Abstract
Recurrent missense mutations in histone H3 were recently reported in pediatric gliomas and soft tissue tumors. Strikingly, these mutations only affected a minority of the total cellular H3 proteins and occurred at or near lysine residues at positions 27 and 36 on the amino-terminal tail of H3 that are subject to well-characterized posttranslational modifications. Here we review recent progress in elucidating the mechanisms by which these mutations perturb the chromatin landscape in cells through their effects on chromatin-modifying machinery, particularly through inhibition of specific histone lysine methyltransferases. One common feature of histone mutations is their ability to arrest cells in a primitive state refractory to differentiation induction, highlighting the importance of studying these mutations in their proper developmental context.
Mutations at key positions in histone H3 are present in some pediatric gliomas and soft-tissue tumors. These histone mutations arrest cells in a primitive state, refractory to differentiation induction.
Chromatin, the combination of DNA and its interacting proteins, is the physiologically relevant form of eukaryotic genomes. The basic repeating unit of chromatin is the nucleosome, comprised of two copies of the core histone proteins H2A, H2B, H3, and H4 that together form an octamer wrapped by two superhelical turns of DNA (Luger et al. 1997). Historically, nucleosomes were thought to mainly provide structural support for genome packaging. However, research from the past two decades has revealed a remarkable role of nucleosome composition, modification, and positioning in virtually all DNA-based processes, including replication, transcription, and damage repair (Jenuwein and Allis 2001; Ernst and Kellis 2010).
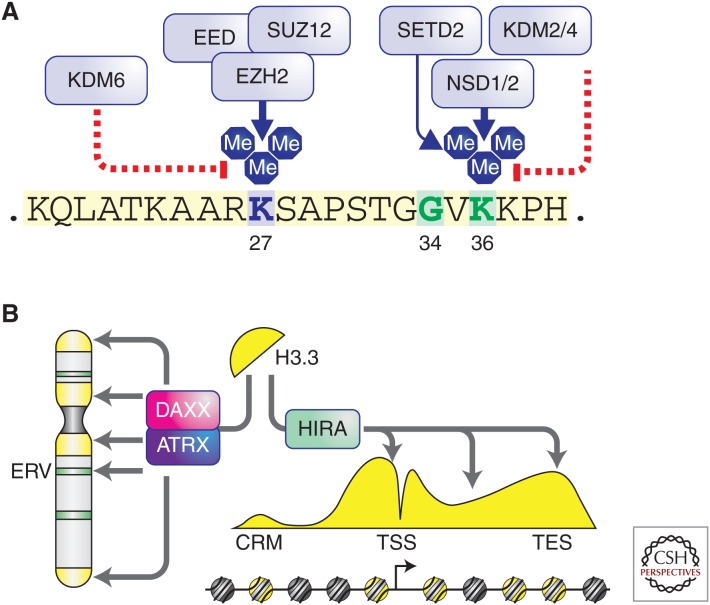
Posttranslational modifications (PTMs) of histones are critically involved in chromatin-mediated gene regulation (Jenuwein and Allis 2001). It is believed that histone PTMs exert their effects through direct physical modulation of nucleosome–DNA contacts and/or recruitment of downstream “reader” protein complexes. To date more than 100 histone PTMs have been identified (Huang et al. 2014), many of which are dynamically controlled by enzymes catalyzing their addition (“writers”) or removal (“erasers”). Among them, PTMs of several lysine residues located at the amino-terminal tail of histone H3 have been extensively characterized (Fig. 1A). For example, H3 lysine 27 (H3K27) can be acetylated by p300/CBP and H3K27ac is preferentially located at promoters and/or enhancers of genes that are actively transcribed (Ogryzko et al. 1996). In contrast, methylation of H3K27, catalyzed by the Polycomb repressive complex 2 (PRC2) and removed by KDM6 family demethylases, is a mark associated with gene silencing (Margueron and Reinberg 2011). Methylation of H3 lysine 36 (H3K36), depending on the context, can regulate transcriptional elongation, RNA processing, and DNA damage sensing (Kuo et al. 2011; Carvalho et al. 2014; Simon et al. 2014; Wen et al. 2014). In mammals, several enzymes targeting H3K36 have been reported (Wagner and Carpenter 2012). Although SETD2 is the only methyltransferase that can generate trimethylation of H3K36 (H3K36me3), multiple methyltransferases catalyze mono- and dimethylation of H3K36 (H3K36me1/2), including NSD1/2/3 and ASH1L. Conversely, members of the KDM2 and KDM4 families act as H3K36-specific demethylases.
Figure 1.
Posttranslational modification of the histone variant H3.3 and its chaperone/deposition machinery. (A) The amino-terminal tail of histone H3.3 (shown here) and other histone H3 proteins are subject to methylation of lysine residue 27 by the Polycomb repressive complex 2 (PRC2), containing core subunits EED, SUZ12, and EZH2. Removal of H3K27 methylation is performed by KDM6 family demethylases, including JMJD3 and UTX. Lysine residue 36 is subject to methylation by multiple enzymes, including the NSD family enzymes and SETD2. Removal of H3K36 methylation is performed by the KDM2 and KDM4 family demethylases. (B) Histone variant H3.3 is deposited at pericentric heterochromatin, telomeres, and certain endogenous retroviral elements (ERV) by the ATRX/DAXX heterodimeric complex. In contrast, H3.3 is deposited at euchromatin regions such as promoters and gene bodies by the histone chaperone HIRA (see Banaszynski et al. 2010 and Maze et al. 2014 for details and references).
