Abstract
Background
Alcohol use typically begins during adolescence and escalates into young adulthood. This represents an important period for the establishment of alcohol use and misuse patterns, which can have psychosocial and medical consequences. Although changes in alcohol use during this time have been phenotypically characterized, their genetic nature is poorly understood.
Methods
Participants of the Avon Longitudinal Study of Parents and Children (ALSPAC) completed the Alcohol Use Disorders Identification Test (AUDIT) four times from age 16–20. We used Mplus to construct a growth model characterizing changes in AUDIT scores across time (N=4545, where data were available for at least two time points). The slope of the model was used as the phenotype in a genome-wide association study (GWAS; N=3380), followed by secondary genetic analyses.
Results
No individual marker met genome-wide significance criteria. Top markers mapped to biologically plausible candidate genes. The slope term was moderately heritable (h2SNP=0.26, p=0.009), and replication attempts using a meta-analysis of independent samples provided support for implicated variants at the aggregate level. Nominally significant (p<0.00001) markers mapped to putatively active genomic regions in brain tissue more frequently than expected by chance.
Conclusions
These results build on prior studies by demonstrating that common genetic variation impacts alcohol misuse trajectories. Influential loci map to genes that merit additional research, as well as to intergenic regions with regulatory functions in the central nervous system. These findings underscore the complex biological nature of alcohol misuse across development.
Keywords: ALSPAC, alcohol problems, genetic influences, heritability, longitudinal model
Introduction
Alcohol use frequently begins in adolescence, with upwards of 50% of US 10th graders (Johnston et al., 2013) and over 40% of UK eleven to fifteen-year-olds (Lifestyle Statistics, 2014) reporting initiation of alcohol use. Consumption typically increases into young adulthood before stabilizing or tapering off (Johnston et al., 2013). The pattern of use established during adolescence into young adulthood can be predictive of later alcohol use problems (Blozis et al., 2007; Duncan et al., 1997; Nixon and McClain, 2010), which are associated with a host of social, economic, and medical costs (McCambridge et al., 2011; Sacks et al., 2015; Secretary of State for the Home Department, 2012a; Secretary of State for the Home Department, 2012b). Thus, clarifying the etiology of alcohol misuse has significant implications for public health.
Genetic factors are known to substantially impact alcohol use phenotypes. A recent meta-analysis of twin and adoption studies reported a heritability estimate of .49 for alcohol use disorders (Verhulst et al., 2015). Other studies have reported heritabilities of .21–.55 for associated phenotypes such as frequency of use, intoxication frequency, and problem drinking (Derks et al., 2014; Edwards et al., 2011a; Edwards et al., 2011b; Geels et al., 2012; Sartor et al., 2013; Wu et al., 2014). Furthermore, evidence suggests that the heritability of alcohol use phenotypes increases from early adolescence into young adulthood (Bergen et al., 2007).
Most prior studies investigating the genetic etiology of alcohol outcomes have focused on either cross-sectional data or lifetime measures. However, abundant evidence suggests that alcohol-related behaviors develop over time and vary across individuals (Casswell et al., 2002; Duncan et al., 1997; Tucker et al., 2003; Wiesner et al., 2007; Windle et al., 2005). Cross-sectional or collapsed (i.e., lifetime) measures fail to capture the dynamic nature of change in alcohol use across time; consequently, genetic studies employing these measures are limited in the extent to which they identify variants or genes that impact the course of alcohol use. Given the complex nature of alcohol use phenotypes, complementary genetic analyses will likely be needed to comprehensively dissect their etiology: Distinct or only partially overlapping genetic factors might impact different aspects such as alcohol use initiation, acceleration of use, alcohol use disorder, recovery, persistence, etc.
While phenotypic analyses of alcohol use/misuse trajectories are common, corresponding genetic analyses are not. To our knowledge, only one prior study has examined genetic influences underlying changes in alcohol outcomes over time. In a study involving three US cohorts longitudinally assessed from childhood into young adulthood, Adkins and colleagues (2015) modeled developmental trajectories of alcohol consumption, followed by genome-wide association analysis of the resulting slope parameter. Results were meta-analyzed across cohorts, and while no marker met genome-wide significance criteria for association with the slope, biologically plausible suggestive markers were identified, and secondary analyses implicated genes involved in axon guidance and development. Parallel to the analysis of the slope, Adkins et al. conducted association tests for mean alcohol consumption across time. Importantly, distinct genetic loci and pathways were implicated across phenotypes, indicating that the genetic factors impacting changes in alcohol consumption differ from those impacting a measure that is effectively cross-sectional.
The current study aims to address the deficit in our understanding of genetic influences on the course of alcohol misuse. We employ data from the Avon Longitudinal Study of Parents and Children (ALSPAC), a prospectively assessed cohort study in the southwest UK, to quantify growth in alcohol misuse from adolescence to emerging adulthood. As only one prior study has subjected a comparable outcome to genetic analysis (Adkins et al., 2015), the current study represents a relatively novel approach to conceptualizing the genetic risk of alcohol use problems.
