In this study, Lahmy et al. used laser UV-assisted zero-length cross-linking to identify a functionally relevant AGO4–DNA interaction at RNA-directed DNA methylation (RdDM) targets in plants. This study provides evidence that AGO4 is targeted to chromatin in RdDM through a protein-based rather than an RNA-based mechanism.
Keywords: Argonaute, DNA methylation, RdDM
Abstract
RNA polymerase V (Pol V) long noncoding RNAs (lncRNAs) have been proposed to guide ARGONAUTE4 (AGO4) to chromatin in RNA-directed DNA methylation (RdDM) in plants. Here, we provide evidence, based on laser UV-assisted zero-length cross-linking, for functionally relevant AGO4–DNA interaction at RdDM targets. We further demonstrate that Pol V lncRNAs or the act of their transcription are required to lock Pol V holoenzyme into a stable DNA-bound state that allows AGO4 recruitment via redundant glycine–tryptophan/tryptophan–glycine AGO hook motifs present on both Pol V and its associated factor, SPT5L. We propose a model in which AGO4–DNA interaction could be responsible for the unique specificities of RdDM.
In fungi, plants, and animals, repeats and transposable elements (TEs) are transcriptionally silenced by packaging of their DNA in a condensed chromatin state (referred to as heterochromatin), established by a set of conserved and specific chromatin-modifying enzymes and characterized by DNA methylation and/or histone modifications (Law and Jacobsen 2010). In addition, small RNAi silencing pathways are universally used by eukaryotes to promote heterochromatin formation and transcriptional gene silencing (TGS) at TEs and repeats (Holoch and Moazed 2015). Central to the activity of these nuclear RNAi pathways are Argonaute (AGO)/PIWI members of the AGO family that bind TE/repeat-derived single-stranded small RNAs to form RNA-induced transcriptional silencing (RITS) complexes that transmit the silencing signal (Holoch and Moazed 2015).
Historically, two models involving base-pairing with target DNA or nascent RNA transcribed at the target loci have been invoked to describe the guiding action of small RNAs in TGS (Matzke and Birchler 2005). However, the detection in fission yeast of RNA polymerase II (Pol II) scaffold RNAs required for RITS recruitment and heterochromatin formation tilted the balance in favor of a RNA-based mode of RITS action known as the “nascent transcript model” (Verdel et al. 2004; Holoch and Moazed 2015). Subsequent studies on RNA-directed DNA methylation (RdDM) in plants and nuclear RNAi in animals, many of them inspired by the yeast model, generalized the notion of a requirement of transcription for the establishment of TGS at target loci (Wierzbicki et al. 2009; Sienski et al. 2012).
RdDM is unique among small RNA-mediated chromatin modification pathways in eukaryotes because it relies on Pol IV and Pol V (two plant-specific homologs of Pol II) for activity (Lahmy et al. 2010). Pol IV long noncoding RNA (lncRNA) precursors are thought to contribute—via the concerted activities of RNA-dependent RNA Pol 2 (RDR2) and DICER-like 3 (DCL3)—to the biogenesis of 24-nucleotide (nt) siRNAs that, upon loading into AGO4-clade proteins, lead to the assembly of an AGO–siRNA RITS-type complex that guides the DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) for DNA methylation of homologous genomic sequences at cytosines in all sequence contexts (CG, CHG, and CHH, where H is A, T, or C) (Zhong et al. 2014; Matzke et al. 2015). Recent studies also revealed that Pol IV lncRNAs can also guide RdDM without being subject to any DICER cleavage (Yang et al. 2016; Ye et al. 2016). Epistasis analysis revealed that Pol V, which does not contribute directly to 24-nt siRNA accumulation, acts downstream from Pol IV to enable RdDM at 24-nt siRNA targeted sites (Kanno et al. 2005; Pontier et al. 2005). The subsequent detection of Pol V lncRNAs and the demonstration of their association with downstream components of the RdDM pathway, including AGO4, led to the proposal of an RNA-based mechanism for RITS guiding to target loci (Wierzbicki et al. 2008, 2009). The generality of a Pol V-based targeting model was further supported by whole-genome studies that revealed a high degree of overlap between the sets of genomic regions bound and transcribed by Pol V and those harboring 24-nt siRNA-dependent DNA methylation (Wierzbicki et al. 2012; Zhong et al. 2012; Böhmdorfer et al. 2016).
