In this study, Luo et al. investigated the regulation of the BRCA2–RAD51 axis in homologous recombination (HR) during DNA double-strand break (DSB) repair. The authors identify UCHL3 as a novel regulator of DNA repair and propose a model in which a phosphorylation–deubiquitination cascade dynamically regulates the BRCA2–RAD51 pathway.
Keywords: BRCA2, DNA damage response, homologous recombination, Rad51, UCHL3, deubiquitination
Abstract
Homologous recombination (HR) is one of the major DNA double-strand break (DSB) repair pathways in mammalian cells. Defects in HR trigger genomic instability and result in cancer predisposition. The defining step of HR is homologous strand exchange directed by the protein RAD51, which is recruited to DSBs by BRCA2. However, the regulation of the BRCA2–RAD51 axis remains unclear. Here we report that ubiquitination of RAD51 hinders RAD51–BRCA2 interaction, while deubiquitination of RAD51 facilitates RAD51–BRCA2 binding and RAD51 recruitment and thus is critical for proper HR. Mechanistically, in response to DNA damage, the deubiquitinase UCHL3 is phosphorylated and activated by ATM. UCHL3, in turn, deubiquitinates RAD51 and promotes the binding between RAD51 and BRCA2. Overexpression of UCHL3 renders breast cancer cells resistant to radiation and chemotherapy, while depletion of UCHL3 sensitizes cells to these treatments, suggesting a determinant role of UCHL3 in cancer therapy. Overall, we identify UCHL3 as a novel regulator of DNA repair and reveal a model in which a phosphorylation–deubiquitination cascade dynamically regulates the BRCA2–RAD51 pathway.
Genome integrity is under constant attack from exogenous and endogenous DNA-damaging factors such as radiation, carcinogens, reactive radicals, and errors in DNA replication. To maintain genomic stability, cells have developed an elaborate DNA damage response (DDR) system to detect, signal, and repair the DNA lesions (Downs et al. 2007; Ciccia and Elledge 2010; Huen and Chen 2010; Lukas et al. 2011). Defects in the DDR pathway lead to genome instability syndromes that are associated with cancer, stem cell exhaustion, developmental defects, infertility, immune deficiency, neurodegenerative disease, and premature aging (Kastan and Bartek 2004; Jackson and Bartek 2009).
In cells, double-strand breaks (DSBs) are typically repaired by end-joining or homologous recombination (HR) (Warmerdam and Kanaar 2010; Chapman et al. 2012). DSB end-joining includes classical nonhomologous end-joining (NHEJ) and alternative NHEJ, which result in quick but error-prone repair (Critchlow and Jackson 1998; Lieber et al. 2004; Chiruvella et al. 2013). Unlike end-joining-mediated DNA repair, HR uses an intact sister chromatid as the template, which makes HR more accurate but limited to the S/G2 phases of the cell cycle. The initial step of HR, DNA resection, is regulated by the MRN complex and CtIP and produces short 3′ overhangs (Paull and Gellert 1998; Sartori et al. 2007; Takeda et al. 2007). The 3′ overhangs are extended through further resection by Exo1 and Dna2 nucleases (Zhu et al. 2008; Mimitou and Symington 2009; Nimonkar et al. 2011). Once ssDNA is generated, it is rapidly bound by the ssDNA-binding protein RPA (replication protein A) and subsequently replaced by RAD51, leading to strand invasion and ensuing HR processes (West 2003; San Filippo et al. 2008). Although the process of HR and end-joining are extensively studied, the regulation of these pathways and their coordination to complete the repair of DSBs remain unclear.
Protein ubiquitination plays an important role in the DDR pathway (Lukas et al. 2011; Jackson and Durocher 2013). For instance, RNF8/RNF168-dependent ubiquitination promotes the recruitment of DSB repair and signaling factors on chromatin surrounding the DNA lesion, which facilitates the DDR process (Huen et al. 2007; Kolas et al. 2007; Mailand et al. 2007; Doil et al. 2009; Mattiroli et al. 2012), while the SUMO targeted ubiquitin (Ub) ligase RNF4 promotes the turnover of MDC1 and RPA from chromatin during the DSB response (Galanty et al. 2012; Luo et al. 2012, 2015; Yin et al. 2012). Multiple recent studies suggest that editing and removal of ubiquitination by deubiquitinases (DUBs) play important roles in regulating ubiquitination events. For example, UCHL5 promotes DNA resection in an EXO-1- and BLM-dependent manner, and USP3 and USP16 are associated with negative regulation of the RNF8 pathway through their ability to oppose H2A ubiquitination (Doil et al. 2009; Shanbhag et al. 2010).
Here we carried out a systematic screen of DUBs for HR and established that UCHL3 interacts with and deubiquitinates RAD51 and promotes binding between RAD51 and BRCA2. Moreover, overexpression of UCHL3 in breast cancer is correlated with poor survival of breast cancer patients and resistance to radiation and chemotherapy. Finally, we clarify a dynamic regulation of the BRCA2–RAD51 axis that is important for HR and may provide new therapeutic targets for overcoming resistance in breast cancer.
Results
UCHL3 regulates RAD51 IR-induced focus formation (IRIF) and HR
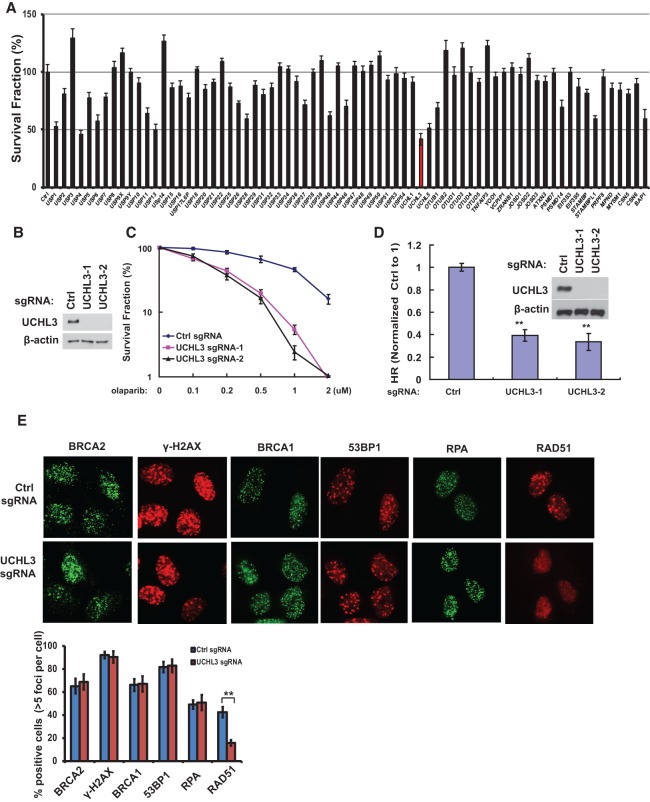
We performed a targeted shRNA library screen of DUBs for their role in HR using PARP inhibitor (PARPi), as PARPi sensitivity is associated with defective HR (Bryant et al. 2005; Farmer et al. 2005). We found that depletion of several DUBs, including UCHL3, resulted in hypersensitivity to olaparib, a PARPi (Fig. 1A). Among them, USP1, USP4, USP11, and UCHL5 have been associated with HR (Wiltshire et al. 2010; Murai et al. 2011; Nishi et al. 2014; Liu et al. 2015; Orthwein et al. 2015; Wijnhoven et al. 2015). However, one of the hits, UCHL3, has not been implicated in DNA repair. UCHL3 is a deubiquitination enzyme that is involved in the processing of both Ub precursors (Grou et al. 2015) and poly-Ub chain from substrates (Roff et al. 1996; Misaghi et al. 2005; Kim et al. 2011) and has been implicated in several cellular processes, such as oocyte maturation, spermatogenesis, osteoblast differentiation, lipogenesis, retinal degeneration, and epithelial-to-mesenchymal transition (Sano et al. 2006; Yi et al. 2007; Suzuki et al. 2009; Kim et al. 2011; Mtango et al. 2012a,b; van Beekum et al. 2012; Song et al. 2014). However, the mechanism of UCHL3's functions remains unclear, as only a limited number of UCHL3 targets were identified. To confirm our screening results, we generated UCHL3 knockout U2OS cells using CRISPR technology and tested cellular sensitivity to PARPi (Fig. 1B,C). Consistent with our initial screening results, UCHL3-deficient cells showed hypersensitivity to olaparib. Because PARPi sensitivity is highly associated with defective HR, we next tested the role of UCHL3 in HR using the well-established DR-GFP reporter system (Pierce et al. 1999). As shown in Figure 1D, UCHL3 deficiency resulted in decreased HR. We did not find significant changes in the cell cycle profile when we deleted the UCHL3 gene (Supplemental Fig. S1A), suggesting that the observed effect was not due to an indirect effect of a change in the cell cycle profile. These results suggest that UCHL3 is important for HR.
Figure 1.
