Abstract
Mammalian cells are commonly employed in screening assays to identify active compounds that could potentially affect the progression of different human diseases including retinitis pigmentosa (RP), a class of inherited diseases causing retinal degeneration with compromised vision. Using transcriptome analysis, we compared NIH3T3 cells expressing wildtype (WT) rod opsin with a retinal disease-causing single P23H mutation. Surprisingly, heterologous expression of WT opsin in NIH3T3 cells caused more than a 2-fold change in 783 out of 16,888 protein coding transcripts. The perturbed genes encoded extracellular matrix proteins, growth factors, cytoskeleton proteins, glycoproteins and metalloproteases involved in cell adhesion, morphology and migration. A different set of 347 transcripts was either up- or down-regulated when the P23H mutant opsin was expressed suggesting an altered molecular perturbation compared to WT opsin. Transcriptome perturbations elicited by drug candidates aimed at mitigating the effects of the mutant protein revealed that different drugs targeted distinct molecular pathways that resulted in a similar phenotype selected by a cell-based high-throughput screen. Thus, transcriptome profiling can provide essential information about the therapeutic potential of a candidate drug to restore normal gene expression in pathological conditions.
Keywords: Rhodopsin, P23H opsin, retinitis pigmentosa, transcriptome, retina, drug discovery, cell-based HTS, transcriptome, RNA-seq
Graphical abstract
INTRODUCTION
Small molecules are the mainstay of pharmacotherapeutics for the treatment of human diseases. Such agents often are easy to administer, well tolerated by patients, and relatively inexpensive. Nonetheless, complex diseases remain difficult to manage, and typically worsen with age. In infectious diseases or cancer, poly-pharmacology aimed at diverse and unrelated targets can effectively deal with the primary problem but often with adverse side effects. This risk can be acceptable for treating terminal diseases or chronic life-threatening infections but not for slowly progressive diseases that are not fatal or overly burdensome during a patient's normal lifespan. To minimize such drug side effects, recent approaches in systems biology can be used to characterize the molecular features of disease models used in both the early and later stages of drug development [1, 2]. Moreover, these technologies can be adapted to improve our understanding of the pharmacology of a starting molecule with respect to its possible off-target effects and therapeutic potential.
Retinitis pigmentosa (RP) is a progressive retinal degenerative disease associated with mutations in more than 50 genes [3, 4]. Effective treatments of RP are unavailable, even though gene therapy [5, 6] and pharmacological intervention with valproic acid [7-11] are currently undergoing clinical trials. The rhodopsin pigment, comprised of the protein opsin bound to a vitamin A chromophore that is regenerated in the endoplasmic reticulum and outer segment disc membranes, is an essential component of rod photoreceptor cells due to its pivotal role in phototransduction and its abundance in the photosensitive outer segments [12]. Thus, it is not surprising that Rho encoding the rod opsin, is the most frequent causal gene among autosomal dominant (ad) RP patients [3, 13]. The P23H mutation, observed in 10% of adRP cases, is a representative Class II mutation that causes opsin misfolding due to its thermal instability [3, 13-16]. Instability of P23H opsin leads to its progressive massive degradation in the rod photoreceptors of P23H knock-in mice [17, 18]. To rescue these photoreceptors, we hypothesized that improving P23H opsin stability could decrease photoreceptor cell death and improve vision. Alternatively, increasing the degradation of mutant opsin and leaving the WT rhodopsin allele to preserve visual function could be an equally viable therapeutic strategy for adRP. Thus, we developed and performed two sets of cell-based, small-molecule high-throughput screens (HTSs) to identify compounds that either improve the stability of the P23H opsin mutant or enhance its degradation [19]. Even though a valid photoreceptor cell line is unavailable, mammalian cells have been commonly used to study the biosynthesis of rhodopsin and to screen for drug candidates because the pre-ciliary biosynthesis of rhodopsin is generally shared by mammalian cells and rod photoreceptor cells [14, 20, 21]. In mammalian cell cultures, heterologously expressed WT opsin is located on the plasma membrane whereas P23H opsin accumulates in the endoplasmic reticulum (ER) due to its structural instability [14-16, 19-21].
For drug discovery, lead compounds identified from HTS in cell models are then tested in an animal model that represents the genetic defect and exhibits pathological signs seen in the corresponding human disease. The challenge of developing drug candidates showing efficacy in both cell and animal models sometimes lies in the dramatic difference of the two model systems. To improve our success rate of drug development, we need a better understanding of our disease models and the lead compounds’ mechanisms of action. Advances in microarray and next generation sequencing (NGS)-based transcriptome profiling (RNA-seq) have already identified novel molecular pathways or key genes associated with the development of disease states in the mouse retina [22-30]. Although transcriptome studies have been applied to low-dose pharmacological treatments in disease models to evaluate drug efficacy and side effects [30-34], RNA-seq studies have rarely been used in the early stages of drug discovery to characterize the cell models used for high throughput screening and to investigate a lead compound's mechanism of action. [30, 35, 36].
Here, using high-throughput RNA-seq technology, we profiled the transcriptomes of three stable cell lines expressing either opsin/green fluorescent protein (GFP), P23Hopsin/GFP or GFP alone, under different treatment conditions. This study addresses three questions related to drug discovery: 1) what are the general transcriptome changes in an established mammalian cell line due to heterologous expression of rod opsin; 2) what transcriptome changes arise from the expression of the P23H opsin mutant; and 3) what genes and associated molecular pathways are affected by treatment with active compounds selected from a HTS?
MATERIALS AND METHODS
Cells
NIH3T3 (WTopsin/GFP), NIH3T3 (P23Hopsin/GFP) and NIH3T3 (GFP) stable cell lines were generated by viral infection with pMiLRO, pMiLRO23 and pMXs-IG constructs, as previously published [19]. Mouse opsin or P23H opsin was co-expressed with GFP. When single clones were selected, no significant differences were observed between them with respect to cell shape, GFP fluorescence or localization of WT opsin (on the plasma membrane) or P23H opsin (in the ER) suggesting the defect of P23H opsin transport is not due to a clonal difference.
Immunostaining and fluorescence imaging
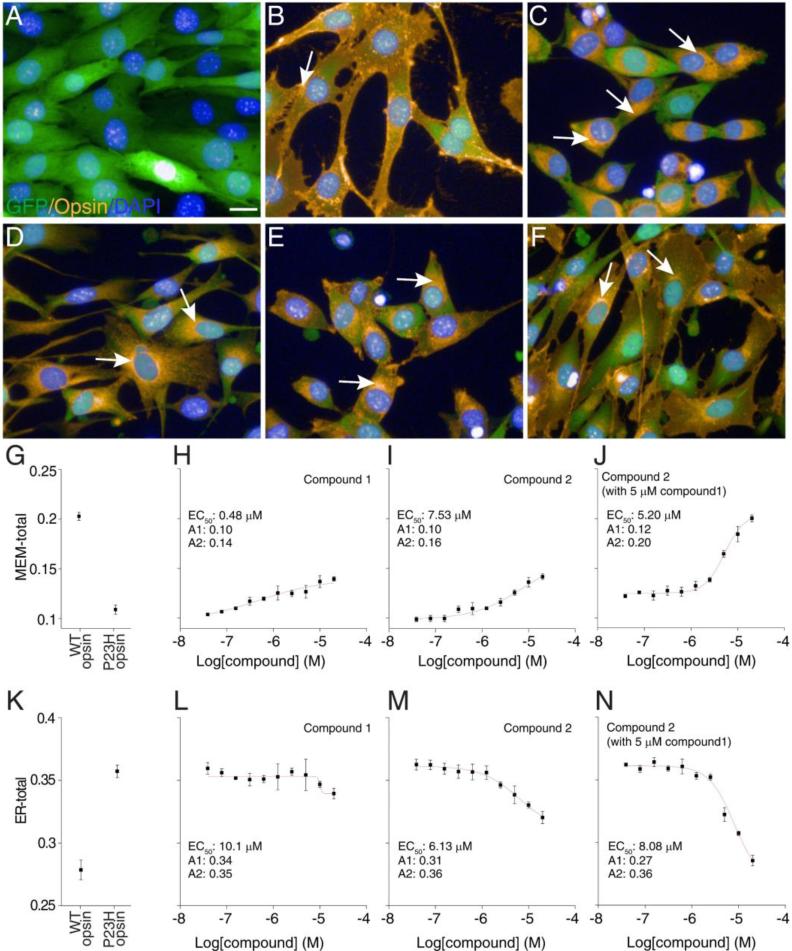
Immunostaining and fluorescence imaging followed published procedures [19]. Briefly, cells were seeded in a 384-well plate at 5,000 cells/well on day 1. These cells were treated with active compounds or a DMSO control on day 2. Cells were fixed with 4% paraformaldehyde 24 h after treatment. Rod opsin was immunostained with 1D4 monoclonal anti-rhodopsin antibody [37] followed by secondary labeling with Cy3-conjugated goat anti-mouse antibody (Jackson Immuno Research, West Grove, PA, USA). Nuclei were stained with DAPI. Fluorescence images were captured with an Operetta High Content Imaging System (Perkin Elmer, Waltham, MA, USA). Five images were taken from each well of the 384-well plate containing a total of 600-1000 cells. Three channels of fluorescence emission were used, Alexa488 (GFP), DAPI, and Cy3. Representative images of immunostained cells are shown in Figure 1A to F.
Figure 1.
Immunofluorescence and image analysis of NIH3T3 cells expressing GFP and opsins. Panel A, only GFP was expressed; panel B, WT opsin and GFP were co-expressed; panels C-F, P23H opsin mutant and GFP were co-expressed; GFP, Cy3 labeling associated with 1D4 anti-rhodopsin antibodies and nuclei stained with DAPI are shown in green, yellow, and blue, respectively. Cells were treated with 0.05% DMSO in A-C; 5 μM of compound 1 in D; 10 μM of compound 2 in E; and 5 μM compound 1 plus 10 μM of compound 2 in F. Arrows in B-F indicate opsin (WT or P23H) staining in the ER region. Scale bar, 20 μm. Quantification of opsin on the plasma membrane (G) and in the ER region (K) by image based analysis. Unlike WT opsin stained on the cell boundary, the P23H opsin mutant accumulated in the perinuclear region of NIH3T3 cells. Treatment with compound 1 reduced the perinuclear accumulation of P23H opsin, whereas treatment with compound 2 induced a significant amount of plasma membrane staining of P23H opsin. Co-treatment with both compounds 1 and 2 demonstrated a synergistic rescue of P23H opsin transport to the plasma membrane. For image analysis, the cytoplasm was defined by GFP fluorescence whereas the nucleus was defined by DAPI fluorescence. The MEM-total was calculated as the mean ratio of Cy3 intensity on the plasma membrane region to that in the entire cell. The ER-total was calculated as the mean ratio of Cy3 intensity in the perinuclear region to that in the entire cell. NIH3T3 cells expressing P23H or WT opsin were treated with 0.1% DMSO as controls (G and K). NIH3T3 cells expressing the P23H opsin were treated with compound 1 (H and L), compound 2 (I and M) or compound 2 with 5 μM compound 1 (J and N); each compound was tested in 10 concentrations starting at 20 μM and followed by 2-fold dilutions. Data points and error bars represent averages and standard deviations of biological replicates.
