Abstract
β-Lactamases, the enzymes that hydrolyze β-lactam antibiotics, remain the greatest threat to the usage of these agents. In this review, the mechanism of hydrolysis is discussed for both those enzymes that use serine at the active site and those that require divalent zinc ions for hydrolysis. The β-lactamases now include >2000 unique, naturally occurring amino acid sequences. Some of the clinically most important of these are the class A penicillinases, the extended-spectrum β-lactamases (ESBLs), the AmpC cephalosporinases, and the carbapenem-hydrolyzing enzymes in both the serine and metalloenzyme groups. Because of the versatility of these enzymes to evolve as new β-lactams are used therapeutically, new approaches to antimicrobial therapy may be required.
β-Lactamase enzymes produced by pathogenic bacteria are the greatest threat to the effectiveness of β-lactam antibiotics (e.g., penicillins). Because of their rapid evolution, new therapeutic approaches may be required.
Despite the tremendous advancements in biomedicine, the production of β-lactam hydrolyzing enzymes, β-lactamases, by Gram-negative and -positive bacteria still remains one of the most significant threats to human health (Hauck et al. 2016). With the introduction of every new class of antibiotics, bacteria have continued to evolve resistance, as they are amazingly capable of responding to environmental pressure via selection of existing mutations and acquisition of new genes. The most significant threat has been faced by β-lactam antibiotics. The rapid evolution of β-lactamases, especially carbapenem hydrolyzing enzymes, makes each new drug obsolete in a very short period of time (Bush 2010a,b, 2014; Drawz and Bonomo 2010).
MECHANISM OF β-LACTAM ACTION
To properly appreciate the mechanisms by which β-lactamases have changed the status of β-lactams in our therapeutic armamentarium, it is important to briefly review how β-lactams kill bacteria. β-Lactam antibiotics show their bactericidal effects by inhibiting enzymes involved in cell-wall synthesis, that is, penicillin-binding proteins (PBPs). The integrity of the bacterial cell wall is essential to maintaining cell shape in a hypertonic and hostile environment such as serum, urine, lung mucus, or gastrointestinal tract. Osmotic stability is preserved by a rigid cell wall comprised of alternating N-acetylmuramic acid (NAM) and N-acetylglucosamine (NAG) units. These glycosidic units are linked by a transglycosidases. A pentapeptide is attached to each NAM unit; the PBPs act as transpeptidases to catalyze the cross-linking of two d-alanine-d-alanine NAM pentapeptides. This cross-linking of adjacent glycan strands confers the rigidity of the cell wall. In the 1960s, Strominger realized that the β-lactam ring is sterically similar to the d-alanine-d-alanine of the NAM pentapeptide (Drawz and Bonomo 2010; Fisher and Mobashery 2014; Fishovitz et al. 2015). As a result, PBPs “mistakenly” use the β-lactam as a substrate “building block” during cell-wall synthesis. This “error” results in acylation of the PBP, which renders the enzyme unable to catalyze further carry out transpeptidation reactions. As cell-wall synthesis slows, constitutive peptidoglycan autolysis continues as a result of amidases, bacterial autolytic enzymes. The breakdown of the murein sacculus, the peptidoglycan net that surrounds the bacterium, leads to cell-wall compromise and increased permeability. In this way, the β-lactam-mediated inhibition of transpeptidation causes cell lysis.
MECHANISMS OF RESISTANCE TO β-LACTAMS
There are four primary mechanisms by which bacteria can overcome β-lactam antibiotics (Drawz and Bonomo 2010; Papp-Wallace et al. 2011). First, changes in the active site of PBPs can lower the affinity for β-lactam antibiotics and subsequently increase resistance to these agents, such as in PBP2x of Streptococcus pneumoniae. In a similar manner, penicillin resistance in Streptococcus sanguis, Streptococcus oralis, and Streptococcus mitis developed from horizontal transfer of a PBP2b gene from S. pneumoniae.
Methicillin resistance in Staphylococcus spp. is another example of an altered PBP. Although the cause for this resistance is heterogeneous, it is often conferred by acquisition of the mec element, the mecA gene, which encodes PBP2a (also denoted PBP2′). This low-affinity transpeptidase can assemble new cell wall in the presence of high concentration of penicillins (i.e., methicillin) and cephalosporins.
Second, to access PBPs on the surface of the inner membrane, β-lactams must either diffuse through or directly traverse porin channels in the outer membrane of Gram-negative bacterial cell walls. Resistance to β-lactams can occur when these porin proteins are modified such that they are not produced in a fully active form. Some Gram-negative bacteria show resistance to carbapenems based on loss and or reduction of these outer membrane proteins, such as the loss of OprD, which is associated with resistance to imipenem and reduced susceptibility to meropenem in Pseudomonas aeruginosa (Papp-Wallace et al. 2011).
Third, multicomponent drug efflux pump systems (mex), as part of either an acquired or intrinsic resistance repertoire, are capable of exporting a wide-range of substrates from the periplasm of Gram-negative bacteria to the surrounding environment (Papp-Wallace et al. 2011). These pumps are an important determinant of multidrug resistance in many Gram-negative pathogens, particularly notable in P. aeruginosa and Acinetobacter spp. Other pumps are found in the enteric bacteria but will not be discussed in detail, as this is beyond the scope of this review. As an example of the role played by efflux pumps, increased production of the MexA–MexB system, in combination with the low intrinsic permeability of P. aeruginosa, can contribute to decreased susceptibility to penicillins, cephalosporins, carbapenems, as well as quinolones, tetracycline, and chloramphenicol.
Last, β-lactamases hydrolyze β-lactams. This is the most common and important mechanism of resistance in Gram-negative bacteria and will be the focus of this review. The description of β-lactamases conferring resistance to penicillins and cephalosporins has been extensively detailed (Drawz and Bonomo 2010; Papp-Wallace et al. 2011). This work will build on those reviews and highlight the role of carbapenemases as clinically important β-lactamases. Emphasis will be placed on select class D oxacillinases as much still needs to be learned about these less popular β-lactamases.
β-LACTAMASE HISTORY
The first β-lactamase was identified in Bacillus (Escherichia) coli before the clinical use of penicillin (Abraham and Chain 1988). Within a decade, a significant clinical problem emerged when S. aureus was observed to be resistant to penicillin owing to the production of the staphylococcal penicillinase, PC1. As more β-lactams were developed by pharmaceutical companies and introduced into the clinic, resistance to each agent was observed; the growing number of β-lactam antibiotics increased the selective pressure on bacteria, promoting the survival of organisms with effective β-lactamases (Massova and Mobashery 1998). As a result, β-lactamases were discovered in a multitude of Gram-negative bacteria including the enteric bacteria such as Klebsiellae spp., Enterobacter spp., and nonfermenters such as P. aeruginosa.