UCHL3 regulates RAD51 IRIF and HR. (A) A panel of DUBs was knocked down in U2OS cells, and cellular response to 1 μM olaparib was assayed using colony formation assay. Error bars represent SEM from three independent experiments. (B) UCHL3 was knocked out in U2OS cells by CRISPR, and Western blot was performed with the indicated antibodies. (C) The sensitivity of control and UCHL3 knockout U2OS cells to olaparib was assessed using colony formation assay. Error bars represent SEM from three independent experiments. (D) The HR-mediated DSB repair capacity of control (Ctrl) and UCHL3 knockout U2OS cells was assessed using a reporter system. Error bars represent SEM from three independent experiments. Statistical significance was determined by ANOVA. (**) P < 0.01. (E) Control or UCHL3 knockout U2OS cells were treated with ionizing radiation (IR), and focus formation of the indicated factors was detected by immunofluorescence. Representative images are shown in the top panel. Quantification of the percentage of cells displaying foci is shown in the bottom panel. Error bars represent SEM from three independent experiments. Statistical significance was determined by two-tailed Student's t-test. (**) P < 0.01.
To identify possible targets of UCHL3 in HR, we first examined focus formation of several DDR factors. Interestingly, we found that UCHL3 deficiency resulted in compromised RAD51 focus formation (Fig. 1E; Supplemental Fig. S1B,C) but did not affect focus formation of upstream regulators of RAD51, such as γ-H2AX, MDC1, RPA, BRCA1, PALB2, and BRCA2 (Fig. 1E; Supplemental Fig. S1C,D). Since focus formation of BRCA1 and 53BP1 also depends on Ub signaling (Jackson and Durocher 2013), these results also suggest that the overall Ub signaling is not affected by UCHL3 deficiency. In addition, we found that UCHL3 itself was recruited to DSBs and colocalized with RAD51 and γ-H2AX (Supplemental Fig. S2A,B). Moreover, we observed accumulation of UCHL3 at the DSB by a chromatin immunoprecipitation (ChIP) assay in which the DSB was introduced by the exogenously expressed I-SceI endonuclease (Supplemental Fig. S2C). Consistent with the immunofluorescence results, ChIP assays revealed that RAD51 recruitment to DSBs was dramatically decreased in UCHL3 knockout cells (Supplemental Fig. S2D), suggesting that UCHL3 is important for RAD51 recruitment to DSBs. Based on these results, we hypothesized that UCHL3 regulates HR through its effect on RAD51.
UCHL3 interacts with and deubiquitinates RAD51
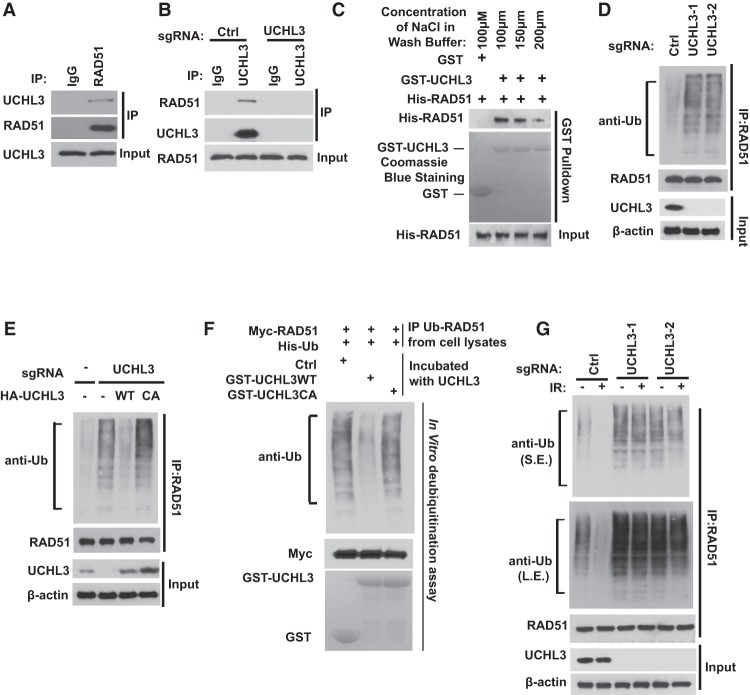
To test our hypothesis, we first examined whether UCHL3 interacts with RAD51. Indeed, we found that endogenous RAD51 coimmunoprecipitated with UCHL3 (Fig. 2A). Reciprocal immunoprecipitation with UCHL3 antibody also brought down RAD51 but failed to do so in UCHL3 knockout cells (Fig. 2B). In addition, UCHL3 interacted with RAD51 when it formed a nucleoprotein filament with ssDNA (Supplemental Fig. S3A). These results suggest a specific interaction between UCHL3 and RAD51 in cells. To determine whether the UCHL3–RAD51 interaction is direct, we generated and purified recombinant UCHL3 and RAD51. Purified His-RAD51 was able to interact with GST-UCHL3 under cell-free conditions even with a relatively high salt concentration in the washing buffer (Fig. 2C), suggesting a direct interaction between UCHL3 and RAD51.
Figure 2.
UCHL3 interacts with and deubiquitinates RAD51. (A) U2OS cell lysates were subjected to immunoprecipitation with control IgG or RAD51 antibodies. The immunoprecipitates were then blotted with the indicated antibodies. (B) Cell lysates from U2OS or UCHL3 knockout cells were subjected to immunoprecipitation with control IgG or UCHL3 antibodies. The immunoprecipitates were then blotted with the indicated antibodies. (C) Purified recombinant GST or GST-UCHL3 on GSH beads were incubated with His-RAD51 in vitro followed by washing with buffers with different salt concentrations. The interaction between UCHL3 and RAD51 was then examined as indicated. (D) U2OS cells or UCHL3-deficient U2OS cells were lysed under denaturing conditions, and RAD51 was immunoprecipitated. Blots were probed with the indicated antibodies. Note that, to make easy comparisons, the loading of immunoprecipitated RAD51 was equalized for D–G. (E) Control cells, UCHL3 knockout cells, and UCHL3 knockout cells reconstituted with the indicated constructs were lysed under denaturing conditions, and RAD51 was immunoprecipitated. Blots were probed with the indicated antibodies. (F) Ubiquitinated Myc-RAD51 was incubated with purified UCHL3 or catalytically inactive UCHL3 (CA) in vitro and then blotted with the indicated antibodies. (G) Control or UCHL3 knockout U2OS cells were treated with 10 Gy of IR. After 1 h, cells were lysed under denaturing conditions, and RAD51 was immunoprecipitated. Blots were probed with the indicated antibodies. (S.E.) Short exposure; (L.E.) long exposure.
Since UCHL3 is a Ub-specific protease that functions to cleave Ub from its substrates, we next tested whether UCHL3 regulates RAD51 ubiquitination. Indeed, we observed increased RAD51 polyubiquitination in UCHL3-deficient cells (Fig. 2D; Supplemental Fig. S3B). Interestingly, RAD51 protein stability did not change when we modulated UCHL3 expression (data not shown). These results suggest that UCHL3 might affect RAD51 function but not stability through its DUB activity. Reconstituting wild-type UCHL3 but not catalytically inactive UCHL3 (CA) reversed the increase in RAD51 ubiquitination induced by UCHL3 deficiency (Fig. 2E), suggesting that UCHL3 regulates RAD51 ubiquitination through its Ub protease activity.
To determine whether UCHL3 directly deubiquitinates RAD51, we performed an in vitro deubiquitination assay. We purified recombinant wild-type UCHL3 and the UCHL3 CA mutant from Escherichia coli and ubiquitinated RAD51 from cells expressing Myc-RAD51 and His-Ub. We then incubated UCHL3 and ubiquitinated RAD51 in a cell-free system. We found that wild-type UCHL3 but not the UCHL3 CA mutant dramatically deubiquitinated RAD51 in vitro (Fig. 2F). Taken together, these results suggest that UCHL3 deubiquitinates RAD51 both in vitro and in vivo. Interestingly, we also found that RAD51 ubiquitination decreased following DNA damage (Fig. 2G, cf. lanes 1 and 2). In contrast, in UCHL3-deficient cells, basal RAD51 ubiquitination increased and did not decrease following DNA damage (Fig. 2G). Collectively, these results suggest that UCHL3 promotes RAD51 deubiquitination following DNA damage.
