Barber et al. show that Drosophila insulin-producing cells (IPCs) are functionally connected to the central circadian clock circuit via DN1 neurons. Insulin mediates circadian output by regulating the rhythmic expression of a metabolic gene (sxe2) in the fat body. The activity of IPCs and the rhythmic expression of sxe2 are additionally regulated by feeding.
Keywords: Drosophila, insulin, circadian, clock, electrophysiology, metabolism
Abstract
Circadian clocks regulate much of behavior and physiology, but the mechanisms by which they do so remain poorly understood. While cyclic gene expression is thought to underlie metabolic rhythms, little is known about cycles in cellular physiology. We found that Drosophila insulin-producing cells (IPCs), which are located in the pars intercerebralis and lack an autonomous circadian clock, are functionally connected to the central circadian clock circuit via DN1 neurons. Insulin mediates circadian output by regulating the rhythmic expression of a metabolic gene (sxe2) in the fat body. Patch clamp electrophysiology reveals that IPCs display circadian clock-regulated daily rhythms in firing event frequency and bursting proportion under light:dark conditions. The activity of IPCs and the rhythmic expression of sxe2 are additionally regulated by feeding, as demonstrated by night feeding-induced changes in IPC firing characteristics and sxe2 levels in the fat body. These findings indicate circuit-level regulation of metabolism by clock cells in Drosophila and support a role for the pars intercerebralis in integrating circadian control of behavior and physiology.
Circadian clocks constitute an endogenous timekeeping system that drives rhythms of behavior and physiology and synchronizes them with respect to each other and the external environment. Anticipation of environmental changes and the coordination of biochemical and physiological processes are critical for optimal physiological function, as evidenced by the association of clock disruption with poor health outcomes, including cancer, cardiovascular disease, and metabolic disorders (Turek et al. 2005; Sahar and Sassone-Corsi 2009; Marcheva et al. 2013).
While circadian clocks are present in almost all body tissues, rhythms of behavior and some physiological processes depend on clock circuits in the brain. In fruit flies (Drosophila melanogaster), the brain clock circuit is comprised of ∼150 neurons that express the core molecular clock components. These neurons are grouped based on their neuroanatomical location and function and include the small and large ventral lateral neurons (LNvs), the dorsal lateral neurons (LNds), and three groups of dorsal neurons (DN1–3) (Nitabach and Taghert 2008; Allada and Chung 2010; Yoshii et al. 2012). The small LNvs (sLNvs) appear to be the master circadian regulators (Allada and Chung 2010; Roberts et al. 2015) and coordinate the activity of the other neurons in the clock network via the release of pigment-dispersing factor (PDF), an LNV-specific neuropeptide (Allada and Chung 2010). Although LNvs are sufficient to drive rest:activity rhythms in constant darkness (DD) and set the period of the clock, robust behavioral and physiological rhythms are an emergent property of the clock network as a whole (Peng et al. 2003; Guo et al. 2014; Yao and Shafer 2014; Roberts et al. 2015; Liang et al. 2016). LNds and DN1 neurons are targets of PDF and play roles in regulating multiple features of circadian behavior, including rhythm strength, phase, and circadian period (Murad et al. 2007; Lear et al. 2009; Zhang et al. 2010; Guo et al. 2014).
In addition to possessing molecular clocks, clock cells show rhythms of electrical activity. This is well documented for clock cells in the mammalian brain clock, the suprachiasmatic nucleus, and has also been demonstrated for some of the Drosophila clock neurons. Although the all-important sLNvs have been difficult to access for electrophysiology, recordings from large LNvs and DN1 neurons show increased action potential firing rates in the early morning that decrease through the day and then begin to rise again during the night (Sheeba et al. 2007, 2008a,b; Cao and Nitabach 2008; Flourakis and Allada 2016). Consistent with their peptidergic identity, LNvs also show rhythms of firing morphology, with higher burst firing during the day and tonic firing or no firing at night (Sheeba et al. 2007, 2008a; Cao and Nitabach 2008). The rhythmic electrical activity of clock cells is required for rest:activity rhythms and is driven by oscillation of the core molecular clock (Nitabach et al. 2006; Park and Griffith 2006; Cao and Nitabach 2008; Sheeba et al. 2008b; Depetris-Chauvin et al. 2011).
A major question in chronobiology is how central clock cells transmit time of day information to other parts of the brain and perhaps even to peripheral tissues to produce rhythms of behavior and physiology. To address this, we needed to identify circadian-relevant cells that act downstream from the clock and determine how these cells encode circadian signals. Recent work identified the pars intercerebralis (PI) as an important output region controlling rest:activity rhythms in Drosophila and additionally showed that the PI is connected to the core clock LNvs via DN1 neurons (Foltenyi et al. 2007; Cavanaugh et al. 2014; Park et al. 2014; Cavey et al. 2016). The PI regulates a number of processes that are under circadian control, including locomotion, metabolism, and sleep (Belgacem and Martin 2002; Rulifson et al. 2002; Broughton et al. 2005; de Velasco et al. 2007; Foltenyi et al. 2007; Crocker and Sehgal 2010). However, PI cells do not express molecular clock machinery, so time of day information from the central clock must propagate directly or indirectly from clock neurons to the PI (Jaramillo et al. 2004; Allada and Chung 2010; Cavanaugh et al. 2014).
Interestingly, the PI is best known as the site of insulin-producing cells (IPCs) in Drosophila. The IPCs have not been implicated in the direct control of circadian rest:activity rhythms, although insulin appears to act during development to modulate rest:activity rhythms in a Drosophila disease model (Cong et al. 2015; Monyak et al. 2016). In mammals, on the other hand, pancreatic β cells contain molecular clocks, and rhythmic secretion of insulin appears to be important for metabolic homeostasis (Van Cauter et al. 1991; Boden et al. 1996; Peschke and Peschke 1998). However, the mechanisms underlying rhythmic insulin secretion are not known. Although less complex and not yet implicated in metabolic rhythms, the Drosophila insulin signaling pathway is similar to that in mammals and is important for metabolism (Rulifson et al. 2002; Haselton and Fridell 2010; Nässel et al. 2013). A major target of insulin in flies is the fat body, a tissue analogous to mammalian liver and adipose tissue with roles in feeding behavior and metabolism (Xu et al. 2008; Arrese and Soulages 2010). The fat body contains a molecular clock that functions together with the brain clock to maintain metabolic homeostasis (Xu et al. 2008). The expression of many genes cycles in the fat body, and ∼20% of these maintain their rhythmicity in the absence of the fat body clock, with cycling of some driven by the central brain clock through the neuropeptide F (NPF)/NPY system (Xu et al. 2011; Erion et al. 2016) However, several fat body transcripts are not modulated by the NPF system in Drosophila but by other, still unidentified clock inputs from other tissues.
Here we demonstrate that the IPCs of the Drosophila PI are functionally connected to the central clock via DN1 neurons and drive rhythmic expression of a lipase transcript in the fat body. This suggests that peptidergic signaling from the PI plays a role in not only modulation of rest:activity rhythms but also coordination of metabolic rhythms in the fat body. Using the IPCs as model clock output neurons, we show that these cells display daily rhythms of electrical activity that are controlled non-cell-autonomously by the circadian clock. In particular, rhythms of burst firing are lost in a clock mutant. Finally, we found that IPC physiology and sxe2 expression are also sensitive to nutritional state, as restricting feeding to the night periods can partially restore “morning-like” firing behavior and increase sxe2 transcript levels. These data are the first to record daily rhythms of neural activity through electrophysiology of nonclock cells as well as of IPCs in any organism. They indicate circuit-level, as opposed to cell-autonomous, circadian control of insulin cells in Drosophila.