Despite recent progress, several aspects of the function of Pol V in RdDM remain to be clarified. In particular, due to the presence of large compositionally conserved glycine–tryptophan/tryptophan–glycine (GW/WG)-rich AGO anchor regions in their C-terminal domains (CTDs), both NRPE1 (the largest subunit of Pol V) and SPT5L (a Pol V auxiliary protein) are likely endowed with high AGO4-binding abilities (El-Shami et al. 2007; Bies-Etheve et al. 2009). In-depth phylogenetic analysis reveals a strict conservation of GW/WG motifs in all NRPE1 and SPT5L orthologs, suggesting that these motifs are functionally relevant (Ma et al. 2015). However, to date, the functional importance of these conserved AGO4-binding platforms in RdDM remains unclear, an issue that we address in this study and that led us to revisit various aspects of Pol V and AGO4 activities in RdDM and propose an alternative model in which AGO4–DNA interactions could be responsible for the highly specific sequence DNA methylation pattern of RdDM.
Results and Discussion
Pol V and SPT5L AGO hook motifs are functionally redundant and essential for RdDM
To assess the role of Pol V/SPT5L AGO hook motifs in vivo, we generated independent plant variants either harboring [NRPE1-WG/Pol V+(1/2) and SPT5L/SPT+(1/2)] or missing [NRPE1-AG/PolV−(1/2) and SPT5Ltr/SPT−(1/2)] these motifs (Supplemental Figs. S1A, S2A) and tested their ability to restore the DNA methylation defects resulting from the loss of either NRPE1 or SPT5L, respectively (Pontier et al. 2005; Bies-Etheve et al. 2009). Using Chop PCR, McrBC-based methylation, and locus-specific bisulfite sequencing analyses, we found that both Pol V and SPT5L variants were able to rescue DNA methylation defects at all RdDM loci tested (Fig. 1A; Supplemental Figs. S1B, S2B). This observation was confirmed and extended by whole-genome bisulfite sequencing analyses (Fig. 1C). Likewise, gene silencing and 24-nt siRNA production at RdDM loci were also restored in plants expressing both Pol V variant types (Supplemental Fig. S1C–E). As expected from their intrinsic capacity to restore RdDM, both Pol V variants were associated with chromatin and displayed a similar capacity to recruit AGO4 at target loci (Supplemental Fig. S1F,G). Taken together, our results indicate that Pol V and SPT5L AGO hook motifs are dispensable for DNA methylation and gene silencing at RdDM loci.
Figure 1.
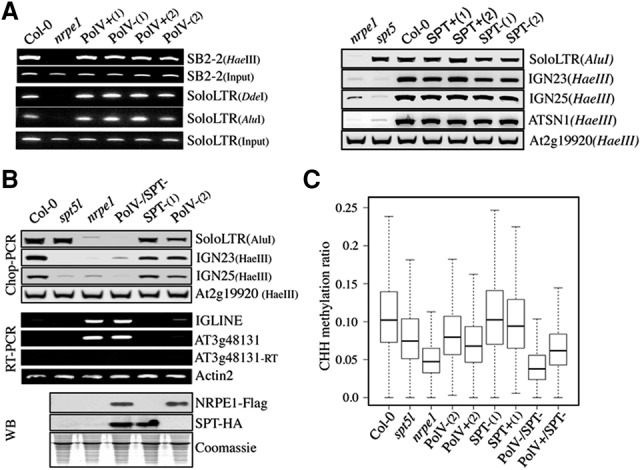
Pol V and SPT5L AGO hook motifs are functionally redundant and essential for RdDM. (A, left panel) Analysis of DNA methylation by Chop PCR at SB2-2 and soloLTR loci in Col-0, nrpe1, Pol V+(1,2), and Pol V−(1,2) lines. Genomic DNA digested with HaeIII, DdeI, and AluI methylation-sensitive enzymes was used as a template for PCR. Undigested DNA (input) was used as a control. (Right panel) Analysis of DNA methylation by Chop PCR at RdDM targets in nrpe1, spt5l, Col-0, SPT+(1,2), and truncated SPT−(1,2) lines. Genomic DNA digested with AluI and HaeIII methylation-sensitive enzymes was used as a template for PCR. The At2g19920 locus was used as a control. (B, top panel) Analysis of DNA methylation by Chop PCR at RdDM targets in Col-0, spt5l, nrpe1, Pol V−/SPT−, SPT−(1), and Pol V−(2) lines. Genomic DNA was digested with HaeIII and AluI methylation-sensitive enzymes. The At2g19920 locus was used as a control. (Middle panel) Transcript levels at two RdDM targets (IGLINE and AT3g48131) in Col-0, spt5l, nrpe1, Pol V−/SPT−, SPT−(1), and Pol V−(2) lines. ACTIN2 was used as a loading control, and −RT reactions show the absence of genomic DNA contamination. (Bottom panel) Detection of Pol V/NRPE1 and SPT variants by Western blot. Coomassie blue staining was used as a loading control. (C) Whole-genome bisulfite analysis of DNA methylation in Col-0, nrpe1, spt5l, Pol V−(2), Pol V+(2), SPT−(1), SPT+(1), Pol V−/SPT−, and Pol V+/SPT− lines. Box plots represent whole-genome CHH methylation ratios at regions previously identified as targeted by Pol V (Zhong et al. 2012). A dependent two-group Wilcoxon signed rank test was run on methylation at the Pol V sites for Pol V−/SPT− versus Pol V+/SPT− and revealed that there is a significant difference in methylation levels. P < 2.2 × 10−16.