UCHL3 regulates HR and radiosensitivity in a RAD51-dependent manner
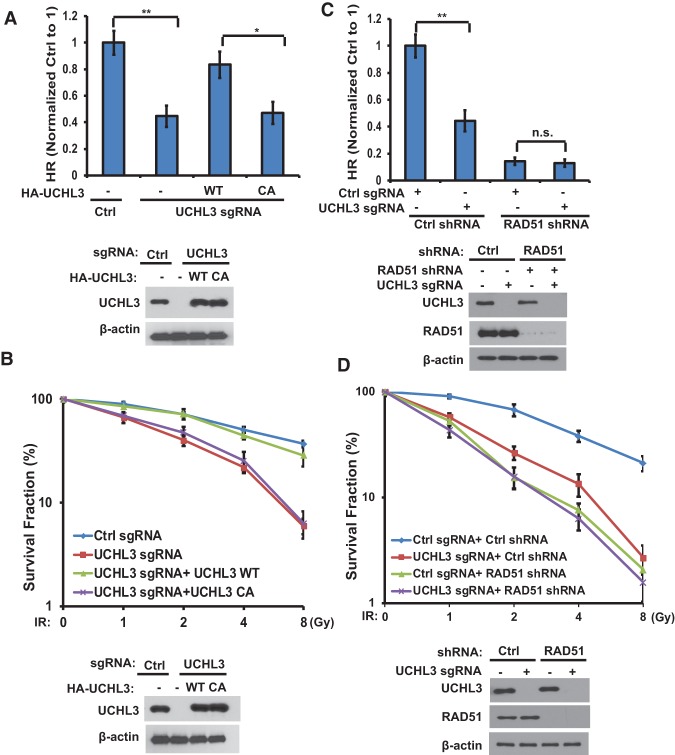
To further confirm that the regulation of HR and DDR by UCHL3 is dependent on its catalytic activity and RAD51, we reconstituted UCHL3-deficient cells with wild-type UCHL3 or UCHL3 CA. As shown in Figure 3, A and B, knocking out UCHL3 in U2OS cells dramatically decreased HR and rendered cells sensitive to ionizing radiation (IR). Reconstitution of wild-type UCHL3 but not the UCHL3 CA mutant rescued these phenotypes, suggesting that the catalytic activity of UCHL3 is important for its regulation of HR and DDR. Consistent with results from human cell lines, mouse embryonic fibroblasts (MEFs) from UCHL3 knockout mice also showed hypersensitivity to IR treatment (Supplemental Fig. S4A). Furthermore, knocking out UCHL3 did not further affect HR and radiosensitivity in RAD51-depleted cells (Fig. 3C,D), suggesting that UCHL3 regulates HR and DDR in a RAD51-dependent manner.
Figure 3.
UCHL3 regulates HR and radiosensitivity through RAD51. (A) The HR-mediated DSB repair capacity of control cells, UCHL3 knockout cells, and UCHL3 knockout cells stably expressing the indicated constructs was assessed using a reporter system. (B) The sensitivity of control cells, UCHL3 knockout cells, and UCHL3 knockout cells stably expressing the indicated constructs to IR was assessed using colony formation assay. (C) Control and UCHL3 knockout U2OS cells stably expressing control or RAD51 shRNA were subjected to HR assay. (D) The sensitivity of control cells and UCHL3 knockout U2OS cells stably expressing control or RAD51 shRNA to IR was assessed using colony formation assay. (A–D) Error bars represent SEM from three independent experiments. Statistical significance was determined by two-tailed Student's t-test, (*) P < 0.05; (**) P < 0.01; (n.s.) no significant difference.
Deubiquitination of RAD51 by UCHL3 is important for HR
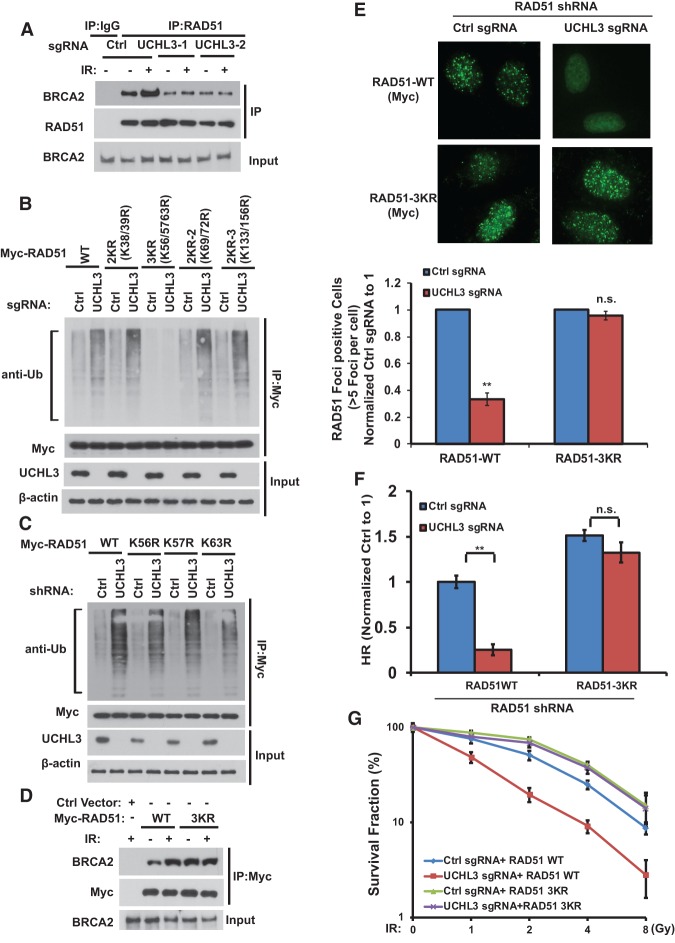
We show that UCHL3 is important for RAD51 deubiquitination and recruitment to DSBs but is not important for BRCA1, PALB2, and BRCA2 recruitment (Figs. 1, 2; Supplemental Fig. S1). In unstressed cells, we observed that ∼40% of RAD51 was ubiquitinated (Supplemental Fig. S5A). Following DNA damage, UCHL3 promotes RAD51 deubiquitination (Figs. 1, 2). It is possible that RAD51 deubiquitination by UCHL3 is important for its recruitment to DSBs. It is well established that the interaction between BRCA2 and RAD51 is critical for the recruitment of RAD51 to the DNA damage sites (Mizuta et al. 1997; Wong et al. 1997; Chen et al. 1998a,b; Moynahan et al. 2001; Galkin et al. 2005). We hypothesized that UCHL3 may regulate the RAD51–BRCA2 interaction. As shown in Figure 4A, the binding between RAD51 and BRCA2 increased after IR. Strikingly, in UCHL3-deficient cells, the basal RAD51–BRCA2 interaction was weak and could not be induced by IR. To directly test how RAD51 deubiquitination affects its binding to BRCA2, we deubiquitinated RAD51 by UCHL3 in vitro and examined its interaction with the BRCA2-BRC4 fragment. We found that deubiquitination of RAD51 increased the binding between RAD51 and BRCA2 (Supplemental Fig. S5B). On the other hand, knocking out UCHL3 in cells did not affect RAD51/RAD52 interaction (Supplemental Fig. S5C). These results suggested that deubiquitination of RAD51 by UCHL3 following DNA damage is important for the interaction between RAD51 and BRCA2.
Figure 4.
Deubiquitination of RAD51 by UCHL3 is important for HR. (A) Control or UCHL3 knockout U2OS cells were treated with 10 Gy of IR. After 1 h, cells were lysed, and lysates were subjected to immunoprecipitation with control IgG or RAD51 antibodies. Blots were probed with the indicated antibodies. (B,C) Control or UCHL3 knockout cells were transfected with the indicated constructs. After 48 h, cells were lysed under denaturing conditions, and Myc-RAD51 was immunoprecipitated. Blots were probed with the indicated antibodies. The loading of immunoprecipitated RAD51 was equalized. (D) U2OS cells stably expressing Myc-RAD51 (wild type or 3KR) constructs were treated with 10 Gy of IR. One hour later, cell lysates were subjected to immunoprecipitation with Myc-tagged antibodies. Blots were probed with the indicated antibodies. (E) Control cells or UCHL3 knockout U2OS cells stably expressing RAD51 shRNA together with shRNA-resistant wild-type or 3KR Myc-RAD51 were treated with IR, and Myc-RAD51 focus formation was detected by immunofluorescence. Representative images are shown in the top panel. Quantification of the percentage of cells displaying foci is shown in the bottom panel. (F) The cells shown in E were subjected to DR-GFP-based HR assay. (G) The sensitivity of cells shown in E to IR was assessed using colony formation assay. (E,G) Error bars represent SEM from three independent experiments. Statistical significance was determined by two-tailed Student's t-test. (**) P < 0.01; (n.s.) no significant difference.
We next mapped potential ubiquitination sites of RAD51 that are regulated by UCHL3. Previous studies have suggested that RAD51 binds to BRCA2 through its N-terminal domain (NTD) and the core domain (Pellegrini et al. 2002; Subramanyam et al. 2013). After analyzing the RAD51 NTD and the core domain sequences, we found nine lysine residues that are conserved in vertebrates, zebrafish, and Xenopus (Supplemental Fig. S6A). We generated combination mutations of RAD51 (mutant K to R) and performed a deubiquitination assay. As shown in Figure 4B, loss of UCHL3 dramatically increased ubiquitination of wild-type RAD51 but not the 3KR (K56/57/63R) mutant with three N-terminal lysines mutated. Combination mutations on other portions of RAD51 had no significant effect on RAD51's ubiquitination in UCHL3-deficient cells (Fig. 4B), suggesting that the three lysines at the N terminus of RAD51 are major ubiquitination sites that are regulated by UCHL3. We next generated single mutations at these sites and found that a single mutation did not significantly affect RAD51 ubiquitination induced by UCHL3 deficiency (Fig. 4C). These results suggested that all three residues at the N terminus of RAD51 are key deubiquitination sites regulated by UCHL3.
