Abstract
CD24 plays an oncogenic role in the onset and progression of various human cancers, including prostate cancer. In the present study, we identified two linkage disequilibrium blocks with four recombination hotspot motifs in human CD24 locus. To elucidate whether genetic variants of CD24 associated with susceptibility to prostate cancer and its disease status, we conducted a case-control association study with two P170 C/T and P-534 A/C polymorphisms of CD24 in 590 patients with prostate cancer and 590 healthy controls. A significant increased risk of prostate cancer was found in men with the P170T/T genotype over the P170C/C genotype (odd ratio=1.74, 95% confidence interval=1.16–2.63, P=0.008), and in men with the P-534C/C genotype over the P-534A/A genotype (odd ratio=1.47, 95% confidence interval=1.18–2.26, P=0.003). Cochran-Armitage trend analysis showed that the P170T allele was significantly correlated with an increased risk of prostate cancer progression (P = 0.029, trend between genotypes and stages) and this observation was also validated in an independent sample cohort. Next, we found that tumors with P170T or P-534C alleles had more 2-fold increased protein expressions of CD24 as compared to those with P170C or P-534A alleles, respectively. Likewise, tumors with a combination of P170T/T and P-534C/C genotypes were associated with a high mRNA level of CD24. Our data suggest a significant association of CD24 genetic variants with prostate cancer onset and progression, which provides new insight into molecular genetics of prostate cancer; however, these findings need to be validated in multiple independent cohorts.
Keywords: CD24, polymorphism, prostate cancer, tumor progression
INTRODUCTION
Prostate cancer (PCa) is one of the most commonly diagnosed malignancies [1]. In clinical practice, PCa is often categorized into different groups primarily based on histological grade (Gleason score), clinical TNM stage, as well as serum prostate-specific antigen (PSA) levels [2]. To date, aggressive, poorly differentiated high-grade PCa remains incurable and potentially lethal, underscoring the urgent need for a better understanding of the molecular basis underlying PCa progression. Whereas 5% to 10% of PCa cases are estimated to be primarily caused by inherited genetic factors or PCa susceptibility genes, large case-control studies and cohort studies suggest that family history is a major genetic determinant in high-risk PCa [3–5]. However, identification of inheritable genetic factors to predict PCa aggressiveness remains fully unknown.
CD24 encodes a glycosylphosphatidylinositol (GPI)-anchored cell surface protein with expression of many tissues and is most abundantly expressed in hematopoietic and immature neuronal cells [6]. Of note, we and three public datasets, such as The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO) and Oncomine, have reported aberrant overexpression of CD24 in human PCa samples, which was associated with aggressive metastasis and poor prognosis for patients with PCa [7–9]. Up to now, CD24 oncogenic function has been well demonstrated in cancer cells by inducing ectopic expression, targeted mutation, gene silencing and antibody blocking [10–12]. Recently, our functional analysis identified that, in PCa cells, intracellular CD24 promoted cell proliferation but inhibited apoptosis, leading to tumor progression and metastasis in both xenogenic and transgenic tumor models [8,9]. This evidence suggests that CD24 as an oncogene appears to be an important determinant for PCa aggressiveness.
Human CD24 messenger RNA (mRNA) contains a 0.24-kb open-reading frame (ORF) and a 1.8kb 3’-untranslated region (UTR). Recently, we first identified the full-length DNA sequence of the human CD24 gene (submitted to NCBI database, accession number FJ226006) [13], as it was wrong to be assembled within the Y chromosome. Thus, while genome-wide association studies analyzed PCa inherited genetic factors or susceptibility genes [14–16], these analyses did not cover CD24 genetic variants due to an unavailable full-length CD24 sequence. Notably, we identified two functional polymorphisms, P170 C/T [11,17] and P1527 TG/deletion [11,18] and a new functional CGC haplotype (P-534C/-492G/-442C in the promoter region) [13] (Fig. 1A). First, P170 C/T at exon 2 in the CD24 putative cleavage site for the GPI anchor [19] results in the replacement of the amino acid alanine with valine. In T cells and hepatocellular cells, the P170T/T genotype expressed higher cell-surface CD24 than the P170C/T or P170C/C genotypes [11,17]. Second, in lymphocyte and hepatocellular cells, the P1527 TG/deletion in the 3’-untranslated region reduced levels of CD24 mRNA by more than two-fold [11,18]. Analysis of RNA decay kinetics revealed that the half-life for the CD24 transcript with the TG deletion was at least 4-fold shorter than that of the TG allele [18]. Third, a haplotype (CGC) consisting of 3 single nucleotide polymorphisms (P-534A/C, P-492G/C, and P-442C/T) specifically binds to a transcription factor, SP1, which is required for promoter activity [13]. Thus, these genetic variants could affect the level of CD24 expression and may be involved in PCa aggressiveness. Given the potential importance of CD24 oncogenic function in PCa aggressiveness, we conducted a case-control association study in 590 unrelated PCa patients and age-matched 590 healthy volunteers in order to assess the genetic susceptibility of CD24 to PCa as well as whether CD24 genetic variants affect tumor progression, which was validated by an independent sample cohort. Likewise, we assessed the association of CD24 genetic variants with its gene expression in prostate cancer.