To further test the hypothesis of a functional redundancy between Pol V and SPT5L AGO hook motifs, we crossed the Pol V−(2) and SPT−(1) lines and identified F2 Pol V−/SPT− offspring lacking both AGO hook platforms. Western blot analysis confirmed that levels of Pol V−(2) and SPT−(1) variants remain almost unchanged in the Pol V−/SPT− line compared with its crossing parents (Fig. 1B, bottom panel). Chop PCR analysis and locus-specific bisulfite sequencing indicated that CHG and CHH DNA methylation levels at all RdDM target loci tested were reduced in Pol V−/SPT− compared with parent lines, to a level similar to that found in the nrpe1-null mutant (Fig. 1B, top panel; Supplemental Fig. S3A). To assess the generality of these findings, we performed whole-genome bisulfite sequencing analyses and confirmed an almost complete loss of CHH DNA methylation at Pol V-binding sites in Pol V−/SPT− compared with parental lines (Fig. 1C), indicating that RdDM is abolished in the AGO hook-minus line. As expected, the decrease in DNA methylation in Pol V−/SPT− was accompanied by a release of gene silencing at various RdDM loci (Fig. 1B, middle panel).
To unambiguously demonstrate that the loss of RdDM in Pol V−/SPT− results from AGO hook motif depletion but not from an indirect effect, we crossed the Pol V+(2) and SPT−(1) lines and identified Pol V+/SPT−, a plant differing from the AGO hook-minus line only by the presence of Pol V AGO hook motifs (Supplemental Fig. S3B, bottom panel). Functional analyses indicated that DNA methylation and gene silencing at RdDM loci were significantly restored in Pol V+/SPT−, confirming that the defects observed in the Pol V−/SPT− line are directly related to the loss of AGO hook motifs (Fig. 1C; Supplemental Fig. S3B,C). Taken together, our results support the model of a compensatory yet essential role of SPT5L and Pol V AGO hook motifs for RdDM, providing a rationale to explain their conservation in all NRPE1 and SPT5L orthologs.
AGO hook motifs are essential determinants of AGO4 recruitment to RdDM loci
To assess the exact contribution of the AGO hook motifs to RdDM, we first compared the chromatin association of Pol V in AGO hook-plus, AGO hook-minus, and control nrpe1 lines by chromatin immunoprecipitation (ChIP) assay using antibodies directed against NRPE5, a Pol V-specific subunit (Lahmy et al. 2009). Despite some variations in ChIP signal strength, all lines showed specific Pol V signals at RdDM loci compared with the nrpe1-null mutant, indicating that Pol V binding is not overly affected by the depletion of the AGO hook motifs (Fig. 2A, top panel). In contrast, a ChIP assay using anti-AGO4 antibodies revealed that the chromatin association of AGO4 was strongly decreased in the AGO hook-minus (Pol V−/SPT−) line compared with the AGO hook-plus [Pol V+(2); SPT−(1) and Pol V+/SPT−] lines, a loss not due to an intrinsic instability of AGO4 in corresponding plant backgrounds (Fig. 2A, bottom panel; Supplemental Fig. S4A,B). Northern blot analyses also indicated that the lack of AGO4 association with chromatin is not due to an indirect loss of siRNA accumulation in the AGO hook-minus line compared with wild type (Supplemental Fig. S4C). Taken together, these results suggest that the RdDM defects observed in the AGO hook-minus line are correlated with a global loss of AGO4 chromatin association at targeted loci.
Figure 2.