Next, we investigated the functional significance of RAD51 ubiquitination/deubiquitination using wild-type RAD51 and the 3KR mutant. As shown in Supplemental Figure S7A, both wild-type RAD51 and the 3KR mutant promoted D-loop formation in vitro, suggesting that RAD51 3KR is still active in vitro. Furthermore, wild-type RAD51 and the 3KR mutant bound equally with UCHL3, suggesting that RAD51 deubiquitination does not affect RAD51/UCHL3 interaction (Supplemental Fig. S7B). As shown in Figure 4D, compared with wild-type RAD51, the 3KR mutant showed higher basal binding efficiency with BRCA2. In addition, unlike wild-type RAD51, IR treatment could not further increase the interaction between the 3KR mutant and BRCA2. Furthermore, the 3KR mutant enhanced the RAD51/BRCA2 interaction in UCHL3-deficient cells (Supplemental Fig. S7C). Collectively, these results suggest that RAD51 deubiquitination is critical for the BRCA2–RAD51 interaction. Next, we examined whether RAD51 deubiquitination regulates its focus formation. As shown in Supplemental Figure S8A, wild-type RAD51 forms foci normally in response to DNA damage. However, the focus formation of wild-type RAD51 was compromised in UCHL3-deficient cells. In contrast, the focus formation of the 3KR mutant was normal in UCHL3-deficient cells in response to DNA damage (Fig. 4E). The RAD51 3KR mutant did not form foci in the absence of DNA damage (Supplemental Fig. S8B), suggesting no premature DDR in the absence of RAD51 ubiquitination. Finally, we found that the 3KR mutant but not wild-type RAD51 was able to rescue HR deficiency and radiosensitivity induced by UCHL3 depletion (Fig. 4F,G). These results suggest that deubiquitination of RAD51 by UCHL3 on Lys56, Lys57, and Lys63 is important for RAD51–BRCA2 interaction, RAD51's recruitment to DSB sites, and DDR.
Regulation of UCHL3 by DDR signaling
Because UCHL3 promotes RAD51 deubiquitination following DNA damage, we further investigated whether and how UCHL3 itself is regulated following DNA damage. ATM and ATR are critical kinases in the DDR pathway and regulate DNA damage signaling by phosphorylating and activating downstream signaling networks. ATM and ATR substrate protein network proteomic data analysis showed that UCHL3 may be a potential ATM/ATR substrate (Matsuoka et al. 2007). To test whether UCHL3 could be phosphorylated by ATM/ATR, we examined UCHL3 phosphorylation using an antibody against the consensus of ATM/ATR phosphorylation sites (anti-phospho-SQ/TQ [anti-pSQ/TQ]) following DNA damage. As shown in Figure 5A, UCHL3 was phosphorylated at the SQ/TQ motif following IR treatment, and this phosphorylation was blocked by an ATM-specific inhibitor (KU55933) and λ phosphatase treatment. Furthermore, UCHL3 was phosphorylated at the SQ/TQ motif in ATM+/+ but not ATM−/− cells (Fig. 5B). These results suggest that UCHL3 is phosphorylated by ATM following DNA damage. Analysis of UCHL3 protein sequence revealed only one SQ/TQ motif: S75. Mutation of S75 (S75A) abolished the pSQ/TQ signal, suggesting that S75 is a major ATM phosphorylation site following DNA damage (Supplemental Fig. S9A). To confirm these results, we generated a site-specific antibody against p-Ser75. As shown in Figure 5C, S75 was phosphorylated after DNA damage, while the S75A mutation inhibited DNA damage-induced UCHL3 phosphorylation. These results suggest that Ser75 is the physiological phosphorylation site for ATM in vivo. To further demonstrate a potential function of UCHL3 phosphorylation, we reconstituted UCHL3-deficient cells with wild-type UCHL3 or the UCHL3 S75A mutant. Re-expression of wild-type UCHL3 restored RAD51 IRIF, while expression of the S75A mutant did not (Fig. 5D; Supplemental Fig. S9B). Furthermore, reconstitution of wild-type UCHL3 but not the S75A mutant rescued HR-mediated DNA repair (Fig. 5E). Finally, re-expression of wild-type UCHL3 but not the S75A mutant was able to reverse hypersensitivity to IR induced by UCHL3 deficiency (Fig. 5F). Together, these results suggest that UCHL3 phosphorylation by ATM is important for RAD51 focus formation, HR, and radiosensitivity.
Figure 5.
Regulation of UCHL3 by DDR signaling. (A) HEK293T cells transfected with HA-UCHL3 were pretreated with DMSO or 25 μM Ku55933 for 2 h followed by mock treatment or 10 Gy of IR. After an additional 1 h, HA-UCHL3 was immunoprecipitated, left untreated or treated with phosphatase, and immunoblotted with pSQ/TQ antibody. (B) ATM+/+ or ATM−/− cells were irradiated (10 Gy) or left untreated. After 1 h, UCHL3 was immunoprecipitated, and blots were probed with the indicated antibodies. (C) HEK293T cells transfected with HA-UCHL3 (wild-type or S75A) were left untreated or irradiated (10 Gy). HA-UCHL3 was immunoprecipitated, and blots were probed with the indicated antibodies. (D) U2OS cells stably expressing HA-UCHL3 (wild-type or S75A) were irradiated, and γ-H2AX and RAD51 focus formation was detected by immunofluorescence. The quantification of cells positive for both γ-H2AX and RAD51 focus formation is shown in the bottom panel. Results were normalized to UCHL3 wild-type cells. Error bars represent SEM from three independent experiments. Statistical significance was determined by two-tailed Student's t-test. (**) P < 0.01. (E) Control cells, UCHL3 knockout cells, and UCHL3 knockout cells stably expressing the indicated constructs were subjected to DR-GFP-based HR assay. UCHL3 expression is shown in the left panel. Error bars represent SEM from three independent experiments. Statistical significance was determined by ANOVA. (*) P < 0.05; (**) P < 0.01. (F) The response of control cells, UCHL3 knockout cells, and UCHL3 knockout U2OS cells stably expressing the indicated constructs to IR was assessed using colony formation assay. UCHL3 expression is shown in the left panel. Error bars represent SEM from three independent experiments. (G) Cells transfected with control, HA-UCHL3 wild type, or the S75A mutant were irradiated (10 Gy). Cell lysates were subjected to immunoprecipitation with HA antibody. Blots were probed with the indicated antibodies. (H) UCHL3 knockout U2OS cells stably expressing the indicated constructs were left untreated or treated with 10 Gy of IR. Cells were lysed under denaturing conditions. RAD51 was immunoprecipitated, and blots were probed with the indicated antibodies. The loading was equalized with immunoprecipitated RAD51. (I) Bacterially expressed GST-UCHL3 wild-type or the S75A mutant was subjected to in vitro kinase reaction with ATM and then used in in vitro deubiquitination reaction with ubiquitinated Myc-RAD51. The reactions were then blotted with the indicated antibodies.
Next, we further investigated how UCHL3 phosphorylation affects its function. We found that S75A mutation did not affect the interaction between UCHL3 and RAD51 (Fig. 5G). Because S75 is located in the catalytic domain of UCHL3, we next examined whether UCHL3 phosphorylation regulates its activation. Wild-type UCHL3 or the S75A mutant was reconstituted in UCHL3-deficient cells, and UCHL3 activity was examined by detecting RAD51 ubiquitination in cells. As shown in Figure 5H, RAD51 ubiquitination was dramatically decreased following IR treatment in cells reconstituted with wild-type UCHL3 but not in those reconstituted with the S75A mutant, suggesting that UCHL3 phosphorylation is important for UCHL3's deubiquitination of RAD51 following DNA damage. RAD51 ubiquitination was equivalent in undamaged cells expressing either wild-type UCHL3 or the S75A mutant, suggesting that the basal activity of UCHL3 is not affected by the S75A mutation. To further test whether UCHL3 phosphorylation is able to activate UCHL3, we performed a two-step assay. We performed an in vitro kinase reaction of UCHL3 by ATM followed by an in vitro deubiquitination assay. Both wild-type UCHL3 and the S75A mutant could deubiquitinate RAD51 in vitro, again suggesting a basal deubiquitination activity of UCHL3 that is not regulated by UCHL3 phosphorylation. This also suggests that the S75 mutation does not abolish the UCHL3 catalytic activity. Interestingly, wild-type UCHL3 after ATM phosphorylation showed increased deubiquitination activity, while the S75A mutant did not have this effect (Fig. 5I). These results demonstrate that UCHL3 phosphorylation by ATM is important for UCHL3 activation following DNA damage. This mechanism can also account for increased RAD51 deubiquitination and BRCA2–RAD51 interaction.
