Abstract
The chemical synthesis of DNA oligonucleotides and their assembly into synthons, genes, circuits, and even entire genomes by gene synthesis methods has become an enabling technology for modern molecular biology and enables the design, build, test, learn, and repeat cycle underpinning innovations in synthetic biology. In this perspective, we briefly review the techniques and technologies that enable the synthesis of DNA oligonucleotides and their assembly into larger DNA constructs with a focus on recent advancements that have sought to reduce synthesis cost and increase sequence fidelity. The development of lower-cost methods to produce high-quality synthetic DNA will allow for the exploration of larger biological hypotheses by lowering the cost of use and help to close the DNA read–write cost gap.
DNA oligonucleotides are typically synthesized using phosphoramidite chemistry methods and then assembled into larger constructs by a variety of methods. Recent advances have sought to reduce cost and increase sequence fidelity.
DNA neither cares nor knows. DNA just is. And we dance to its music.
—Richard Dawkins
River Out of Eden: A Darwinian Life
It has been said that the 20th century was the “century of the atom” in which discoveries on the physical and chemical properties of the elements led to breakthroughs such as atomic energy (and weaponry), medical diagnostics, computers, and the microchip to name just a few. These advances had a dramatic effect on our way of life and helped shape the promise and possibilities of science and technology. In the early part of the 21st century, we are witnessing what could very likely become known as the “century of DNA.” As the score to life’s intricate symphony, DNA provides the blueprint for biological function. Advances over the last few decades in both reading (sequencing) and writing (synthesis) DNA sequences have made marked changes in our ability to understand and engineer biological systems. These advancements have led to the development of groundbreaking technologies for the design, assembly, and manipulation of DNA encoded genes, materials, circuits, and metabolic pathways, which are allowing for an ever greater manipulation of biological systems and even entire organisms.
Thanks to next-generation sequencing (NGS) technologies capable of generating an estimated 15 petabases of sequence data per year worldwide (Schatz and Phillippy 2012), the current megagenomics era has led to the swelling of biological sequence repositories with DNA sequences isolated from every organism and environment imaginable. Associated improvements in bioinformatics techniques and software allow researchers to obtain, analyze, and manipulate these DNA sequences in ways easier than ever. The ever-increasing availability of biological sequence information from all branches of the “tree of life” has deepened our understanding of biological systems and the interrelated nature of organisms at the genetic level. NGS technologies have led to a deeper understanding of human diseases, the microbiome, and the genetic diversity of organisms in our environment. This sequence boom is also allowing for the expansion of scientific disciplines such as metabolic engineering and synthetic biology as researchers seek to use novel sequences in the manipulation of biological systems for anthropocentric means. In addition, this wealth of information is leading to the development of a variety of diagnostics and therapeutics, which will contribute to the long-term improvement of human health.
This biological sequence data bonanza has been aided by a stream of innovations in instrumentation and techniques for generating sequencing data with high fidelity, increased throughput, and decreased cost, which has contributed to making NGS a go-to technology for many applications in biology. The ability to generate large DNA sequence data sets has allowed researchers to create and test large scoped biological hypotheses using NGS technologies that were not possible before their development. Beyond analyzing biological systems at the DNA sequence level, the need to construct DNA from designed sequences has driven the parallel development of methods and instrumentation to produce synthetic DNA at scale to enable the testing of engineered biological components. DNA reading by NGS when combined with modern DNA synthesis technologies form the two foundational technologies driving synthetic biology efforts and will eventually instill the predictability and reliability to engineered biological systems that chemical engineering has brought to chemical systems. This being said, our ability to sequence DNA is currently better than our ability to synthesize DNA de novo, although new technologies are helping to close the gap.
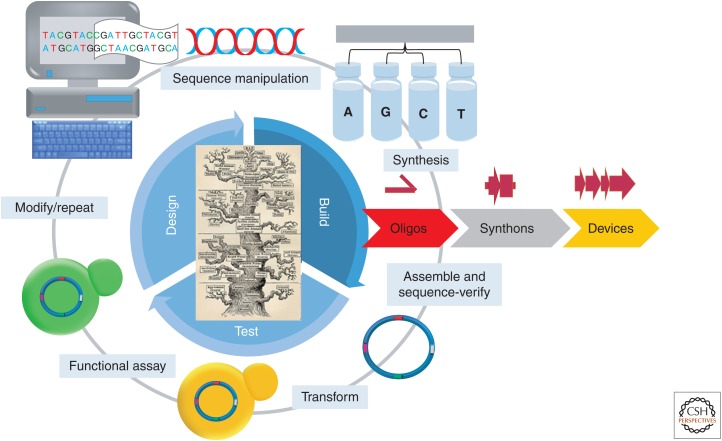
Synthetic biology is emerging as an important discipline with the potential to impact a number of academic and industrial applications including the creation of novel therapeutics, materials, biosensing, and manufacturing capabilities. Although our current understanding of biological systems is vast, it is still far from complete. Adapting exogenous DNA sequences and the functional components they encode that have naturally evolved within a network of interrelated sequences remains a principle challenge of engineering biology. Currently, the engineering of biological systems requires a heavy dose of empirical trial and error to evaluate novel enzymes, expression systems, and pathways for the desired function. Typically, this process is accomplished by first designing a desired synthetic biological circuit or pathway using computer-aided design tools (Fig. 1). Next, the resulting construct DNA is divided into smaller overlapping pieces (typically 200–1500-bp segments) that are easier to synthesize. These DNA components are then synthesized from a set of overlapping single-stranded oligonucleotides in-house (or by commercial vendors). The resulting overlapping synthons are then assembled into larger pieces of DNA and cloned into an expression vector and the sequence of the resulting construct is then verified. The sequence-verified constructs are then transformed into a cell and assayed for function. Depending on the results, changes to the construct design can be made and further iterations of the test cycle repeated. This design, build, test, learn, and repeat process has become the backbone of synthetic biology (Fig. 1), which in turn has put a premium on automated processes and methods that can shorten the development cycle and increase throughput.
Figure 1.
The synthetic biology test cycle. (From top, clockwise) Synthetic DNA constructs are designed and manipulated using computer-aided design software. The designed DNA is then divided into synthesizable pieces (synthons) up to 1–1.5 kbp. The synthons are then broken up into overlapping single-stranded oligonucleotide sequences and chemically synthesized. The oligonucleotides are then assembled together into the designed synthons using gene synthesis techniques. If necessary, multiple synthons can be assembled together into larger DNA assemblies or devices. The assembled DNAs are then typically cloned into an expression vector and sequence-verified. Once verified, the synthetic constructs are transformed into a cell and the function of the synthetic construct is assayed. Depending on the results the constructs can then be modified or refined and the test cycle is repeated until a DNA construct is obtained that produces the desired function.
One of the attributes that makes biological systems attractive from an engineering perspective is the fact that biological functions are encoded to a large part in DNA. Therefore, a gross simplification of biological engineering can be reduced to the design, production, and testing of DNA sequences. As researchers seek to engineer biological systems with novel DNA sequences, the need for custom synthetic DNA sequences has grown. This is particularly true when the sequences to be engineered are derived from metagenomic sequences in which no organism may be available from which to isolate the DNA via other methods. The synthesis of synthetic DNA is often referred to generically as “gene synthesis,” which specifically is the synthesis of gene-length pieces of DNA (250–2000 bp) directly from single-stranded synthetic DNA oligonucleotides. Unfortunately, whereas the cost of sequencing has decreased precipitously over time, the cost of gene synthesis and oligonucleotide synthesis in general has not kept pace, although technological innovation and market forces are progressively lowering the cost of synthetic DNA. The cost of gene synthesis is typically directly tied to the cost of oligonucleotide synthesis from which the genes are made. The cost of oligonucleotide synthesis has not decreased appreciably in more than a decade, generally ranging from $0.05 to $0.17 per base depending on the synthesis scale, the length of the oligonucleotide, and the supplier (Kosuri and Church 2014). Traditionally, this cost floor has been carried through to the production of gene synthesis products. These synthetic genes currently range in cost from $0.10 to $0.30 per bp ($100–$300 for a 1-kb gene) from traditional commercial suppliers, although companies exploiting newer lower-cost synthesis methods are starting to bring the cost down.
