN slice (Pauls et al. 2014). Spatiotemporal gradients in VIP-mediated signaling may therefore be a core feature of circuit-level function in the SCN. The development of real-time reporters of neuropeptidergic function would enable significant advances to be made in this area.
Figure 5.
Vasoactive intestinal peptide (VIP) as a circadian synchronizer and de-synchronizer of suprachiasmatic nucleus (SCN) circuits. (A) Aggregate PER2 bioluminescence from VIP-null SCN recorded before and after graft with wild-type SCN (arrow). In the absence of VIP, SCN organotypic slices lose circadian amplitude and the coherence of cell-autonomous molecular oscillations, and these can be restored by coculture with a wild-type, VIP-competent SCN. (B) Raster plots of cellular PER2 bioluminescence recorded by CCD, before and after grafting (different slice from A). (C) Rayleigh plots of cellular phase determined in relation to grafting. (D) In wild-type SCN, treatment with exogenous VIP can desynchronize the previously tightly coupled circuit. (E, F) Raster and Rayleigh plots as in B and C. (Redrawn from data in Maywood et al. 2011b and An et al. 2013.)
It is thus easy to refer to VIP as the master synchronizer of circadian rhythms. Recent results, however, provide evidence that VIP can also reduce synchrony (An et al. 2013). When applied in vitro or in vivo to the SCN, VIP phase shifts some cells more than others. This is especially apparent at higher concentrations of VIP (e.g., 150 nM) in which the transient desynchronization has been termed “phase tumbling” (Fig. 5D–F). This observation led to the hypothesis that low concentrations of VIP entrain circadian cells to each other, whereas higher levels of release, which may result from light-induced activation of VIP neurons by RHT input, weaken the synchrony. It is possible that this is a design feature of the SCN to allow it to entrain to environmental lighting cues, loosening up the clock circuit when it needs to be reset. Consistent with this, administration of VIP before a shift in the light cycle can accelerate reentrainment in vivo. Future experiments are required to resolve how VIP release is regulated by environmental and circadian signals to allow the SCN to be a robust coordinated circadian clock that nevertheless remains sufficiently sensitive to adjust to changes in the environment.
Role of GABA: Modulating Synchrony with the Seasons
Is it as simple as “VIP sets synchrony in the SCN”? The only neurotransmitter produced and received by all SCN neurons is γ-aminobutyric acid (GABA) (Moore and Speh 1993; Moldavan et al. 2015), which also modulates the phase relationships among SCN cells from day to day and across the seasons. Until recently, the role of GABA within the SCN was unclear. Although daily addition of GABA can synchronize SCN rhythms (Liu and Reppert 2000), GABA is not required for SCN cells to maintain their daily synchrony (Aton et al. 2006; Evans et al. 2013; Freeman et al. 2013). Yet, blocking GABA signaling impairs phase shifts in response to light (Ralph and Menaker 1989; Gillespie et al. 1997) and synchronization of the dorsal and ventral SCN following a shift in the light cycle (Albus et al. 2005). Finally, GABA receptor antagonists enhance synchrony and precision in the SCN (Aton et al. 2006), rescue coordinated circadian rhythms in VIP-deficient SCN (Freeman et al. 2013), and abolish stochastic circadian oscillations in BMAL1-deficient SCN (Ko et al. 2010). These results led to the hypothesis that GABA actively opposes synchrony in the SCN by injecting “jitter” into rhythms. Mechanistically, it could be proposed that VIP and GABA exert opposing effects on SCN neurons: loss of VIP causes hyperpolarization, whereas loss of GABAergic tone leads to depolarization. Under normal circumstances, the coordinated actions of VIP and GABA leave the membrane potential of SCN cells in an operating range that supports the molecular clockwork and interneuronal communication, and this is perturbed when either the VIP or GABAergic input is attenuated. Thus, loss of GABA can restore synchronized rhythms in a VIP-defective SCN by reversing the associated hyperpolarization. This proposed relationship between the effectiveness of the TTFL and the “permissive operating range” of membrane potential is an unexplored feature of cell-autonomous and circuit-level timekeeping in the SCN and warrants greater attention.
Three recent studies have placed the role of SCN GABA into an intriguing ecological context—photoperiodism. Many organisms adjust their behavior and physiology (e.g., reproduction) to longer days of summer. In all organisms, this seasonal (photoperiodic) induction depends on a circadian clock and in mammals the SCN is necessary to respond to seasonal cues. Part of this response includes changes within the SCN. After several weeks of exposure to short days (long nights), cells within the SCN compress their times of daily peak firing and Per gene expression to peak around midday. This coincides with a dramatic decrease in GABA-dependent postsynaptic potentials during the day (Farajnia et al. 2014). During long days, however, SCN cells distribute their phase relationships more widely, so that they are less synchronized in their time of daily peak activity. This coincides with a depolarization in the equilibrium potential for GABA (i.e., chloride) (Myung et al. 2012; Farajnia et al. 2014). Consequently, GABA signaling has the potential to switch from low-frequency, inhibitory communication to higher-frequency, often excitatory, communication. These results complement the observation that GABA becomes important on long days for the dorsal and ventral SCN to resynchronize, raising the possibility that GABA might oppose synchrony at some times of year—and promote synchrony at others (Evans et al. 2013; DeWoskin et al. 2015). In support of this hypothesis, experiments and computational modeling argue that, during the long days of summer, GABA acts as an excitatory neurotransmitter and phase-repulsive (desynchronizing) coupling factor (Myung et al. 2015).
Observations from these previous studies have raised some additional questions that will help in elucidating the relative roles of VIP and GABA as synchronizing and desynchronizing agents, respectively. For example, does VIP signaling modulate resting membrane potential (e.g., with the changing day length) to determine GABA’s actions? Second, does GABA modulate the actions of VIP? Finally, what are the epigenetic steps that normally mediate the photoperiodic changes in GABA and VIP signaling? Of the many possible, proximal targets for day-length-dependent changes in the SCN, synaptic plasticity has perhaps attracted the most attention. Recent advances in our ability to map connections and synaptic strength in the nervous system have revealed that VIP neurons, for example, make sparse, GABAergic synapses with some preference for neurons that do not express VIP (Fan et al. 2015). Simultaneous recordings from >100 SCN neurons in dispersed cultures revealed that GABAergic connections are sparse, stable for days, change in strength with time of day, and are important for reducing the cycle-to-cycle precision of the SCN (Freeman et al. 2013). It will be informative to map other connections in the SCN (e.g., VIP, AVP) and monitor their changes in response to seasonal cues. More speculatively, it is intriguing to consider that disruption of photoperiodically induced changes in the balance between VIP and GABA might underlie mood and metabolic disorders, including seasonal affective disorder.