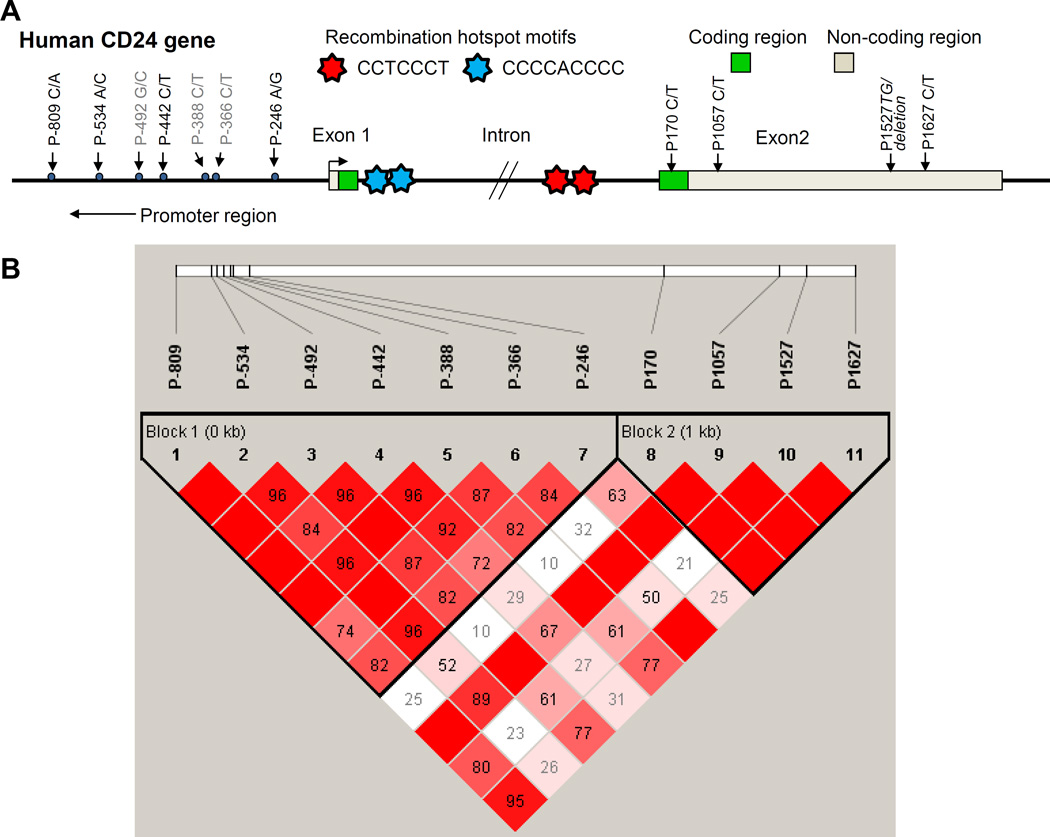
Figure 1. Genetic variants and LD map in human CD24.
A, The genomic exon–intron structure of the human CD24. The polymorphisms are numbered relative to position + 1 representing the ATG translation start site. The vertical black arrows indicate the position of polymorphisms that have been reported in our previous studies [13,17,18]. The recombination hotspot motifs are shown by seven-point stars. The transverse black arrows indicate the transcription start site. B, LD map of human CD24 was analyzed by Haploview [21] software using seven polymorphisms in the proximal promoter with four polymorphisms in exon 2. These polymorphisms are shown as a vertical line within white box. The box with bold black line indicates a LD block.
MATERIALS AND METHODS
Study participants
A total of 1,200 male participants from the Second Hospital of Jilin University were enrolled in our study. Six hundred PCa patients were registered from December 2000 to March 2014. All PCa patients were diagnosed by histological examination of specimens obtained by transrectal sextant needle biopsy, by transurethral resection of the prostate, or by surgical resections. The clinical or pathological stage of PCas at the time of diagnosis was determined by reviewing the medical records based on the American Joint Committee on Cancer (AJCC) Cancer stage grouping system [20]. Pathological grading was based on specimens corresponding to Gleason scores of 2–10. In five patients, no definitive Gleason scores could be determined due to inadequate information. Male controls comprised 600 volunteers without any apparent urinary voiding symptoms or any history of prostatic disease. They were mainly selected randomly from the native Northern Chinese population attending a medical check-up in the Second Hospital of Jilin University between December 2000 and March 2014. Serum total PSA was tested in all male controls with normal levels (defined as 4.0 ng/mL or less). Written informed consent was obtained from all control subjects. The present study was approved by the Ethics Committee of the Second Hospital of Jilin University and written informed consents from all participants were obtained.
Two hundred and six formalin-fixed paraffin-embedded (FFPE) PCa tissue microarray (TMA) specimens were purchased from US Biomax, Inc. (Rockville, MD). Twenty six frozen PCa tissue specimens were collected at the University of Alabama at Birmingham (UAB) Hospital between May 2012 and November 2015. Brief clinical and pathological characteristics of the subjects are presented in Table 1. All patients were diagnosed by histological examination of specimens obtained from surgical resections. The pathological stage of PCa at the time of diagnosis was determined based on the Tumor-Node-Metastasis (TNM) system for US Biomax and UAB specimens. Pathological grading was based on specimens corresponding to Gleason scores of 2–10. In the UAB cohort, all subjects are Caucasians, but there are only Asian subjects in the US Biomax cohort. For all UAB specimens, informed consent was obtained from all subjects in accordance with the requirements of the Institutional Review Board of UAB.
Table 1.
Human Subject Characteristics
| Jilin Universitya | US Biomaxb | UABc | ||
|---|---|---|---|---|
| Control | Case | Case | Case | |
| Total number | n=590 | n=590 | n=206 | n=26 |
| Age, mean±SD | 71.18±7.9 | 70.99±7.7 | 69.21±7.2 | 72.06±9.9 |
| Race | Chinese | Chinese | Asian | Caucasian |
| PSA (ng/ml), median (range) | - | 15.8 (0–152) | 17.5 (0–161) | 16.1 (0–146) |
| Tumor stage (AJCC)d | ||||
| I | - | 187 | - | - |
| II | - | 163 | - | - |
| III | - | 152 | - | - |
| IV | - | 88 | - | - |
| Tumor stage (TNM) | ||||
| T1–2 | - | - | 72 | - |
| T3 | - | - | 56 | 11 |
| T4 | - | - | 18 | 5 |
| N+ or M+ | - | - | 60 | 10 |
| Gleason score | ||||
| G2–6 | - | 117 | 40 | 5 |
| G7 | - | 257 | 67 | 9 |
| G8–10 | - | 216 | 94 | 12 |
Blood DNA samples
Formalin-fixed TMA samples from US Biomax, Inc.
Frozen PCa samples from University of Alabama at Birmingham (UAB)
the American Joint Committee on Cancer (AJCC) TNM system
Linkage disequilibrium (LD) analysis
All CD24 genetic variants were carried out using Haploview software [21] to study the LD pattern at the CD24 locus using the 301 Caucasian healthy control data from our previous studies [13,18]. Specifically, pairwise LD measured r2 was calculated for each of the eleven pairs of genetic variants, and their results were displayed in a tilted matrix.
