Westphal et al. demonstrate a role of lysosomal trafficking regulator Lyst that couples the regulation of endolysosomal trafficking to inflammatory responses by the control of toll-like receptor–mediated endosomal TRIF signaling pathways.
Abstract
Subcellular compartmentalization of receptor signaling is an emerging principle in innate immunity. However, the functional integration of receptor signaling pathways into membrane trafficking routes and its physiological relevance for immune responses is still largely unclear. In this study, using Lyst-mutant beige mice, we show that lysosomal trafficking regulator Lyst links endolysosomal organization to the selective control of toll-like receptor 3 (TLR3)– and TLR4-mediated proinflammatory responses. Consequently, Lyst-mutant mice showed increased susceptibility to bacterial infection and were largely resistant to endotoxin-induced septic shock. Mechanistic analysis revealed that Lyst specifically controls TLR3- and TLR4-induced endosomal TRIF (TIR domain–containing adapter-inducing interferon β) signaling pathways. Loss of functional Lyst leads to dysregulated phagosomal maturation, resulting in a failure to form an activation-induced Rab7+ endosomal/phagosomal compartment. This specific Rab7+ compartment was further demonstrated to serve as a major site for active TRIF signaling events, thus linking phagosomal maturation to specific TLR signaling pathways. The immunoregulatory role of Lyst on TLR signaling pathways was confirmed in human cells by CRISPR/Cas9-mediated gene inactivation. As mutations in LYST cause human Chédiak-Higashi syndrome, a severe immunodeficiency, our findings also contribute to a better understanding of human disease mechanisms.
Introduction
Microbial detection receptors sample different subcellular compartments (Akira et al., 2006), and the sites of microbial detection by innate immune receptors are thought to be spatially separated from the sites of receptor signal transduction (Kagan, 2012). In this context, recent work has begun to appreciate that receptor trafficking routes can not only control timing and amplitude of signaling, but also confer signaling specificity by enabling the assembly of activated receptors with downstream signaling molecules that are recruited to specific subcellular compartments (Sorkin and von Zastrow, 2009). Thus, the cellular membrane trafficking machinery and endosomal routes appear to be intimately linked to receptor signal transduction. However, although this concept of a regulatory nexus between endosomal trafficking and signaling has mainly been developed from in vitro cell culture experiments, the impact of membrane trafficking networks on immune cell signaling under physiological in vivo conditions is still largely unclear.
Defects in endosomal trafficking have been linked to human disease and are frequently associated with impaired immune function (Huizing et al., 2008; Krzewski and Cullinane, 2013). Human Chédiak-Higashi syndrome (CHS) and its orthologous mouse disorder beige are characterized by defects in endolysosomal biogenesis that result in enlarged lysosome-related organelles caused by mutations in the lysosomal trafficking regulator (LYST) gene (Barbosa et al., 1996; Kaplan et al., 2008). This highly conserved gene encodes for a large cytosolic protein of poorly defined biochemical function. CHS is inherited as an autosomal recessive trait, and absence of functional LYST protein in individuals with CHS is associated with severe immune defects (Introne et al., 1999; Kaplan et al., 2008). CHS patients suffer from persistent and recurrent infections of the skin, respiratory tract, and mucous membranes. Other manifestations of CHS include partial albinism, mild bleeding, and progressive neurological dysfunction. Moreover, patients with CHS frequently develop a lethal accelerated phase of the disease, characterized by lymphoproliferative infiltration of major organs (Lozano et al., 2014). However, the mechanistic link between altered lysosomal organization in LYST-mutant cells and the immunological pathophysiology of CHS is still only poorly understood.