Lowering the cost of gene synthesis would enable the generation of larger data sets by making the cost of gene construction less expensive, meaning that more constructs could be generated for the same cost investment. This would allow researchers the ability to sample a greater amount of the design landscape. With a greater understanding of what does and does not work in a given construct design, a set of general design rules could be created that would improve the success of the design process and eventually shorten the design–build–test–learn process. Because the major cost of gene synthesis is the reagents that are needed for oligonucleotide synthesis, approaches that reduce reagent consumption, improve the robustness and accuracy of the gene assembly process, and enable increased throughput have become valuable tools for advancing the usage of synthetic DNA by allowing for lower cost of use. In this perspective, a brief overview of the methods and technologies that have contributed to the production of synthetic DNA via gene synthesis methods will be highlighted as well as some of the challenges that have had to be overcome to reduce the cost, increase the throughput, and ensure the fidelity of synthetic DNA.
OLIGONUCLEOTIDE SYNTHESIS
Modern de novo gene synthesis techniques can trace their beginnings to the mid-1960s and Gobind Khorana and colleagues’ efforts to decipher the genetic code and to reconstitute biological function synthetically (Scheuerbrandt et al. 1963; Khorana et al. 1968). These early efforts focused on chemically synthesizing and ligating together 17 oligonucleotides, which encoded the gene for a 77-nucleotide alanine tRNA using synthetic chemistries still in their infancy in an effort that took more than 5 years to complete (Agarwal et al. 1970; Khorana et al. 1972). Within 7 years of this work, the first reported synthetic gene encoding the 14-amino-acid hormone, somatostatin, was expressed in recombinant Escherichia coli (Itakura et al. 1977). Subsequent improvements to oligonucleotide synthesis chemistries and techniques in the early 1980s (reviewed in Caruthers 2011; Roy and Caruthers 2013) led to the development of solid-phase phosphoramidite chemistries whose robustness and fidelity allowed for automated methods to be developed to enable scalable oligonucleotide synthesis that is used to this day for the commercial synthesis of oligonucleotides. Synthetic oligonucleotides for gene synthesis applications are generally synthesized using variations of the phosphoramidite chemistry methods either on traditional column-based synthesizers or microarray-based synthesizers that will be discussed in the following sections.
COLUMN-BASED OLIGONUCLEOTIDE SYNTHESIS
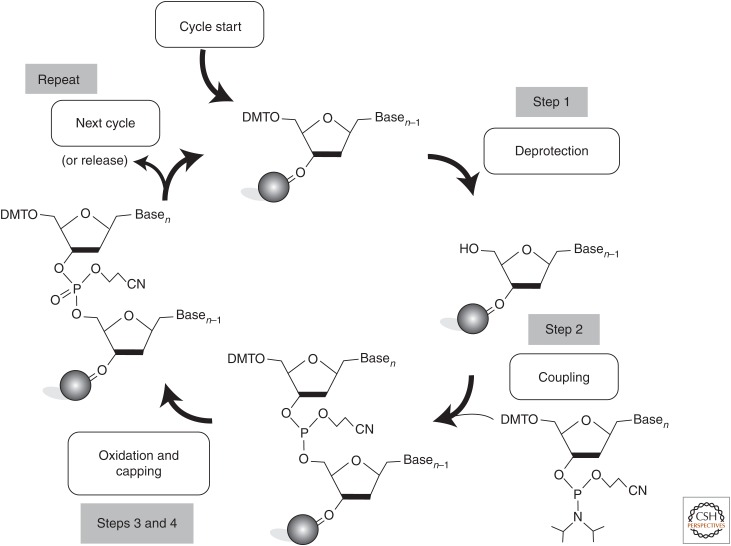
The commonly used phosphoramidite synthesis chemistry consists of a four-step chain elongation cycle that adds one base per cycle onto a growing oligonucleotide chain attached to a solid support matrix (Fig. 2). In the first step, a dimethoxytrityl (DMT)-protected nucleoside phosphoramidite that is attached to a solid support (usually contained within a synthesis column) is deprotected by the addition of trichloroacetic acid. This activates the support-attached phosphoramidite for chain elongation with the next phosphoramidite monomer. In the second step, the next base in the sequence is added in the form of a DMT-protected phosphoramidite and is coupled to the 5′-hydroxyl group of the previous nucleoside phosphoramidite in the sequence forming a phosphite triester. Third, any unreacted 5′-hydroxyl groups are capped by acylation to render any unextended sequences inert in subsequent rounds of the chain elongation cycle and thus reducing deletion errors in the finished oligonucleotide sequences. In the fourth step, the phosphite triester linkage between the monomers is converted to a phosphate linkage via oxidation with an iodine solution to produce a cyanoethyl-protected phosphate backbone. The synthesis cycle then repeats for the next base in the sequence via the removal of the 5′-terminal DMT protecting group. After the desired sequence has been synthesized from the 3′ to 5′, the oligonucleotide is chemically cleaved from the solid synthesis support and the protecting groups on the bases and the backbone are removed. This process is highly amenable to automation and forms the basis for oligonucleotide synthesizers, which can synthesize 96–1536 distinct oligonucleotides simultaneously (Fig. 3A) (Rayner et al. 1998; Cheng et al. 2002). These synthesizers typically synthesize oligonucleotides at scales ranging from 10 to 1000 nmol with a cost ranging between $0.05 and $0.17 per base. These oligonucleotides can generally be synthesized up to ∼100 nt with error rates at or below 0.5%. Column-based coupling yields are generally quite efficient (typically >99% per coupling); however, the yield of full-length oligonucleotides typically decrease with increasing oligonucleotide length. Yields of full-length oligonucleotides can be affected by spurious depurination of the synthesized oligomer, especially at adenosines during the acid deprotection steps of the synthesis cycle and becomes especially problematic for longer oligonucleotide sequences (Efcavitch and Heiner 1985; Septak 1996; LeProust et al. 2010). These spurious abasic sites can reduce the yields of full-length oligonucleotides by promoting the cleavage of the oligonucleotide phosphate backbone during the removal of the remaining nucleobase and backbone protecting groups following the final synthesis cycle. An additional reduction of synthesis quality is caused by the introduction of synthesis-related errors into the synthesized sequences, primarily in the form of single-base deletions. The main source of this type of error is the incomplete removal of the DMT protecting group or the combined inefficiencies of the coupling and capping steps during the synthesis cycle. Although complete removal of synthesis-related errors will likely be impossible because of less than 100% reaction efficiencies in a step-wise multiple reaction synthesis, recent improvements in the synthesis process and the chemistries used will likely lead to gains in the length and quality of oligonucleotides synthesized in the not-too-distant future (LeProust et al. 2010). For gene synthesis applications, because every oligonucleotide synthesized is made on individual synthesis columns the oligonucleotides necessary to assemble a gene must be added together into an assembly pool postsynthesis, which inherently reduces the oligonucleotide synthesis pool complexity because only the oligonucleotides needed for a given synthesis are included in the assembly mixture.
Figure 2.
Phosphoramidite-based synthesis of oligonucleotides. This synthesis process is the most commonly used for the synthesis of DNA oligonucleotides for gene synthesis.