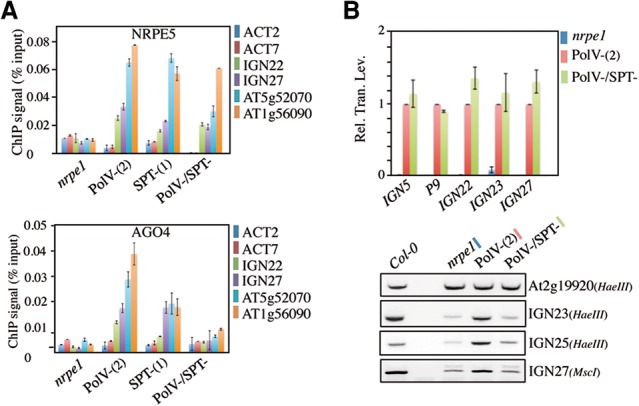
AGO hook motifs are essential determinants of AGO4 recruitment to RdDM loci. (A) ChIP analysis of Pol V (top panel) and AGO4 (bottom panel) binding in nrpe1, Pol V−(2), and SPT−(1) complemented lines and a Pol V−/SPT− cross line. The tested targets are indicated at the right. Actin2 and Actin7 were used as negative controls. After formaldehyde cross-link, Pol V complexes were immunoprecipitated using either the anti-NRPE5 or anti-AGO4 antibodies. Values are means ± SD from two independent amplifications. (B, top panel) Quantitative RT–PCR (qRT–PCR) analysis of Pol V lncRNA levels. IGN5, P9, IGN22, IGN23, and IGN27 transcript accumulation was tested in nrpe1, Pol V−(2), and Pol V−/SPT− lines. (Rel. Tran. Lev.) Relative transcript level normalized to Actin and Pol V−(2) using the ΔΔCt method. (Bottom panel) Analysis of DNA methylation by Chop PCR at IGN23, IGN25, and IGN27 loci. Genomic DNA was digested with HaeIII or MscI methylation-sensitive enzymes and used as a template for PCR. The At2g19920 locus has no HaeIII site and was used as a control (cont). DNA methylation was assessed in Col-0, nrpe1, Pol V−(2), and Pol V−/SPT− lines.
Although our previous data support the idea of a direct role for AGO hook motifs in AGO4 recruitment to chromatin, the loss of AGO4 binding in the AGO minus line could result from a global decrease of Pol V lncRNA levels due to a Pol V transcriptional dysfunction incurred by AGO hook motif depletion. To address this point, we assessed Pol V lncRNA accumulation in AGO hook-plus, AGO hook-minus, and control nrpe1 mutant lines using quantitative RT–PCR (qRT–PCR). Interestingly, Pol V lncRNAs accumulate to closely similar levels in both AGO hook-plus and AGO hook-minus lines, indicating that the loss of DNA methylation and AGO4 chromatin binding is not directly attributable to a general impairment of Pol V-mediated transcription (Fig. 2B top panel; Supplemental Fig. S4D, top panel). More interestingly, Chop PCR analyses performed directly on Pol V lncRNA-generating loci confirm that the AGO hook motif mutation causes the functional uncoupling of Pol V transcription and AGO4-dependent DNA methylation activities (Fig. 2B, bottom panel; Supplemental Fig. S4D, bottom panel), indicating that AGO hook motifs are essential determinants of AGO4 recruitment to RdDM loci.
Pol V lncRNAs or the act of their transcription mediate the association of Pol V to chromatin
Since our results point out the importance of the Pol V/SPT5L AGO hook motifs in the binding of AGO4 to chromatin, we investigated their contribution in securing AGO4 recruitment to chromatin with Pol V lncRNAs and focused our analysis on the nrpe1-3/drd3-3 catalytic mutant allele of Pol V (Fig. 3A; Kanno et al. 2005), a mutant line in which the loss of RdDM was associated with a general defect of Pol V lncRNA production at RdDM loci (Wierzbicki et al. 2008). In line with these previous results, the nrpe1-3 mutation abolishes Pol V lncRNA production and AGO4 chromatin binding (Fig. 3B; Supplemental Fig. S5A). However, ChIP analyses performed with anti-NRPE1 and anti-NRPE5 antibodies also revealed that Pol V binding to RdDM loci is abrogated in the catalytic nrpe1-3 mutant (Fig. 3C), an effect that is not due to a general decrease in the stability of Pol V subunits or associated factors in nrpe1-3 (Supplemental Fig. S5B). Although this observation precluded us from providing any definitive conclusion regarding the concerted implication of both Pol V AGO hook motifs and lncRNAs into AGO4 recruitment to chromatin, our data provide new insights into Pol V function during RdDM, indicating that transcription initiation and productive elongation are essential to lock the Pol V holoenzyme into a stable and productive DNA-bound state. Although the underlying mechanism remains unclear, the RNA–DNA hybrid formed within the transcription bubble as Pol V elongates could directly stabilize Pol V via the establishment of extensive RNA–DNA hybrid/polymerase contacts, as shown previously in the structure of the yeast Pol II elongation complex (Kettenberger et al. 2004).
Figure 3.