Results
IPCs represent a metabolic clock output region
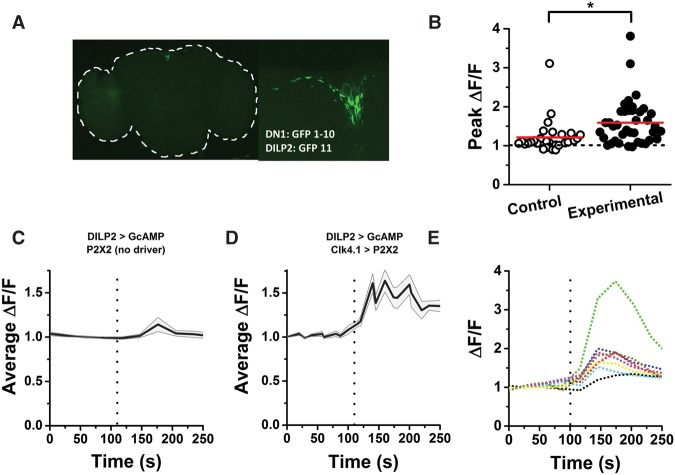
The PI comprises a circadian output region required for rhythmic rest:activity. Of the different groups of peptidergic cells in the PI, those that secrete DH44 and SIFamide are relevant for rest:activity rhythms. Manipulation of IPCs does not disrupt circadian locomotor rhythms (Cavanaugh et al. 2014), but we found through GFP reconstitution across synaptic partners (GRASP) experiments that, like the DH44 cells, IPCs physically connect to DN1 cells of the clock network (Fig. 1A). To test for functional connectivity between DN1 neurons and IPCs, we used the mammalian purinergic receptor P2X2, which can be stimulated with ATP application, to specifically depolarize the DN1 neurons (Yao et al. 2012) and simultaneously expressed GCaMP6m in IPCs to allow monitoring of IPC calcium levels before and after stimulation of DN1 neurons (Chen et al. 2013). Stimulation of P2X2-expressing DN1 neurons with 2 mM ATP increased GCaMP fluorescence in IPCs to an average of 60% above baseline (Fig. 1B,D,E). GCaMP-expressing IPCs in brains that did not express a DN1 driver for P2X2 had a greatly reduced response to the application of ATP (Fig. 1 B,C). From these data, we conclude that IPCs are both physically and functionally connected to DN1 neurons. The response of IPCs to DN1 stimulation was heterogeneous within a single brain, with some cells showing a large response, while others had only moderate or no response to ATP application (Fig. 2E). This suggests heterogeneity in the connections between DN1 neurons and IPCs such that some IPCs may not receive any direct input from DN1 neurons.
Figure 1.
IPCs are functionally connected to the circadian clock. (A) GRASP between DN1 neurons and IPCs (+; DILP2-Gal4/LexAOP-GFP11; Clk4.1-LexA/UAS-GFP1-10). Whole brain (left), outlined in white, and zoomed view of the PI region (right) showing the GFP signal in the PI and along the length of the projection from the DN1 to the PI. (B) Peak-normalized GCaMP6m fluorescence from individual IPCs after application of 2.5 mM ATP to stimulate control brains lacking P2X2 expression (control [open symbols]; mean ΔF/F = 1.2 ± 0.1) or DN1 cells expressing P2X2 receptors (experimental [filled symbols]; mean ΔF/F = 1.6 ± 0.1). Each point represents a single cell, and horizontal lines indicate the mean of all cells. The peak GCaMP excitation upon ATP application is significantly higher in the experimental group. P < 0.001 by Mann-Whitney test. Experimental fly genotype: +;DILP2-Gal4/UAS-mCherry.NLS;Clk4.1-LexA/LexAop-P2X2,UAS-GCaMP6m; control genotype: +;DILP2-Gal4/UAS-mCherry.NLS;+/LexAop-P2X2,UAS-GCaMP6m. (C) Averaged time course of GCaMP fluorescence in control flies. ATP was applied at the time indicated by the dashed line. Shading shows SEM. n = 29 cells from nine brains (five male and four female). (D) Averaged time course of GCaMP fluorescence in experimental flies as in C. n = 39 cells from eight brains (four male and four female). (E) Normalized time course of GCaMP6m fluorescence from eight IPCs in the same experimental brain shows the heterogeneity of the response to ATP applied at the time indicated by the dashed line.
Figure 2.
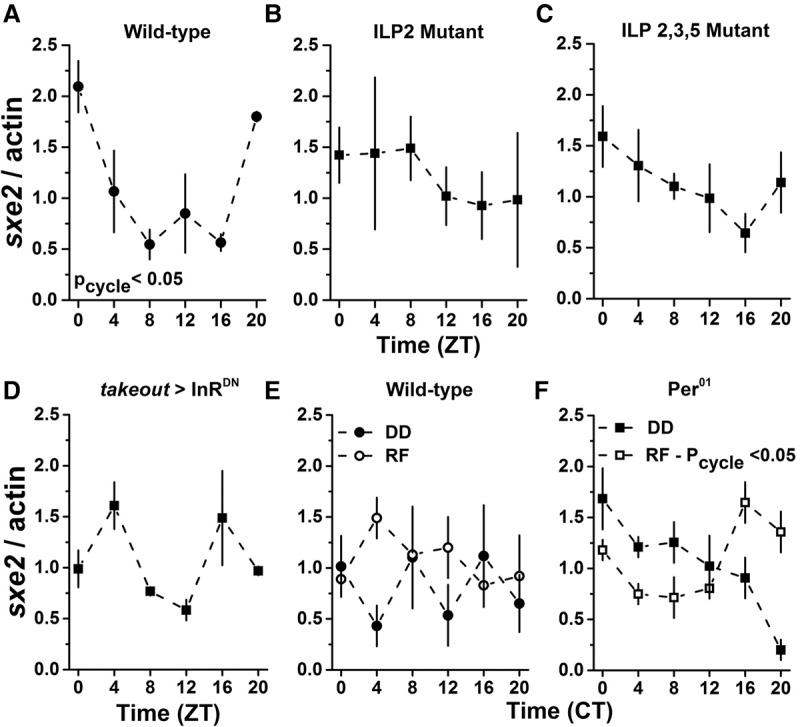
sxe2 transcript rhythms in the fat body are regulated by insulin and feeding in the absence of a clock. (A) Wild-type (Iso31) flies displayed rhythmic sxe2 transcript expression in the fat body (P = 0.047) as assessed by JTK_Cycle analysis of quantitative RT–PCR (qRT–PCR) data. (B,C) Loss of DILP2 (B) or DILP2, DILP3, and DILP5 (C) resulted in loss of sxe2 transcript rhythm. P = 1.0 and P = 0.42, respectively. (D) Expression of a dominant-negative insulin receptor in the fat body (takeout-Gal4 > UAS-InRDN) resulted in a loss of sxe2 transcript rhythm. P = 0.88. (E) On the fourth day in DD, wild-type flies do not maintain rhythmic sxe2 transcript expression (filled symbols). P = 1.00. Restricted feeding (RF; open symbols) from circadian time (CT) 9–15 does not restore rhythmic sxe2 transcript expression. P = 0.11. (F) On the fourth day in DD, period mutant (per01) flies do not show rhythmic sxe2 transcript expression (filled symbols). P = 0.35. Restricted feeding (RF; open symbols) from CT 9–15 restores a circadian rhythm with a peak at CT 16. P = 2.0 × 10−4. n = 3 biological replicates per time point for A–C, E, and F. n = 2 biological replicates per time point for D.
The functional connectivity between the clock network and IPCs suggested that IPCs play a clock output role, perhaps for outputs other than rest:activity behavior. Given their well-known role in metabolism, we investigated whether the PI IPCs might be clock output cells that regulate the timing of metabolic processes.