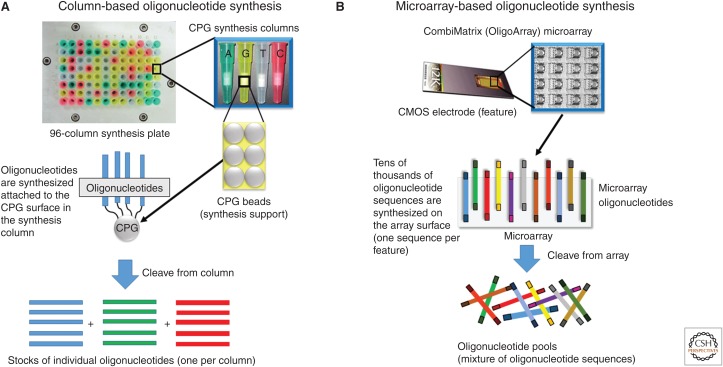
Figure 3.
Methods for solid-phase synthesis of oligonucleotides. (A) Column-based oligonucleotide synthesis. This is the traditional method of synthesizing DNA using solid-phase phosphoramidite chemistry. The synthesis of a unique oligonucleotide sequence is done on the surface of controlled-porosity glass beads (CPG) contained with a synthesis column. During the synthesis process, the reagents flow through the column and across the packed CPG matrix and the oligonucleotide “grows” off from the bead surface. Only one sequence can be synthesized per column, although high-throughput synthesizers exist that can synthesize on multiple columns at once. A 96-column synthesis plate is shown as an example. (B) Microarray-based oligonucleotide synthesis. In this method, microarray chips containing tens of thousands of distinct features synthesize unique oligonucleotide sequences at once with one unique oligonucleotide sequence synthesized per chip feature. On standard arrays, there are no physical barriers between features, so following cleavage of the synthesized oligonucleotides from the chip surface the end product is a pool of sequences containing every oligonucleotide synthesized on the array. Subsequent processing steps are required to “fish” the desired oligonucleotide sequences out of the synthesis pool for subsequent gene synthesis. The OligoArray CMOS microarray chip is shown as an example, although a handful of different array formats exist for oligonucleotide synthesis.
MICROARRAY-BASED OLIGONUCLEOTIDE SYNTHESIS
An alternative to traditional column-based oligonucleotide synthesis has emerged from the use of microarray oligonucleotide synthesis (Fig. 3B). These technologies, which were originally developed for producing oligonucleotide microarrays for diagnostic applications, have emerged as a promising low-cost alternative to column-based oligonucleotide synthesis. Early versions of these synthesizers synthesized the oligonucleotides attached to a microchip surface using a modification of the phosphoramidite synthesis process. Affymetrix was one of the early pioneers in this field and developed light-activated chemistries to control the spatially separated synthesis of oligonucleotides on the chip surface using standard mask-based photolithography techniques to selectively deprotect special photolabile nucleoside phosphoramidites (Fodor et al. 1991; Pease et al. 1994). Companies such as NimbleGen and LC Sciences further simplified the light-directed synthesis procedures by eliminating the photolithography masks by using programmable micromirror devices to precisely control the light-based chemistries (Singh-Gasson et al. 1999; Gao et al. 2001). Alternatively, ink-jet printing-based technologies developed by Agilent and semiconductor-based chip technologies by CombiMatrix (CustomArray) allow for the use of standard phosphoramidites and reagents without the need for expensive micromirror controllers or photomasking. In particular, these two synthesis technologies currently provide the best combination of cost, oligonucleotide synthesis length, and accuracy, which has led to their use in several recent gene synthesis applications (Cleary et al. 2004; Warner et al. 2010; Patwardhan et al. 2012). Using microarray-based synthesizers, oligonucleotides are typically synthesized at femtomolar synthesis scales, that is, typically two to four orders of magnitude lower than that used in column-based synthesis. This lower synthesis scale leads to a concomitant decrease in the quantity of reagents needed for the synthesis. The reduction of reagent use in the synthesis process leads to dramatically lower costs. In addition to the reduced synthesis scale, chip-based oligonucleotide synthesizers offer levels of multiplexing not possible on traditional column-based synthesizers that allows for a greater number of unique oligonucleotide sequences to be synthesized in a given synthesis run. With possible feature densities in the tens of thousands per chip and the capability for some synthesizers to synthesize multiple chips simultaneously, the ability of microarray-based synthesis methods far outstrip the capacity of even the largest column-based instruments. The combined reduced-synthesis scale and massive multiplexing capability means that oligonucleotide synthesis costs from array-based platforms can range from $0.00001 to $0.0001 per nucleotide depending on the platform, oligonucleotide length and synthesis scale. When compared with the $0.05–$0.10 cost per base attainable for column-synthesized oligonucleotides, the appeal of chip-synthesized oligonucleotides for gene synthesis applications becomes obvious.
Although array-based platforms offer superior synthesis capabilities in terms of multiplexing and cost there are some challenges with using these platforms for making oligonucleotides for gene synthesis applications. One problem is that planar array synthesized oligonucleotides tend to be relatively low quality, which leads to more synthesis-related errors when compared with column-synthesized oligonucleotides (Kosuri and Church 2014; Wan et al. 2014). Despite this, recent improvements in chip design and refinements to the synthesis process have led to an increase in the quality of array-synthesized oligonucleotides. One such source of reduced synthesis quality is the depurination of oligonucleotide sequences on chips, which can occur spontaneously because of prolonged exposure of the nascent oligonucleotides to the deprotecting reagents during the synthesis cycle. Optimizations to the reaction conditions and the flow of reagents to the chip during the synthesis cycle has been shown to effectively improve the quality of the oligonucleotides synthesized on chips by reducing depurination and increasing the overall synthesis yields, which in turn enables the synthesis of oligonucleotides up to 200 nts in length (LeProust et al. 2010). Another source of reduced synthesis quality with chip produced oligonucleotides is because of “edge effects,” which is caused by misalignment of the reagent droplets with the selected chip feature, or inaccurate selective deprotection of the proper synthesis features caused by improper reagent sequestration or light beam drifts in synthesizers using light-activated synthesis chemistries. Edge effects reduce oligo quality by reducing the selective control of the synthesis reactions in adjacent oligonucleotides, which can lead to substitution or deletion errors in oligonucleotides synthesized on adjacent features. Work to reduce edge effects during the synthesis process has shown that refinements to the chip design can improve the fidelity of array-synthesized oligonucleotides to rival column-synthesized oligonucleotides (Saaem et al. 2010). Continued improvement in array design and refinements to synthesis reagents and processes used will lead to routine robust synthesis of high-quality long oligonucleotides from arrays, which will further their use as the go to source for low-cost oligonucleotides for gene synthesis applications.
GENE SYNTHESIS FROM OLIGONUCLEOTIDES
Because DNA is a polymer made up of four different nucleotide monomers, gene synthesis and DNA assembly methods are in effect a form of hierarchical polymer synthesis. For synthetic DNA, individual phosphoramidite monomers are combined together to create individual oligonucleotides 60–100 nt in length. To facilitate the assembly of a synthetic double-stranded DNA (synthon) from single-stranded oligonucleotides, adjacent oligonucleotides are designed to contain overlapping sequences between the oligonucleotides encoded on the opposing strands of the DNA duplex and are assembled together during the gene synthesis process to make double-stranded synthons from 200 to 2000 bp in length. Sequence-verified synthons can then be assembled together by numerous methods to make larger synthetic DNA constructs encoding entire metabolic pathways or genomes.
