Abstract
Introduction
The aim was to examine the influence of the SCN1A gene polymorphism IVS5-91 rs3812718 G>A on the response to antiepileptic drugs (AEDs) in monotherapy or polytherapy.
Material and methods
Two hundred epilepsy patients and 200 healthy subjects were genotyped for SCN1A IVS5-91 rs3812718 G>A polymorphism using TaqMan assay. Patients were divided into drug-responsive and drug-resistant patients. The drug-responsive group was further studied, comparing monotherapy in maximum and minimum doses and monotherapy-responsive and -resistant groups.
Results
There were no statistically significant differences in the allelic frequencies and genotype distributions between patients and controls (p = 0.178). The distribution of SCN1A IVS5-91 rs3812718 G>A genotypes was similar between drug-responsive and drug-resistant patients (p = 0.463). The differences in genotype distributions (A/A or A/G vs. G/G) between monotherapy-responsive and -resistant groups were statistically significant (p = 0.021). Within the monotherapy-responsive group, patients with the A/A or A/G genotype needed higher dose AEDs than patients with the G/G genotype (p = 0.032). The relative risk for generalized epilepsy due to A-containing genotypes was of marginal statistical significance when compared with the G/G genotype (p = 0.05).
Conclusions
Overall, our findings demonstrate an association of SCN1A IVS5-91 rs3812718 G>A polymorphism with AED responsiveness in monotherapy without evidence of an effect on drug-resistant epilepsy.
Keywords: antiepileptic drugs, gene polymorphism, SCN1A, responsiveness, resistant epilepsy
Introduction
Genetic factors have been implicated in epilepsy and other disorders [1–4]. Epilepsy is a common and very heterogeneous neurological disorder in which genetics play an important etiological role, either because the underlying cause of epilepsy is primarily genetic or because genes modulate susceptibility to an epileptogenic insult [5]. Genetic factors also play a role in the response to antiepileptic drugs (AEDs), affecting their efficacy.
Several single nucleotide polymorphisms (SNPs) have been reported to be associated with AED dosage and drug resistance [6, 7].
The majority of AEDs exhibit anticonvulsant effects by blocking voltage gated sodium channels (Nav). Sodium channels play an essential role in the generation and propagation of action potentials in neurons and other excitable cells [8]. Their composition as heteromultimeric complexes comprise a large (approximately 260 kDa), pore-forming α-subunit, responsible for sodium channel function, and smaller accessory β-subunits [9, 10]. Architecture of the α-subunit consists of four domains (D1–D4), each containing six α-helical transmembrane segments (S1–S6). The positively charged amino acid residues in the S4 segments (voltage sensors) are involved in action potentials and membrane potential changes. Eleven genes (SCN1A–SCN11A) encode the α-subunit, which is a target for commonly used sodium channel blockers such as carbamazepine (CBZ), oxcarbamazepine (OXZ), phenytoin (PHT), and lamotrigine (LTG) [11, 12]. Genetic variations in the α-subunit may affect the electro-physiological properties of sodium channels in drug-resistant epilepsy patients [13].
The SCN1A gene is widely investigated. Over 500 mutations have been identified in the SCN1A gene, some of which correlated with the generation of epileptic seizures as febrile seizures, generalized epilepsy with febrile seizures plus (GEFS+), or severe myoclonic epilepsy of infancy (SMEI) [14–17]. Furthermore, pharmacogenetic studies revealed that genetic polymorphisms in SCN1A and SCN2A influence the clinical response to AEDs [18, 19].
A SCN1A functional intronic polymorphism (IVS5-91 rs3812718 G>A) located in the 5ʹ splice donor site of a highly conserved, alternatively spliced exon (exon 5N) has been reported to be a possible modifying factor for AED responsiveness. The polymorphism was shown to be significantly associated with maximum doses of CBZ and PHT in a cohort of patients with various types of epilepsy [20]. In that study Tate and his colleagues demonstrated a strong relationship between the A/A genotype and the highest required doses of CBZ and PHT. In a separate study by the same authors a similar relationship was detected between PHT serum levels at maintenance dose and genotypes [21]. However, subsequent studies have failed to identify an association between this polymorphism and dosages of sodium channel blocker AEDs [22], although in a Japanese study Abe et al. demonstrated an association of SCN1A IVS5-91 rs3812718 G>A polymorphism with CBZ-resistant epilepsy [23].
The aim of the present study was to examine the effect of SCN1A gene polymorphism IVS5-91 rs3812718 G>A on the response to sodium channel blocker AEDs in monotherapy or polytherapy in a population in Thrace, in northern Greece.
Material and methods
Patients and controls
This was a multicenter study involving patients of the Democritus University of Thrace in Alexandroupolis, the General Hospital of Komotini, the General Hospital of Didimoticho and the General Hospital of Xanthi, all located in Thrace, northern Greece. The present study included 200 epilepsy patients. Patients were followed up as outpatients in the Epilepsy Clinic of Democritus University of Thrace (Greece) for at least 2 years. During the same period 200 healthy subjects with no evidence of epilepsy, individually matched to epilepsy patients based on gender and age, who visited the Democritus University Hospital of Thrace in Alexandroupolis for a routine health checkup, were recruited as controls.
Seizures and epilepsy syndromes were classified according to the International League Against Epilepsy (ILAE) (1989) [24]. The inclusion criteria for the study comprised: 1) sex: male and female, 2) diagnosis of generalized or partial epilepsy, in accordance with ILAE criteria, 3) known clinical response to the AEDs administered, 4) neuroimaging performed (MRI). The exclusion criteria comprised: 1) idiopathic partial epilepsy, 2) previous history of non-epileptic seizures, 3) poor compliance with AEDs, 4) severe adverse drug reactions, 5) alcohol and/or drug abuse, 6) significant mental disorders, 7) medical conditions that might affect liver and renal metabolism.
Before the beginning of the study, an informed consent form was obtained from each participant. The study protocol was approved by the Ethics Committee of the Faculty of Medicine, Democritus University of Thrace, Alexandroupolis, Greece.
Demographic and clinical data for each patient were collected from questionnaires. At the first visit we recorded sex, age, age at onset of epilepsy, family history of epilepsy, medical and neurological history, type of epileptic seizures, disease classification, frequency and duration of seizures, electro-encephalographic findings, imaging data and the AED treatment. Medical records were subsequently collected every three months until the end of the study, and recordings in each follow-up visit included doses of AEDs on monotherapy or polytherapy, patient compliance, responsiveness to AED treatment and possible side effects.