The fat body is a major regulator of metabolism in Drosophila. Like mammalian livers and adipose tissue, the fat body mobilizes nutrients from energy stores in response to starvation (Arrese and Soulages 2010). Also, as noted above, the fat body contains a clock that drives rhythmic expression of many genes, but some cycling genes in the fat body are regulated by clocks in other tissues, with at least two controlled by clock cells that release NPF (Erion et al. 2016). The sxe2 gene, which encodes a lipase, cycles robustly independently of both the fat body clock and NPF. To determine whether cycling of sxe2 is regulated by IPCs, we initially partially ablated IPCs using the cell death gene reaper. This manipulation caused a moderate loss of sxe2 rhythm, but, as reaper ablation of IPCs is associated with other deleterious phenotypes (Rulifson et al. 2002), we examined insulin-like peptide (ILP) signaling directly. The IPCs produce three ILPs—ILP2, ILP3, and ILP5—of which ILP2 is most analogous to mammalian insulin (Nässel et al. 2013). To determine whether the effect on sxe2 cycling was due to loss of any of these peptides, we examined mutants lacking ILP2 as well as those lacking ILP2, ILP3, and ILP5 together. These mutants abrogated cycling of sxe2 (Fig 2 A–C), suggesting that IPCs act as clock output cells that modulate the rhythm of a metabolic transcript in the fat body via insulin. To determine whether insulin signals directly to the fat body to regulate sxe2, we examined sxe2 cycling in flies expressing a dominant-negative insulin receptor (InRDN) in the fat body. We found that loss of functional insulin receptor in the fat body results in loss of the sxe2 transcript rhythm (Fig. 2D). This finding supports the conclusion that insulin is a clock output signaling molecule that regulates the rhythmic expression of sxe2.
IPCs display rhythms of electrical activity
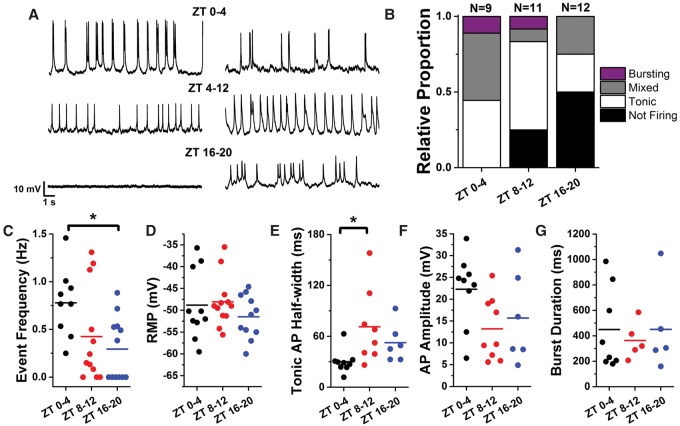
While rhythms of physiology have been found for clock cells (Sheeba et al. 2007, 2008a; Cao and Nitabach 2008; Flourakis and Allada 2016) and predicted in clock output cells based on calcium oscillation and response to stimulation (Cavey et al. 2016), the rhythmic electrical activity of clock output cells has never been directly assessed. Using IPCs as model clock output cells, we characterized their circadian electrophysiology using whole-cell patch clamp analysis (Fig. 3). Flies were entrained to 12:12 light:dark (LD) cycles and collected in the morning (Zeitgeber time [ZT] 0–4), evening (ZT 8–12), or night (ZT 16–20). We then acutely dissected whole brains and patched the IPCs to assess multiple electrophysiological parameters, including firing phenotype, event frequency, resting membrane potential, tonic action potential half-width, mean burst duration, and event amplitude.
Figure 3.
IPCs display circadian patterns of event frequency and morphology. (A) Fifteen-second representative traces from whole-cell patch clamp of GFP-labeled DILP2+ neurons from acutely dissected female Drosophila brains. Flies were entrained to a 12:12 LD cycle and sacrificed and recorded within the time window indicated. (B) Relative proportions of firing phenotypes from IPCs at different windows of the circadian day. (C) Event frequency of tonic and bursting events for three circadian time windows. Each point represents a single cell, and horizontal lines indicate the mean of all cells. Event frequency in the morning (ZT 0–4) is significantly different from night (ZT 16–20) by one-way ANOVA. P < 0.05. (D) Resting membrane potential for three circadian time windows. There was no time of day difference detected by one-way ANOVA. (E) Mean half-width of tonic action potentials for three circadian time windows. Tonic action potential (AP) half-width is significantly larger in the evening (ZT 8–12) than in the morning (ZT 0–4) by one-way ANOVA. P < 0.05. (F) Mean action potential amplitude for three circadian time windows. There was no time of day difference detected by one-way ANOVA. (G) Mean burst duration for three circadian time windows. There was no time of day difference detected by one-way ANOVA.
We detected a range of firing phenotypes of IPCs—including nonfiring cells, tonically firing cells, bursting cells, and cells with mixed tonic and burst firing (Fig. 3A)—similar to that seen for core peptidergic pacemaker neurons. IPC firing frequency varied by time of day (Fig. 3B), being highest in the morning (ZT 0–4). At this time, all recorded cells showed firing events, with an equal proportion of cells exhibiting tonic and burst firing. In the night (ZT 16–20), half of the cells assayed showed no firing events, although the firing cells displayed an equal proportion of tonic and burst firing. In the late day (ZT 8–12), the distribution of bursting, tonic, and silent cells was intermediate between the morning and night distributions. Event frequency of all events considered together showed time of day variation, with higher firing frequency in the morning and lower firing frequency in the evening, when many cells were silent (Fig. 3C). This temporal pattern of firing frequency and temporal distribution of burst/tonic firing is very similar to that seen for core pacemaker neurons (Sheeba et al. 2007, 2008a). The difference in event frequency was significant between ZT 0–4 and ZT 16–20.
Despite the difference in firing rates around the circadian day, resting membrane potential showed no significant differences across time points (Fig. 3D). For tonic action potentials, the half-width varied, with wider action potentials later in the day; this difference was significant between ZT 0–4 and ZT 8–12, with a trend toward longer action potentials at ZT 16–20 that could not be statistically confirmed due to the small number of cells that were active at this time point (Fig. 3E). Action potential amplitude showed no circadian variation across time points. Action potential amplitude varied within any one time point from ∼5 mV to ∼35 mV, but the amplitude within any individual cell was highly consistent across long periods of recording (Fig. 3F). While the proportion of bursting cells varied across time points, mean burst duration did not show significant time of day differences (Fig. 3G). Overall, these data indicate time of day differences in firing frequency (including burst firing) and action potential width.
Electrical characteristics of IPCs are clock-controlled
To test whether the observed physiological rhythms in IPCs are controlled by the circadian clock, we assessed whether these rhythms are maintained in DD in the absence of a functional core clock. After 18–20 h of DD, physiological rhythms were not detected (Fig. 4). During the subjective morning (circadian time [CT] 0–4), the proportions of nonfiring, tonic, and bursting cells resembled the evening time point in LD cycles (ZT 8–12), with a small proportion of silent cells and both mixed and tonic firing in the active cells (Fig. 4A). On the other hand, the proportions of firing cells during subjective evening (CT 8–12) were nearly identical to the LD morning time point (ZT 0–4), with no silent cells and more than half of the active cells exhibiting mixed or predominantly burst firing (Fig. 4A). These data likely reflect loss of timing/synchrony of different parameters rather than an alteration in phase. Importantly, no difference was observed in event frequency, (Fig. 4B) between CT 0–4 and CT 8–12. There was a slight trend toward increased tonic action potential half-width at CT 8–12 (Fig. 4F), similar to the wider action potentials observed during the evening under LD conditions (ZT 8–12). As in LD conditions, resting membrane potential, action potential amplitude, and burst duration did not show circadian variation (Fig. 4C,E,F).
Figure 4.