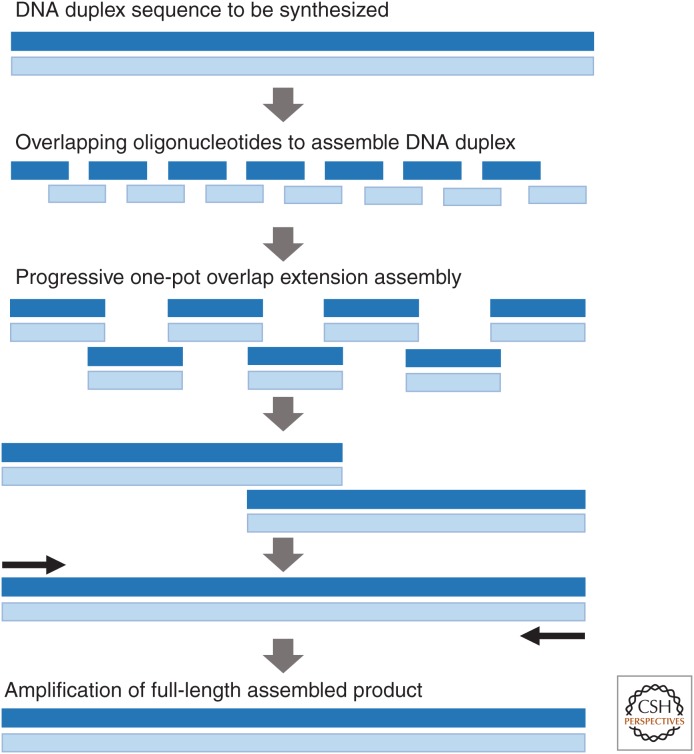
Early oligonucleotide assembly methods relied on the ligation of adjacent oligonucleotides via the use of a DNA ligase (Agarwal et al. 1970; Sekiya et al. 1976, 1979). Initially, the ligation of adjacent oligonucleotides was done sequentially, but as assembly methods and the quality of the synthetic oligonucleotides improved and the use of thermostable ligases were used to improve the stringency of the assembly reaction the one-pot assembly of DNA synthons via ligation became possible (Au et al. 1998; Xiong et al. 2008b). Shortly after the discovery of the polymerase chain reaction (PCR) in the 1980s ligation-free methods for using synthetic single-stranded oligonucleotides were developed to make completely synthetic pieces of DNA without the need for a naturally derived DNA template (Dillon and Rosen 1990; Jayaraman et al. 1991; Stemmer et al. 1995). Since this time, dozens of different methods have been created to assemble double-stranded DNA from single-stranded oligonucleotides via PCR-like methods. A summary of many of these methods has been reviewed in the literature (Xiong et al. 2008a; Czar et al. 2009; Hughes et al. 2011; Ma et al. 2012) and will not be reviewed extensively here. All of these methods use single-stranded synthetic oligonucleotides with complementary overlapping sequences between adjacent oligonucleotides to assemble double-stranded DNA synthons using a thermostable DNA polymerase and PCR. The only differences between the myriad of PCR-based DNA assembly methods is in how the substituent oligonucleotides are designed to be assembled together and the reaction conditions under which they are assembled, the end product of all of these methods is the same, a double-stranded DNA (dsDNA) synthon. Of these assembly methods, variations of the polymerase cycle assembly (PCA) method is by far the most commonly used to assemble DNA synthons from oligonucleotides (Fig. 4) (Stemmer et al. 1995). Using PCA, a dsDNA sequence is divided into oligonucleotide sequences (typically 60–80 nt), which encode both strands of the DNA duplex with overlaps between adjacent oligonucleotides that range from 15 to 25 nt in length. Typically, adjacent oligonucleotides are designed with gaps between the forward and the reverse overlapping regions of the assembly oligonucleotides to reduce the amount of oligonucleotide synthesis required to synthesize a given sequence. Once designed and synthesized, the substituent oligonucleotides for an assembly are pooled together in equimolar concentrations and cycled in a one-pot “assembly” reaction in which adjacent oligonucleotides are randomly extended in a nonexponential manner by a DNA polymerase to produce a mixture of oligonucleotide extension products of various lengths. This mixture is then used as a template to seed a second PCR reaction, in which the desired “full-length” product is amplified from the assembly mixture in the presence of an excess of the outermost assembly primers. In addition to ligation and PCA-based assembly methods, Gibson and colleagues, as part of their pioneering work in synthetic genomics, have developed both in vitro (Gibson et al. 2009; Gibson 2011a), and in vivo (Gibson 2009, 2011b, 2012) one-pot, single-step methods for simultaneously assembling DNA synthons from oligonucleotides and cloning them directly into plasmids. In some variation, these three assembly methods are used in most commercial and academic gene synthesis applications reported to date.
Figure 4.
Polymerase chain assembly. Overlapping oligonucleotides encoding a DNA duplex are assembled together via progressive overlap extension assembly in a one-pot reaction. Following assembly the full-length assembled synthon is amplified out of the assembly mixture by polymerase chain reaction (PCR) with the outermost primers.
GENE SYNTHESIS FROM ARRAY-DERIVED OLIGONUCLEOTIDE POOLS
Over the last decade, the desire for cheaper sources of synthetic DNA has fueled an interest in array-synthesized DNA as a way to produce oligonucleotides inexpensively. Beyond early demonstrations of gene synthesis from array produced oligonucleotides (Richmond et al. 2004; Tian et al. 2004; Zhou et al. 2004), recent innovations have made substantial improvements to the quality, efficiency, and scalability of microarray-based oligonucleotide synthesis and gene assembly. To use microarray-synthesized DNA for gene synthesis, several hurdles have had to be overcome to make this source of DNA an attractive alternative to oligonucleotides synthesized by conventional column-based approaches. Some of the properties that make array-synthesized oligonucleotides attractive as a low-cost source of DNA for gene synthesis present challenges for actually using oligonucleotides synthesized in this way for gene synthesis. The reduced synthesis scale of array-produced oligonucleotides is attractive from a cost perspective because of the concomitant reduction in reagent consumption needed to synthesize a given DNA sequence. However, the amount of DNA produced (fmol) is generally much less than what is needed (pmol) to reliably assemble them into larger synthons. Similarly, the massive multiplexing capabilities of array-based oligonucleotide synthesis means that tens of thousands of unique oligonucleotide sequences can be synthesized simultaneously on the array surface. However, the sequence complexity of the oligonucleotide pools produced on the array can lead to problems with assembling them into larger constructs due to the interference between oligonucleotides that contain even modest sequence homology. Last, as mentioned previously, array-synthesized oligonucleotides are in general lower quality as compared with their column-synthesized counterparts, which manifests itself as sequence errors in the synthetic DNA assembled from them. So, in general, the use of array-synthesized oligonucleotides requires the use of some sort of error reduction strategy to reduce the work required to obtain sequence-verified constructs.
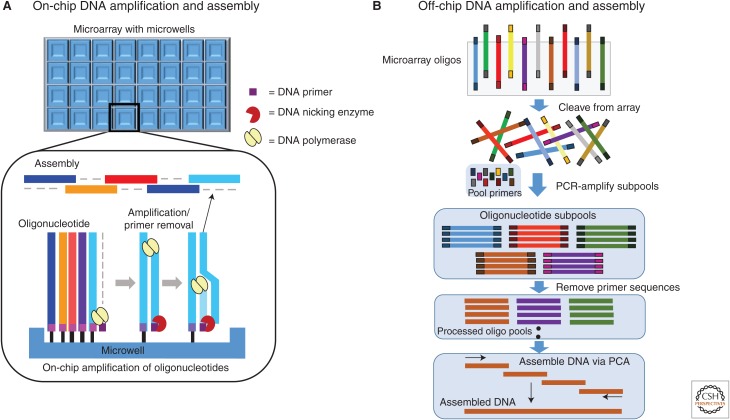
To overcome these limitations, a couple of different methods have been developed to increase the effective oligonucleotide pool concentration to ensure reliable assembly and reproducibility (Tian et al. 2004; Kosuri et al. 2010; Quan et al. 2011). Tian et al. (2004) addressed the concentration issue by including a common priming sequence followed by a nicking endonuclease recognition site on the 3′-end of every oligonucleotide synthesized on the array (Fig. 5A). The array-synthesized oligonucleotides serve as the template strand for a primer extension reaction using the common priming sequence incorporated into the array-attached oligonucleotide pool. Following primer extension, a nicking endonuclease included in the reaction mixture removes the common primer sequences to release a single-stranded DNA oligonucleotide that is complementary to each oligonucleotide synthesized on the array. This process amplifies the array-synthesized oligonucleotide pool to increase the concentration of the oligonucleotides to levels suitable for gene synthesis. In this method, because the array-synthesized oligonucleotides remain attached to the array, spatial control of oligonucleotide assembly can occur on chip. To exploit this feature and to improve the multiplexing capabilities of chip-based oligonucleotide synthesis, Quan et al. (2011) used a custom ink-jet synthesizer that synthesized oligonucleotides on specialized chips. Within these specialized chips, micro-compartmentalized assembly wells can be designed into the synthesis array with the oligonucleotides necessary to assemble a unique synthon, synthesized, amplified, and assembled within individual microwells (Fig. 5A). Using such a method effectively reduces the sequence complexity of the localized oligonucleotide pool, which in turn increases the robustness of assembly for gene synthesis while also allowing for synthesis multiplexing that can occur in each of the many microwells designed into the chip.
