Abstract
Chromosomal aberrations during cell division represent one of the first recognized features of human cancer cells, and modern detection methods have revealed the pervasiveness of aneuploidy in cancer. The ongoing karyotypic changes brought about by chromosomal instability (CIN) contribute to tumor heterogeneity, drug resistance, and treatment failure. Whole-chromosome and segmental aneuploidies resulting from CIN have been proposed to allow “macroevolutionary” leaps that may contribute to profound phenotypic change. In this review, we will outline evidence indicating that aneuploidy and CIN contribute to cancer evolution.
Chromosome segregation errors during cell division are some of the first recognized features of cancer cells. The resulting karyotypic changes contribute to tumor heterogeneity, drug resistance, and treatment failure.
Intratumor heterogeneity is increasingly recognized as a ubiquitous feature across human tumors. Contributing to this heterogeneity are an increased mutation rate because of various defects in DNA damage detection and repair (genomic instability or GIN), along with gross chromosomal rearrangements and deviations from the euploid chromosome number (aneuploidy). Cancer cells derived from solid tumors almost invariably display a high rate of chromosome segregation errors, called chromosomal instability (CIN), which leads to aneuploidy. This feature was recognized early on in human tumors (see Boveri 2008, for an English translation), but its importance has been obscured by the complexity of the cancer phenotype. Multiple mechanisms have been described to account for the high rate of whole-chromosome segregation errors (wCIN) and structural aberrations (sCIN) in cancer and have been comprehensively discussed elsewhere (Lengauer et al. 1998; Ganem et al. 2007; Holland and Cleveland 2009; Burrell et al. 2013b; Duijf and Benezra 2013). Here, we will briefly summarize some of the key defects leading to CIN, emphasize the importance of phenotypes allowing the expansion of highly aneuploid cells, and discuss its impact on tumor progression and treatment.
CAUSES OF ANEUPLOIDY
Mitotic Checkpoint Defects
The spindle assembly checkpoint (SAC) or mitotic checkpoint monitors kinetochore attachment to the mitotic spindle and delays sister chromatid separation until biorientation is achieved at metaphase (Fig. 1) (Musacchio and Salmon 2007). Heterozygous mice for SAC genes, such as BUB1, BUBR1 (BUB1B), BUB3, MAD1, MAD2, and CENPE, all display aneuploidy to various degrees (reviewed in Giam and Rancati 2015; Simon et al. 2015). SAC impairment has been linked to increased aneuploidy and in many mouse models associated with increased tumor development (either spontaneous, following drug-induced tumorigenesis or when crossed into a sensitized background). Biallelic mutations in BUB1B are linked to mosaic variegated aneuploidy (MVA) syndrome (Hanks et al. 2004). Nevertheless, it is under debate whether SAC defects represent a common CIN driver because SAC gene mutations are rarely found in cancer (discussed in the section Chromosome Cohesion Defects). In fact, some SAC proteins are frequently overexpressed in cancer, such as Mad2 (MAD2L1), which is an E2F transcriptional target aberrantly expressed in Rb mutant cells (Hernando et al. 2004). MAD2 transgenic mice develop aneuploid tumors and it was therefore proposed that SAC hyperactivation was causing CIN in this model (Sotillo et al. 2007; Kato et al. 2011). However, tetraploidization and hyperstabilization of kinetochore–microtubule (k-MT) attachments are observed upon Mad2 overexpression, which could explain how it drives CIN (discussed in the next two sections) (Hernando et al. 2004; Sotillo et al. 2007; Kabeche and Compton 2012). Another SAC protein, namely, Mad1 (MAD1L1), is overexpressed in cancer but in this case produces a dominant-negative effect and impairs SAC signaling (Ryan et al. 2012). Many SAC proteins also have roles outside mitosis that may also contribute to tumorigenesis.
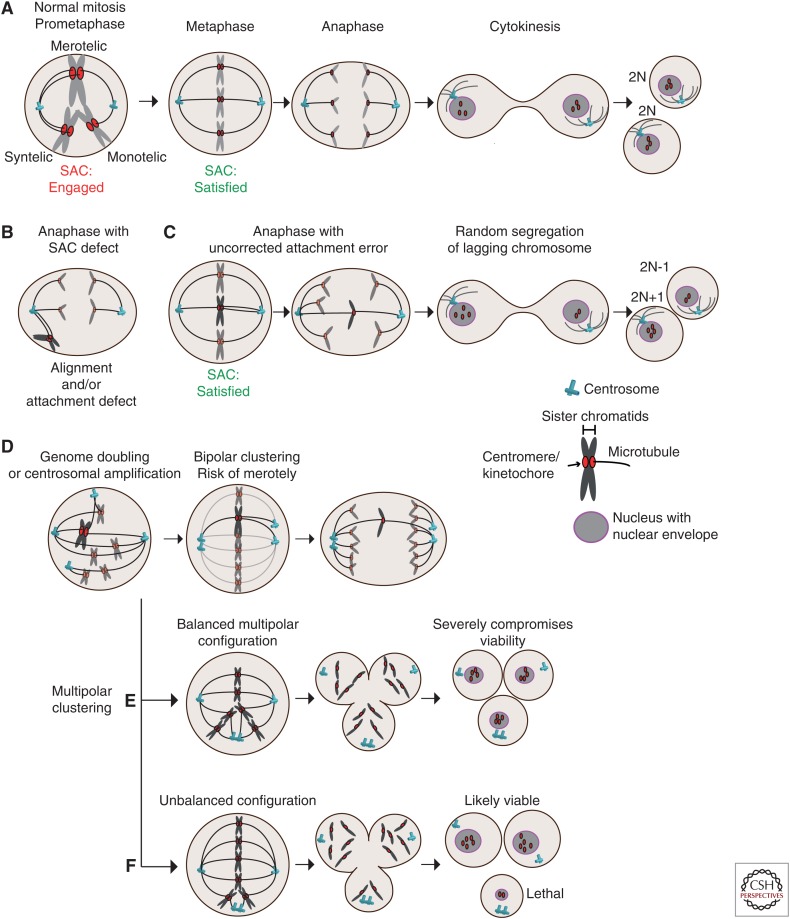
Figure 1.
Mitotic defects leading to aneuploidy. (A) Kinetochore–microtubule attachment configurations and stages of mitosis. In early mitosis, improper attachments (syntelic and monotelic) activate the spindle assembly checkpoint (SAC) to delay anaphase onset. Merotelic misattachments poorly activate the checkpoint, but error correction mechanisms contribute to the formation of correct (amphiletic) bipolar attachments prior to chromosome separation at anaphase. (B) Cells with compromised SAC activity can enter anaphase with various attachment defects, here shown with syntelic chromosomes that did not congress to the metaphase plate. (C) Uncorrected merotelic attachments lead to lagging chromosomes at anaphase onset, causing random segregation, which can result in aneuploid daughter cells. (D) Cells with centrosome amplification and tetraploid cells enter mitosis with more than two centrosomes, creating a transient multipolar spindle. Bipolar clustering creates merotelic attachments because of the random clustering of the extra centrosomes. (E) Extra centrosome can result in a multipolar anaphase creating daughter cells lacking several chromosomes in a random fashion. (F) Random clustering can create a functional spindle pole that attaches only to few chromosomes, potentially resulting in viable aneuploidy in two out of three cells. Examples in D and E are especially well tolerated by tetraploid cells.