Materials and Methods
Sample
The Avon Longitudinal Study of Parents and Children (ALSPAC) is a cohort-based sample recruited in southwest England. ALSPAC recruited 14,541 pregnant women resident in Avon, UK with expected dates of delivery 1st April 1991 to 31st December 1992. 14,541 is the initial number of pregnancies for which the mothers enrolled in the ALSPAC study and had either returned at least one questionnaire or attended a “Children in Focus” clinic by 19 July 1999. Of these initial pregnancies, there was a total of 14,676 fetuses, resulting in 14,062 live births and 13,988 children who were alive at 1 year of age. Subsequent phases of enrolment increased the sample size over time. The phases of enrolment are described in more detail elsewhere (Boyd et al., 2013; Fraser et al., 2013). For the current analyses, full or partial phenotypic data were available for 4545 participants (see below). The study website contains details of all the data that is available through a fully searchable data dictionary (http://www.bris.ac.uk/alspac/researchers/data-access/data-dictionary/). Ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees.
Measures
Participants completed the 10 items of the Alcohol Use Disorders Identification Test (AUDIT; (Babor and Grant, 1989)) at approximately ages 16.5, 17.5, 18.75, and 20.75. At age 17.5, data was collected at a clinic, at which participants completed the AUDIT on a computer. Otherwise, data was collected via postal or online questionnaire. Scores for each AUDIT item range from 0–4 as a function of how frequently (e.g., “Never” to “Daily or almost daily”) the respondent has experienced that item. Total scores could range from 0–40.
Genotyping
Samples were genotyped on the Illumina HumanHap550 quad SNP genotyping platform by the Wellcome Trust Sanger Institute (Cambridge, UK) and Laboratory Corporation of America (Burlington, NC, US). Individuals were excluded on the basis of gender mismatch; minimal or excessive heterozygosity; individual missingness >3%; and insufficient sample replication (IBD<0.8). Population stratification was assessed using multidimensional scaling analysis and compared to Hapmap II populations; individuals not of European ancestry were excluded. Markers with MAF<1%, call rate <95%, and violations of Hardy-Weinberg equilibrium (p<5e–7) were removed. Individuals with evidence of cryptic relatedness were removed (IBD>0.1). After quality control filters were applied, data were available for 9,048 subjects and 526,688 SNPs. Haplotypes were estimated using ShapeIT (v2.r644), and imputation was conducted using a phased version of the 1000 Genomes reference panel (Phase 1, Version 3), using Impute V2.2.2 and all reference haplotypes to maximize imputation quality.
Statistical Analyses
Growth model
In Mplus version 7.11, we fit a latent growth model (Muthén and Muthén, 2012) for the four waves of AUDIT scores. We estimated an intercept (I), slope (S), quadratic (Q), and cubic (C) term, and conducted a series of tests to identify the most parsimonious model that provided a good fit to the data, using Root Mean Square Error of Approximation (RMSEA), the Tucker-Lewis Index (TFI), and Comparative Fit Index (CFI) to interpret fit.
Genetic analyses
We conducted GWAS on the imputed (dosage) genetic data using Plink v1.07 (Purcell et al., 2007). Only autosomal markers were analyzed. We included sex and 10 ancestry-informative principal components as covariates. Slope was the focal phenotype; other relevant growth factors (I and Q, see Results) as covariates.
We used VEGAS2 (Mishra and Macgregor, 2015) to perform gene-based analyses. Such tests represent a biology-based approach to interpreting marker-level results, as they consider aggregate effects across a functionally meaningful genomic region. VEGAS2 mapped markers falling within 50kb to known genes; this flanking region was selected in order to capture potentially influential regulatory regions. VEGAS2 accounts for linkage disequilibrium among markers mapping to a given gene based on the selected 1000 Genomes (The 1000 Genomes Project Consortium et al., 2012) reference population (in this case, the European subset). We considered both VEGAS2’s gene-based p-values and Benjamini-Hochberg false discovery rates (FDR); the latter were derived using p. adjust in R version 3.2.3.
We used Genome-wide Complex Trait Analysis (GCTA; (Yang et al., 2011)) to calculate the proportion of phenotypic variance attributable to genetic variation (h2SNP), using only observed variants. The genetic relationship matrix was constructed using a relatedness cut-off of 0.025. We included sex, non-focal growth parameters, and 10 ancestry-informative principal components as covariates.
Replication
We selected promising (p<0.0001) markers for individual SNP-based replication attempts using the only available published study of a similar phenotype (Adkins et al., 2015). P-values were combined across the current study and the meta-analytic results from Adkins et al., using Fisher’s method. In addition, we conducted sign tests, using a subset of markers: markers were first selected for their availability in the replication sample; redundant markers were removed, and reference alleles were aligned across the current results and the replication sample. Because the replication sample was a meta-analysis, combined t-statistics were used rather than betas. Markers were next selected for independence using Plink’s --clump option in two stages: first, we applied a LD threshold of r2=0.5 within a 250kb window; we then applied a threshold of r2=0.2 within a 5mb window. Selected markers were binned by p-value for sign tests.
Epigenomic annotations
SNPs were annotated for various epigenetic and functional features using data obtained from HaploReg (v4.1) (Ward and Kellis, 2016). These data included DNaseI hypersensitive sites (DHS), histone modification sites and chromatin state segmentations (15-state) produced by the RoadMap Epigenomics Project for various cell types and biological samples (Roadmap Epigenomics et al., 2015). For each of these cell type-specific features we calculated the frequencies with which they overlapped any of the assayed SNPs. We then compared the overlap among SNPs nominally associated with growth in alcohol misuse to the background frequencies in order to calculate a cell type-specific fold enrichment score. The background for this analysis comprised a set of 8,887,107 markers, which represents the overlap of markers assayed in this study and included in dbSNP release b137. Enrichment relative to this background was calculated using a binomial statistic. Within each class, we limited our assessment to feature-threshold combinations to which at least 3 markers mapped.