More recently, it was appreciated that changes to the cellular epigenetic state could also result from expression and incorporation of histone variants (Banaszynski et al. 2010; Maze et al. 2014), further increasing the complexity of chromatin regulation. With the exception of H4, all histones are expressed in variant forms differing in primary amino acid sequence, leading to minor or major structural dissimilarities. Importantly, such variations were shown to have functional relevance. For example, in metazoans, variants of H3 include H3.1, H3.2, H3.3, and CENPA (Hake and Allis 2006). CENPA is centromere-specific and structurally dissimilar to the other H3 variants, whereas H3.3 differs from the “canonical” H3.1 and H3.2 by only a few amino acids. However, several lines of evidence suggest that H3.3 plays a distinct role in chromatin biology from canonical H3 (Fig. 1B). First, although canonical H3 is expressed and incorporated into chromatin in a DNA replication-dependent manner, the expression, assembly, and deposition of H3.3-containing nucleosomes are cell-cycle-independent (Tagami et al. 2004). Second, in proliferating cells, H3.3 is enriched at selected genomic regions, including promoters and gene bodies of highly transcribed or transcriptionally “poised” genes in euchromatin (McKittrick et al. 2004), pericentric heterochromatin and telomeres (Goldberg et al. 2010), and certain classes of endogenous retroviral elements (ERVs) (Elsässer et al. 2015). Third, compared with canonical nucleosomes, distinct sets of factors facilitate the assembly and deposition of H3.3-containing nucleosomes. Deposition of H3.3 to euchromatic regions is mainly mediated by the HIRA complex (Tagami et al. 2004), whereas ATRX/DAXX were identified as the complex that specifically incorporates H3.3 to heterochromatic regions (Lewis et al. 2010; Elsässer et al. 2012, 2015). Depletion of H3.3 in embryonic stem cells leads to aberrant PRC2 binding at developmentally regulated genes, derepression of ERVs, and abnormal cell differentiation (Banaszynski et al. 2013; Elsässer et al. 2015). In mammals, two genes (H3F3A and H3F3B) encode H3.3 and mice deficient for either H3.3 gene show postnatal death, growth retardation, and infertility (Couldrey et al. 1999; Bush et al. 2013). Therefore, although the specific mechanisms remain to be fully elucidated, it appears that H3.3 is required to establish the proper chromatin states at specific genomic regions to maintain cell identity during development.
As the precise regulation of chromatin is essential for many cellular events, including proliferation and differentiation, not surprisingly chromatin misregulation has been linked to various human diseases, notably cancer. A major finding from recent tumor genome sequencing studies was the discovery that chromatin regulators, including writers, erasers, and readers of histone and DNA modifications and nucleosome remodelers, are frequently altered in malignancies (Shen and Laird 2013). Although aberrant chromatin states are increasingly recognized as an emerging hallmark of cancer, few researchers in the cancer epigenetics field anticipated the reports of recurrent mutations in histone H3 themselves. As detailed in the following sections, these mutations are highly clustered missense mutations of residues at or near well-studied PTM sites. Furthermore, the mutations are always monoallelic and affect only one of the 16 genes encoding H3 in humans. These interesting features have attracted considerable attention from the oncology and chromatin biology communities and recently the mechanisms underlying these so-called “oncohistones” have begun to be unraveled. In this review, we will summarize and discuss efforts to identify and understand histone H3 mutations.
HISTONE MUTATIONS IN CANCER
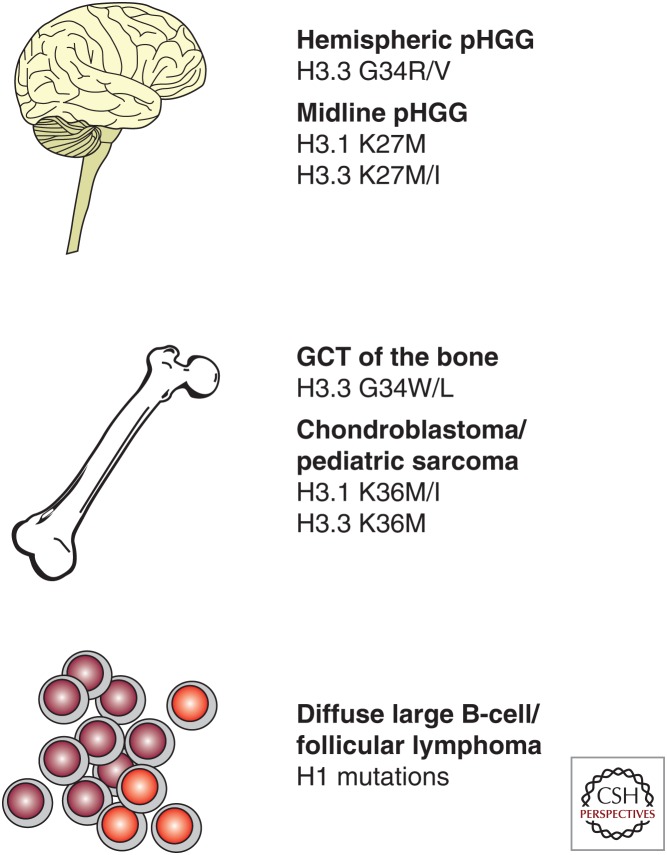
Recurrent mutations in histone H3 were first reported in pediatric high-grade gliomas (pHGGs) (Fig. 2). Simultaneous reports described H3K27M mutations in the majority of diffuse intrinsic pontine gliomas (DIPGs), a type of pHGG associated with dismal prognosis owing to its location in the brainstem, as well as in thalamic gliomas (Schwartzentruber et al. 2012; Wu et al. 2012). H3K27M mutations were not detected in other pediatric brain tumors including medulloblastomas and ependymomas (Wu et al. 2012) and seemed to be specific for midline pHGGs as they were subsequently found in pHGGs involving the cerebellum and the spinal cord (Sturm et al. 2012). Overall, pHGGs were characterized by additional mutations affecting growth factor signaling (e.g., RAS and PI3K) and the RB1 and TP53 pathways for cell-cycle regulation (Buczkowicz et al. 2014; Fontebasso et al. 2014b; Taylor et al. 2014; Wu et al. 2014). Evolutionary reconstruction of individual DIPG tumors revealed that H3K27M mutations arose early and were accompanied by an obligate partner mutation in a member of either of these pathways (e.g., PIK3R1 or TP53), suggesting that both are needed for tumorigenesis (Nikbakht et al. 2016). H3K27M mutations were also reported in rare cases of pediatric low-grade gliomas (Jones et al. 2013; Zhang et al. 2013) as well as in thalamic gliomas in adults under the age of 50 (Aihara et al. 2014). Notably, DIPG patients carrying the H3K27M mutation had worse overall survival compared with patients lacking the mutation (Khuong-Quang et al. 2012; Feng et al. 2015). Because of its prevalence and association with patient outcomes, the detection of H3K27M mutations by immunohistochemistry is increasingly being considered for diagnostic purposes in pediatric gliomas. Antibodies directed against H3K27M have been developed and were shown to be 100% sensitive and specific for the mutation (Bechet et al. 2014; Venneti et al. 2014).
Figure 2.
Recurrent histone mutations in human cancer (see text and Fontebasso et al. 2014a; Kallappagoudar et al. 2015). pHGG, Pediatric high-grade glioma; GCT, giant cell tumor.