Treatment protocol
In the present study, the AEDs phenytoin, carbamazepine, oxcarbamazepine, and lamotrigine (sodium channel blockers) were used in monotherapy. Patients were treated initially with a single AED. If seizures persisted, the AED was replaced by a different single AED. In the monotherapy group doses of sodium channel blocker AEDs, such as PHT > 200 mg, CBZ > 800 mg, OXC > 1200 mg, LTG > 200 mg were considered as maximum doses. In case of resistance to monotherapy, a combination of AEDs (polytherapy) was applied. In polytherapy, in addition to sodium channel blocker AEDs we administered AEDs with a different or multiple mechanism of action (topiramate, valproate, lacosamide, levetiracetam, benzodiazepines).
Patients were divided into two groups according to drug response: drug-responsive and drug-resistant patients. The drug-responsive group was further studied, comparing monotherapy in maximum and minimum doses and monotherapy to polytherapy (monotherapy-resistant group). The definitions of drug responsiveness and drug resistance proposed by the Task Force of the ILAE Commission on Therapeutic Strategies (2009) were applied in the current study [25].
DNA analysis
Genomic DNA was extracted from venous blood using a PureLink DNA extraction kit and the procedure recommended by the manufacturer (Qiagen AS, Oslo, Norway). Extracted DNA was quantified using a NanoDrop Analyzer (ND-1000) spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA). The SCN1A IVS5-91 rs3812718 G>A polymorphism was analyzed by allelic discrimination TaqMan assay according to the manufacturer’s protocols (Applied Biosystems, Foster City, CA, USA). Genotyping was performed using real-time PCR (polymerase chain reaction) on a MxPro 3005P, Stratagene (MxPro 3005P, Stratagene, La Jolla, CA, USA).
Statistical analysis
Statistical analysis was performed using the Statistical Package for the Social Sciences (SPSS), version 19.0 (IBM). The χ2 test was used to assess differences of genotype and allele frequencies between patients with epilepsy and controls. Unconditional logistic regression analysis was used to estimate gender, age and type of epilepsy-adjusted odds ratios (aOR) and 95% confidence intervals (CI) as the measure of association of SCN1A IVS5-91G>A polymorphism with several characteristics of epilepsy patients. Student’s t test and χ2 test were used to assess age and gender differences between drug-resistant and drug-responsive epilepsy, respectively. All tests were two-tailed and statistical significance was considered for p-values less than 0.05.
Results
Characteristics of study participants
The demographic characteristics of the 200 enrolled patients are shown in Table I. The study included 101 females and 99 males, with a mean age of 41.74 ±16.5 years (range: 16 to 73 years). Drug-resistant epilepsy was diagnosed in 70 (35%) patients and drug-responsive epilepsy (monotherapy- and polytherapy-responsive epilepsy) in 130 (65%) patients. There were no statistically significant differences in gender (p = 0.486) or age (p = 0.254) between these two groups of patients (Table I). Moreover, idiopathic generalized epilepsy was more frequent among drug-responsive patients, while cryptogenic partial epilepsy was more frequent among drug-resistant patients (p < 0.001). Of the 200 participants, 65 (32.5%) suffered generalized epilepsy, 69 (34.5%) symptomatic partial epilepsy and 66 (33%) cryptogenic partial epilepsy.
Table I.
Demographic and clinical characteristics of patients
| Parameter | Patients | Drug-responsive | Drug-resistant | P-value |
|---|---|---|---|---|
| Gender, n (%): | 0.486 | |||
| Females | 101 (50.5) | 68 (52.3) | 33 (47.1) | |
| Males | 99 (49.5) | 62 (47.7) | 37 (52.9) | |
| Age, mean ± SD [years] | 41.74 ±16.50 | 40.76 ±16.55 | 43.56 ±16.37 | 0.254 |
| Type of epilepsy, n (%): | < 0.001 | |||
| Idiopathic generalized | 56 (28.0) | 55 (42.3) | 1 (1.4) | |
| Symptomatic generalized | 9 (4.5) | 7 (5.4) | 2 (2.9) | |
| Symptomatic partial | 69 (34.5) | 44 (33.8) | 25 (35.7) | |
| Cryptogenic partial | 66 (33.0) | 24 (18.5) | 42 (60.0) | |
| Drug response | 130 (65.0) | 70 (35.0) | ||
| Therapy, n (%): | ||||
| Monotherapy minimum doses | 37 (18.5) | |||
| Monotherapy maximum doses | 58 (29.0) | |||
| Polytherapy | 105 (52.5) |
Correlation between IVS5-91 rs3812718 G>A polymorphism and generalized epilepsy
The distribution of genotypes and alleles of SCN1A IVS5-91 rs3812718 G>A polymorphism was compared between generalized and partial epilepsy. Although the genotypic and allelic frequencies were similar in generalized and partial epilepsy (p = 0.135 and p = 0.371, respectively), the A-containing genotypes were marginally more prevalent in generalized compared to partial epilepsy (p = 0.055). Moreover, multivariate logistic regression analysis, adjusting for gender and age, revealed that patients with generalized epilepsy were almost 3 times more likely to have an A-containing genotype than patients with partial epilepsy (OR = 2.62, 95% CI: 1.00–6.94, p = 0.050) (Table II).
Table II.
Distribution of genotypes and alleles of SCN1A IVS5-91 rs3812718 G>A polymorphism in relation to type of epilepsy
| Variable | Generalized epilepsy* | Partial epilepsy* | P-value | aOR (95% CI) | P-value |
|---|---|---|---|---|---|
| Genotype: | 0.135 | ||||
| G/G | 6 (9.2) | 27 (20.0) | Reference | ||
| A/G | 55 (84.6) | 98 (72.6) | 2.60 (0.98–6.91) | 0.055 | |
| A/A | 4 (6.2) | 10 (7.4) | 2.97 (0.64–13.80) | 0.165 | |
| Genotype: | 0.055 | ||||
| G/G | 6 (9.2) | 27 (20.0) | Reference | ||
| A/G or A/A | 59 (90.8) | 108 (80.0) | 2.62 (1.00–6.94) | 0.050 | |
| Allele: | 0.371 | ||||
| G | 67 (51.5) | 152 (56.3) | Reference | ||
| A | 63 (48.5) | 118 (43.7) | 1.31 (0.84–2.04) | 0.231 |
Data are expressed as number of patients and percentage (%); aOR – adjusted odds ratio for gender and age. Marginal statistical significance of SCN1A IVS5-91 rs3812718 G>A polymorphism and generalized epilepsy in A-containing genotypes (p = 0.050). Significant differences are bolded.