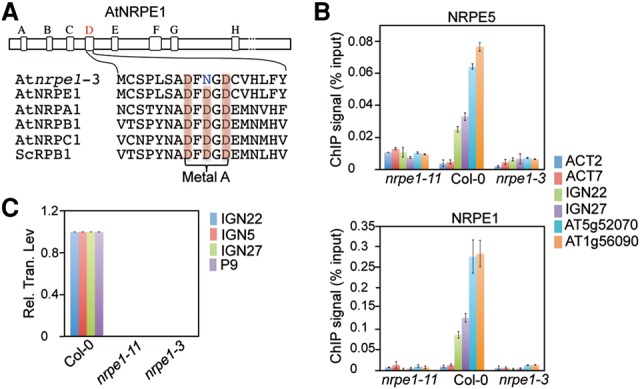
Pol V lncRNAs or the act of their transcription primarily mediate the association of Pol V to chromatin. (A) Amino acid sequences of the catalytic sites (metal A) of different polymerase subunits from Arabidopsis thaliana, Saccharomyces cerevisiae, and Escherichia coli. (B) qRT–PCR analysis of Pol V lncRNA levels in Col-0 and nrpe1-11 and nrpe1-3 mutants. (Rel. Tran. Lev.) Relative transcript level. Values represent the means ± SD of three independent experiments. (C) ChIP analysis of Pol V binding in nrpe1-11 and nrpe1-3 mutants at different RdDM targets, as indicated at the right. Actin2 and Actin7 were used as controls. Values are means ± SD of two independent amplifications. After formaldehyde cross-link, Pol V complexes were immunoprecipitated using either the anti-NRPE5 or anti-NRPE1 antibody.
Evidence for AGO4–DNA interactions at RdDM loci
Despite its wide acceptance, the “nascent transcript” model does not easily explain the strand-biased nature and exquisite degree of specificity displayed by siRNA-guided DNA methylation in RdDM, and other models involving AGO4/siRNA–DNA interaction have been invoked to describe the guiding action of RITS in RdDM (Zhong et al. 2014; Matzke et al. 2015; Wang et al. 2015). Assessing the interaction between AGO4 and DNA using conventional formaldehyde-based cross-linking approaches would be difficult given the existence of the intricate network of molecular interactions among components of the active Pol V holoenzyme revealed by this study. Indeed, these approaches generate extensive protein–protein and protein–nucleic acid cross-links that reveal both direct and indirect interactions (Zentner and Henikoff 2014; Hoffman et al. 2015). To unambiguously assess the DNA-binding specificity of AGO4 in vivo, we performed ChIP on UV laser cross-linked chromatin (LChIP) (Fig. 4A). The benefit of laser UV light lies in the fact that it is a zero-length cross-linking agent that generates only direct DNA–protein cross-links when performed under “single-hit” conditions (i.e., no more than 5%–10% of DNA–protein noncovalent complexes are cross-linked) and does not form protein–protein cross-links (Mutskov et al. 1997; Altintas et al. 2011). Because Pol V has been shown to transcribe bipartite synthetic DNA templates in vitro (Haag et al. 2012), we first developed LChIP by looking at Pol V/DNA interaction at both control and RdDM loci. Under optimal conditions for photochemical cross-linking, LChIP analysis using Flag tag antibodies indicated a specific and reproducible enrichment of Flag-NRPE1 on RdDM targets versus control loci (Fig. 4B). As expected for a specific LChIP signal, DNA was not significantly enriched in the nonirradiated samples (Fig. 4B). These data confirm that Pol V contacts DNA at RdDM loci and provide proof of concept data for the use of a UV laser device in assessing protein–DNA interaction at RdDM targets in vivo.
Figure 4.
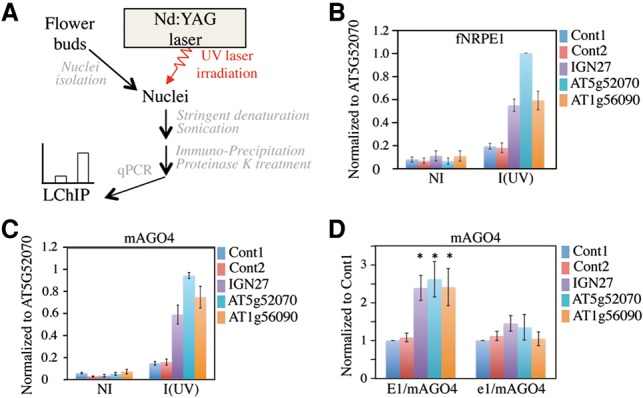
Detection of AGO4–DNA interactions at RdDM loci by LChIP. (A) Scheme of LChIP. (B) LChIP analysis of Pol V binding in a Pol V+-complemented line. Nonirradiated (NI) or irradiated [I (UV)] nuclei were used to prepare chromatin. ChIP was then performed using anti-Flag antibodies. The RdDM target loci tested are indicated at the right. Cont1 and Cont2 correspond to two independent regions located 2 kb from the At4g04920 gene and represent negative controls. Values of DNA enrichment were calculated as percentage of input and were normalized to At5g52070. Error bars are SEM of three independent ChIP experiments. (C) LChIP analysis of mAGO4 binding in an E1/mAGO4-complemented line. Nonirradiated (NI) or irradiated [I (UV)] nuclei were used to prepare chromatin. ChIP was then performed using anti-myc antibodies. The RdDM target loci tested are indicated at the right. Cont1 and Cont2 represent negative controls. Values of DNA enrichment were calculated as percentage input and were normalized to At5g52070. Error bars are SEM of three independent ChIP experiments. (D) LChIP analysis of mAGO4 binding in E1/mAGO4-complemented versus e1/mAGO4-complemented lines. ChIP was then performed using anti-myc antibodies on irradiated nuclei. The RdDM target loci tested are indicated at the right. Cont1 and Cont2 correspond to two independent regions located 2 kb from the At4g04920 gene and represent negative controls. Values of DNA enrichment were calculated as the percentage of input and were normalized to Cont1. Error bars are SEM of three independent ChIP experiments. (*) P < 0.05 compared with Cont1. n = 3.