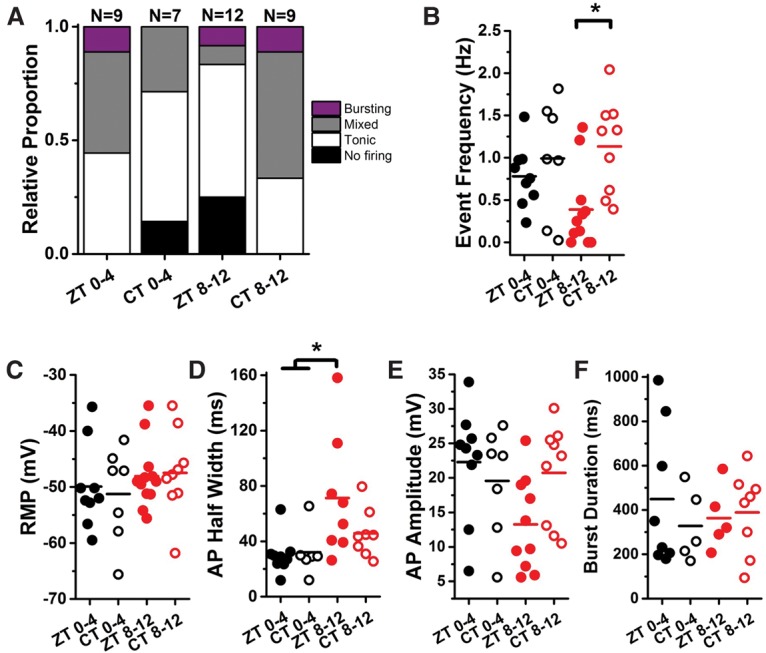
IPC electrophysiological rhythms are not maintained in DD. (A) Relative proportions of firing phenotypes from IPCs at different windows of the circadian day when entrained to a light cycle (ZT 0–4 and ZT 8–12) (data replotted from Fig. 1) compared with flies maintained in DD for 18–22 h prior to recording. CT 0–4, n = 7; CT 8–12, n = 9. (B) Event frequency was not different between ZT 0–4 and CT 0–4; however, we observed significantly higher event frequency at CT 8–12 compared with ZT 8–12. (C) Resting membrane potential was constant across all conditions. (D) Mean half-width of tonic action potentials was nearly identical between ZT 0–4 and CT 0–4 and also between ZT 8–12 and CT 8–12. Although the trend toward longer action potentials was maintained at CT 8–12, the half-width difference was not significant between CT 8–12 and morning time points. (E,F) Mean action potential amplitude (E) and mean burst duration (F) were similar across all conditions.
To assess the role of the molecular clock more directly, we used period mutant (per01) flies, which are arrhythmic under constant conditions but show rhythmic rest:activity patterns in a LD cycle (Konopka and Benzer 1971). As wild-type flies do not display significant rhythms of IPC firing in DD, we assayed per01 mutants in a 12:12 LD cycle. We reasoned that if the electrophysiological rhythms of IPC activity were light-driven, they would persist in per01 mutants under LD conditions, whereas if the rhythms were clock-driven, they would be eliminated in the absence of period.
Recordings of per01 mutants showed a nearly complete loss of burst firing in IPCs at all times of day (Fig. 5A). In the morning (ZT 0–4), when a large proportion of control IPCs exhibit burst firing, we found no burst firing in per01 mutants. In the evening (ZT 8–12) and night (ZT 16–20), we observed only one cell at each of these time points that displayed “mixed” firing phenotypes, with 41% and 46% of events being bursts, respectively. Additionally, the proportions of firing and nonfiring cells across the day were nearly identical in per01 mutants (Fig. 5A). Thus, the rhythm of event frequency found in wild-type neurons (Fig. 3C) is lost in per01 mutants (Fig. 5B), as is the rhythm in tonic action potential half-width (Fig. 5D). Similar to recordings from wild-type control IPCs, there was no rhythm in resting membrane potential or action potential amplitude (Fig. 5C,E).
Figure 5.
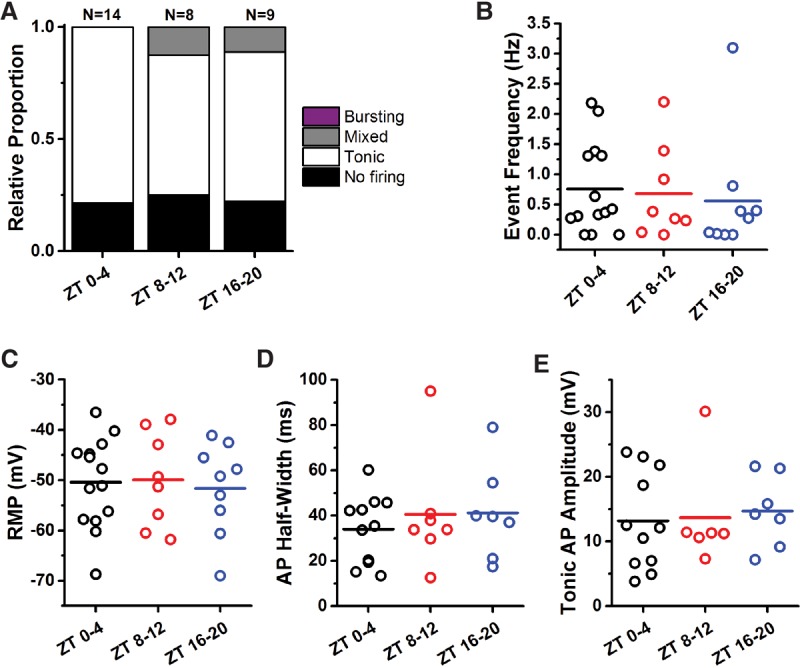
Ablation of the molecular clock results in a loss of electrophysiological rhythm. (A) Relative proportions of firing phenotypes from IPCs in per01 flies in the morning (ZT 0–4), evening (ZT 8–12), and night (ZT 16–20) showed nearly identical proportions of firing versus nonfiring cells and a loss of cells displaying burst firing events. (B–E) Event frequency, resting membrane potential (RMP), tonic action potential (AP) half-width, and action potential (AP) amplitude from per01 flies at ZT 0–4 (black), ZT 8–12 (red), and ZT 16–20 (blue). Each point represents a single cell, and horizontal lines indicate the mean. There were no significant differences between time points for any parameter.
IPCs and their rhythmic output are also modulated by feeding
Drosophila display feeding rhythms, with the majority of food intake in the presence of LD cycles occurring during the morning peak of locomotor activity and a second smaller peak in the evening (Supplemental Fig. S1). This feeding rhythm is maintained in wild-type flies in DD, albeit with reduced amplitude (Supplemental Fig. S1), and is regulated by the clock in the fat body (Xu et al. 2008; Seay and Thummel 2011). Because of the role of IPCs in sensing metabolic state, we assessed whether feeding contributes to the observed physiological rhythms of IPCs. Thus, we used a food restriction paradigm to force flies to feed during the night. Flies in 12:12 LD conditions were starved for 18 h from ZT 18 on day 1 to ZT 12 on day 2. Starved flies were subsequently refed from ZT 12 to ZT 15 prior to electrophysiological recording from ZT 15 to ZT 19. Capillary feeder (CAFE) assays (Ja et al. 2007) demonstrated that refeeding at night after starvation mimics the food intake pattern observed during the morning feeding peak (Supplemental Fig. S2).
Night-fed flies showed a mixed IPC electrophysiological profile, with some properties shifted to more morning-like phenotypes, while other properties remained similar to the night time point (ZT 16–20). Similarities to the normal control morning time point (Fig. 6A) consisted of fewer nonfiring cells and a larger proportion of bursting cells than typically seen at the control ZT 16–20 time point. Event frequency also increased with night feeding and was similar to the firing event frequency observed at ZT 0–4; i.e., significantly different from the lower firing event frequency at the control ZT 16–20 time point (Fig. 6B). Not all IPC electrophysiological features recorded from night-fed flies shifted to resemble the control morning phenotype though. Notably, the tonic action potential half-width remained widened even with night feeding, similar to the control ZT 16–20 time point, and was significantly different from the control morning tonic action potential half-width (Fig. 6C). This may be due to preserved expression of a common repertoire of voltage-gated ion channels for both conditions, as spike width is determined largely by membrane ion channel composition (Bean 2007). As in all other conditions, there was no rhythm in resting membrane potential or action potential amplitude in IPCs from night-fed flies (Fig. 6D,E). Together, these data indicate that firing of IPCs is modulated dually by the circadian clock and feeding.