No correlation in distribution of IVS5-91 rs3812718 G>A polymorphism among patients and controls
Regarding the genotypic frequencies of SCN1A IVS5-91 rs3812718 G>A polymorphism, G/G, A/G and A/A genotypes were found in 16.5%, 76.5% and 7% of patients and 23.5%, 71.5% and 5% of the control group, respectively. Allelic frequency was 54.7% for the G allele and 45.3% for the A allele in patients and 59.3% and 40.7% in the control group. There were no statistically significant differences in genotypic or allelic frequencies between patients and controls (p = 0.178 and p = 0.199, respectively), although A-containg genotypes were more frequent in patients than in controls (83.5% vs. 76.5%, p = 0.080); epilepsy patients were 55% more likely to have an A-containing genotype than healthy controls (OR = 1.55, 95% CI: 0.95–2.55) (Table III).
Table III.
Distribution of genotypes and alleles of SCN1A IVS5-91 rs3812718 G>A polymorphism among patients and controls
| Variable | Controls* | Patients* | P-value | aOR (95% CI) | P-value |
|---|---|---|---|---|---|
| Genotype: | 0.178 | ||||
| G/G | 47 (23.5) | 33 (16.5) | Reference | ||
| A/G | 143 (71.5) | 153 (76.5) | 1.52 (0.92–2.51) | 0.098 | |
| A/A | 10 (5.0) | 14 (7.0) | 1.99 (0.79–5.03) | 0.144 | |
| Genotype: | 0.080 | ||||
| G/G | 47 (23.5) | 33 (16.5) | Reference | ||
| A/G or A/A | 153 (76.5) | 167 (83.5) | 1.55 (0.95–2.55) | ||
| Allele: | 0.199 | ||||
| G | 237 (59.3) | 219 (54.7) | Reference | ||
| A | 163 (40.7) | 181 (45.3) | 1.20 (0.91–1.59) |
Data are expressed as number of patients-controls and percentage (%); aOR – adjusted odds ratio for gender and age. No significant association of SCN1A IVS5-91 rs3812718 G>A polymorphism among patients and controls.
No correlation of IVS5-91 rs3812718 G>A polymorphism and drug-resistant epilepsy
The distribution of SCN1A IVS5-91 rs3812718 G>A genotypes did not differ significantly between drug-resistant and drug-responsive epilepsy patients (p = 0.463, p = 0.328 and p = 0.776 for genotype, A-containing genotype and allelic frequencies, respectively) (Table IV).
Table IV.
Distribution of genotypes and alleles of SCN1A IVS5-91 rs3812718 G>A polymorphism in relation to AED responsiveness
| Variable | Drug responder* | Drug-resistant* | P-value | aOR (95% CI) | P-value |
|---|---|---|---|---|---|
| Genotype: | 0.463 | ||||
| G/G | 19 (14.6) | 14 (20.0) | Reference | ||
| A/G | 103 (79.2) | 50 (71.4) | 0.93 (0.41–2.16) | 0.874 | |
| A/A | 8 (6.2) | 6 (8.6) | 1.45 (0.35–6.07) | 0.608 | |
| Genotype: | 0.328 | ||||
| G/G | 19 (14.6) | 14 (20.0) | Reference | ||
| A/G or A/A | 111 (85.4) | 56 (80.0) | 0.97 (0.42–2.22) | 0.946 | |
| Allele: | 0.776 | ||||
| G | 141 (54.2) | 78 (55.7) | Reference | ||
| A | 119 (45.8) | 62 (44.3) | 0.93 (0.61–1.41) | 0.730 |
Data are expressed as number of patients and percentage (%); aOR, adjusted odds ratio for gender and age. No significant association of SCN1A IVS5-91 rs3812718 G>A polymorphism among drug-responsive and drug-resistant patients.
Positive correlation between IVS5-91 rs3812718 G>A polymorphism and monotherapy-resistant patients
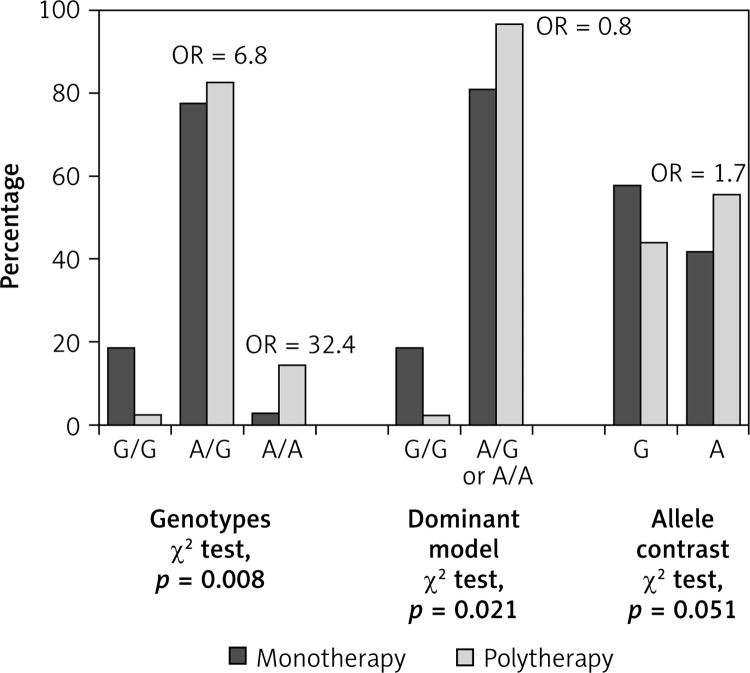
Among the entire cohort of patients, 95 (47.5%) of them were on monotherapy and 105 (52.5%) were on polytherapy. The distribution of SCN1A IVS5-91 rs3812718 G>A genotypes was compared between polytherapy-responsive (monotherapy-resistant) and monotherapy-responsive patients (Table V, Figure 1); the presence of A/A genotype was associated with a more than 30-fold (aOR = 32.43, 95% CI: 2.68–272.73, p = 0.006) and with a more than 5-fold (aOR = 5.07, 95% CI: 1.05–24.40, p = 0.043) increase in risk of resistance in monotherapy compared to the G/G and A/G genotypes, respectively. Heterozygous A/G genotype also showed a risk of 6.83 (95% CI: 0.87–53.73) compared to G/G genotype, which did not reach the statistical significance level (p = 0.068). The presence of the A-containing genotypes (A/G or A/A) and presence of the A allele yielded adjusted odds ratios of 7.96 (95% CI: 1.02–62.26, p = 0.048) and 1.73 (95% CI: 1.00–3.02, p = 0.050) for resistance in monotherapy compared to G/G genotype and G allele, respectively.