Microtubule Attachment Defects
In mitosis, spindle microtubules capture chromosomes at the kinetochore, a complex protein structure built around the centromeric region. Correct k-MT attachment is crucial for faithful chromosome segregation, and kinetochores from sister chromatids must be attached to opposite poles (Fig. 1). Aurora kinase B (AURKB) activity at the inner centromere (along with the microtubule depolymerizing protein KIF2B and KIF2C [MCAK]) plays a central role in the establishment of correct (amphitelic) attachments (Fig. 1A,B). This pathway functions from prometaphase to metaphase and destabilize syntelic attachments (in which both sister kinetochores are attached to the same pole) and merotelic attachments (in which one of the kinetochores is attached to both pole) (Fig. 1A). Merotelic attachments in particular are thought to constitute a major cause of CIN because this configuration satisfies the requirements of the SAC (MT attachment and interkinetochore tension), resulting in lagging chromosomes at anaphase (Fig. 1C). Cyclin A was also shown to destabilize k-MT and promote error correction during mitotic progression, giving rise to a model whereby cyclin A degradation as cells advance from prometaphase to metaphase gradually stabilizes k-MT attachments (Kabeche and Compton 2013; Godek et al. 2015). The rate of cyclin A degradation during prometaphase might thus establish a window during which erroneous attachments can be corrected, and variations in mitotic duration could potentially compromise error correction. Impaired k-MT dynamics (stabilization) appears to contribute to CIN following deregulation of several mitotic proteins, and was also proposed to underpin CIN in adenomatous polyposis coli (APC) mutant cells (reviewed in Godek et al. 2015). Enhanced microtubule assembly rates is also a feature shared by CIN colorectal cancer cell lines and also results in microtubule hyperstabilization and chromosome segregation errors (Ertych et al. 2014). Reducing microtubule polymerization rates using a low dose of taxol or by RNAi against the microtubule stabilizer Ch-TOG reduced classical features of CIN such as lagging chromosomes and karyotypic variation. Proper activity of the CHK2-BRCA1 pathways seems important to limit AURKA activity in controlling MT assembly rates (Ertych et al. 2014, 2016).
Centrosome and Mitotic Spindle Defects
Centrosome amplification is a widespread feature of human tumors associated with CIN and is thought to contribute to tumorigenesis through several mechanisms (Pihan et al. 1998; Ghadimi et al. 2000; Nigg and Stearns 2011; Godinho et al. 2014). Several defects result in supernumerary centrosomes, either directly through centriole overduplication (Nigg and Stearns 2011) or indirectly following an aborted mitosis resulting in genome doubling (tetraploidization). Extra centrosomes randomly attach to chromosomes early in mitosis, creating a transient multipolar spindle, and tend to cluster into two poles before division (Fig. 1D). Random clustering consequently increases the probability of creating merotelic attachments, and lagging chromosomes are frequent in cells with supernumerary centrosomes (Kwon et al. 2008; Ganem et al. 2009). Tetraploid cells resulting from cytokinesis failure display CIN and spontaneously generate tumors when injected in mice, whereas tetraploid cells that have lost their extra centrosomes are karyotipically stable (Fujiwara et al. 2005; Ganem et al. 2009). Genome-doubling events are also common in cancer and appear to be a precursor of CIN (Zack et al. 2013; Dewhurst et al. 2014).
Failure to cluster extra centrosomes into two poles results in multipolar divisions, likely to generate severe karyotypic changes in daughter cells. Balanced multipolar divisions are therefore lethal and very unlikely to efficiently propagate CIN, whereas an unbalanced multipolar division cell could result in the missegregation or loss of few chromosomes (Fig. 1E,F) (Ganem et al. 2007, 2009; Kwon et al. 2008; Gisselsson et al. 2010). Knockdown of the kinesin KIFC1 (HSET) by siRNA prevents clustering and results in multipolar divisions and lethality specifically in cells with multiple centrosomes. HSET small molecule inhibitors therefore represent an attractive avenue to target cancer cells (Kwon et al. 2008; Raab et al. 2012; Watts et al. 2013; Johannes et al. 2015). Interestingly, multipolarity can also occur without centrosome amplification. For example, defects in mitotic spindle maintenance can lead to an aberrant yet functional spindle pole (Logarinho et al. 2012).
Chromosome Cohesion Defects
The cohesin protein complex maintains centromeric cohesion between sister chromatids until anaphase onset, and unscheduled loss of cohesion was shown to cause CIN (Jallepalli et al. 2001; Barber et al. 2008). Truncating mutations in the cohesin complex subunit STAG2 were identified in cancer cell lines and tumor samples and account for ∼39% of STAG2 mutations reported in The Cancer Genome Atlas (TCGA) (Fig. 2) (Solomon et al. 2011). Introduction of a STAG2 inactivating mutations in a karyotypically stable cell line was sufficient to induce CIN, whereas correction of naturally occurring mutations reduced CIN in glioblastoma cell lines (Solomon et al. 2011). Because STAG2 is located on the X chromosome, complete gene inactivation can be achieved through a single mutation without the need for loss of heterozygosity (LOH). Although most but not all STAG2 truncating mutations lead to CIN and aneuploidy, the consequence of missense mutations causes various phenotypes, and the impact of cohesion complex mutations in cancer might also involve their general role of cohesion in transcription (Taylor et al. 2014; Kim et al. 2016).
Figure 2.
Mutations identified in genes important for chromosome segregation fidelity. Lollipop plots depicting the location and nature of mutations identified in 6807 tumors (32 cancer types) sequenced by The Cancer Genome Atlas (TCGA). Pathway analyses might shed new light on the frequency of chromosomal instability (CIN)-causing defects and the underlying mutations driving CIN, which will require a deeper understanding of the consequence of missense mutations. Many mutations occur in functional domains: for example, 33/67 (49%) of missense mutations in TTK (Mps1) occur in the kinase domain. Green dot, missense mutation; red dot, truncating mutation (nonsense, splice site, and frameshifts); purple dot, various mutation types. (Data and diagrams modified from cBioportal [Cerami et al. 2012; Gao et al. 2013].)
Centromeric cohesion maintenance during mitosis also requires the action of Shugoshin 1 (SGO1) at the inner centromere (Watanabe 2005; Marston 2015). Recently, it was shown that SGO1 maintenance at the inner centromere was impaired in CIN cells when centromeres come under tension at metaphase. Interestingly, SGO1 recruitment at the centromere required both cohesin and the histone mark H3K9me3, and several CIN cell lines were found to be defective in one or both pathways (Tanno et al. 2015). Owing to its interaction with AURKB-containing chromosomal passenger complex, SGO1 mislocalization impairs error correction. This supports the notion that cohesion defects might cause CIN through stabilization of k-MT attachment errors (Kleyman et al. 2014; Tanno et al. 2015).
CIN-Causing Mutations
The general view when considering the underlying genetic causes of wCIN is that direct mutations in any single gene responsible for segregation fidelity are rare. This conclusion is based on studies in which the mutational status of CIN genes were analyzed independently in relatively small cohorts. This finding is perhaps expected for such a complex process in which mild disruptions in the activity of most proteins involved could elevate chromosome missegregation rates. Pan-cancer analyses based on whole-genome or whole-exome sequencing data will enable the identification of low-frequency drivers, and allow pathway-driven rather than gene-centric analyses. This might reveal that tumors bear a mutation in a CIN-causing gene more frequent than previously anticipated (Leiserson et al. 2015). In Figure 2, we summarize the mutational data for some genes involved in error correction/cohesion and the SAC collected from 6807 tumors of diverse origins sequenced by TCGA (Cerami et al. 2012; Gao et al. 2013). As discussed further below (the section on Determinants of CIN Propagation), mildly disrupting mutations may be more potent CIN drivers than deleterious mutations, by causing low levels of CIN more likely to be viable and tolerated. A deeper understanding of the impact of missense mutations in candidate CIN drivers is also required, and genome-editing techniques will now enable us to explore the functional impact of these mutations in unprecedented detail. Epigenetic silencing may also emerge as a frequent CIN driver, as has been reported for BUB1B (Park et al. 2007; Haruta et al. 2008; Landau et al. 2014).
Mutations in common oncogenes and tumor suppressors are also associated with CIN. For example, deregulation of the RB-E2F and MYC pathways, or oncogenic Ras signaling has been reported to cause to aberrant expression of SAC genes, perturb microtubule dynamics, and cause centrosome amplification and cohesion defects (Hernando et al. 2004; Adon et al. 2010; Zeng et al. 2010; Kamata and Pritchard 2011; Schvartzman et al. 2011; Orr and Compton 2013; Manning et al. 2014; reviewed in Duijf and Benezra 2013; Orr and Compton 2013). Mutations in numerous tumor-suppressor genes have been shown to cause CIN, such as APC and WNT signaling pathway components (CHK1 and CHK2-BRCA1) (Zachos et al. 2007; Stolz et al. 2010; Orr and Compton 2013). The convergence of these various pathways on processes directly controlling segregation fidelity lead to the concept of oncogene-induced mitotic stress (Malumbres 2011; Duijf and Benezra 2013).