Image analysis
Only intact cell images were selected for analysis. The cytoplasm was defined by the GFP fluorescence of each cell with the cell boundary as the 0% line. Nuclei were visualized by DAPI fluorescence, and each nucleus, as defined by its edges, was assumed to comprise 50% of each cell. The membrane region was defined within −5 to 5% around the cell boundary, and the ER region was defined within 25 to 50% at the perinuclear region. Opsin staining on the plasma membrane was represented by the ratio of Cy3 intensity in the membrane region to that in the entire cell (MEM-total). Opsin staining in the ER region was denoted by the ratio of Cy3 intensity in the ER region to that in the entire cell (ER-total). MEM-total and ER-total were averaged from all the imaged intact cells in each well. Each condition was repeated in 3 or 8 wells, for treatments or controls, respectively. MEM-total and ER-total were then averaged from those biological repeats and presented as data points in Figure 1G to N. Error bars were standard deviations from those biological repeats.
Identification of active compounds by HTS
Compound 1 (an isoquinoline-2(3H)-hexanamide) and compound 2 (4-(5-chlorothiophen-2-yl)furan-2(5H)-one) were selected by a cell-based HTS of small molecules from the University of Cincinnati 2.5 K Diversity Set of Small-Molecules Library [19]. To identify small molecular compounds which rescue the P23H opsin mutant from ER retention, we performed a HTS with a β-galactosidase fragment complementation assay as described in reference [19]. Briefly, two complementary subunits of beta-galactosidase were individually fused with a plasma membrane-anchored peptide, the PH domain of phospholipase C delta (PLC), and the mouse opsin P23H mutant, respectively. A U2OS stable cell line was generated that consistently expresses these two fusion proteins. Due to its inherent instability, the P23H opsin fusion protein accumulates in the ER, whereas the PLC fusion protein remains anchored on the plasma membrane of cells. The separation of the two fusion proteins led to a complete lack of beta-galactosidase activity. Upon treatment with an active compound that stabilizes P23H opsin folding and transport to plasma membrane, two complementary beta-galactosidase subunits were colocalized on the plasma membrane resulting in a recovery of beta-galactosidase activity read by its luminescence. Using this cell-based assay in 384-well format, we screened the University of Cincinnati 2.5 K Diversity Set of small-molecules library and identified four hit compounds. An immunostaining and high-content imaging assay then was used to identify lead compounds with true activity. Both compound 1 and compound 2 increased the transport of the P23H opsin mutant to the plasma membrane, despite sharing little chemical or structural similarity.
Treatment of cell culture with active compounds
Each cell line was cultured at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS, Hyclone), 100 units/mL penicillin, 100 μg/mL streptomycin, and 2.92 μg/mL L-glutamine (Hyclone). The three cell lines were cultured to the same passage to achieve 90% confluence in a 50 mm culture dish before subsequent treatment. Five cell assay conditions were used: 1) NIH3T3(GFP) cells were treated with 0.05% DMSO, denoted as Control; 2) NIH3T3(WTopsin/GFP) cells were treated with 0.05% DMSO, denoted as Opsin; 3) NIH3T3(P23Hopsin/GFP) cells were treated with 0.05% DMSO, denoted as P23H; 4) NIH3T3(P23Hopsin/GFP) cells were treated with 5 μM of compound 1, denoted as T1; 5) NIH3T3(P23Hopsin/GFP) cells were treated with 10 μM of compound 2, denoted as T2. Each treatment condition was replicated in three 50 mm dishes. After 24 h of each treatment, the medium was aspirated, and cells were washed with phosphate-buffered saline (PBS, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4, 137 mM NaCl, and 2.7 mM KCl) before being lysed.
RNA extraction
Cells were lysed with TRIzol (1 mL/dish, Thermo Fisher Scientific, Grand Island, NY, USA), and total RNA was extracted from each sample according to the TRIzol reagent manual [38, 39]. Extracted RNA samples were analyzed with a 2100 Bioanalyzer Nano chip (Agilent Technologies Genomics, Santa Clara, CA, USA) to assess total RNA quality (RIN: 10). RNA from each sample then was labeled and stored at −80 °C before RNA-seq analysis.
RNA-seq and data analyses
All transcript libraries were made with the TruSeq Stranded mRNA Sample Prep Kit (Illumina, San Diego, CA, USA). The 100 base paired samples were run on 2 lanes of a Hiseq 2500 Sequencing system (Illumina) with Rapid mode running RTAv1.18.61 software. Illumina adapter trimming was performed with Trimmomatic v0.33. Quality control (QC) was analyzed with FastQCv0.11.2 software. Reads were processed in parallel by both qualitative and quantitative analyses with multiple types of software including Bowtie2 v2.2.1, TopHat2 v2.0.11, Samtools v0.1.19, and eXpress v1.3.1, as previously described in Ref. [40] using the GRCm38 assembly and ENSEMBL v78 annotation. Expression analyses were performed with R v3.11. Principal component analysis was conducted to validate the reproducibility of replication (Table 1). TMM (trimmed mean of M-values) normalization was done with EdgeR v3.6.8 to normalize the count data in different samples that varied in depth. Differential expression (DE) was analyzed by Iimma v3.22.3 that employed the Iimma voom function for estimating dispersion [41]. For DE analysis, only transcripts wherein all replicates of any sample group were greater than 1 fragment per kilobase of transcript per million mapped reads (FPKM) were kept in the data set. FPKM values rather than count values were used to reduce length bias in transcript filtering. DE statistics were obtained by moderated t-test, Enhanced Bayes, and Benjamini-Hochberg calculations, yielding P values, F and B-statistics, and false discovery rates (FDRs), respectively. The DE filter was set for transcripts with FPKM fold changes of more than 2 and an FDR of less than 5%. Hierarchical clustering heatmaps were generated by Affinity Propagation employing the APcluster package in R using the Z-score of normalized FPKM values.
Table 1.
The number of sequencing reads generated for each sample and their alignment statistics. The alignment statistics refer to the number and percentage of reads that align to the genome and to the annotation used in the analysis. Reads mapping to the genome assembly but not the annotation are from intergenic, intronic, or novel transcripts not present in the annotation.
| Samples | Total Reads | Assembly | Annotation | % Genome | % Assembly |
|---|---|---|---|---|---|
| Control.1 | 30,452,046 | 27,923,900 | 25,058,876 | 91.7 | 82.3 |
| Control.2 | 26,566,098 | 24,353,828 | 21,365,196 | 91.7 | 80.4 |
| Control.3 | 24,620,210 | 23,212,126 | 20,288,806 | 94.3 | 82.4 |
| Opsin.1 | 41,396,766 | 37,892,165 | 34,548,292 | 91.5 | 83.5 |
| Opsin.2 | 26,478,896 | 24,207,557 | 21,314,236 | 91.4 | 80.5 |
| Opsin.3 | 46,300,468 | 43,618,647 | 37,763,672 | 94.2 | 81.6 |
| P23H.1 | 30,715,732 | 28,228,632 | 25,307,996 | 91.9 | 82.4 |
| P23H.2 | 40,926,422 | 37,480,508 | 34,042,686 | 91.6 | 83.2 |
| P23H.3 | 32,057,578 | 30,168,399 | 26,602,622 | 94.1 | 83.0 |
| T1.1 | 21,895,902 | 19,480,527 | 17,761,142 | 89.0 | 81.1 |
| T1.2 | 23,551,414 | 21,054,805 | 18,928,522 | 89.4 | 80.4 |
| T1.3 | 33,157,974 | 30,313,499 | 26,632,680 | 91.4 | 80.3 |
| T2.1 | 31,953,284 | 29,212,898 | 26,769,282 | 91.4 | 83.8 |
| T2.2 | 45,090,996 | 41,511,166 | 37,576,600 | 92.1 | 83.3 |
| T2.3 | 46,026,166 | 43,463,234 | 37,740,812 | 94.4 | 82.0 |
Gene ontology and biological process pathway analyses
Gene ontology (GO) analysis was initially performed with DAVID Bioinformatics Resources 6.7 (The Database for Annotation, Visualization and Integrated Discovery, http://david.abcc.ncifcrf.gov/home.jsp) [42, 43]. Functional Annotation Clustering was used only for the “Biological Processes” category at a GO level of 5 and greater. GO terms that had the lowest P values in each cluster as well as low FDR values were listed. The total number of genes included in all of the listed GO terms was considered to be 100%, and the number of genes in each GO term was calculated as a percentage in a pie chart for data visualization.