Table V.
Distribution of genotypes and alleles among monotherapy-responsive and monotherapy-resistant group
| Variable | Monotherapy responder* | Monotherapy resistant* | P-value | aOR (95% CI) | P-value |
|---|---|---|---|---|---|
| Genotype: | 0.008 | ||||
| G/G | 18 (18.9) | 1 (2.9) | Reference | ||
| A/G | 74 (77.9) | 29 (82.9) | 6.83 (0.87–53.73) | 0.068 | |
| A/A | 3 (3.2) | 5 (14.3) | 32.43 (2.68–272.73) | 0.006 | |
| A/A vs. A/G | 5.07 (1.05–24.40) | 0.043 | |||
| Genotype: | 0.021 | ||||
| G/G | 18 (18.9) | 1 (2.9) | Reference | ||
| A/G or A/A | 77 (81.1) | 34 (97.1) | 7.96 (1.02–62.26) | 0.048 | |
| Allele: | 0.051 | ||||
| G | 110 (57.9) | 31 (44.3) | Reference | ||
| A | 80 (42.1) | 39 (55.7) | 1.73 (1.00–3.02) | 0.050 |
Data are expressed as number of patients and percentage (%); aOR – adjusted odds ratio for gender and age. Statistical significant association of IVS5-91 rs 3812718 G>A polymorphism and monotherapy-resistant epilepsy. Significant differences are bolded.
Figure 1.
Distribution of genotypes and alleles of SCN1A IVS5-91 rs3812718 G>A polymorphism in relation to responsiveness to AEDs in monotherapy among epilepsy patients
Positive correlation between IVS5-91 rs 3812718 G>A polymorphism and maximum doses of AEDs in monotherapy-responsive patients
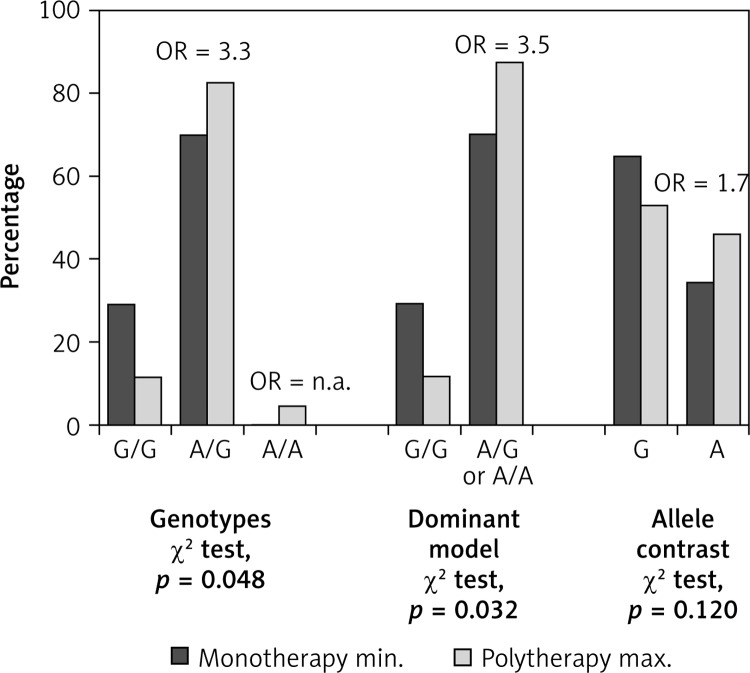
In monotherapy-responsive group the presence of A/A or A/G genotypes was associated with higher dose of AEDs compared to G/G genotype (p = 0.048). In this regard, an increased risk of higher dose of AEDs with an adjusted odds ratio of 3.54 (95% CI: 1.18–10.64, p = 0.024) was associated with the A-containing genotypes among monotherapy-responsive patients. However, the allelic association with the response to different doses of AEDs in monotherapy was of marginal statistical significance (aOR = 1.66, 95% CI: 0.90–3.06, p = 0.097) (Table VI, Figure 2).
Table VI.
Distribution of genotypes and alleles in monotherapy-responsive group
| Variable | Monotherapy minimum doses* | Monotherapy maximum doses* | P-value | aOR (95% CI) | P-value |
|---|---|---|---|---|---|
| Genotype: | 0.048 | ||||
| G/G | 11 (29.7) | 7 (12.1) | Reference | ||
| A/G | 26 (70.3) | 48 (82.8) | 3.33 (1.11–9.99) | 0.031 | |
| A/A | 0 (0.0) | 3 (5.2) | – | – | |
| Genotype: | 0.032 | ||||
| G/G | 11 (29.7) | 7 (12.1) | Reference | ||
| A/G or A/A | 26 (70.3) | 51 (87.9) | 3.54 (1.18–10.64) | 0.024 | |
| Allele: | 0.120 | ||||
| G | 48 (64.9) | 62 (53.4) | Reference | ||
| A | 26 (35.1) | 54 (46.6) | 1.66 (0.90–3.06) | 0.097 |
Data are expressed as number of patients and percentage (%); aOR – adjusted odds ratio for gender and age. Statistically significant association of IVS5-91 rs 3812718 G>A polymorphism and maximum doses of AEDs in monotherapy-responsive patients. Significant differences are bolded.
Figure 2.
Distribution of genotypes and alleles of SCN1A IVS5-91 rs3812718 G>A polymorphism in relation to doses (min. vs. max.) of AEDs in monotherapy-responsive epilepsy patients
Discussion
In the present multicenter study, we investigated whether the SCN1A IVS5-91 rs3812718 G>A polymorphism influences the response to antiepileptic treatment. We demonstrated that the rs3812718 SNP in the SCN1A gene was significantly associated with monotherapy-resistant epilepsy and with maximum doses of AEDs effective in monotherapy. The presence of the A-containing genotype was a risk factor in the monotherapy-resistant patients, and carriers of the A allele (A/A or A/G genotype) required significantly higher doses of AEDs than those with the G/G genotype, whereas a correlation with multidrug resistance phenotype was not detectable. In addition, a possible positive correlation was revealed in carriers of the A-containing genotype and generalized epilepsy.