Figure 5.
Gene synthesis from microarray-synthesized oligonucleotides. (A) Synthons can be assembled using on-chip synthesis and assembly by including a single priming site into the 3′-end of every oligonucleotide synthesized on the microarray. The oligonucleotides can then be amplified within microwells designed into the array by incubating with a common primer and a DNA polymerase. The primer sequence is removed from the assembly oligonucleotides using a nicking endonuclease, freeing the oligonucleotides to be assembled together via polymerase chain assembly within the same well. (B) Synthons can also be synthesized off-chip by first cleaving the oligonucleotide pools from the array. The cleaved synthesis oligonucleotide pool can be amplified into subpools by PCR using priming sites incorporated into both ends of the synthesized oligonucleotide. The amplified subpools can then be further subdivided into assembly oligonucleotide pools by additional unique priming sites included in the oligonucleotide flanking sequences. Following segregation by amplification the priming sequences are removed from the assembly oligonucleotides by restriction enzyme digestion. The processed oligonucleotides can then be assembled together by polymerase cycle assembly.
Another method first introduced by Kosuri et al. (2010) solves the oligonucleotide concentration and pool complexity problems by including “bar-coded” priming sequences into each of the oligonucleotides synthesized on the array (Fig. 5B). In this method, the oligonucleotides required to synthesize a desired synthon is flanked on both ends by sets of predefined primer sequences. The outermost priming sequences allow for amplification of the array-synthesized oligonucleotide pool, which has been cleaved from the chip surface following synthesis. This step allows for one set of unique primers to amplify the entire oligonucleotide pool such that a portion of the pool can be used for subsequent assembly or archival purposes. Inboard of the pool amplification primers is a set of flanking primer-binding sequences that allow for selective amplification of assembly subpools from the synthesis pool. Each unique synthon would be given a unique set of flanking subpool primer sequences such that the oligonucleotides necessary to assemble any given synthon would be selectively amplified from the synthesis pool for subsequent assembly. Following amplification of the subpool oligonucleotides, the priming sequences are removed by restriction digestion of the amplified dsDNA oligonucleotides using a unique enzyme recognition sequence flanking the assembly oligonucleotide sequence. This oligonucleotide design scheme simultaneously solves the oligonucleotide concentration and pool complexity problems associated with array-derived oligonucleotides and does not require specialized chips or array synthesizers. This method is also highly amenable to automation as each subpool can be assembled in unique wells in a multiwell plate format using PCA assembly techniques.
ERROR CORRECTION AND SEQUENCE VALIDATION
One of the chief challenges in the synthesis and assembly of synthetic DNA from unpurified oligonucleotides is dealing with sequence errors introduced into the sequences by synthesis by-products and the assembly process itself. Following assembly, a synthetic DNA can be thought of as a population of sequences containing a mixture of correct and incorrect sequences. Traditionally, the synthetic DNA would be cloned into a vector and individual clones isolated and sequenced by Sanger sequencing. However, the probability of any given sequence containing an error increases with the length of DNA, which in turn means that more clones must be sequenced to obtain a completely correct clone. This procedure is time-consuming and the setup work required adds significantly to the cost of the DNA being produced. To reduce the number of clones necessary to obtain a correct sequence, some postsynthesis method must be applied to the assembled DNA to sieve correct sequences from incorrect ones. As a result, methods have been developed that approximate the error-correcting process that cells use to maintain sequence fidelity during DNA replication. In general, most methods for removing DNA sequence errors from synthetic DNA start with the formation of a DNA heteroduplex. This is done by heating up the synthetic DNA to denature and disassociate the strands followed by cooling the sample to reanneal the strands together. For any given position within a DNA sequence, most of the duplexes in the synthetic mixture will contain the correct base at that position with errors sprinkled throughout the population. Because sequence errors occur randomly within an assembled DNA sequence this denaturation and reannealing process leads to the formation of heteroduplexed DNA at positions that contain errors. Positions that contain mutations within these heteroduplexes can be acted on by proteins, which specifically recognize sequence mutations in DNA. One such group of methods relies on the sequence mismatch recognition capabilities of the MutS protein to specifically bind to sequence mismatches in synthetic DNA duplexes. Selective binding of MutS to error-containing DNA can be used to sieve error-free sequences from those that contain errors (Carr et al. 2004; Binkowski et al. 2005; Wan et al. 2014). These methods usually immobilize MutS to a solid matrix material and then purify column-bound (error-containing) DNA sequences from unbound material (error-reduced). Error-containing heteroduplex DNA can also be sieved using enzymes that recognize and cut the DNA duplex at the site of the base mismatch (Young and Dong 2004; Fuhrmann et al. 2005; Kosuri et al. 2010; Saaem et al. 2012; Dormitzer et al. 2013). The use of endonuclease enzymes (or enzyme cocktails), which recognize and cleave DNA heteroduplexes at the sites of mismatches, has been shown to be highly effective at reducing synthesis-related errors in synthetic genes allowing for time and material savings such that in some cases the treated genes can be used directly in functional assays without cloning and sequence verification (Dormitzer et al. 2013). These methods are a relatively straightforward and cost-effective approach for sieving error-containing sequences from synthetic DNA and can reduce the observed error rate from 1/100 bases to 1/10,000 bases (Dormitzer et al. 2013; Kosuri and Church 2014).
An alternative to enzyme-mediated error correction techniques is the direct sequencing of oligonucleotides and assembled synthons using NextGen sequencing techniques. Although more expensive on a per-sequence basis than enzyme-based error correction techniques, NextGen sequencing techniques offers tremendous multiplexing capabilities in which thousands of sequence reads can be taken on multiple oligonucleotides or synthons simultaneously. In one such application, Matzas et al. (2010) used Roche 454 sequencing combined with a bead-picking robot to selectively remove oligonucleotide sequences (attached to beads) from the sequencing array that showed perfect sequences that were then used for gene synthesis. Kim et al. (2012) used random sequence tags to mark individual synthetic sequences in a 454 sequencing reaction and used these tags to selectively amplify correct sequences from the sequenced pool. In a similar approach, Schwartz et al. (2012) used Illumina sequencing barcodes to “dial-out” correct sequences via selective amplification of the sequence-verified bar-coded sequences. Recently, they have refined this approach to improve the multiplexing capacity of microarray-synthesized DNA to pairwise assemble 2271 131–250 bp synthons from a single oligonucleotide pool and used bar-coded primers to selectively isolate sequence-verified individuals (Klein et al. 2016). All of these methods point to ways in which NextGen technologies can be used to improve the quality of synthetic DNA. In addition, because sequence-based approaches evaluate collections of individual molecules of DNA, they are suitable for the sequence verification of synthetic DNA, which may contain regions of sequence degeneracy such as libraries for directed evolution. Currently, each of these methods are limited by the sequence read-length capabilities of the NextGen instrument used and by sequencing-related errors. However, as NextGen instrumentation and techniques continue to improve, these limitations will become less significant and allow for the accurate verification of longer pieces of synthetic DNA.