Role of AVP-Rs: Weaker Synchronizers
In contrast to the dramatic effects of loss of VIP signaling on the SCN clock, impairments of AVP and GRP signaling have until recently not been viewed as significant. For example, AVP has been considered an E-box-regulated target of the TTFL, acting principally as an output factor but with little influence within the SCN (Jin et al. 1999). Nevertheless, AVP and GRP receptors are broadly expressed in the SCN and appear able to mediate weak coordination between circadian cells, especially in the absence of VIP (Maywood et al. 2011b). Loss of the AVP receptors in vivo accelerates the reentrainment of mice exposed to shifts in their lighting cycle (Yamaguchi et al. 2013). One interpretation of these results is that loss of this coupling signal loosens up the circuit and thereby allows the population of SCN cells to adjust more readily to environmental cues. This observation echoes the biphasic effects of VIP and the phenomenon of “phase-tumbling,” because in both cases “loosening” of the network is associated with more rapid resetting. Given that VIP and AVP cells are anatomically segregated to the core and shell regions of the SCN, this suggests that they exert reciprocal effects on synchrony, both as subpopulations and also across the entire circuit. VIP release by the core maintains synchrony in the shell, but equally the shell in turn regulates the response of the core to retinal input. Together, these results lead to a model in which there is a hierarchy of partially redundant peptidergic signals mediating synchrony in the SCN with VIP most potent and AVP (and possibly GRP) providing weaker effects, but their overall reciprocity generates a very stable and precise oscillatory network.
Role of ZFHX3: Bridging Cells and Circuits
The emerging model, therefore, is that neuropeptides engage the TTFL of SCN neurons and thereby maintain synchrony and amplitude of cellular oscillations across the circuit. Recently, a mutagenesis screen has identified an additional transcriptional axis in the SCN. The Short-circuit (Sci) mutation of the transcription factor ZFHX3 causes a short period of behavioral and SCN molecular rhythms in the heterozygote mouse (the homozygous mutation is embryonic lethal) (Parsons et al. 2015). It is associated with reduced expression of various SCN neuropeptides and neuropeptide receptors, including VIP and VPAC2. ZFHX3 controls gene expression via enhancer sequences rich in A and T called AT-boxes (its alternative name is AT-binding factor, ATBF1), and several of the peptidergic genes dysregulated in the SCN of Zfhx3Sci mice carry AT-boxes. Furthermore, their transcriptional activation in vitro is compromised by the mutation. The additional feature, however, is that by using a lentiviral bioluminescent reporter carrying concatenated AT-boxes, it was possible to show that AT-dependent transcription in the SCN is circadian and directed by the TTFL. Moreover, the period of the AT-box rhythm was lengthened by inhibition of CK1δ/ɛ, confirming that the core TTFL drives transcriptional activity at AT-boxes. This may, however, be dependent on circadian changes in the expression of cofactors because Zfhx3 expression is not rhythmic. Importantly, the coherence of the rhythm of AT-box expression is compromised in SCN from Zfhx3Sci mice and the period shortened. Conversely, hemizygous deletion of Zfhx3 lengthened SCN period, indicating that Zfhx3Sci is a gain-of-function, rather than a null, mutation. Thus, the ZFHX3/AT axis is a circadian transcriptional output of the TTFL, and, because it regulates neuropeptidergic signaling, it also provides an input to the TTFL. The ZFHX3/AT axis therefore bridges across cell-autonomous and circuit-level circadian functions (Parsons et al. 2015). Other developmentally relevant transcription factors are also known to regulate specific neuropeptidergic expression in the SCN, suggesting that the motif of combined cell-autonomous and circuit-level activity seen in ZFHX3 may be more prevalent (Bedont et al. 2014). In the particular case of ZFHX3, the effects on period of heterozygotes deletion and in vivo knockdown occur after maturation of the SCN and therefore are independent of any developmental role regulated by this transcription factor.
Are There “Pacemaker Cells” in the SCN?
In addition to its cellular synchrony and spatiotemporal wave of gene expression, another emergent property of the SCN circuit is ensemble period. When synaptic communication is blocked with TTX, the molecular clocks of individual SCN neurons express their intrinsic cell-autonomous periods, which can deviate considerably from the ensemble period in the absence of TTX. This raises the question of whether ensemble period is determined by a subset of “pacemaker” cells, or by a circuit-level computation to which all of the individual SCN neurons contribute equally? One way to address this is to create temporally chimeric SCN, consisting of neurons with a range of distinct, genetically specified intrinsic periods and then examine how the circuit behaves. In embryonic chimeras carrying WT and long-period ClockΔ19 mutant cells in an untargeted mix, strong evidence was provided for a simple averaging model for the determination of period (Low-Zeddies and Takahashi 2001): the period of circadian behavior of individual mice was in direct proportion to the relative abundance of mutant and WT cells in their SCN. This led to the conclusion that the “complex integration of cellular phenotypes determines the generation and expression of coherent circadian rhythms at the organismal level” (Low-Zeddies and Takahashi 2001).