Figure 6.
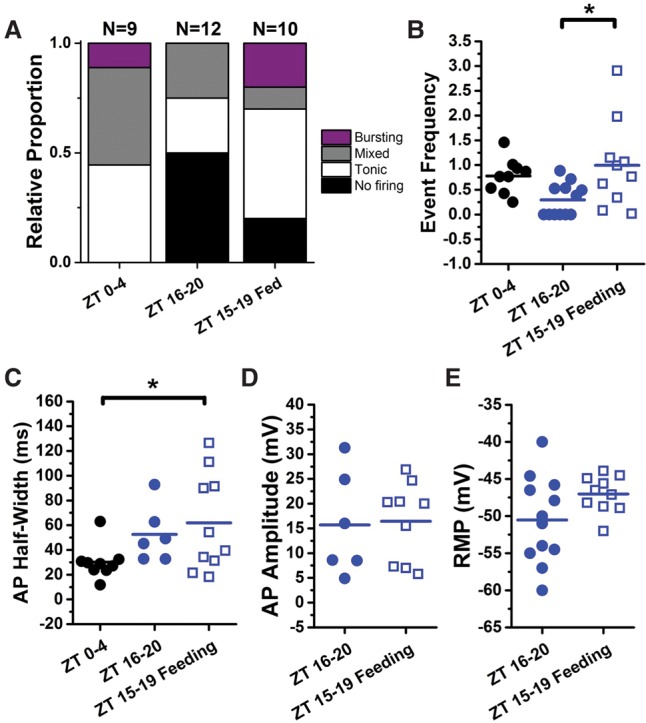
Restricting feeding during the night period partially restores “morning-like” firing properties. (A) Relative proportions of firing phenotypes from ZT 0–4 and ZT 16–20 (data replotted from Fig. 1) compared with flies starved for 18–22 h, fed for 2 h from ZT 12–14, and sacrificed for recording from ZT 15–19. (B) The event frequency for flies fed at night (open blue circles) is similar to the morning event frequency (black) and significantly higher than for control flies from the same time window fed ad lib (filled blue circles). (C) The mean tonic action potential half-width for flies fed at night (open blue circles) still resembles the night phenotype (filled blue circles) and is significantly different from the morning time window. (D,E) The mean action potential amplitude (D) and resting membrane potential (E) for flies fed ad lib versus flies with night-restricted feeding are not significantly different.
To determine how feeding affects the rhythmic output of the IPCs, specifically rhythmic sxe2 expression, we sought to determine whether cyclic expression of sxe2 was controlled directly by the clock, perhaps through the DN1–IPC circuit, or indirectly through clock modulation of feeding rhythms. Previous work in our laboratory identified food-entrainable genes in the fat body, some of which are food-entrainable only in the absence of a functional clock, and some of which are food-entrainable through the clock (Xu et al. 2011). To test whether sxe2 is a food-entrainable gene, we kept both wild-type and per01 flies in DD either with ad lib food or under restricted feeding conditions, with food supplied only from CT 9–15, a time when wild-type flies normally have low food intake (Supplemental Fig. S1). sxe2 transcript rhythms dampened in DD under ad lib conditions, as is typical for cyclically expressed fat body genes (Xu et al. 2011), and restricted feeding did not restore a rhythm (Fig. 2E).
per01 flies show arrhythmic rest:activity patterns under free-running conditions and have dampened to no feeding rhythms in LD and DD conditions, respectively (Supplemental Fig. S1). per01 flies also lack sxe2 transcript rhythms when fed ad lib in DD, but restricted feeding from CT 9–15 drove rhythms in the expression of sxe2, producing an elevated transcript level in the hours after feeding (Fig. 2F). We conclude that the predominant driver of sxe2 transcript rhythms in the fat body is the central brain clock via circuitry that includes, but may not be limited to, IPCs. Restricted feeding can entrain an sxe2 rhythm only in the absence of a clock, suggesting a secondary role for feeding, which, under normal conditions, would coordinate with more directly clock-driven rhythms to support robust sxe2 cycling.
Discussion
We present here the first direct evidence of clock-controlled electrophysiological rhythms in an identified class of circadian circuit-driven output cells. Functional imaging demonstrated that PI IPCs are functionally connected to the DN1 neurons of the Drosophila brain clock. The fine resolution achieved by patch clamp electrophysiology allowed us to determine that not only does firing rate show circadian rhythmicity, but event morphology also shows time of day changes in spite of the fact that these cells do not express a cell-autonomous clock. Additionally, we show that insulin-producing neurons of the PI comprise part of a circuit that modulates rhythmic gene expression in the fat body and can be modulated by nutrient intake. Integration of circadian and metabolic inputs in the PI likely permits fine-tuned homeostatic regulation of physiological processes downstream from the circadian clock. Together with our previous findings that demonstrate the control of rest:activity rhythms by the PI, these data underscore the importance of the PI as a relay station for multiple outputs from the brain clock circuit.
The circadian system controls physiology in large part through regulation of circadian gene transcription. While many tissue-specific transcript rhythms are generated by tissue-autonomous clocks, the clock in the fat body does not drive the cycling of all circadian fat body genes (Xu et al. 2011); rhythmic expression of several transcripts is regulated by clocks in other tissues, such as in NPF/NPY neurons (Erion et al. 2016). In this study, we found that another neuropeptide signal from the brain regulates rhythmic expression of a different fat body gene (sxe2) (Fig. 1B,C). However, unlike NPF neurons, IPCs do not possess a cell-autonomous molecular clock; instead, time of day is encoded at the circuit level via connections to DN1 neurons of the core clock system. We show that IPC regulation of sxe2 transcript rhythm is dependent on the presence of both insulin and functional insulin receptors in the fat body, suggesting that insulin may transmit time of day signals from IPCs directly to the fat body. Interestingly, while sxe2 is not a known target for FOXO, the best-known insulin-responsive transcription factor, a few other genes that cycle independently of the fat body clock—including the clock genes tim and vri as well as sas, α-man-IIb, ebony, CG17562, and CG5156—are potential FOXO targets (Alic et al. 2011; Xu et al. 2011; Bai et al. 2013). We note, however, that other insulin-responsive transcription factors could also be involved in the regulation of cyclically expressed fat body transcripts.
We report a circadian rhythm of IPC firing, with a higher firing rate and a larger proportion of bursting events in the morning. During the night, cells show a lower firing rate, with wider tonic action potentials and a higher likelihood of being electrically silent (Fig. 2). This is consistent with firing patterns observed in lLNv and DN1p neurons of the clock circuit, both of which show high morning firing and a nighttime firing trough (Sheeba et al. 2007, 2008a; Cao and Nitabach 2008; Flourakis and Allada 2016). Like IPCs, lLNv neurons show an increased proportion of burst firing during the day, while DN1p neurons display a strictly tonic firing morphology. The circadian distribution of burst versus tonic firing morphology is interesting, as peptidergic corelease requires burst firing, while the release of classical small molecule neurotransmitters without peptide corelease is coded by tonic firing (Whim and Lloyd 1989; Tallent 2008). In mammalian pancreatic β cells, synchronous glucose-dependent bursting gives rise to widespread changes in calcium concentration that lead to pulsatile insulin release (Santos et al. 1991; Gilon and Henquin 1992). Thus, the presence of burst firing may be associated with insulin release in IPCs, which would suggest an increased likelihood of insulin release early in the day, when food consumption is high. Such circadian “priming” of the metabolic system for food intake could maximize metabolic efficiency.