To follow up on this observation, we assessed AGO4–DNA interaction by performing LChIP experiments on an epitope-tagged cmyc-AGO4 (mAGO4) line whose functionality was confirmed previously by its ability to restore DNA methylation of RdDM targets in the ago4-1 mutant line (Li et al. 2006). We found that the mAGO4 LChIP signal was reproducibly enriched over the RdDM loci compared with control loci, as was the Pol V LChIP signal, suggesting that AGO4 also contacts DNA at these loci (Fig. 4C). To assess the specificity of the AGO4 LChIP signal, we crossed the mAGO4 and nrpe1 lines and retrieved both E1/mAGO4 and e1/mAGO4 offspring expressing similar levels of epitope-tagged AGO4 in either a wild-type or nrpe1 mutant background (Supplemental Fig. S6A). Chop PCR analysis confirmed that DNA methylation at RdDM loci was abolished in e1/mAGO4 compared with the E1/mAGO4 parent line, in keeping with the idea that Pol V is essential to guide AGO4 to RdDM loci (Supplemental Fig. S6B). While LChIP experiments performed on the parent line showed a statistically significant enrichment of mAGO4 over the RdDM loci, there was no statistical difference in the binding of mAGO4 on RdDM versus control loci in e1/mAGO4 lines (Fig. 4D), confirming that the AGO4 LChIP signal is specific and that AGO4 contacts DNA in a Pol V-dependent manner.
Our results shed new light on the previously underappreciated contribution of the conserved AGO hook motifs present on Pol V and SPT5L CTDs, emphasizing the redundant yet essential implication of these motifs in RdDM. In particular, our results suggest that AGO4 is recruited to the Pol V holoenzyme via a protein-based mechanism (Fig. 5) involving multiple AGO hook motifs that are likely to contribute to DNA methylation by the mass action effect of concentrating AGO4 to its site of action. The AGO hook motif-mediated mechanism of AGO4 recruitment proposed in our study provides an explanation for the emergence of a unique transcriptional machinery dedicated to RdDM in plants. We also show that Pol V transcription and/or elongation is essential to lock this enzyme into a productive DNA-bound state (Fig. 5). This result may explain why the Pol V catalytic mutant fails to display the characteristic punctate immunostaining pattern shown by the wild-type enzyme in Arabidopsis nuclei (Haag et al. 2009). These data suggest an intrinsic lack of stability of Pol V on DNA that could be linked to the single-stranded nature of the Pol V template, as proposed recently in a transcription fork model (Pikaard et al. 2012; Matzke et al. 2015). Whatever the mechanism, a functional consequence of transcription-dependent stabilization of Pol V would be to increase the local concentration of AGO4 through the AGO hook platforms, thus potentiating interactions with DNA targets. Consistent with this idea, we provide, through laser UV-assisted cross-linking experiments, the first evidence for specific AGO4–DNA interaction at RdDM loci. We propose a model (Fig. 5A) in which, as Pol V proceeds through transcription elongation, AGO4 would interact with Pol V lncRNAs, likely generating a metastable multimeric complex that would serve as an intermediate for the transfer of AGO4 to the DNA template. Following the interaction with the complementary ssDNA, AGO4–siRNA complexes would recruit DRM2 to the opposite siRNA-like DNA strand for DNA methylation. Such a mechanism, suggesting a possible role for DNA in specifying AGO4-dependent DNA methylation, could easily account for the strand-biased nature and high degree of specificity of DNA methylation in RdDM. One could imagine that the pool of AGO4–siRNA effector complexes associated with the AGO hook platforms would directly inspect both DNA strands for complementary base pairing as Pol V proceeds through transcription elongation (Fig. 5B). Further studies will help to answer these fundamental questions.
Figure 5.