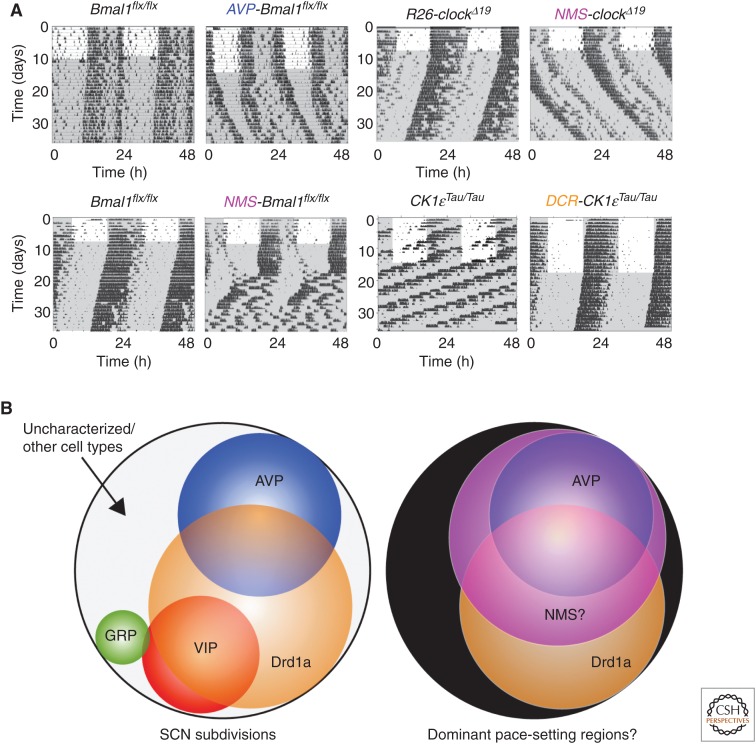
More recently, intersectional genetics has been used to address the same question, but now with the advantage that specific subpopulations can be addressed, creating targeted chimeras (Fig. 6A) (Lee et al. 2015; Mieda et al. 2015; Smyllie et al. 2016). Two types of strategies have been used to generate temporally chimeric animals. First, either reversible (“TET-off”) overexpression of CLOCKΔ19 or deletion of BMAL1 was used to disrupt TTFL clock function in targeted cell populations. Overexpression of CLOCKΔ19 in the bulk of VIP and AVP cells (characterized by their coexpression of the peptide neuromedin S [NMS]) led to reversible lengthening of the period of behavioral and SCN molecular rhythms (Lee et al. 2015). Given that the targeted neurons represent only 40% of the total SCN, the extension of period by ca. 1 h in vivo and 2 h in vitro suggests that the NMS neurons can exert a disproportionate effect on ensemble period. This argues against the simple averaging model of period determination. In addition, constitutive deletion of BMAL1 from the NMS neurons rendered the mice arrhythmic, as did conditional and reversible overexpression of PER2. For the SCN to function as a clock at the circuit level, therefore, the NMS neurons have to have functional cell-autonomous clocks. Moreover, the period of the SCN is determined by the period of the NMS cells, identifying them as a “pacemaker” population. Finally, conditional expression of tetanus toxin in NMS cells was used to compromise synaptic vesicle release, blocking their communication with the rest of the SCN circuit. Consequently, circadian behavior was dramatically and reversibly disrupted and, at the SCN level, ensemble oscillation of bioluminescence was damped and a subset of cells became arrhythmic, showing the necessity of synaptic signaling from these cells to maintain function across the SCN. The NMS subpopulation therefore fulfills the criteria to be defined as a “pacemaker.” Ensemble circadian timing requires a functional cell-autonomous clock in these cells; the ensemble period is determined by the period of these cells, and to sustain circadian timekeeping these cells need to be able to communicate with the rest of the SCN. This elegant, groundbreaking work carries, nevertheless, a caveat—given that NMS cells constitute almost all of the VIP and AVP cells, and that loss of NMS itself has no effect on SCN timekeeping, it is not possible to assign specific roles for these two, well-studied cell types of the SCN that constitute elements of both core and shell. It is likely that a “pacemaker” population constitutes a subset of NMS neurons, and thus further subdivision is required.
Figure 6.
Dissection of suprachiasmatic nucleus (SCN) circuit-level pacemaking by intersectional genetic approaches. (A) By manipulating components of the transcriptional and posttranslational feedback loop (TTFL) (BMAL, CLOCK) or the TTFL regulator, CK1ɛ, in defined subpopulations of SCN neurons (AVP, NMS, D1aR), it is possible to alter the period and/or the coherence of mouse activity/rest behavioral rhythms. (Redrawn from data in Lee et al. 2015, Mieda et al. 2015, and Smyllie et al. 2016.) (B) Schematic representation (left) of the partially overlapping subpopulations of SCN neurons, and a meta-analysis (right) of putative pace-setting regions. (Redrawn from data in Smyllie et al. 2016.)
This issue has been addressed by using an AVP-Cre mouse to achieve selective deletion of BMAL1 in AVP cells (Mieda et al. 2015). This did not lead to arrhythmia, as might have been anticipated if the AVP neurons were dominant drivers to the circuit. Rather, the mice showed long periods of circadian behavior and SCN bioluminescence rhythms. At an operational level, therefore, AVP neurons can be considered pacemakers because manipulation of their cell-autonomous clock affects ensemble period. Furthermore, CCD imaging revealed that cells in the dorsal SCN, a principal location of AVP neurons, had low amplitude and unstable molecular oscillations of variable periods, whereas at a behavioral level the mice showed more rapid reentrainment to altered lighting cycles. This is reminiscent of the rapid resetting of mice deficient in AVP receptors, again emphasizing that a “loosened” clock network is more responsive to altered lighting cycles (Yamaguchi et al. 2013). Importantly, the effects of genomic deletion of BMAL1 were rescued by targeted AAV-mediated expression of recombinant BMAL1 in the SCN (Mieda et al. 2015), but again there are caveats. The most significant is that deletion of BMAL1 disrupted the expression of a large number of neuropeptides in the SCN. Therefore, the observed phenotype, as with the comparable manipulations of NMS neurons, is likely a result of both cell-autonomous and circuit-level effects. For example, expression of AVP and other neuropeptides (but not VIP) was dramatically reduced, and the retinal induction of Per1 expression in the core SCN was attenuated, even though resetting was accelerated. Assignment of particular circadian functions to AVP and non-AVP cells is therefore difficult.
A complement to these transcription factor-based approaches therefore requires an alternative strategy that preserves neuropeptide-dependent, circuit-level communication within the SCN. This has been achieved using Cre-mediated deletion of the CK1ɛTau mutation in SCN subpopulations. The SCN from such mice contain cells that are either Tau-competent or Tau-deleted, and so express periods of either ca. 20 h or 24 h, respectively (Smyllie et al. 2016). Tau deletion was targeted by the gene encoding the dopamine 1a receptor (Drd1a), a population that covers 60% of SCN neurons, spanning both core and shell, and likely overlapping with the NMS cell population. Importantly, these chimeric SCN circuits can sustain stable, high-amplitude circadian oscillations of behavior and gene expression, despite incorporating cells with very divergent cell-autonomous periods. This highlights the power of neuropeptidergic signaling pathways that are not compromised in these SCN. In the majority (ca. 60%) of mice and SCN slices, the emergent period was 24 h, identifying the Drd1a cells as circuit pacemakers. There was, however, some variability and a significant minority (ca. 35%) of mice and SCN had periods closer to 20 h, suggesting that non-Drd1a cells can also act as pacesetters on occasion. Indeed, a few mice and SCN (ca. 10%) had unstable periods that flipped between ca. 20 h and ca. 24 h, suggesting some sort of ongoing competition between cell populations in specifying the emergent period. CCD imaging of SCN with stable 24 h or 20 h periods treated with TTX revealed the range of cell-autonomous periods in the slice. Importantly, the ensemble period observed in the absence of TTX was not the arithmetic mean of the individual cell-autonomous periods. In 24 h SCN, the ensemble period was longer than the mean, whereas in 20 h SCN it was shorter. The computation of ensemble period, therefore, is not linear: in both cases, one subpopulation of cells exerted greater influence.