TLRs recognize conserved microbial components and are key regulators of the innate immune response to pathogens (Kawai and Akira, 2010). As such, TLRs are highly expressed by macrophages and dendritic cells, specialized phagocytic immune cells. Different TLRs recognize their ligands at distinct subcellular compartments. Whereas nucleic acid–specific TLRs (TLR3, TLR7, TLR8, and TLR9) are expressed within endolysosomal compartments, other TLRs (TLR1, TLR4, TLR5, and TLR6) are localized to the cell surface (Blasius and Beutler, 2010; Kawai and Akira, 2010). Localization and trafficking of TLRs appears not only to be important for ligand accessibility, but is also thought to be critical for their signaling function (Barton and Kagan, 2009; McGettrick and O’Neill, 2010). Upon ligand binding, TLRs initiate a series of signaling events that, dependent on the usage of distinct adaptor molecules, are classified in MyD88- and TRIF (TIR domain–containing adapter-inducing IFN-β)-dependent pathways to induce the production of proinflammatory cytokines and type I IFNs (Kawai and Akira, 2006; Moresco et al., 2011). Although TLR3 signals exclusively via the TRIF pathway, all other TLRs use the adaptor MyD88. The LPS receptor TLR4 is unique in that it assembles a MyD88-signaling complex at the plasma membrane to induce MAPKs and NF-κB activation and, upon receptor internalization to endosomal compartments, signals via a TRIF-dependent pathway to induce type I IFNs (Yamamoto et al., 2003; Kagan et al., 2008). Although much has been learned on the biochemical and cell biological characteristics of TLR signaling, the integration of TLR signaling pathways into the intracellular membrane trafficking network under inflammatory conditions and its physiological relevance for protective immunity still remain to be defined. The elucidation of these issues is also of clinical relevance, as dysregulated signaling via TLR family members is involved in the immunopathogenesis of infectious diseases, chronic inflammatory disorders, and autoimmunity (Moresco et al., 2011). Excessive inflammatory responses to TLR ligands can also induce life-threatening conditions, such as endotoxin-mediated septic shock. Thus, to provide immune protection against infection and also avoid harmful overreactions, TLR signals need to be tightly regulated.
Here, we explored the idea that TLR signaling and function can be controlled by the regulation of the endosomal trafficking network. Our data demonstrate that lysosomal trafficking regulator Lyst selectively controls TLR3- and TLR4-mediated proinflammatory responses. Mechanistic in vitro studies revealed that Lyst regulates phagosomal trafficking/maturation upon microbial encounter and specifically affects TRIF/IRF3 (IFN regulatory factor 3)-mediated endosomal TLR signaling. In vivo, loss of functional Lyst results in impaired immune responses against infection and prevents uncontrolled inflammation during septic shock. The identification of Lyst as a physiological regulator of TLR function reveals how the regulation of the cellular membrane trafficking network can affect specific immune receptor signaling pathways and inflammatory responses.
RESULTS
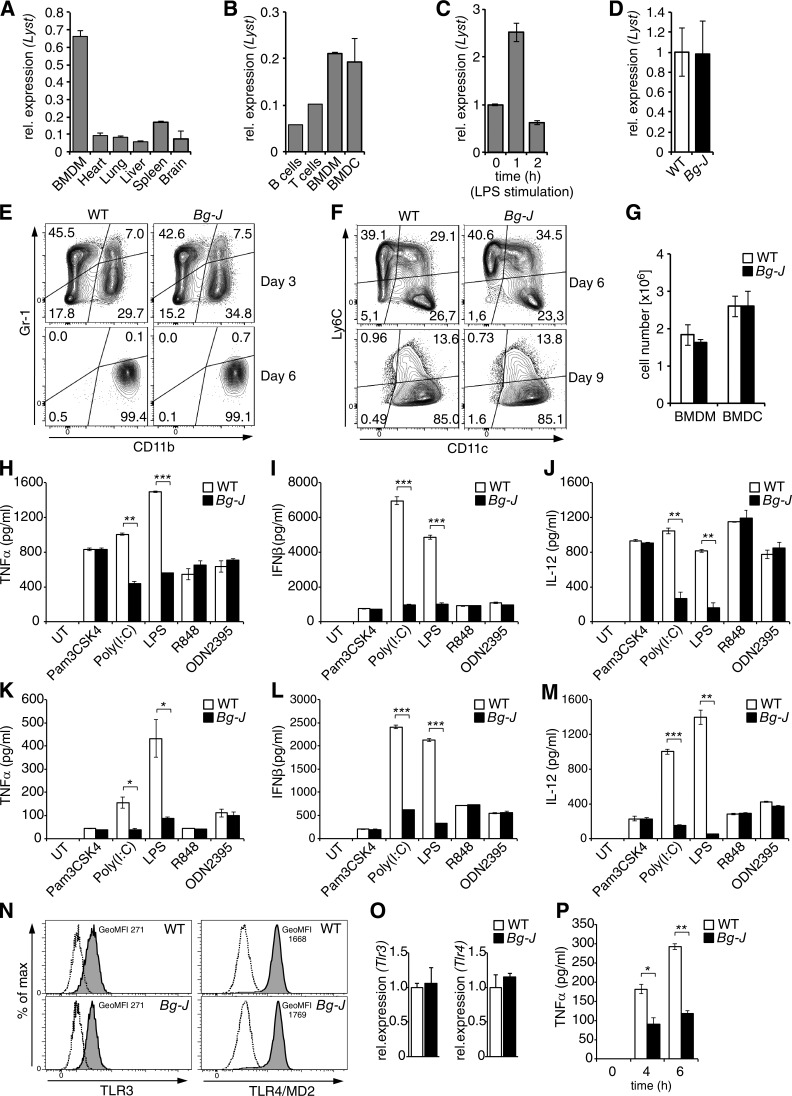
Lyst selectively controls TLR3- and TLR4-mediated production of proinflammatory cytokines