The majority of pHGG-associated H3K27M mutations affect the variant histone H3.3, whereas others are found in canonical histone H3.1/2. Intriguingly, H3.3K27M mutations differ from H3.1/2K27M mutations in several clinical features. H3.1K27M-mutant gliomas were restricted to the brainstem, unlike H3.3K27M-mutant gliomas, which were found in the brainstem in addition to other midline locations (Fontebasso et al. 2014b; Castel et al. 2015). DIPG patients with H3.1K27M were, on average, 2 years younger than those with H3.3K27M (Castel et al. 2015) and had a distinctive set of co-occurring mutations. Missense mutations in ACVR1, which encodes the bone morphogenetic protein (BMP) type I receptor ALK2, were significantly associated only with H3.1K27M but not H3.3K27M (Buczkowicz et al. 2014; Fontebasso et al. 2014b; Taylor et al. 2014; Wu et al. 2014). Many of the ACVR1 mutations were identical to those found in the autosomal dominant syndrome fibrodysplasia ossificans progressiva in which gain-of-function mutations in ACVR1 lead to heterotopic ossification (Shore et al. 2006; Chaikuad et al. 2012). Phosphorylation of SMAD1/5/8, downstream events of ALK2 activation, were increased in ACVR1-mutant DIPG tumors (Buczkowicz et al. 2014; Fontebasso et al. 2014b), and expression of mutant ACVR1 transgenes was sufficient to increase phosphorylation of SMAD1/5/8 in astrocytes (Wu et al. 2014) and DIPG cell lines (Taylor et al. 2014). Therefore, ALK2 inhibitors currently in development to treat fibrodysplasia ossificans progressiva could be therapeutically effective against a subset of pHGGs. In contrast, amplification of PDGFRA was significantly associated only with H3.3K27M (Buczkowicz et al. 2014; Castel et al. 2015). Other H3.3K27M-mutant gliomas, particularly in the thalamus, were associated with mutations in both FGFR1 and NF1 (Jones et al. 2013). H3.1K27M and H3. 3K27M DIPGs also have distinct gene expression profiles, although it remains unclear to what extent this is attributable to differences in the H3 variants as opposed to the unique co-occurring mutations in each subgroup or the possibility that each subgroup arises from a distinct cell of origin (Castel et al. 2015).
Unlike midline pHGGs, many pHGGs located in the cerebral hemispheres carried H3.3G34R or H3.3G34V mutations (Fig. 2) (Schwartzentruber et al. 2012; Wu et al. 2012). H3.3G34R/V-mutant tumors were typically diagnosed during adolescence, rather than in early childhood like most H3K27M-mutant tumors. They had distinct gene expression and DNA methylation signatures from H3K27M-mutant tumors and H3 mutational status could be predicted based on the expression of FOXG1 exclusively in H3.3G34-mutant gliomas and OLIG2 exclusively in H3K27M-mutant gliomas (Sturm et al. 2012). Mutations in ATRX, which heterodimerizes with the histone chaperone DAXX to specifically incorporate H3.3 at pericentric heterochromatin and telomeres, were significantly associated with H3.3G34-mutant pHGGs although they were also observed at a lower frequency in wild-type (WT) and H3K27M-mutant pHGGs. Similar to other cancer types, ATRX-mutant pHGGs show alternative lengthening of telomeres (Schwartzentruber et al. 2012). Notably, mutations in the H3K36 methyltransferase SETD2 were also seen in high-grade gliomas involving the cerebral hemispheres in adolescents and were mutually exclusive with H3 mutations (Fontebasso et al. 2013).
Recurrent mutations in histone H3 were subsequently reported in specific types of bone and cartilage tumors (Fig. 2). H3.3K36M mutations were found in nearly all chondroblastomas, a benign tumor of the active growth plate of bones, and rarely in malignant conventional and clear-cell chondrosarcomas (Behjati et al. 2013). Antibodies directed against H3K36M were 100% specific and sensitive for the mutation and may prove useful for diagnosis (Amary et al. 2016; Lu et al. 2016). Interestingly, although H3.3K27M mutations occurred exclusively in H3F3A, H3.3K36M mutations predominately were found in H3F3B despite similar expression levels and identical amino acid sequence of both isoforms (Behjati et al. 2013). In addition, nearly all giant cell tumors of the bone had H3.3G34 mutations mostly to tryptophan (and in one case, to leucine), whereas osteosarcomas infrequently had the H3.3G34R mutation originally identified in pHGGs (Behjati et al. 2013; Joseph et al. 2014; Sarungbam et al. 2016). Two patients with postzygotic H3.3G34W mutations presented with paragangliomas and recurrent giant cell tumors of the bone, suggesting that somatic mosaic mutation of H3.3G34 may be the basis for a new nonhereditary cancer syndrome (Toledo et al. 2015). No H3 mutations were detected in other types of bone and cartilage tumors including chondromyxoid fibromas, chordomas, and chondromas (Behjati et al. 2013).
Recurrent mutations in the linker histone H1 have also been reported in diffuse large B-cell lymphomas (Lohr et al. 2012) and follicular lymphomas (Okosun et al. 2014). Many of these mutations are believed to be loss-of-function (Okosun et al. 2014), but their contribution to oncogenesis remains unexplored and therefore will not be discussed in this review.