Surprisingly, chimeric 24-h animals were nevertheless able to entrain to both 20 h and 24 h lighting schedules, and maintained the entrained period in subsequent free-running conditions: the computation of SCN period was therefore reprogrammed in vivo. When SCN rhythmicity was subsequently assessed in vitro, the initial period also reflected the period seen in vivo. Interestingly, there appeared to be a “relaxation” effect in which the period gradually changed back to 24 h. This suggests that period setting is complex, in which additional inputs (e.g., from other brain areas) may be able to alter the computation of circadian period. This remarkable flexibility thus begs the question of whether “absolute” pacemaker cells exist, or whether in fact pace setting reflects both the environmental and circuit context. Nevertheless, under normal 24 h environmental conditions, given that the NMS cells constitute both AVP and VIP neurons, and deletion of BMAL1 in VIP cells had very little effect on circadian behavior and cellular rhythms, the combination of these investigations (Lee et al. 2015; Mieda et al. 2015; Smyllie et al. 2016) points toward an important role for both AVP neurons and non-VIP NMS and Drd1a neurons as the dominant pacemaking elements in a nonlinear, hierarchical SCN circuit (Fig. 6B).
Taken together, these studies lead to the intriguing conclusion that circadian rhythms are generated within cells and sustained and synchronized by diverse cell–cell interactions. Efforts to map the connections that mediate circadian synchrony require the development of new methods. One recent investigation took advantage of the resynchronization of circadian cells within the SCN following removal of TTX to infer the underlying functional connections (Abel et al. 2016). Based on fluctuations in PER2 expression recorded during resynchronization, they applied the maximal information coefficient statistic to identify pairs of neurons that share a high degree of mutual information. They found that individual cells tend to have few connections, but those in the SCN core have more connections than those in the shell. Strikingly, they found strong connections between the left and right SCN core regions. Future studies should focus on further tests to validate these inferred functional connections and to understand how this architecture supports the self-sustained, entrainable rhythms in the SCN.
Thus, the interplay between cell-autonomous and circuit-level functions confers on the SCN the emergent properties that enable it to operate as the principal clock in the brain. Nevertheless, even though the composition of the TTFL seems to be well understood, its relationship with neuronal electrical activity, the computations performed by the SCN circuit and the cellular mechanisms that serve those computations are poorly understood. Likely they involve changes at all levels. For example, changes in ensemble period following entrainment of mice to non-24-h lighting cycles are mediated by epigenetic modification, specifically DNA methylation (Azzi et al. 2014). Thus, what might be viewed as an effect on a neural network may also have consequences at the level of genomic networks.
CONCLUDING REMARKS: BRAIN CLOCKS BEYOND THE SCN
Beyond the SCN, most brain regions have local TTFL-based circadian clocks that direct rhythms of gene expression appropriate to the functions of that region, and how those functions are integrated into the daily rhythm of sleep and wakefulness (Chaudhury et al. 2005; Wardlaw et al. 2014). Ordinarily, the SCN coordinates these local clocks to ensure optimum mental function, but in the absence of effective SCN outflow, brain function can be compromised. For example, environmentally induced arrhythmia in hamsters compromises hippocampal-dependent memory, but lesioning the SCN rescues this effect (Fernandez et al. 2014), indicating that an aberrant SCN signal is worse than no signal at all. Equally, compromise of local clock functions can also disrupt brain function. For example, selective deletion of BMAL1 in histaminergic (HA) neurons in the posterior hypothalamus of mice does not affect their behavioral activity rhythms, but it does deregulate circadian histamine rhythms across the brain and this is associated with fragmented sleep and compromised hippocampus-dependent memory (Yu et al. 2014). The principal effect appears to be a failure in the tuberomamillary nucleus-BMAL (TMN-BMAL)-deficient mice to enforce the normal circadian down-regulation of HA activity during the circadian day, leading to an inability to sustain consolidated sleep. Thus, the role of the local clock in TMN neurons is to generate a daytime decline on HA activity appropriate to the greater overall SCN-defined sleep–wake cycle. A major challenge, therefore, will be to tease apart the interdependent nexus of SCN timing, local clock functions, and sleep/wakefulness. Indeed, the field has arrived at a model in which the entire nervous system can be viewed as a temporally resonant system, pretuned to oscillate on a circadian basis, each brain region locked into a pattern that complements and supports those of other regions. Therefore, events that have an impact on this circadian structure would be expected to disturb normal brain functions. This might be seen at the level of compromised sleep-dependent clearance of neurotoxic aggregates (Roh et al. 2012; Xie et al. 2013) or at the level of psychiatric illness (Frank et al. 2013). In both cases, future investigation of the close associations between “the clock” and disease, and their mechanistic basis will yield dramatic and unanticipated translational opportunities (Hastings and Goedert 2013; Frank et al. 2014).
Footnotes
Editors: Paolo Sassone-Corsi, Michael W. Young, and Akhilesh B. Reddy
Additional Perspectives on Circadian Rhythms available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abel JH, Meeker K, Granados-Fuentes D, St John PC, Wang TJ, Bales BB, Doyle FJ III, Herzog ED, Petzold LR. 2016. Functional network inference of the suprachiasmatic nucleus. Proc Natl Acad Sci 113: 4512–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albus H, Bonnefont X, Chaves I, Yasui A, Doczy J, van der Horst GT, Meijer JH. 2002. Cryptochrome-deficient mice lack circadian electrical activity in the suprachiasmatic nuclei. Curr Biol 12: 1130–1133. [DOI] [PubMed] [Google Scholar]
- Albus H, Vansteensel MJ, Michel S, Block GD, Meijer JH. 2005. A GABAergic mechanism is necessary for coupling dissociable ventral and dorsal regional oscillators within the circadian clock. Curr Biol 15: 886–893. [DOI] [PubMed] [Google Scholar]