To study the biological relevance of endosomal trafficking for TLR signaling, we generated BMDMs and BMDCs from WT mice and Lyst-mutant beige (Bg-J) mice that carry a 3-bp deletion in exon 54 of the Lyst gene (Trantow et al., 2009). BMDMs and BMDCs express high levels of Lyst (Fig. 1, A–D), but development of these innate immune cells is not affected by the Bg-J mutation (Fig. 1, E–G). As activation of intracellular TLRs occurs within endolysosomal compartments (Blasius and Beutler, 2010), we initially studied the potential involvement of lysosomal trafficking regulator Lyst in the signaling of endosomal TLRs, such as TLR3, TLR7, TLR8, and TLR9. Interestingly, the analysis of TLR-induced cytokine production revealed that Lyst-mutant Bg-J BMDCs and BMDMs were selectively impaired in cellular cytokine responses to the endosomal receptor TLR3 and the predominantly cell surface–localized TLR4, whereas Lyst was dispensable for cytokine production via other cell-surface or intracellular TLRs (Fig. 1, H–M). Concentrations of IFN-β and proinflammatory cytokines such as TNF and IL-12 were substantially reduced in supernatants from cultures of Bg-J BMDCs and BMDMs compared with respective WT cells upon stimulation with polyinosinic-polycytidylic acid (Poly[I:C]; TLR3 agonist) or LPS (TLR4 agonist; Fig. 1, H–M). In contrast, secretion of IFN-β, TNF, and IL-12 was similar between WT and Bg-J cells in response to triggering with Pam3CSK4 (TLR1/TLR2 agonist), R848 (TLR7/TLR8 agonist), and ODN2395 (TLR9 agonist; Fig. 1, H–M). Reduced cytokine production by Lyst-mutant Bg-J cells upon TLR3 and TLR4 triggering was not secondary to reduced expression of these TLRs, as normal levels of TLR3 and TLR4 were detected in Bg-J cells (Fig. 1, N and O). Next, to test the functional significance of these findings, we examined cytokine responses upon infection of cells with live bacteria. BMDMs from Bg-J mice showed reduced release of TNF in response to in vitro infection with Salmonella enterica sv Typhimurium (S. Typhimurium; Fig. 1 P).
Figure 1.
Lyst is selectively involved in TLR3- and TLR4-mediated cytokine production. (A and B) Quantitative real-time PCR analysis of Lyst mRNA expression in different tissues (A) and different immune cell types (purified B and T lymphocytes, BMDMs, and BMDCs; B). (C) Lyst mRNA expression in BMDMs stimulated with 500 ng/ml LPS for the indicated times. The expression level of Lyst in unstimulated BMDMs was set as 1. (A–C) Error bars represent mean ± SD from duplicate samples. rel., relative. (D) Lyst mRNA expression levels in WT and Bg-J BMDMs. Data (mean ± SD) are pooled from three independent experiments. (E and F) BMDMs and BMDCs were differentiated by culturing bone marrow cells from WT and Bg-J mice in the presence of either M-CSF (BMDMs; E) or GM-CSF (BMDCs; F). The progress of cell differentiation was monitored at the indicated days of culture by flow cytometric analysis. (E) For BMDM differentiation cultures, CD11b and Gr-1 cell-surface expression is shown. (F) For BMDC differentiation cultures, CD11c and Ly6C cell-surface expression is shown. (G) Mean cell number in BMDM and BMDC cultures obtained from 106 bone marrow cells from WT and Bg-J mice. BMDMs: day 7; BMDCs: day 9. WT, n = 3; Bg-J, n = 4. Data are mean ± SD. (H–M) BMDCs (H–J) or BMDMs (K-M) from WT and Bg-J mice were stimulated for 6 h with the indicated TLR ligands. Release of TNF (H and K), IFN-β (I and L), and IL-12 (J and M) into culture supernatants was measured by ELISA. Graphs show means ± SD from two cultures derived from different mice. UT, untreated. (N) BMDMs from WT and Bg-J mice were analyzed by flow cytometry for expression of TLR3 (intracellular staining) and TLR4/MD2 (cell-surface staining). Shaded histograms show staining with specific antibodies. Dashed lines depict matching isotype controls. GeoMFI, geometric mean fluorescence intensity. (O) Quantitative real-time PCR analysis of Tlr3 and Tlr4 mRNA expression levels in WT and Bg-J BMDMs. Data are mean ± SD from duplicate samples. (P) BMDMs form WT and Bg-J mice were infected with S. Typhimurium at a multiplicity of infection of 5:1. Supernatants were collected at the indicated times after infection, and TNF levels were determined by ELISA. Data are mean ± SD from two cultures derived from different mice. All data are representative of at least three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test).