INTERPLAY BETWEEN GENOMIC INSTABILITY AND CHROMOSOMAL INSTABILITY
Cancer cells almost invariably display a combination of numerical (aneuploidy) and structural aberrations (translocations, deletions), as well as defects affecting accurate replication or repair of damaged DNA (GIN). There is now accumulating evidence describing how the various types of instabilities can impact on each other, explaining at least in part how any of these triggers can pave the way for cancer cells to acquire such complex genetic compositions.
Genomic Instability Fuels CIN
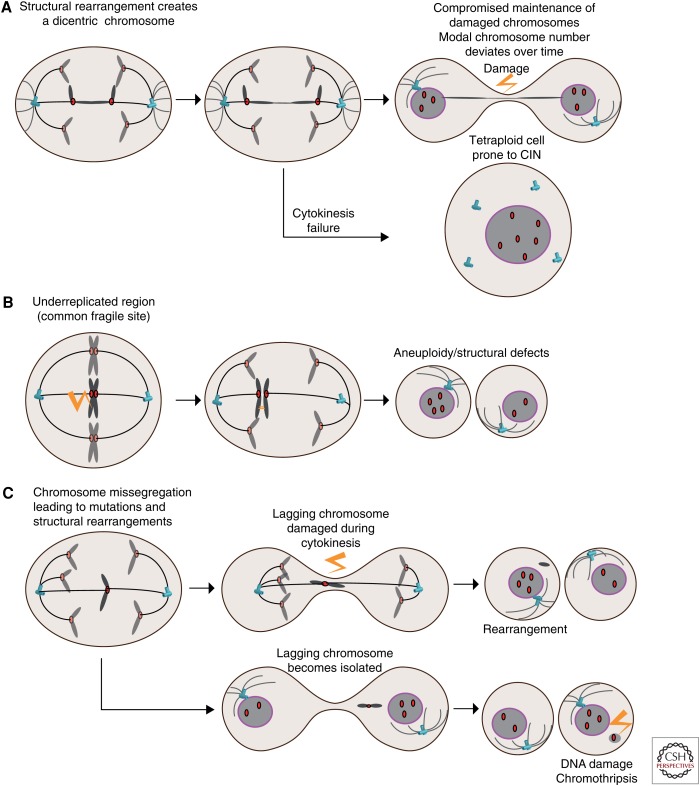
Premitotic defects arising from DNA replication fork collapse (replication stress) can result in double strand breaks (DSBs) especially at common fragile sites, which are late-replicating loci in the genome. Replication stress leads to rearrangement and chromosomes breakage during the repair process especially when coupled to DNA repair defects (Halazonetis et al. 2008). As a result, replication-stress-induced defects can result in acentric chromosome fragments that will not be captured by the mitotic spindle or in dicentric chromosomes leading to chromosome bridges at anaphase (Fig. 3A) (Gisselsson 2008). These structural defects constitute a large fraction of mitotic aberrations during anaphase found in CIN colorectal cancer cells, and alleviation of replication stress using nucleoside complementation reduced their frequency indicative of their de novo formation during each phase of DNA replication (Burrell et al. 2013a). Transient induction of replication stress with aphidicolin also caused a deviation in chromosome number, a defining feature of CIN, when analyzed by clonal fluorescence in situ hybridization (FISH) using centromeric probes (Burrell et al. 2013a). Remarkably, it was recently shown that DNA replication occurs at common fragile sites during mitosis, presumably in a last attempt to complete DNA replication before cell division, and failure to do so causes chromosome missegregation as revealed by centromeric FISH (Minocherhomji et al. 2015). Replication stress can therefore lead to the missegregation of an entire, yet probably damaged, chromosome (Fig. 3B). Replication stress in precancerous lesions might therefore allow the onset of CIN in some tumors (Gorgoulis et al. 2005).
Figure 3.
The interplay between whole-chromosome instability (wCIN), structural CIN (sCIN), and genomic instability (GIN). (A) Structural rearrangements caused by replication stress before mitosis created dicentric chromosomes. These can result in ultrafine bridges devoid of nuclear membrane and subject to extensive DNA damage and rearrangements akin to chromothripsis or hypermutation through kateagis and severing of the chromosome by exonucleases. Dicentric chromosomes can also prevent cytokinesis, as observed following telomere dysfunction. (B) Replication stress at common fragile sites triggers DNA replication during mitosis. Failing to complete DNA replication can lead to nondysjunction and aneuploidy. (C) Whole-chromosome missegregation can result in the trapping of the lagging chromosome during cytokinesis, causing DNA damage and structural aberrations. Alternatively, lagging chromosomes can become isolated in a micronucleus, in which the lack of nuclear membrane renders it susceptible to extensive DNA damage and chromothripsis. Damaged micronuclei can reintegrate the other chromosomes in the following division and be propagated.
Apart from common fragile sites, telomeres are also intrinsically sensitive to replication stress. Telomere maintenance is crucial for ongoing cell proliferation, and failure to do so results in telomere attrition and eventually activation of a DNA damage response, which halts cell proliferation (telomere crisis). Evidence of transient telomere crisis during early stages of breast cancer, followed by stabilization of shorter telomeres through telomerase reactivation, was shown for several cancers including prostate (Meeker et al. 2002), colon (Rudolph et al. 2001), and pancreas (Chin et al. 2004; Feldmann et al. 2007). Interestingly, telomere dysfunction can result in genome-doubling through cytokinesis failure or rereplication (endoreduplication) (Fig. 3A). The resulting tetraploid cells displayed hallmarks of CIN and were tumorigenic (Davoli et al. 2010; Davoli and de Lange 2012). Dysfunctional telomeres also cause chromosomal rearrangements, including dicentric chromosomes, which result in chromosome bridges during division (Fig. 3A). Dicentric chromosomes were shown to be prone to extensive localized mutagenesis (kateagis, bearing a mutation signature characteristic of the APOBEC family of cytidine deaminase enzymes) and chromothripsis (the random rearrangement of an entire chromosome or chromosome segment) (Stephens et al. 2011; Leibowitz et al. 2015; Maciejowski et al. 2015). Chromosome bridges were found to be coated with RPA proteins, indicative of single-stranded (ssDNA) exposure (the substrate for APOBEC enzymes), and the bridge actively processed by an exonuclease (Maciejowski et al. 2015). This is consistent with the detection of DNA damage markers on chromosome bridges that can form following SAC dysfunction (Janssen et al. 2011b). Segregation error defects, by physically isolating chromosomes or part thereof, can thus provide an explanation for the observation of mutational clusters, and for segmental structural rearrangements.
Chromosome Missegregation and Genomic Instability
A large body of evidence suggests that whole-chromosome missegregation and aneuploidy can in turn drive further GIN. Studies in yeast revealed aneuploid strains are prone to further segregation errors, display a mutator phenotype, and acquire chromosomal rearrangements (Pavelka et al. 2010; Sheltzer et al. 2011). A single chromosome missegregation event can thus trigger hallmarks of full-blown GIN. Similarly, engineered human cell lines bearing an extra chromosome acquire segmental aneuploidy and rearrangements, an event most likely driven by replication stress (Passerini et al. 2016). In diploid cells, lagging chromosomes caused by SAC inhibition or microtubule attachment defects can also get trapped and damaged during cytokinesis, resulting in translocations (Janssen et al. 2011b). Whole-chromosome missegregation (wCIN) and aneuploidy can therefore lead to structural chromosomal aberrations (Fig. 3C). Consequently, CIN allows cells to rapidly explore various compositions of whole-chromosome and segmental aneuploidies, and the combination of oncogenes and tumor-suppressor genes on each chromosomal fragment most likely drives karyotypic selection (Davoli et al. 2013).