- *.Allen CN, Nitabach MN, Colwell CS. 2016. Membrane currents, gene expression, and circadian clocks. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Irwin RP, Allen CN, Tsai C, Herzog ED. 2011. Vasoactive intestinal polypeptide requires parallel changes in adenylate cyclase and phospholipase C to entrain circadian rhythms to a predictable phase. J Neurophysiol 105: 2289–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S, Harang R, Meeker K, Granados-Fuentes D, Tsai CA, Mazuski C, Kim J, Doyle FJ III, Petzold LR, Herzog ED. 2013. A neuropeptide speeds circadian entrainment by reducing intercellular synchrony. Proc Natl Acad Sci 110: E4355–E4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand SN, Maywood ES, Chesham JE, Joynson G, Banks GT, Hastings MH, Nolan PM. 2013. Distinct and separable roles for endogenous CRY1 and CRY2 within the circadian molecular clockwork of the suprachiasmatic nucleus, as revealed by the Fbxl3Afh mutation. J Neurosci 33: 7145–7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson SE, Maywood ES, Chesham JE, Wozny C, Colwell CS, Hastings MH, Williams SR. 2011. Cyclic AMP signaling control of action potential firing rate and molecular circadian pacemaking in the suprachiasmatic nucleus. J Biol Rhythms 26: 210–220. [DOI] [PubMed] [Google Scholar]
- Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. 2005. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci 8: 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aton SJ, Huettner JE, Straume M, Herzog ED. 2006. GABA and Gi/o differentially control circadian rhythms and synchrony in clock neurons. Proc Natl Acad Sci 103: 19188–19193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzi A, Dallmann R, Casserly A, Rehrauer H, Patrignani A, Maier B, Kramer A, Brown SA. 2014. Circadian behavior is light-reprogrammed by plastic DNA methylation. Nat Neurosci 17: 377–382. [DOI] [PubMed] [Google Scholar]
- Bedont JL, LeGates TA, Slat EA, Byerly MS, Wang H, Hu J, Rupp AC, Qian J, Wong GW, Herzog ED, et al. 2014. Lhx1 controls terminal differentiation and circadian function of the suprachiasmatic nucleus. Cell Rep 7: 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard S, Gonze D, Cajavec B, Herzel H, Kramer A. 2007. Synchronization-induced rhythmicity of circadian oscillators in the suprachiasmatic nucleus. PLoS Comput Biol 3: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besing RC, Paul JR, Hablitz LM, Rogers CO, Johnson RL, Young ME, Gamble KL. 2015. Circadian rhythmicity of active GSK3 isoforms modulates molecular clock gene rhythms in the suprachiasmatic nucleus. J Biol Rhythms 30: 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenstein C, Gosak M, Schuster S, Marhl M, Perc M. 2012. Modeling the seasonal adaptation of circadian clocks by changes in the network structure of the suprachiasmatic nucleus. PLoS Comput Biol 8: e1002697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancaccio M, Maywood ES, Chesham JE, Loudon AS, Hastings MH. 2013. A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron 78: 714–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr ED, Yoo SH, Takahashi JS. 2010. Temperature as a universal resetting cue for mammalian circadian oscillators. Science 330: 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Robinson B, Xu HY, Gkogkas C, Khoutorsky A, Alain T, Yanagiya A, Nevarko T, Liu AC, Amir S, et al. 2013. Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E-BP1 signaling. Neuron 79: 712–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Gkogkas CG, de Zavalia N, Blum ID, Yanagiya A, Tsukumo Y, Xu H, Lee C, Storch KF, Liu AC, et al. 2015. Light-regulated translational control of circadian behavior by eIF4E phosphorylation. Nat Neurosci 18: 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Guarente L. 2013. SIRT1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 153: 1448–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury D, Wang LM, Colwell CS. 2005. Circadian regulation of hippocampal long-term potentiation. J Biol Rhythms 20: 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yoo SH, Park YS, Kim KH, Wei S, Buhr E, Ye ZY, Pan HL, Takahashi JS. 2012. Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci 109: 101–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS. 2011. Linking neural activity and molecular oscillations in the SCN. Nat Rev Neurosci 12: 553–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X, Waschek JA. 2003. Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am J Physiol Regul Integr Comp Physiol 285: R939–R949. [DOI] [PubMed] [Google Scholar]
- Deery MJ, Maywood ES, Chesham JE, Sladek M, Karp NA, Green EW, Charles PD, Reddy AB, Kyriacou CP, Lilley KS, et al. 2009. Proteomic analysis reveals the role of synaptic vesicle cycling in sustaining the suprachiasmatic circadian clock. Curr Biol 19: 2031–2036. [DOI] [PubMed] [Google Scholar]
- Delogu A, Sellers K, Zagoraiou L, Bocianowska-Zbrog A, Mandal S, Guimera J, Rubenstein JL, Sugden D, Jessell T, Lumsden A. 2012. Subcortical visual shell nuclei targeted by ipRGCs develop from a Sox14+-GABAergic progenitor and require Sox14 to regulate daily activity rhythms. Neuron 75: 648–662. [DOI] [PubMed] [Google Scholar]
- DeWoskin D, Myung J, Belle MD, Piggins HD, Takumi T, Forger DB. 2015. Distinct roles for GABA across multiple timescales in mammalian circadian timekeeping. Proc Natl Acad Sci 112: E3911–E3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. 2010. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu Rev Physiol 72: 517–549. [DOI] [PubMed] [Google Scholar]
- Doi M, Ishida A, Miyake A, Sato M, Komatsu R, Yamazaki F, Kimura I, Tsuchiya S, Kori H, Seo K, et al. 2011. Circadian regulation of intracellular G-protein signalling mediates intercellular synchrony and rhythmicity in the suprachiasmatic nucleus. Nat Commun 2: 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, et al. 2012. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485: 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards MD, Brancaccio M, Chesham JE, Maywood ES, Hastings MH. 2016. Rhythmic expression of cryptochrome induces the circadian clock of arrhythmic suprachiasmatic nuclei through arginine vasopressin signaling. Proc Natl Acad Sci 113: 2732–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoki R, Kuroda S, Ono D, Hasan MT, Ueda T, Honma S, Honma K. 2012. Topological specificity and hierarchical network of the circadian calcium rhythm in the suprachiasmatic nucleus. Proc Natl Acad Sci 109: 21498–21503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JP, Yu EA, Indic P, Dallmann R, Weaver DR. 2010. Casein kinase 1δ (CK1δ) regulates period length of the mouse suprachiasmatic circadian clock in vitro. PLoS ONE 5: e10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. 2013. Dynamic interactions mediated by nonredundant signaling mechanisms couple circadian clock neurons. Neuron 80: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Zeng H, Olson DP, Huber KM, Gibson JR, Takahashi JS. 2015. Vasoactive intestinal polypeptide (VIP)-expressing neurons in the suprachiasmatic nucleus provide sparse GABAergic outputs to local neurons with circadian regulation occurring distal to the opening of postsynaptic GABAA ionotropic receptors. J Neurosci 35: 1905–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farajnia S, van Westering TL, Meijer JH, Michel S. 2014. Seasonal induction of GABAergic excitation in the central mammalian clock. Proc Natl Acad Sci 111: 9627–9632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez F, Lu D, Ha P, Costacurta P, Chavez R, Heller HC, Ruby NF. 2014. Circadian rhythm. Dysrhythmia in the suprachiasmatic nucleus inhibits memory processing. Science 346: 854–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flourakis M, Kula-Eversole E, Hutchison AL, Han TH, Aranda K, Moose DL, White KP, Dinner AR, Lear BC, Ren D, et al. 2015. A conserved bicycle model for circadian clock control of membrane excitability. Cell 162: 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forger DB, Peskin CS. 2003. A detailed predictive model of the mammalian circadian clock. Proc Natl Acad Sci 100: 14806–14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank E, Sidor MM, Gamble KL, Cirelli C, Sharkey KM, Hoyle N, Tikotzky L, Talbot LS, McCarthy MJ, Hasler BP. 2013. Circadian clocks, brain function, and development. Ann NY Acad Sci 1306: 43–67. [DOI] [PubMed] [Google Scholar]