MECHANISM OF H3K27M-MEDIATED ONCOGENESIS
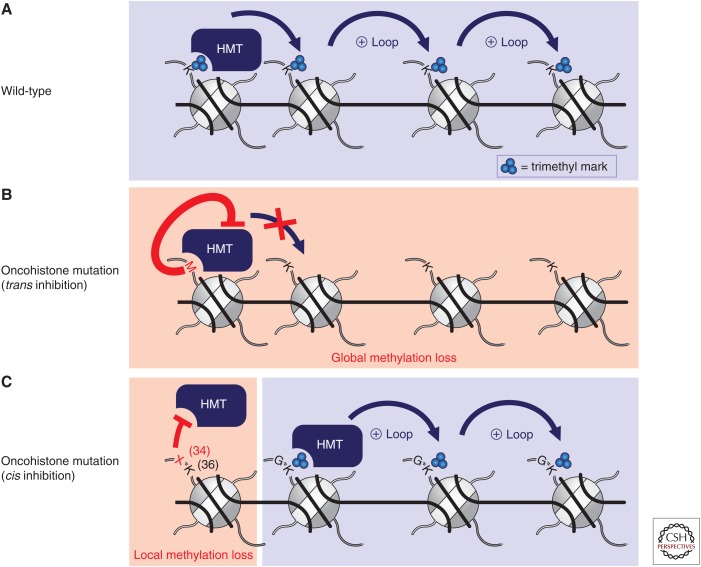
Initial characterization of the histone PTMs in H3K27M-mutant DIPG patient tumors and cell lines revealed a global reduction of H3K27me3 compared with H3 WT DIPG tumors (Chan et al. 2013; Lewis et al. 2013; Venneti et al. 2013), despite similar expression of EZH2 (Venneti et al. 2013). K27M-mutant H3 contributed to only approximately 3%–17% of the total H3 proteins in DIPG samples (Lewis et al. 2013), which is consistent with the fact that only one of the 32 H3-encoding alleles harbors the mutation and suggests that the global reduction in H3K27me3 was caused by a dominant effect of the H3K27M mutation. Indeed, expression of an H3K27M transgene in 293T cells was sufficient to reduce H3K27me2 and H3K27me3 globally despite the mutant histone accounting for only 1% of total H3 protein (Lewis et al. 2013). Similar effects were seen regardless of whether the transgene was H3.1 or H3.3 (Chan et al. 2013) and were consistent across a range of different cell types including astrocyte, fibroblast, and glioma cell lines (Bender et al. 2013; Chan et al. 2013). This effect was restricted to the mutation of the lysine residue at position 27 to methionine (and to a lesser extent isoleucine), but not to other amino acids (Lewis et al. 2013). The dominant nature of the mutation was further shown by the loss of H3K27me3 on endogenous WT H3 proteins (Bender et al. 2013; Lewis et al. 2013).
In-depth biochemical and molecular work has revealed that the H3K27M mutation achieves global reductions in H3K27 methylation through inhibition of the PRC2 complex in trans (Fig. 3A,B). Addition of H3K27M peptides (Lewis et al. 2013) or mononucleosomes (Brown et al. 2014), or heterotypic nucleosome arrays containing three WT nucleosomes and one spatially defined H3K27M mutant nucleosome (Brown et al. 2015), inhibited recombinant PRC2 activity in a dose-dependent and competitive manner. The IC50 for H3K27M inhibition of PRC2 calculated in these in vitro assays is less than the estimated nuclear concentration of a 3%–5% fraction of the total H3 pool (Lewis and Allis 2013), suggesting that such a mechanism could be responsible for the global reduction of H3K27me3 in cells. Although nucleosomes isolated from H3K27M-expressing cells were able to inhibit the activity of recombinant PRC2, their effect on the activity of H3K27 demethylases JMJD3 and UTX was relatively minimal (Bender et al. 2013).
Figure 3.
Inhibition of histone lysine methyltransferases (HMTs) by histone H3 oncohistone mutations. (A) Unimpeded activity of HMTs in the absence of a mutation (wild-type). (B) H3K9M, H3K27M, and H3K36M mutations dominantly inhibit their respective HMTs in trans. Consequently, nonmutant nucleosomes are hypomethylated (global methylation loss). (C) H3G34 mutations inhibit H3K36 HMTs in cis such that only nucleosomes containing the mutation are unable to be methylated (localized methylation loss).
Insights into the interaction between H3K27M and PRC2 have begun to elucidate the structural basis of the inhibition. H3K27M peptides with photo-cross-linkers inserted at or near the mutated residue both pulled down EZH2 from the recombinant PRC2 (Lewis et al. 2013). EZH2 is also enriched on mononucleosomes containing H3K27M isolated from cells (Bender et al. 2013; Chan et al. 2013). Screening of H3K27-mutant peptides found that mutation of K27 to amino acids that have a long, hydrophobic side chain with minimal branching such as methionine, isoleucine, and the unnatural amino acid norleucine were among the most potent inhibitors of PRC2 activity (Lewis et al. 2013; Brown et al. 2014). The inhibitory properties of isoleucine led to the prediction of H3K27I mutation in pHGGs (Lewis and Allis 2013), which was indeed recently identified in a DIPG patient tumor (Castel et al. 2015). These three side chains are thought to be compatible with binding to the active site of the SET domain of EZH2, which normally uses an aromatic cage to bind the unbranched, hydrophobic side chain of lysine. Mutation of one of these aromatic residues in EZH2 rendered the protein less sensitive to inhibition by H3K27M (Lewis et al. 2013). Recently, the crystal structure of Chaetomium thermophilum PRC2 bound to a H3K27M peptide in the presence of S-adenosyl-l-homocysteine (SAH) was solved and suggests that a residue adjacent to the missense mutation, H3 arginine 26 (H3R26), occupies the active site and prevents substrate binding (Jiao and Liu 2015). Interestingly, in the human PRC2:H3K27M complex the methionine 27 side chain was positioned in the lysine 27 access channel (Justin et al. 2016). Further structural and biochemical studies of the PRC2-H3K27M interaction will be important to uncover potential ways to reverse the inhibition of PRC2 (Brown et al. 2014).
Based on the strong degree of conservation among SET domain–containing proteins, the widely adaptable inhibition of histone methyltransferases by H3 “K-to-M” mutations was appreciated even before reports surfaced of H3K36M mutations in patients (Behjati et al. 2013; Lewis et al. 2013). Analogous to H3K27M, the introduction of exogenous H3K9M and H3K36M into 293T cells was able to reduce global levels of H3K9me3 and H3K36me3, respectively (Lewis et al. 2013). Recombinant H3K9-specific methyltransferases, SUV39H1 and G9a, were also each shown to be inhibited by the addition of H3K9M peptides in trans and in an S-adenosyl methionine (SAM)-dependent manner (Lewis et al. 2013; Jayaram et al. 2016; Justin et al. 2016). In contrast, introduction of H3K4M had minimal effect on global H3K4me3 levels in cells, a finding Lewis and colleagues suggest may be a result of inhibition of the H3K4 demethylases LSD1 and LSD2 by H3K4M counterbalancing any potential inhibition of H3K4 methyltransferases by H3K4M (Karytinos et al. 2009).