Reverse transcription and quantitative real-time polymerase chain reaction (qPCR)
RNA was extracted from cells and reverse transcribed into cDNA using the high capacity RNA-to-cDNA kit (Applied Biosystems, Foster City, CA, USA) following the manufacturer's protocol. These cDNA samples were used as templates for qPCR. The qPCRs were performed using CYBR Green Super Mix (Bio-Rad Laboratories, Hercules, CA, USA). A total of 2 ng of cDNA was added to 10 μL of CYBR Green Super Mix, 8 μL of purified H2O and 1 μL of primers including forward and reverse primers at 10 μM each. Each reaction therefore contained a 20 μL solution in a single well of a 96-well PCR plate. We quantified 9 transcripts by qPCR using Gapdh as the control transcript. The 9 transcripts included: Txnip, Nqo1, Gstp1, Pfdn6, Itch, Dnajb14, Vdr, Clec16a, and Masp1. Specific bands of qPCR products were confirmed by DNA gel electrophoresis, stained with ethidium bromide and visualized by UV fluorescence. Primers for the 9 transcripts were: Txnip (forward 5’-GTTGCGTAGACTACTGGGTGAAG-3’; reverse: 5’-CTCCTTTTTGGCAGACACTGGTG-3’); Nqo1 (forward: 5’-GCCGAACACAAGAAGCTGGAAG-3’; reverse: 5’-GGCAAATCCTGCTACGAGCACT-3’); Gstp1 (forward: 5’-TGGAAGGAGGAGGTGGTTACCA-3’; reverse: 5’-GGTAAAGGGTGAGGTCTCCATC-3’); Pfdn6 (forward: 5’-GATTACAGCGGGTAGAGCGT-3’; reverse: 5’-TACTCAAGTCCTTCTGCAGCTGT-3’); Itch (forward: 5’-CTCGGATTACTCAGTGGGAAGAC-3’; reverse: 5’-GTTGCTCTTCTATTGTGGTCCAC-3’); Vdr (forward: 5’-GCTCAAACGCTGCGTGGACATT-3’; reverse: 5’-GGATGGCGATAATGTGCTGTTGC-3’) Dnajb14 (Forward: 5’-GCGCGCGCGTTATTGG-3’; reverse: 5’-CAGAAGTGCCGTCCTTTCCA-3’); Dedd (forward: 5’-CACACTTGGGAGCCAGCGAAAA-3’; reverse: 5’-AGCCGTCTCATGCTGGCAGTAT-3’); Clec16a (forward: 5’-GAACACCACAGACGAGGAGAAG-3’; reverse: 5’-CATACAGGAGGCAGAGCACGAA-3’); Masp1 (forward: 5’-CCTTCAAAGACCAAGTGCTCGTC-3’; reverse: 5’-ACTCCATGCACCGTCCTTCAGA-3’); Coro1b (forward: 5’-CAGCCCGAAATGTGCTTCTCAG-3’; reverse: 5’-GCAAAAGAGGCTGCCATTGTGG-3’); Ttll1 (forward: 5’-ACCGGCTATTCTAGTGACGC-3’; reverse: 5’-GGGACACTGGTCAACACTCC-3’). Primers for Gapdh were: forward: 5’-TGGTGAAGCAGGC-3’; reverse: 5’-TGAAGTCGCAGGAGACAACC-3’. The fold change for each transcript was first normalized by Gapdh, and then compared to Control (NIH3T3 cells expressing GFP only). Fold changes were averaged from three biological replicates and error bars were from standard deviations of those replicates. P values comparing two conditions were calculated with the two-sample Student's t-test.
RESULTS
To discover effective small molecular drugs for retinitis pigmentosa associated with the rhodopsin P23H mutation, we previously undertook a cell-based HTS to identify small molecules rescuing the P23H opsin from misfolding and ER retention, using a beta-galactosidase complementation assay [19]. Briefly, using the complemented beta-galactosidase activity as a reporter, the amount P23H opsin successfully transported to the plasma membrane was quantified by its luminescence read from a microplate reader. In 384-well plates, we screened the University of Cincinnati 2.5 K Diversity Set of small-molecule library and identified compound 1 and compound 2 that featured outstanding potency as well as efficacy. Compound 1 (an isoquinoline-2(3H)-hexanamide) showed its EC50 at 3.1 μM with an efficacy score of 87% whereas compound 2 ((4-(5-chlorothiophen-2-yl)furan-2(5H)-one)) had an EC50 at 5.5 μM with an efficacy score of 166%. Both activity scores were normalized to 100% as exhibited by the effect of treatment with 5 μM 9-cis-retinal.
To confirm the activity of these two lead compounds for rescuing the transport of P23H opsin mutant from the ER to the plasma membrane, three stable NIH3T3 cell lines were generated for the immunostaining and high-content imaging assay [19]. GFP was expressed, either with the opsin protein or by itself, as a positive selection marker to help generate the stable cell lines. In agreement with reports describing other mammalian cell models [14, 21, 44-49], the WT opsin protein localized to the plasma membrane of NIH3T3 cells as demonstrated by the staining of cellular boundaries, whereas the P23H opsin mutant accumulated in the perinuclear region with little plasma membrane staining indicating its predominant retention in the endoplasmic reticulum (ER) (Figure 1A-C). This mislocalization was noted in all clones of stable cells, suggesting the defect of P23H opsin transport is due to its structural instability, rather than an artificial clonal difference. To quantitatively compare the cellular localization of the WT and P23H opsins, we undertook an image-based analysis to quantify the immunostaining of opsin on the plasma membrane versus that in the ER relative to the total opsin staining in each cell (MEM-total versus ER-total, Figure 1). For controls, quantitative analysis of 8 biological repeats, each calculated from images of 600-1000 cells, provided reliable baselines representing “normal” and “aberrant” conditions. In agreement with what was observed from the images in Figure 1B and C, relative staining of WT opsin was significantly higher on the plasma membrane (MEM-total=0.20, Figure 1G), and lower in the ER (ER-total=0.27, Figure 1K), as compared to P23H opsin staining.
Two small molecular compounds were found from the HTS which rescued transport of P23H opsin from the ER to the plasma membrane. Treatment with compound 1 resulted in a reproducible redistribution of P23H opsin throughout the cell body (Figure 1D), with a dose-dependent increase in cell boundary staining (MEM-total increased from 0.10 up to 0.14, EC50=0.48 μM, Figure 1H), but little change in the ER region (Figure 1,L). Comparably, treatment with compound 2 also increased the plasma membrane staining of P23H opsin (MEM-total increased from 0.10 to 0.16, EC50=7.5 μM, Figure 1E and I), whereas its perinuclear staining was reduced in a dose dependent manner (ER-total decreased from 0.36 to 0.31, EC50=6.1 μM, Figure 1M). Interestingly, co-treatment with both compounds produced a significant improvement of P23H opsin transport to the plasma membrane (MEM-total increased from 0.12 up to 0.20, EC50=5.2 μM, Figure 1J), which also dramatically reduced the perinuclear accumulation of the opsin mutant (ER-total decreased from 0.36 to 0.27, EC50=8.1 μM, Figure 1N). The differing cellular distribution of P23H opsin upon treatment with these two compounds and their synergic effect suggest that they utilize distinct cellular targets.
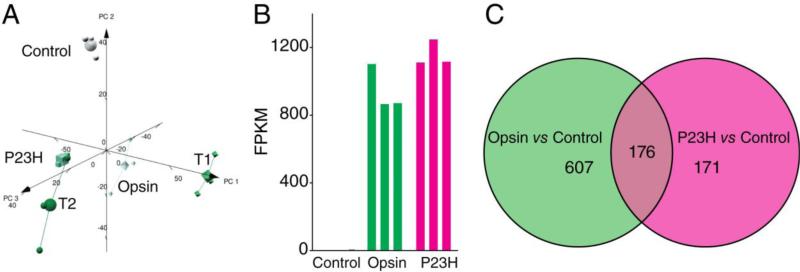
To understand the effect of heterologous expression of opsin or the P23H mutant and the molecular events altered by treatment with compounds 1 and 2, we profiled the transcriptomes of different sample groups. Principle component analysis showed the consistency between repeats of each sample group (Figure 2A and Table 1). Protein-coding transcripts with fold changes of more than 2 and a false discovery rate (FDR) less than 5% were collected in each DE profile of two cellular conditions. FPKM reads of Rho transcripts from NIH3T3 cells expressing WT opsin and NIH3T3 cells expressing the P23H opsin mutant were similar (Figure 2B), suggesting that the changes in transcripts between the two cell lines were not due to differential expression of the Rho transgenes.
Figure 2.
Transcriptome overview of NIH3T3 cells expressing GFP and opsin under different treatment conditions: Control, only GFP was expressed; Opsin, opsin and GFP were co-expressed; P23H, P23H opsin mutant and GFP were co-expressed. Cells were treated with 0.05% DMSO as a vehicle control. T1 or T2, the P23H opsin and GFP expressing cells were treated with 5 μM of compound 1 or 10 μM of compound 2, respectively. A. Principle component analysis plot of the five sample groups. B. FPKM reads of Rho transcripts from three NIH3T3 stable cell lines revealed similar heterologous expression levels of WT opsin or the P23H opsin mutant in each cell line. Each sample group contained three biological replicates. C. Comparison of differential expression (DE) profiles of Opsin versus Control and P23H versus Control. The two circles represent each of the two DE profiles. The number of transcripts located in both DE profiles is shown in the overlapping area of the two circles, whereas the number of transcripts detected in only one DE profile is placed in the corresponding circle. The DE filter was set for fold changes of more than 2 and an FDR smaller than 5%.
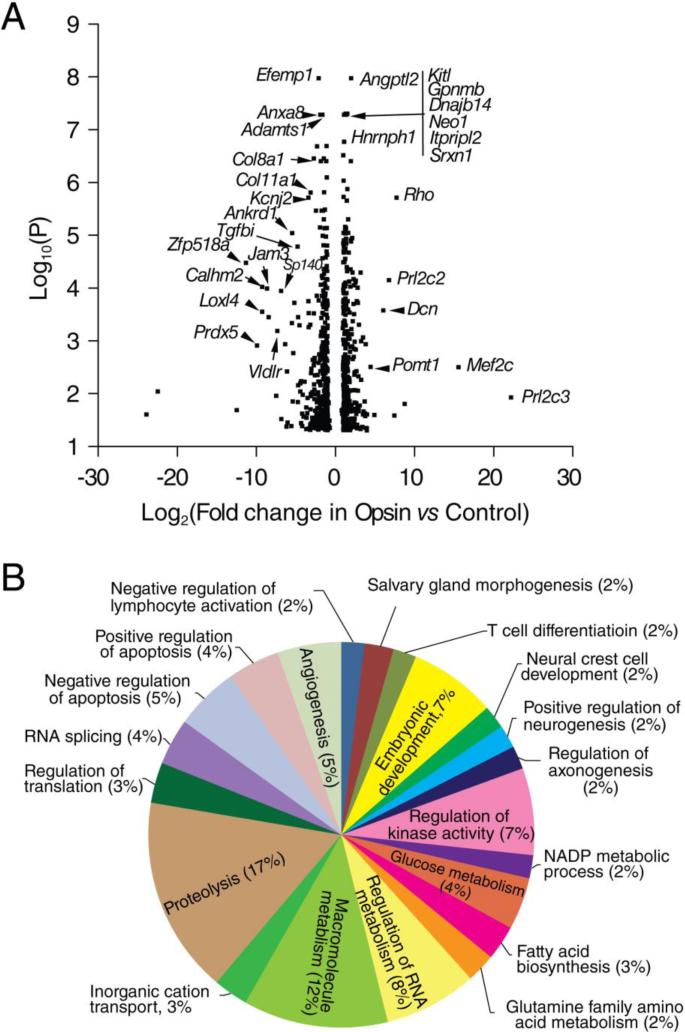
Heterologous expression of opsin and its mutants in mammalian cells has been used extensively to study rhodopsin biochemistry and identify drug candidates [14, 20, 21, 44-48]. However, it is unknown whether expression of opsin produces a transcriptome change within the host cell line and if analysis of changes in the transcriptome could be used for validation of lead compounds from HTS. Here, a total of 783 out of 16,888 protein coding transcripts revealed differential expression in Opsin versus Control cell lines (Figure 2C). Notably, many of the perturbed genes are relevant to pathways involved in cell adhesion, morphology and migration and encode extracellular matrix proteins, growth factors, cytoskeleton proteins, glycoproteins or metalloproteases (Efemp1, Adamts1, Col8a1, Col11a1, Calhm2, Loxl4, Tgfbi, Jam3, Angptl2, Kitl, Gpnmb, Neo1, Dcn, Prl2c2 and Prl2c3) (Figure 3A). These changes in expression could represent a stress response due to the heterologous expression of opsin in NIH3T3 cells, which are fibroblasts that feature elevated expression of extracellular matrix proteins [35, 50, 51]. Among other significantly altered genes, some are involved in transcriptional regulation or RNA splicing (Zfp518a, Hnmph1, Sp140, Ankrd1 and Mef2c), and others participate in cellular responses to oxidative stress (Srxn1, Prdx5), protein glycosylation (Prmt1), and lipid transport (Vldlr). Their changes could be due to cellular adaptation required to support the biosynthesis of the opsin protein, a G protein-coupled receptor (GPCR). Alternatively, the DE profile of Opsin versus Control cells includes many genes containing cAMP responsive elements (CREs) [52] which could be regulated by the leaky activity of free opsin [53-55] coupled to endogenous Gi/o protein signaling [7, 16, 56]. Such genes include but are not limited to Gsta4, Pcx, and Pdk4 involved in metabolic processes; Per1, Ppargc1a and Egfr1 that regulate gene transcription; the growth factor Prl2c2; the neurotransmitter Inhba; and Ppp1r15a involved in DNA repair (Table 2). A more complete evaluation of biological processes affected by the expression of opsin in NIH3T3 cells is presented in Figure 3B.