The role of UCHL3 in response to PARP inhibition and irradiation
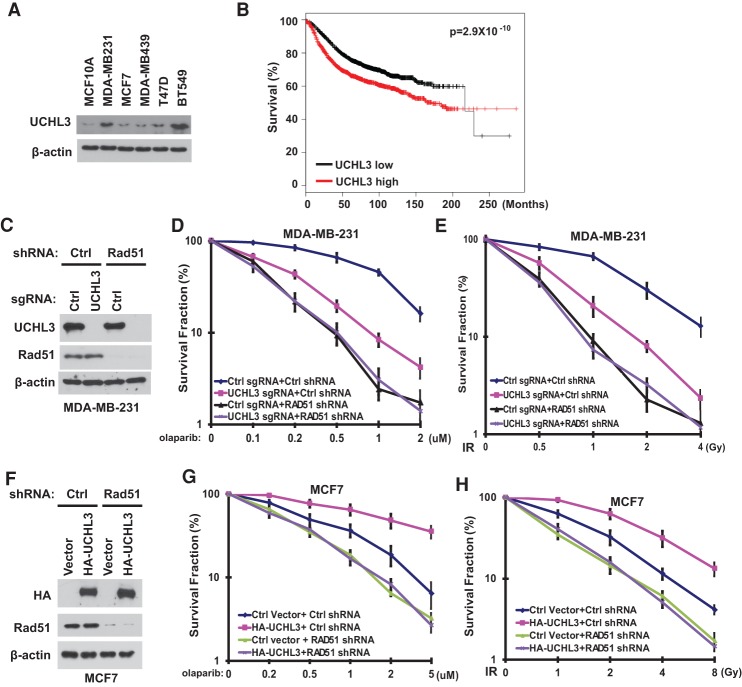
The DDR pathway is important for DNA repair. In addition, many studies suggest that the status of the DDR pathway affects cancer cells’ response to radiation and chemotherapy. Deficiency in the HR pathway (i.e., BRCA1 mutation) renders cells sensitive to cross-linking agents and PARPi. In contrast, enhanced DNA repair capability (e.g., RAD51 overexpression in breast cancer) renders cells resistant to these treatments (Klein 2008). Since UCHL3 deubiquitinates RAD51 and regulates HR, we next investigated the role of UCHL3 in cancer. We blotted for UCHL3 protein levels in normal breast epithelial and breast cancer cell lines and found that UCHL3 is overexpressed in several breast cancer cell lines (Fig. 6A). To examine whether UCHL3 overexpression can affect patient response to cancer therapy, we first explored a public database (Kaplan-Meier Plotter, http://kmplot.com; Gyorffy et al. 2013). Interestingly, we found that increased expression of UCHL3 correlates with poor survival of breast cancer patients (Fig. 6B). We hypothesized that increased UCHL3 expression in breast cancer cells would increase DNA repair capability and render cells resistant to cancer therapy. To test this hypothesis, we generated UCHL3 knockout MDA-MB-231 and BT549 cell lines (Fig. 6C; Supplemental Fig. S10A), which highly express UCHL3. We found that disruption of the UCHL3 gene rendered both MDA-MB-231 and BT549 cells sensitive to PARPi (Fig. 6D; Supplemental Fig. S10B). However, knocking out UCHL3 in RAD51 knockdown cells did not further sensitize cells to PARPi (Fig. 6D; Supplemental Fig. S10B). Furthermore, we examined radiosensitivity of breast cancer cells, as radiation is used as an adjuvant therapy for breast cancer. As shown in Figure 6E, UCHL3 deficiency rendered MDA-MB-231 cells more sensitive to IR in parental MDA-MB-231 cells but not in RAD51-depleted cells. Similar results were obtained when we used BT549 cells (Supplemental Fig. S10C). These results demonstrate that UCHL3 regulates breast cancer cell response to radiation and chemotherapy in a RAD51-dependent manner. Conversely, we overexpressed UCHL3 in UCHL3-low MCF7 cells (Fig. 6F). As shown in Figure 6, G and H, overexpression of UCHL3 in MCF7 cells rendered cells resistant to radiation and PARPi treatment. However, overexpression of UCHL3-mediated resistance to radiation and PARPi was abolished in RAD51-depleted cells (Fig. 6G,H). Taken together, our results demonstrate that UCHL3 may be a causal factor of cancer cell response to radiation and chemotherapy.
Figure 6.
The role of UCHL3 in response to PARP inhibition and irradiation. (A) Expression of UCHL3 in human breast epithelial cell lines and breast carcinoma cell lines. (B) Correlation between 3554 cases of breast cancer patient survival and UCHL3 expression. (C–E) Survival assays for control and UCHL3 knockout MDA-MB-231 cells stably expressing control or RAD51 shRNA exposed to olaparib (D) and IR (E). (C) UCHL3 and RAD51 expression was detected by Western blot. (F–H) Survival assays for control or RAD51-depleted MCF7 cells stably expressing HA-UCHL3 exposed to olaparib (G) and IR (H). (F) HA-UCHL3 and RAD51 expression was detected by Western blot. (D,E,G,H) Error bars represent SEM from three independent experiments.
Discussion
RAD51, a strand exchange protein, is a central player in HR (San Filippo et al. 2008). The loading of RAD51 to ssDNA is a critical step during HR and is regulated by BRCA2 and RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3) (Cromie et al. 2001). Recent studies have provided more mechanisms for how BRCA2 interacts with and affects RAD51 function (Roy et al. 2012). BRCA2 interacts with RAD51 through two domains: the BRC repeat domain and C-terminal domain (Roy et al. 2012). The BRC repeat domain exhibits multiple capacities for RAD51 interaction (Carreira and Kowalczykowski 2011). Besides facilitating the recruitment of RAD51 to ssDNA and nucleoprotein filament formation (Pellegrini et al. 2002), the BRC repeat domain of BRCA2 also accelerates RPA displacement from ssDNA by RAD51 and blocks RAD51 nucleation at dsDNA (Wooster et al. 1995; Carreira et al. 2009; Shivji et al. 2009). Besides the BRC domain, BRCA2 also interacts with RAD51 through its C-terminal domain (Davies and Pellegrini 2007; Esashi et al. 2007). The binding of RAD51 by the C terminus of BRCA2 is dependent on CDK activity in a cell cycle-dependent fashion (Esashi et al. 2005). However, this interaction appears to be important for the rapid focus disassembly of RAD51 complexes and mitotic entry (Ayoub et al. 2009). A previous study also suggested that the interaction between BRCA2 and RAD51 is regulated by localization, as DNA damage can induce redistribution of soluble nucleoplasmic BRCA2 available for RAD51 binding (Jeyasekharan et al. 2010). However, whether and how the BRCA2–RAD51 interaction is regulated following DDR is still unclear. We found that, in unstressed cells, ubiquitination of RAD51 hinders the interaction between BRCA2 and RAD51. Following DNA damage, a DUB, UCHL3, deubiquitinates RAD51 and promotes binding between RAD51 and BRCA2 and the recruitment of RAD51 to DSBs, which in turn facilitates HR. This reveals a new regulatory mechanism for activating RAD51.
Post-translational modification (PTM) of RAD51 is important for its function in DNA repair and tumor radiosensitivity (Flott et al. 2011; Krejci et al. 2012). For example, Chk1-mediated phosphorylation of RAD51 has been shown to be important for efficient HR (Sorensen et al. 2005), while Mec1-mediated phosphorylation of RAD51 is required for its ATPase and DNA-binding activities (Flott et al. 2011). Phosphorylation of Tyr315 by BCR/ABL is important for DSB repair and drug resistance, while phosphorylation of Tyr54 by c-Abl inhibits RAD51 binding to DNA and its ATP-dependent DNA strand exchange reaction (Yuan et al. 1998; Slupianek et al. 2011). FBH1-mediated RAD51 monoubiquitination influences DNA replication fork stability and plays an important role in DNA replication stress (Chu et al. 2015). However, the role of ubiquitination/deubiquitination of RAD51 in DSBs is unclear. Here, we identified polyubiquitination of RAD51 as a negative regulatory mechanism not through the regulation of protein stability but through the regulation of its interaction with BRCA2. Ubiquitination/deubiquitination regulates multiple cellular functions in DDR, such as signaling transduction and proteasome-mediated protein degradation (Jackson and Durocher 2013; Brown and Jackson 2015). During signal transduction, it is generally thought that ubiquitination acts as a dock to mediate protein–protein interaction. However, ubiquitination-mediated inhibition of protein–protein interaction is less studied. Recently, it was reported that PALB2 polyubiquitination blocks its interaction with BRCA1 (Orthwein et al. 2015). Our study suggests a parallel mode of regulation for the modulation of BRCA2–RAD51 interaction. We found that polyubiquitination of the NTD of RAD51 blocks its interaction with BRCA2. Previous structural studies suggested that both the NTD and the core domain of RAD51 bind with the BRC4 domain of BRCA2 (Pellegrini et al. 2002; Conway et al. 2004; Spies and Kowalczykowski 2006; Nomme et al. 2010; Scott et al. 2013; Subramanyam et al. 2013). This interaction between RAD51 and BRCA2 guides positioning of the BRC4 peptide within a cavity between the core domain and the NTD, separates the two domains, and restricts the internal dynamics of RAD51 protomers. Three amino acids (E42, E59, and E237) within the NTD and core domains are critical for RAD51–BRCA2 binding. Our results suggested that three lysine sites (56, 57, and 63) on RAD51 that are close to E59 are deubiquitinated by UCHL3. The deubiquitination is important for RAD51–BRCA2 interaction. These results raise a hypothesis that ubiquitination on the three lysine sites may physically block RAD51–BRCA2 interaction. Alternatively, the ubiquitination may change the structure of RAD51 and restrict the position of the BRC4 peptide within the cavity between the core domain and the NTD. Further structural studies and conformational analyses of RAD51 will be necessary to better understand the precise mechanism. Given that RAD51 3KR can rescue UCHL3 deficiency-induced DDR phenotypes, it suggested that RAD51 may be the major DDR substrate for UCHL3.