Presently, >2000 naturally occurring β-lactamases are now identified and each possesses a unique amino acid sequence and characteristic hydrolysis profile (K Bush, pers. comm.). It is speculated that the rapid replication rate and high mutation frequency permit bacteria to adapt to novel β-lactams by evolution of these β-lactamases. Because we also enjoy the benefits of rapid whole genome sequencing methods, novel variants of a particular family of β-lactamases are constantly being discovered, for example, Acinetobacter-derived cephalosporinases, ADCs, and species-specific families of OXAs.
NAMING OF β-LACTAMASES
Jacoby (2006) has provided a very nice summary of this interesting practice of naming the β-lactamases. β-Lactamases have been named on the basis of molecular characteristics or functional properties. Earlier, β-lactamases were initially designated by the name of the bacteria or plasmid that produced them (e.g., PC1 or P99). Since these original descriptions, β-lactamases have been named after substrates that are hydrolyzed (FOX), discovery location (OHIO), patient’s names (TEM), or the names of the discoverers (HMS). Notably, a few β-lactamases have been given more than one name (e.g., ARI-1 and OXA-23; YOU-1 and TEM-26; YOU-2 and TEM-12; PIT-2 and SHV-1). The interested reader should refer to Jacoby (2006).
CLASSIFICATION
Two major classification schemes exist for categorizing β-lactamase enzymes (the Ambler and Bush–Jacoby systems). Ambler classes A through D use amino acid sequence homology to categorize β-lactamases (Bush and Jacoby 2010). The Bush–Jacoby system groups 1 through 4 are based on substrate hydrolysis profiles (penicillin, cephalosporin, extended-spectrum cephalosporin, carbapenem) and inhibitor profile (inhibition by β-lactamase inhibitors clavulanate and tazobactam) (shown in Table 1).
Table 1.
Comparison of nomenclature systems
| Bush–Jacoby | Ambler | Defining substrates | Inhibited by EDTA | Inhibited by clavulanaic acid or tazobactam | Representatives |
|---|---|---|---|---|---|
| 1 | Class C | Cephalosporinases | (–) | No | P99 |
| Cephamycinases | FOX-4 | ||||
| 2 | Class A | (–) | Yes | ||
| 2a | Penicillins | Yes | PC1 | ||
| 2b | Yes | TEM-1, SHV-1 | |||
| 2be | Cephalosporins | Yes | TEM-10, SHV-2 | ||
| 2br | No | TEM-30 | |||
| 2ber | No | TEM-50 | |||
| 2ce | Yes | RTG-4 | |||
| 2d | Class D | Penicillins | (–) | Variable | OXA-1 |
| 2de | Cephalosporins | OXA-11 | |||
| 2df | Carbapenems | OXA-23 | |||
| 2e | Yes | CepA | |||
| 2f | Carbapenems | Variable | KPC-2 | ||
| 3 | Class B | Carbapenems | (+) | No | |
| 3a | B1 | NDM-1, VIM-2,IMP-1 | |||
| 3b | B2 | CphA | |||
| 3a | B3 | L1 |
Based on data from Bush and Jacoby (2010).
EDTA, ethylenediaminetetraacetate.
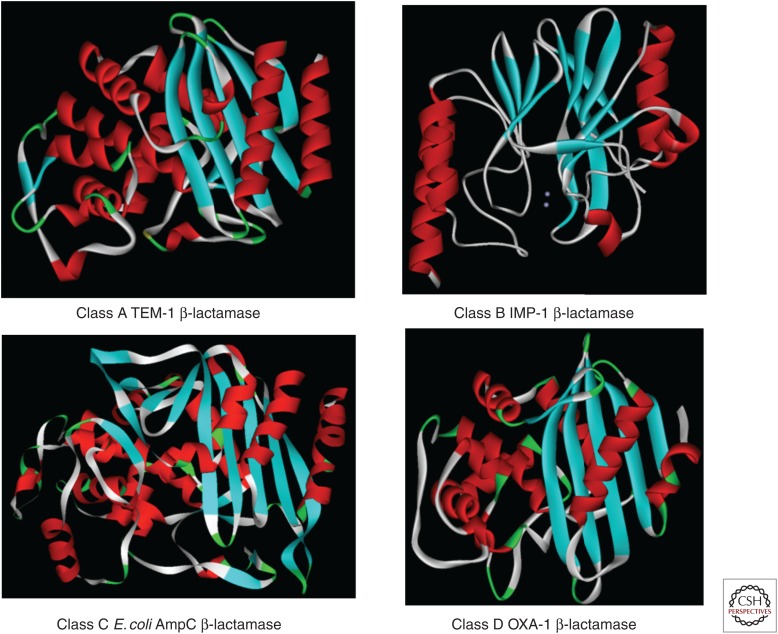
A “family portrait” reveals the structural similarity of class A, C, and D serine β-lactamases (Fig. 1). Class B β-lactamases (“a class apart”) are metallo-β-lactamases (MBLs). These proteins possess either a single or pair of Zn2+ ions coordinated in their active sites. More details will be discussed below.
Figure 1.
The structural similarity of class A, C, and D serine β-lactamases.
CLASS A HYDROLYTIC MECHANISM
β-Lactamases are bacterial hydrolases that bind and acylate β-lactam antibiotics, much like PBPs, and then use strategically positioned water molecules to hydrolyze and inactivate the antibiotic before it can reach its target (Drawz and Bonomo 2010). In this manner, the β-lactamase is regenerated and can inactivate additional antibiotic molecules. This reaction may be schematically represented by the following equation:
| (1) |
In this scheme, E is a β-lactamase, S is a β-lactam substrate, E:S is the Henri–Michaelis complex, E–S is the acyl enzyme, and P is the product devoid of antibacterial activity. The rate constants for each step are represented by: k1, k−1, k2, and k3; k1 and k−1 are association and dissociation rate constants for the preacylation complex, respectively; k2 is acylation rate constant; and k3 is deacylation rate constant.
Serine β-lactamases, for example, TEM-1, SHV-1, P99, and KPC-2, actually use a multistep process to inactivate β-lactams. First, after penicillin or cephalosporin binding, nucleophilic attack by the active site serine on the carbonyl group of the β-lactam antibiotic results in a high-energy acylation intermediate. Next, this intermediate “transitions” into a lower energy covalent acyl enzyme. Following this, a catalytic water molecule attacks the covalent complex and leads to a high-energy deacylation intermediate, with subsequent hydrolysis of the bond between the β-lactam carbonyl and the serine oxygen. Last, deacylation regenerates the active enzyme and renders the β-lactam inactive.
Both acylation and deacylation require the activation of the nucleophilic serine and hydrolytic water, respectively. In class A enzymes, the ultra-high-resolution (0.85 Å) structure of TEM-1 in complex with an acylation transition state analogue revealed the protonated state of Glu166, supporting the hypothesis that Glu166 acts as the activating base for both acylation and deacylation.