Figure 3.
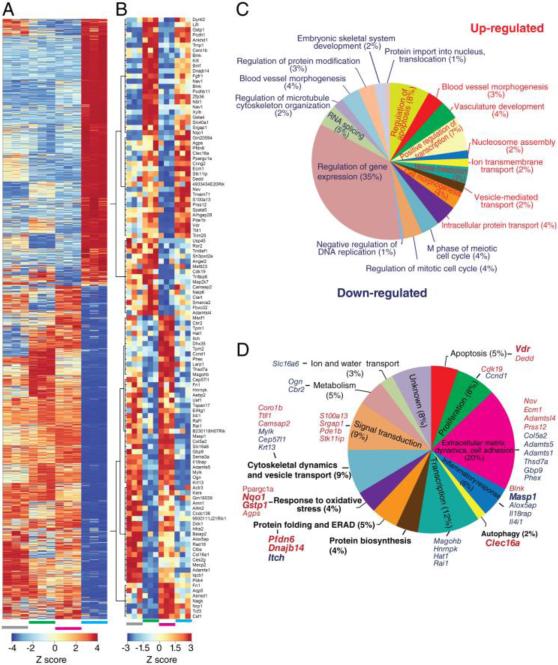
Transcriptome changes in NIH3T3 cells from heterologous expression of mouse opsin. Control, NIH3T3 cells expressing only GFP; Opsin, NIH3T3 cells expressing both opsin and GFP. A. Volcano plot of transcripts which showed DE in Opsin versus Control. P values are plotted in a −log10 format as y-values, and fold changes of FPKM reads in P23H versus Opsin are plotted in a Log2 format as x-values. Representative transcripts that showed significant differential expression are labeled with gene names. B. Pie chart showing biological processes (BP) enriched in the DE profile of Opsin versus Control. The size of each pie is correlated with number of genes included in the corresponding BP as a percentage of the total number of genes counted in all BPs.
Table 2.
List of cAMP responsive elements (CRE)-containing genes with their transcripts differentially expressed in NIH3T3 cells expressing opsin and GFP compared to those expressing GFP only (Opsin versus Control). Cutoff: Fold change >2 or <0.5, P value<0.05.
| Gene name | Transcript name | Description | Log2(Fold Opsin versus Control) | P value (Opsin versus Control) |
|---|---|---|---|---|
| Gsta4 | Gsta4-201 | glutathione S-transferase, alpha 4 | 1.08 | 7.76E-08 |
| Pcx | Pcx-201 | pyruvate carboxylase | −1.31 | 2.65E-05 |
| Pdk4 | Pdk4-001 | pyruvate dehydrogenase kinase, isoenzyme 4 | −1.61 | 5.73E-08 |
| Per1 | Per1-001 | period circadian clock 1 | −1.15 | 2.26E-04 |
| Per1 | Per1-003 | period circadian clock 1 | −1.06 | 1.86E-03 |
| Pparg c1a | Ppargc1a-001 | peroxisome proliferative activated receptor, gamma, coactivator 1 alpha | 1.85 | 7.63E-06 |
| Egfr | Egfr-001 | epidermal growth factor receptor | −1.19 | 5.98E-08 |
| Prl2c2 | Prl2c2-201 | prolactin family 2, subfamily c, member 2 | 7.27 | 9.85E-07 |
| Inhba | Inhba-202 | inhibin beta-A | −1.08 | 2.34E-03 |
| Ppp1r15a | Ppp1r15a-001 | protein phosphatase 1, regulatory (inhibitor) subunit 15A | −1.10 | 6.93E-05 |
P23H opsin revealed a transcriptome shift compared to WT opsin expressed in NIH3T3 cells. We identified 347 differentially expressed genes from the DE profile of P23H versus control, indicating that far fewer genes were affected by expression of the mutant opsin (Figure 2C). A total of 176 of these genes showed differential expression in both Opsin versus Control and P23H versus Control, and among these, 159 genes evidenced the same trend. These similar changes could result from common structural and biochemical properties shared by P23H opsin and WT opsin. GO analysis of these genes revealed a significant enrichment of genes involved in the vesicle transport pathway, such as Klc1, Stx16, Kif1c, Myo7a, Dlg4 and Lrp8 (Table 3). Transcripts involved in metabolism, apoptosis, and transcription pathways were similarly changed in both DE profiles of Opsin versus Control and P23H versus Control.
Table 3.
List of transcripts enriched in the vesicle transport pathway that revealed the same trends of differential expression when comparing P23H versus Control and Opsin versus Control. Cutoff: Fold change >2 or <0.5, P value<0.05.
| Gene name | Transcript name | Description | Log2(Fold P23H versus Control) | P value (P23H versus Control) | Log2(Fold Opsin versus Control) | P value (Opsin versus control) |
|---|---|---|---|---|---|---|
| Klc1 | Klc1-004 | kinesin light chain 1 | 1.50 | 2.74E-03 | 1.33 | 1.04E-02 |
| Stx16 | Stx16-001 | syntaxin 16 | −1.06 | 3.54E-03 | −1.22 | 1.40E-02 |
| Stx16 | Stx16-201 | syntaxin 16 | 1.36 | 5.32E-03 | 1.34 | 3.22E-02 |
| Kif1c | Kif1c-005 | kinesin family member 1C | 1.83 | 1.31E-04 | 1.30 | 2.36E-02 |
| Myo7a | Myo7a-001 | myosin VIIA | 1.14 | 5.00E-05 | 1.03 | 9.53E-03 |
| Dlg4 | Dlg4-001 | discs, large homolog4 (Drosophila) | −1.19 | 2.36E-05 | −1.50 | 2.44E-02 |
| Lrp8 | Lrp8-003 | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor | 1.36 | 2.43E-04 | 1.54 | 2.82E-03 |
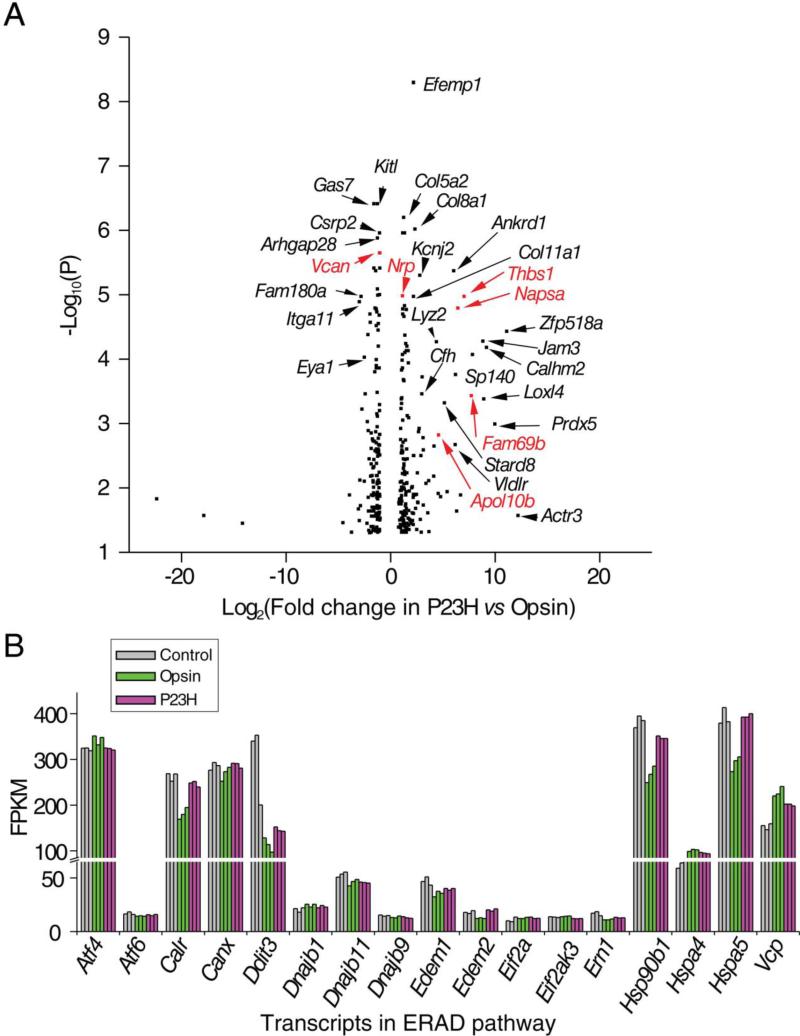
To assess transcriptome differences between the disease and control models, we compared the DE profiles of P23H versus Opsin that identified 330 transcripts (Figure 4A). Among these transcripts, 271 were not seen in the DE profile of P23H versus Control, but did show up in the profile of Opsin versus Control. This result suggests that 84% of the transcripts that displayed differential expression in P23H versus Opsin are altered due to molecular perturbations induced by the expression of WT opsin but not P23H opsin. This result could be due to the significant reduction of folded P23H opsin protein as a result of its ER accumulation and its enhanced degradation in these NIH3T3 cells [19]. GO analysis showed significant enrichment in genes involved in cell adhesion (P value = 0.00004 and Benjamini = 0.06), including Col8a1, Col16a1, Col11a1, Cdh18, Cdh11a, Pcdhb1, Pcdhb17, Pcdhb21, Pcdh1, Alcam, Ncam, Fn1, and Jam3 (Figure 4A, genes denoted in black).