Although much interest has been directed toward understanding the inhibition of methyltransferases by H3 “K-to-M” mutations, these analyses may overlook additional cellular targets of these mutations. Indeed, histone demethylases have alternatively been implicated as being responsible for the alteration of H3K9 methylation in cells expressing H3K9M. Unbiased mass-spectrometry analysis of immunoprecipitated mononucleosomes containing H3K9M revealed an enrichment for the K9 demethylase KDM3B but a depletion for HP1, a reader of H3K9me3, in native conditions (Herz et al. 2014). In this model, H3K9M serves to recruit KDM3B to genomic loci where it demethylates adjacent WT H3K9, thereby releasing HP1 and decompacting the surrounding chromatin (Herz et al. 2014). It will be important to further validate this model by directly testing the ability of H3K9M to potentiate the activity of KDM3B in cis on nucleosomal templates as well as to assess whether deletion of KDM3B can blunt local and/or global changes in H3K9 methylation induced by the H3K9M mutation. Nonetheless, these findings highlight that multiple non–mutually exclusive mechanisms may shape how H3 “K-to-M” mutations alter the chromatin landscape in cells.
Despite global depletion of H3K27 methylation in H3K27M-expressing cells, genome-wide profiling of the chromatin landscape with high-throughput sequencing revealed a striking gain of H3K27me3 at selected genomic loci. The existence of these regions was first observed when comparing H3K27M-mutant DIPG cell lines with human neural stem cells (NSCs) (Chan et al. 2013) and also when comparing H3K27M and H3 WT DIPG tumor samples (Bender et al. 2013). Although H3K27me3 was reduced in mutant DIPG cell lines at every type of genetic element examined (including promoters, 5′ UTRs, coding exons, introns, 3′ UTRs, and intergenic regions), ∼60% of the H3K27me3 peaks that remained were not present in NSCs (Chan et al. 2013). A similar reduction in overall H3K27me3 peaks was observed in H3K27M-mutant DIPG tumors compared with H3 WT tumors, with gain of H3K27me3 at limited loci that were significantly enriched for intergenic regions (Bender et al. 2013). Integrated RNA-seq and ChIP-seq analyses in both cell lines and tumors revealed that expression of genes that gained H3K27me3 in H3K27M-mutant samples was down-regulated (e.g., the long isoform of CDK6, p16Ink4a, MICA). Conversely, genes that lost H3K27me3 showed enhanced expression (Bender et al. 2013; Chan et al. 2013). In addition, H3K27M-mutant DIPG tumors showed global DNA hypomethylation, which was associated with the loss of H3K27me3 at promoter regions (Bender et al. 2013). A better understanding of the mechanism through which H3K27me3 is established and maintained at certain regions will likely require assessing how EZH2 localization and H3K27 methylation change genome-wide following the introduction and removal of H3K27M in the same cell type.
New models to study DIPG biology have been developed in light of the discovery of H3K27M mutations and the recognition of their ability to perturb the chromatin landscape in cells. Human embryonic stem cell–derived NSCs have been used to elucidate the oncogenic nature of H3K27M based on their role as the presumed cells-of-origin for DIPG. Introduction of H3K27M selectively increased cell proliferation in NSCs, but not in embryonic stem cells (ESCs), ESC-derived astrocytes, or human fibroblasts (Funato et al. 2014). In the setting of TP53 knockdown and the presence of a constitutively active form of PDGFRa, addition of the H3K27M mutation enabled NSCs to acquire several neoplastic properties (Funato et al. 2014). These included the ability to suppress apoptosis on growth factor withdrawal, sustained proliferation following irradiation, increased invasiveness, impaired differentiation into astrocytes and oligodendrocytes, and formation of tumors when transplanted in vivo (Funato et al. 2014). None of these attributes were observed with H3K27M expression alone, or when the combination of PDGFRa activation and p53 knockdown was not paired with H3K27M, consistent with the observations that these mutations tend to co-occur in patient tumors. Leveraging this model, a small molecule screen was performed, which identified the menin inhibitor MI-2 to selectively decrease the proliferation of transformed H3K27M-expressing NSCs and an H3K27M DIPG cell line, but not H3 WT NSCs (Funato et al. 2014). Treatment with MI-2 also led to the removal of the differentiation block and slowed in vivo growth of orthotopic tumor xenografts, indicating that menin inhibition might be a potential therapeutic option for DIPG patients (Funato et al. 2014).
Other approaches taken to therapeutically target H3K27M-mutant DIPG focused on reversing the reduction of H3K27me3 through manipulating various forms of chromatin-modifying machinery. For example, a small molecule inhibitor of the H3K27 demethylases JMJD3 and UTX known as GSKJ4 was able to increase global levels of H3K27me2/3 in pediatric glioma cell lines expressing H3K27M (Hashizume et al. 2014). GSKJ4 more potently inhibited the growth of these lines both in vitro and in vivo compared with other glioma cell lines that were H3 WT (Hashizume et al. 2014). The efficacy of GSKJ4 may be largely through its effect on JMJD3, as knockdown of JMJD3 but not UTX slowed the growth of H3K27M-expressing cell lines (Hashizume et al. 2014). An alternative method of restoring H3K27me3 levels in cells involves “detoxifying” the H3K27M nucleosomes by weakening their interaction with PRC2 (Brown et al. 2014). PRC2 interacts with the entire H3 N-terminal tail, and its inhibition by peptides containing K27Nle was substantially weakened when residues next to position 27 were deleted or altered (Brown et al. 2014). PTMs, including K4 methylation, polyacetylation, and S28 phosphorylation, all reduced inhibition of PRC2 activity by H3K27Nle peptides in vitro (Brown et al. 2014). Furthermore, the expression of an H3K27M, S28E (mimicking a phosphoserine) double mutant transgene partially rescued the global reduction of H3K27me2/3 in cells (Brown et al. 2014). This strategy has now been taken one step closer to the clinic as H3K27M DIPG cell lines were found to be selectively sensitive to histone deacetylase (HDAC) inhibitors (Grasso et al. 2015). Knockdown of either HDAC1 or HDAC2 showed a similar effect. The clinically approved multi-HDAC inhibitor panobinostat led to a dose-dependent increase in both global H3 acetylation and H3K27me3, likely reflecting diminished inhibition of PRC2 and thereby contributing to a normalization of the H3K27M-induced aberrant gene expression signature (Grasso et al. 2015). Several clinical trials are ongoing to test the safety and efficacy of panobinostat and other HDAC inhibitors in patients with DIPG.