IPCs do not maintain a clear pattern of circadian firing after 18–20 h of DD. We note that, even in clock cells, it has been technically challenging to detect firing rhythms in DD (Sheeba et al. 2007; Cao and Nitabach 2008). Drosophila IPCs do not express the core molecular clock components and hence must receive time of day input from upstream clock neurons. These circumstances may promote an even more rapid desynchrony and drift from circadian firing patterns under constant conditions. The firing in IPCs is clearly under circadian control, as per01 flies show a loss of firing frequency rhythm as well as an absence of burst firing at all recorded time points. These data demonstrate that PERIOD is required for not only circadian variation in IPC firing frequency but also burst firing at any time of day.
Food intake is expected to affect the activity of insulin cells, and, indeed, we observed feeding-dependent changes in IPC firing. In fact, feeding at night overrides clock control to increase firing at a time when it would normally be low. This increase likely drives insulin release to mediate nutrient absorption. Coregulation of IPCs by both the clock and starvation response systems would serve to maximize metabolic homeostasis. However, as the IPCs are also implicated in promoting wakefulness (Crocker and Sehgal 2010), we cannot rule out the possibility that increased firing arouses the flies to allow eating at a time when they would normally be asleep. We also found that restricted feeding can entrain the sxe2 transcript rhythm in the fat body but only in the absence of a functional clock. We surmise that feeding cannot override circadian control to drive sxe2 cycling, suggesting that feeding cannot sustain rhythms in the system; alternatively, IPC-independent clock signals may contribute to sxe2 rhythms
As seen in electrophysiological analysis of clock neurons (and most other neuronal classes), IPC properties show heterogeneity between cells even within a time point. We also observed cell-to-cell heterogeneity in the GCaMP response to stimulation of DN1 neurons. This is not due to experimental variation but likely reflects real heterogeneity of cellular behavior. Although we define all 14 DILP2+ cells of the PI as “IPCs,” these cells differ from each other in terms of several properties, such as secretion of drosulfakinin and the expression of SLOB protein (Jaramillo et al. 2004; Söderberg et al. 2012). Such heterogeneity is increasingly recognized in the nervous system and elsewhere, leading to the rapid advent of approaches targeted toward single-cell analysis (Baslan and Hicks 2014).
We propose a model in which IPCs are signal integrators that can be regulated dually by the clock and nutrient intake (Fig. 7) and whose firing modulates multiple downstream physiological processes in a coordinated fashion. In this model, the clock may both directly and indirectly regulate IPC firing. Our functional imaging data show that DN1 clock neurons directly regulate IPCs, although the clock may also indirectly regulate IPC activity by influencing the timing of food intake. Loss of IPC firing rhythms in per01 flies also supports the role of direct circadian input in rhythmic IPC firing. This model also shows that, under starvation conditions, altered food intake patterns can drive IPC firing activity, presumably to allow insulin secretion to facilitate nutrient uptake and preserve metabolic homeostasis. We suggest that IPC firing is both a circadian and metabolic output in Drosophila that leads to insulin-dependent signaling to the fat body and other regions. Together with previous work, these findings indicate that the Drosophila PI is a clock output region and signal integration center that receives a diverse array of environmental and endogenous inputs to regulate multiple behavioral and physiological processes via both local and circulating peptide signals.
Figure 7.
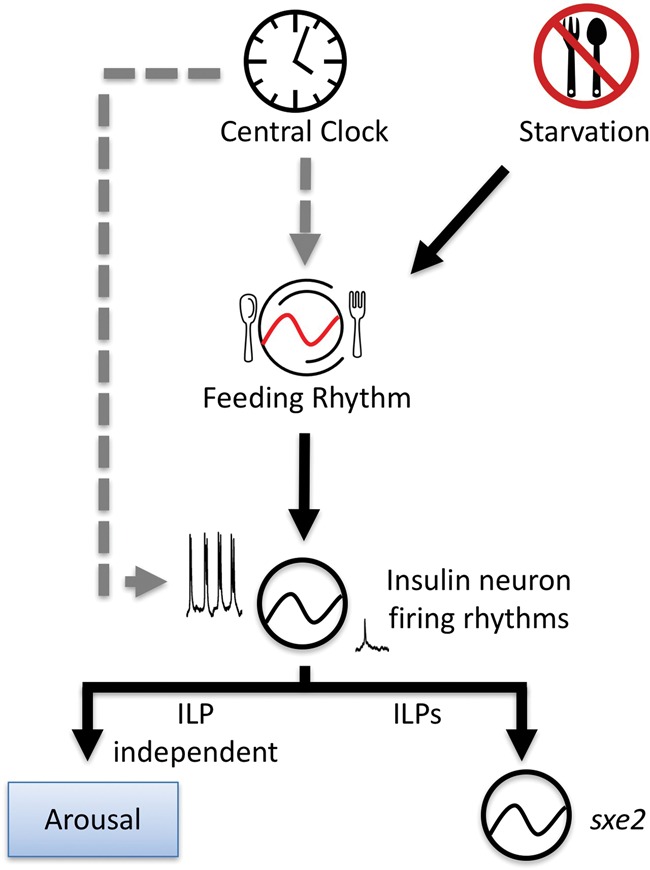
Model for dual modulation of IPCs by the circadian clock and feeding. Inputs from the brain clock can modulate IPC activity both directly via inputs from DN1 neurons and indirectly via clock control of feeding rhythms. Starvation can alter feeding rhythms, leading to altered IPC activity. IPC firing rhythms control insulin-dependent and insulin-independent downstream processes, which include metabolic transcript rhythms in the fat body and behavioral arousal.
Materials and methods
Fly lines
Flies were maintained on standard cornmeal/molasses food at 25°C in 12:12 LD conditions unless otherwise specified. The following fly lines were used: Iso31 (isogenic w1118 stock), Dilp2mCherry, per01, DILP2-Gal4, takeout-Gal4, Clk4.1-LexA, UAS-dORK-NC1, UAS-InRDN (K1409A), UAS-mCherry.NLS, UAS-GCaMP6m, UAS-GFP1-10, LexAop-GFP11, LexAop-P2X2 (III), ILP2, and ILP2,3,5 knockout mutants (Grönke et al. 2010).
GRASP
Brains expressing GFP1-10 in IPCs under the control of the Gal4-UAS system (DILP2-Gal4>UAS-GFP1-10) and GFP11 in DN1 neurons under the control of the LexAop system (Clk4.1-LexA>LexAop-GFP-11) were imaged via confocal microscopy to assess GFP fluorescence (Cavanaugh et al. 2014).
GCaMP imaging and analysis
We constructed +;DILP2-Gal4/UAS-mCherry.NLS;Clk4.1-LexA/LexAop-P2X2,UAS-GCaMP6m flies. These flies express mCherry with a nuclear localization sequence and GCaMP6m in IPCs under the control of the Gal4-UAS system (DILP2-Gal4> UAS-mCherry.NLS, UAS-GCaMP6m) and P2X2 receptor in DN1 neurons under the control of the LexAop system (Clk4.1-LexA>LexAop-P2X2). GCaMP and mCherry fluorescence in acutely dissected brains from both male and female flies was imaged via confocal microscopy. We acquired z-stacks of the PI region while perfusing control artificial hemolymph (AHL) and after establishing a stable GCaMP baseline, switching to perfusion of 2 mM ATP in AHL. AHL consisted of 70 mM NaCl, 5 mM KCl, 1.5 mM CaCl2, 4 mM MgCl2, 10 mM NaHCO3, 5 mM trehalose, 115 mM sucrose, and 5 mM HEPES (pH 7.1).
ImageJ was used to create sum projections of GCaMP and mCherry fluorescence for each z-stack. Regions of interest were drawn around individual IPCs that showed no overlap with other IPCs to measure fluorescence intensity. Only four to eight cells per brain were suitable for analysis, as many cells were too tightly clustered to allow single-cell analysis. From this point, analysis was conducted on each cell independently. Maximum GCaMP fluorescence intensity was normalized to mCherry fluorescence intensity at each time step. To observe changes in GCaMP fluorescence intensity over time, the GCaMP/mCherry value was normalized to baseline fluorescence by dividing by the average GCaMP/mCherry fluorescence for the first four time steps of each experiment for each cell; this measurement is reported as ΔF/F.