Figure 4.
Transcriptome differences between NIH3T3 cells expressing the P23H opsin mutant versus those expressing WT opsin. Control, NIH3T3 cells expressing only GFP; P23H, NIH3T3 cells expressing both the P23H opsin mutant and GFP; Opsin, NIH3T3 cells expressing both opsin and GFP. A. Volcano plot of transcripts which showed DE in P23H versus Opsin. P values are plotted in a −log10 format as y-values, and fold changes of FPKM reads in P23H versus Opsin are plotted in a Log2 format as x-values. Representative transcripts that showed DE in P23H versus Opsin but not in P23H versus Control are in black print, suggesting loss-of-opsin-properties in the P23H opsin mutant. Those that showed DE in both P23H versus Opsin and P23H versus Control are indicated in red, suggesting gain-of-abnormal-properties in the P23H opsin. B. FPKM reads of transcripts in the endoplasmic reticulum associated protein degradation (ERAD) pathway of the three stable cell lines. The results suggest no significant activation of this pathway due to expression of WT or P23H opsin. Each sample group had three biological replicates.
Apart from genes revealing the lost transcriptional perturbation by the P23H opsin mutant, there were 59 genes observed in DE profiles of both P23H versus Control and P23H versus Opsin, suggesting that these changes resulted from the aberrant properties of the P23H opsin mutant. These genes can be classified as participating in: 1) cell adhesion (Thbs1, Napsa, Vcan); 2) cell differentiation (Vdr, Eya1, Sprr1a, Nrp, Egln3); 3) membrane protein biosynthesis (Galnt13, Fam69b); 4) lipid transport (Apol10b); and 5) glycoprotein degradation (Lyz2) (Figure 4A, genes denoted in red). Activation of the ER-associated degradation (ERAD) pathway has been reported in the P23H knock-in mouse model [57] and in mammalian cell models transfected with P23H opsin[21, 58]. However, genes related to ERAD did not change significantly in NIH3T3 cells upon continuous expression of either WT opsin or the P23H opsin mutant (Table 4). Molecular chaperones including calnexin, BIP/Grp78 and GRP94 were reported to directly associate with P23H opsin in cell cultures [49, 59], and EDEM1 was reported to be responsible for the initiation of the ERAD of P23H opsin [60]. Here we observed a slight upregulation of molecular chaperones including Hspa5 (Bip), Hsp90b1 (Grp94), Edem2 and Calr (encoding calreticulin) but not Edem1 or Canx (encoding calnexin) in cells expressing P23H opsin compared to those expressing WT opsin. These data suggest that the instability of P23H opsin could have moderately induced molecular chaperones to maintain protein homeostasis (Figure 4B). However, no significant changes of other ERAD genes (Atf4, Atf6, Ern1(Ire1), Eif2ak3, Eif2a or Ddit (Chop)) were observed in DE profiles of P23H versus Opsin (Figure 4B), suggesting that NIH3T3 cells can be intrinsically adapted to protein misfolding. Moreover, no significant induction of caspase expression was seen in DE profiles of P23H versus Opsin. Due to differences between immortalized NIH3T3 cells and mouse photoreceptors [61, 62], transcriptome changes in a cell line should be interpreted with caution and cannot be directly correlated with the molecular events that occur in a mouse bearing the P23H mutation. Rather, to understand the molecular events involved in photoreceptor death, transcriptomes derived from mouse retina at an early age before photoreceptor death occurs should be compared between wild type and heterozygous P23H knock-in animals. But here, our goal of comparing transcriptomes between these cell lines was to obtain information pertinent to drug discovery through early stage analyses of molecular events following various treatments.
Table 4.
Mean FPKM reads of transcripts involved in the endoplasmic reticulum associated degradation (ERAD) pathway of NIH3T3 cells expressing GFP (Control), Opsin/GFP (Opsin), or P23Hopsin/GFP (P23H). Mean FPKM reads were calculated from 3 biological repeats.
| Gene name | Transcript name | Description | Mean FPKM reads | ||
|---|---|---|---|---|---|
| Control | Opsin | P23H | |||
| Atf4 | Atf4-201 | activating transcription factor 4 | 322.7 | 343.6 | 323.2 |
| Atf6 | Atf6-001 | activating transcription factor 6 | 16.9 | 14.4 | 15.5 |
| Calr | Calr-001 | calreticulin | 262.9 | 181.4 | 246.8 |
| Canx | Canx-001 | calnexin | 285.3 | 269.4 | 287.8 |
| Ddit3 | Ddit3-001 | DNA-damage inducible transcript 3 | 297.8 | 113.3 | 146.7 |
| Dnajb11 | Dnajb11-001 | DnaJ (Hsp40) homolog, subfamily B, member 11 | 53.3 | 46.0 | 45.8 |
| Dnajb9 | Dnajb9-201 | DnaJ (Hsp40) homolog, subfamily B, member 9 | 15.0 | 13.5 | 13.1 |
| Edem1 | Edem1-201 | ER degradation enhancer, mannosidase alpha-like 1 | 47.1 | 35.3 | 39.8 |
| Edem2 | Edem2-001 | ER degradation enhancer, mannosidase alpha-like 2 | 18.1 | 12.4 | 20.1 |
| Eif2a | Eif2a-001 | eukaryotic translation initiation factor 2A | 38.7 | 41.7 | 33.3 |
| Eif2ak3 | Eif2ak3-001 | eukaryotic translation initiation factor 2 alpha kinase 3 | 13.5 | 14.2 | 12.1 |
| Ernl | Ernl-001 | endoplasmic reticulum (ER) to nucleus signaling 1 (IRE1) | 16.8 | 11.2 | 13.0 |
| Hsp90b1 | Hsp90b1-001 | heat shock protein 90, beta (GRP94), member 1 | 382.8 | 267.3 | 347.5 |
| Hspa4 | Hspa4-001 | heat shock protein 4 | 64.8 | 101.7 | 95.2 |
| Hspa5 | Hspa5-001 | heat shock protein 5 | 391.6 | 292.0 | 394.9 |
| Vcp | Vcp-001 | valosin containing protein | 153.7 | 228.5 | 201.1 |
Thus, we identified two active compounds through a cell-based, small molecule HTS, both of which rescued the transport of P23H opsin back from the ER to the plasma membrane. Each compound produced a different distribution pattern for the P23H opsin and co-treatment with both resulted in a synergistic rescue of P23H opsin homeostasis (Figure 1). These results suggest that these two active compounds target different cellular pathways which culminate in a similar cellular effect. Such affected pathways can be revealed by cellular transcriptome changes that are sensitive to perturbations of cellular conditions. To delineate the molecular events initiated by each of these compounds, we compared the DE profiles of T1 versus P23H and T2 versus P23H (Figure 5). The DE profile of T1 versus P23H recorded 2,715 changed transcripts, whereas that of T2 versus P23H showed no significant changes. These transcriptome changes were captured 24 h after treatments when rescue of P23H opsin transport was first observed (Figure 1D and E). Therefore, this finding suggests that the rescue of P23H opsin transport by T1 could primarily result from transcriptional regulation, whereas compound 2 could directly target the P23H opsin protein or other proteins that improve the folding or transport of the opsin mutant without directly affecting transcription.
Figure 5.
Transcriptome shift of NIH3T3 cells expressing P23H opsin and GFP upon treatment with compound 1 (T1 versus P23H). A. Heat map of transcripts showing DE in T1 versus P23H. Four sample groups are shown, each with three biological replicates: grey bar, Control - NIH3T3 cells expressing only GFP; green bar, Opsin - NIH3T3 cells expressing both opsin and GFP; magenta bar, P23H - NIH3T3 cells expressing both P23H opsin mutant and GFP; blue bar, T1 - NIH3T3 cells expressing both P23H opsin mutant and GFP that were treated with 5 μM of compound 1. The heat map legend is displayed at the bottom. B. Heat map of transcripts in DE of T1 versus P23H that were reversed as compared to their changes in DE of P23H versus Opsin. C. Pie chart of biological processes (BP) which were enriched with up-regulated genes are denoted in red and BPs associated with down-regulated genes are denoted in blue. Numbers of genes changed in each BP are shown as a percentage of the total gene number of all BPs represented by the size of the corresponding pie. D. Pie chart of BPs enriched among the 117 genes that were reversed in DE of T1 versus P23H compared to their changes in P23H versus Opsin. The percentage numbers in parenthesis following each BP represent the percentage of transcripts among the 117 genes classified in that BP. Gene names of the top 25 up- and down-regulated transcripts are labeled at the side of each BP. BPs that are most relevant to opsin biosynthesis are labeled in bold. Transcripts that were selected and confirmed by qPCR assay were labeled in bold.