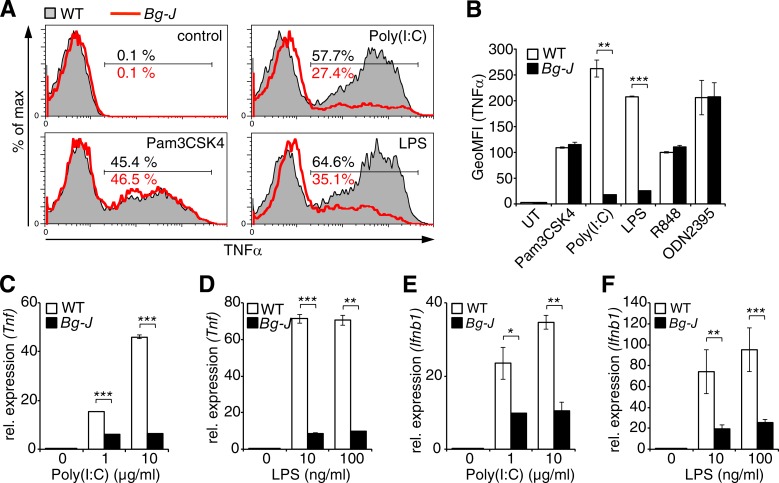
To dissect whether reduced cytokine levels in supernatants from Lyst-mutant cells were caused by defects in exocytic processes or caused by impaired de novo cytokine production, cells were treated with the Golgi inhibitor brefeldin A, which blocks protein secretion, and subsequently, total cellular cytokine production was examined by intracellular flow cytometry. The data revealed reduced intracellular TNF protein levels in Bg-J BMDCs in response to stimulation with TLR3 and TLR4 ligands (Poly[I:C] and LPS, respectively; Fig. 2, A and B). However, cytokine production was not generally impaired, as TNF production induced via other TLRs was not affected (Fig. 2, A and B). Thus, Lyst mutation specifically affects TLR3- and TLR4-induced cytokine production upstream of the secretion process. Furthermore, reduced cytokine production upon triggering of TLR3 and TLR4 was associated with reduced cytokine gene transcription, as reduced levels of Tnf and Ifnb1 mRNA were detected in Bg-J versus WT BMDCs stimulated with either TLR3 or TLR4 ligands (Fig. 2, C–F). Together, these data indicate regulatory effects of Lyst on TLR-induced cytokine gene induction that are restricted to TLR3- and TLR4-specific pathways but are not caused by defects on the actual secretion process.
Figure 2.
Lyst controls TLR3- and TLR4-induced inflammatory cytokine gene expression. (A and B) BMDCs from WT and Bg-J mice were stimulated for 6 h with the indicated TLR ligands in the presence of brefeldin A. Cells were subsequently subjected to intracellular staining for TNF and analyzed by flow cytometry. Representative flow cytometric histograms (WT, shaded; Bg-J, red lines; A) and the geometric mean fluorescence intensity (GeoMFI; B) ± SD from two cultures derived from different mice are shown. UT, untreated. (C–F) Quantitative real-time PCR analysis of Tnf (C and D) and Ifnb1 (E and F) mRNA levels in BMDCs from WT and Bg-J mice stimulated with the indicated concentrations of poly(I:C) (C and E) and LPS (D and F). Data are means ± SD from two cultures derived from different mice. rel., relative. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test). All data are representative of at least three independent experiments.