Results
Descriptive statistics
Individuals who did not endorse alcohol use initiation are coded as missing as they did not satisfy the screening condition for administration of the AUDIT items. A total of 1332 individuals participated in all 4 waves; N=1681 participated in 3 waves; N=1532 participated in 2 waves; and N=2026 participated in only 1 wave. AUDIT scores at each wave of data collection are provided in Table 1; scores increased gradually across time.
Table 1.
Mean (SD) AUDIT scores and number of participants with valid data at each age.
| Age | N | % Male | Mean (SD) | Skewness | Cronbach’s α |
|---|---|---|---|---|---|
| 16.50 | 4660 | 41.2 | 6.29 (5.29) | 1.09 | 0.78 |
| 17.50 | 3929 | 43.9 | 6.94 (4.89) | 1.10 | 0.76 |
| 18.75 | 3100 | 36.2 | 7.80 (5.06) | 0.78 | 0.78 |
| 20.75 | 3772 | 39.4 | 8.88 (5.45) | 0.81 | 0.77 |
Growth model fitting
We fit a model based on the AUDIT scores across 4 waves, specifying within Mplus the average temporal lag between waves of data collection. We began with a multi-group model where males were group 1 and females were group 2; we estimated an intercept (I), slope (S), quadratic (Q), and cubic (C) term. The variance of C was constrained to 0 to facilitate model convergence; in addition, within sex, the residual variances of the AUDIT scores were constrained to be equal across waves (Grimm and Widaman, 2010). The model provided an excellent fit to the data (CFI=1.000, TLI=0.999, RMSEA=0.012). We next assessed sex differences by testing whether the means of I, S, Q, and C differed across sexes, and found that they did not (p>0.1); this model also provided an excellent fit (CFI=0.998, TLI=0.998, RMSEA=0.017). Therefore, subsequent models combined the sexes. We next restricted the model to individuals for whom at least 2 AUDIT scores were available (N=4545) (CFI=0.995, TFI=0.991, RMSEA=0.029). Within this sample, we tested models that dropped the C and/or Q growth parameters or restricted Q variance to 0, all of which resulted in a decrement in fit (e.g., RMSEA>0.05, CFI/TLI<0.99). Therefore, we selected the model with all four growth parameters, with C variance constrained to 0, as our final model. Individual scores on each parameter were exported for use in the subsequent GWAS.
Genomewide association analysis
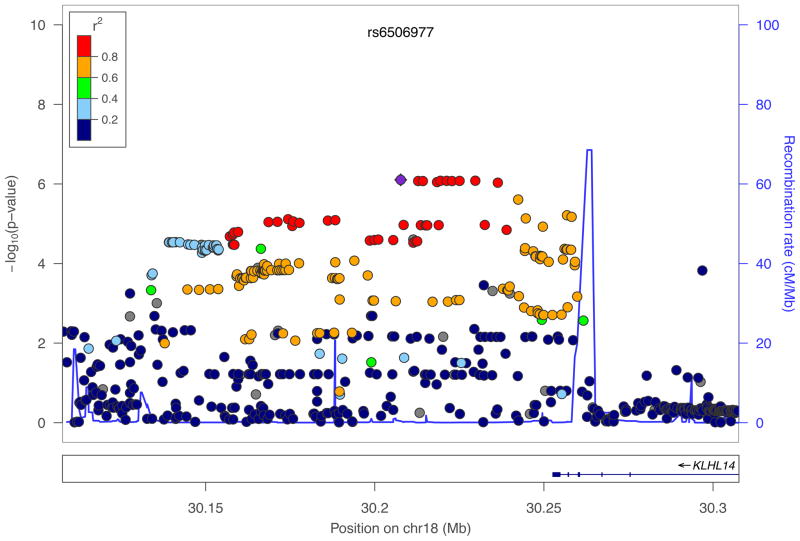
We used the slope term (Grimm et al., 2013) from the growth model as the focal phenotype in a GWAS. Of the 4545 individuals with phenotypic data, genotypic data were available for N=3380 individuals. After restricting to markers with INFO≥0.50 and minor allele frequency (MAF) ≥0.01, results were available for 8,972,921 markers. There was no evidence of inflation (λ=1.01, SE=1.11e-6). No marker met genome-wide significance criteria (p<5e-8). The top marker, rs143795029, is located on chromosome 2p12 in an intronic region of LRRTM4 (leucine rich repeat transmembrane neuronal 4). However, low MAF (0.01) and local linkage disequilibrium suggest that this association is potentially artifactual. The next marker, rs6506977 (p=7.88e-7), along with 9 neighboring markers with p<1e-7, maps to the region 45kb 3′ of KLHL14 (kelch-like family member 14) on chromosome 18q12.1 (Figure 1). The next most strongly implicated region, with top marker rs17397108, maps to 66kb 3′ of LMX1A (LIM homeobox transcription factor 1, alpha) on chromosome 1q24.1.
Figure 1.
Regional association plot of the top marker (rs6506977, in purple) mapping to the region 3′ of KLHL14, created by LocusZoom (Pruim et al., 2010).