Based on mechanisms of pharmacoresistant epilepsy, alterations in drug targets (genetic or acquired) make them less sensitive to AEDs according to the target hypothesis [26]. Antiepileptic drugs targets include voltage-gated channels and neurotransmitter receptors associated with neuronal excitation. Most AEDs act by blocking sodium channels in their resting phase (tonic block), which prevents channel opening and ion conductance. It has been suggested that altered composition, function and/or expression of different subunits of sodium channels may affect the clinical response to AEDs [27].
SCN1A IVS5-91 rs3812718 G>A polymorphism alter the sensitivity to AEDs. The functional SNP rs3812718 contributes to the alternative splicing of exon 5 of SCN1A observed in brain tissue. In the genomic DNA, two alternative spliced forms of exon 5 have been described, a neonatal (5N) and an adult form (5A), that are coexpressed in the adult brain. Recent reports revealed that the SCN1A IVS5-91 rs3812718 G>A polymorphism affects the proportions of adult and neonatal mRNA transcripts in adults with epilepsy. Therefore, the G allele allows expression of both forms while the A allele significantly reduces the expression of the 5N form in relation to the 5A form. Furthermore, in individuals with the G/G genotype, up to 50% of the transcripts include the 5N form, whereas the A/A genotype allows an undetectable level of the 5N form to be expressed. The difference between 5N and 5A form consists in three amino acids and may modify the electrophysiological properties of the sodium channels (S4 sensors) and their susceptibility to AEDs. Sodium channels (Nav1.1) containing exon 5N are more sensitive to AEDs, unlike channels containing exon 5A [28]. The expression of these two alternatively spliced products is tightly controlled by the regulatory protein Nova-2. A larger Nova-2 mediated effect was detected in the A/A genotype, which was associated with increased doses of AEDs [29].
In acquired drug resistance, the disease state can promote transcriptional or post-transcriptional changes that induce structural changes in voltage-gated sodium channels and modify their sensitivity to AEDs [30]. Similarly, the subunit composition of GABAA receptors is altered in epileptic tissue [31, 32]. A decrease of a1 subunits and an increase of a4 subunits in the GABA receptor may underlie the decrease in GABA receptor response to benzodiazepines [33].
Another mechanism of pharmacoresistance, the transporter hypothesis, proposed that levels of AEDs to the epileptogenic focus are decreased because of overexpression of multi-drug transporter proteins, most notably P-glycoprotein (P-gp), encoded by the ABCB1 gene. The multi-drug transporters, P-gp and multi-drug resistance associated protein (MRP) are expressed in the apical parts of endothelial cells in the brain tissue and act as efflux pumps of lipophilic molecules, being a protection against xenobiotics. It has been found that their concentration is increased in epileptogenic foci from drug-resistant patients, thus preventing the penetration of AEDs to the brain tissue [34, 35]. After many years of research, there remains a lack of convincing evidence that AEDs are high-affinity substrates for P-gp [36, 37]. In addition, genetic association studies have failed to replicate early reports of an association between polymorphisms in the ABCB1 gene and epilepsy pharmacoresistance [38–40].
A more recent approach – the inherent severity model of epilepsy – was proposed by Rogawski [41]. According to this hypothesis, there is a continuum in severity of the disease, which determines its relative response to medication. The frequency of seizures in the early phase of epilepsy is the single most important factor that determines responsiveness to AED treatment. High frequency of seizures prior to onset of treatment is a prognostic signal of increased severity and future drug failure. Although high seizure frequency is often associated with drug resistance, a subgroup of epilepsy patients becomes seizure free after a change of AED treatment. A model alone is not able to fully explain or predict pharmacoresistance. The majority of patients are likely to have an overlap of drug resistance mechanisms.
New data suggest that inflammation might constitute a central key factor in drug-resistant epilepsy of various etiologies. Several clinical studies have demonstrated increased levels of inflammatory mediators such as interleukin (IL)-6, tumor necrosis factor α (TNF-α), IL-1β, IL-1 receptor antagonist in serum or cerebrospinal fluid (CSF) in patients with refractory epilepsy [42]. Inflammatory mediators induce changes in the expression rates, subunit composition and phosphorylation state of neurotransmitter receptors AMPA, NMDA, and GABAA and might affect drug sensitivity [43]. A pathological role of protein high-mobility group box 1 (HMGB1) has been demonstrated in microglia and astrocytes in temporal lobe epilepsy and focal cortical dysplasia (FCD) [44]. A major challenge for the future is to define specific biomarkers in serum or CSF of drug-resistant patients who might benefit from anti-inflammatory therapies.
In our study a comparison of the distribution of genotypes for the rs3812718 G>A SNP between the monotherapy-responsive and the monotherapy-resistant group showed that the carriers of A/G or A/A genotype were more likely to be resistant to monotherapy. At the allele level the association was of marginal significance. These findings indicated the A allele as a probable risk factor for monotherapy resistance to sodium channel blocker AEDs. Similarly, Abe et al. and Ma et al. [23, 45] demonstrated that the frequency of the SCN1A IVS5-91 rs3812718 A/A genotype was significantly higher in CBZ-resistant patients in contrast to the results reported by Manna et al. [46] and Sterjev et al. [47]. Further studies of Haerian et al. examining SCN1A IVS5-91 rs3812718 G>A polymorphism and clinical response to valproate (VPA) or CBZ failed to demonstrate any association [48, 49]. Moreover, regarding the effect of the SCN1A IVS5-91 rs 3812718 G>A polymorphism on the multidrug resistance phenotype, we failed to show any association with drug-resistant epilepsy patients. The lack of influence of this polymorphism on drug-resistant epilepsy confirmed the results of recent reports [46, 50, 51].
According to our data, in the monotherapy-responsive group A/A homozygotes and heterozygotes required higher doses of sodium channel blocker AEDs for effective treatment than wild-type G/G homozygotes. Using the dominant model (A/G + A/A vs. G/G) there was a more significant correlation between the maximum doses of AEDs and the carriers of the A allele (A/G or A/A genotype). Our data are partially consistent with the findings of Tate et al. [20]. In this large pharmacogenetic study involving patients with various types of epilepsy, a significant association was reported in the intronic SNP rs3812718 and maximum tolerated dose of CBZ and PHT. A/A homozygote patients had to be prescribed higher doses of CBZ and PHT as compared to G/G homozygotes. Although only two additional studies by Hung et al. and Zhou et al. confirmed that carriers of the rs3812718 A allele required higher maintenance doses of CBZ [52, 53], most investigations failed to reproduce the association between the SCN1A IVS5-91 rs3812718 G>A genotypes and dosages of sodium channel blocking AEDs [22, 46]. SCN1A IVS5-91 rs3812718 G>A polymorphism was also associated with changes in cortical excitability. In a pharmacogenetic transcranial magnetic stimulation study, Menzler et al. suggested that the rs3812718 SNP is associated with altered cortical excitability after administration of CBZ. G/G homozygotes showed increased cortical inhibition (increase in cortical silent period – CSP) after administration of CBZ as compared to A/A homozygotes [54].