MAKE IT BIGGER: SYNTHESIS OF LONGER DNA ASSEMBLIES
Recent advancements in oligonucleotide synthesis and techniques for gene synthesis have largely focused on reducing costs, increasing throughput, and the quality of synthetic DNA. Concomitant advances in assembling DNA into longer and longer pieces have led to methods to construct large enzyme complexes (Kodumal et al. 2004), entire metabolic pathways (Temme et al. 2012), and even entire genomes (Smith et al. 2003; Gibson et al. 2008a; Hutchison et al. 2016) synthetically. Methods to assemble synthons have been developed to assemble synthons and larger DNA assemblies in both in vitro (Gibson et al. 2010) and in vivo (Gibson et al. 2008b). Although restriction enzyme-based cloning techniques have been the de rigueur choice for manipulating DNA constructs for a couple of decades and were the basis of early BioBrick and similar assembly methods (Shetty et al. 2008; Lee et al. 2011), the need to simplify the cloning/assembly process while reducing the limitations on sequence design has led to the development of scar-less restriction-enzyme-free cloning and assembly techniques. Seam-less assembly and cloning methods available to the modern DNA jockey include Gibson assembly (Gibson et al. 2009; Gibson 2011a), Golden Gate assembly (Engler et al. 2008), sequence and ligation-independent cloning (SLIC) (Li and Elledge 2007), ligation cycling reaction (de Kok et al. 2014), paper-clip assembly (Trubitsyna et al. 2014), yeast assembly (Gibson et al. 2008b; Gibson 2011b), and circular polymerase extension cloning (CPEC) (Quan and Tian 2009), to name just a few. Which DNA assembly technique to use is largely a matter of choice, and multiple approaches are often applied in parallel. Of these methods, Gibson assembly is probably the most commonly used to assemble multiple pieces of DNA together into larger constructs. This method uses a one-pot isothermal technique, which uses an enzyme mixture containing a thermostable DNA polymerase, DNA ligase, and exonuclease to chew-back, anneal and repair adjacent overlapping DNA sequences to assemble the desired construct. Recent innovations in the design of unique overlapping sequences to direct the assembly process has further expanded the usage of the Gibson assembly method for combinatorial assembly of large DNA sequences (Guye et al. 2013; Torella et al. 2014).
One of the hallmarks of synthetic biology is the application of rational design principles to the design and assembly of biological components; however, because it is often difficult to know a priori how well a given DNA construct will work once introduced into a cell, it is often necessary to try several versions of the construct to find which one will work best. Therefore, a greater emphasis on the modular design of DNA parts enables the assembly of a greater variety of potential constructs through mix-and-match combinatorial assembly of DNA components. In addition to simplifying the overall assembly process, modular design and assembly of DNA components makes automation of the process possible, which can reduce the time, labor, and cost of making and testing multiple constructs. Most of the aforementioned assembly methods can be used for the assembly in vitro, and Gibson assembly has been applied to the assembly of DNA segments multiple kilobases in length (Gibson et al. 2009). In another such example, Gibson assembly was used to assemble the 16.3-kb mouse mitochondrial genome directly from 60 mer oligonucleotides (Gibson et al. 2010). Efficient assembly of even larger synthetic DNA segments can also be performed in vivo by using the homologous recombination capabilities of the yeast Saccharomyces cerevisiae. In an example of the exceptional ability of yeast to assemble exogenous DNA into larger assemblies from overlapping synthons or subassemblies, researchers at the J. Craig Venter Institute have successfully used yeast to assemble multiple 0.5–1 Mbp bacterial genomes (Gibson et al. 2008a; Hutchison et al. 2016) and even assembled synthons directly from overlapping oligonucleotides (Gibson 2009, 2011b, 2012). Each of the aforementioned assembly techniques could be automated to further increase the throughput for constructing larger synthetic DNAs and enable the exploration and testing of large biological hypotheses.
CONCLUDING REMARKS
The development of automatable robust chemistries for chemical DNA synthesis over the last 40 years has contributed to the advancement of our understanding of biology and has laid the groundwork for the predictable engineering of biological systems. Synthetic DNA is central to the development of methods to engineer biology and when combined with the massive amounts of sequence data being generated by NGS efforts will contribute to the advancement of synthetic biology toward applications heretofore unimaginable. To date, there have been a handful of moonshot demonstrations such as the complete synthesis of an entire yeast chromosome (Annaluru et al. 2014), an entire bacterial genome (Gibson et al. 2008a), and the subsequent synthesis of a minimal bacterial genome (Hutchison et al. 2016), which illustrate the use of synthetic DNA and the capabilities of existing gene synthesis methods to accomplish large-scale synthetic biology efforts. These examples, when combined with numerous projects in which synthetic DNA has been used to evaluate functional biological components (Salis et al. 2009; Callura et al. 2012; Brophy and Voigt 2016), create synthetic vaccines (Dormitzer et al. 2013), and construct synthetic genetic circuits (Salis et al. 2009; Sowa et al. 2015; Nielsen et al. 2016; Rubens et al. 2016) and when applied to wholesale genomic editing (Wang et al. 2009; Doudna and Charpentier 2014), point to a future where the nuances of biological function can in part be understood by the design, synthesis, and assay of interchangeable synthetic components akin to synthetic drug development.
To enable larger-scale engineering efforts and to democratize the use of synthetic biology techniques and design principles, recent innovations have sought to lower the cost of synthesizing DNA oligonucleotides and their subsequent assembly into synthetic constructs ready for use. The use of microarray/microfluidic oligonucleotide synthesis techniques, when combined with automated gene synthesis protocols and improvements in synthetic DNA quality, have made great strides toward this goal. In response to market pressures, commercialization of these technologies and the continued development of other efficient DNA synthesis techniques will further enhance the availability of increasingly inexpensive synthetic DNA.
Although the synthesis of relatively small synthons (<1 kb) can often be straightforward and accomplished at the undergraduate level the assembly of larger pieces of synthetic DNA can still pose several technical challenges to even the most experienced DNA jockey. Problems created by a high degree of secondary structure in the target sequence, as well as repetitive DNA sequences, can pose challenges to their assembly. Sequences that are unknowingly toxic to the host cells used for intermediate manipulations of the DNA (e.g., cloning for sequence verification) or assay of synthetic DNA constructs can also pose problems for the successful assembly and use of some synthetic DNA constructs. Here to, further improvements in synthetic construct design (Tang and Chilkoti 2016) and in the DNA assembly process will likely enable the successful synthesis of even the most difficult target sequences in the not-to-distant future. What started with a synthetic tRNA gene more than 40 years ago has led to the recent synthesis of the first minimal bacterial genome (Hutchison et al. 2016) and a deep appreciation of the complexities and challenges of engineering biology. Much of which still remains to be explored. In this, the century of DNA, the question remains. What is next?