CLASS C, B, AND D HYDROLYTIC MECHANISM
The mechanistic symmetry seen in the class A discussion above is mirrored in class C enzymes, in which Tyr150 likely behaves as the general base for both acylation and deacylation, increasing the nucleophilicity of Ser64 and the catalytic water, respectively (Chen et al. 2009). The hydrolysis reaction catalyzed by class C β-lactamases consists of two steps: acylation and deacylation. In the acylation half of the reaction, Ser64 attacks the β-lactam ring carbon and forms a covalent acyl-enzyme complex. Current structural and conformational evidence suggests that during acylation the catalytic nucleophile, Ser64, is deprotonated and a proton is also transferred to the leaving group, the β-lactam ring nitrogen. In deacylation, it is believed that a general base activates the structurally conserved deacylating water, whereas a general acid may be needed to reprotonate Ser64. In the deacylation step, the catalytic water reacts with the covalent linkage between the enzyme and the substrate, leading to the release of the hydrolyzed product. Both acylation and deacylation reactions proceed through a high-energy tetrahedral transition state.
Despite this similarity, debate exists about the role of Lys67 (Chen et al. 2009). It is generally agreed that many mechanisms may contribute to hydrolysis in class C β-lactamases, depending on the enzyme and the substrate, explaining why different variants and substrates seem to support different pathways and mechanisms, that is, substrate assisted or conjugate base. For the wild-type enzyme itself, the conjugate base mechanism may be well favored. Substrate assisted catalysis may also occur; in this mechanism, the proton from the catalytic water is transferred to the substrate ring nitrogen, whereas Tyr150 stabilizes the water molecule. It is the opinion of this researcher that the two different hypotheses are not mutually exclusive.
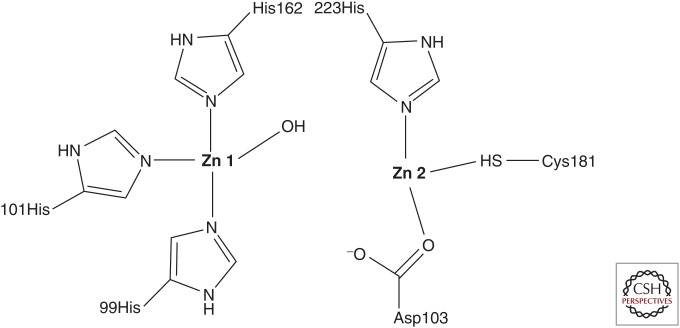
Class B includes Zn2+-dependent enzymes that follow a different hydrolytic mechanism. These MBLs use the OH group from a water molecule that is coordinated by Zn2+ to hydrolyze the scissile amide bond of a β-lactam (Fig. 2). The importance of a high-energy anionic intermediate is currently favored as being essential to the reaction coordinate.
Figure 2.
The active site of a metallo-β-lactamase.
Class D enzymes hydrolyze β-lactams using a slightly different scheme by featuring a carbamylated lysine. OXA β-lactamases use this carbamylated lysine to activate the nucleophilic serine used for β-lactam hydrolysis (like Glu166 in class A β-lactamases). The deacylating water molecule approaches the acyl-enzyme species, anchored at the nucleophilic serine using the carbamylated lysine as a chemical anchor. Carbapenem hydrolyzing class D β-lactamases have evolved the ability to hydrolyze imipenem, an important carbapenem in clinical use, by subtle structural changes in the active site. These changes may contribute to tighter binding of imipenem to the active site and removal of steric hindrances from the path of the deacylating water molecule as was shown in class A (Verma et al. 2011).
CLINICALLY IMPORTANT β-LACTAMASES
Class A—Serine Penicillinases TEM, SHV, and CTX-M and the Carbapenemases KPC, etc.
Class A β-lactamases are often plasmid-encoded, but can also be located on the bacterial chromosome. For example, blaSHV-1 is a chromosomal gene in Klebsiella pneumoniae, but may also be found on plasmics; penA from Burkholderia pseudomallei is chromosomally encoded (Papp-Wallace et al. 2015). In general, class A enzymes are usually susceptible to inactivation by the clinically available β-lactamase inhibitors: clavulanate, sulbactam, tazobactam, and avibactam. TEM, SHV, and CTX-M β-lactamases are mostly found in E. coli and Klebsiellae spp. Many class A β-lactamases have substrate profiles that include expanded-spectrum cephalosporins, and these extended-spectrum β-lactamases (ESBLs) have been discussed extensively (Paterson and Bonomo 2005; Perez et al. 2007). The widespread distribution of CTX-M β-lactamases, especially CTX-M-14 and CTX-M-15, in E. coli is responsible for the large part of the global advanced generation cephalosporin resistance seen in many clinical isolates. Other class A enzymes are encoded on integrons, for example, GES-1 from K. pneumoniae and VEB-1 in P. aeruginosa and Acinetobacter baumannii (Poirel et al. 2012).
Few Ambler class A β-lactamases show carbapenem-hydrolyzing activity. The major class A carbapenemases include KPC, GES, Nmc-A/IMI, and SME β-lactamases, with SME carbapenemases identified, to date, only in Serratia marcescens. With the notable exception of KPCs and GES, the clinical distribution of the types of carbapenemases is relatively limited (Bush 2010a).
Currently, most carbapenem resistance among Enterobacteriaceae in the United States is attributed to plasmid-mediated expression of a KPC-type (K. pneumoniae carbapenemase) carbapenemase. KPC-producing Enterobacteriaceae are considered endemic in many places, for example, in Greece, along with other carbapenemases, specifically VIM-type metallo-β-lactamases. KPC β-lactamases efficiently hydrolyze carbapenems as well as penicillins, cephalosporins, and aztreonam and are not overcome in vitro by clinically available β-lactamase inhibitors; in fact, clavulanic acid, sulbactam, and tazobactam are hydrolyzed. Avibactam inhibits KPC enzymes, but is hydrolyzed slowly (Nguyen et al. 2016).
Carbapenem resistance secondary to KPC production was first described in a K. pneumoniae isolate recovered in North Carolina in 1996 (Yigit et al. 2003). The blaKPC gene has been mapped to a highly conserved Tn3-based transposon, Tn4401, and different isoforms of Tn4401 are described. Plasmids carrying blaKPC are of various sizes and many carry additional genes conferring resistance to fluoroquinolones and aminoglycosides, thus limiting the antibiotics available to treat infections with KPC-producing pathogens; blaKPC has rarely been mapped to a chromosomal location (Chen et al. 2009).
A predominant strain of K. pneumoniae appears responsible for outbreaks and the international spread of KPC-producing K. pneumoniae. A specific MLST sequence type (ST), ST258, has spread in the United States. Now there is widespread acknowledgment that two clades of ST258 exist (Deleo et al. 2014). A second sequence type, ST14, was also common in institutions in the midwestern region of the United States. These findings implied that certain strains of K. pneumoniae may be more apt to obtain and retain the blaKPC gene.