Selection of haplotype-tagging CD24 genetic variants
In the International HapMap Project, CD24 is wrong to be assembled within the Y chromosome, thus LD information is unavailable at CD24 locus. However, we used our identified two LD blocks to select two potentially functional genetic variants P170 C/T and P-534 A/C as haplotype-tagging single nucleotide polymorphisms (htSNPs) for each LD block, respectively. P170 C/T and P-534 A/C htSNPs with minor allele frequency (MAF) > 0.05 and their potential function have been identified in our previous studies [13,18].
PCR-restriction fragment length polymorphism analysis
CD24 genotypes were determined by PCR-restriction fragment length polymorphism (PCR-RFLP). We observed the following two CD24 loci: the P170 C/T SNP located at exon 2 in the coding region and the P-534 A/C SNP located in the proximal promoter region. Genomic DNA was isolated using the Wizard genomic DNA purification kit (Promega Corp, Fitchburg, WI, USA) for white blood cells and the PicoPure DNA isolation kit (Molecular Devices, Sunnyvale, CA, USA) for PCa cells according to the manufacturer’s instructions. The P170 C/T was amplified by PCR from intron to the end of exon 2 by using a forward primer (5’-CTA AAG AGA ATG ACC TTG GTG GGT TGA-3’) and a reverse primer (5’-GGA TTG GGT TTA GAA GAT GGG GAA A-3’). The PCR conditions were as follows: 95°C for 2 min, 94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec, for 35 cycles, followed by 72°C for 5 min. The PCR products were digested overnight with BstXI (50°C) (New England Biolabs, Ipswich, MA, USA) and then electrophoresed on 3.0% agarose gels. The genotypes were designated as C when the restriction sites of BstXI were absent (404bp) and as T when each restriction site was present (274bp + 136bp) as described in our previous study [18]. For P-534: forward primer (5’-AGA GAT AAC CCT GCC CGA G-3’) and a reverse primer (5’-CCA AGT TTC CTT TGT TTC CC-3’), and the PCR conditions were as follows: 95°C for 2 min, 94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec, for 35 cycles, following 72°C for 5 min. The PCR products were digested overnight with BsrFI (37°C) (New England Biolabs, Ipswich, MA, USA) and were electrophoresed on 3.0% agarose gels. The genotypes were designated as A when the restriction sites of BsrFI were absent (209bp) and as C when each restriction site was present (128bp + 85bp) as described in our previous study [13].
Immunohistochemistry (IHC) and immunofluorescence
The ABC detection system (Vectastain Elite ABC) was used for immunostaining according to the manufacturer’s protocol as described previously [9,22]. Specific primary antibodies were used to detect CD24 (BD Pharmingen, ML5, 1:100, Cat #:555426) and GAPDH (Cell Signaling, 1:500, Cat#: 2118). Protein expressions of CD24 in the plasma membrane and cytoplasm were classified as negative or positive. The results were determined to be negative if <10% of cells within tumor areas were stained or positive if 10%–100% were stained. All slides were examined by two pathologists in a blinded fashion. Mouse anti-CD24 (BD Pharmingen, ML5, 1:200, Cat #:555426) was used for immunofluorescence staining in the pTracer CMV2-CD24C and pTracer CMV2-CD24T [18]-transfected PC3 cells. Prostate cancer cell line PC3 was obtained from the American Type Culture Collection, authenticated by examination of morphology and growth characteristics, and confirmed to be mycoplasma-free. Cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, and cultured for less than 6 months.
Laser capture microdissection
Frozen tissue sections were used for laser capture microdissection to obtain prostate epithelial and cancer cells, as described previously [23,24]. For analysis of gene expression, 5000 cells were microdissected from target tissues.
Quantitative real-time PCR (qPCR)
mRNA expression was analyzed using qPCR on the Real-Time PCR System (Roche) with SYBR Green dye (Promega) in accordance with the manufacturer’s protocol. Relative expression levels were determined using the comparative method (2−ΔΔCt) against endogenous GAPDH controls. The primer sequences for human CD24 expression analysis are a forward primer (5’-TTC TCC AAG CAC CCA GCA-3’) and a reverse primer (5’-TGG AAT AAA TCT GCG TGG GTA-3’).
Statistical analysis
In the case-control association study, cases and controls were examined for any significant differences in their genotype distributions in each of the CD24 genetic variants at the population level. The Hardy-Weinberg equilibrium assumption was confirmed for each genetic variant in the control group. We performed the Pearson Chi-square test for each genetic variant by comparing the distribution of the genotypes of the cases to those of the controls. The Odd ratios (ORs) for the remaining genotype variants were computed using the counts of one of the genotypes as a reference. The associated 95% confidence intervals (CIs) for the ORs were obtained through a multivariate logistic regression model adjusted by age as a potential confounding factor. Patient disease characteristics at diagnosis were summarized as the number and percentage of patients or as the ages, tumor stages, and Gleason scores. Associations of tumor progression with CD24 genotypes were evaluated using both the Pearson Chi-square test and the Cochran-Armitage trend test. For quantitative data, the distribution of data for each group was evaluated using a one-sample Kolmogorov-Smirnov test. In samples with normal distributions, the means of the variables were compared using a two-tailed t test between two groups. In samples with non-normal distributions, the medians of the variable between two groups were compared using a Mann-Whitney test. One-way analysis of variance (ANOVA) was used to test for overall differences followed by Fisher’s least significant difference (PLSD) test. Statistical analyses were performed using the Microsoft Excel version 2010 and SPSS version 22.0 software (IBM/SPSS Inc., Chicago, IL, USA), and P < 0.05 (two sided) was considered statistically significant.