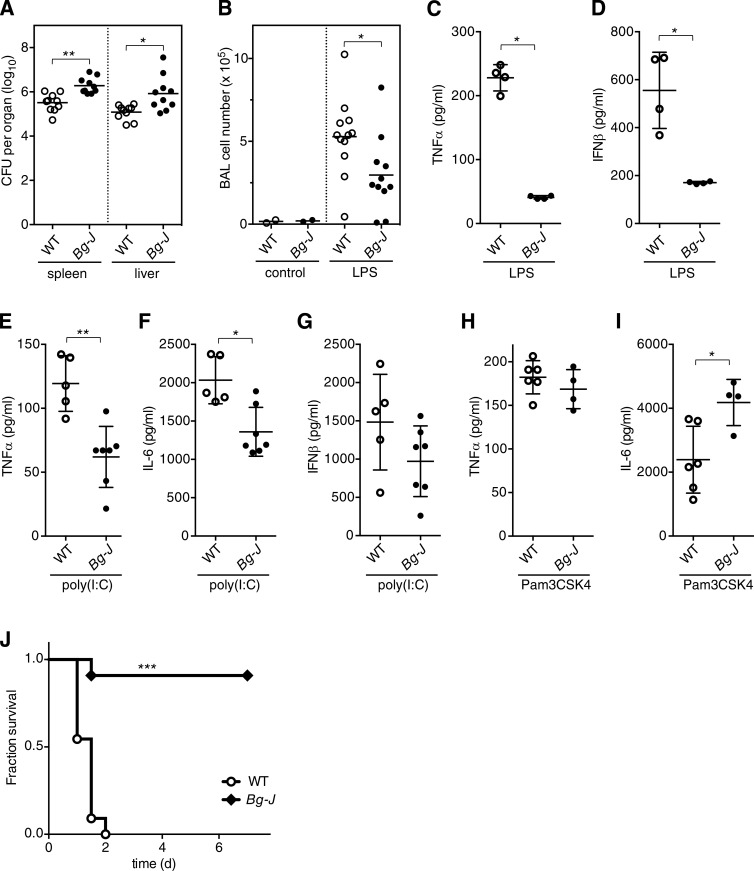
Lyst-mutant Bg-J mice show impaired inflammatory responses in vivo and are protected from endotoxin-induced septic shock
Defective pathogen-induced TNF production and the enhanced susceptibility of CHS patients to bacterial infection prompted us to assess the effect of Lyst on colonization/clearance of S. Typhimurium in vivo. Upon oral infection with S. Typhimurium, significantly higher bacterial loads were detected in spleen and liver of Bg-J mice compared with WT mice (Fig. 3 A), indicating that Lyst contributes to the host defense against Salmonella infection in vivo. Mechanistic dissection of the role of Lyst in the relative resistance to S. Typhimurium is however complicated by the complexity of the host–pathogen interaction in vivo, which involves not only TLR4-mediated defense mechanisms, but also other TLRs (Arpaia et al., 2011; Mathur et al., 2012). Thus, to evaluate the pathophysiological relevance of the Bg-J mutation in a more defined in vivo system, we next analyzed acute inflammatory reactions by directly challenging Bg-J and WT mice with specified TLR ligands. In a model of pulmonary inflammation induced via intranasal inoculation of LPS, Bg-J mice showed significantly decreased numbers of infiltrating inflammatory cells in the bronchoalveolar lavage (BAL; Fig. 3 B), indicating a reduced inflammatory response. However, immune cell recruitment was not generally impaired in Bg-J mice, as TNF-induced peritoneal infiltration of granulocytes is not affected by the Bg-J mutation (not depicted). Furthermore, upon systemic administration of LPS, serum levels of TNF and IFN-β were strongly reduced in Bg-J mice compared with WT controls (Fig. 3, C and D). Also, when injected i.p. with poly(I:C), Bg-J mice exhibited reduced serum levels of TNF and IL-6 (Fig. 3, E and F). Poly(I:C)-induced serum levels of IFN-β were not significantly affected by the Lyst mutation (Fig. 3 G), which has also been reported for TRIF−/− mice (Kato et al., 2006). Upon in vivo administration of the TLR1/2 ligand Pam3CSK4, serum levels of TNF and IL-6 were normal or increased, respectively (Fig. 3, H and I). Thus, in vivo cytokine production in Bg-J mice is specifically impaired in response to TLR3 and TLR4 pathways.