Fat body quantitative RT–PCR (qRT–PCR)
Abdominal fat bodies were collected from 5- to 10-d-old male flies. For experiments in a LD cycle, flies were entrained to a 12:12 LD light cycle for at least 3 d. For experiments in DD, entrained flies were moved to DD and dissected on the fourth day of DD. For food restriction experiments, on the first day in DD, flies were switched from standard food to 1% agar. Starting the following day, flies were fed standard food for a specific 6-h period each day and maintained in 1% agar the rest of the time. Flies were again dissected on the fourth day of food restriction and DD.
For each time point, 16–20 fat bodies were collected for each genotype, and each time point was replicated in three independent experiments. The abdominal fat body was dissected in 100% ethanol by separating the abdomen from the rest of the body and removing internal organs, leaving the fat body attached to the cuticle. Fat bodies were then placed in RNeasy lysis buffer (Qiagen, Inc.) on dry ice for RNA extraction according to the RNeasy manufacturer's protocol. RNA was reverse-transcribed using the SuperScript II reverse transcription kit (ThermoFisher). qRT–PCR was performed on a ViiA 7 real-time PCR system (Applied Biosystems) using SYBR Green (Life Technologies). The following primer sequences were used for qPCR: sxe2 (forward, 5′-TGCGGTACGATCTTTATACGCC-3′, and reverse, 5′-CTAACTGGCCATTTCGGATTGA-3′) and β-actin (forward, 5′-GGACCGGACTCGTCATACTC-3′, and reverse 5′-CTGGCGGCACTACCATGTATC-3′). sxe2 transcript levels were normalized to β-actin to control for variations in total RNA content between samples. Significant rhythmicity of transcript levels was determined using the JTK_Cycle algorithm (Hughes et al. 2010).
Capillary feeder (CAFE) assays
For each assay, 10 groups of four female flies were housed in standard vials on 1% agar with a calibrated capillary tube containing 5% sucrose for 1 d prior to the assay to familiarize them with the food source. On the day of the assay, capillaries were refilled, and the meniscus level was marked for each capillary every 2 h. These measurements were converted to microliters and normalized to single-fly consumption. Circadian rhythmicity was assessed and determined using the JTK_Cycle algorithm (Hughes et al. 2010).
Electrophysiology
Whole brains were dissected from 5- to 10-d-old female flies immobilized on ice in cold external recording solution. Brains from time points between ZT 0 and ZT 12 were dissected under white light. Dissections from ZT 16 to ZT 20 were illuminated by weak 600-nm LEDs. Dissected whole brains were placed in a chamber and secured using a nylon fiber holder. After finding cells in white light, daytime recordings were conducted using only room lighting. For ZT 16–20, white light was filtered to allow only 550- to 600-nm light for finding cells, and recordings were conducted in darkness. To visualize DILP2+ neurons in wild-type flies, expression of the GFP-tagged physiologically neutral nonconducting open rectifier channel (dORK-NC1-eGFP) was driven using the Gal4-UAS system in Iso31 flies (Nitabach et al. 2002). In per01 mutant flies, DILP2+ neurons were visualized by expression of mCherry under the DILP2 promoter. Brief illumination of brains at excitatory wavelengths for GFP (480 nm) or mCherry (560 nm) was required at all time points to identify DILP2-positive cells.
Electrophysiological recordings were made using an Axon Multiclamp 700B patch clamp amplifier (Molecular Devices), digitized with a Digidata 1550 (Molecular Devices), and acquired using pClamp 10 software (Molecular Devices). Recordings were low-pass Bessel-filtered at 2 kHz and digitized at 10 kHz. Borosilicate glass patch pipettes (10–15 mΩ resistance) were filled with an internal solution consisting of 102 mM K-gluconate, 0.085 mM CaCl2, 1.7 mM MgCl2, 17 mM NaCl, 0.94 mM EGTA, and 8.5 mM HEPES with an osmolality of 235 mOsm and a pH of 7.2. External solution consisted of 101 mM NaCl, 1 mM CaCl2, 4 mM MgCl2, 3 mM KCl, 5 mM glucose, 1.25 mM NaH2PO4, and 20.7 mM NaHCO3 with osmolality 250 mOsm and a pH of 7.2 (Sheeba et al. 2007).
Data analysis was performed using pClamp10 and OriginPro9.1. Neuronal firing was analyzed over a 2-min window acquired from the midpoint of the stable recording time window to avoid biasing data sampling. Firing events were identified and characterized using the Event Detection feature of pClamp10 and validated manually. Events that consisted of a single action potential that repolarized to the resting membrane potential before another firing event was initiated were classified as tonic. Events comprised of multiple spikes that did not repolarize to the resting membrane potential between spikes were classified as bursting. Neurons were classified based on the behavior within the analysis window. Nonfiring neurons had no firing events within the analysis window. Neurons were classified as tonic if >80% of events were tonic and as bursting if >80% of events were bursts. Neurons were classified as mixed when neither tonic nor burst events accounted for 80% of events; i.e., one phenotype did not predominate (Sheeba et al. 2007).
Supplementary Material
Acknowledgments
We thank Keri Fogle for electrophysiological technique training; Anna King, Jack Jacobs, and Shirley Zhang for technical and analytical assistance; Michael Gulledge, Christine Quake, Han Wang, and Zhifeng Yue for tissue dissection work; and members of the Sehgal laboratory for helpful discussions. This work was supported by National Institutes of Health grants R37 NS048471 (to A.S.), GM102965 (to T.C.H.), GM107405 (to T.C.H.), and F32 NS090811 (to A.F.B.). A.S. is a Howard Hughes Medical Institute investigator. A.F.B. and A.S. conceived the study; A.F.B., T.C.H., and A.S. supplied the methodology; A.F.B. and R.E. performed the investigation; A.F.B. wrote the original draft; A.F.B., T.C.H., and A.S. reviewed and edited the manuscript; A.F.B. visualized the study; T.C.H. and A.S. supervised the work; and A.F.B., T.C.H., and A.S. acquired funding.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.288258.116.