MECHANISM OF H3K36M-MEDIATED ONCOGENESIS
The discovery of H3.3K36M mutations in the vast majority of chondroblastomas and in rare cases of chondrosarcomas (Behjati et al. 2013) prompted the initial description of global reduction in H3K36me3 in 293T cells (Lewis et al. 2013) to be revisited in a more developmentally relevant context. The introduction of an H3K36M transgene into murine mesenchymal progenitor cells blocked their ability to differentiate into chondrocytes and promoted tumor formation when the cells were subcutaneously transplanted into mice (Lu et al. 2016). The tumors did not appear by histology as chondroblastomas or chondrosarcomas but rather as undifferentiated sarcomas and indeed H3K36M was found to also block the differentiation of these cells into adipocytes and osteocytes. Screening a small panel of 10 pediatric undifferentiated soft tissue sarcomas identified one tumor with an H3.1K36M mutation and another with an H3.1K36I mutation. H3K36I similarly blocked the differentiation of murine mesenchymal progenitor cells and its presence in patient tumors mirrors the existence of H3K27I mutations in pediatric gliomas (Castel et al. 2015). Consistent with these findings, CRISPR-Cas9-mediated knock-in of the K36M mutation to the endogenous H3F3B allele in immortalized human chondrocytes bestowed similar oncogenic properties, including impaired differentiation and suppressed alkaloid-induced apoptosis (Fang et al. 2016).
Expression of either H3K36M or H3K36I resulted in global reductions in H3K36me2/3 in murine mesenchymal progenitor cells, an effect that appeared to be independent of whether the mutation occurred in H3.1 or H3.3 (Lu et al. 2016). Interestingly, reductions in H3K36me2/3 levels were positively correlated to enrichment of H3.3K36M, suggesting that in addition to its global effects, H3K36M may also exert a localized inhibitory influence (Fang et al. 2016). Nucleosomes containing H3K36M inhibited purified SETD2 and NSD2 activity in vitro, and knockdown of H3K36 methyltransferases recapitulated the impact of H3K36M on the epigenome, transcriptome, and cellular differentiation (Fang et al. 2016; Lu et al. 2016). These results lend support for a critical role of methyltransferase inhibition downstream of the H3K36M mutation.
Profiling the perturbations in the chromatin landscape induced by expression of H3K36M revealed one mechanism by which differentiation blockade is achieved. H3K36me2 was found to be significantly reduced at intergenic domains, which was associated with an increase in H3K27me3 at the same regions. This is consistent with the known inhibition of PRC2 methyltransferase activity by nucleosomes carrying H3K36me2/3 in vitro and the mutual exclusivity of H3K27me3 and H3K36me2/3 genome-wide (Schmitges et al. 2011; Yuan et al. 2011). The increase in intergenic H3K27me3 served to compete with gene-associated H3K27me3 for the recruitment of H3K27me3 “readers” and led to a dilution of the H3K27me3-binding canonical PRC1 complex away from its target genes where it normally represses gene expression. As a consequence, genes that were normally silenced by PRC1 became derepressed on expression of H3K36M. These genes were highly enriched for mesenchymal tissue development regulators and consistently, knockdown of PRC1 components significantly blocked mesenchymal progenitor cell differentiation (Lu et al. 2016).
As H3K36 methylation has also been implicated in the regulation of transcriptional elongation (Wen et al. 2014), RNA processing (Simon et al. 2014), and the DNA damage response (Carvalho et al. 2014), the H3K36M mutation may rely on additional oncogenic mechanisms besides dilution of PRC1. Indeed, defective homologous recombination was noted in chondrocytes expressing H3K36M (Fang et al. 2016). Further study is needed to address whether dysregulation of these processes contributes to H3K36M-mediated oncogenesis.
MECHANISM OF H3G34R/V-MEDIATED ONCOGENESIS
Less is known about H3G34 mutations in cancer, but they appear to work through a different mechanism than H3 “K-to-M” mutations. Expression of H3G34R or H3G34V in 293T cells both had no effect on global H3K27me2/3 or H3K36me3 levels (Lewis et al. 2013). Instead, H3K36me2/3 was exclusively reduced on exogenous H3G34-mutant nucleosomes but not on endogenous WT nucleosomes (Lewis et al. 2013). Consistent with this finding, recombinant SETD2 was less efficient at methylating H3K36 using H3G34-mutant nucleosomes in vitro (Lewis et al. 2013). Thus, H3G34 mutations appear to exert their effect in cis, in contrast to H3K27M and H3K36M mutations, which are able to inhibit SET-domain methyltransferases in trans (Fig. 3C). Of note, whereas H3K27M and H3K36M mutations have been reported in H3.1 and H3.3, H3G34 mutations have only been identified in H3.3, further suggesting that they may result in local chromatin changes in genomic regions enriched for H3.3. How H3G34 mutations contribute to oncogenesis remains largely unexplored, although an initial report points to MYCN as being the most highly differentially expressed gene in a H3G34-mutant glioma cell line (Bjerke et al. 2013). Knockdown of MYCN decreased viability of H3G34-mutant cells and expression of H3G34-mutant transgenes in astrocyte and glial cells was sufficient to upregulate MYCN (Bjerke et al. 2013). Further work is needed to assess how H3G34 mutations act to regulate MYCN expression as well as other genes associated with early brain development (Bjerke et al. 2013).
CONCLUDING REMARKS
The application of next-generation sequencing to an increasing number and array of human cancers is revealing mutations where no one expected—perhaps most surprisingly in histones, the fundamental building blocks of chromatin. Strikingly, these mutations occur at or near residues on the amino-terminal tail of histone H3 that have well-characterized writers, readers, and erasers. As more types of cancer are sequenced and as appreciation for the dominant nature of these “oncohistone” mutations grows, it will be interesting to determine if new histone mutations, “K-to-M” or otherwise, are identified. One common feature of histone mutations is their ability to arrest cells in a primitive state refractory to differentiation induction. This is in line with the exquisite tissue and lineage specificity of these mutations and highlights the importance of modeling and studying oncohistones in the appropriate developmental context.
Despite rapid progress in elucidating the molecular mechanism of H3K27M, H3K36M, and H3G34R/V mutations, many questions remain. Although the effects of H3K27M and H3K36M mutations seemed to be independent of the H3 isoforms in cell culture models, patient tumors carrying H3.1/2 “K-to-M” mutations show clear pathological and clinical distinctions from H3.3 “K-to-M” mutant tumors. Further research is warranted to resolve this discrepancy. In addition, more work is needed to understand the persistence of H3K27me3 enrichment at limited genomic loci in H3K27M-mutant gliomas and whether or how it may contribute to oncogenesis. This will likely require a renewed focus on changes to the chromatin landscape beyond genic regions to include intergenic loci. Why specific oncogenes and tumor suppressors co-occur with different oncohistones, such as H3K27M with specific oncogenic signaling pathways or H3G34R/V with ATRX mutations, is also unknown. At another level, the remarkable anatomical and age specificity of oncohistone-associated cancers begs for studies aimed at determining underlying cells-of-origin and developmental timing issues. If our current knowledge of histone mutations is any indication, answering these questions and making further progress in developing new therapies for patients will require integrating our understanding of chromatin structure and biochemistry with developmental pathways and the hallmarks of cancer.