Gene-based analysis
Markers with MAF≥0.01 and INFO≥0.50 were submitted to VEGAS2 for gene-based analysis. These markers mapped to 24,233 defined genes. No gene survived corrected significance criteria (q<0.05). The top gene was NSMCE3 (NSE3 homolog, SMC5-SMC6 complex component; alias NDNL2; p=0.000037; q=0.51), on chromosome 15, which plays a role in chromatin remodeling. The 2nd ranking gene, DHX34 (DEAH box polypeptide 34; p=0.000105; q=0.51), maps to chromosome 19 and is involved in RNA regulation (Longman et al., 2013).
Genomewide Complex Trait Analysis
We used GCTA to investigate whether, in aggregate, common genetic variants account for a significant proportion of the phenotypic variance of the slope term. The h2SNP estimate was 0.26 (SE=0.11, p=0.009), indicating moderate heritability that differed significantly from 0. Although slope was the primary phenotype of interest, we also computed the heritability of the intercept and quadratic terms, which were h2SNP=0.11 (SE=0.11, p=0.1) and h2SNP=0 (SE=0.11, p=0.5), respectively. We further tested the genetic correlation between I and S (Q was excluded due to its 0 estimate), and obtained an estimate of 0.05 (SE=0.60, ns).
Replication
We selected markers with p<0.0001 (a threshold chosen to balance statistical support and sample size) for comparison to results reported by Adkins and colleagues (Adkins et al., 2015). Of 9397 markers selected, target sample results were available for 1141 due to the use of different imputation reference panels across studies. The Fisher’s combined p-values were <0.05 for 1115 of these markers; however, we observed little support for specific variants across the current study and the 3 component studies of the Adkins et al. meta-analysis (Supplementary Table 1).
We conducted sign tests using independent markers selected for their availability in both the current study and the replication study (N=465,265). We then binned according to discovery sample p-value and determined the number of markers within each bin with the same direction of effect across studies. Results are provided in Table 2. At stringent significance thresholds (p≤0.0001) we observed modest evidence of agreement across studies, though these results are based on small numbers of markers. At more inclusive thresholds (p≤0.10 to p≤0.50), results indicated more consistently significant overlap.
Table 2.
Sign test results.
| P-value Bin | # with Consistent Sign | Total # in Bin | P-value |
|---|---|---|---|
| p≤0.00001 | 9 | 13 | 0.1334 |
| p≤0.0001 | 69 | 114 | 0.01539 |
| p≤0.001 | 411 | 821 | 0.5 |
| p≤0.01 | 4105 | 8240 | 0.6336 |
| p≤0.10 | 36312 | 71856 | 0.002109 |
| p≤0.25 | 81360 | 161723 | 0.00663 |
| p≤0.50 | 144472 | 286659 | 9.95E-06 |
Localization of implicated markers to putative functional regions
Enrichment analyses were conducted for three classes of epigenetic features: i) chromatin marks, which are subdivided into 15 classes; ii) DNase Hypersensitivity Sites (DHS); and iii) histone mark peaks. Table 3 provides the 5 highest enrichment scores for each class; Supplementary Table 2 provides complete results. For DHS, the top categories included markers with p<0.00001; the two top categories correspond to putatively active genomic regions in fetal brain tissue. Top active chromatin mark categories, all of which included markers with p<0.0001, correspond to transcription start sites or enhancers across a variety of tissues, including hippocampal cells, ovary, and hematopoietic stem cells. Finally, top enrichment scores for histone marks correspond to markers with p<0.00001, and map to brain, lung, spleen, blood, and embryonic stem cells.
Table 3.
Top five enrichment scores for each class of epigenetic mark.
| Class | P-value Threshold | Type (if applicable) | # Markers | Enrichment Score | p-value | q-value | Tissue |
|---|---|---|---|---|---|---|---|
| DNase Hypersensitivity Site | 0.00001 | NA1 | 7 | 3.2476 | 1.48E-03 | 2.94E-01 | Fetal Brain Female |
| 0.00001 | NA | 7 | 3.0431 | 2.22E-03 | 2.94E-01 | Fetal Brain Male | |
| 0.00001 | NA | 5 | 2.6031 | 1.29E-02 | 5.15E-01 | H1 Derived Neuronal Progenitor Cultured Cells | |
| 0.00001 | NA | 7 | 2.2289 | 1.36E-02 | 5.15E-01 | Fetal Lung | |
| 0.00001 | NA | 4 | 2.1409 | 3.99E-02 | 7.69E-01 | HepG2 Hepatocellular Carcinoma Cell Line | |
|
| |||||||
| Chromatin Mark | 0.0001 | Bivalent/Poised TSS2 | 3 | 21.9606 | 1.29E-05 | 2.44E-03 | Ovary |
| 0.0001 | Bivalent/Poised TSS | 5 | 14.1634 | 1.96E-06 | 7.12E-04 | H1 BMP4 Derived Trophoblast Cultured Cells | |
| 0.0001 | Bivalent Enhancer | 7 | 13.8871 | 6.46E-08 | 7.33E-05 | Primary T cells from cord blood | |
| 0.0001 | Bivalent/Poised TSS | 3 | 13.3484 | 8.84E-05 | 9.17E-03 | Primary hematopoietic stem cells | |
| 0.0001 | Bivalent Enhancer | 11 | 11.6911 | 4.00E-10 | 1.02E-06 | Brain Hippocampus Middle | |
|
| |||||||
| Histone Mark Peaks | 0.00001 | H3K4me1_Enh | 21 | 2.4863 | 2.48E-05 | 1.01E-03 | Fetal Brain Female |
| 0.00001 | H3K4me3_Pro | 9 | 2.3770 | 4.66E-03 | 7.67E-02 | Fetal Lung | |
| 0.00001 | H3K4me1_Enh | 27 | 2.1450 | 2.74E-05 | 1.07E-03 | Spleen | |
| 0.00001 | H3K4me1_Enh | 15 | 2.1009 | 1.88E-03 | 3.67E-02 | ES-WA7 Cells | |
| 0.00001 | H3K4me1_Enh | 15 | 2.0685 | 2.20E-03 | 4.17E-02 | Primary T cells effector/memory enriched from peripheral blood | |
DNase Hypersensitivity Sites are not categorized into genomic locations by the Roadmap Project.