KPC-production can confer variable levels of carbapenem resistance with reported minimum inhibitory concentrations (MICs) ranging from ≤1 µg/mL (susceptible) to ≥16 µg/mL. Analysis of isolates displaying high-level carbapenem resistance showed that increased phenotypic resistance may be caused by increased blaKPC gene copy number or the loss of an outer membrane porin, Omp K35 and/or Omp K36. The highest level of imipenem resistance was seen with isolates lacking both porins and with augmented KPC enzyme production (Chen et al. 2009).
Nmc-A (non-metallo-carbapenemase-A) is a chromosomal carbapenemase originally isolated from Enterobacter cloacae in France. Currently, reports of this particular β-lactamase are still rare. IMI-1 was initially recovered from the chromosome of an E. cloacae isolate in the southwestern United States. A variant of IMI-1, IMI-2, has been identified on plasmids isolated from environmental strains of Enterobacter absuriae in U.S. rivers.
SME-1 (S. marcescens enzyme) was originally identified in an isolate of S. marcescens from a patient in London in 1982. SME-2 and SME-3 were subsequently isolated in the United States, Canada, and Switzerland. Chromosomally encoded SME-type carbapenemases continue to be isolated at a low frequency in North America (Naas et al. 2016). Although infrequent, it is currently recommended to screen for SME enzymes in carbapenem-resistant S. marcescens isolates (Bush et al. 2013).
The GES-type (Guiana extended-spectrum) β-lactamases are acquired β-lactamases recovered from P. aeruginosa, Enterobacteriaceae, and A. baumannii. The genes encoding these β-lactamase have often, but not exclusively, been identified within class 1 integrons residing on transferrable plasmids. GES-1 has a similar hydrolysis profile to other ESBLs, although they essentially spare monobactams. Several GES β-lactamases are described with six (i.e., GES-2, GES-4, GES-5, GES-6, GES-11, and GES-14), showing detectable carbapenemase activity in the setting of amino acid substitutions at their active sites (specifically at residue 104 and 170). These GES-type carbapenemases have been described in Europe, South Africa, Asia, and the Middle East.
Class B Metallo-β-Lactamases
Class B β-lactamases are bacterial enzymes that degrade β-lactam antibiotics with the help of a metal cofactor (divalent zinc in the natural form). Class B enzymes are Zn2+-dependent β-lactamases that follow a different hydrolytic mechanism than the serine β-lactamases of classes A, C, and D. Although catalyzing the same overall reaction as serine-β-lactamases, that is, breaking the amide bond, class B MBLs are structurally and mechanistically unrelated to the former enzymes, and their common function apparently represents an example of convergent evolution of different protein lineages within the bacterial domain (Mojica et al. 2015).
Organisms producing MBLs usually show resistance to penicillins, cephalosporins, carbapenems, and the clinically available β-lactamase inhibitors. MBL bla genes are located on the chromosome, plasmid, and integrons. Until a few years ago, the clinically important class B enzymes included those found in the nosocomial pathogen Stenotrophomonas maltophilia. The rapid emergence of NDM MBLs in the Enterobacteriaceae has changed this (Walsh et al. 2011).
Because of the dependence on Zn2+, catalysis is inhibited in the presence of metal-chelating agents like EDTA. MBLs are not inhibited by the presence of commercially available β-lactamase inhibitors; however, hydrolytic stability of monobactams (i.e., aztreonam) leading to susceptibility of the producing organism appears to be preserved in the absence of concomitant expression of other resistance mechanisms (e.g., ESBL production). The more geographically widespread MBLs include IMP, VIM, and NDM (Mojica et al. 2015).
NDM-1 (New Delhi MBL) was first identified in 2008. NDM-1 was first discovered in Sweden in a patient of Indian descent previously hospitalized in India (Yong et al. 2009; Kumarasamy et al. 2010). The patient was colonized with a K. pneumoniae strain and an E. coli–carrying blaNDM-1 on transferrable plasmids. In the United Kingdom, an increase in the number of clinical isolates of carbapenem-resistant Enterobacteriaceae was also seen in both 2008 and 2009. A U.K. reference laboratory reported that at least 17 of 29 patients found to be harboring NDM-1 expressing Enterobacteriaceae had a history of recent travel to the Indian subcontinent with the majority having been hospitalized in those countries.
NDM-1 shares the most homology (32.4%) with VIM-1 and VIM-2 (Yong et al. 2009). It is a 28-kDa monomeric protein that shows tight binding to both penicillins and cephalosporins. Binding to carbapenems does not appear to be as avid as other MBLs, but catalytic efficiencies appear to be similar. Using ampicillin as a substrate allowed for detailed characterization of the interactions between NDM’s active site and β-lactams as well as improved evaluation of MBLs unique mechanism of β-lactam hydrolysis. More recent crystal structures of NDM-1 reveal the molecular details of how carbapenem antibiotics are recognized by dizinc-containing metallo-β-lactamases (King et al. 2012).
Because of its rapid international dissemination and its ability to be expressed by numerous Gram-negative pathogens, NDM is poised to become the most commonly isolated and distributed carbapenemase worldwide. Initial reports frequently showed an epidemiologic link to the Indian subcontinent where these MBLs are endemic. Indeed, retrospective analyses of stored isolates suggest that NDM-1 may have been circulating in the subcontinent as early as 2006. Despite initial controversy, the Balkans may be another area of endemicity for NDM-1. Sporadic recovery of NDM-1 in the Middle East suggests that this region may be an additional reservoir.
Like KPCs, the conveniences of international travel and medical tourism have quickly propelled this relatively novel MBL into a formidable public health threat. Gram-negative bacilli harboring blaNDM have been identified worldwide (Nordmann et al. 2011).
European reports suggest that horizontal transfer of blaNDM-1 exists within hospitals outside of endemic areas. Of overwhelming concern are the reported cases without specific contact with the healthcare system locally or in endemic areas, suggesting autochthonous acquisition.
Surveillance of public water supplies in India indicates that exposure to NDM-1 may be environmental. Walsh et al. (2011) analyzed samples of public tap water and seepage water from sites around New Delhi. The results were disheartening in that blaNDM-1 was detected by polymerase chain reaction (PCR) in 4% of drinking water samples and 30% of seepage samples. In this survey, carriage of blaNDM-1 was noted in 11 species of bacteria not previously described, including virulent ones like Shigella boydii and Vibrio cholerae.
The rapid spread of NDM-1 highlights the fluidity and rapidity of gene transfer among bacterial species. Although blaNDM-1 was initially and repeatedly mapped to plasmids isolated from carbapenem-resistant E. coli and K. pneumoniae, reports of both plasmid and chromosomal expression of blaNDM-1 has been noted in other species of Enterobacteriaceae as well as Acinetobacter spp. and P. aeruginosa (Jones et al. 2014). Recently, bacteremia with an NDM-1 expressing V. cholerae has been described in a patient previously hospitalized in India colonized with a variety of Enterobacteriaceae previously known to be capable of carrying plasmids with blaNDM-1 (Darley et al. 2012).