Our data showed that the SCN1A IVS5-91 rs3812718 G>A polymorphism might be associated with generalized epilepsy. The presence of A/G or A/A genotype could be a risk factor for generalized epilepsy. A recent study by Tang et al. suggested that SCN1A IVS5-91 rs3812718 G>A polymorphism is a risk factor of epilepsy with febrile seizures, especially in Caucasians [55]. In a case-control association study of polymorphisms in the voltage-gated sodium channel genes SCN1A, SCN2A, SCN3A, SCN1B, SCN2B and epilepsy, SNP rs3812718 in the SCN1A gene showed the strongest correlation with all types of epilepsy (with or without febrile seizures). A meta-analysis confirmed the correlation and demonstrated a protective effect of the G allele on epilepsy risk [56].
Inconsistencies observed between studies may be explained by various factors including genetic heterogeneity, sample size, population substructure, duration of treatment, variation in definition of treatment outcomes, type of used AEDs (sodium channel blockers or AEDs with multiple mechanisms of action), interaction of AEDs in polytherapy and their competition for binding to common targets, differences in etiology of epilepsy or differences in dosing regimens. Duration of treatment should not be less than 12 months according to the new definition of treatment outcome from the ILAE [25]. As the majority of previous studies included polytherapy regimens, the possibility of drug interactions cannot be ignored. In addition, etiology of epilepsy could affect the responsiveness to AEDs and the results of studies. Most reports were performed in patients with different types of epilepsy. Evidence showed that pharmacoresistance was more frequent in patients with symptomatic or cryptogenic epilepsy than in patients with idiopathic epilepsy [57]. Further studies are needed to clarify these issues.
The present study has some limitations. First, only a single SNP in the sodium channel SCN1A gene was screened in the study population without estimating the presence of other SNPs in the same gene or other neighboring genes in influencing the response to AED treatment [18, 51]. Interaction of SCN1A IVS5-91 rs3812718 G>A polymorphism with various loci in SCN1A and other genes among different populations needs to be taken into account. SNPs in the SCN1A gene such as rs1813502, rs1461195, rs1020853, rs6432860, rs10188577, rs11686142, rs7607543, rs1972445, and rs1461197, neighboring genes such as SCN2A (rs17183814, rs2304016) and SCN3A, and the more distant genes SCN1B, SCN7A, SCN8A, and SCN9A might be the best candidates to explain the differences of AED responsiveness in the population of Thrace. Moreover, the inherited component of the response to drugs is polygenic, and environmental factors cannot be ignored [13]. Secondly, the small sample size of the epilepsy population may have led to possible false positive results. Thirdly, in pharmacoresistant epilepsy, in addition to genetic factors, other factors could be taken in account, such as past treatment history, etiology of epilepsy, duration of seizures, and number of seizures prior to initiation of therapy [58]. Finally, in polytherapy we used sodium blockers and AEDs with different or multiple mechanisms of action.
In conclusion, this study suggests that the presence of the A allele of the SCN1A IVS5-91 rs3812718 G>A polymorphism is correlated with responsiveness to AEDs in monotherapy but not with resistance to drug polytherapy. These results provide evidence that the α-subunit of the SCN1A gene may play a role in the modified drug response in epilepsy, underlying the benefit of pharmacogenetic tests in clinical practice for the individualization of AED treatment.
Acknowledgments
The authors gratefully acknowledge all the participants in this study, as well as the medical staff of Xanthi General Hospital for their assistance in recruiting patients.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Monastiriotis C, Papanas N, Veletza S, Maltezos E. APOE gene polymorphisms and diabetic peripheral neuropathy. Arch Med Sci. 2012;8:583–8. doi: 10.5114/aoms.2012.30279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nowak A, Scaflik J, Gacek M, et al. BDNF and HSP gene polymorphisms and their influence on the progression of primary open-angle glaucoma in a Polish population. Arch Med Sci. 2013;10:1206–13. doi: 10.5114/aoms.2014.45089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alkharfy K, Aghamdi A, Bagulb K, et al. Distribution of selected gene polymorphisms of UGT1A1 in a Saudi population. Arch Med Sci. 2013;9:731–8. doi: 10.5114/aoms.2013.37012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lesiak A, Zakrzewski M, Przybyłowska K, et al. Atopic dermatitis patients carrying G allele in -1082 G/A IL-10 polymorphism are predisposed to higher serum concentration of IL-10. Arch Med Sci. 2014;10:1239–43. doi: 10.5114/aoms.2014.47833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poduri A, Lowenstein D. Epilepsy genetics – past, present, and future. Curr Opin Genet Dev. 2011;21:325–32. doi: 10.1016/j.gde.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldstein DB, Need AC, Singh R, Sisodiya SM. Potential genetic causes of heterogeneity of treatment effects. Am J Med. 2007;120:S21–5. doi: 10.1016/j.amjmed.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Chmielewska B, Lis K, Rejdak K, et al. Pattern of adverse events of antiepileptic drugs: results of the aESCAPE study in Poland. Arch Med Sci. 2013;9:858–64. doi: 10.5114/aoms.2013.38679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hille B. Ion channels of excitable membranes. 3rd ed. Sunderland, MA: Sinauer Associates Inc; 2001. [Google Scholar]