Previous studies suggested that knockout of RAD51 or BRCA2 in mice leads to embryonic lethality (Lim and Hasty 1996; Ludwig et al. 1997; Sharan et al. 1997; Suzuki et al. 1997); however, UCHL3 knockout mice did not show such a severe phenotype (Kurihara et al. 2000). One possibility is the adaptive response in mice during development. Other factors, such as UCHL1, might compensate for UCHL3, as suggested by Tilghman and colleagues (Kurihara et al. 2000). The mouse UCHL3 protein displays 52% identity to its mouse paralog, UCHL1, and several studies suggest that UCHL3 and UCHL1 have redundant as well as distinct functions in oocyte and sperm development (Kwon et al. 2004a,b; Mtango et al. 2012a,b; Yi et al. 2015). Another complimentary possibility is that, unlike BRCA2/RAD51 knockout, UCHL3 knockout only decreases Rad51 function. The remaining Rad51 function might be sufficient for normal development of mice without external stress. Our results also suggest that UCHL3 deficiency causes HR defect but does not affect cell proliferation (Fig. 1D; Supplemental Fig. S1A). Importantly, when we used MEFs from UCHL3−/− mice, we were able to show that these cells have defective DDR in response to external genotoxic stress (Supplemental Fig. S4A). In the future, more extensive characterization of UCHL3 knockout mice, especially under stress, would be necessary.
Many studies suggest that HR-based DNA repair affects the outcome of cancer treatment and drug resistance (Helleday 2010). For instance, better response of primary ovarian cancers to platinum-based therapy is correlated with decreased expression of HR proteins, such as BRCA1 or FANCF (Taniguchi et al. 2003; Teodoridis et al. 2005), and mutations in BRCA1 or BRCA2 (Edwards et al. 2008; Sakai et al. 2008). However, cisplatin resistance in ovarian cancer cells is correlated with re-expression of FANCF (Taniguchi et al. 2003) or genetic reversion of BRCA1 or BRCA2 mutations (Edwards et al. 2008; Sakai et al. 2008; Swisher et al. 2008). Similarly, high levels of the HR protein RAD51 in cancer result in enhanced DNA repair capability, rendering cells resistant to chemotherapy (Klein 2008). We found that UCHL3 is overexpressed in breast cancer cell lines. Furthermore, overexpression of UCHL3 in breast cancer is correlated with poor survival of breast cancer patients. In addition, disruption of the UCHL3 gene renders breast cancer cells sensitive to PARPi and radiotherapy. Taken together, our results suggest that UCHL3 may be a new therapeutic target for overcoming resistance to standard therapy, and therapeutic interventions targeting UCHL3 may improve outcome in combination with existing therapies.
Materials and methods
Cell culture, plasmids, and antibodies
HEK293T, U2OS, and human breast cancer cell lines MDA-MB-231, MCF7, MDA-MB-439, T47D, and BT549 were purchased from American Type Culture Collection, and the identities of all cell lines were confirmed by the medical genome facility at the Mayo Clinic Center (Rochester, MN) using short tandem repeat profiling on receipt. The cell lines were maintained in the appropriate media with 10% FBS.
HA-FLAG-UCHL3 was purchased from Addgene (plasmid #22564, provided by Dr. Wade Harper) and subcloned into pGEX-4T-2 vector (Clontech). UCHL3C94A and S75A mutants were generated by site-directed mutagenesis (Stratagene). pCMV-Myc-RAD51 was a gift from Dr. Junjie Chen (University of Texas MD Anderson Cancer Center).
Anti-UCHL3 antibody was purchased from ProteinTech (12384-1-AP). Anti-Ub (P4D1), anti-RPA32 (9H8), and anti-BRCA1 (D9) antibodies were purchased from Santa Cruz Biotechnology. Anti-RAD51 (N1C2) was purchased from GeneTex. Anti-γH2AX (05-636), anti-BRCA2 (OP95), and anti-MDC1 (05-1572) were purchased from Millipore. Anti-BRCA2 (A303-435A) and anti-γH2AX (A300-081A) were purchased from Bethyl Laboratories. Anti-53BP1 (NB100-304) was purchased from Novus Biologicals. Anti-Flag (m2), anti-HA, and anti-β-actin antibodies were purchased from Sigma. Anti-phospho-(Ser/Thr) ATM/ATR substrate antibody (2851) was purchased from Cell Signaling Technology. Anti-PALB2 was a gift from Dr. Bing Xia (Rutgers, The State University of New Jersey)
CRISPR/Cas9 knockout
For CRISPR/Cas9 knockout of human UCHL3 in BT-549, U2OS, and MDA-MB-231 cells, the following small guide RNAs (sgRNAs) were used: sgUCHL3-1 (5′-GCCGCTGGAGGCCAATCCCGAGG-3′) and sgUCHL3-2 (5′-GCCCCGAAGCGCGCCCACCTCGG-3′). The gRNA sequences were cloned into the vector LentiCRISPR-V2-puro. Cells were infected with Lenti-UCHL3-sgRNA-puro followed by extensive selection with 2 μg/mL puromycin, and single colonies were obtained by serial dilution and amplification. Clones were identified by immunoblotting with anti-UCHL3 antibody and were verified by DNA sequencing.
Recombinant protein expression and pull-down assay
To construct the plasmids that express His-RAD51 and GST-UCHL3 proteins, the coding sequences of RAD51 and UCHL3 were subcloned into pET-32a and pGEX-4T-2. To produce recombinant proteins, each expression construct was transformed into E. coli BL21 (DE3). In brief, bacteria were cultured in LB medium at 37°C to reach 1.2 (OD600) and cooled for 30 min on ice. The cells were then continuously cultured for 15 h at 18°C after adding 0.8 mM isopropyl-b-D-thiogalactopyranoside (IPTG). Cells were then harvested and lysed by sonication. The cell lysates were centrifuged at 16,000g for 30 min, and the resulting supernatants were incubated with His Mag Sepharose Ni (GE Healthcare) to purify His-RAD51 proteins and GSH beads (Promega) to purify GST or GST-UCHL3 proteins, respectively. The proteins were then purified according to the manufacturer's instructions.
For the in vitro GST-UCHL3 pull-down assay, 50 µg of recombinant GST and GST-UCHL3 proteins bound to GSH beads was blocked by 5% BSA for 2 h at 4°C followed by incubation with 50 µg of His-UCHL3 for an additional 2 h at 4°C. The beads were then washed five times with NETN buffer with different salt concentration. The bound proteins were eluted by 1× SDS-PAGE buffer with heating for 10 min at 95°C. His-RAD51 was detected by a monoclonal anti-His antibody (MBL Life Science), and GST-UCHL3 was stained by Coomassie blue.
Denatured deubiquitination assay in vivo and deubiquitination assay in vitro
For the in vivo deubiquitination assay, control U2OS cells, UCHL3 knockout U2OS cells, or UCHL3 knockout U2OS cells stably expressing HA-UCHL3 wild-type or mutant Cys95 to Ala (CA mutant) were lysed in 120 μL of 62.5 mM Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, 20 mM NEM, and 1 mM iodoacetamide; boiled for 15 min; diluted 10 times with NETN buffer containing protease inhibitors, 20 mM NEM, and 1 mM iodoacetamide; and centrifuged to remove cell debris. The cell extracts were subjected to immunoprecipitation with the indicated antibodies and blotted with anti-Ub antibody.
For the preparation of a large amount of ubiquitinated proteins as the substrate for the in vitro deubiquitination assay, HEK293T cells were transfected together with the Myc-RAD51 and His-Ub expression vectors. Ubiquitinated proteins were purified from the cell extracts with His beads in a denatured condition. Next, the Ub-RAD51 proteins were purified from the cell extracts with anti-Myc-agarose beads in lysis buffer (50 mM Tris-HCl at pH 7.8, 137 mM NaCl, 10 mM NaF, 1 mM EDTA, 1% Triton X-100, 0.2% Sarkosyl, 1 mM DTT, 10% glycerol, fresh proteinase inhibitors). The recombinant GST-UCHL3 and UCHL3 CA were expressed in BL21 cells and purified following the standard protocol. For the deubiquitination assay in vitro, ubiquitinated proteins were incubated with recombinant UCHL3 in a deubiquitination buffer (50 mM Tris-HCl at pH 8.0, 50 mM NaCl, 1 mM EDTA, 10 mM DTT, 5% glycerol) for 4 h at 30°C.