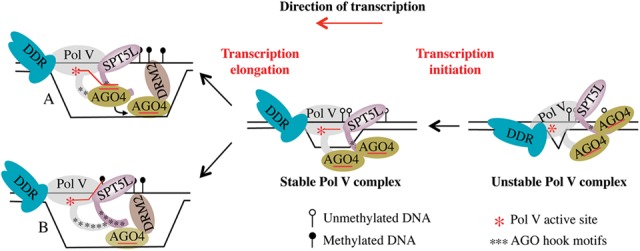
Revisited model for Pol V− and AGO4-dependent RNA-mediated DNA methylation. The Pol V and SPT5L AGO hook platforms are essential determinants of AGO4 recruitment to chromatin at RdDM loci. (Right) The DDR complex interacts with Pol V and triggers a local DNA unwinding that is proposed to facilitate Pol V recruitment at RdDM targets. (Middle) Further stabilization of Pol V on single-stranded target DNA requires active transcription and the formation of a DNA–RNA hybrid in the transcription bubble. (Model A) As Pol V elongates, AGO4 would interact with Pol V lncRNAs, likely generating a metastable multimeric complex that would serve as the intermediate for the transfer of AGO4 to the DNA template. (Model B) Alternatively, one could imagine that the pool of AGO4–siRNA effector complexes associated with the AGO hook platforms would directly inspect both DNA strands for complementary base pairing as Pol V proceeds through transcription elongation. Following the interaction with the complementary ssDNA, AGO4–siRNA complexes would recruit DRM2 to the opposite siRNA-like DNA strand for DNA methylation.
Materials and methods
Establishment of NRPE1 and SPT5L gene constructs, plant material, and growth conditions
NRPE1-WG (Pol V+) and NRPE1-AG (POL V−) were obtained by swapping the amino acid 1258–1708 region of NRPE1 with multimers of wild-type WG or mutant AG consensus motifs (Pontier et al. 2005) in a 2xFlag-containing pCambia 1300 vector. SPT+ and SPT− constructs were based on a genomic fragment, including the SPT5L promoter and ORF with or without the AGO hook platform. Further details and plant material are described in the Supplemental Material.
Protein extraction, immunodetection, and immunoprecipitation
Conventional methods for protein extraction, immunodetection, and immunoprecipitation were used as described in the Supplemental Material.
DNA methylation analysis and whole-genome bisulfite sequencing
Genomic DNA used in the Chop PCR, locus-specific, and whole-genome bisulfite sequencing experiments was extracted from flowers buds. Further details are in the Supplemental Material.
Sequencing data were deposited to the NCBI Gene Expression Omnibus with accession number GSE67216.
ChIP on formaldehyde cross-linked samples
For each plant line, 2 g of inflorescences was used for conventional ChIP experiments. The ChIP procedure was adapted from Wierzbicki et al. (2008) and is detailed in the Supplemental Material.
LChIP
The LChIP procedure was adapted from the conventional ChIP protocol described above. The washed nuclei were resuspended in 25 mL of Honda buffer without Triton. UV laser cross-linking was performed as described in Mutskov et al. (1997). Briefly, nuclei were subjected to a single UV laser pulse (E = 0.2 J/cm2) provided by the fourth harmonic generation (λ = 266 nm) of a nanosecond Nd:YAG laser (Surelite II, Continuum) operating at a 10-Hz repetition rate. To ensure homogenous irradiation, nuclei were irradiated in a laminar flowthrough quartz cuvette at a flow rate of 2 mL/min, corresponding to 35 μL/sec (3.5 μL per pulse), provided by a peristaltic pump. They were then pelleted by centrifugation at 1500g for 15 min, lysed in nucleus lysis buffer, and processed as described above. Immunoprecipitation was performed as for conventional ChIP with slight modifications as described in the Supplemental Material.
Supplementary Material
Acknowledgments
We are grateful to Françoise Moneger and the Laboratoire de Reproduction et Developpement des Plantes at Ecole Normale Supérieure Lyon for providing us with facilities to prepare material for UV cross-link. We also thank C. Picart and J. Azevedo for fruitful discussions and technical help, and Jean-René Pagès for technical assistance. Lagrange laboratory research was supported by the Agence Nationale de la Recherche (ANR; grants 08-BLAN-0206 and 12-BSV6-0010), Centre National de la Recherche Scientifique (CNRS), and Université de Perpignan Via Domitia (UPVD). This work was supported by the European Union's Seventh Framework Programme FP7/2007-2013/COST Action CM1201 “Biomimetic Radical Chemistry” to D.A. This work was supported by National Institutes of Health grant GM60398 (to S.E.J.). S.E.J. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.289553.116.