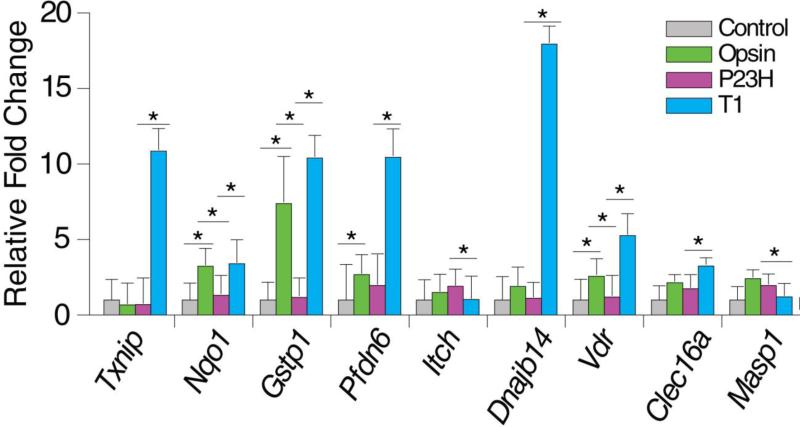
The transcriptional impact of treatment with compound 1 (T1) was profound, causing a greater than 16% change in the transcriptome (Figure 5A). Due to the large number of transcripts affected by T1, these transcripts were initially grouped as either up- or down-regulated before the GO analysis. Biological processes affected by T1 then were classified (Figure 5C). Among the enriched pathways, genes involved in vesicle-mediated transport (2%), regulation of microtubule cytoskeleton organization (2%), and regulation of protein modifications (3%) correlated with the improved P23H opsin transport observed by immunostaining (compare Figure 1D with 5C). Notably, 117 genes were reversed in the DE profile of T1 versus P23H compared to the DE profile of P23H versus Opsin (Figure 5B), suggesting these affected genes by P23H opsin were somehow “corrected” by T1. Correlating with the observation that T1 partially rescued the transport of the P23H opsin, changes of these transcripts by T1 could involve pathways either causing or being effected by the rescued P23H opsin transport. These genes were manually classified according to their biological functions documented in the Gene Cards Human Gene Database (http://www.genecards.org) (Figure 5D). The top 25 up- and down-regulated transcripts with known functions are listed next to their classifications. The most relevant biological processes to P23H opsin biosynthesis include, but are not limited to those regulating cytoskeleton dynamics, vesicle transport, protein folding and ERAD, as well as a response to oxidative stress. Among transcripts responding to oxidative stress, Gstp1 and Nqo1 were down-regulated in P23H versus Opsin and up-regulated in T1 versus P23H. These genes protect cells from reactive oxygen species generated during protein biosynthesis by transferring thiol groups for disulfide bonds [63]. Up-regulation of Gstp1 and Nqo1 could represent an elevated response to reactive oxygen species that is frequently correlated with protein misfolding. Notably, molecular chaperones Dnajb14 and Pfdn6 (prefoldin), both involved in protein folding and ERAD, were down-regulated in P23H and reversed by T1. Expression of Dnajb14 was reported to accelerate the degradation of misfolded membrane proteins such as the cystic fibrosis transmembrane conductance regulator Δ508 (CFTRΔ508) [64]. Prefoldins were previously reported to prevent ubiquitinated-protein aggregation under ER-stress [65-67]. Up-regulation of these two molecular chaperones can be correlated with the improved homeostasis of P23H opsin in NIH3T3 cells. The same trend of changes was confirmed by qPCR (Figure 6; Table 5). Collectively, this study identified candidate genes for further investigation to attain a better understanding of the molecular targets of T1 and its affected pathways that can improve the homeostasis of misfolded opsin mutants.
Figure 6.
Fold changes of 9 transcripts in NIH3T3 cells expressing GFP only (Control, grey), Opsin and GFP (Opsin, green), or P23H opsin and GFP treated with DMSO (P23H, magenta) or compound 1 (DMSO), quantified by qPCR. Fold changes of transcripts were first normalized by Gapdh as a control transcript, and then compared to Control in the qPCR assay. The fold change of each transcript was averaged from three biological replicates, and error bars show the standard deviations of those replicates. * indicates a P value smaller than 0.05. Transcriptional regulation of the 9 transcripts by T1 was in agreement with what was found by RNA-seq.
Table 5.
Fold change of transcripts comparing NIH3T3 cells expressing the P23H opsin and GFP that was treated with compound 1 and those treated with DMSO (T1 versus P23H).
| RNA-seq | qPCR | ||||
|---|---|---|---|---|---|
| Gene name | Description | Fold change (T1 versus P23H) | P value (T1 versus P23H) | Fold change (T1 versus P23H) | P value (T1 versus P23H) |
| Txnip | thioredoxin interacting protein | 9.44 | 4.12E-13 | 15.63 | 2.32E-03 |
| Nqo1 | NAD(P)H dehydrogenase, quinone 1 | 2.55 | 1.68E-06 | 2.60 | 4.14E-02 |
| Gstp1 | glutathione S-transferase, pi 1 | 2.43 | 4.98E-06 | 8.92 | 8.29E-03 |
| Pfdn6 | prefoldin subunit 6 | 4.25 | 4.20E-03 | 5.36 | 1.89E-02 |
| Itch | itchy, E3 ubiquitin protein ligase | 0.48 | 1.29E-03 | 0.54 | 4.81E-02 |
| Vdr | vitamin D receptor | 3.33 | 5.86E-09 | 4.44 | 7.59E-04 |
| Dnajb14 | DnaJ (Hsp40) homolog, subfamily B, member 14 | 1.49 | 2.19E-09 | 16.09 | 3.00E-02 |
| Clec16a | C-type lectin domain family 16, member A | 2.46 | 2.00E-03 | 1.86 | 2.93E-02 |
| Masp1 | mannan-binding lectin serine peptidase 1 | 0.22 | 3.34E-07 | 0.61 | 4.98E-02 |
In contrast to the profound transcriptome shift by T1, the lack of transcriptome alterations by treatment with compound 2 (T2) suggests that, in addition to transcriptome analysis, other systems biology techniques such as proteomics [68] are also needed for compound-target profiling. Recent work on a truncated mutant of CFTR (F508) clearly demonstrates that a global interactome remodeling occurs to CFTR interacting partners which are crucial for membrane localization [69].
DISCUSSION
We have identified two small molecular compounds that rescued the transport of a disease-causing P23H opsin mutant in a mammalian stable cell line. Though the synergistic effect of these two compounds supports their distinctive molecular targets, it is still unclear as to which molecular pathways were affected by their treatment. In attempting to answer this question, we report several observations after analyzing the transcriptomes of stable cell lines expressing WT or P23H opsin that we had used for discovering drugs for the treatment of retinitis pigmentosa [19]. First, we have shown that cell lines used for drug discovery should be profiled to appreciate their advantages and limitations as disease models. Second, we demonstrated that techniques associated with approaches in systems biology such as transcriptome profiling can be developed as a standard procedure for compound profiling at early stages of drug discovery to guide further investigation of lead compounds’ mechanisms of action. Whereas treatment with compound 1 reversed 117 transcripts that were “abnormally” expressed due to aberrant P23H opsin, treatment with compound 2 did not affect the transcriptome of cells expressing the P23H opsin. This finding confirmed that these two compounds have distinct mechanisms of action, even though treatment with each compound resulted in a partially rescued transport of P23H opsin. Transcriptome analysis already is used routinely to study molecular events during both the development of the normal retina and the onset and progression of degenerative retinal diseases. It will not be too long before this technique is broadly employed during drug discovery for treatment of such diseases.
Treatment with two active compounds caused distinct transcriptome changes. RNA-seq-based transcriptome analysis has been used to examine the efficacy of potential drugs for diabetic retinopathy in a mouse model [30]. A recent study used microarray analysis to examine the pathways changed by small molecular agents that could prevent staurosporine-induced apoptosis of NIH3T3 cells [35]. The transcriptome analysis of control and treated cells identified changes in gene expression affected by apoptotic conditions that could be reversed by treatment with drugs used in the screen.
Modern genetic profiling can enhance systems pharmacology in the endeavor to improve the design of drugs and their testing in models of complex human diseases. First, genetic profiling can provide a global assessment of the alterations in signaling systems caused by disease. Any perturbation of a native state, even resulting from a single mutation, can alter many cellular networks to induce a new steady-state that then is manifestd as a disease phenotype. For example, removal of the retina-specific leucine zipper gene, Nrl [70] alters hundreds of transcripts that change the photoreceptor population [28, 29, 71]. Similarly, mice with a genetic A/J background exhibit variations in hundreds of transcripts compared to C57Bl/6J WT mice, driving complex alterations associated with cone photoreceptor cell degeneration [72]. Modern genetic profiling also can determine how well a therapy returns an abnormal gene expression landscape toward more normal conditions. In this study, we showed that treatment with compound 1 regulated transcripts involved in multiple pathways related to membrane protein biosynthesis and transport. These effects can be viewed as desirable activities of compound 1. In contrast, transcripts affected by compound 1 that relate to cell cycle and apoptosis could be counted as risk factors that should be tested in animal models for potential side effects. Connections between various intra- and inter-cellular pathways must be considered when evaluating drug safety and efficacy and even the duration of a successful therapy. Thus, the initial specificity of a drug does not guarantee its long-term effectiveness or safety. Rather, genetic profiling can assess the molecular consequences in multiple cell-types associated with a particular drug treatment and thereby improve ultimate drug design [73].
The transcriptome changes observed in NIH3T3 cells with rod opsin expression could be specific to this cell line and have little direct correlation with free rod opsin signaling in photoreceptor cells in vivo 37, 58, 59. However, the DE profile of Opsin versus Control is a transcriptional measure of the molecular properties induced by WT opsin in NIH3T3 cells that can be compared to the DE profile of P23H versus Control (Table 3). This finding could link the cause, namely the P23H single mutation, with its consequences, i.e. ER retention of P23H opsin in these cells. Notably, even though NIH3T3 cells lack the photoreceptor cell's tertiary outer segment ciliary structure, the pre-ciliary biosynthesis of opsin is similar in both types of cells. The defect of P23H opsin transport is due to its structural instability and ER retention, a pre-ciliary defect. Therefore, the transcriptome difference between the two stable cell lines expressing the WT or P23H opsin, in parallel to the transcriptome difference between WT or P23H knock-in mouse models, provides an essential set of data to understand the advantages and limitations of cells used for drug discovery .
In short, uncommonly used in small molecular screening, we employed NGS to evaluate a simple cellular system as a disease model for drug discovery. This strategy can then be tailored to identify novel and effective compounds or combinations of compounds applicable to the treatment of complex diseases as recently reported in animal models [74] and eventually in humans.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Leslie T. Webster, Jr., and all members of the Palczewski laboratory (Case Western Reserve University) for valuable comments regarding this manuscript. This work utilized the computational resources of the HPC Biowulf cluster at NIH (http://hpc.nih.gov). K.P. is the John H. Hord Professor of Pharmacology.
Funding Information
This research was supported in part by grants from the National Institutes of Health (NIH) (EY022326 and EY R24024864 to K.P and K99 EY024992 to Y.C.), the Arnold and Mabel Beckman Foundation, Foundation Fighting Blindness, and the Intramural Research Program of the National Eye Institute (EY000474 and EY000546 to A.S.).
ABBREVIATIONS
- ad
autosomal dominant
- BP
biological process
- CHOP
C/EBP homologous protein
- CRE
the cAMP-responsive element
- CREB
the cAMP-responsive element-binding protein
- CREM
the cAMP-responsive element modulator
- ATF-1
activating transcription factor-1
- DAVID
the database for annotation, visualization and integrated discovery
- DE
differential expression
- DMEM
Dulbecco's modified Eagle's medium
- ER
endoplasmic reticulum
- ERAD
ER-associated protein degradation
- FDA
Food and Drug Administration
- FDR
false discovery rate
- FPKM
fragments per kilobase of transcript per million mapped reads
- GFP
green fluorescence protein
- GPCR
G protein-coupled receptor
- GO
gene ontology
- HTS
high-throughput screen or screening
- NGS
next-generation sequencing
- qPCR
quantitative real-time polymerase chain reaction
- RP
retinitis pigmentosa
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ASSOCIATED CONTENT
Data access
The data presented in this publication have been deposited in NCBI's Gene Expression Omnibus [75] and are accessible through GEO Series accession number GSE77449 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE77449).