ChIP
Induction of a single DSB in U2OS DR-GFP cells was performed through transfection of the I-SceI expression plasmid. Twenty-four hours after transfection, cells were fixed with 1% formaldehyde for 10 min at room temperature to cross-link proteins to DNA. Glycine (0.125 M) was added for 5 min to stop the cross-linking. Cells were harvested, and the pellets were resuspended in cell lysis buffer (5 mM PIPES [KOH] at pH 8.0, 85 mM KCl, 0.5% NP-40) with 1 μg/mL leupeptin, 1 μg/mL aprotinin, and 1 mM PMSF. Nuclei were pelleted by centrifugation at 2,000g for 5 min. Nuclei were then resuspended in nuclear lysis buffer (50 mM Tris at pH 8.1, 10 mM EDTA, 1% SDS with1 μg/mL leupeptin, 1 μg/mL aprotinin, 1 mM PMSF) and sonicated to shear chromatin to an average size of 0.6 kb. The lysates were precleared overnight with salmon sperm DNA/protein-A agarose slurry. Ten percent of each supernatant was used as input control and processed with the cross-linking reversal step. The rest of the supernatant was incubated with 5 μg of the indicated antibody overnight at 4°C. The beads were washed four times: once in high-salt buffer (50 mM Tris-HCl at pH 8.0, 500 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% NP-40, 1 mM EDTA), once in LiCl buffer (50 mM Tris-HCl at pH 8.0, 250 mM LiCl, 1% NP-40, 0.5% deoxycholate, 1 mM EDTA), and twice in TE buffer (10 mM Tris-HCl at pH 8.0, 1 mM EDTA, pH 8.0). Beads were then resuspended in elution buffer (1% SDS, 0.1 M NaHCO3) and rotated for 20 min at room temperature. Eluted samples were incubated with 0.2 M NaCl overnight at 65°C to reverse cross-link. The samples were incubated with RNase A for 30 min at 37°C followed by proteinase K for 1 h at 37°C. DNA was then purified using PCR Cleanup kit and used for quantitative PCR analysis. The PCR primers for ChIP, ∼220 base pairs away from the I-SceI cut site, were as follows: forward, 5′-GAGCAAGGGCGAGGAGCTGT -3′; and reverse, 5′-CCGTAGGTCAGGGTGGTCAC-3′.
HR assay
We generated control or UCHL3 knockout U2OS DR-GFP cell lines by the CRISPR system using the U2OS DR-GFP cells from Dr. Maria Jasin (Memorial Sloan Kettering Cancer Center). I-SceI expression vector (pCBA-I-SceI) was transfected into the cells. Cells were harvested 2 d after I-SceI transfection and subjected to analysis using flow cytometry to examine recombination induced by I-SceI digestion. The parallel transfection with pEGFP-C1 was used to normalize for transfection efficiency.
Statistics
For the cell survival assay and HR assay, data are presented as the mean ± SEM of three independent experiments. For the focus formation assay, data are presented as the mean ± SEM of three independent experiments. More than 200 cells were counted per experiment. Statistical analyses were performed with the Student's t-test, ANOVA, or log-rank test. Statistical significance is represented in figures by one asterisk for P < 0.05 and two asterisks for P < 0.01.
Supplementary Material
Acknowledgments
We thank Dr. Keji Wade (National Institute of Neuroscience, Tokyo) for providing the UCHL3+/+ and UCHL3−/− MEF cells, Dr. Junjie Chen (University of Texas MD Anderson Cancer Center) for providing the Myc-RAD51 constructs, and Dr. Bing Xia (Rutgers, The State University of New Jersey) for providing the PALB2 antibody. This work was supported by the National Basic Research Program of China (973 Program, grant no. 2013CB530700), the International S&T Cooperation Program of China (2015DFA30610), the National Natural Science Foundation of China (31270806, 81322031, 81572770, and 31371367), the National Institutes of Health (grants CA203971, CA130996, CA189666, and CA203561), the Mayo Clinic Ovarian Cancer Specialized Program of Research Excellence (SPORE) (P50CA136393), the Mayo Clinic Breast Cancer SPORE (P50CA116201), and the Mayo Clinic Fraternal Order of Eagles Cancer Research Fund-Pilot Project Award Program.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.289439.116.
References
- Ayoub N, Rajendra E, Su X, Jeyasekharan AD, Mahen R, Venkitaraman AR. 2009. The carboxyl terminus of Brca2 links the disassembly of Rad51 complexes to mitotic entry. Curr Biol 19: 1075–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JS, Jackson SP. 2015. Ubiquitylation, neddylation and the DNA damage response. Open Biol 5: 150018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917. [DOI] [PubMed] [Google Scholar]
- Carreira A, Kowalczykowski SC. 2011. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc Natl Acad Sci 108: 10448–10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira A, Hilario J, Amitani I, Baskin RJ, Shivji MK, Venkitaraman AR, Kowalczykowski SC. 2009. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell 136: 1032–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell 47: 497–510. [DOI] [PubMed] [Google Scholar]
- Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM, et al. 1998a. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell 2: 317–328. [DOI] [PubMed] [Google Scholar]
- Chen PL, Chen CF, Chen Y, Xiao J, Sharp ZD, Lee WH. 1998b. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci 95: 5287–5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiruvella KK, Liang Z, Wilson TE. 2013. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol 5: a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WK, Payne MJ, Beli P, Hanada K, Choudhary C, Hickson ID. 2015. FBH1 influences DNA replication fork stability and homologous recombination through ubiquitylation of RAD51. Nat Commun 6: 5931. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. 2010. The DNA damage response: making it safe to play with knives. Mol Cell 40: 179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway AB, Lynch TW, Zhang Y, Fortin GS, Fung CW, Symington LS, Rice PA. 2004. Crystal structure of a Rad51 filament. Nat Struct Mol Biol 11: 791–796. [DOI] [PubMed] [Google Scholar]
- Critchlow SE, Jackson SP. 1998. DNA end-joining: from yeast to man. Trends Biochem Sci 23: 394–398. [DOI] [PubMed] [Google Scholar]
- Cromie GA, Connelly JC, Leach DR. 2001. Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell 8: 1163–1174. [DOI] [PubMed] [Google Scholar]
- Davies OR, Pellegrini L. 2007. Interaction with the BRCA2 C terminus protects RAD51–DNA filaments from disassembly by BRC repeats. Nat Struct Mol Biol 14: 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446. [DOI] [PubMed] [Google Scholar]
- Downs JA, Nussenzweig MC, Nussenzweig A. 2007. Chromatin dynamics and the preservation of genetic information. Nature 447: 951–958. [DOI] [PubMed] [Google Scholar]
- Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. 2008. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451: 1111–1115. [DOI] [PubMed] [Google Scholar]
- Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. 2005. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature 434: 598–604. [DOI] [PubMed] [Google Scholar]
- Esashi F, Galkin VE, Yu X, Egelman EH, West SC. 2007. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol 14: 468–474. [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921. [DOI] [PubMed] [Google Scholar]
- Flott S, Kwon Y, Pigli YZ, Rice PA, Sung P, Jackson SP. 2011. Regulation of Rad51 function by phosphorylation. EMBO Rep 12: 833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Jackson SP. 2012. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev 26: 1179–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Esashi F, Yu X, Yang S, West SC, Egelman EH. 2005. BRCA2 BRC motifs bind RAD51–DNA filaments. Proc Natl Acad Sci 102: 8537–8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grou CP, Pinto MP, Mendes AV, Domingues P, Azevedo JE. 2015. The de novo synthesis of ubiquitin: identification of deubiquitinases acting on ubiquitin precursors. Sci Rep 5: 12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorffy B, Surowiak P, Budczies J, Lanczky A. 2013. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 8: e82241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T. 2010. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis 31: 955–960. [DOI] [PubMed] [Google Scholar]
- Huen MS, Chen J. 2010. Assembly of checkpoint and repair machineries at DNA damage sites. Trends Biochem Sci 35: 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. 2009. The DNA-damage response in human biology and disease. Nature 461: 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Durocher D. 2013. Regulation of DNA damage responses by ubiquitin and SUMO. Mol Cell 49: 795–807. [DOI] [PubMed] [Google Scholar]
- Jeyasekharan AD, Ayoub N, Mahen R, Ries J, Esposito A, Rajendra E, Hattori H, Kulkarni RP, Venkitaraman AR. 2010. DNA damage regulates the mobility of Brca2 within the nucleoplasm of living cells. Proc Natl Acad Sci 107: 21937–21942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. 2004. Cell-cycle checkpoints and cancer. Nature 432: 316–323. [DOI] [PubMed] [Google Scholar]
- Kim JY, Lee JM, Cho JY. 2011. Ubiquitin C-terminal hydrolase-L3 regulates Smad1 ubiquitination and osteoblast differentiation. FEBS Lett 585: 1121–1126. [DOI] [PubMed] [Google Scholar]
- Klein HL. 2008. The consequences of Rad51 overexpression for normal and tumor cells. DNA Repair 7: 686–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci L, Altmannova V, Spirek M, Zhao X. 2012. Homologous recombination and its regulation. Nucleic Acids Res 40: 5795–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara LJ, Semenova E, Levorse JM, Tilghman SM. 2000. Expression and functional analysis of Uch-L3 during mouse development. Mol Cell Biol 20: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J, Wang YL, Setsuie R, Sekiguchi S, Sakurai M, Sato Y, Lee WW, Ishii Y, Kyuwa S, Noda M, et al. 2004a. Developmental regulation of ubiquitin C-terminal hydrolase isozyme expression during spermatogenesis in mice. Biol Reprod 71: 515–521. [DOI] [PubMed] [Google Scholar]
- Kwon J, Wang YL, Setsuie R, Sekiguchi S, Sato Y, Sakurai M, Noda M, Aoki S, Yoshikawa Y, Wada K. 2004b. Two closely related ubiquitin C-terminal hydrolase isozymes function as reciprocal modulators of germ cell apoptosis in cryptorchid testis. Am J Pathol 165: 1367–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K. 2004. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 3: 817–826. [DOI] [PubMed] [Google Scholar]