Figure 3.
Enhanced bacterial dissemination but protection from LPS-induced septic shock in Lyst-mutant Bg-J mice. (A) Dissemination of S. Typhimurium in orally infected WT and Bg-J mice was detected on day 4 after infection by determining CFU in spleen and liver. Data shown are combined results from three independent experiments. (B) Bronchoalveolar cell infiltration 24 h after intranasal inoculation of WT and Bg-J mice with LPS from S. Typhimurium or vehicle control. Data are pooled from three independent experiments. (C–G) WT and Bg-J mice were injected i.p. with LPS from S. Typhimurium (C and D) or with poly(I:C) (E–G). Blood was taken 1 h after LPS injection and 4 h after poly(I:C) injection. Serum concentrations of TNF (C and E), IFN-β (D and G), and IL-6 (F) were determined by ELISA. (H and I) WT and Bg-J mice were injected i.p. with Pam3CSK4. Blood was taken 2 h after injection, and serum concentrations of TNF (H) and IL-6 (I) were determined by ELISA. (C–I) Data are representative of at least two independent experiments. (A–I) Circles indicate values for individual animals (n ≥ 4), and bars indicate means. *, P < 0.05; **, P < 0.005 (Mann-Whitney U test). (J) Kaplan-Meier analysis of the survival of WT and Bg-J mice injected i.p. with LPS from S. Typhimurium. Mice were monitored over 7 d (n = 11 per genotype). ***, P < 0.0001 (log-rank test). Data are pooled from three independent experiments.
As massive release of proinflammatory cytokines, including TNF, contributes to the toxic effects of LPS during sepsis, we analyzed susceptibility to endotoxin-mediated lethal shock. Upon challenge with a lethal dose of LPS, WT mice died within 2 d, whereas Bg-J mice were largely protected from LPS-induced lethality (Fig. 3 J). These data are consistent with substantially reduced inflammatory cytokine production in Bg-J mice upon in vivo LPS administration (Fig. 3, C and D). Thus, Lyst is specifically involved in poly(I:C)- and LPS-induced proinflammatory responses and is a physiological mediator of excessive inflammation during endotoxin-mediated septic shock.
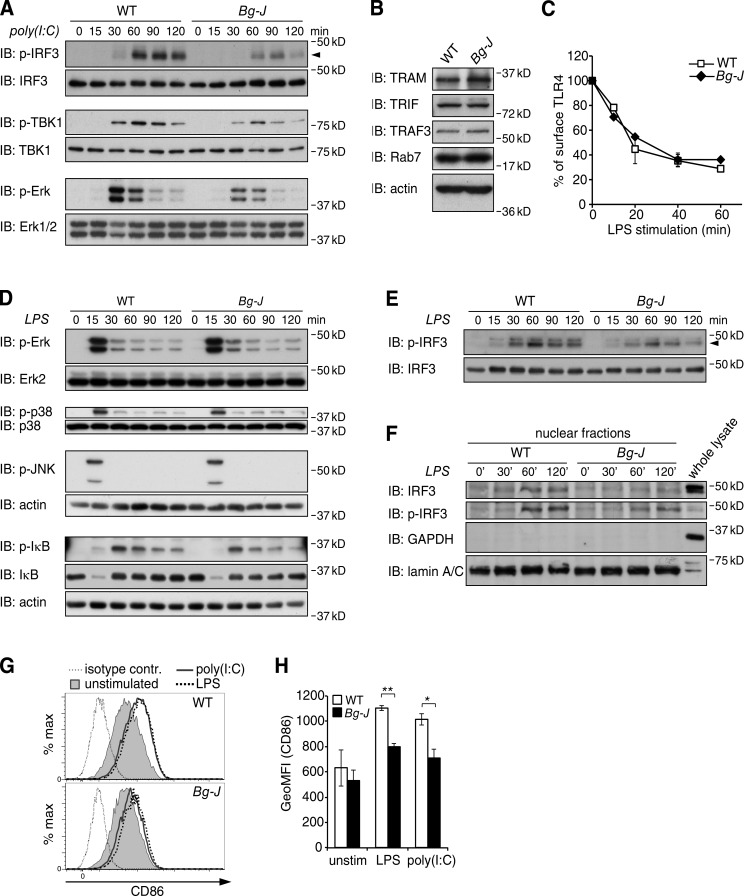
Lyst is involved in the regulation of endosomal TRIF signaling
To define the mechanism underlying defective reactivity to poly(I:C) and LPS in Lyst-mutant Bg-J mice, we analyzed the effects of the Bg-J mutation on TLR signaling. The selective defect in TLR3- and TLR4-induced cytokine production suggests that Lyst affects a signaling pathway common to both of these TLR systems but not shared with other TLRs. TLR3 and TLR4 are unique among TLRs in their ability to signal via a specialized endosomal pathway involving the adaptor protein TRIF (Yamamoto et al., 2003). Key components of the TRIF pathway, such as TRIF, TRAF3, IRF3, and TBK1, were expressed at comparable levels between WT and Bg-J BMDMs (Fig. 4, A and B). Importantly, however, poly(I:C)-induced phosphorylation of both TBK1 and the transcription factor IRF3, two critical TRIF-mediated signaling events leading to the induction of type I IFNs (Doyle et al., 2002; Fitzgerald et al., 2003; Sato et al., 2003; Kagan et al., 2008), were substantially impaired in Bg-J versus WT BMDMs (Fig. 4 A). Consistent with defective TRIF signaling (Hoebe et al., 2003; Yamamoto et al., 2003), poly(I:C)-induced Erk activation, which is detected within 30–60 min, was also reduced in Bg-J cells (Fig. 4 A).