TSS=transcription start site
Discussion
Genetic factors are known to influence alcohol use phenotypes including initiation, frequency of use, and alcohol use disorder. Prior studies have illustrated that there is variation in the development of alcohol use over time, with adolescence into young adulthood comprising a critical period for the establishment of alcohol use patterns that can be predictive of later problems. We demonstrate that growth in alcohol misuse is moderately heritable (h2SNP=0.26), and that genes contributing to growth represent biologically plausible candidate genes. Furthermore, the low co-heritability between initial (age 16.5) alcohol misuse and slope indicates that genetic factors influencing distinct aspects of the development of alcohol misuse are largely, though not entirely, independent. Indeed, risk factors, genetic or otherwise, impacting alcohol misuse may only partially overlap with risk factors for alcohol consumption, initiation, etc. We observed support for implicated variants, in aggregate, in a US replication sample, suggesting that genetic factors influencing changes in alcohol misuse in the UK are generalizable to an ethnically similar population. Exploratory in silico molecular analyses indicate that implicated variants map to genomic locations transcriptionally active during brain development.
The growth model demonstrates that AUDIT scores increase from age 16.5 through 20.75 in a non-linear fashion, with the inclusion of both quadratic and cubic terms contributing to model fit. Previous studies of samples in a similar age range have also shown that alcohol use/misuse increases during this period (Casswell et al., 2002; Jackson et al., 2008; Tucker et al., 2003). Though not all prior studies have included a non-linear growth term in their longitudinal models, Marmorstein (2009) reported that a quadratic term improved model fit when examining alcohol use-related problems in a large sample of individuals in their adolescence into early adulthood. In that sample, alcohol problems peaked at 22, followed by a gradual decline. Similarly, Walden et al. (2007) reported nonlinear growth in alcohol consumption and alcohol dependence symptom counts. We included the quadratic term as a covariate in our analyses (the cubic term was excluded as its variance was fixed to 0), but focused on the slope term as the outcome of interest given its interpretable nature. Future studies may benefit from analysis of genetic influences on the non-linear components of growth in alcohol misuse, and we note that the current study’s focus on the slope represents a limited perspective on genetic influences on change in alcohol misuse. Joint consideration of the growth factors (e.g., a multivariate GWAS) may have had an impact on our results and their interpretation.
Walden et al. (2007), using a sample of adolescent twins, found that a parental history of alcohol problems and/or higher parental alcohol consumption predicted greater rates of change in offspring alcohol outcomes. Given that a proportion of the risk conferred by parental problems is genetic, those results are conceptually consistent with the current findings, in that genetic liability contributes to the course of alcohol misuse during this critical developmental period.
The heritability estimate of the slope obtained from observed variants is consistent with, though modestly lower than, estimates for other alcohol-related phenotypes. Evidently, and unsurprisingly, environmental factors account for a substantial proportion of risk in course of alcohol problems during this time frame. Such factors might include peer behavior (Barnett et al., 2014; Bertholet et al., 2013) or college attendance (Carter et al., 2010; Slutske, 2005). Though outside the scope of the current analyses, future studies might assess whether genetic liability for increasing alcohol problems over time is moderated by specific environmental protective or risk factors.
Our findings suggest two genes, KLHL14 and LMX1A, warrant further investigation based on SNP-level findings. KLHL14 is expressed in neuronal cell bodies, and the protein interacts with the TOR1A protein (encoded by torsin family 1, member A), which is expressed in the substantia nigra pars compacta, a region that is responsive to ethanol infusion (Asyyed et al., 2006) and ethanol withdrawal (Kozell et al., 2005) in mice. LMX1A, proximal to the set of SNPs nominally implicated (p≤1.19e-6) on chromosome 1, is of potential interest based on its known function: in addition to its role in insulin gene transcription, it is involved in the embryonic development of dopaminergic neurons (Doucet-Beaupre et al., 2015), which are central to the neurobiology of drugs of abuse (Korpi et al., 2015). In addition, preliminary evidence suggests an association between variation in LMX1A and cognitive functioning and psychiatric disorders (Bergman et al., 2010; Rolstad et al., 2015). Thus, although these genes have not been directly associated with alcohol-related outcomes in prior studies, they represent biologically plausible candidates for follow-up. Molecular follow-up of implicated genes and putatively functional polymorphisms is ongoing.