Footnotes
Editors: Daniel G. Gibson, Clyde A. Hutchison III, Hamilton O. Smith, and J. Craig Venter
Additional Perspectives on Synthetic Biology available at www.cshperspectives.org
REFERENCES
- Agarwal KL, Buchi H, Caruthers MH, Gupta N, Khorana HG, Kleppe K, Kumar A, Ohtsuka E, Rajbhandary UL, Van de Sande JH, et al. 1970. Total synthesis of the gene for an alanine transfer ribonucleic acid from yeast. Nature 227: 27–34. [DOI] [PubMed] [Google Scholar]
- Annaluru N, Muller H, Mitchell LA, Ramalingam S, Stracquadanio G, Richardson SM, Dymond JS, Kuang Z, Scheifele LZ, Cooper EM, et al. 2014. Total synthesis of a functional designer eukaryotic chromosome. Science 344: 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au LC, Yang FY, Yang WJ, Lo SH, Kao CF. 1998. Gene synthesis by an LCR-based approach: High-level production of leptin-l54 using synthetic gene in Escherichia coli. Biochem Biophys Res Commun 248: 200–203. [DOI] [PubMed] [Google Scholar]
- Binkowski BF, Richmond KE, Kaysen J, Sussman MR, Belshaw PJ. 2005. Correcting errors in synthetic DNA through consensus shuffling. Nucleic Acids Res 33: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy JA, Voigt CA. 2016. Antisense transcription as a tool to tune gene expression. Mol Syst Biol 12: 854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callura JM, Cantor CR, Collins JJ. 2012. Genetic switchboard for synthetic biology applications. Proc Natl Acad Sci 109: 5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr PA, Park JS, Lee YJ, Yu T, Zhang S, Jacobson JM. 2004. Protein-mediated error correction for de novo DNA synthesis. Nucleic Acids Res 32: e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruthers MH. 2011. A brief review of DNA and RNA chemical synthesis. Biochem Soc Trans 39: 575–580. [DOI] [PubMed] [Google Scholar]
- Cheng JY, Chen HH, Kao YS, Kao WC, Peck K. 2002. High throughput parallel synthesis of oligonucleotides with 1536 channel synthesizer. Nucleic Acids Res 30: e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary MA, Kilian K, Wang Y, Bradshaw J, Cavet G, Ge W, Kulkarni A, Paddison PJ, Chang K, Sheth N, et al. 2004. Production of complex nucleic acid libraries using highly parallel in situ oligonucleotide synthesis. Nat Methods 1: 241–248. [DOI] [PubMed] [Google Scholar]
- Czar MJ, Anderson JC, Bader JS, Peccoud J. 2009. Gene synthesis demystified. Trends Biotechnol 27: 63–72. [DOI] [PubMed] [Google Scholar]
- de Kok S, Stanton LH, Slaby T, Durot M, Holmes VF, Patel KG, Platt D, Shapland EB, Serber Z, Dean J, et al. 2014. Rapid and reliable DNA assembly via ligase cycling reaction. ACS Synth Biol 3: 97–106. [DOI] [PubMed] [Google Scholar]
- Dillon PJ, Rosen CA. 1990. A rapid method for the construction of synthetic genes using the polymerase chain reaction. BioTechniques 9: 298–300. [PubMed] [Google Scholar]
- Dormitzer PR, Suphaphiphat P, Gibson DG, Wentworth DE, Stockwell TB, Algire MA, Alperovich N, Barro M, Brown DM, Craig S, et al. 2013. Synthetic generation of influenza vaccine viruses for rapid response to pandemics. Sci Transl Med 5: 185ra168. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Efcavitch JW, Heiner C. 1985. Depurination as a yield decreasing mechanism in oligodeoxynucleotide synthesis. Nucleosides Nucleotides 4: 267–267. [Google Scholar]
- Engler C, Kandzia R, Marillonnet S. 2008. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3: e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor SP, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. 1991. Light-directed, spatially addressable parallel chemical synthesis. Science 251: 767–773. [DOI] [PubMed] [Google Scholar]
- Fuhrmann M, Oertel W, Berthold P, Hegemann P. 2005. Removal of mismatched bases from synthetic genes by enzymatic mismatch cleavage. Nucleic Acids Res 33: e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, LeProust E, Zhang H, Srivannavit O, Gulari E, Yu P, Nishiguchi C, Xiang Q, Zhou X. 2001. A flexible light-directed DNA chip synthesis gated by deprotection using solution photogenerated acids. Nucleic Acids Res 29: 4744–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG. 2009. Synthesis of DNA fragments in yeast by one-step assembly of overlapping oligonucleotides. Nucleic Acids Res 37: 6984–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG. 2011a. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498: 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG. 2011b. Gene and genome construction in yeast. In Current protocols in molecular biology (ed. Ausubel FM et al. ), Chapter 3, Unit 3.22 Wiley, Hoboken, NJ. [DOI] [PubMed] [Google Scholar]
- Gibson DG. 2012. Oligonucleotide assembly in yeast to produce synthetic DNA fragments. Methods Mol Biol 852: 11–21. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. 2008a. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319: 1215–1220. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Axelrod KC, Zaveri J, Algire MA, Moodie M, Montague MG, Venter JC, Smith HO, Hutchison CA III. 2008b. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci 105: 20404–20409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Method 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Smith HO, Hutchison CA III, Venter JC, Merryman C. 2010. Chemical synthesis of the mouse mitochondrial genome. Nat Method 7: 901–903. [DOI] [PubMed] [Google Scholar]
- Guye P, Li Y, Wroblewska L, Duportet X, Weiss R. 2013. Rapid, modular and reliable construction of complex mammalian gene circuits. Nucleic Acids Res 41: e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes RA, Miklos AE, Ellington AD. 2011. Gene synthesis: Methods and applications. Methods Enzymol 498: 277–309. [DOI] [PubMed] [Google Scholar]
- Hutchison CA 3rd, Chuang RY, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, et al. 2016. Design and synthesis of a minimal bacterial genome. Science 351: aad6253. [DOI] [PubMed] [Google Scholar]
- Itakura K, Hirose T, Crea R, Riggs AD, Heyneker HL, Bolivar F, Boyer HW. 1977. Expression in Escherichia coli of a chemically synthesized gene for the hormone somatostatin. Science 198: 1056–1063. [DOI] [PubMed] [Google Scholar]
- Jayaraman K, Fingar SA, Shah J, Fyles J. 1991. Polymerase chain reaction–mediated gene synthesis: Synthesis of a gene coding for isozyme c of horseradish peroxidase. Proc Natl Acad Sci 88: 4084–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorana HG, Buchi H, Caruthers MH, Chang SH, Gupta NK, Kumar A, Ohtsuka E, Sgaramella V, Weber H. 1968. Progress in the total synthesis of the gene for ala-tRNA. Cold Spring Harb Symp Quant Biol 33: 35–44. [DOI] [PubMed] [Google Scholar]
- Khorana HG, Agarwal KL, Buchi H, Caruthers MH, Gupta NK, Kleppe K, Kumar A, Otsuka E, RajBhandary UL, Van de Sande JH, et al. 1972. Studies on polynucleotides. 103. Total synthesis of the structural gene for an alanine transfer ribonucleic acid from yeast. J Mol Biol 72: 209–217. [DOI] [PubMed] [Google Scholar]
- Kim H, Han H, Ahn J, Lee J, Cho N, Jang H, Kim H, Kwon S, Bang D. 2012. “Shotgun DNA synthesis” for the high-throughput construction of large DNA molecules. Nucleic Acids Res 40: e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JC, Lajoie MJ, Schwartz JJ, Strauch EM, Nelson J, Baker D, Shendure J. 2016. Multiplex pairwise assembly of array-derived DNA oligonucleotides. Nucleic Acids Res 44: e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV. 2004. Total synthesis of long DNA sequences: Synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proc Natl Acad Sci 101: 15573–15578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri S, Church GM. 2014. Large-scale de novo DNA synthesis: Technologies and applications. Nat Method 11: 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri S, Eroshenko N, Leproust EM, Super M, Way J, Li JB, Church GM. 2010. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat Biotechnol 28: 1295–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TS, Krupa RA, Zhang F, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD. 2011. BglBrick vectors and datasheets: A synthetic biology platform for gene expression. J Biol Eng 5: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeProust EM, Peck BJ, Spirin K, McCuen HB, Moore B, Namsaraev E, Caruthers MH. 2010. Synthesis of high-quality libraries of long (150mer) oligonucleotides by a novel depurination controlled process. Nucleic Acids Res 38: 2522–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. 2007. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Method 4: 251–256. [DOI] [PubMed] [Google Scholar]