Figure 4.
Defective TLR3- and TLR4-induced activation of IRF3 in Lyst-mutant Bg-J cells. (A) WT and Bg-J BMDMs were stimulated with 10 µg/ml poly(I:C) for the indicated times. Whole cellular lysates were immunoblotted (IB) for phosphorylated IRF3 (p-IRF3), p-TBK1, or p-Erk1/2. Reprobing with anti-IRF3, TBK1, and Erk1/2, respectively, served as the loading control. The arrowhead indicates p-IRF3. (B) Whole cellular lysates from WT and Bg-J BMDMs were immunoblotted with antibodies against TRAM, TRIF, TRAF3, and Rab7. Immunoblotting with actin-specific antibodies served as a loading control. (C) WT and Bg-J BMDMs were stimulated with 100 ng/ml LPS for the indicated times. TLR4 endocytosis was measured by flow cytometric assessment of the loss of TLR4 cell-surface expression. Data are mean ± SD from duplicate cultures. (D–F) BMDMs from WT and Bg-J mice were stimulated with 100 ng/ml LPS for the indicated times. (D) Whole cellular lysates were immunoblotted for p-Erk1/2, p-p38, p-JNK, p-IκBα, and IκBα. Blots were reprobed for total Erk2, p38, and actin to verify equal protein loading. (E) Immunoblot analysis as in D using antibodies specific for p-IRF3. p-IRF3 is indicated by an arrowhead. Reprobing with anti-IRF3 served as the loading control. (F) Nuclear extracts were prepared and analyzed by immunoblotting for p-IRF3 and total IRF3. Blots were reprobed for GAPDH (cytoplasmic marker) and lamin A/C (nuclear marker). Whole cellular lysate from WT BMDMs served as a control. (G and H) WT and Bg-J BMDMs were treated with 10 µg/ml poly(I:C) or 100 ng/ml LPS or left unstimulated (unstim.). After 4 h of stimulation, cell-surface expression of CD86 was determined by flow cytometry. (G) Representative flow cytometric histograms. Thin dotted line: isotype control; shaded: CD86 unstimulated; continuous line: CD86 poly(I:C) stimulated; thick dotted line: CD86 LPS stimulated. contr., control. (H) Geometric mean fluorescence intensity (GeoMFI) of CD86 staining ± SD of n = 2 independent cultures. *, P < 0.05; **, P < 0.01 (Student’s t test). All data are representative of at least three independent experiments.
As TLR4-mediated TRIF signaling requires dynamin-dependent endocytosis of TLR4 (Husebye et al., 2006; Kagan et al., 2008), we next analyzed LPS-induced TLR4 endocytosis. The data revealed comparable LPS-induced TLR4 endocytosis rates between WT and Bg-J BMDMs (Fig. 4 C). In contrast to poly(I:C) stimulation, LPS-induced phosphorylation of the MAPKs Erk, p38, and JNK, which peaked early (∼15 min) upon LPS triggering and are largely TRIF independent, was similar between WT and Bg-J cells (Fig. 4 D). TLR4-induced phosphorylation and degradation of IκBα was also not affected by the Lyst mutation (Fig. 4 D). However, TRIF-dependent IRF3 phosphorylation in response to LPS stimulation was reduced and less sustained in Bg-J BMDMs (Fig. 4 E). Also, LPS-induced nuclear translocation of IRF3, an indicator of IRF3 activation, was impaired in Bg-J BMDMs (Fig. 4 F).