- Frank E, Benabou M, Bentzley B, Bianchi M, Goldstein T, Konopka G, Maywood E, Pritchett D, Sheaves B, Thomas J. 2014. Influencing circadian and sleep-wake regulation for prevention and intervention in mood and anxiety disorders: What makes a good homeostat? Ann NY Acad Sci 1334: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman GM Jr, Krock RM, Aton SJ, Thaben P, Herzog ED. 2013. GABA networks destabilize genetic oscillations in the circadian pacemaker. Neuron 78: 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al. 2013. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155: 793–806. [DOI] [PubMed] [Google Scholar]
- Gillespie CF, Mintz EM, Marvel CL, Huhman KL, Albers HE. 1997. GABAA and GABAB agonists and antagonists alter the phase-shifting effects of light when microinjected into the suprachiasmatic region. Brain Res 759: 181–189. [DOI] [PubMed] [Google Scholar]
- Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, et al. 2007. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316: 897–900. [DOI] [PubMed] [Google Scholar]
- Gonze D, Bernard S, Waltermann C, Kramer A, Herzel H. 2005. Spontaneous synchronization of coupled circadian oscillators. Biophys J 89: 120–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Fuentes D, Norris AJ, Carrasquillo Y, Nerbonne JM, Herzog ED. 2012. IA channels encoded by Kv1.4 and Kv4.2 regulate neuronal firing in the suprachiasmatic nucleus and circadian rhythms in locomotor activity. J Neurosci 32: 10045–10052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Fuentes D, Hermanstyne TO, Carrasquillo Y, Nerbonne JM, Herzog ED. 2015. IA channels encoded by Kv1.4 and Kv4.2 regulate circadian period of PER2 expression in the suprachiasmatic nucleus. J Biol Rhythms 30: 396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. 2008. The meter of metabolism. Cell 134: 728–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Koeppl H, Gonze D. 2012. Effect of network architecture on synchronization and entrainment properties of the circadian oscillations in the suprachiasmatic nucleus. PLoS Comput Biol 8: e1002419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, et al. 2002. The VPAC2 receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 109: 497–508. [DOI] [PubMed] [Google Scholar]
- Hastings MH, Goedert M. 2013. Circadian clocks and neurodegenerative diseases: Time to aggregate? Curr Opin Neurobiol 23: 8807–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings MH, Field MD, Maywood ES, Weaver DR, Reppert SM. 1999. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: New insights into a core clock mechanism J Neurosci 19: RC11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S, Kumar M, Park A, Tong P, Tung J, Yau KW, Berson DM. 2006. Central projections of melanopsin-expressing retinal ganglion cells in the mouse. J Comp Neurol 497: 326–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog ED, Huckfeldt RM. 2003. Circadian entrainment to temperature, but not light, in the isolated suprachiasmatic nucleus. J Neurophysiol 90: 763–770. [DOI] [PubMed] [Google Scholar]
- Herzog ED, Takahashi JS, Block GD. 1998. Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nat Neurosci 1: 708–713. [DOI] [PubMed] [Google Scholar]
- Hirano A, Yumimoto K, Tsunematsu R, Matsumoto M, Oyama M, Kozuka-Hata H, Nakagawa T, Lanjakornsiripan D, Nakayama KI, Fukada Y. 2013. FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152: 1106–1118. [DOI] [PubMed] [Google Scholar]
- Hirota T, Lewis WG, Liu AC, Lee JW, Schultz PG, Kay SA. 2008. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3β. Proc Natl Acad Sci 105: 20746–20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lee JW, Lewis WG, Zhang EE, Breton G, Liu X, Garcia M, Peters EC, Etchegaray JP, Traver D, et al. 2010. High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIα as a clock regulatory kinase. PLoS Biol 8: e1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Lee JW, St John PC, Sawa M, Iwaisako K, Noguchi T, Pongsawakul PY, Sonntag T, Welsh DK, Brenner DA, et al. 2012. Identification of small molecule activators of cryptochrome. Science 337: 1094–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Sugiyama T, Wallace CS, Gompf HS, Yoshioka T, Miyawaki A, Allen CN. 2003. Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron 38: 253–263. [DOI] [PubMed] [Google Scholar]
- Jagannath A, Butler R, Godinho SI, Couch Y, Brown LA, Vasudevan SR, Flanagan KC, Anthony D, Churchill GC, Wood MJ, et al. 2013. The CRTC1-SIK1 pathway regulates entrainment of the circadian clock. Cell 154: 1100–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. 1999. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 96: 57–68. [DOI] [PubMed] [Google Scholar]
- Jones CR, Huang AL, Ptacek LJ, Fu YH. 2013. Genetic basis of human circadian rhythm disorders. Exp Neurol 243: 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JR, Tackenberg MC, McMahon DG. 2015. Manipulating circadian clock neuron firing rate resets molecular circadian rhythms and behavior. Nat Neurosci 18: 373–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Yamada YR, Welsh DK, Buhr ED, Liu AC, Zhang EE, Ralph MR, Kay SA, Forger DB, Takahashi JS. 2010. Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol 8: e1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. 2012. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338: 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinuma S, Asakawa T, Nagano M, Furukawa K, Sujino M, Masumoto KH, Nakajima Y, Hashimoto S, Yagita K, Shigeyoshi Y. 2013. Regional circadian period difference in the suprachiasmatic nucleus of the mammalian circadian center. Eur J Neurosci 38: 2832–2841. [DOI] [PubMed] [Google Scholar]
- Korencic A, Kosir R, Bordyugov G, Lehmann R, Rozman D, Herzel H. 2014. Timing of circadian genes in mammalian tissues. Sci Rep 4: 5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman SJ, McMahon DG. 2004. Rhythmic regulation of membrane potential and potassium current persists in SCN neurons in the absence of environmental input. Eur J Neurosci 20: 1113–1117. [DOI] [PubMed] [Google Scholar]
- Kuhlman SJ, Quintero JE, McMahon DG. 2000. GFP fluorescence reports Period 1 circadian gene regulation in the mammalian biological clock. Neuroreport 11: 1479–1482. [PubMed] [Google Scholar]