RESULTS
CD24 haplotype structure and recombination hotspot motifs
In the International HapMap Project, CD24 is incorrectly assembled within the Y chromosome, thus the LD information in CD24 locus is unavailable for us. We have identified several genetic variants in the ORF and promoter regions, but did not find any polymorphisms in the intron of CD24 in 301 normal healthy Caucasians (Fig. 1A) [13]. Using this sample database, we constructed the LD map of human CD24 with Haploview software with seven polymorphisms in the proximal promoter and four polymorphisms in exon 2 (Fig. 1B). Of note, two LD blocks are located in the promoter and in exon 2, respectively. Since the P-534C/-492G/-442C haplotype in the promoter region and P170 and P1527 polymorphisms in exon 2 are functional genetic variants [13,17,18] and MAF in P-534 A/C and P170 C/T polymorphisms are more 0.05, we selected these two polymorphisms to determine the genetic susceptibility of CD24 to PCa in our case-control association study.
CD24 genetic variants associated with risk and progression of PCa
The patient clinic characteristics, including age, race, PSA level, tumor stage and Gleason score, are summarized in Table 1. No significant difference in the mean age was found between the PCa group and the control group in the Jilin University sample cohort (P > 0.05). The CD24 genotypes were successfully obtained from 98.3% of blood DNA samples in the PCa cases (590/600) and the controls (590/600). The distribution of CD24 genotypes was in Hardy-Weinberg equilibrium in the control group (P170, P=0.932 and P-534, P=0.280). To assess the risk of PCa according to CD24 genotypes, logistic regression analysis was conducted with adjustment for age at the time of diagnosis. As shown in Table 2, the P170T/T genotype was more commonly observed in PCa patients (12.0%) than in controls (7.8%), and men with the P170T/T genotype had a 1.74-fold increased risk of PCa compared to those with the P170C/C genotype. However, no significant increased risk of PCa (OR = 1.18) was observed when men with the P170C/T genotype were compared to those with the P170C/C genotype (Table 2). Due to a small number of those with the P170T/T genotype, the P170T/T and P170C/T genotypes were combined and then compared to the P170C/C genotype, and a marginal significant association was observed (OR=1.27, 95% CI=1.01–1.59, P=0.045), but OR was dramatically reduced as compared to P170T/T genotype alone, suggesting that the P170T allele is a recessive genetic risk factor for PCa onset. Likewise, men with the P-534C/C genotype had a 1.47-fold increased risk of PCa compared to those with the P-534A/A genotype, but no significant increased risk of PCa (OR = 1.11) was observed in men with the P534A/C genotype (Table 2), suggesting that the P-534C allele is also a recessive genetic risk factor for PCa onset.
Table 2.
CD24 Genotype Frequencies and PCa Risk Against Controls
| Jilin University | ||||||
|---|---|---|---|---|---|---|
| Control | Case | |||||
| SNP | Genotype | N(%) | N(%) | ORa | 95% C.I. | Pb |
| P170 | C/C | 308(52.5) | 273(46.3) | 1.00 (ref)c | - | - |
| C/T | 236(40.0) | 246(41.7) | 1.18 | 0.92–1.50 | 0.189 | |
| T/T | 46(7.8) | 71(12.0) | 1.74 | 1.16–2.63 | 0.008 | |
| P534 | A/A | 147(24.9) | 122(20.7) | 1.00 (ref)c | - | - |
| A/C | 308(52.2) | 285(48.3) | 1.11 | 0.83–1.48 | 0.477 | |
| C/C | 135(22.9) | 183(31.0) | 1.47 | 1.18–2.26 | 0.003 | |
Adjustment for age at the time of diagnosis
Pearson Chi-square test
ref, reference
To further assess the relationship of CD24 genetic variants with tumor stage or Gleason grade at the time of diagnosis, we performed both the Pearson Chi-square test and the Cochran-Armitage trend test in the case group. As shown in Table 3, frequency of the P170C/C genotype was gradually reduced with tumor stages from I to IV (P-trend = 0.029, Cochran-Armitage trend test), whereas those of P170T/T genotype was gradually increased with tumor stages (P-trend = 0.220, Cochran-Armitage trend test). While total genotype distribution was not significant in the Pearson Chi-square test (P = 0.280), the Cochran-Armitage trend test showed a significant trend across all three P170 genotypes with tumor stages (P-trend = 0.029), indicating that P170T allele carrier status influences tumor progression. However, this observation was not reproduced in the P-534 polymorphism with similar analyses (Table 3). Likewise, no significant relationships were observed between CD24 genetic variants and Gleason grades (Table 4).
Table 3.
Association of CD24 Genotypes with Tumor Stages in Patients with PCa
| Jilin University | |||||||
|---|---|---|---|---|---|---|---|
| Tumor stage, N(%) | |||||||
| SNP | Genotype | I | II | III | IV | Pa | Pb |
| P170 | C/C | 92(49.2) | 81(49.7) | 70(46.1) | 30(34.1) | 0.052, trend within genotypes | |
| C/T | 76(40.6) | 64(39.3) | 61(40.1) | 45(51.1) | 0.085, trend within stages | ||
| T/T | 19(10.2) | 18(11.0) | 21(13.8) | 13(14.8) | 0.280 | 0.029, trend between genotypes and stages | |
| P534 | A/A | 41(21.9) | 28(17.2) | 37(24.3) | 16(18.2) | 0.254, trend within genotypes | |
| A/C | 93(49.7) | 80(49.1) | 69(45.4) | 43(48.9) | 0.824, trend within stages | ||
| C/C | 53(28.3) | 55(33.7) | 46(30.3) | 29(33.0) | 0.719 | 0.645, trend between genotypes and stages | |
Pearson Chi-square test
Cochran-Armitage trend test
Table 4.