In contrast to KPCs, the presence of a dominant clone among blaNDM-1-carrying isolates remains elusive. NDM-1 expression in E. coli has been noted among sequence types previously associated with the successful dissemination of other β-lactamases, including ST101 and ST131. Mushtaq et al. analyzed a relatively large group of blaNDM-1 expressing E. coli from the United Kingdom, Pakistan, and India to potentially identify a predominant strain responsible for the rapid and successful spread of NDM-1 (Mushtaq et al. 2011). The most frequent sequence type identified was ST101. Another study examining a collection of carbapenem-resistant Enterobacteriaceae from India shows the diversity of strains capable of harboring blaNDM-1. Carriage of blaNDM-1 was confirmed in ten different sequence types of K. pneumoniae and five sequence types of E. coli. This multiplicity was confirmed in a study looking at a collection of blaNDM-1 expressing Enterobacteriaceae from around the world (Poirel et al. 2011). Of most concern is that NDM-1 has been identified in E. coli ST131, the strain of E. coli credited with the global propagation of CTX-M-15 ESBLs. Similar to KPCs, NDM-1 expression portends variable levels of carbapenem resistance, and there is often concomitant carriage of a myriad of resistance determinants including other β-lactamases and carbapenemases as well as genes associated with resistance to fluoroquinolones and aminoglycosides.
To date, NDM-1 remains the most common NDM variant isolated. It is currently believed that blaNDM-1 is a chimeric gene that may have evolved from A. baumannii. Contributing to this theory is the presence of complete or variations of the insertion sequence, ISAba125, upstream of the blaNDM-1 gene in both Enterobacteriaceae and A. baumannii. This insertion sequence has primarily been found in A. baumannii.
A recent evaluation of the genetic construct associated with blaNDM-1 has led to the discovery of a new bleomycin resistance protein, BRPMBL. Evaluation of 23 isolates of blaNDM-1/2 harboring Enterobacteriaceae and A. baumannii noted that the overwhelming majority of them possessed a novel bleomycin resistance gene, bleMBL. Coexpression of blaNDM-1 and bleMBL appears to be mediated by a common promoter (PNDM-1), which includes portions of ISAba125. It is postulated that BRPMBL expression may contribute some sort of selective advantage allowing NDM-1 to persist in the environment.
A contemporary evaluation of recently recovered NDM-1 producing A. baumannii isolates from Europe shows that blaNDM-1 and blaNDM-2 genes are situated on the same chromosomally located transposon, Tn125. Dissemination of blaNDM in A. baumannii seems be caused by different strains carrying Tn125 or derivatives of Tn125, rather than plasmid-mediated or clonal (Poirel et al. 2012).
Before the description of NDM-1, frequently detected MBLs include IMP-type (imipenem-resistant) and VIM-type (Verona integron-encoded MBL) with VIM-2 being the most prevalent. These MBLs are embedded within a variety of genetic structures, most commonly integrons. When these integrons are associated with transposons or plasmids, they can readily be transferred among species (Mojica et al. 2015).
A more commonly recovered MBL is the VIM-type enzyme. VIM-1 was first described in Italy in 1997 in P. aeruginosa. VIM-2 was next discovered in southern France in P. aeruginosa cultured from a neutropenic patient in 1996. Although originally thought to be limited to nonfermenting Gram-negative bacilli, VIM-type MBLs are being increasingly identified in Enterobacteriaceae as well. Many variants of VIM have been described with VIM-2 being the most common MBL recovered worldwide (Mojica et al. 2015).
Other more geographically restricted MBLs include (1) SPM-1, Sao Paulo MBL, which has been associated with hospital outbreaks in Brazil; (2) GIM-1, German imipenemase, isolated in carbapenem-resistant P. aeruginosa isolates in Germany; (3) SIM-1, Seoul imipenemase, isolated from A. baumannii isolates in Korea; (4) KHM-1, Kyorin Health Science MBL, isolated from a Citrobacter freundii isolate in Japan (Sekiguchi et al. 2008); (5) AIM-1, Australian imipenemase, isolated from P. aeruginosa in Australia; (6) DIM-1, Dutch imipenemase, isolated from a clinical Pseudomonas stutzeri isolate in the Netherlands; (7) SMB-1, S. marcescens MBL, in S. marcescens in Japan; (8) TMB-1, Tripoli MBL, in Achromobacter xylosoxidans in Libya; and (9) FIM-1, Florence imipenemase, from a clinical isolate of P. aeruginosa in Italy. With the notable exception of SPM-1, which has been introduced into European hospitals by a Brazilian pediatric patient, these MBLs have remained confined to their countries/cities of origin.
Class C Cephalosporinases
Class C enzymes include the AmpC β-lactamases, which are usually encoded by bla genes located on the bacterial chromosome, although plasmid-borne AmpC enzymes have become more prevalent. Organisms expressing the AmpC β-lactamase are typically resistant to penicillins, β-lactamase inhibitors (clavulanate and tazobactam), and most cephalosporins including cefoxitin, cefotetan, ceftriaxone, and cefotaxime. AmpC enzymes poorly hydrolyze cefepime, an expanded-spectrum cephalosporin, and are readily inactivated by carbapenems. Notably, AmpC cephalosporinases are very susceptible to inactivation by avibactam. These enzymes are absent in K. pneumoniae, Klebsiella oxytoca, Proteus mirabilis, and Salmonella spp. as well as other bacteria (Perez et al. 2016).
Benzylpenicillin, ampicillin, amoxicillin, and cephalosporins such as cefazolin and cephalothin are very good inducers and good substrates for AmpC β-lactamase. Cefoxitin and imipenem are also strong inducers but are much more stable to hydrolysis. Cefotaxime, ceftriaxone, ceftazidime, cefepime, cefuroxime, piperacillin, and aztreonam are weak inducers and weak substrates but can be hydrolyzed if enough enzyme is expressed. Consequently, MICs of weakly inducing oxyimino-β-lactams are dramatically increased with AmpC hyperproduction. Conversely, MICs of agents that are strong inducers show little change with regulatory mutations because the level of induced ampC expression is already high.
Some β-lactamase inhibitors are also inducers, especially clavulanate, which has little inhibitory effect on AmpC β-lactamase activity, but can paradoxically appear to increase AmpC-mediated resistance in an inducible organism. The inducing effect of clavulanate is especially important for P. aeruginosa, in which clinically achieved concentrations of clavulanate by inducing AmpC expression have been shown to antagonize the antibacterial activity of ticarcillin (Papp-Wallace et al. 2014).
Production of AmpC enzymes in clinically important Gram-negative bacteria is normally at a low level (“repressed”), but can be “derepressed” by induction with certain β-lactams, particularly cefoxitin and imipenem. Sulbactam, but not tazobactam, is also a good inducer of AmpC β-lactamases. The genetic underpinnings of this regulation have been the subject of intense investigation, but are not the subject of this review. Members of the Enterobacteriaceae family, such as Citrobacter, Salmonella, and Shigella, are clinically relevant producers of AmpC enzymes that resist inhibition by clavulanate and sulbactam (Bauvois and Wouters 2007).