- 9.Yu FH, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Biol. 2003;4:207. doi: 10.1186/gb-2003-4-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George AL. Inherited disorders of voltage-gated sodium channels. J Clin Invest. 2005;115:1990–9. doi: 10.1172/JCI25505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plummer NW, Meisler MH. Evolution and diversity of mammalian sodium channel genes. Genomics. 1999;57:323–31. doi: 10.1006/geno.1998.5735. [DOI] [PubMed] [Google Scholar]
- 12.Kuo CC. A common anticonvulsant binding site for phenytoin, carbamazepine and lamotrigine in neuronal Na+ channels. Mol Pharmacol. 1998;54:712–21. [PubMed] [Google Scholar]
- 13.Löscher W, Klotz U, Zimprich F, Schmidt D. The clinical impact of pharmacogenetics on the treatment of epilepsy. Epilepsia. 2009;50:1–23. doi: 10.1111/j.1528-1167.2008.01716.x. [DOI] [PubMed] [Google Scholar]
- 14.Schlachter K, Gruber-Sedlmayr U, Stogmann E, et al. A splice site variant in the sodium channel gene SCN1A confers risk of febrile seizures. Neurology. 2009;72:974–8. doi: 10.1212/01.wnl.0000344401.02915.00. [DOI] [PubMed] [Google Scholar]
- 15.Escayg A, Heils A, MacDonald BT, et al. A novel SCN1A mutation associated with generalized epilepsy with febrile seizures plus: and prevalence of variants in patients with epilepsy. Am J Hum Genet. 2001;68:866–73. doi: 10.1086/319524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meisler MH, Kearney JA. Sodium channel mutations in epilepsy and other neurological disorders. J Clin Invest. 2005;115:2010–7. doi: 10.1172/JCI25466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lossin C. A catalog of SCN1A variants. Brain Dev. 2009;31:114–30. doi: 10.1016/j.braindev.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 18.Lakhan R, Kumari R, Misra U, et al. Differential role of sodium channels SCN1A and SCN2A gene polymorphisms with epilepsy and multiple drug resistance in the north Indian population. BJCP. 2009;68:214–20. doi: 10.1111/j.1365-2125.2009.03437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumari R, Lakhan R, Garg RK, et al. Pharmacogenomic association study on the role of drug metabolizing, drug transporters and drug target gene polymorphisms in drug resistant epilepsy in a North Indian population. IJHG. 2011;17:32–40. doi: 10.4103/0971-6866.80357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tate SK, Depondt C, Sisodiya SM, et al. Genetic predictors of the maximum doses patients receive during clinical use of the anti-epileptic drugs carbamazepine and phenytoin. Proc Natl Acad Sci USA. 2005;102:5507–12. doi: 10.1073/pnas.0407346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tate SK, Singh R, Hung CC, et al. A common polymorphism in the SCN1A gene associates with phenytoin serum levels at maintenance dose. Pharmacogenet Genom. 2006;16:721–6. doi: 10.1097/01.fpc.0000230114.41828.73. [DOI] [PubMed] [Google Scholar]
- 22.Zimprich F, Stogmann E, Bonelli S, et al. A functional polymorphism in the SCN1A gene is not associated with carbamazepine dosages in Austrian patients with epilepsy. Epilepsia. 2008;49:1108–9. doi: 10.1111/j.1528-1167.2008.01549_4.x. [DOI] [PubMed] [Google Scholar]
- 23.Abe T, Seo T, Ishitsu T, et al. Association between SCN1A polymorphism and carbamazepine resistant epilepsy. Br J Clin Pharmacol. 2008;66:304–7. doi: 10.1111/j.1365-2125.2008.03203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Commission on Classification and Terminology of the International League Against Epilepsy. Proposal for revised classification of epilepsies and epileptic syndromes. Epilepsia. 2008;30:389–99. doi: 10.1111/j.1528-1157.1989.tb05316.x. [DOI] [PubMed] [Google Scholar]
- 25.Kwan P, Arzimanoglou A, Berg A, et al. Definition of drug resistant epilepsy: consensus proposal by the ad hoc Task Force of the ILAE Commision on Therapeutic Strategies. Epilepsia. 2010;51:1069–77. doi: 10.1111/j.1528-1167.2009.02397.x. [DOI] [PubMed] [Google Scholar]
- 26.Remy S, Beck H. Mollecular and cellular mechanisms of pharmacoresistance in epilepsy. Brain. 2006;129:18–35. doi: 10.1093/brain/awh682. [DOI] [PubMed] [Google Scholar]
- 27.Ragsdale DS, Avoli M. Sodium channels as molecular targets for antiepileptic drugs. Brain Res Rev. 1998;26:16–28. doi: 10.1016/s0165-0173(97)00054-4. [DOI] [PubMed] [Google Scholar]
- 28.Heinzen EL, Yoon W, Tate SK, et al. Nova2 interacts with a cis-acting polymorphism to influence the proportions of drug-responsive splice variants of SCN1A. Am J Hum Genet. 2007;80:876–83. doi: 10.1086/516650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomson CH, Kahlig KM, George AL. SCN1A splice variants exhibit divergent sensitivity to commonly used antiepileptic drugs. Epilepsia. 2011;52:1000–9. doi: 10.1111/j.1528-1167.2011.03040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beck H. Plasticity of antiepileptic drug targets. Epilepsia. 2007;48(Suppl. 1):14–8. doi: 10.1111/j.1528-1167.2007.00994.x. [DOI] [PubMed] [Google Scholar]
- 31.Volk HA, Arabadzisc D, Fritschy JM, et al. Antiepileptic drug-resistant rats differ from drug-responsive rats in hippocampal neurodegeneration and GABA (A) receptor ligand binding in a model of temporal lobe epilepsy. Neurobiol Dis. 2006;21:633–6. doi: 10.1016/j.nbd.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Ferando I, Mody I. GABAA receptor modulation by neurosteroids in models of temporal lobe epilepsies. Epilepsia. 2012;53(Suppl 9):89–101. doi: 10.1111/epi.12038. [DOI] [PubMed] [Google Scholar]
- 33.Brooks-Kayal AR, Shumate MD, Jin H, et al. Selective changes in single cell GABA(A) receptor subunit expression and function in temporal lobe epilepsy. Nat Med. 1998;4:1166–72. doi: 10.1038/2661. [DOI] [PubMed] [Google Scholar]
- 34.Löscher W, Potschka H. Drug resistance in brain diseases and the role of drug efflux transporters. Nature Rev Neurosci. 2005;6:591–602. doi: 10.1038/nrn1728. [DOI] [PubMed] [Google Scholar]