Association of CD24 Genotypes with Tumor Grades in Patients with PCa
| Jilin University | ||||||
|---|---|---|---|---|---|---|
| Gleason grades, N(%) | ||||||
| SNP | Genotype | G2–6 | G7 | G8–10 | Pa | Pb |
| P170 | C/C | 50(42.7) | 119(46.3) | 104(48.1) | 0.526 Trend within genotypes | |
| C/T | 50(42.7) | 108(42.0) | 88(40.7) | 0.556 trend within grades | ||
| T/T | 17(14.5) | 30(11.7) | 24(11.1) | 0.853 | 0.281 trend between genotypes and grades | |
| P534 | A/A | 26(22.2) | 47(18.3) | 49(22.7) | 0.508 trend within genotypes | |
| A/C | 52(44.4) | 128(31.9) | 105(48.6) | 0.645 trend within grades | ||
| C/C | 39(33.3) | 82(31.9) | 62(28.7) | 0.664 | 0.419 trend between genotypes and grades | |
Pearson Chi-square test
Cochran-Armitage trend test
Validation of the association of CD24 genotypes with tumor progression of PCa
To further validate the potential association of the CD24 genotype with tumor progression, we assessed the CD24 genotypes in an independent sample cohort from 206 Asian patients with PCa (US Biomax, Inc.). All CD24 genotypes were obtained from TMA PCa DNA samples. As shown in Table 5, the frequency of the P170C/C genotype appeared to be gradually reduced with tumor stages from T1 to T4 or N+/M+, but no significance was found (P-trend = 0.197, Cochran-Armitage trend test), whereas that of the P170T/T genotype was gradually increased with tumor stages (P-trend = 0.035, Cochran-Armitage trend test). Total genotype distribution was not significant in the Pearson Chi-square test (P = 0.090), but the Cochran-Armitage trend test showed a significant trend across either three P170 genotypes (P-trend = 0.010) or four tumor stages (P-trend = 0.009) or all P170 genotypes with tumor stages (P-trend = 0.002), validating P170T allele as a genetic risk factor for tumor progression of PCa. However, this significant trend was not observed in the P-534 polymorphism with the similar analyses (Table 5).
Table 5.
Association of CD24 Genotypes with Tumor Stages in Patients with PCa
| US Biomax | |||||||
|---|---|---|---|---|---|---|---|
| Tumor stage, N(%) | |||||||
| SNP | Genotype | T1–2 | T3 | T4 | N+ or M+ |
Pa | Pb |
| P170 | C/C | 41(56.9) | 28(50.0) | 9(50.0) | 21(35.0) | 0.010, trend within genotypes | |
| C/T | 26(36.1) | 22(39.3) | 5(27.8) | 26(43.3) | 0.009, trend within stages | ||
| T/T | 5(6.9) | 6(10.7) | 4(22.2) | 13(21.7) | 0.090 | 0.002, trend between genotypes and stages | |
| P534 | A/A | 19(26.4) | 14(25.0) | 5(27.8) | 14(23.3) | 0.634, trend within genotypes | |
| A/C | 36(50.0) | 29(51.8) | 8(44.4) | 27(45.0) | 0.532, trend within stages | ||
| C/C | 17(23.6) | 13(23.2) | 5(27.8) | 19(31.7) | 0.951 | 0.369, trend between genotypes and stages | |
Pearson Chi-square test
Cochran-Armitage trend test
In addition, as shown in Table 6, frequency of the P170C/C genotype was gradually reduced with Gleason grades from G2–6 to G8–10 (P-trend = 0.017, Cochran-Armitage trend test), whereas that of the P170T/T genotype was not significantly associated with Gleason grades (P-trend = 0.358, Cochran-Armitage trend test). Total genotype distribution was not significant in the Pearson Chi-square test (P = 0.090), but the Cochran-Armitage trend test showed a significant trend across all P170 genotypes with Gleason grades (P-trend = 0.034) (Table 6), indicating that the P170T allele may also be associated with an increased tumor grade of PCa. However, this significant trend was not observed in the P-534 polymorphism with similar analyses (Table 6).
Table 6.
Association of CD24 Genotypes with Tumor Grades in Patients with PCa
| US Biomax | ||||||
|---|---|---|---|---|---|---|
| Gleason grades, N(%) | ||||||
| SNP | Genotype | G2–6 | G7 | G8–10 | Pa | Pb |
| P170 | C/C | 24(60.0) | 36(53.7) | 37(39.4) | 0.104 Trend within genotypes | |
| C/T | 13(32.5) | 20(29.9) | 43(45.7) | 0.058 trend within grades | ||
| T/T | 3(7.5) | 11(16.4) | 14(14.9) | 0.106 | 0.034 trend between genotypes and grades | |
| P534 | A/A | 12(30.0) | 13(19.4) | 27(28.7) | 0.387 trend within genotypes | |
| A/C | 18(45.0) | 34(50.7) | 44(46.8) | 0.959 trend within grades | ||
| C/C | 10(25.0) | 20(29.9) | 23(24.5) | 0.678 | 0.775 trend between genotypes and grades | |
Pearson Chi-square test
Cochran-Armitage trend test
Association of CD24 genetic variants with CD24 gene expression
Our recent IHC analysis has identified CD24 protein expression in 48% of human PCa samples [22]. Of note, CD24 protein expression is associated with tumor progression of PCa, especially tumor metastasis [22]. In the present study, we determined the effect of CD24 genetic variants on CD24 protein expression in the US Biomax sample cohort. As shown in Table 7, the P170T/T genotype was significantly associated with a 2.61-fold increase of CD24 protein expression compared with the P170C/C genotype. While the P170C/T genotype was also associated with a 1.85-fold increase of CD24 protein expression, no statistical significance was found (Table 7). Likewise, the P-534C/C genotype was associated with a 2.53-fold increase of CD24 protein expression compared with the P-534A/A genotype (Table 7). While a 1.73-fold increase of CD24 protein expression was also observed in tumors with P-534A/C genotype, no statistical significance was found (Table 7).
Table 7.