Class D Serine Oxacillinases
Class D β-lactamases were initially categorized as “oxacillinases” because of their ability to hydrolyze oxacillin at a rate of at least 50% that of benzylpenicillin, in contrast to the relatively slow hydrolysis of oxacillin by classes A and C. Different enzymes in this diverse class can also confer resistance to penicillins, cephalosporins, extended-spectrum cephalosporins (OXA-type ESBLs), and carbapenems (OXA-type carbapenemases) (Leonard et al. 2013).
Oxacillinases comprise a heterogeneous group of class D β-lactamases, which are able to hydrolyze amino- and carboxypenicillins. The majority of class D β-lactamases are not inhibited by commercially available β-lactamase inhibitors but are inhibited in vitro by NaCl. OXA enzymes are insensitive to inhibition by clavulanate, sulbactam, and tazobactam, with some exceptions; for example, OXA-2 and OXA-32 are inhibited by tazobactam, but not sulbactam and clavulanate; OXA-53 is inhibited by clavulanate. Interestingly, sodium chloride at concentrations >50–75 mm inhibits some carbapenem-hydrolyzing oxacillinases (e.g., OXA-25 and OXA-26). Site-directed mutagenesis studies suggest that susceptibility to inhibition by sodium chloride is related to the presence of a Tyr residue at position 144, which may facilitate sodium chloride binding better than the Phe residue found in resistant oxacillinases. Examples of OXA enzymes include those rapidly emerging in A. baumannii (e.g., OXA-23) and P. aeruginosa (e.g., OXA-50).
Carbapenem-Hydrolyzing Class D β-Lactamases
More than 490 types of oxacillinases are reported with a minority showing low levels of carbapenem-hydrolyzing activity (lahey.org/Studies/other.asp#table1). This select group of enzymes is also referred to as the carbapenem-hydrolyzing class D β-lactamases (CHDLs). CHDLs have been identified most frequently in Acinetobacter spp.; however, there has been increasing isolation among Enterobacteriaceae, specifically OXA-48 (Patel and Bonomo 2013).
With the exception of OXA-163, CHDLs efficiently inactivate penicillins, early cephalosporins, and β-lactam/β-lactamase inhibitor combinations, but spare expanded-spectrum cephalosporins. Carbapenem hydrolysis efficiency is lower than that of other carbapenemases, including the MBLs, and often additional resistance mechanisms are expressed in organisms showing higher levels of phenotypic carbapenem resistance. These include expression of other carbapenemases, alterations in outer membrane proteins (e.g., CarO, OmpK36), increased transcription mediated by IS elements functioning as promoters, increased gene copy number, and amplified drug efflux. Many subgroups of CHDLs have been described. We will focus on those found in A. baumannii and Enterobacteriaceae: OXA-23 and -27; OXA-24/40, -25, and -26; OXA-48 variants; OXA-51, -66, and -69; OXA-58 and OXA-143.
CHDLs can be intrinsic or acquired (Patel and Bonomo 2013). A. baumannii does have naturally occurring but variably expressed chromosomal CHDLs, OXA-51, OXA-66, and OXA-69. For the most part, in isolation the phenotypic carbapenem resistance associated with these oxacillinases is low. However, levels of carbapenem resistance appear to be increased in the presence of specific insertion sequences promoting gene expression. Additional resistance to extended-spectrum cephalosporins can be seen in the setting of coexpression of ESBLs and/or other carbapenemases (Leonard et al. 2013).
The first reported acquired oxacillinase with appreciable carbapenem-hydrolyzing activity was OXA-23. OXA-23, or ARI-1, was identified from an A. baumannii isolate in Scotland in 1993, although the isolate was first recovered in 1985 (Paton et al. 1993). Subsequently, OXA-23 expression has been reported worldwide and both plasmid and chromosomal carriage of blaOXA-23 are described. The OXA-23 group includes OXA-27, found in a single A. baumannii isolate from Singapore. With the exception of an isolate of P. mirabilis identified in France in 2002, this group of β-lactamases has been exclusively recovered from Acinetobacter species. Increased expression of OXA-23 has been associated with the presence of upstream insertion sequences (e.g., ISAba1 and ISAba4) acting as strong promoters.
Another group of CHDLs includes OXA-24/40, OXA-25, and OXA-26. OXA-24 and OXA-40 differ by a few amino acid substitutions, and OXA-25 and OXA-26 are point mutation derivatives of OXA-40. Although primarily linked with clonal outbreaks in Spain and Portugal, OXA-24/40 β-lactamases has been isolated in other European countries and the United States. OXA-40 was in fact the first CHDL documented in the United States. OXA-58 has also only been detected in Acinetobacter spp. Initially identified in France, OXA-58 has been associated with institutional outbreaks and has been recovered from clinical isolates of A. baumannii worldwide (Leonard et al. 2013).
As civilian and military personnel began returning from Afghanistan and the Middle East, practitioners noted increasing recovery of A. baumannii from skin and soft tissue infections. Drug resistance was associated with expression of both OXA-23 and OXA-58. Many isolates carrying the blaOXA-58 gene concurrently carry insertion sequences (e.g., ISaba1, ISAba2, or ISAba3) associated with increased carbapenemase production and, thus, higher levels of carbapenem resistance. In one report, increased gene copy number was also associated with a higher level of enzyme production and increased phenotypic carbapenem resistance (Hujer et al. 2006).
Spread of OXA-type carbapenemases among A. baumannii appears to be clonal, and in-depth reviews of the molecular epidemiology and successful dissemination of these clones has been published. Two MLST schemes with three loci in common exist for A. baumannii—the PubMLST scheme and the Pasteur scheme. Both schemes assign different sequence types into clonal complexes. Sequence types and clonal complexes (CCs) from both schemes can be further categorized into the international (European) clones I, II, and III. It should be noted, however, that the molecular taxonomy of A. baumannii continues to evolve. OXA-23 producing A. baumannii predominantly belong to international clones I and II with a notable proportion being part of CC92 (PubMLST). Similarly, A. baumannii isolates associated with epidemic spread of OXA-24/40 in Portugal and Spain appear to be incorporated in international clone II and ST56 (PubMLST). OXA-58 expressing A. baumannii have been associated with international clones I, II, and III and a variety of unrelated sequence types (Hujer et al. 2006).
OXA-48 was originally identified in a carbapenem-resistant isolate of K. pneumoniae in Turkey. Early reports suggested that this enzyme was geographically restricted to Turkey. In the past few years, however, the enzyme has been recovered from variety of Enterobacteriaceae and has successfully circulated outside of Turkey with reports of isolation in the Middle East, North Africa, Europe, and, most recently, the United States. The Middle East and North Africa may be secondary reservoirs for these CHDLs. Indeed, the introduction of OXA-48 expressing Enterobacteriaceae in some countries has been from patients from the Middle East or Northern Africa. In the United States, the first clinical cases were associated with ST199 and ST43.