References
- Alic N, Andrews TD, Giannakou ME, Papatheodorou I, Slack C, Hoddinott MP, Cochemé HM, Schuster EF, Thornton JM, Partridge L. 2011. Genome-wide dFOXO targets and topology of the transcriptomic response to stress and insulin signalling. Mol Syst Biol 7: 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Chung BY. 2010. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol 72: 605–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Soulages JL. 2010. Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol 55: 207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H, Kang P, Hernandez AM, Tatar M. 2013. Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila. PLoS Genet 9: e1003941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baslan T, Hicks J. 2014. Single cell sequencing approaches for complex biological systems. Curr Opin Genet Dev 26: 59–65. [DOI] [PubMed] [Google Scholar]
- Bean BP. 2007. The action potential in mammalian central neurons. Nat Rev Neurosci 8: 451–465. [DOI] [PubMed] [Google Scholar]
- Belgacem YH, Martin JR. 2002. Neuroendocrine control of a sexually dimorphic behavior by a few neurons of the pars intercerebralis in Drosophila. Proc Natl Acad Sci 99: 15154–15158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boden G, Ruiz J, Urbain JL, Chen X. 1996. Evidence for a circadian rhythm of insulin secretion. Am J Physiol Endocrinol Metab 271: E246–E252. [DOI] [PubMed] [Google Scholar]
- Broughton SJ, Piper MDW, Ikeya T, Bass TM, Jacobson J, Driege Y, Martinez P, Hafen E, Withers DJ, Leevers SJ, et al. 2005. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci 102: 3105–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G, Nitabach MN. 2008. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci 28: 6493–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh DJ, Geratowski JD, Wooltorton JRA, Spaethling JM, Hector CE, Zheng X, Johnson EC, Eberwine JH, Sehgal A. 2014. Identification of a circadian output circuit for rest:activity rhythms in Drosophila. Cell 157: 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavey M, Collins B, Bertet C, Blau J. 2016. Circadian rhythms in neuronal activity propagate through output circuits. Nat Neurosci 19: 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. 2013. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499: 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong X, Wang H, Liu Z, He C, An C, Zhao Z. 2015. Regulation of sleep by insulin-like peptide system in Drosophila melanogaster. Sleep 38: 1075–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker A, Sehgal A. 2010. Genetic analysis of sleep. Genes Dev 24: 1220–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depetris-Chauvin A, Berni J, Aranovich EJ, Muraro NI, Beckwith EJ, Ceriani MF. 2011. Adult-specific electrical silencing of pacemaker neurons uncouples molecular clock from circadian outputs. Curr Biol 21: 1783–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Velasco B, Erclik T, Shy D, Sclafani J, Lipshitz H, McInnes R, Hartenstein V. 2007. Specification and development of the pars intercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev Biol 302: 309–323. [DOI] [PubMed] [Google Scholar]
- Erion R, King AN, Wu G, Hogenesch JB, Sehgal A. 2016. Neural clocks and neuropeptide F/Y regulate circadian gene expression in a peripheral metabolic tissue. Elife 5: e13552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flourakis M, Allada R. 2016. And the beat goes on: from clock to behavior. Nat Neurosci 19: 529–530. [DOI] [PubMed] [Google Scholar]
- Foltenyi K, Greenspan RJ, Newport JW. 2007. Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat Neurosci 10: 1160–1167. [DOI] [PubMed] [Google Scholar]
- Gilon P, Henquin JC. 1992. Influence of membrane potential changes on cytoplasmic Ca2+ concentration in an electrically excitable cell, the insulin-secreting pancreatic β-cell. J Biol Chem 267: 20713–20720. [PubMed] [Google Scholar]
- Grönke S, Clarke D-F, Broughton S, Andrews TD, Partridge L. 2010. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet 6: e1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Cerullo I, Chen X, Rosbash M. 2014. PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. Elife 3: e02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselton AT, Fridell YWC. 2010. Adult Drosophila melanogaster as a model for the study of glucose homeostasis. Aging (Albany NY) 2: 523–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. 2010. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms 25: 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ja WW, Carvalho GB, Mak EM, de la Rosa NN, Fang AY, Liong JC, Brummel T, Benzer S. 2007. Prandiology of Drosophila and the CAFE assay. Proc Natl Acad Sci 104: 8253–8256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo AM, Zheng X, Zhou Y, Amado DA, Sheldon A, Sehgal A, Levitan IB. 2004. Pattern of distribution and cycling of SLOB, Slowpoke channel binding protein, in Drosophila. BMC Neurosci 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka RJ, Benzer S. 1971. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci 68: 2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear BC, Zhang L, Allada R. 2009. The neuropeptide PDF acts directly on evening pacemaker neurons to regulate multiple features of circadian behavior. PLoS Biol 7: e1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Holy TE, Taghert PH. 2016. Synchronous Drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science 351: 976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Peek CB, Affinati A, Maury E, Bass J. 2013. Circadian clocks and metabolism. Handb Exp Pharmacol 127–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monyak RE, Emerson D, Schoenfeld BP, Zheng X, Chambers DB, Rosenfelt C, Langer S, Hinchey P, Choi CH, McDonald TV, et al. 2016. Insulin signaling misregulation underlies circadian and cognitive deficits in a Drosophila fragile X model. Mol Psychiatry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad A, Emery-Le M, Emery P. 2007. A subset of dorsal neurons modulates circadian behavior and light responses in Drosophila. Neuron 53: 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel DR, Kubrak OI, Liu Y, Luo J, Lushchak OV. 2013. Factors that regulate insulin producing cells and their output in Drosophila. Front Physiol 4: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. 2008. Organization of the Drosophila circadian control circuit. Curr Biol 18: R84–R93. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Blau J, Holmes TC. 2002. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell 109: 485–495. [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, White BH, Holmes TC. 2006. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci 26: 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Griffith LC. 2006. Electrophysiological and anatomical characterization of PDF-positive clock neurons in the intact adult Drosophila brain. J Neurophysiol 95: 3955–3960. [DOI] [PubMed] [Google Scholar]
- Park S, Sonn JY, Oh Y, Lim C, Choe J. 2014. SIFamide and SIFamide receptor defines a novel neuropeptide signaling to promote sleep in Drosophila. Mol Cells 37: 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Stoleru D, Levine JD, Hall JC, Rosbash M. 2003. Drosophila free-running rhythms require intercellular communication. PLoS Biol 1: E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke E, Peschke D. 1998. Evidence for a circadian rhythm of insulin release from perifused rat pancreatic islets. Diabetologia 41: 1085–1092. [DOI] [PubMed] [Google Scholar]
- Roberts L, Leise TL, Noguchi T, Galschiodt AM, Houl JH, Welsh DK, Holmes TC. 2015. Light evokes rapid circadian network oscillator desynchrony followed by gradual phase retuning of synchrony. Curr Biol 25: 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson EJ, Kim SK, Nusse R. 2002. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296: 1118–1120. [DOI] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. 2009. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer 9: 886–896. [DOI] [PubMed] [Google Scholar]
- Santos RM, Rosario LM, Nadal A, Garcia-Sancho J, Soria B, Valdeolmillos M. 1991. Widespread synchronous [Ca2+]i oscillations due to bursting electrical activity in single pancreatic islets. Pflugers Arch 418: 417–422. [DOI] [PubMed] [Google Scholar]
- Seay DJ, Thummel CS. 2011. The circadian clock, light, and cryptochrome regulate feeding and metabolism in Drosophila. J Biol Rhythms 26: 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Gu H, Sharma VK, O'Dowd DK, Holmes TC. 2007. Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J Neurophysiol 99: 976–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Kaneko M, Rashid S, Chou Y-T, Sharma VK, Holmes TC. 2008a. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr Biol 18: 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Sharma VK, Gu H, Chou Y-T, O'Dowd DK, Holmes TC. 2008b. Pigment dispersing factor-dependent and -independent circadian locomotor behavioral rhythms. J Neurosci 28: 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg JA, Carlsson MA, Nässel DR. 2012. Insulin-producing cells in the Drosophila brain also express satiety-inducing cholecystokinin-like peptide, Drosulfakinin. Front Endocrinol (Lausanne) 3: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallent MK. 2008. Presynaptic inhibition of glutamate release by neuropeptides: use-dependent synaptic modification. Results Probl Cell Differ 44: 177–200. [DOI] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. 2005. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308: 1043–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cauter E, Blackman JD, Roland D, Spire JP, Refetoff S, Polonsky KS. 1991. Modulation of glucose regulation and insulin secretion by circadian rhythmicity and sleep. J Clin Invest 88: 934–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whim MD, Lloyd PE. 1989. Frequency-dependent release of peptide cotransmitters from identified cholinergic motor neurons in Aplysia. Proc Natl Acad Sci 86: 9034–9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zheng X, Sehgal A. 2008. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab 8: 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, DiAngelo JR, Hughes ME, Hogenesch JB, Sehgal A. 2011. The circadian clock interacts with metabolic physiology to influence reproductive fitness. Cell Metab 13: 639–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Shafer OT. 2014. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 343: 1516–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Macara AM, Lelito KR, Minosyan TY, Shafer OT. 2012. Analysis of functional neuronal connectivity in the Drosophila brain. J Neurophysiol 108: 684–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T, Rieger D, Helfrich-Förster C. 2012. Two clocks in the brain: an update of the morning and evening oscillator model in Drosophila. Prog Brain Res 199: 59–82. [DOI] [PubMed] [Google Scholar]
- Zhang L, Chung BY, Lear BC, Kilman VL, Liu Y, Mahesh G, Meissner R-A, Hardin PE, Allada R. 2010. DN1(p) circadian neurons coordinate acute light and PDF inputs to produce robust daily behavior in Drosophila. Curr Biol 20: 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.