- Ma S, Tang N, Tian J. 2012. DNA synthesis, assembly and applications in synthetic biology. Curr Opin Chem Biol 16: 260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzas M, Stahler PF, Kefer N, Siebelt N, Boisguerin V, Leonard JT, Keller A, Stahler CF, Haberle P, Gharizadeh B, et al. 2010. High-fidelity gene synthesis by retrieval of sequence-verified DNA identified using high-throughput pyrosequencing. Nat Biotechnol 28: 1291–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AA, Der BS, Shin J, Vaidyanathan P, Paralanov V, Strychalski EA, Ross D, Densmore D, Voigt CA. 2016. Genetic circuit design automation. Science 352: aac7341. [DOI] [PubMed] [Google Scholar]
- Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, May D, Lee C, Andrie JM, Lee SI, Cooper GM, et al. 2012. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol 30: 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease AC, Solas D, Sullivan EJ, Cronin MT, Holmes CP, Fodor SP. 1994. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc Natl Acad Sci 91: 5022–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan J, Tian J. 2009. Circular polymerase extension cloning of complex gene libraries and pathways. PLoS ONE 4: e6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan J, Saaem I, Tang N, Ma S, Negre N, Gong H, White KP, Tian J. 2011. Parallel on-chip gene synthesis and application to optimization of protein expression. Nat Biotechnol 29: 449–452. [DOI] [PubMed] [Google Scholar]
- Rayner S, Brignac S, Bumeister R, Belosludtsev Y, Ward T, Grant O, O’Brien K, Evans GA, Garner HR. 1998. MerMade: An oligodeoxyribonucleotide synthesizer for high throughput oligonucleotide production in dual 96-well plates. Genome Res 8: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond KE, Li MH, Rodesch MJ, Patel M, Lowe AM, Kim C, Chu LL, Venkataramaian N, Flickinger SF, Kaysen J, et al. 2004. Amplification and assembly of chip-eluted DNA (AACED): A method for high-throughput gene synthesis. Nucleic Acids Res 32: 5011–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Caruthers M. 2013. Synthesis of DNA/RNA and their analogs via phosphoramidite and H-phosphonate chemistries. Molecules 18: 14268–14284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubens JR, Selvaggio G, Lu TK. 2016. Synthetic mixed-signal computation in living cells. Nat Commun 7: 11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saaem I, Ma KS, Marchi AN, LaBean TH, Tian J. 2010. In situ synthesis of DNA microarray on functionalized cyclic olefin copolymer substrate. ACS Appl Mater Interfaces 2: 491–497. [DOI] [PubMed] [Google Scholar]
- Saaem I, Ma S, Quan J, Tian J. 2012. Error correction of microchip synthesized genes using Surveyor nuclease. Nucleic Acids Res 40: e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA. 2009. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol 27: 946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz MC, Phillippy AM. 2012. The rise of a digital immune system. GigaScience 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuerbrandt G, Duffield AM, Nussbaum AL. 1963. Stepwise synthesis of deoxyribo-oligonucleotides. Biochem Biophys Res Commun 11: 152–155. [DOI] [PubMed] [Google Scholar]
- Schwartz JJ, Lee C, Shendure J. 2012. Accurate gene synthesis with tag-directed retrieval of sequence-verified DNA molecules. Nat Method 9: 913–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya T, Besmer P, Takeya T, Khorana HG. 1976. Total synthesis of the structural gene for the precursor of a tyrosine suppressor transfer RNA from Escherichia coli. 7. Enzymatic joining of the chemically synthesized segments to form a DNA duplex corresponding to the nucleotide sequence 1-26. J Biol Chem 251: 634–641. [PubMed] [Google Scholar]
- Sekiya T, Takeya T, Brown EL, Belagaje R, Contreras R, Fritz HJ, Gait MJ, Lees RG, Ryan MJ, Khorana HG. 1979. Total synthesis of a tyrosine suppressor transfer RNA gene. XVI: Enzymatic joinings to form the total 207-base pair-long DNA. J Biol Chem 254: 5787–5801. [PubMed] [Google Scholar]
- Septak M. 1996. Kinetic studies on depurination and detritylation of CPG-bound intermediates during oligonucleotide synthesis. Nucleic Acids Res 24: 3053–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty RP, Endy D, Knight TF Jr. 2008. Engineering BioBrick vectors from BioBrick parts. J Biol Eng 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh-Gasson S, Green RD, Yue Y, Nelson C, Blattner F, Sussman MR, Cerrina F. 1999. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat Biotechnol 17: 974–978. [DOI] [PubMed] [Google Scholar]
- Smith HO, Hutchison CA III, Pfannkoch C, Venter JC. 2003. Generating a synthetic genome by whole genome assembly: φX174 bacteriophage from synthetic oligonucleotides. Proc Natl Acad Sci 100: 15440–15445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa SW, Gelderman G, Contreras LM. 2015. Advances in synthetic dynamic circuits design: Using novel synthetic parts to engineer new generations of gene oscillations. Curr Opin Biotechnol 36: 161–167. [DOI] [PubMed] [Google Scholar]
- Stemmer WP, Crameri A, Ha KD, Brennan TM, Heyneker HL. 1995. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene 164: 49–53. [DOI] [PubMed] [Google Scholar]
- Tang NC, Chilkoti A. 2016. Combinatorial codon scrambling enables scalable gene synthesis and amplification of repetitive proteins. Nat Mater 15: 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme K, Zhao D, Voigt CA. 2012. Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proc Natl Acad Sci 109: 7085–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Gong H, Sheng N, Zhou X, Gulari E, Gao X, Church G. 2004. Accurate multiplex gene synthesis from programmable DNA microchips. Nature 432: 1050–1054. [DOI] [PubMed] [Google Scholar]
- Torella JP, Lienert F, Boehm CR, Chen JH, Way JC, Silver PA. 2014. Unique nucleotide sequence-guided assembly of repetitive DNA parts for synthetic biology applications. Nat Protoc 9: 2075–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trubitsyna M, Michlewski G, Cai Y, Elfick A, French CE. 2014. PaperClip: Rapid multi-part DNA assembly from existing libraries. Nucleic Acids Res 42: e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W, Li L, Xu Q, Wang Z, Yao Y, Wang R, Zhang J, Liu H, Gao X, Hong J. 2014. Error removal in microchip-synthesized DNA using immobilized MutS. Nucleic Acids Res 42: e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. 2009. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460: 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, Reeder PJ, Karimpour-Fard A, Woodruff LB, Gill RT. 2010. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat Biotechnol 28: 856–862. [DOI] [PubMed] [Google Scholar]
- Xiong AS, Peng RH, Zhuang J, Gao F, Li Y, Cheng ZM, Yao QH. 2008a. Chemical gene synthesis: Strategies, softwares, error corrections, and applications. FEMS Microbiol Rev 32: 522–540. [DOI] [PubMed] [Google Scholar]
- Xiong AS, Peng RH, Zhuang J, Liu JG, Gao F, Chen JM, Cheng ZM, Yao QH. 2008b. Non-polymerase-cycling-assembly-based chemical gene synthesis: Strategies, methods, and progress. Biotechnol Adv 26: 121–134. [DOI] [PubMed] [Google Scholar]
- Young L, Dong Q. 2004. Two-step total gene synthesis method. Nucleic Acids Res 32: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Cai S, Hong A, You Q, Yu P, Sheng N, Srivannavit O, Muranjan S, Rouillard JM, Xia Y, et al. 2004. Microfluidic PicoArray synthesis of oligodeoxynucleotides and simultaneous assembling of multiple DNA sequences. Nucleic Acids Res 32: 5409–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]