Our attempts at replication using meta-analytic results, which were derived from three longitudinally assessed samples of comparable ages (Adkins et al., 2015), provide additional support for our results, and suggest that the current findings are generalizable. Although comparisons of significance of individual markers did not strongly implicate specific loci, sign tests indicate that the direction of effect is consistent across studies more frequently than expected by chance. This was true for markers meeting relatively stringent significance thresholds as well as for markers meeting higher p-value thresholds. This is consistent with the polygenic nature of alcohol use phenotypes: we expect many genetic variants to incrementally impact the trajectory of alcohol use from adolescence into emerging adulthood. We note that the strength of the replication using this method is modest: only a small number of markers meet the more stringent thresholds, thereby reducing statistical power, and while the large number of markers meeting less stringent thresholds conveys higher power, the proportion of markers with the same direction of effect at those thresholds only slightly exceeds 50%. We therefore caution against over-interpretation of the sign test results.
Our exploratory enrichment analysis provides insight as to the potential functional role of markers implicated at various p-value thresholds. Results suggest that markers with p<0.00001 disproportionately map to open chromatin (indicated by DHS or H3K4me1 marks) in brain tissue. Fetal brain tissue is of particular relevance, implying that these markers play a role in CNS development. Further analyses are planned to investigate whether genotypes at the implicated loci have potential functional consequences, such as differential expression of proximal genes, in the interim the current findings should be considered preliminary.
Limitations
The refined phenotype limited our ability to attempt replication in other datasets. While the AUDIT does include items related to alcohol consumption, the total score used in the current analyses is designed to capture problems/misuse, and in particular our analysis of growth in this construct was relatively novel. Though comparable phenotypic analyses (i.e., growth models of alcohol misuse) exist, there is, to our knowledge, only one published report of a genetic analysis of the slope of a longitudinally assessed alcohol outcome (Adkins et al., 2015). We observed inconsistent evidence of replication, with top markers failing to replicate across studies. However, sign tests, which investigate replication at the aggregate level, did provide modest support for common SNP effects across samples. These findings warrant confirmation in additional studies. However, we note that phenotype definition is critical for valid replication attempts: the low (and non-significant) coheritability between the intercept and slope terms within the current sample suggests that replication using a more standard phenotype (e.g., a cross-sectional measure of alcohol misuse) would not be informative, as variants would not be expected to impact both measures. Furthermore, this may have impacted the somewhat modest level of replication we observed, as Adkins and colleagues employed a measure of alcohol consumption rather than problems: there is likely incomplete overlap among the factors – genetic or otherwise – that impact various alcohol-related outcomes such as problems vs. consumption.
The ALSPAC sample is homogenous with respect to ethnicity. While this makes the sample ideal for genetic analysis, it does present limitations with respect to generalizability to other samples. Environmental factors that impact alcohol outcomes differ across cultures; for example, the legal drinking age in the UK is younger than in the US, and light alcohol consumption among early adolescents is sanctioned within some cultures but not others. One straightforward consequence of cross-sample comparability is that such environmental influences affect heritability estimates. However, the genetic etiology of alcohol problems is not known to differ substantially across populations, with the important exception of variation within alcohol metabolizing genes being far more common within East Asian populations (Edenberg, 2007).
Despite these concerns, the results presented herein represent a contribution to emergent evidence that the course of alcohol use phenotypes has a substantial genetic component. While phenotypic studies have long acknowledged the essential developmental aspect of alcohol problems, the application of genetic approaches to this aspect of etiology is relatively nascent. Further clarification of these genetic factors is critical to improving our understanding of how alcohol misuse unfolds during development.
Supplementary Material
Acknowledgments
Funding: NIH: AA021399, AA018333, 1P50AA022537, R21AA022749-01, UL1TR000058 and R37AA011408. MRC and ESRC: MR/L022206/1 & ES/L015471/1.
We are grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council and the Wellcome Trust (Grant ref: 092731) and the University of Bristol provide core support for ALSPAC. This publication is the work of the authors and ACE will serve as guarantor for the contents of this paper. This research was specifically funded by the National Institutes of Health (AA021399, AA018333, 1P50AA022537, R21AA022749-01, UL1TR000058 and R37AA011408). We acknowledge additional support from MRC and ESRC (MR/L022206/1 & ES/L015471/1). The authors have no conflicts of interest to declare.