Chromosome missegregation can result in the physical isolation of one or few chromosomes into micronulei, in which they are prone to extensive DNA damage because of severe nuclear envelope defects (Fig. 3C) (Crasta et al. 2012; Hatch et al. 2013). DNA from micronuclei can be reincorporated into the main chromosome mass and transmitted during the following division (Crasta et al. 2012). This provides a mechanism whereby a single chromosome segregation error results in the acquisition of mutations without prior defects in genome maintenance, and can therefore be a precursor of GIN. By combining live-cell imaging and single-cell sequencing, Pellman and colleagues showed that chromothripsis can occur in micronuclei, providing a simple mechanism to massively rearrange entire chromosomes while leaving the others intact (Zhang et al. 2015). Interestingly, chromothripsis can also explain the formation of double-minute chromosomes, short circular acentric DNA fragments that are often amplified and contain oncogenes. Pan-cancer studies have detected evidence of chromothripsis in 2%–5% of samples, with higher prevalence in some cancer types such as glioblastoma and bone cancers (Stephens et al. 2011; Zack et al. 2013; Rode et al. 2016). CIN thus creates at least two situations, namely, chromosome bridges (including ultra-fine bridges) (Liu et al. 2014) and micronuclei, in which loss of nuclear envelope integrity renders chromosomes prone to GIN (Hatch et al. 2013).
DETERMINANTS OF CIN PROPAGATION
Sustainable versus Lethal Levels of CIN
CIN is generally linked to poor outcomes in cancer (McGranahan et al. 2012). However, there is mounting evidence that the level of CIN determines its impact on tumor progression, in which a trade off at each division must be reached between the selective advantages that may be acquired versus the risk of acquiring an unviable karyotype. The concept of a “just right” level of CIN was crystallized by Lengauer and colleagues who proposed that CIN can create a breath of clones, which may by chance, possess the right characteristics to cross various selection barriers. However, this is only if CIN itself does not impair cell viability (Cahill et al. 1999). This idea is now supported by evidence from sequencing data and experimental models.
Genomic analysis of human data revealed a nonlinear relationship between the extent of chromosomal rearrangements and patient outcome. Patients whose tumor displayed either the lowest or highest instability (measured as the weighed Genome Instability Index, wGII) had better outcome than patients with intermediate levels of GIN in breast, ovarian, NSCLC, and gastric adenocarcinoma (Birkbak et al. 2011; Roylance et al. 2011). Extreme CIN was also linked to improved outcome in a multivariate analysis performed on a cohort of more than 1100 ER-negative breast cancer patients in which CIN was derived from centromeric FISH analysis (Jamal-Hanjani et al. 2015). More recently, similar conclusions were reached in a pan-cancer analysis exploiting exome data from 1165 tumors across 12 tumor types and further validated with SNP-array data sets from more than 2000 samples (Andor et al. 2016). By examining the correlation between survival and the genome fraction affected by copy number alterations, the investigators found better outcome in patients when either <25% or >75% of the tumor genome was affected by copy-number variants. Tumor progression is thus more likely to occur with moderate levels of karyotypic variations.
Mouse models of CIN also support the hypothesis that only moderate CIN levels are viable and can promote cancer, whereas extreme CIN is unviable. Homozygous deletion of many CIN-causing genes is embryonically lethal in mice (reviewed in Simon et al. 2015), whereas heterozygous mice show moderate levels of aneuploidy with a higher incidence of cancer (either spontaneous, in a sensitized background or with carcinogens). In a landmark study using CENPE+/– mice, Cleveland and colleagues showed that CIN promotes tumorigenesis in tissues with low endogenous levels of aneuploidy (lung and lymphocytes), but is tumor suppressive in the liver, which displays high levels of endogenous segregation errors (Weaver et al. 2007). Exploiting the tumor suppressive function of excessive CIN is the basis for using SAC inhibitors to target cancer cells and could contribute to the cytotoxic effect of paclitaxel (Janssen et al. 2011a; Zasadil et al. 2014; Drost et al. 2015; Lee et al. 2016).
Aneuploidy Tolerance Mechanisms
Aneuploidy has been well documented to be detrimental to overall organismal development and cell fitness in virtually every species studied, from yeast, plants, and mammals (Siegel and Amon 2012). Studies in yeast and mouse showed that the presence of an extra chromosome impairs cell proliferation regardless of the extra chromosome involved and was associated with altered metabolism (Torres et al. 2007; Williams et al. 2008). The presence of an extra chromosome and segmental aneuploidies results in a proportional perturbation in transcript and protein levels for most encoded genes (Rancati et al. 2008; Pavelka et al. 2010; Torres et al. 2010; Stingele et al. 2012; Dephoure et al. 2014), although some gene dosage compensation has been reported. Increased pressure on the translation machinery, protein quality control mechanisms, and stoichiometric imbalances brought about by aneuploidy, contribute to a global stress response believed to hamper cell proliferation (Santaguida and Amon 2015). Aneuploid cells displayed greater sensitivity to drugs that further impair these pathways, exposing vulnerabilities of aneuploid cells that could be exploited therapeutically (Tang et al. 2011).
TP53 status appears to be an important determinant of CIN propagation, and analysis of TCGA data revealed it is the only gene significantly associated with CIN (Burrell et al. 2013a). In Barrett’s eosophagus, TP53 LOH precedes tetraploidization and aneuploidy (Barrett et al. 1999; Lai et al. 2007). TP53 is stabilized following chromosome missegregation and results in a p21(CDKN1A)-dependent cell-cycle arrest (Thompson and Compton 2008, 2010). Interestingly, mice expressing a p53 mutant (R175P in human) defective for apoptotic induction but that retains cell-cycle arrest functions, showed delayed lymphomagenesis compared with TP53−/− mice and only gave rise to diploid tumors, an effect which was entirely dependent on p21 induction (Liu et al. 2004; Barboza et al. 2006). The mechanistic basis linking chromosome segregation errors and p53 stabilization is still elusive, but it is likely that several upstream signaling pathways are necessary, one of which being the p38 MAPK pathway (Thompson and Compton 2010; Kumari et al. 2014). p53-independent pathways might also act in parallel. Mutational timing analysis in a pan-cancer dataset revealed that TP53 mutations are predominantly clonal and as such tumors might be primed to be CIN tolerant early in the etiology of cancer (McGranahan et al. 2015). TP53 mutations are also significantly associated with genome doubled tumors and occur before genome duplication in >90% of tumors (McGranahan et al. 2015). 17p deletions encompassing the TP53 locus affect ∼70% of human cancers and result in co-loss of an AURKB allele, potentially facilitating CIN and tolerance in a single event (Fernandez-Miranda et al. 2011; Liu et al. 2016). The importance of TP53 in blocking proliferation of tetraploid cells, by imposing a G1 arrest, has been well documented (Andreassen et al. 2001; Fujiwara et al. 2005; Davoli and de Lange 2012; Ganem et al. 2014). TP53 disruption is therefore thought to render cells permissive for CIN by relieving the stress associated with acute segregation errors and by allowing the expansion of genome doubled cells, which are intrinsically more prone to CIN.
Genome-doubling represents arguably one of the most underestimated precursor of CIN in human cancers. Newly formed tetraploidization cells display CIN leading to extensive karyotypic heterogeneity and chromosomal rearrangements in various systems, and can initiate tumor formation in mice (Fujiwara et al. 2005; Davoli and de Lange 2012; Dewhurst et al. 2014). Tetraploidization and CIN are also early events in esophageal cancer, such as in Barrett’s esophagus (Barrett et al. 1999; Lai et al. 2007; Davoli and de Lange 2011; Murugaesu et al. 2015). Recent large-scale copy number analyses of human cancers revealed that an ancestral whole-genome doubling event is detected in ∼37% of tumors (11% to 64% depending on cancer type) (Cibulskis et al. 2012; Zack et al. 2013; Dewhurst et al. 2014). Such tumors display a much higher rate of copy-number gain and losses, most of which is acquired post genome-doubling (Zack et al. 2013; Dewhurst et al. 2014). Experiments with various cell lines showed that tetraploid cells are also much more tolerant to segregation errors than their diploid precursors, without prior mutations (Dewhurst et al. 2014; Kuznetsova et al. 2015). Deleterious genetic alterations are most likely buffered in a tetraploid background, providing a genetic environment into which a wider range of genetic makeups can be explored.