A temporary private link to give to reviewers and authors to access the submission is: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=qhgvkswelzwbhil&acc=GSE77449
The Authors declare that they have no conflicts of interest with the contents of this article.
SUPPORTING INFORMATION
Quality control parameters of the RNA-seq data set and complete lists of transcripts that are differentially expressed among different sample groups.
REFERENCES
- 1.Boland MR, Jacunski A, Lorberbaum T, Romano JD, Moskovitch R, Tatonetti NP. Systems biology approaches for identifying adverse drug reactions and elucidating their underlying biological mechanisms. Wiley interdisciplinary reviews. Systems biology and medicine. 2015 doi: 10.1002/wsbm.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Westerhoff HV, Nakayama S, Mondeel TD, Barberis M. Systems Pharmacology: An opinion on how to turn the impossible into grand challenges. Drug discovery today. Technologies. 2015;15:23–31. doi: 10.1016/j.ddtec.2015.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Daiger SP, Sullivan LS, Bowne SJ. Genes and mutations causing retinitis pigmentosa. Clinical genetics. 2013;84(2):132–41. doi: 10.1111/cge.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ratnapriya R, Swaroop A. Genetic architecture of retinal and macular degenerative diseases: the promise and challenges of next-generation sequencing. Genome Med. 2013;5(10):84. doi: 10.1186/gm488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LaVail MM, Yasumura D, Matthes MT, Yang H, Hauswirth WW, Deng WT, Vollrath D. Gene Therapy for MERTK-Associated Retinal Degenerations. Advances in experimental medicine and biology. 2016;854:487–93. doi: 10.1007/978-3-319-17121-0_65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conlon TJ, Deng WT, Erger K, Cossette T, Pang JJ, Ryals R, Clement N, Cleaver B, McDoom I, Boye SE, Peden MC, Sherwood MB, Abernathy CR, Alkuraya FS, Boye SL, Hauswirth WW. Preclinical potency and safety studies of an AAV2-mediated gene therapy vector for the treatment of MERTK associated retinitis pigmentosa. Human gene therapy. Clinical development. 2013;24(1):23–8. doi: 10.1089/humc.2013.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar A, Midha N, Gogia V, Gupta S, Sehra S, Chohan A. Efficacy of oral valproic acid in patients with retinitis pigmentosa. Journal of ocular pharmacology and therapeutics : the official journal of the Association for Ocular Pharmacology and Therapeutics. 2014;30(7):580–6. doi: 10.1089/jop.2013.0166. [DOI] [PubMed] [Google Scholar]
- 8.Bhalla S, Joshi D, Bhullar S, Kasuga D, Park Y, Kay CN. Long-term follow-up for efficacy and safety of treatment of retinitis pigmentosa with valproic acid. The British journal of ophthalmology. 2013;97(7):895–9. doi: 10.1136/bjophthalmol-2013-303084. [DOI] [PubMed] [Google Scholar]
- 9.Shanmugam PM, Minija CK, Ramanjulu R, Tekwani P, Saxena M. Effect of short-term oral valproic Acid on vision and visual field in retinitis pigmentosa. Ophthalmology and therapy. 2012;1(1):6. doi: 10.1007/s40123-012-0006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sisk RA. Valproic acid treatment may be harmful in non-dominant forms of retinitis pigmentosa. The British journal of ophthalmology. 2012;96(8):1154–5. doi: 10.1136/bjophthalmol-2012-301950. [DOI] [PubMed] [Google Scholar]
- 11.Clemson CM, Tzekov R, Krebs M, Checchi JM, Bigelow C, Kaushal S. Therapeutic potential of valproic acid for retinitis pigmentosa. The British journal of ophthalmology. 2011;95(1):89–93. doi: 10.1136/bjo.2009.175356. [DOI] [PubMed] [Google Scholar]
- 12.Palczewski K. Chemistry and biology of vision. The Journal of biological chemistry. 2012;287(3):1612–9. doi: 10.1074/jbc.R111.301150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sung CH, Davenport CM, Hennessey JC, Maumenee IH, Jacobson SG, Heckenlively JR, Nowakowski R, Fishman G, Gouras P, Nathans J. Rhodopsin mutations in autosomal dominant retinitis pigmentosa. Proc. Natl. Acad. Sci. U. S. A. 1991;88(15):6481–5. doi: 10.1073/pnas.88.15.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sung CH, Schneider BG, Agarwal N, Papermaster DS, Nathans J. Functional heterogeneity of mutant rhodopsins responsible for autosomal dominant retinitis pigmentosa. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(19):8840–4. doi: 10.1073/pnas.88.19.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noorwez SM, Kuksa V, Imanishi Y, Zhu L, Filipek S, Palczewski K, Kaushal S. Pharmacological chaperone-mediated in vivo folding and stabilization of the P23H-opsin mutant associated with autosomal dominant retinitis pigmentosa. The Journal of biological chemistry. 2003;278(16):14442–50. doi: 10.1074/jbc.M300087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Jastrzebska B, Cao P, Zhang J, Wang B, Sun W, Yuan Y, Feng Z, Palczewski K. Inherent instability of the retinitis pigmentosa P23H mutant opsin. The Journal of biological chemistry. 2014;289(13):9288–303. doi: 10.1074/jbc.M114.551713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakami S, Maeda T, Bereta G, Okano K, Golczak M, Sumaroka A, Roman AJ, Cideciyan AV, Jacobson SG, Palczewski K. Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. The Journal of biological chemistry. 2011;286(12):10551–67. doi: 10.1074/jbc.M110.209759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakami S, Kolesnikov AV, Kefalov VJ, Palczewski K. P23H opsin knock-in mice reveal a novel step in retinal rod disc morphogenesis. Human molecular genetics. 2014;23(7):1723–41. doi: 10.1093/hmg/ddt561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Tang H, Seibel W, Papoian R, Li X, Lambert NA, Palczewski K. A High-Throughput Drug Screening Strategy for Detecting Rhodopsin P23H Mutant Rescue and Degradation. Investigative ophthalmology & visual science. 2015;56(4):2553–67. doi: 10.1167/iovs.14-16298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendes HF, Cheetham ME. Pharmacological manipulation of gain-of-function and dominant-negative mechanisms in rhodopsin retinitis pigmentosa. Human molecular genetics. 2008;17(19):3043–54. doi: 10.1093/hmg/ddn202. [DOI] [PubMed] [Google Scholar]
- 21.Saliba RS, Munro PM, Luthert PJ, Cheetham ME. The cellular fate of mutant rhodopsin: quality control, degradation and aggresome formation. Journal of cell science. 2002;115(Pt 14):2907–18. doi: 10.1242/jcs.115.14.2907. [DOI] [PubMed] [Google Scholar]
- 22.Uren PJ, Lee JT, Doroudchi MM, Smith AD, Horsager A. A profile of transcriptomic changes in the rd10 mouse model of retinitis pigmentosa. Molecular vision. 2014;20:1612–28. [PMC free article] [PubMed] [Google Scholar]
- 23.Sundermeier TR, Vinberg F, Mustafi D, Bai X, Kefalov VJ, Palczewski K. R9AP overexpression alters phototransduction kinetics in iCre75 mice. Investigative ophthalmology & visual science. 2014;55(3):1339–47. doi: 10.1167/iovs.13-13564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alves CH, Bossers K, Vos RM, Essing AH, Swagemakers S, van der Spek PJ, Verhaagen J, Wijnholds J. Microarray and morphological analysis of early postnatal CRB2 mutant retinas on a pure C57BL/6J genetic background. PloS one. 2013;8(12):e82532. doi: 10.1371/journal.pone.0082532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J, Farjo R, MacNee SP, Baehr W, Stambolian DE, Swaroop A. Annotation and analysis of 10,000 expressed sequence tags from developing mouse eye and adult retina. Genome Biol. 2003;4(10):R65. doi: 10.1186/gb-2003-4-10-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roger JE, Hiriyanna A, Gotoh N, Hao H, Cheng DF, Ratnapriya R, Kautzmann MA, Chang B, Swaroop A. OTX2 loss causes rod differentiation defect in CRX-associated congenital blindness. The Journal of clinical investigation. 2014;124(2):631–43. doi: 10.1172/JCI72722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang HJ, Ratnapriya R, Cogliati T, Kim JW, Swaroop A. Vision from next generation sequencing: multi-dimensional genome-wide analysis for producing gene regulatory networks underlying retinal development, aging and disease. Progress in retinal and eye research. 2015;46:1–30. doi: 10.1016/j.preteyeres.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akimoto M, Cheng H, Zhu D, Brzezinski JA, Khanna R, Filippova E, Oh EC, Jing Y, Linares JL, Brooks M, Zareparsi S, Mears AJ, Hero A, Glaser T, Swaroop A. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc. Natl. Acad. Sci. U. S. A. 2006;103(10):3890–5. doi: 10.1073/pnas.0508214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brooks MJ, Rajasimha HK, Roger JE, Swaroop A. Next-generation sequencing facilitates quantitative analysis of wild-type and Nrl(−/−) retinal transcriptomes. Mol. Vis. 2011;17:3034–54. [PMC free article] [PubMed] [Google Scholar]
- 30.Kandpal RP, Rajasimha HK, Brooks MJ, Nellissery J, Wan J, Qian J, Kern TS, Swaroop A. Transcriptome analysis using next generation sequencing reveals molecular signatures of diabetic retinopathy and efficacy of candidate drugs. Mol. Vis. 2012;18:1123–46. [PMC free article] [PubMed] [Google Scholar]
- 31.Das A, Chai JC, Yang CS, Lee YS, Das ND, Jung KH, Chai YG. Dual transcriptome sequencing reveals resistance of TLR4 ligand-activated bone marrow-derived macrophages to inflammation mediated by the BET inhibitor JQ1. Scientific reports. 2015;5:16932. doi: 10.1038/srep16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakuma K, Komatsu H, Maruyama M, Imaichi S, Habata Y, Mori M. Temporal and spatial transcriptional fingerprints by antipsychotic or propsychotic drugs in mouse brain. PloS one. 2015;10(2):e0118510. doi: 10.1371/journal.pone.0118510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giordano E, Davalos A, Crespo MC, Tome-Carneiro J, Gomez-Coronado D, Visioli F. Soy isoflavones in nutritionally relevant amounts have varied nutrigenomic effects on adipose tissue. Molecules. 2015;20(2):2310–22. doi: 10.3390/molecules20022310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tavares R, Scherer NM, Ferreira CG, Costa FF, Passetti F. Splice variants in the proteome: a promising and challenging field to targeted drug discovery. Drug discovery today. 2015;20(3):353–60. doi: 10.1016/j.drudis.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Wu J, Wang Y, Liang S, Ma H. Cytoprotective effect of selective small-molecule caspase inhibitors against staurosporine-induced apoptosis. Drug design, development and therapy. 2014;8:583–600. doi: 10.2147/DDDT.S60283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palfreyman MG, Hook DJ, Klimczak LJ, Brockman JA, Evans DM, Altar CA. Novel directions in antipsychotic target identification using gene arrays. Curr. Drug Targets CNS Neurol. Disord. 2002;1(2):227–38. doi: 10.2174/1568007024606203. [DOI] [PubMed] [Google Scholar]