References
- Adkins DE, Clark SL, Copeland WE, Kennedy M, Conway K, Angold A, Maes H, Liu Y, Kumar G, Erkanli A, Patkar AA, Silberg J, Brown TH, Fergusson DM, Horwood LJ, Eaves L, Van Den Oord EJ, Sullivan PF, Costello EJ. Genome-Wide Meta-Analysis of Longitudinal Alcohol Consumption Across Youth and Early Adulthood. Twin Res Hum Genet. 2015;18:335–47. doi: 10.1017/thg.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asyyed A, Storm D, Diamond I. Ethanol activates cAMP response element-mediated gene expression in select regions of the mouse brain. Brain Res. 2006;1106:63–71. doi: 10.1016/j.brainres.2006.05.107. [DOI] [PubMed] [Google Scholar]
- Babor T, Grant M. From clinical research to secondary prevention: International collaboration in the development of the Alcohol Use Disorders Identification Test (AUDIT) Alcohol Health and Research World. 1989;13:371–374. [Google Scholar]
- Barnett NP, Ott MQ, Clark MA. The relevance of network prominence and reciprocity of relationships for alcohol use and alcohol-related problems in a college residence hall network. Psychol Addict Behav. 2014;28:980–9. doi: 10.1037/a0038354. [DOI] [PubMed] [Google Scholar]
- Bergen SE, Gardner CO, Kendler KS. Age-related changes in heritability of behavioral phenotypes over adolescence and young adulthood: a meta-analysis. Twin Research and Human Genetics. 2007;10:423–33. doi: 10.1375/twin.10.3.423. [DOI] [PubMed] [Google Scholar]
- Bergman O, Westberg L, Nilsson LG, Adolfsson R, Eriksson E. Preliminary evidence that polymorphisms in dopamine-related transcription factors LMX1A, LMX1B and PITX3 are associated with schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. 2010;34:1094–7. doi: 10.1016/j.pnpbp.2010.05.032. [DOI] [PubMed] [Google Scholar]
- Bertholet N, Faouzi M, Studer J, Daeppen JB, Gmel G. Perception of tobacco, cannabis, and alcohol use of others is associated with one’s own use. Addict Sci Clin Pract. 2013;8:15. doi: 10.1186/1940-0640-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blozis SA, Feldman B, Conger RD. Adolescent alcohol use and adult alcohol disorders: a two-part random-effects model with diagnostic outcomes. Drug Alcohol Depend. 2007;88(Suppl 1):S85–96. doi: 10.1016/j.drugalcdep.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, Molloy L, Ness A, Ring S, Davey Smith G. Cohort Profile: the ‘children of the 90s’--the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013;42:111–27. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AC, Brandon KO, Goldman MS. The college and noncollege experience: a review of the factors that influence drinking behavior in young adulthood. J Stud Alcohol Drugs. 2010;71:742–50. doi: 10.15288/jsad.2010.71.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casswell S, Pledger M, Pratap S. Trajectories of drinking from 18 to 26 years: identification and prediction. Addiction. 2002;97:1427–37. doi: 10.1046/j.1360-0443.2002.00220.x. [DOI] [PubMed] [Google Scholar]
- Derks EM, Vink JM, Willemsen G, Van Den Brink W, Boomsma DI. Genetic and environmental influences on the relationship between adult ADHD symptoms and self-reported problem drinking in 6024 Dutch twins. Psychol Med. 2014;44:2673–83. doi: 10.1017/S0033291714000361. [DOI] [PubMed] [Google Scholar]
- Doucet-Beaupre H, Ang SL, Levesque M. Cell fate determination, neuronal maintenance and disease state: The emerging role of transcription factors Lmx1a and Lmx1b. FEBS Lett. 2015 doi: 10.1016/j.febslet.2015.10.020. [DOI] [PubMed] [Google Scholar]
- Duncan SC, Alpert A, Duncan TE, Hops H. Adolescent alcohol use development and young adult outcomes. Drug Alcohol Depend. 1997;49:39–48. doi: 10.1016/s0376-8716(97)00137-3. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism. Role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Research & Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- Edwards AC, Larsson H, Lichtenstein P, Kendler KS. Early environmental influences contribute to covariation between internalizing symptoms and alcohol intoxication frequency across adolescence. Addict Behav. 2011a;36:175–82. doi: 10.1016/j.addbeh.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AC, Sihvola E, Korhonen T, Pulkkinen L, Moilanen I, Kaprio J, Rose RJ, Dick DM. Depressive symptoms and alcohol use are genetically and environmentally correlated across adolescence. Behav Genet. 2011b;41:476–87. doi: 10.1007/s10519-010-9400-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Davey Smith G, Henderson J, Macleod J, Molloy L, Ness A, Ring S, Nelson SM, Lawlor DA. Cohort Profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol. 2013;42:97–110. doi: 10.1093/ije/dys066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geels LM, Bartels M, Van Beijsterveldt TC, Willemsen G, Van Der Aa N, Boomsma DI, Vink JM. Trends in adolescent alcohol use: effects of age, sex and cohort on prevalence and heritability. Addiction. 2012;107:518–27. doi: 10.1111/j.1360-0443.2011.03612.x. [DOI] [PubMed] [Google Scholar]
- Grimm K, Widaman K. Residual Structures in Latent Growth Curve Modeling. Structural Equation Modeling: A Multidisciplinary Journal. 2010;17:424–442. [Google Scholar]
- Grimm K, Zhang Z, Hamagami F, Mazzocco M. Modeling Nonlinear Change via Latent Change and Latent Acceleration Frameworks: Examining Velocity and Acceleration of Growth Trajectories. Multivariate Behav Res. 2013;48:117–43. doi: 10.1080/00273171.2012.755111. [DOI] [PubMed] [Google Scholar]