Sporadic CIN Episodes and Compensation
CIN is unlikely to follow a linear trajectory in which the level of instability may fluctuate dramatically during tumor development. As a result, CIN may itself be dynamic during tumor evolution and subject to various compensatory mechanisms, an area that remains poorly explored. Although genomic imbalances globally translate into proportional differences on transcript and protein abundance, as much as 20% of the genes on an extra chromosome may not show aberrant expression (Torres et al. 2007; Williams et al. 2008; Pavelka et al. 2010; Stingele et al. 2012). This regulation might take place on a gene-specific basis in aneuploid cells, and could involve translational compensation (Dassi et al. 2015). Cells could also compensate for the sudden loss of CIN inducing genes, especially for those whose loss would lead to excessive CIN, by developing alternative coping mechanisms. This phenomenon has been described following Securin (PTTG1) homozygous deletion, which causes extensive CIN, followed by adaptation to the lack of securin and genome stabilization (Pfleghaar et al. 2005). SAC activity is also dispensable after the combined loss of at least two E2 ubiquitin ligases involved in mitotic progression (Wild et al. 2016). Cells can thus respond to intensive CIN episodes through buffering mechanisms, to promote survival.
EXPLORING THE FITNESS LANDSCAPE THROUGH CIN
Aneuploidy Drives Adaptation
Elegant experiments in yeast have provided evidence that aneuploidy can drive phenotypic adaptation. They revealed that although aneuploidy hampers cell proliferation under homeostatic conditions, it gives rises to clones with growth-promoting properties when exposed to chemotherapeutic and antifungal drugs, and drives the acquisition of entirely new traits on deletion of a gene thought to be essential for cell division (Rancati et al. 2008; Pavelka et al. 2010). Aneuploidies involving random combinations of chromosomes offer further possibilities to modulate independent pathways through additive and synergistic effects, producing complex phenotypes (Potapova et al. 2013). A striking observation is that aneuploid cells invariably show a greater phenotypic variance in response to cytotoxic agents, even when aneuploid cells were overall more sensitive to a drug than euploid cells, rare aneuploid clones showed greater viability (Chen et al. 2015). Similarly, increased polyploidy greatly accelerates adaptation in budding yeast grown in raffinose as sole carbon source (Selmecki et al. 2015). Tetraploid cells not only acquired significantly more mutations than diploid or haploid cells, but only tetraploid clones almost inevitably displayed whole-chromosome and segmental aneuploidies (Selmecki et al. 2015). Remarkably, copy gain of chromosome XIII accelerated adaptation in tetraploid clones but was detrimental when occurring in diploid cells. Tetraploidy thus provides a more permissive background in which a greater breath of karyotypic and mutational variations can be explored, increasing the probability of acquiring growth-promoting properties.
CIN in Tumor Progression
CIN remains underestimated when analyzing mechanisms of tumor progression and relapse. Indeed, copy-number analyses on single biopsies poorly capture the extent of cell-to-cell variations and rather reflect pervasive copy number states. High degree of intratumor heterogeneity results in neutral copy number being observed for most chromosomes. In addition, analyses of highly aneuploid tumors (Sotillo et al. 2010) or mixtures of distinct aneuploid populations (Chen et al. 2015) create an illusion of diploidy. Multiregion sampling provides a better resolution and revealed in several studies that CIN onset preceded subclonal expansion and metastatic dispersion in several cancers, because chromosomal aberrations were shared between all regions, or occurred after genome-doubling (Campbell et al. 2010; de Bruin et al. 2014; Kim et al. 2015). Single-cell analysis of two paired primary and metastatic breast carcinomas revealed ongoing CIN in the primary tumor with the predominance of a highly aneuploidy clone, which seeded the metastasis (Navin et al. 2011). Multiregion sequencing also revealed that most metastatic copy number and structural variants arose from a primary tumor subclone, but with evidence of persistent CIN during the metastatic process (Yates et al. 2015).
CIN Drives Recurrence and Metastasis
Seminal experiments using an inducible K-Ras-driven model of lung cancer showed that concomitant inducible CIN induction (through MAD2 overexpression) during tumor development greatly increases recurrence following K-Ras oncogene withdrawal (Sotillo et al. 2010). In this system, K-Ras/MAD2 tumors displayed CIN and enhanced the aggressiveness, but tumor regression was just as profound following oncogene withdrawal. However, 11/24 of tumors from K-Ras/MAD2 mice recurred, as opposed to 0/25 for K-Ras tumors after oncogene withdrawal. As pointed out by the investigators, this situation is reminiscent of Bcr-Abl-driven CML tumors, which invariably respond to imatinib; however, recurrence is significantly higher in tumors initially showing CIN (Cortes and O’Dwyer 2004). The fact that MAD2 transgene expression is dispensable for tumor maintenance reflects an important feature of CIN in general. Once initiated, CIN provides a pool of highly diversified cells, which are prone to acquire further numerical and structural aberrations. This relieves the selective pressure to maintain the initial genetic lesion, and is therefore not subject to oncogene addiction. Mouse models of skin and pancreatic cancers have similarly shown that CIN drives metastatic progression (Hingorani et al. 2005; McCreery et al. 2015). In human tumors, increase in CIN has been linked to metastatic progression in prostate cancer, pancreatic cancer, breast cancer, colorectal cancer, and renal cell carcinoma (reviewed in Turajlic and Swanton 2016).
CIN Contributes to Drug Resistance
CIN has long been associated with multidrug resistance (Duesberg et al. 2007; Lee et al. 2011; McGranahan et al. 2012). Tetraploid cells, by virtue of their higher CIN rate and tolerance (Dewhurst et al. 2014), also display greater resistance to a broad range of compounds (Lee et al. 2011; Dewhurst et al. 2014; Kuznetsova et al. 2015). One explanation for this phenomenon is that CIN allows the maintenance of various karyotypes, and the one(s) conferring a proliferative advantage expand on selection. For example, in a longitudinal study of CLL, increased genomic complexity on recurrence was observed more frequently amongst patients who received therapy (Ouillette et al. 2013). In medulloblastoma, profound divergence was observed posttherapy, yet the clone representing <5% of the primary tumor driving recurrence featured aneuploidy and GIN (Morrissy et al. 2016). Detailed phylogenetic reconstruction of chemotherapy resistant breast cancer also uncovered the presence of the resistant subclones before treatment, showing that chemotherapy did not drive the acquisition of mutations (Yates et al. 2015). Instead, resistance could be traced back to the expansion of a preexisting clone with persistent GIN.
CONCLUDING REMARKS
Although wCIN is a pervasive feature of human tumors, it is challenging to identify the genetic and/or epigenetic cause(s) of wCIN in individual tumors. This might be predictable given that CIN can be brought about by a multitude of defects and its propagation does not require maintenance of the CIN initiating lesion. An alternative explanation is that mutations that cause a high level of CIN might be excessively deleterious, even in a genetic background permissive to aneuploidy, such that CIN might be driven mainly by partially deleterious mutations. For similar reasons, known oncogenes might be potent CIN drivers because they moderately affect the expression and function of genes involved in genome maintenance. Nevertheless, CIN enables the exploration of a phenotypic landscape in a way that cannot be achieved with an increased mutation rate alone (Mroz et al. 2015). A deep understanding of the underlying causes of CIN and the context enabling its propagation is still lacking and needs thorough longitudinal studies to fully understand its importance. Stochastic chromosome missegregation in normal cells, although rare (one error per 100–500 divisions), explains the presence of aneuploid cells in various healthy tissues (Knouse et al. 2014) and could represent early precursors of CIN. Understanding genome dynamics during therapy response and resistance might also offer new strategies to ambush cancer cells by forcing them through an “evolutionary trap” that could make them exquisitely sensitive to a second treatment (Chen et al. 2015). The development of sensitive approaches for copy-number analysis should now prompt study design aimed at tracking more precisely the onset of CIN in tumors and targeting of CIN for therapeutic benefit.
ACKNOWLEDGMENTS
L.S. is supported by a grant from the Novo Nordisk Foundation, the Canadian Institutes of Health Research (CIHR), and the European Molecular Biology Organization (EMBO). C.S. is a senior Cancer Research UK clinical research fellow and is funded by Cancer Research UK (TRACERx), the Royal Society (Napier Professorship), the CRUK Lung Cancer Centre of Excellence, Stand Up 2 Cancer (SU2C), the Rose-trees Trust, the Novo Nordisk Foundation (ID 16584), EU FP7 (projects PREDICT and RESPONSIFY, ID: 259303), the Prostate Cancer Foundation, the Breast Cancer Research Foundation, the European Research Council (THESEUS), and the National Institute for Health Research/University College London Hospitals Biomedical Research Centre.