- Kuhlman SJ, Silver R, Le Sauter J, Bult-Ito A, McMahon DG. 2003. Phase resetting light pulses induce Per1 and persistent spike activity in a subpopulation of biological clock neurons. J Neurosci 23: 1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. 2009. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326: 437–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HM, Chen R, Kim H, Etchegaray JP, Weaver DR, Lee C. 2011. The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc Natl Acad Sci 108: 16451–16456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IT, Chang AS, Manandhar M, Shan Y, Fan J, Izumo M, Ikeda Y, Motoike T, Dixon S, Seinfeld JE, et al. 2015. Neuromedin S-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron 85: 1086–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGates TA, Fernandez DC, Hattar S. 2014. Light as a central modulator of circadian rhythms, sleep and affect. Nat Rev Neurosci 15: 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Holy TE, Taghert PH. 2016. Synchronous Drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science 351: 976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, Guttler T, Davis F, Asara JM, Sahin M. 2015. The circadian protein BMAL1 regulates translation in response to S6K1-mediated phosphorylation. Cell 161: 1138–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Reppert SM. 2000. GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron 25: 123–128. [DOI] [PubMed] [Google Scholar]
- Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, et al. 2007. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell 129: 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke JC, Westermark PO, Kramer A, Herzel H. 2008. Global parameter search reveals design principles of the mammalian circadian clock. BMC Syst Biol 2: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low-Zeddies SS, Takahashi JS. 2001. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell 105: 25–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundkvist GB, Kwak Y, Davis EK, Tei H, Block GD. 2005. A calcium flux is required for circadian rhythm generation in mammalian pacemaker neurons. J Neurosci 25: 7682–7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Reddy AB, Wong GK, O’Neill JS, O’Brien JA, McMahon DG, Harmar AJ, Okamura H, Hastings MH. 2006. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol 16: 599–605. [DOI] [PubMed] [Google Scholar]
- Maywood ES, O’Neill JS, Chesham JE, Hastings MH. 2007. Minireview: The circadian clockwork of the suprachiasmatic nuclei—Analysis of a cellular oscillator that drives endocrine rhythms. Endocrinology 148: 5624–5634. [DOI] [PubMed] [Google Scholar]
- Maywood ES, Chesham JE, Meng QJ, Nolan PM, Loudon AS, Hastings MH. 2011a. Tuning the period of the mammalian circadian clock: Additive and independent effects of CK1ɛTau and Fbxl3Afh mutations on mouse circadian behavior and molecular pacemaking. J Neurosci 31: 1539–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Chesham JE, O’Brien JA, Hastings MH. 2011b. A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc Natl Acad Sci 108: 14306–14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maywood ES, Drynan L, Chesham JE, Edwards MD, Dardente H, Fustin JM, Hazlerigg DG, O’Neill JS, Codner GF, Smyllie NJ, et al. 2013. Analysis of core circadian feedback loop in suprachiasmatic nucleus of mCry1-luc transgenic reporter mouse. Proc Natl Acad Sci 110: 9547–9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill DS, Sheely CJ, Ecker JL, Badea TC, Morhardt D, Guido W, Hattar S. 2011. Development of melanopsin-based irradiance detecting circuitry. Neural Dev 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker K, Harang R, Webb AB, Welsh DK, Doyle FJ 3rd, Bonnet G, Herzog ED, Petzold LR. 2011. Wavelet measurement suggests cause of period instability in mammalian circadian neurons. J Biol Rhythms 26: 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng QJ, Logunova L, Maywood ES, Gallego M, Lebiecki J, Brown TM, Sladek M, Semikhodskii AS, Glossop NR, Piggins HD, et al. 2008. Setting clock speed in mammals: The CK1ɛ tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 58: 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng QJ, Maywood ES, Bechtold DA, Lu WQ, Li J, Gibbs JE, Dupre SM, Chesham JE, Rajamohan F, Knafels J, et al. 2010. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci 107: 15240–15245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieda M, Ono D, Hasegawa E, Okamoto H, Honma K, Honma S, Sakurai T. 2015. Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron 85: 1103–1116. [DOI] [PubMed] [Google Scholar]
- Militi S, Maywood ES, Sandate CR, Chesham JE, Barnard AR, Parsons MJ, Vibert JL, Joynson GM, Partch CL, Hastings MH, et al. 2016. Early doors (Edo) mutant mouse reveals the importance of period 2 (PER2) PAS domain structure for circadian pacemaking. Proc Natl Acad Sci 113: 2756–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsky HP, Liu AC, Welsh DK, Kay SA, Doyle FJ III. 2009. A model of the cell-autonomous mammalian circadian clock. Proc Natl Acad Sci 106: 11107–11112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldavan M, Cravetchi O, Williams M, Irwin RP, Aicher SA, Allen CN. 2015. Localization and expression of GABA transporters in the suprachiasmatic nucleus. Eur J Neurosci 42: 3018–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RY, Speh JC. 1993. GABA is the principal neurotransmitter of the circadian system. Neurosci Lett 150: 112–116. [DOI] [PubMed] [Google Scholar]
- Myung J, Hong S, Hatanaka F, Nakajima Y, De Schutter E, Takumi T. 2012. Period coding of Bmal1 oscillators in the suprachiasmatic nucleus. J Neurosci 32: 8900–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung J, Hong S, DeWoskin D, De Schutter E, Forger DB, Takumi T. 2015. GABA-mediated repulsive coupling between circadian clock neurons in the SCN encodes seasonal time. Proc Natl Acad Sci 112: E3920–E3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M, Adachi A, Nakahama K, Nakamura T, Tamada M, Meyer-Bernstein E, Sehgal A, Shigeyoshi Y. 2003. An abrupt shift in the day/night cycle causes desynchrony in the mammalian circadian center. J Neurosci 23: 6141–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. 2004. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 119: 693–705. [DOI] [PubMed] [Google Scholar]
- Nakamura W, Honma S, Shirakawa T, Honma K. 2002. Clock mutation lengthens the circadian period without damping rhythms in individual SCN neurons. Nat Neurosci 5: 399–400. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Michihata T, Nakamura W, Takumi T, Shimizu R, Yamamoto M, Ikeda M, Ohmiya Y, Nakajima Y. 2010. Dual-color luciferase mouse directly demonstrates coupled expression of two clock genes. Biochemistry 49: 8053–8061. [DOI] [PubMed] [Google Scholar]
- Oakhill JS, Steel R, Chen ZP, Scott JW, Ling N, Tam S, Kemp BE. 2011. AMPK is a direct adenylate charge-regulated protein kinase. Science 332: 1433–1435. [DOI] [PubMed] [Google Scholar]
- *.Ode K. 2016. Synthetic biology of circadian clocks. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a028357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono D, Honma S, Honma K. 2013. Cryptochromes are critical for the development of coherent circadian rhythms in the mouse suprachiasmatic nucleus. Nat Commun 4: 1666. [DOI] [PubMed] [Google Scholar]