- 35.Sisodiya SM. Mechanisms of antiepileptic drug resistance. Curr Opin Neurol. 2003;16:197–201. doi: 10.1097/01.wco.0000063771.81810.6c. [DOI] [PubMed] [Google Scholar]
- 36.Anderson GD, Shen DD. Where is the evidence that p-glycoprotein limits brain uptake of antiepileptic drug and contributes to drug resistance in epilepsy. Epilepsia. 2007;48:2372–4. doi: 10.1111/j.1528-1167.2007.01260_3.x. [DOI] [PubMed] [Google Scholar]
- 37.Dickens D, Yosuf SR, Abbott NJ, et al. A multi-system approach assessing the interaction of anticonvulsantes. PLoS One. 2013;8:e64854. doi: 10.1371/journal.pone.0064854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leschziner G, Jorgensen AL, Andrew T, et al. Clinical factors and ABCB1 polymorphisms in prediction of antiepileptic drug response: a prospective cohort study. Lancet Neurol. 2006;5:668–76. doi: 10.1016/S1474-4422(06)70500-2. [DOI] [PubMed] [Google Scholar]
- 39.Leschziner G, Andrew T, Leach JP, et al. Common ABCB1 polymorphisms are not associated with multidrug resistance in epilepsy using a gene-wide tagging approach. Pharmacogenet Genom. 2007;17:217–20. doi: 10.1097/01.fpc.0000230408.23146.b1. [DOI] [PubMed] [Google Scholar]
- 40.Balan S, Bharathan SP, Vellichiramal N, et al. Genetic association analysis of ATP binding cassette protein family reveals a novel association of ABCB1 genetic variants with epilepsy risk, but not with drug resistance. PLoS One. 2014;8:e64854. doi: 10.1371/journal.pone.0089253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogawski MA. The intrinsic severity hypothesis of pharmacoresistance to antiepileptic drugs. Epilepsia. 2013;54(Suppl 2):33–40. doi: 10.1111/epi.12182. [DOI] [PubMed] [Google Scholar]
- 42.Aronica E, Crino PB. Inflammation in epilepsy: clinical observations. Epilepsia. 2011;52(Suppl. 3):26–32. doi: 10.1111/j.1528-1167.2011.03033.x. [DOI] [PubMed] [Google Scholar]
- 43.Vezzani A, Aronica E, Mazarati A, et al. Epilepsy and brain inflammation. Exp Neurol. 2014;244:11–21. doi: 10.1016/j.expneurol.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 44.Maroso M, Balosso S, Ravizza T, et al. Toll-like receptor 4 and high-mobility group box-1 are involved in ictogenesis and can be targeted to reduce seizures. Nat Med. 2010;16:413–9. doi: 10.1038/nm.2127. [DOI] [PubMed] [Google Scholar]
- 45.Ma CL, Wu XY, Zheng J, et al. Association of SCN1A, SCN2A and ABCC2 gene polymorphisms with the response to antiepileptic drugs in Chinese Han patients with epilepsy. Pharmacogenomics. 2014;15:1323–36. doi: 10.2217/pgs.14.89. [DOI] [PubMed] [Google Scholar]
- 46.Manna I, Gambardella A, Bianchi A, et al. A functional polymorphism in the SCN1A gene does not influence the antiepileptic drug responsiveness in Italian patients with focal epilepsy. Epilepsia. 2011;52:40–4. doi: 10.1111/j.1528-1167.2011.03097.x. [DOI] [PubMed] [Google Scholar]
- 47.Sterjev Z, Kiteva G, Cvetkovska E, et al. Influence of the SCN1A IVS5N+5G>A polymorphism on therapy with carbamazepine for epilepsy. BJMG. 2012;15:19–24. doi: 10.2478/v10034-012-0003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haerian BS, Baum L, Tan HJ, et al. SCN1A IVS5N+5 polymorphism and response to sodium valproate: a multicenter study. Pharmacogenetics. 2012;13:1477–85. doi: 10.2217/pgs.12.127. [DOI] [PubMed] [Google Scholar]
- 49.Haerian BS, Baum L, Kwan P, et al. SCN1A, SCN2A and SCN3A gene polymorphisms and responsiveness to antiepileptic drugs: a multicenter cohort study and meta-analysis. Pharmacogenetics. 2013;14:1153–66. doi: 10.2217/pgs.13.104. [DOI] [PubMed] [Google Scholar]
- 50.Kumari R, Lakhan R, Kumar S, et al. SCN1A IVS5-91G>A polymorphism is associated with susceptibility to epilepsy but not with drug responsiveness. Biochimie. 2013;95:1350–3. doi: 10.1016/j.biochi.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 51.TSC Yip, O’Doherty C, Tan NCK, et al. SCN1A variations and response to multiple antiepileptic drugs. Pharmacogenomics. 2014;14:385–9. doi: 10.1038/tpj.2013.43. [DOI] [PubMed] [Google Scholar]
- 52.Hung CC, Chang WL, Ho JL, et al. Association of polymorphisms in EPHX1, UGT2B7, ABCB1, ABCC2, SCN1A and SCN2A genes with carbamazepine therapy optimization. Pharmacogenomics. 2012;13:159–69. doi: 10.2217/pgs.11.141. [DOI] [PubMed] [Google Scholar]
- 53.Zhou BT, Zhou QH, Yin JY, et al. Comprehensive analysis of the association of SCN1A gene polymorphisms with the retention rate of carbamazepine following monotherapy for new-onset focal seizures in the Chinese Han population. Clin Exp Pharmacol Physiol. 2012;39:379–84. doi: 10.1111/j.1440-1681.2012.05680.x. [DOI] [PubMed] [Google Scholar]
- 54.Menzler K, Hermsen A, Balkenhol K, et al. A common SCN1A splice- site polymorphism modifies the effect of carbamazepine on cortical excitability – a pharmacogenetic transcranial magnetic stimulation study. Epilepsia. 2014;55:362–3. doi: 10.1111/epi.12515. [DOI] [PubMed] [Google Scholar]
- 55.Tang L, Lu X, Tao Y, et al. SCN1A rs3812718 polymorphism and susceptibility to epilepsy with febrile seizures: a meta-analysis. Gene. 2014;533:26–31. doi: 10.1016/j.gene.2013.09.071. [DOI] [PubMed] [Google Scholar]
- 56.Baum L, Haerian BS, Ng HK, et al. Case-control association study of polymorphisms in the voltage-gated sodium channel genes SCN1A, SCN2A, SCN3A, SCN1B, and SCN2B and epilepsy. Hum Genet. 2014;133:651–9. doi: 10.1007/s00439-013-1405-1. [DOI] [PubMed] [Google Scholar]
- 57.Kwan P, Brodie MJ. Early identification of refractory epilepsy. N Engl J Med. 2000;342:314–9. doi: 10.1056/NEJM200002033420503. [DOI] [PubMed] [Google Scholar]
- 58.Sanchez MB, Herranz JL, Leno C, et al. Genetic factors associated with drug-resistance of epilepsy: relevance of stratification by patient age and aetiology of epilepsy. Seizure. 2010;19:93–101. doi: 10.1016/j.seizure.2009.12.004. [DOI] [PubMed] [Google Scholar]