Footnotes
Editors: Charles Swanton, Alberto Bardelli, Kornelia Polyak, Sohrab Shah, and Trevor A. Graham
Additional Perspectives on Cancer Evolution available at www.perspectivesinmedicine.org
REFERENCES
- Adon AM, Zeng X, Harrison MK, Sannem S, Kiyokawa H, Kaldis P, Saavedra HI. 2010. Cdk2 and Cdk4 regulate the centrosome cycle and are critical mediators of centrosome amplification in p53-null cells. Mol Cell Biol 30: 694–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, Ji HP, Maley CC. 2016. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 22: 105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreassen PR, Lohez OD, Lacroix FB, Margolis RL. 2001. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol Biol Cell 12: 1315–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber TD, McManus K, Yuen KWY, Reis M, Parmigiani G, Shen D, Barrett I, Nouhi Y, Spencer F, Markowitz S., et al. 2008. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci 105: 3443–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barboza JA, Liu G, Ju Z, El-Naggar AK, Lozano G. 2006. p21 delays tumor onset by preservation of chromosomal stability. Proc Natl Acad Sci 103: 19842–19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett MT, Sanchez CA, Prevo LJ, Wong DJ, Galipeau PC, Paulson TG, Rabinovitch PS, Reid BJ. 1999. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet 22: 106–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkbak NJ, Eklund AC, Li Q, McClelland SE, Endesfelder D, Tan P, Tan IB, Richardson AL, Szallasi Z, Swanton C. 2011. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res 70: 3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T. 2008. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J Cell Sci 121 (Suppl 1): 1–84. [DOI] [PubMed] [Google Scholar]
- Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N, Domingo E, Kanu N, Dewhurst SM, Gronroos E, et al. 2013a. Replication stress links structural and numerical cancer chromosomal instability. Nature 494: 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, McGranahan N, Bartek J, Swanton C. 2013b. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501: 338–345. [DOI] [PubMed] [Google Scholar]
- Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. 1999. Genetic instability and Darwinian selection in tumours. Trends Cell Biol 9: M57–M60. [PubMed] [Google Scholar]
- Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, et al. 2010. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467: 1109–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Mulla WA, Kucharavy A, Tsai HJ, Rubinstein B, Conkright J, McCroskey S, Bradford WD, Weems L, Haug JS, et al. 2015. Targeting the adaptability of heterogeneous aneuploids. Cell 160: 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, de Solorzano CO, Knowles D, Jones A, Chou W, Rodriguez EG, Kuo WL, Ljung BM, Chew K, Myambo K, et al. 2004. In situ analyses of genome instability in breast cancer. Nat Genet 36: 984–988. [DOI] [PubMed] [Google Scholar]
- Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, Beroukhim R, et al. 2012. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 30: 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes J, O’Dwyer ME. 2004. Clonal evolution in chronic myelogenous leukemia. Hematol Oncol Clin North Am 18: 671–684. [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassi E, Greco V, Sidarovich V, Zuccotti P, Arseni N, Scaruffi P, Tonini GP, Quattrone A. 2015. Translational compensation of genomic instability in neuroblastoma. Sci Rep 5: 14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, de Lange T. 2011. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol 27: 585–610. [DOI] [PubMed] [Google Scholar]
- Davoli T, de Lange T. 2012. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell 21: 765–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Denchi EL, de Lange T. 2010. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell 141: 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Xu Andrew W, Mengwasser Kristen E, Sack Laura M, Yoon John C, Park Peter J, Elledge Stephen J. 2013. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155: 948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, Jamal-Hanjani M, Shafi S, Murugaesu N, Rowan AJ, et al. 2014. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346: 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Hwang S, O’Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM. 2014. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. eLife 3: e03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhurst SM, McGranahan N, Burrell RA, Rowan AJ, Grönroos E, Endesfelder D, Joshi T, Mouradov D, Gibbs P, Ward RL, et al. 2014. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov 4: 175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, et al. 2015. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521: 43–47. [DOI] [PubMed] [Google Scholar]
- Duesberg P, Li R, Sachs R, Fabarius A, Upender MB, Hehlmann R. 2007. Cancer drug resistance: The central role of the karyotype. Drug Resist Updat 10: 51–58. [DOI] [PubMed] [Google Scholar]
- Duijf PHG, Benezra R. 2013. The cancer biology of whole-chromosome instability. Oncogene 32: 4727–4736. [DOI] [PubMed] [Google Scholar]
- Ertych N, Stolz A, Stenzinger A, Weichert W, Kaulfuß S, Burfeind P, Aigner A, Wordeman L, Bastians H. 2014. Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nat Cell Biol 16: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertych N, Stolz A, Valerius O, Braus GH, Bastians H. 2016. CHK2-BRCA1 tumor-suppressor axis restrains oncogenic Aurora-A kinase to ensure proper mitotic microtubule assembly. Proc Natl Acad Sci 113: 1817–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann G, Beaty R, Hruban RH, Maitra A. 2007. Molecular genetics of pancreatic intraepithelial neoplasia. J Hepatobiliary Pancreat Surg 14: 224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Miranda G, Trakala M, Martin J, Escobar B, Gonzalez A, Ghyselinck NB, Ortega S, Canamero M, De Castro IP, Malumbres M. 2011. Genetic disruption of aurora B uncovers an essential role for aurora C during early mammalian development. Development 138: 2661–2672. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. 2005. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 138: 1043–1047. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D. 2007. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev 17: 157–162. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature 460: 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Cornils H, Chiu SY, O’Rourke Kevin P, Arnaud J, Yimlamai D, Théry M, Camargo Fernando D, Pellman D. 2014. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell 158: 833–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadimi BM, Sackett DL, Difilippantonio MJ, Schröck E, Neumann T, Jauho A, Auer G, Ried T. 2000. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosomes Cancer 27: 183–190. [PMC free article] [PubMed] [Google Scholar]
- Giam M, Rancati G. 2015. Aneuploidy and chromosomal instability in cancer: A jackpot to chaos. Cell Div 10: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D. 2008. Classification of chromosome segregation errors in cancer. Chromosoma 117: 511–519. [DOI] [PubMed] [Google Scholar]
- Gisselsson D, Jin Y, Lindgren D, Persson J, Gisselsson L, Hanks S, Sehic D, Mengelbier LH, Ora I, Rahman N, et al. 2010. Generation of trisomies in cancer cells by multipolar mitosis and incomplete cytokinesis. Proc Natl Acad Sci 107: 20489–20493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godek KM, Kabeche L, Compton DA. 2015. Regulation of kinetochore-microtubule attachments through homeostatic control during mitosis. Nat Rev Mol Cell Biol 16: 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SA, Picone R, Burute M, Dagher R, Su Y, Leung CT, Polyak K, Brugge JS, Théry M, Pellman D. 2014. Oncogene-like induction of cellular invasion from centrosome amplification. Nature 510: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LVF, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Kastrinakis NG, Levy B, et al. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434: 907–913. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. 2008. An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355. [DOI] [PubMed] [Google Scholar]
- Hanks S, Coleman K, Reid S, Plaja A, Firth H, Fitzpatrick D, Kidd A, Méhes K, Nash R, Robin N, et al. 2004. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet 36: 1159. [DOI] [PubMed] [Google Scholar]
- Haruta M, Matsumoto Y, Izumi H, Watanabe N, Fukuzawa M, Matsuura S, Kaneko Y. 2008. Combined BubR1 protein down-regulation and RASSF1A hypermethylation in Wilms tumors with diverse cytogenetic changes. Mol Carcinog 47: 660–666. [DOI] [PubMed] [Google Scholar]