- Parsons MJ, Brancaccio M, Sethi S, Maywood ES, Satija R, Edwards JK, Jagannath A, Couch Y, Finelli MJ, Smyllie NJ, et al. 2015. The regulatory factor ZFHX3 modifies circadian function in SCN via an AT motif-driven axis. Cell 162: 607–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch CL, Green CB, Takahashi JS. 2014. Molecular architecture of the mammalian circadian clock. Trends Cell Biol 24: 90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls S, Foley NC, Foley DK, LeSauter J, Hastings MH, Maywood ES, Silver R. 2014. Differential contributions of intra-cellular and inter-cellular mechanisms to the spatial and temporal architecture of the suprachiasmatic nucleus circadian circuitry in wild-type, cryptochrome-null and vasoactive intestinal peptide receptor 2-null mutant mice. Eur J Neurosci 40: 2528–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilorz V, Cunningham PS, Jackson A, West AC, Wager TT, Loudon AS, Bechtold DA. 2014. A novel mechanism controlling resetting speed of the circadian clock to environmental stimuli. Curr Biol 24: 766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power A, Hughes AT, Samuels RE, Piggins HD. 2010. Rhythm-promoting actions of exercise in mice with deficient neuropeptide signaling. J Biol Rhythms 25: 235–246. [DOI] [PubMed] [Google Scholar]
- Ralph MR, Menaker M. 1989. GABA regulation of circadian responses to light. I: Involvement of GABAA-benzodiazepine and GABAB receptors. J Neurosci 9: 2858–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph MR, Foster RG, Davis FC, Menaker M. 1990. Transplanted suprachiasmatic nucleus determines circadian period. Science 247: 975–978. [DOI] [PubMed] [Google Scholar]
- *.Reddy AB. 2016. Redox oscillations and posttranslational modifications. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a027698. [DOI] [Google Scholar]
- Reischl S, Vanselow K, Westermark PO, Thierfelder N, Maier B, Herzel H, Kramer A. 2007. β-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms 22: 375–386. [DOI] [PubMed] [Google Scholar]
- Roh JH, Huang Y, Bero AW, Kasten T, Stewart FR, Bateman RJ, Holtzman DM. 2012. Disruption of the sleep-wake cycle and diurnal fluctuation of β-amyloid in mice with Alzheimer’s disease pathology. Sci Transl Med 4: 150ra122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini C, Liani A, Curie T, Gos P, Kreppel F, Emmenegger Y, Bonacina L, Wolf JP, Poget YA, Franken P, et al. 2013. Real-time recording of circadian liver gene expression in freely moving mice reveals the phase-setting behavior of hepatocyte clocks. Genes Dev 27: 1526–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Norona FE, Alzate-Correa D, Scarberry D, Hoyt KR, Obrietan K. 2013. Clock and light regulation of the CREB coactivator CRTC1 in the suprachiasmatic circadian clock. J Neurosci 33: 9021–9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. 2004. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43: 527–537. [DOI] [PubMed] [Google Scholar]
- Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. 2007. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of Cryptochrome and Period gene expression. Cell 129: 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyllie NJ, Chesham JE, Hamnett R, Maywood ES, Hastings MH. 2016. Temporally chimeric mice reveal flexibility of circadian period-setting in the suprachiasmatic nucleus. Proc Natl Acad Sci 113: 3657–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John PC, Hirota T, Kay SA, Doyle FJ III. 2014. Spatiotemporal separation of PER and CRY posttranslational regulation in the mammalian circadian clock. Proc Natl Acad Sci 111: 2040–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. 2002. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci 99: 7728–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um JH, Pendergast JS, Springer DA, Foretz M, Viollet B, Brown A, Kim MK, Yamazaki S, Chung JH. 2011. AMPK regulates circadian rhythms in a tissue- and isoform-specific manner. PLoS ONE 6: e18450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasalou C, Herzog ED, Henson MA. 2011. Multicellular model for intercellular synchronization in circadian neural networks. Biophys J 101: 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virshup DM, Eide EJ, Forger DB, Gallego M, Harnish EV. 2007. Reversible protein phosphorylation regulates circadian rhythms. Cold Spring Harb Symp Quant Biol 72: 413–420. [DOI] [PubMed] [Google Scholar]
- Walton KM, Fisher K, Rubitski D, Marconi M, Meng QJ, Sladek M, Adams J, Bass M, Chandrasekaran R, Butler T, et al. 2009. Selective inhibition of casein kinase 1ɛ minimally alters circadian clock period. J Pharmacol Exp Ther 330: 430–439. [DOI] [PubMed] [Google Scholar]
- Wardlaw SM, Phan TX, Saraf A, Chen X, Storm DR. 2014. Genetic disruption of the core circadian clock impairs hippocampus-dependent memory. Learn Mem 21: 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver DR. 1998. The suprachiasmatic nucleus: A 25-year retrospective. J Biol Rhythms 13: 100–112. [DOI] [PubMed] [Google Scholar]
- Webb AB, Angelo N, Huettner JE, Herzog ED. 2009. Intrinsic, nondeterministic circadian rhythm generation in identified mammalian neurons. Proc Natl Acad Sci 106: 16493–16498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AB, Taylor SR, Thoroughman KA, Doyle FJ III, Herzog ED. 2012. Weakly circadian cells improve resynchrony. PLoS Comput Biol 8: e1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermark PO, Welsh DK, Okamura H, Herzel H. 2009. Quantification of circadian rhythms in single cells. PLoS Comput Biol 5: e1000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt JP, Montgomery JR, Meredith AL. 2016. BK channel inactivation gates daytime excitability in the circadian clock. Nat Commun 7: 10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, O’Donnell J, Christensen DJ, Nicholson C, Iliff JJ, et al. 2013. Sleep drives metabolite clearance from the adult brain. Science 342: 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Gustafson CL, Sammons PJ, Khan SK, Parsley NC, Ramanathan C, Lee HW, Liu AC, Partch CL. 2015. Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nat Struct Mol Biol 22: 476–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. 2003. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302: 1408–1412. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Suzuki T, Mizoro Y, Kori H, Okada K, Chen Y, Fustin JM, Yamazaki F, Mizuguchi N, Zhang J, et al. 2013. Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science 342: 85–90. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. 2004. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci 101: 5339–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, Kornblum I, Kumar V, Koike N, Xu M, et al. 2013. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152: 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Zecharia A, Zhang Z, Yang Q, Yustos R, Jager P, Vyssotski AL, Maywood ES, Chesham JE, Ma Y, et al. 2014. Circadian factor BMAL1 in histaminergic neurons regulates sleep architecture. Curr Biol 24: 2838–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. 2014. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci 111: 16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Kim JK, Eng GW, Forger DB, Virshup DM. 2015. A Period2 phosphoswitch regulates and temperature compensates circadian period. Mol Cell 60: 77–88. [DOI] [PubMed] [Google Scholar]