Effect of CD24 Genotypes on CD24 Protein Expression in PCa Tumors
| US Biomax | ||||||
|---|---|---|---|---|---|---|
| CD24− | CD24+ | |||||
| SNP | Genotype | N(%) | N(%) | ORa | 95% C.I. | Pb |
| P170 | C/C | 62(56.4) | 37(38.5) | 1.00 (ref)c | ||
| C/T | 37(33.6) | 42(43.8) | 1.85 | 0.58–3.38 | 0.456 | |
| T/T | 11(10.0) | 17(17.7) | 2.61 | 1.10–6.17 | 0.029 | |
| P534 | A/A | 33(30.0) | 19(19.8) | 1.00 (ref)c | ||
| A/C | 55(50.0) | 45(46.9) | 1.73 | 0.88–3.40 | 0.111 | |
| C/C | 22(20.0) | 32(33.3) | 2.53 | 1.15–5.53 | 0.020 | |
Adjustment for age at the time of diagnosis
Pearson Chi-square test
ref, reference
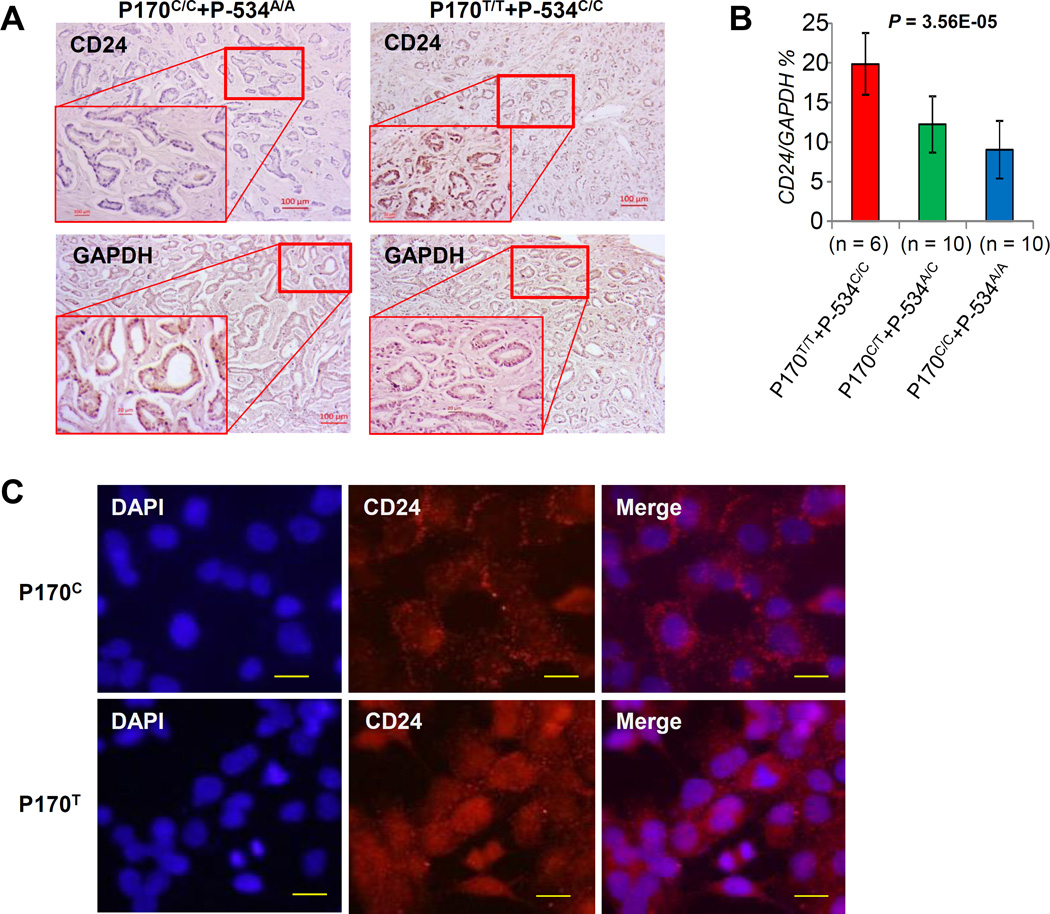
Our recent Analysis of the TCGA and GEO databases demonstrated that CD24 mRNA levels significantly increase with tumor stages, particularly for tumor metastasis in human PCas [22]. In the present study, we used a laser capture microdissection to extract the prostate cancer cells from frozen human PCa samples. After the CD24 genotyping, we obtained 6 PCa cases with both P170T/T and P-534C/C genotypes, 10 PCa cases with both P170C/T and P-534A/C genotypes, and 10 PCa cases with both P170C/C and P-534A/A genotypes (Fig. 2A). Notably, tumor cells with P170T/T and P-534C/C genotypes were associated with the highest levels of CD24 mRNA expression, whereas the lowest levels were found in tumor cells with P170C/C and P-534A/A genotypes (Fig. 2B). In addition, to determine whether the P170 at cleavage site of the GPI anchor [19] affects CD24 expression on plasma membrane, we transfected the exogenous CD24 P170C and P170T, respectively, into PC3 cells. PC3 cells do not express CD24 on the plasma membrane and express small amount of intracellular CD24 [9]. As shown in Figure 2C, the CD24 P170C was obviously transferred on plasma membrane, but the majority of the CD24 P170T was expressed intracellularly.
Figure 2. CD24 gene expressions associated with CD24 genetic variants in human PCa cells.
A, IHC representative CD24 staining (top panels) and GAPDH staining (low panels) of PCa samples with CD24 genotypes. B, mRNA expression levels of CD24 in human PCa cells with CD24 genotypes. The relative amounts are expressed as percentages of GAPDH. Data are presented as the means and SD. Differences in mRNA expression levels among groups were calculated by one-way ANOVA tests. n, sample size. C, immunofluorescence representative CD24 staining of PC3 cells after transfection with pTracer CMV2-CD24-P170C allele (top panels) and pTracer CMV2-CD24-P170T allele (low panels). Scale bar, 100 µm. DAPI, 4',6-diamidino-2-phenylindole.
DISCUSSION
Human CD24 is located at 6q21 [13]. Our haplotype analysis identified two LD blocks on the human CD24 locus in the Caucasian population. One LD block in the promoter region covers the P170 C/T genetic variant and another LD block in exon 2 includes the P-534 A/C genetic variant. Of note, we identified four recombination hotspot motifs in the CD24 intron [25–27]. Thus, these may be the recombination hot spots on the CD24 intron that potentially influence the haplotype patterns and the genetic recombination and sequence evolution of this gene, which may explain the identification of several intronless CD24 pseudogenes on chromosomes 1, 15 and Y in the human genome [28]. In addition, the P170 C/T SNP in the coding region has been cited as rs52812045 in recent publications [29,30], but this reference SNP (refSNP) identification number is still recorded on the Y chromosome in the Database of SNP (dbSNP). P-534 A/C SNP is identified in our previous study [13] and still not issued with a refSNP identification number in refSNP. Thus, P170 C/T and P-534 A/C were used for the two genetic variants of human CD24.