- Lim DS, Hasty P. 1996. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol 16: 7133–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang H, Wang X, Tian Q, Hu Z, Peng C, Jiang P, Wang T, Guo W, Chen Y, et al. 2015. The deubiquitylating enzyme USP4 cooperates with CtIP in DNA double-strand break end resection. Cell Rep 13: 93–107. [DOI] [PubMed] [Google Scholar]
- Ludwig T, Chapman DL, Papaioannou VE, Efstratiadis A. 1997. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev 11: 1226–1241. [DOI] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J. 2011. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol 13: 1161–1169. [DOI] [PubMed] [Google Scholar]
- Luo K, Zhang H, Wang L, Yuan J, Lou Z. 2012. Sumoylation of MDC1 is important for proper DNA damage response. EMBO J 31: 3008–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K, Deng M, Li Y, Wu C, Xu Z, Yuan J, Lou Z. 2015. CDK-mediated RNF4 phosphorylation regulates homologous recombination in S-phase. Nucleic Acids Res 43: 5465–5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166. [DOI] [PubMed] [Google Scholar]
- Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. 2012. RNF168 ubiquitinates K13–15 on H2A/H2AX to drive DNA damage signaling. Cell 150: 1182–1195. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. 2009. Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci 34: 264–272. [DOI] [PubMed] [Google Scholar]
- Misaghi S, Galardy PJ, Meester WJ, Ovaa H, Ploegh HL, Gaudet R. 2005. Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate. J Biol Chem 280: 1512–1520. [DOI] [PubMed] [Google Scholar]
- Mizuta R, LaSalle JM, Cheng HL, Shinohara A, Ogawa H, Copeland N, Jenkins NA, Lalande M, Alt FW. 1997. RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein. Proc Natl Acad Sci 94: 6927–6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Pierce AJ, Jasin M. 2001. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell 7: 263–272. [DOI] [PubMed] [Google Scholar]
- Mtango NR, Sutovsky M, Susor A, Zhong Z, Latham KE, Sutovsky P. 2012a. Essential role of maternal UCHL1 and UCHL3 in fertilization and preimplantation embryo development. J Cell Physiol 227: 1592–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mtango NR, Sutovsky M, Vandevoort CA, Latham KE, Sutovsky P. 2012b. Essential role of ubiquitin C-terminal hydrolases UCHL1 and UCHL3 in mammalian oocyte maturation. J Cell Physiol 227: 2022–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J, Yang K, Dejsuphong D, Hirota K, Takeda S, D'Andrea AD. 2011. The USP1/UAF1 complex promotes double-strand break repair through homologous recombination. Mol Cell Biol 31: 2462–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. 2011. BLM–DNA2–RPA–MRN and EXO1–BLM–RPA–MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25: 350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi R, Wijnhoven P, le Sage C, Tjeertes J, Galanty Y, Forment JV, Clague MJ, Urbe S, Jackson SP. 2014. Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat Cell Biol 16: 1016–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomme J, Renodon-Corniere A, Asanomi Y, Sakaguchi K, Stasiak AZ, Stasiak A, Norden B, Tran V, Takahashi M. 2010. Design of potent inhibitors of human RAD51 recombinase based on BRC motifs of BRCA2 protein: modeling and experimental validation of a chimera peptide. J Med Chem 53: 5782–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, Munro M, Pinder J, Salsman J, Dellaire G, et al. 2015. A mechanism for the suppression of homologous recombination in G1 cells. Nature 528: 422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. 1998. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell 1: 969–979. [DOI] [PubMed] [Google Scholar]
- Pellegrini L, Yu DS, Lo T, Anand S, Lee M, Blundell TL, Venkitaraman AR. 2002. Insights into DNA recombination from the structure of a RAD51–BRCA2 complex. Nature 420: 287–293. [DOI] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 13: 2633–2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roff M, Thompson J, Rodriguez MS, Jacque JM, Baleux F, Arenzana-Seisdedos F, Hay RT. 1996. Role of IκBα ubiquitination in signal-induced activation of NFκB in vivo. J Biol Chem 271: 7844–7850. [DOI] [PubMed] [Google Scholar]
- Roy R, Chun J, Powell SN. 2012. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 12: 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ, et al. 2008. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451: 1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. 2008. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–257. [DOI] [PubMed] [Google Scholar]
- Sano Y, Furuta A, Setsuie R, Kikuchi H, Wang YL, Sakurai M, Kwon J, Noda M, Wada K. 2006. Photoreceptor cell apoptosis in the retinal degeneration of Uchl3-deficient mice. Am J Pathol 169: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. 2007. Human CtIP promotes DNA end resection. Nature 450: 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DE, Ehebauer MT, Pukala T, Marsh M, Blundell TL, Venkitaraman AR, Abell C, Hyvonen M. 2013. Using a fragment-based approach to target protein–protein interactions. Chembiochem 14: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. 2010. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141: 970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. 1997. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386: 804–810. [DOI] [PubMed] [Google Scholar]
- Shivji MK, Mukund SR, Rajendra E, Chen S, Short JM, Savill J, Klenerman D, Venkitaraman AR. 2009. The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange. Proc Natl Acad Sci 106: 13254–13259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupianek A, Dasgupta Y, Ren SY, Gurdek E, Donlin M, Nieborowska-Skorska M, Fleury F, Skorski T. 2011. Targeting RAD51 phosphotyrosine-315 to prevent unfaithful recombination repair in BCR–ABL1 leukemia. Blood 118: 1062–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song HM, Lee JE, Kim JH. 2014. Ubiquitin C-terminal hydrolase-L3 regulates EMT process and cancer metastasis in prostate cell lines. Biochem Biophys Res Commun 452: 722–727. [DOI] [PubMed] [Google Scholar]
- Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. 2005. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol 7: 195–201. [DOI] [PubMed] [Google Scholar]
- Spies M, Kowalczykowski SC. 2006. The RecA binding locus of RecBCD is a general domain for recruitment of DNA strand exchange proteins. Mol Cell 21: 573–580. [DOI] [PubMed] [Google Scholar]
- Subramanyam S, Jones WT, Spies M, Spies MA. 2013. Contributions of the RAD51 N-terminal domain to BRCA2–RAD51 interaction. Nucleic Acids Res 41: 9020–9032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, de la Pompa JL, Hakem R, Elia A, Yoshida R, Mo R, Nishina H, Chuang T, Wakeham A, Itie A, et al. 1997. Brca2 is required for embryonic cellular proliferation in the mouse. Genes Dev 11: 1242–1252. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Setsuie R, Wada K. 2009. Ubiquitin carboxyl-terminal hydrolase l3 promotes insulin signaling and adipogenesis. Endocrinology 150: 5230–5239. [DOI] [PubMed] [Google Scholar]
- Swisher EM, Sakai W, Karlan BY, Wurz K, Urban N, Taniguchi T. 2008. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res 68: 2581–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Nakamura K, Taniguchi Y, Paull TT. 2007. Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol Cell 28: 351–352. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Tischkowitz M, Ameziane N, Hodgson SV, Mathew CG, Joenje H, Mok SC, D'Andrea AD. 2003. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med 9: 568–574. [DOI] [PubMed] [Google Scholar]
- Teodoridis JM, Hall J, Marsh S, Kannall HD, Smyth C, Curto J, Siddiqui N, Gabra H, McLeod HL, Strathdee G, et al. 2005. CpG island methylation of DNA damage response genes in advanced ovarian cancer. Cancer Res 65: 8961–8967. [DOI] [PubMed] [Google Scholar]
- van Beekum O, Gao Y, Berger R, Koppen A, Kalkhoven E. 2012. A novel RNAi lethality rescue screen to identify regulators of adipogenesis. PLoS One 7: e37680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmerdam DO, Kanaar R. 2010. Dealing with DNA damage: relationships between checkpoint and repair pathways. Mutat Res 704: 2–11. [DOI] [PubMed] [Google Scholar]
- West SC. 2003. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol 4: 435–445. [DOI] [PubMed] [Google Scholar]
- Wijnhoven P, Konietzny R, Blackford AN, Travers J, Kessler BM, Nishi R, Jackson SP. 2015. USP4 auto-deubiquitylation promotes homologous recombination. Mol Cell 60: 362–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltshire TD, Lovejoy CA, Wang T, Xia F, O'Connor MJ, Cortez D. 2010. Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair. J Biol Chem 285: 14565–14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. 1997. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem 272: 31941–31944. [DOI] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G. 1995. Identification of the breast cancer susceptibility gene BRCA2. Nature 378: 789–792. [DOI] [PubMed] [Google Scholar]
- Yi YJ, Manandhar G, Sutovsky M, Li R, Jonakova V, Oko R, Park CS, Prather RS, Sutovsky P. 2007. Ubiquitin C-terminal hydrolase-activity is involved in sperm acrosomal function and anti-polyspermy defense during porcine fertilization. Biol Reprod 77: 780–793. [DOI] [PubMed] [Google Scholar]
- Yi YJ, Sutovsky M, Song WH, Sutovsky P. 2015. Protein deubiquitination during oocyte maturation influences sperm function during fertilisation, antipolyspermy defense and embryo development. Reprod Fertil Dev 27: 1154–1167. [DOI] [PubMed] [Google Scholar]
- Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT. 2012. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev 26: 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan ZM, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Kharbanda S, Wang R, Sung P, Shinohara A, Weichselbaum R, et al. 1998. Regulation of Rad51 function by c-Abl in response to DNA damage. J Biol Chem 273: 3799–3802. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.