References
- Altintas DM, Vlaeminck V, Angelov D, Dimitrov S, Samarut J. 2011. Cell cycle regulated expression of NcoR in prostate cells might control cyclic expression of androgen responsive genes. Mol Cell Endocrinol 332: 149–162. [DOI] [PubMed] [Google Scholar]
- Bies-Etheve N, Pontier D, Lahmy S, Picart C, Vega D, Cooke R, Lagrange T. 2009. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep 10: 649–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhmdorfer G, Sethuraman S, Rowley MJ, Krzyszton M, Rothi MH, Bouzit L, Wierzbicki AT. 2016. Long non-coding RNA produced by RNA polymerase V determines boundaries of heterochromatin. Elife 5: e19092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi MA, Jacobsen SE, Cooke R, Lagrange T. 2007. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev 21: 2539–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Pontes O, Pikaard CS. 2009. Metal A and Metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing. PLoS One 4: e4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Ream TS, Marasco M, Nicora CD, Norbeck AD, Pasa-Tolic L, Pikaard CS. 2012. In vitro transcription activities of PolIV, PolV, and RDR2 reveal coupling of PolIV and RDR2 for dsRNA synthesis in plants RNA silencing. Mol Cell 48: 811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman EA, Frey BL, Smith LM, Auble DT. 2015. Formaldehyde crosslinking: a tool for the study of chomatin complexes. J Biol Chem 290: 26404–26411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D, Moazed D. 2015. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJM. 2005. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37: 761–765. [DOI] [PubMed] [Google Scholar]
- Kettenberger H, Armache KJ, Cramer P. 2004. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell 16: 955–965. [DOI] [PubMed] [Google Scholar]
- Lahmy S, Pontier D, Cavel E, Vega D, El-Shami M, Kanno T, Lagrange T. 2009. PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit. Proc Natl Acad Sci 106: 941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahmy S, Bies-Etheve N, Lagrange T. 2010. Plant-specific multisubunit RNA polymerase in gene silencing. Epigenetics 5: 4–8. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SWL, Lagrange T, Pikaard CS, Jacobsen CS. 2006. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126: 93–106. [DOI] [PubMed] [Google Scholar]
- Ma L, Hatlen A, Kelly LJ, Becher H, Wang W, Kovarik A, Leitch IJ, Leitch AR. 2015. Angiosperms are unique among land plant lineage in the occurrence of key genes in the RNA-directed DNA methylation (RdDM) pathway. Genome Biol Evol 7: 2648–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA. 2005. RNAi-mediated pathways in the nucleus. Nat Rev Genet 6: 24–35. [DOI] [PubMed] [Google Scholar]
- Matzke MA, Kanno T, Matzke AJ. 2015. RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu Rev Plant Biol 66: 9.1–9.25. [DOI] [PubMed] [Google Scholar]
- Mutskov V, Angelov D, Pashev I. 1997. Crosslinking proteins to DNA in nuclei by single-pulse UV laser using a flow cuvette. Photochem Photobiol 66: 42–45. [DOI] [PubMed] [Google Scholar]
- Pikaard CS, Haag JR, Pontes OM, Blevins T, Cocklin R. 2012. A transcription fork model for Pol IV and Pol V-dependent RNA-directed DNA methylation. Cold Spring Harb Symp Quant Biol 77: 205–212. [DOI] [PubMed] [Google Scholar]
- Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T. 2005. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerase IV in Arabidopsis. Genes Dev 19: 2030–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, Brennecke J. 2012. Transcriptional silencing of transposons by Piwi and Maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, Moazed D. 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Polydore S, Axtell MJ. 2015. More than meets the eye? Factors that affect target selection by plant miRNAs and heterochromatic siRNAs. Curr Opin Plant Biol 27: 118–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. 2008. Noncoding transcription by RNA polymerase PolIVb/PolV mediates transcriptional silencing of overlapping and adjacent genes. Cell 135: 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. 2009. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet 41: 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Cocklin R, Mayampurath A, Lister R, Rowley MJ, Gregory BD, Ecker JR, Tang H, Pikaard CS. 2012. Spatial and functional relationships among Pol V-associated loci, Pol IV-dependent siRNAs, and cytosine methylation in the Arabidopsis epigenome. Genes Dev 26: 1825–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DL, Zhang G, Tang K, Li J, Yang L, Huang H, Zhang H, Zhu JK. 2016. Dicer-independent RNA-directed DNA methylation in Arabidopsis. Cell Res 26: 66–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Chen Z, Lian B, Rowley MJ, Xia N, Chai J, Li Y, He XJ, Wierzbicki AT, Qi Y. 2016. A dicer-independent route for biogenesis of siRNAs that direct DNA methylation in Arabidopsis. Mol Cell 61: 22–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Henikoff S. 2014. High-resolution digital profiling of the epigenome. Nat Rev Genet 15: 814–827. [DOI] [PubMed] [Google Scholar]
- Zhong X, Hale CJ, Law JA, Johnson LM, Feng S, Tu A, Jacobsen SE. 2012. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol 19: 870–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Du J, Hale CJ, Gallego-Bartolome J, Feng S, Vashisht AA, Chory J, Wohlschlegel JA, Patel DJ, Jacobsen SE. 2014. Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157: 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