While the genetic effect of CD24 has been investigated in a population-based case–control study with a total of 2,514 patients and 4,858 controls from Caucasian women using TaqMan custom genotyping assays, the association of CD24 genetic variants with risk of breast cancer was not found. However, the P170T/T genotype was significantly associated with breast cancer prognosis for all-cause and breast cancer-specific mortality (hazard ratio = 1.52 and 1.83, respectively) [30]. Also, the P170T/T genotype is suggested to be a significant predictor of pathologic complete response to neoadjuvant chemotherapy for primary breast cancer [31]. Since several intronless CD24 pseudogenes have been identified in the human genome [28], at least one primer in the intron is required for P170 C/T genotyping, but the primer information was not described in the above studies. In addition, a few studies estimated the association of CD24 genetic variants with risk of breast cancer [32] and esophageal cancer [33], but the small sample size of studies limited the power of the statistical analyses to reflect a true effect. The present study is the first to assess the association of CD24 genetic variants with susceptibility to PCa and its disease progression in Asian men. We identified that both the P170T and P-534C alleles are likely to be genetic risk factors for PCa onset, but only the P170T allele appears to be associated with more aggressive PCa, which is also validated in an independent sample cohort. Notably, the P170T and P-534C alleles are related to an increased expression of CD24 in both protein and mRNA levels.
Accumulating data have demonstrated the central role of CD24 in tumor formation, progression, and metastasis in human cancers [34], including PCa [22]. Our previous studies also suggest that the P170T allele appears to be associated with an increased expression of CD24 in T cells [11,17] and the P-534C allele may increase SP1-induced transcription of CD24 in breast cancer cells [13]. Given CD24 is sufficient to promote PCa progression [9], both the P170T and P-534C alleles may also be related to tumor progression through alterations of CD24 expression and protein function. However, as a result, our Cochran-Armitage trend analyses identified a significant association of CD24 P170 genotypes with tumor stages or Gleason grades. More rapid progression of PCa was observed in the patients who carry the P170T allele compared to those patients carrying the P170C allele. Though the P-534 genotypes were significantly associated with risk of PCa onset, no significant relationship between the P-534 genotypes and tumor progression was found in the present study. The underlying reason for the different effects of two genetic variants on tumor progression remains unknown. Since P170 C/T is located at cleavage site for the GPI anchor [19], this genetic variant appears to be associated with trafficking CD24 protein to the cell membrane. In fact, the P170T allele results in the substitution of the amino acid alanine with valine, and our data revealed that this change reduces CD24 protein on cell membrane. Because intracellular CD24 has an oncogenic function in prostate cancer cells [9], the P170 C/T change may enhance CD24 protein function as well as its oncogenic function during tumor progression.
In conclusion, our data suggest the significant association of CD24 genetic susceptibility to PCa onset and progression. P170T and P-534C alleles increase the risk of PCa in Asian men, and patients with the P170C allele had more aggressive disease progression. Our results provide new insights into the molecular genetics of PCa; however, these findings need to be validated by multiple independent cohorts.
Acknowledgments
This work is supported in the grants from the Natural Science Foundation of China (No. 81272472 and No.31571342 for X.Z. and R.L.), the National Institutes of Health/National Cancer Institute (CA179282, CA118948, and CA199586 for L.W. and CA013148 for R.L.), National Institutes of Health/National Center for Advancing Translational Sciences (part of UL1TR001417 for L.W.), the Department of Defense (PC140308 for R.L. and PC130594 for L.W.), and the UAB Pittman Scholar Award (for L.W.).
Abbreviations
- AJCC
American Joint Committee on Cancer
- ANOVA
analysis of variance
- CI
confidence interval
- dbSNP
database of single nucleotide polymorphism
- FFPE
formalin-fixed paraffin-embedded
- GEO
Gene Expression Omnibus
- GPI
glycosylphosphatidylinositol
- GWAS
genome-wide association study
- htSNP
haplotype-tagging single nucleotide polymorphism
- IHC
immunohistochemistry
- LD
linkage disequilibrium
- MAF
minor allele frequency
- mRNA
messenger RNA
- OR
odd ratio
- ORF
open-reading frame
- PCa
prostate cancer
- PCR
polymerase chain reaction
- PCR-RFLP
PCR-restriction fragment length polymorphism
- PLSD
Fisher’s least significant difference
- PSA
prostate-specific antigen
- refSNP
reference single nucleotide polymorphism
- SNP
single nucleotide polymorphism
- SNP
single-nucleotide polymorphism
- TCGA
The Cancer Genome Atlas
- TMA
tissue microarray
- TNM
Tumor-Node-Metastasis
- UAB
University of Alabama at Birmingham
- UTR
untranslated region
Footnotes
Conflict of interest: The authors state no conflict of interest.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA: a cancer journal for clinicians. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Partin AW, Kattan MW, Subong EN, et al. Combination of prostate-specific antigen, clinical stage, and Gleason score to predict pathological stage of localized prostate cancer. A multi-institutional update. Jama. 1997;277(18):1445–1451. [PubMed] [Google Scholar]