At least six OXA-48 variants (e.g., OXA-48, OXA-162, OXA-163, OXA-181, OXA-204, and OXA-232) have been identified. OXA-48 is by far the most globally dispersed and its epidemiology has been recently reviewed. Unlike KPCs and NDM-1, which have been associated with a variety of plasmids, a single 62-kb self-conjugative IncL/M-type plasmid has contributed to a large proportion of the distribution of blaOXA-48 in Europe. Sequencing of this plasmid (pOXA-48a) notes that blaOXA-48 had been integrated through the acquisition of a Tn1999 composite transposon. blaOXA-48 appears to be associated with a specific insertion sequence, IS1999. A variant of Tn1999, Tn1999.2, has been identified among isolates from Turkey and Europe. Tn1999.2 harbors an IS1R element within the IS1999. OXA-48 appears to have the highest affinity for imipenem of the CHDLs specifically those harboring blaOXA-48 within a Tn1999.2 composite transposon. Three isoforms of the Tn1999 transposon have been described.
Although much of the spread of OXA-48 is attributed to a specific plasmid, outbreak evaluations show that a variety of strains have contributed to dissemination of this emerging carbapenemase in K. pneumoniae. The same K. pneumoniae sequence type, ST395, harboring blaOXA-48 was identified in Morocco, France, and the Netherlands. ST353 was associated with an outbreak of OXA-48 producing K. pneumoniae in London (Woodford et al. 2011) and ST221 with an outbreak of OXA-48 in Ireland. OXA-48 production in K. pneumoniae, like KPC-expressing K. pneumoniae, has also been associated with ST14 (Poirel et al. 2004a) and a recent outbreak in Greece was associated with ST11. blaOXA-48 is remarkably similar to blaOXA-54, a β-lactamase gene intrinsic to Shewanella oneidensis (Poirel et al. 2004b). Shewanella spp. are relatively ubiquitous waterborne Gram-negative bacilli and are proving to be a potential environmental reservoir for OXA-48 like carbapenemases as well as other resistance determinants.
OXA-163, a single amino acid variant of OXA-48, was identified in isolates of K. pneumoniae and E. cloacae from Argentina and is unique in that it has activity against expanded-spectrum cephalosporins (Poirel et al. 2011). OXA-163 also has been identified in Egypt, which has a relatively prevalence of OXA-48, in patients without epidemiologic links to Argentina (Abdelaziz et al. 2012).
OXA-181 was initially identified among carbapenem-resistant Enterobacteriaceae collected from India (Castanheira et al. 2011). OXA-181 differs from OXA-48 by four amino acids; however, it appears to be nestled in an entirely different genetic platform. The blaOXA-181 gene has been mapped to a different group of plasmids, the ColE family, and has been associated with an alternative insertion sequence, ISEcp1. The latter insertion sequence has been associated with the acquisition of other β-lactamases including CTX-M-like ESBLs. Like, OXA-48, it appears that OXA-181 may have evolved from a waterborne environmental species Shewanella xiamenensis. OXA-204 differs from OXA-48 by two amino acid substitution. It was recently identified in a clinical K. pneumoniae isolate from Tunisia. Its genetic construct appears to be similar to that of OXA-181. OXA-232 was recently identified among K. pneumoniae isolates in France. OXA-143 is a novel plasmid-borne carbapenem-hydrolyzing oxacillinase recovered from clinical A. baumannii isolates in Brazil. Information regarding its significance and prevalence continues to evolve.
CONCLUSIONS
This review briefly recapitulates our understanding of the history of this complex family of enzymes, reviews the mechanisms by which these hydrolases inactivate β-lactams, and highlights the current challenges that threaten our β-lactam armamentarium, especially the carbapenemases KPCs, NDMs, and OXAs. In 80 years, we have learned that the constant evolution of substrate specificity meets each new β-lactam introduced. The “long view” predicts these enzymes will continue to evolve in novel forms. Perhaps, we will eventually understand why this occurs and how to prevent this. The future of research in this arena needs to be open and mindful to new approaches. We have still not defined all the correlates of activity and resistance or have answered why novel structural variants emerge. The current challenges will have to be faced with new technologies.
Footnotes
Editors: Lynn L. Silver and Karen Bush
Additional Perspectives on Antibiotics and Antibiotic Resistance available at www.perspectivesinmedicine.org
REFERENCES
- Abdelaziz MO, Bonura C, Aleo A, El-Domany RA, Fasciana T, Mammina C. 2012. OXA-163-producing Klebsiella pneumoniae in Cairo, Egypt, in 2009 and 2010. J Clin Microbiol 50: 2489–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham EP, Chain E. 1988. An enzyme from bacteria able to destroy penicillin. Rev Infect Dis 10: 677–678. [PubMed] [Google Scholar]
- Bauvois C, Wouters J. 2007. Crystal structures of class C β-lactamases: Mechanistic implications and perspectives in drug design. In Enzyme-mediated resistance to antibiotics: Mechanisms, dissemination, and prospects for inhibition (ed. Bonomo RA, Tolmasky ME), pp. 145–161. ASM, Washington DC. [Google Scholar]
- Bush K. 2010a. Alarming β-lactamase-mediated resistance in multidrug-resistant Enterobacteriaceae. Curr Opin Microbiol 13: 558–564. [DOI] [PubMed] [Google Scholar]
- Bush K. 2010b. Bench-to-bedside review: The role of β-lactamases in antibiotic-resistant Gram-negative infections. Crit Care 14: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K. 2014. Introduction to Antimicrobial therapeutics reviews: Infectious diseases of current and emerging concern. Ann NY Acad Sci 1323: v–vi. [DOI] [PubMed] [Google Scholar]
- Bush K, Jacoby GA. 2010. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 54: 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K, Pannell M, Lock JL, Queenan AM, Jorgensen JH, Lee RM, Lewis JS, Jarrett D. 2013. Detection systems for carbapenemase gene identification should include the SME serine carbapenemase. Int J Antimicrob Agents 41: 1–4. [DOI] [PubMed] [Google Scholar]
- Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. 2011. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: Report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob Agents Chemother 55: 1274–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, McReynolds A, Shoichet BK. 2009. Re-examining the role of Lys67 in class C β-lactamase catalysis. Protein Sci 18: 662–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darley E, Weeks J, Jones L, Daniels V, Wootton M, MacGowan A, Walsh T. 2012. NDM-1 polymicrobial infections including Vibrio cholerae. Lancet 380: 1358. [DOI] [PubMed] [Google Scholar]