- 37.Hodges RS, Heaton RJ, Parker JM, Molday L, Molday RS. Antigen-antibody interaction. Synthetic peptides define linear antigenic determinants recognized by monoclonal antibodies directed to the cytoplasmic carboxyl terminus of rhodopsin. J. Biol. Chem. 1988;263(24):11768–75. [PubMed] [Google Scholar]
- 38.Chomczynski P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. BioTechniques. 1993;15532-4(3):536–7. [PubMed] [Google Scholar]
- 39.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanatephenol-chloroform extraction. Analytical biochemistry. 1987;162(1):156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 40.Kaewkhaw R, Kaya KD, Brooks M, Homma K, Zou J, Chaitankar V, Rao M, Swaroop A. Transcriptome Dynamics of Developing Photoreceptors in Three-Dimensional Retina Cultures Recapitulates Temporal Sequence of Human Cone and Rod Differentiation Revealing Cell Surface Markers and Gene Networks. Stem Cells. 2015;33(12):3504–18. doi: 10.1002/stem.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 43.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krebs MP, Holden DC, Joshi P, Clark CL, 3rd, Lee AH, Kaushal S. Molecular mechanisms of rhodopsin retinitis pigmentosa and the efficacy of pharmacological rescue. Journal of molecular biology. 2010;395(5):1063–78. doi: 10.1016/j.jmb.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 45.Mendes HF, Zaccarini R, Cheetham ME. Pharmacological manipulation of rhodopsin retinitis pigmentosa. Advances in experimental medicine and biology. 2010;664:317–23. doi: 10.1007/978-1-4419-1399-9_36. [DOI] [PubMed] [Google Scholar]
- 46.Ohgane K, Dodo K, Hashimoto Y. Retinobenzaldehydes as proper-trafficking inducers of folding-defective P23H rhodopsin mutant responsible for retinitis pigmentosa. Bioorganic & medicinal chemistry. 2010;18(19):7022–8. doi: 10.1016/j.bmc.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 47.Zhu L, Imanishi Y, Filipek S, Alekseev A, Jastrzebska B, Sun W, Saperstein DA, Palczewski K. Autosomal recessive retinitis pigmentosa and E150K mutation in the opsin gene. The Journal of biological chemistry. 2006;281(31):22289–98. doi: 10.1074/jbc.M602664200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang H, Xiong S, Xia X. Retinitis pigmentosaassociated rhodopsin mutant T17M induces endoplasmic reticulum (ER) stress and sensitizes cells to ER stress-induced cell death. Molecular medicine reports. 2014;9(5):1737–42. doi: 10.3892/mmr.2014.1987. [DOI] [PubMed] [Google Scholar]
- 49.Anukanth A, Khorana HG. Structure and function in rhodopsin. Requirements of a specific structure for the intradiscal domain. J. Biol. Chem. 1994;269(31):19738–44. [PubMed] [Google Scholar]
- 50.Leibiger C, Kosyakova N, Mkrtchyan H, Glei M, Trifonov V, Liehr T. First molecular cytogenetic high resolution characterization of the NIH 3T3 cell line by murine multicolor banding. The journal of histochemistry and cytochemistry : official journal of the Histochemistry Society. 2013;61(4):306–12. doi: 10.1369/0022155413476868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. The Journal of cell biology. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim HJ, Son ED, Jung JY, Choi H, Lee TR, Shin DW. Violet light down-regulates the expression of specific differentiation markers through Rhodopsin in normal human epidermal keratinocytes. PloS one. 2013;8(9):e73678. doi: 10.1371/journal.pone.0073678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cornwall MC, Fain GL. Bleached pigment activates transduction in isolated rods of the salamander retina. The Journal of physiology. 1994;480(Pt 2):261–79. doi: 10.1113/jphysiol.1994.sp020358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cornwall MC, Matthews HR, Crouch RK, Fain GL. Bleached pigment activates transduction in salamander cones. The Journal of general physiology. 1995;106(3):543–57. doi: 10.1085/jgp.106.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buczylko J, Saari JC, Crouch RK, Palczewski K. Mechanisms of opsin activation. The Journal of biological chemistry. 1996;271(34):20621–30. doi: 10.1074/jbc.271.34.20621. [DOI] [PubMed] [Google Scholar]
- 56.Jastrzebska B, Chen Y, Orban T, Jin H, Hofmann L, Palczewski K. Disruption of Rhodopsin Dimerization with Synthetic Peptides Targeting an Interaction Interface. The Journal of biological chemistry. 2015;290(42):25728–44. doi: 10.1074/jbc.M115.662684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chiang WC, Kroeger H, Sakami S, Messah C, Yasumura D, Matthes MT, Coppinger JA, Palczewski K, LaVail MM, Lin JH. Robust Endoplasmic Reticulum-Associated Degradation of Rhodopsin Precedes Retinal Degeneration. Molecular neurobiology. 2015;52(1):679–95. doi: 10.1007/s12035-014-8881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mendes HF, van der Spuy J, Chapple JP, Cheetham ME. Mechanisms of cell death in rhodopsin retinitis pigmentosa: implications for therapy. Trends Mol. Med. 2005;11(4):177–85. doi: 10.1016/j.molmed.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 59.Noorwez SM, Sama RR, Kaushal S. Calnexin improves the folding efficiency of mutant rhodopsin in the presence of pharmacological chaperone 11-cis-retinal. J. Biol. Chem. 2009;284(48):33333–42. doi: 10.1074/jbc.M109.043364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kosmaoglou M, Kanuga N, Aguila M, Garriga P, Cheetham ME. A dual role for EDEM1 in the processing of rod opsin. Journal of cell science. 2009;122(Pt 24):4465–72. doi: 10.1242/jcs.055228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sizova OS, Shinde VM, Lenox AR, Gorbatyuk MS. Modulation of cellular signaling pathways in P23H rhodopsin photoreceptors. Cellular signalling. 2014;26(4):665–72. doi: 10.1016/j.cellsig.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318(5852):944–9. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Espinosa-Diez C, Miguel V, Mennerich D, Kietzmann T, Sanchez-Perez P, Cadenas S, Lamas S. Antioxidant responses and cellular adjustments to oxidative stress. Redox biology. 2015;6:183–97. doi: 10.1016/j.redox.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sopha P, Kadokura H, Yamamoto YH, Takeuchi M, Saito M, Tsuru A, Kohno K. A novel mammalian ER-located J-protein, DNAJB14, can accelerate ERAD of misfolded membrane proteins. Cell structure and function. 2012;37(2):177–87. doi: 10.1247/csf.12017. [DOI] [PubMed] [Google Scholar]
- 65.Tashiro E, Zako T, Muto H, Itoo Y, Sorgjerd K, Terada N, Abe A, Miyazawa M, Kitamura A, Kitaura H, Kubota H, Maeda M, Momoi T, Iguchi-Ariga SM, Kinjo M, Ariga H. Prefoldin protects neuronal cells from polyglutamine toxicity by preventing aggregation formation. The Journal of biological chemistry. 2013;288(27):19958–72. doi: 10.1074/jbc.M113.477984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takano M, Tashiro E, Kitamura A, Maita H, Iguchi-Ariga SM, Kinjo M, Ariga H. Prefoldin prevents aggregation of alpha-synuclein. Brain research. 2014;1542:186–94. [PubMed] [Google Scholar]
- 67.Abe A, Takahashi-Niki K, Takekoshi Y, Shimizu T, Kitaura H, Maita H, Iguchi-Ariga SM, Ariga H. Prefoldin plays a role as a clearance factor in preventing proteasome inhibitor-induced protein aggregation. The Journal of biological chemistry. 2013;288(39):27764–76. doi: 10.1074/jbc.M113.476358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar A, Baycin-Hizal D, Shiloach J, Bowen MA, Betenbaugh MJ. Coupling enrichment methods with proteomics for understanding and treating disease. Proteomics. Clinical applications. 2015;9(1-2):33–47. doi: 10.1002/prca.201400097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pankow S, Bamberger C, Calzolari D, Martinez-Bartolome S, Lavallee-Adam M, Balch WE, Yates JR., 3rd F508 CFTR interactome remodelling promotes rescue of cystic fibrosis. Nature. 2015;528(7583):510–6. doi: 10.1038/nature15729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat. Genet. 2001;29(4):447–52. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 71.Mustafi D, Kevany BM, Genoud C, Okano K, Cideciyan AV, Sumaroka A, Roman AJ, Jacobson SG, Engel A, Adams MD, Palczewski K. Defective photoreceptor phagocytosis in a mouse model of enhanced S-cone syndrome causes progressive retinal degeneration. FASEB J. 2011;25(9):3157–76. doi: 10.1096/fj.11-186767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mustafi D, Maeda T, Kohno H, Nadeau JH, Palczewski K. Inflammatory priming predisposes mice to age-related retinal degeneration. The Journal of clinical investigation. 2012;122(8):2989–3001. doi: 10.1172/JCI64427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Y, Palczewski K. Systems Pharmacology Links GPCRs with Retinal Degenerative Disorders. Annu Rev Pharmacol Toxicol. 2016;56:273–98. doi: 10.1146/annurev-pharmtox-010715-103033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Y, Palczewska G, Masuho I, Gao S, Jin H, Dong Z, Gieser L, Brooks MJ, Kiser PD, Kern TS, Martemyanov KA, Swaroop A, Palczewski K. Synergistically acting agonists and antagonists of G protein-coupled receptors prevent photoreceptor cell degeneration. Sci Signal. 2016;9(438):ra74. doi: 10.1126/scisignal.aag0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.