- Jackson KM, Sher KJ, Schulenberg JE. Conjoint developmental trajectories of young adult substance use. Alcohol Clin Exp Res. 2008;32:723–37. doi: 10.1111/j.1530-0277.2008.00643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LD, O’malley PM, Bachman JG, Schulenberg JE. Monitoring the Future National Survey Results on Drug Use, 1975–2012: Vol. 1. Secondary School Students. Bethesda, MD: National Institute of Drug Abuse; 2013. [Google Scholar]
- Korpi ER, Den Hollander B, Farooq U, Vashchinkina E, Rajkumar R, Nutt DJ, Hyytia P, Dawe GS. Mechanisms of Action and Persistent Neuroplasticity by Drugs of Abuse. Pharmacol Rev. 2015;67:872–1004. doi: 10.1124/pr.115.010967. [DOI] [PubMed] [Google Scholar]
- Kozell LB, Hitzemann R, Buck KJ. Acute Alcohol Withdrawal is Associated with c-Fos Expression in the Basal Ganglia and Associated Circuitry: C57BL/6J and DBA/2J Inbred Mouse Strain Analyses. Alcoholism: Clinical & Experimental Research. 2005;29:1939–1948. doi: 10.1097/01.alc.0000187592.57853.12. [DOI] [PubMed] [Google Scholar]
- Lifestyle Statistics HSCIC. Statistics on Alcohol, England, 2014. Health & Social Care Information Centre; 2014. [Google Scholar]
- Longman D, Hug N, Keith M, Anastasaki C, Patton EE, Grimes G, Caceres JF. DHX34 and NBAS form part of an autoregulatory NMD circuit that regulates endogenous RNA targets in human cells, zebrafish and Caenorhabditis elegans. Nucleic Acids Res. 2013;41:8319–31. doi: 10.1093/nar/gkt585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein NR. Longitudinal associations between alcohol problems and depressive symptoms: early adolescence through early adulthood. Alcohol Clin Exp Res. 2009;33:49–59. doi: 10.1111/j.1530-0277.2008.00810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccambridge J, Mcalaney J, Rowe R. Adult consequences of late adolescent alcohol consumption: a systematic review of cohort studies. PLoS Medicine. 2011;8:e1000413. doi: 10.1371/journal.pmed.1000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A, Macgregor S. VEGAS2: Software for More Flexible Gene-Based Testing. Twin Res Hum Genet. 2015;18:86–91. doi: 10.1017/thg.2014.79. [DOI] [PubMed] [Google Scholar]
- Muthén BO, Muthén LK. Mplus User’s Guide. 7. Los Angeles, CA: 2012. [Google Scholar]
- Nixon K, Mcclain JA. Adolescence as a critical window for developing an alcohol use disorder: current findings in neuroscience. Curr Opin Psychiatry. 2010;23:227–32. doi: 10.1097/YCO.0b013e32833864fe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, Mcmanus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolstad S, Palsson E, Ekman CJ, Eriksson E, Sellgren C, Landen M. Polymorphisms of dopamine pathway genes NRG1 and LMX1A are associated with cognitive performance in bipolar disorder. Bipolar Disord. 2015 doi: 10.1111/bdi.12347. [DOI] [PubMed] [Google Scholar]
- Sacks JJ, Gonzales KR, Bouchery EE, Tomedi LE, Brewer RD. 2010 National and State Costs of Excessive Alcohol Consumption. Am J Prev Med. 2015;49:e73–9. doi: 10.1016/j.amepre.2015.05.031. [DOI] [PubMed] [Google Scholar]
- Sartor CE, Nelson EC, Lynskey MT, Madden PA, Heath AC, Bucholz KK. Are there differences between young African-American and European-American women in the relative influences of genetics versus environment on age at first drink and problem alcohol use? Alcohol Clin Exp Res. 2013;37:1939–46. doi: 10.1111/acer.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secretary of State for the Home Department. The Government’s Alcohol Strategy. HM Government; 2012a. [Google Scholar]
- Secretary of State for the Home Department. Impact Assessment: A minimum unit price for alcohol. 2012b. [Google Scholar]
- Slutske WS. Alcohol use disorders among US college students and their non-college-attending peers. Arch Gen Psychiatry. 2005;62:321–7. doi: 10.1001/archpsyc.62.3.321. [DOI] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, Depristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, Mcvean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker JS, Orlando M, Ellickson PL. Patterns and correlates of binge drinking trajectories from early adolescence to young adulthood. Health Psychol. 2003;22:79–87. doi: 10.1037//0278-6133.22.1.79. [DOI] [PubMed] [Google Scholar]
- Verhulst B, Neale MC, Kendler KS. The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychol Med. 2015;45:1061–72. doi: 10.1017/S0033291714002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden B, Iacono WG, Mcgue M. Trajectories of change in adolescent substance use and symptomatology: impact of paternal and maternal substance use disorders. Psychol Addict Behav. 2007;21:35–43. doi: 10.1037/0893-164X.21.1.35. [DOI] [PubMed] [Google Scholar]
- Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44:D877–81. doi: 10.1093/nar/gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesner M, Weichold K, Silbereisen RK. Trajectories of alcohol use among adolescent boys and girls: identification, validation, and sociodemographic characteristics. Psychol Addict Behav. 2007;21:62–75. doi: 10.1037/0893-164X.21.1.62. [DOI] [PubMed] [Google Scholar]
- Windle M, Mun EY, Windle RC. Adolescent-to-young adulthood heavy drinking trajectories and their prospective predictors. J Stud Alcohol. 2005;66:313–22. doi: 10.15288/jsa.2005.66.313. [DOI] [PubMed] [Google Scholar]
- Wu SH, Guo Q, Viken RJ, Reed T, Dai J. Heritability of usual alcohol intoxication and hangover in male twins: the NAS-NRC Twin Registry. Alcohol Clin Exp Res. 2014;38:2307–13. doi: 10.1111/acer.12487. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.