- Deleo FR, Chen L, Porcella SF, Martens CA, Kobayashi SD, Porter AR, Chavda KD, Jacobs MR, Mathema B, Olsen RJ, et al. 2014. Molecular dissection of the evolution of carbapenem-resistant multilocus sequence type 258 Klebsiella pneumoniae. Proc Natl Acad Sci 111: 4988–4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drawz SM, Bonomo RA. 2010. Three decades of β-lactamase inhibitors. Clin Microbiol Rev 23: 160–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JF, Mobashery S. 2014. The sentinel role of peptidoglycan recycling in the β-lactam resistance of the Gram-negative Enterobacteriaceae and Pseudomonas aeruginosa. Bioorg Chem 56: 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishovitz J, Taghizadeh N, Fisher JF, Chang M, Mobashery S. 2015. The Tipper–Strominger hypothesis and triggering of allostery in penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus (MRSA). J Am Chem Soc 137: 6500–6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck C, Cober E, Richter SS, Perez F, Salata RA, Kalayjian RC, Watkins RR, Scalera NM, Doi Y, Kaye KS, et al. 2016. Spectrum of excess mortality due to carbapenem-resistant Klebsiella pneumoniae infections. Clin Microbiol Infect 10.1016/j.cmi.2016.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, Ecker DJ, Massire C, Eshoo MW, Sampath R, et al. 2006. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother 50: 4114–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby GA. 2006. β-Lactamase nomenclature. Antimicrob Agents Chemother 50: 1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LS, Toleman MA, Weeks JL, Howe RA, Walsh TR, Kumarasamy KK. 2014. Plasmid carriage of bla NDM-1 in clinical Acinetobacter baumannii isolates from India. Antimicrob Agents Chemother 58: 4211–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DT, Worrall LJ, Gruninger R, Strynadka NC. 2012. New Delhi metallo-β-lactamase: Structural insights into β-lactam recognition and inhibition. J Am Chem Soc 134: 11362–11365. [DOI] [PubMed] [Google Scholar]
- Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, et al. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study. Lancet Infect Dis 10: 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard DA, Bonomo RA, Powers RA. 2013. Class D β-lactamases: A reappraisal after five decades. Acc Chem Res 46: 2407–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massova I, Mobashery S. 1998. Kinship and diversification of bacterial penicillin-binding proteins and β-lactamases. Antimicrob Agents Chemother 42: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica MF, Bonomo RA, Fast W. 2015. B1-metallo-β-lactamases: Where do we stand? Curr Drug Targets 17: 1029–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, Livermore DM, Woodford N. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J Antimicrob Chemother 66: 2002–2005. [DOI] [PubMed] [Google Scholar]
- Naas T, Dortet L, Iorga BI. 2016. Structural and functional aspects of class A carbapenemases. Curr Drug Targets 17: 1006–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NQ, Krishnan NP, Rojas LJ, Prati F, Caselli E, Romagnoli C, Bonomo RA, van den Akker F. 2016. Crystal structures of KPC-2 and SHV-1 β-lactamases in complex with the boronic acid transition state analog S02030. Antimicrob Agents Chemother 60: 1760–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol 19: 588–595. [DOI] [PubMed] [Google Scholar]
- Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. 2011. Carbapenems: Past, present, and future. Antimicrob Agents Chemother 55: 4943–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp-Wallace KM, Winkler ML, Gatta JA, Taracila MA, Chilakala S, Xu Y, Johnson JK, Bonomo RA. 2014. Reclaiming the efficacy of β-lactam-β-lactamase inhibitor combinations: Avibactam restores the susceptibility of CMY-2-producing Escherichia coli to ceftazidime. Antimicrob Agents Chemother 58: 4290–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp-Wallace KM, Becka SA, Taracila MA, Winkler ML, Gatta JA, Rholl DA, Schweizer HP, Bonomo RA. 2015. Exposing a β-lactamase “twist”: The mechanistic basis for the high level of ceftazidime resistance in the C69F variant of the Burkholderia pseudomallei PenI β-lactamase. Antimicrob Agents Chemother 60: 777–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel G, Bonomo RA. 2013. “Stormy waters ahead”: Global emergence of carbapenemases. Front Microbiol 4: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson DL, Bonomo RA. 2005. Extended-spectrum β-lactamases: A clinical update. Clin Microbiol Rev 18: 657–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton R, Miles RS, Hood J, Amyes SG. 1993. ARI 1: β-lactamase-mediated imipenem resistance in Acinetobacter baumannii. Int J Antimicrob Agents 2: 81–87. [DOI] [PubMed] [Google Scholar]
- Perez F, Endimiani A, Hujer KM, Bonomo RA. 2007. The continuing challenge of ESBLs. Curr Opin Pharmacol 7: 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F, El Chakhtoura NG, Papp-Wallace KM, Wilson BM, Bonomo RA. 2016. Treatment options for infections caused by carbapenem-resistant Enterobacteriaceae: Can we apply “precision medicine” to antimicrobial chemotherapy? Expert Opin Pharmacother 17: 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Heritier C, Tolun V, Nordmann P. 2004a. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother 48: 15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Heritier C, Nordmann P. 2004b. Chromosome-encoded ambler class D β-lactamase of Shewanella oneidensis as a progenitor of carbapenem-hydrolyzing oxacillinase. Antimicrob Agents Chemother 48: 348–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Castanheira M, Carrer A, Rodriguez CP, Jones RN, Smayevsky J, Nordmann P. 2011. OXA-163, an OXA-48-related class D β-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob Agents Chemother 55: 2546–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L, Bonnin RA, Nordmann P. 2012. Genetic support and diversity of acquired extended-spectrum β-lactamases in Gram-negative rods. Infect Genet Evol 12: 883–893. [DOI] [PubMed] [Google Scholar]
- Sekiguchi JI, Morita K, Kitao T, Watanabe N, Okazaki M, Miyoshi-Akiyama T, Kanamori M, Kirikae T. 2008. KHM-1, a novel plasmid-mediated metallo-β-lactamase from a Citrobacter freundii clinical isolate. Antimicrob Agents Chemother 52: 4194–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V, Testero SA, Amini K, Wei W, Liu J, Balachandran N, Monoharan T, Stynes S, Kotra LP, Golemi-Kotra D. 2011. Hydrolytic mechanism of OXA-58 enzyme, a carbapenem-hydrolyzing class D β-lactamase from Acinetobacter baumannii. J Biol Chem 286: 37292–37303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: An environmental point prevalence study. Lancet Infect Dis 11: 355–362. [DOI] [PubMed] [Google Scholar]
- Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: The role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35: 736–755. [DOI] [PubMed] [Google Scholar]
- Yigit H, Queenan AM, Rasheed JK, Biddle JW, Domenech-Sanchez A, Alberti S, Bush K, Tenover FC. 2003. Carbapenem-resistant strain of Klebsiella oxytoca harboring carbapenem-hydrolyzing β-lactamase KPC-2. Antimicrob Agents Chemother 47: 3881–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-β-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53: 5046–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]