- Hatch Emily M, Fischer Andrew H, Deerinck Thomas J, Hetzer Martin W. 2013. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154: 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando E, Nahlé Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, et al. 2004. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430: 797–802. [DOI] [PubMed] [Google Scholar]
- Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA. 2005. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 7: 469–483. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. 2009. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 10: 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli PV, Waizenegger IC, Bunz F, Langer S, Speicher MR, Peters JM, Kinzler KW, Vogelstein B, Lengauer C. 2001. Securin is required for chromosomal stability in human cells. Cell 10: 445–457. [DOI] [PubMed] [Google Scholar]
- Jamal-Hanjani M, A’Hern R, Birkbak NJ, Gorman P, Grönroos E, Ngang S, Nicola P, Rahman L, Thanopoulou E, Kelly G, et al. 2015. Extreme chromosomal instability forecasts improved outcome in ER-negative breast cancer: A prospective validation cohort study from the TACT trial. Ann Oncol 26: 1340–1346. [DOI] [PubMed] [Google Scholar]
- Janssen A, Kops GJ, Medema RH. 2011a. Targeting the mitotic checkpoint to kill tumor cells. Horm Cancer 2: 113–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, Van Der Burg M, Szuhai K, Kops GJPL, Medema RH. 2011b. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 333: 1895–1898. [DOI] [PubMed] [Google Scholar]
- Johannes JW, Almeida L, Daly K, Ferguson AD, Grosskurth SE, Guan H, Howard T, Ioannidis S, Kazmirski S, Lamb ML, et al. 2015. Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering. Bioorg Med Chem Lett 25: 5743–5747. [DOI] [PubMed] [Google Scholar]
- Kabeche L, Compton DA. 2012. Checkpoint-independent stabilization of kinetochore-microtubule attachments by Mad2 in human cells. Curr Biol 22: 638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeche L, Compton DA. 2013. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature 502: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata T, Pritchard C. 2011. Mechanisms of aneuploidy induction by RAS and RAF oncogenes. Am J Cancer Res 1: 955–971. [PMC free article] [PubMed] [Google Scholar]
- Kato T, Daigo Y, Aragaki M, Ishikawa K, Sato M, Kondo S, Kaji M. 2011. Overexpression of MAD2 predicts clinical outcome in primary lung cancer patients. Lung Cancer 74: 124–131. [DOI] [PubMed] [Google Scholar]
- Kim TM, Jung SH, An CH, Lee SH, Baek IP, Kim MS, Park SW, Rhee JK, Lee SH, Chung YJ. 2015. Subclonal genomic architectures of primary and metastatic colorectal cancer based on intratumoral genetic heterogeneity. Clin Cancer Res 21: 4461–4472. [DOI] [PubMed] [Google Scholar]
- Kim JS, He X, Orr B, Wutz G, Hill V, Peters JM, Compton DA, Waldman T. 2016. Intact cohesion, anaphase, and chromosome segregation in human cells harboring tumor-derived mutations in STAG2. PLoS Genet 12: e1005865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleyman M, Kabeche L, Compton DA. 2014. STAG2 promotes error correction in mitosis by regulating kinetochore-microtubule attachments. J Cell Sci 127: 4225–4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouse KA, Wu J, Whittaker CA, Amon A. 2014. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc Natl Acad Sci 111: 13409–13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari G, Ulrich T, Krause M, Finkernagel F, Gaubatz S. 2014. Induction of p21CIP1 protein and cell cycle arrest after inhibition of Aurora B kinase is attributed to aneuploidy and reactive oxygen species. J Biol Chem 289: 16072–16084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsova AY, Seget K, Moeller GK, de Pagter MS, de Roos JADM, Dürrbaum M, Kuffer C, Müller S, Zaman GJR, Kloosterman WP, et al. 2015. Chromosomal instability, tolerance of mitotic errors and multidrug resistance are promoted by tetraploidization in human cells. Cell Cycle 14: 2810–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D. 2008. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev 22: 2189–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai LA, Paulson TG, Li X, Sanchez CA, Maley C, Odze RD, Reid BJ, Rabinovitch PS. 2007. Increasing genomic instability during premalignant neoplastic progression revealed through high resolution array-CGH. Genes Chromosomes Cancer 46: 532–542. [DOI] [PubMed] [Google Scholar]
- Landau DA, Clement K, Ziller MJ, Boyle P, Fan J, Gu H, Stevenson K, Sougnez C, Wang L, Li S, et al. 2014. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell 26: 813–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AJX, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, Downward J, Szallasi Z, Tomlinson IPM, Howell M, et al. 2011. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res 71: 1858–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Lee NCO, Kouprina N, Kim JH, Kagansky A, Bates S, Trepel JB, Pommier Y, Sackett D, Larionov V. 2016. Effects of anticancer drugs on chromosome instability and new clinical implications for tumor-suppressing therapies. Cancer Res 76: 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz ML, Zhang CZ, Pellman D. 2015. Chromothripsis: A new mechanism for rapid karyotype evolution. Annu Rev Genet 49: 183–211. [DOI] [PubMed] [Google Scholar]
- Leiserson MDM, Vandin F, Wu HT, Dobson JR, Eldridge JV, Thomas JL, Papoutsaki A, Kim Y, Niu B, McLellan M, et al. 2015. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet 47: 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. 1998. Genetic instabilities in human cancers. Nature 396: 643–649. [DOI] [PubMed] [Google Scholar]
- Liu G, Parant JM, Lang G, Chau P, Chavez-Reyes A, El-Naggar AK, Multani A, Chang S, Lozano G. 2004. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat Genet 36: 63–68. [DOI] [PubMed] [Google Scholar]
- Liu Y, Nielsen CF, Yao Q, Hickson ID. 2014. The origins and processing of ultra fine anaphase DNA bridges. Curr Opin Genet Dev 26: 1–5. [DOI] [PubMed] [Google Scholar]
- Liu Y, Chen C, Xu Z, Scuoppo C, Rillahan CD, Gao J, Spitzer B, Bosbach B, Kastenhuber ER, Baslan T, et al. 2016. Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature 531: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarinho E, Maffini S, Barisic M, Marques A, Toso A, Meraldi P, Maiato H. 2012. CLASPs prevent irreversible multipolarity by ensuring spindle-pole resistance to traction forces during chromosome alignment. Nat Cell Biol 14: 1–10. [DOI] [PubMed] [Google Scholar]
- Maciejowski J, Li Y, Bosco N, Campbell Peter J, De Lange T. 2015. Chromothripsis and kataegis induced by telomere crisis. Cell 163: 1641–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M. 2011. Oncogene-induced mitotic stress: p53 and pRb get mad too. Cancer Cell 19: 691–692. [DOI] [PubMed] [Google Scholar]
- Manning Amity L, Yazinski Stephanie A, Nicolay B, Bryll A, Zou L, Dyson Nicholas J. 2014. Suppression of genome instability in pRB-deficient cells by enhancement of chromosome cohesion. Mol Cell 53: 993–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL. 2015. Shugoshins: Tension-sensitive pericentromeric adaptors safeguarding chromosome segregation. Mol Cell Biol 35: 634–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCreery MQ, Halliwill KD, Chin D, Delrosario R, Hirst G, Vuong P, Jen KY, Hewinson J, Adams DJ, Balmain A. 2015. Evolution of metastasis revealed by mutational landscapes of chemically induced skin cancers. Nat Med 21: 1514–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. 2012. Cancer chromosomal instability: Therapeutic and diagnostic challenges. EMBO Rep 13: 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, Swanton C. 2015. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med 7: 283ra254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeker AK, Hicks JL, Platz EA, March GE, Bennett CJ, Delannoy MJ, De Marzo AM. 2002. Telomere shortening is an early somatic DNA alteration in human prostate tumorigenesis. Cancer Res 62: 6405–6409. [PubMed] [Google Scholar]
- Minocherhomji S, Ying S, Bjerregaard VA, Bursomanno S, Aleliunaite A, Wu W, Mankouri HW, Shen H, Liu Y, Hickson ID. 2015. Replication stress activates DNA repair synthesis in mitosis. Nature 528: 286–290. [DOI] [PubMed] [Google Scholar]
- Morrissy AS, Garzia L, Shih DJH, Zuyderduyn S, Huang X, Skowron P, Remke M, Cavalli FMG, Ramaswamy V, Lindsay PE, et al. 2016. Divergent clonal selection dominates medulloblastoma at recurrence. Nature 529: 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroz EA, Tward AD, Tward AM, Hammon RJ, Ren Y, Rocco JW. 2015. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: Analysis of data from the Cancer Genome Atlas. PLoS Med 12: e1001786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugaesu N, Wilson GA, Birkbak NJ, Watkins TBK, McGranahan N, Kumar S, Abbassi-Ghadi N, Salm M, Mitter R, Horswell S, et al. 2015. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov 5: 821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. 2007. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393. [DOI] [PubMed] [Google Scholar]
- Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. 2011. Tumour evolution inferred by single-cell sequencing. Nature 742: 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. 2011. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol 13: 1154–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr B, Compton DA. 2013. A double-edged sword: How oncogenes and tumor suppressor genes can contribute to chromosomal instability. Front Oncol 3: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouillette P, Saiya-Cork K, Seymour E, Li C, Shedden K, Malek SN. 2013. Clonal evolution, genomic drivers, and effects of therapy in chronic lymphocytic leukemia. Clin Cancer Res 19: 2893–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HY, Jeon YK, Shin HJ, Kim IJ, Kang HC, Jeong SJ, Chung DH, Lee CW. 2007. Differential promoter methylation may be a key molecular mechanism in regulating BubR1 expression in cancer cells. Exp Mol Med 39: 195–204. [DOI] [PubMed] [Google Scholar]
- Passerini V, Ozeri-Galai E, de Pagter MS, Donnelly N, Schmalbrock S, Kloosterman WP, Kerem B, Storchová Z. 2016. The presence of extra chromosomes leads to genomic instability. Nat Commun 7: 10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. 2010. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468: 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfleghaar K, Heubes S, Cox J, Stemmann O, Speicher MR. 2005. Securin is not required for chromosomal stability in human cells. PLoS Biol 3: e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihan GA, Purohit A, Wallace J, Knecht H, Woda B, Quesenberry P, Doxsey SJ. 1998. Centrosome defects and genetic instability in malignant tumors. Cancer Res 58: 3974–3985. [PubMed] [Google Scholar]
- Potapova TA, Zhu J, Li R. 2013. Aneuploidy and chromosomal instability: A vicious cycle driving cellular evolution and cancer genome chaos. Cancer Metastasis Rev 32: 377–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab MS, Breitkreutz I, Anderhub S, Rønnest MH, Leber B, Larsen TO, Weiz L, Konotop G, Hayden PJ, Podar K, et al. 2012. GF-15, a novel inhibitor of centrosomal clustering, suppresses tumor cell growth in vitro and in vivo. Cancer Res 72: 5374–5385. [DOI] [PubMed] [Google Scholar]
- Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, Perera A, Staehling-Hampton K, Seidel CW, Li R. 2008. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell 135: 879–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode A, Maass KK, Willmund KV, Lichter P, Ernst A. 2016. Chromothripsis in cancer cells: An update. Int J Cancer 138: 2322–2333. [DOI] [PubMed] [Google Scholar]
- Roylance R, Endesfelder D, Gorman P, Burrell RA, Sander J, Tomlinson I, Hanby AM, Speirs V, Richardson AL, Birkbak NJ, et al. 2011. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol Biomarkers Prev 20: 2183–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph KL, Millard M, Bosenberg MW, DePinho RA. 2001. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat Genet 28: 155–159. [DOI] [PubMed] [Google Scholar]
- Ryan SD, Britigan EMC, Zasadil LM, Witte K, Audhya A, Roopra A, Weaver BA. 2012. Up-regulation of the mitotic checkpoint component Mad1 causes chromosomal instability and resistance to microtubule poisons. Proc Natl Acad Sci 109: E2205–E2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Amon A. 2015. 16: Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol 473–485. [DOI] [PubMed] [Google Scholar]
- Schvartzman JM, Duijf Pascal HG, Sotillo R, Coker C, Benezra R. 2011. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell 19: 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki AM, Maruvka YE, Richmond PA, Guillet M, Shoresh N, Sorenson AL, De S, Kishony R, Michor F, Dowell R, et al. 2015. Polyploidy can drive rapid adaptation in yeast. Nature 519: 349–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A. 2011. Aneuploidy drives genomic instability in yeast. Science 333: 1026–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JJ, Amon A. 2012. New insights into the troubles of aneuploidy. Annu Rev Cell Dev Biol 28: 189–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JE, Bakker B, Foijer F. 2015. CINcere modelling: What have mouse models for chromosome instability taught us? Recent Results Cancer Res 200: 39–60. [DOI] [PubMed] [Google Scholar]
- Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M, et al. 2011. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 333: 1039–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Díaz-Rodríguez E, Teruya-Feldstein J, Cordón-Cardo C, Lowe SW, Benezra R. 2007. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell 11: 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Schvartzman JM, Socci ND, Benezra R. 2010. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature 464: 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. 2012. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol 8: 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Ertych N, Kienitz A, Vogel C, Schneider V, Fritz B, Jacob R, Dittmar G, Weichert W, Petersen I, et al. 2010. The CHK2–BRCA1 tumour suppressor pathway ensures chromosomal stability in human somatic cells. Nat Cell Biol 12: 492–499. [DOI] [PubMed] [Google Scholar]
- Tang YC, Williams BR, Siegel JJ, Amon A. 2011. Identification of aneuploidy-selective antiproliferation compounds. Cell 144: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno Y, Susumu H, Kawamura M, Sugimura H, Honda T, Watanabe Y. 2015. The inner centromere-shugoshin network prevents chromosomal instability. Science 349: 1237–1240. [DOI] [PubMed] [Google Scholar]
- Taylor CF, Platt FM, Hurst CD, Thygesen HH, Knowles MA. 2014. Frequent inactivating mutations of STAG2 in bladder cancer are associated with low tumour grade and stage and inversely related to chromosomal copy number changes. Hum Mol Genet 23: 1964–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. 2008. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol 180: 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. 2010. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol 188: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924. [DOI] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A. 2010. Identification of aneuploidy-tolerating mutations. Cell 143: 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turajlic S, Swanton C. 2016. Metastasis as an evolutionary process. Science 352: 169–175. [DOI] [PubMed] [Google Scholar]
- Watanabe Y. 2005. Shugoshin: Guardian spirit at the centromere. Curr Opin Cell Biol 17: 590–595. [DOI] [PubMed] [Google Scholar]
- Watts CA, Richards FM, Bender A, Bond PJ, Korb O, Kern O, Riddick M, Owen P, Myers RM, Raff J, et al. 2013. Design, synthesis, and biological evaluation of an allosteric inhibitor of HSET that targets cancer cells with supernumerary centrosomes. Chem Biol 20: 1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BAA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. 2007. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11: 25–36. [DOI] [PubMed] [Google Scholar]
- Wild T, Larsen Marie Sofie Y, Narita T, Schou J, Nilsson J, Choudhary C. 2016. The spindle assembly checkpoint is not essential for viability of human cells with genetically lowered APC/C activity. Cell Rep 14: 1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. 2008. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322: 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, Aas T, Alexandrov LB, Larsimont D, Davies H, et al. 2015. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 21: 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachos G, Black EJ, Walker M, Scott MT, Vagnarelli P, Earnshaw WC, Gillespie DAF. 2007. Chk1 is required for spindle checkpoint function. Dev Cell 12: 247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang CZ, Wala J, Mermel CH, et al. 2013. Pan-cancer patterns of somatic copy number alteration. Nat Genet 45: 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasadil LM, Andersen KA, Yeum D, Rocque GB, Wilke LG, Tevaarwerk AJ, Raines RT, Burkard ME, Weaver BA. 2014. Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation on multipolar spindles. Sci Transl Med 6: 229ra243–229ra243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Shaikh FY, Harrison MK, Adon AM, Trimboli AJ, Carroll KA, Sharma N, Timmers C, Chodosh LA, Leone G, et al. 2010. The Ras oncogene signals centrosome amplification in mammary epithelial cells through cyclin D1/Cdk4 and Nek2. Oncogene 29: 5103–5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. 2015. Chromothripsis from DNA damage in micronuclei. Nature 522: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]