- 3.Carter BS, Beaty TH, Steinberg GD, Childs B, Walsh PC. Mendelian inheritance of familial prostate cancer. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(8):3367–3371. doi: 10.1073/pnas.89.8.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gronberg H, Damber L, Damber JE. Familial prostate cancer in Sweden. A nationwide register cohort study. Cancer. 1996;77(1):138–143. doi: 10.1002/(SICI)1097-0142(19960101)77:1<138::AID-CNCR23>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 5.Dean M, Lou H. Genetics and genomics of prostate cancer. Asian journal of andrology. 2013;15(3):309–313. doi: 10.1038/aja.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang X, Zheng P, Tang J, Liu Y. CD24: from A to Z. Cell Mol Immunol. 2010;7(2):100–103. doi: 10.1038/cmi.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kristiansen G, Pilarsky C, Pervan J, et al. CD24 expression is a significant predictor of PSA relapse and poor prognosis in low grade or organ confined prostate cancer. Prostate. 2004;58(2):183–192. doi: 10.1002/pros.10324. [DOI] [PubMed] [Google Scholar]
- 8.Zhang W, Yi B, Wang C, et al. Silencing of CD24 enhances the PRIMA-1-induced restoration of mutant p53 in prostate cancer cells. Clin Cancer Res. 2015 doi: 10.1158/1078-0432.CCR-15-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Liu R, Ye P, et al. Intracellular CD24 disrupts the ARF-NPM interaction and enables mutational and viral oncogene-mediated p53 inactivation. Nat Commun. 2015;6:5909. doi: 10.1038/ncomms6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith SC, Oxford G, Wu Z, et al. The Metastasis-Associated Gene CD24 Is Regulated by Ral GTPase and Is a Mediator of Cell Proliferation and Survival in Human Cancer. Cancer Res. 2006;66(4):1917–1922. doi: 10.1158/0008-5472.CAN-05-3855. [DOI] [PubMed] [Google Scholar]
- 11.Li D, Zheng L, Jin L, et al. CD24 polymorphisms affect risk and progression of chronic hepatitis B virus infection. Hepatology. 2009;50(3):735–742. doi: 10.1002/hep.23047. [DOI] [PubMed] [Google Scholar]
- 12.Sagiv E, Kazanov D, Arber N. CD24 plays an important role in the carcinogenesis process of the pancreas. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2006;60(6):280–284. doi: 10.1016/j.biopha.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Liu R, Li D, et al. A hypermorphic SP1-binding CD24 variant associates with risk and progression of multiple sclerosis. Am J Transl Res. 2012;4(3):347–356. [PMC free article] [PubMed] [Google Scholar]
- 14.Kote-Jarai Z, Olama AA, Giles GG, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;43(8):785–791. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berndt SI, Wang Z, Yeager M, et al. Two susceptibility loci identified for prostate cancer aggressiveness. Nat Commun. 2015;6:6889. doi: 10.1038/ncomms7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann TJ, Van Den Eeden SK, Sakoda LC, et al. A large multiethnic genome-wide association study of prostate cancer identifies novel risk variants and substantial ethnic differences. Cancer Discov. 2015;5(8):878–891. doi: 10.1158/2159-8290.CD-15-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Q, Rammohan K, Lin S, et al. CD24 is a genetic modifier for risk and progression of multiple sclerosis. Proc Natl Acad Sci U S A. 2003;100(25):15041–15046. doi: 10.1073/pnas.2533866100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Lin S, Rammohan KW, et al. A dinucleotide deletion in CD24 confers protection against autoimmune diseases. PLoS Genet. 2007;3(4):e49. doi: 10.1371/journal.pgen.0030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zarn JA, Jackson DG, Bell MV, et al. The small cell lung cancer antigen cluster-4 and the leukocyte antigen CD24 are allelic isoforms of the same gene (CD24) on chromosome band 6q21. Cytogenet Cell Genet. 1995;70(1–2):119–125. doi: 10.1159/000134075. [DOI] [PubMed] [Google Scholar]
- 20.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W, Yi B, Wang C, et al. Silencing of CD24 enhances the PRIMA-1-induced restoration of mutant p53 in prostate cancer cells. Clinical Cancer Research. 2016 doi: 10.1158/1078-0432.CCR-15-1927. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Liu R, Li W, et al. Somatic single hits inactivate the X-linked tumor suppressor FOXP3 in the prostate. Cancer Cell. 2009;16(4):336–346. doi: 10.1016/j.ccr.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu R, Yi B, Wei S, et al. FOXP3-miR-146-NF-kappaB Axis and Therapy for Precancerous Lesions in Prostate. Cancer Res. 2015;75(8):1714–1724. doi: 10.1158/0008-5472.CAN-14-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310(5746):321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 26.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40(9):1124–1129. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 27.Jeffreys AJ, Neumann R. The rise and fall of a human recombination hot spot. Nat Genet. 2009;41(5):625–629. doi: 10.1038/ng.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hough MR, Rosten PM, Sexton TL, Kay R, Humphries RK. Mapping of CD24 and homologous sequences to multiple chromosomal loci. Genomics. 1994;22(1):154–161. doi: 10.1006/geno.1994.1356. [DOI] [PubMed] [Google Scholar]
- 29.Yan S, Xu D, Jiang T, et al. CD24 single nucleotide polymorphisms and cancer risk. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35(9):8927–8932. doi: 10.1007/s13277-014-2127-2. [DOI] [PubMed] [Google Scholar]
- 30.Buck K, Hug S, Seibold P, et al. CD24 polymorphisms in breast cancer: impact on prognosis and risk. Breast cancer research and treatment. 2013;137(3):927–937. doi: 10.1007/s10549-012-2325-9. [DOI] [PubMed] [Google Scholar]
- 31.Marme F, Werft W, Walter A, et al. CD24 Ala57Val polymorphism predicts pathologic complete response to sequential anthracycline- and taxane-based neoadjuvant chemotherapy for primary breast cancer. Breast cancer research and treatment. 2012;132(3):819–831. doi: 10.1007/s10549-011-1759-9. [DOI] [PubMed] [Google Scholar]
- 32.Zhou X, Cao Y, Luo J, Zeng X. [Association between CD24 polymorphism and genetic susceptibility to breast cancer: a case-control study] Zhong nan da xue xue bao Yi xue ban = Journal of Central South University Medical sciences. 2013;38(11):1122–1129. doi: 10.3969/j.issn.1672-7347.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 33.Sadot E, Kraus S, Stein M, et al. CD24 gene polymorphism--a novel prognostic factor in esophageal cancer. The International journal of biological markers. 2014;29(1):e49–e54. doi: 10.5301/jbm.5000071. [DOI] [PubMed] [Google Scholar]
- 34.Lee JH, Kim SH, Lee ES, Kim YS. CD24 overexpression in cancer development and progression: a meta-analysis. Oncol Rep. 2009;22(5):1149–1156. doi: 10.3892/or_00000548. [DOI] [PubMed] [Google Scholar]