ACKNOWLEDGMENTS
We apologize to colleagues whose work could not be cited because of space limitations. We thank Alexey Soshnev for help in the preparation of the figures. C.D.A. is supported by funding from The Rockefeller University and the National Institutes of Health (NIH) Grant P01CA196539. C.L. is the Kandarian Family Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2195-14). D.N.W. is supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the NIH under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan Kettering Tri-Institutional MD-PhD Program.
Footnotes
Editors: Scott A. Armstrong, Steven Henikoff, and Christopher R. Vakoc
Additional Perspectives on Chromatin Deregulation in Cancer available at www.perspectivesinmedicine.org
REFERENCES
- Aihara K, Mukasa A, Gotoh K, Saito K, Nagae G, Tsuji S, Tatsuno K, Yamamoto S, Takayanagi S, Narita Y, et al. 2014. H3F3A K27M mutations in thalamic gliomas from young adult patients. Neuro Oncol 16: 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amary MF, Berisha F, Mozela R, Gibbons R, Guttridge A, O’Donnell P, Baumhoer D, Tirabosco R, Flanagan AM. 2016. The H3F3 K36M mutant antibody is a sensitive and specific marker for the diagnosis of chondroblastoma. Histopathology 69: 121–127. [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Allis CD, Lewis PW. 2010. Histone variants in metazoan development. Dev Cell 19: 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N, Elsässer SJ, Chapgier A, Goldberg AD, Canaani E, et al. 2013. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES Cells. Cell 155: 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechet D, Gielen G, Korshunov A, Pfister SM, Rousso C, Faury D, Fiset PO, Benlimane N, Lewis PW, Lu C, et al. 2014. Specific detection of methionine 27 mutation in histone 3 variants (H3K27M) in fixed tissue from high-grade astrocytomas. Acta Neuropathol 128: 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behjati S, Tarpey PS, Presneau N, Scheipl S, Pillay N, Van Loo P, Wedge DC, Cooke SL, Gundem G, Davies H, et al. 2013. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet 45: 1479–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT, Kool M, Zapatka M, Northcott PA, Sturm D, Wang W, et al. 2013. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 24: 660–672. [DOI] [PubMed] [Google Scholar]
- Bjerke L, Mackay A, Nandhabalan M, Burford A, Jury A, Popov S, Bax DA, Carvalho D, Taylor KR, Vinci M, et al. 2013. Histone H3.3 mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov 3: 512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown ZZ, Müller MM, Jain SU, Allis CD, Lewis PW, Muir TW. 2014. Strategy for “fetoxification” of a cancer-derived histone mutant based on mapping its interaction with the methyltransferase PRC2. J Am Chem Soc 136: 13498–13501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown ZZ, Müller MM, Kong HE, Lewis PW, Muir TW. 2015. Targeted histone peptides: Insights into the spatial regulation of the methyltransferase PRC2 by using a surrogate of heterotypic chromatin. Angew Chem Int Ed Engl 54: 6457–6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buczkowicz P, Hoeman C, Rakopoulos P, Pajovic S, Letourneau L, Dzamba M, Morrison A, Lewis P, Bouffet E, Bartels U, et al. 2014. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet 46: 451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush KM, Yuen BT, Barrilleaux BL, Riggs JW, O’Geen H, Cotterman RF, Knoepfler PS. 2013. Endogenous mammalian histone H3.3 exhibits chromatin-related functions during development. Epigenet Chrom 6: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S, Vítor AC, Sridhara SC, Filipa BM, Ana CR, Desterro JMP, Ferreira J, de Almeida SF. 2014. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. eLife 2014: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel D, Philippe C, Calmon R, Le Dret L, Truffaux N, Boddaert N, Pagès M, Taylor KR, Saulnier P, Lacroix L, et al. 2015. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol 130: 815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaikuad A, Alfano I, Kerr G, Sanvitale CE, Boergermann JH, Triffitt JT, Von Delft F, Knapp S, Knaus P, Bullock AN. 2012. Structure of the bone morphogenetic protein receptor ALK2 and implications for fibrodysplasia ossificans progressiva. J Biol Chem 287: 36990–36998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, et al. 2013. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 27: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couldrey C, Carlton MB, Nolan PM, Colledge WH, Evans MJ. 1999. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum Mol Genet 8: 2489–2495. [DOI] [PubMed] [Google Scholar]
- Elsässer SJ, Huang H, Lewis PW, Chin JW, Allis CD, Patel DJ. 2012. DAXX envelops a histone H3.3–H4 dimer for H3.3-specific recognition. Nature 491: 560–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsässer SJ, Noh KM, Diaz N, Allis CD, Banaszynski LA. 2015. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522: 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kellis M. 2010. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol 28: 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D, Gan H, Lee JH, Han J, Wang Z, Riester SM, Jin L, Chen J, Zhou H, Wang J, et al. 2016. The histone H3.3K36M mutation reprograms the epigenome of chondroblastomas. Science 352: 1344–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Hao S, Pan C, Wang Y, Wu Z, Zhang J, Yan H, Zhang L, Wan H. 2015. The H3.3 K27M mutation results in a poorer prognosis in brainstem gliomas than thalamic gliomas in adults. Hum Pathol 46: 1626–1632. [DOI] [PubMed] [Google Scholar]
- Fontebasso AM, Schwartzentruber J, Khuong-Quang DA, Liu XY, Sturm D, Korshunov A, Jones DT, Witt H, Kool M, Albrecht S, et al. 2013. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol 125: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontebasso AM, Gayden T, Nikbakht H, Neirinck M, Papillon-Cavanagh S, Majewski J, Jabado N. 2014a. Epigenetic dysregulation: A novel pathway of oncogenesis in pediatric brain tumors. Acta Neuropathol 128: 615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, Nikbakht H, Gerges N, Fiset PO, Bechet D, Faury D, De Jay N, Ramkissoon LA, et al. 2014b. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet 46: 462–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funato K, Major T, Lewis PW, Allis CD, Tabar V. 2014. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science 346: 1529–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsässer SJ, Stadler S, Dewell S, Law M, Guo X, Li X, et al. 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140: 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily MA, Quist MJ, Davis LE, Huang EC, Woo PJ, et al. 2015. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med 21: 555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake SB, Allis CD. 2006. Histone H3 variants and their potential role in indexing mammalian genomes: The “H3 barcode hypothesis.” Proc Natl Acad Sci 103: 6428–6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume R, Andor N, Ihara Y, Lerner R, Gan H, Chen X, Fang D, Huang X, Tom MW, Ngo V, et al. 2014. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med 20: 1394–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Morgan M, Gao X, Jackson J, Rickels R, Swanson SK, Florens L, Washburn MP, Eissenberg JC, Shilatifard A. 2014. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science 345: 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Sabari BR, Garcia BA, Allis CD, Zhao Y. 2014. SnapShot: Histone modifications. Cell 159: 458–458.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaram H, Hoelper D, Jain SU, Cantone N, Lundgren SM, Poy F, Allis CD, Cummings R, Bellon S, Lewis PW. 2016. S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by lysine 9 to methionine mutation on histone H3. Proc Natl Acad Sci 113: 6182–6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jiao L, Liu X. 2015. Structural basis of histone H3K27 trimethylation by an active Polycomb repressive complex 2. Science 350: 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Hutter B, Jäger N, Korshunov A, Kool M, Warnatz HJ, Zichner T, Lambert SR, Ryzhova M, Quang DA, et al. 2013. Recurrent somatic alterations of FGFR1 and NTRK2 in pilocytic astrocytoma. Nat Genet 45: 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph CG, Hwang H, Jiao Y, Wood LD, Kinde I, Wu J, Mandahl N, Luo J, Hruban RH, Diaz LA Jr, et al. 2014. Exomic analysis of myxoid liposarcomas, synovial sarcomas, and osteosarcomas. Genes Chromosomes Cancer 53: 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justin N, Zhang Y, Tarricone C, Martin SR, Chen S, Underwood E, De Marco V, Haire LF, Walker PA, Reinberg D, et al. 2016. Structural basis of oncogenic histone H3K27M inhibition of human Polycomb repressive complex 2. Nat Commun 7: 11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallappagoudar S, Yadav RK, Lowe BR, Partridge JF. 2015. Histone H3 mutations—A special role for H3.3 in tumorigenesis? Chromosoma 124: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A. 2009. A novel mammalian flavin-dependent histone demethylase. J Biol Chem 284: 17775–17782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, Bartels U, Albrecht S, Schwartzentruber J, Letourneau L, et al. 2012. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol 124: 439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J, Xi Y, Park BH, Shi X, Garcia BA, et al. 2011. NSD2 links fimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell 44: 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Allis CD. 2013. Poisoning the “histone code” in pediatric gliomagenesis. Cell Cycle 12: 3241–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Elsässer SJ, Noh KM, Stadler SC, Allis CD. 2010. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci 107: 14075–14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD. 2013. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340: 857–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, et al. 2012. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci 109: 3879–3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Jain SU, Hoelper D, Bechet D, Molden R, Ran L, Murphy D, Venneti S, Hameed M, Pawel B, et al. 2016. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science 352: 844–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Noh KM, Soshnev AA, Allis CD. 2014. Every amino acid matters: Essential contributions of histone variants to mammalian development and disease. Nat Rev Genet 15: 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick E, Gafken PR, Ahmad K, Henikoff S. 2004. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci 101: 1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikbakht H, Panditharatna E, Mikael LG, Li R, Gayden T, Osmond M, Ho CY, Kambhampati M, Hwang EI, Faury D, et al. 2016. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nat Commun 7: 11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko V, Schiltz RL, Russanova V, Howard BH, Nakatani Y. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87: 953–959. [DOI] [PubMed] [Google Scholar]
- Okosun J, Bödör C, Wang J, Araf S, Yang CY, Pan C, Boller S, Cittaro D, Bozek M, Iqbal S, et al. 2014. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet 46: 176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarungbam J, Agaram N, Hwang S, Lu C, Wang L, Healey J, Hameed M. 2016. Symplastic/pseudoanaplastic giant cell tumor of the bone. Skeletal Radiol 10.1007/s00256-016-2373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341. [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tönjes M, et al. 2012. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482: 226–231. [DOI] [PubMed] [Google Scholar]
- Shen H, Laird PW. 2013. Interplay between the cancer genome and epigenome. Cell 153: 38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore EM, Xu M, Feldman GJ, Fenstermacher DA, Cho TJ, Choi IH, Connor JM, Delai P, Glaser DL, LeMerrer M, et al. 2006. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat Genet 38: 525–527. [DOI] [PubMed] [Google Scholar]
- Simon JM, Hacker KE, Singh D, Brannon AR, Parker JS, Weiser M, Ho TH, Kuan PF, Jonasch E, Furey TS, et al. 2014. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res 24: 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C, Pfaff E, Tönjes M, Sill M, Bender S, et al. 2012. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22: 425–437. [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116: 51–61. [DOI] [PubMed] [Google Scholar]
- Taylor KR, Mackay A, Truffaux N, Butterfield YS, Morozova O, Philippe C, Castel D, Grasso CS, Vinci M, Carvalho D, et al. 2014. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet 46: 457–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo RA, Qin Y, Cheng ZM, Gao Q, Iwata S, Silva GM, Prasad ML, Ocal IT, Rao S, Aronin N, et al. 2015. Recurrent mutations of chromatin-remodeling genes and kinase receptors in pheochromocytomas and paragangliomas. Clin Cancer Res 22: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, Judkins AR. 2013. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol 23: 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti S, Santi M, Felicella MM, Yarilin D, Phillips JJ, Sullivan LM, Martinez D, Perry A, Lewis PW, Thompson CB, et al. 2014. A sensitive and specific histopathologic prognostic marker for H3F3A K27M mutant pediatric glioblastomas. Acta Neuropathol 128: 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. 2012. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 13: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Li Y, Xi Y, Jiang S, Stratton S, Peng D, Tanaka K, Ren Y, Xia Z, Wu J, et al. 2014. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 508: 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, et al. 2012. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44: 251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, Zhu X, Qu C, Chen X, Zhang J, et al. 2014. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 46: 444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. 2011. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 286: 7983–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B, Orisme W, Punchihewa C, Parker M, Qaddoumi I, et al. 2013. Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet 45: 602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]