Together, our data suggest that Lyst is specifically involved in TLR3- and TLR4-induced production of proinflammatory cytokines by regulating the endosomal TRIF/IRF3 signaling pathway. Further support for an involvement of the TRIF pathway came from the observation that TRIF-controlled up-regulation of CD86 (Yamamoto et al., 2003) upon stimulation with LPS and poly(I:C) was clearly reduced in Lyst-mutant Bg-J cells (Fig. 4, G and H).
Lyst controls endolysosomal trafficking and phagosomal maturation
Given our finding that Lyst regulates endosomal TRIF signaling and the characteristic enlargement of lysosome-related organelles in cells from CHS patients (Kaplan et al., 2008), we hypothesized that endolysosomal trafficking of activated TLR4, which is critical for TRIF signaling, might be dysregulated in Lyst-mutant cells. Thus, we next investigated the effects of the Bg-J mutation on the organization and dynamics of the endosomal/lysosomal system. Immunofluorescence staining for endogenous EEA1 and Rab5 (early endosomal structures), Rab4 and Rab11 (early sorting and recycling endosomes), and Rab7 (late endosomes) showed similar structures in WT and Bg-J BMDCs (Fig. 5 A). However, consistent with enlarged lysosome-related organelles in CHS and beige cells (Burkhardt et al., 1993; Kaplan et al., 2008), staining for the lysosomal marker Lamp1 or acidotropic staining with LysoTracker and Lysosensor indicated abnormally large lysosome-related, acidic vesicles in Lyst-mutant Bg-J BMDCs (Fig. 5, A and B). In addition, Rab9+ vesicles (late endosome to trans-Golgi network) were modestly enlarged in Bg-J BMDCs (Fig. 5 A). Thus, in addition to confirming the well-known defects in lysosome-related structures in Bg-J cells, the data also indicate that the endosomal system is not generally affected by the Lyst mutation, and particularly, structural organization of early endosomal compartments appeared to be normal. Upon phagocytic uptake by innate immune cells, bacteria are progressively transported via an endosomal trafficking route to lysosomal compartments for final degradation. Because of the observed lysosomal defects in Bg-J cells, we next investigated the role of Lyst on phagosomal maturation. Overall, phagocytic capacity was normal in Bg-J cells, as measured by the uptake of fluorescence-labeled Escherichia coli particles (Fig. 5 C). As phagocytosis is associated with a progressive decrease in phagosomal pH, we monitored phagocytosis of E. coli particles conjugated with pHrodo, a pH-sensitive reporter dye. Comparison of pHrodo fluorescence in WT and Lyst-mutant Bg-J cells indicated normal phagosomal acidification in Bg-J cells (Fig. 5, D–F). During maturation, phagosomes sequentially acquire different sets of proteins. An essential step in this maturation process is the loss of early endosomal markers, such as Rab5 and its effector EEA1, and recruitment of Rab7 (Rink et al., 2005). Thus, to gain further insights into the role of Lyst in phagosomal trafficking, we compared the acquisition of phagosomal maturation markers in WT and Bg-J cells by exposing BMDCs to LPS-coated beads (LPS beads) as surrogates for bacteria. 15 min upon phagocytosis of LPS beads, phagosomes of both WT and Bg-J BMDCs were positive for the early endosomal marker EEA1 (Fig. 6 A). However, the recruitment of Rab7, which was observed on phagosomes of WT BMDCs at later stages of the maturation process (30–60 min), was largely absent in Bg-J cells (Fig. 6, B and C).
Figure 5.
Enlarged lysosome-related compartments but normal phagocytic uptake in Lyst-mutant cells. (A, left) WT and Bg-J BMDCs were stained for the indicated endosomal/lysosomal marker proteins. Localization of endogenous proteins was analyzed by confocal microscopy. Bars, 5 µm. (Right) The mean size of the indicated particles was quantified by using ImageJ software (1.50c). In the graphs, each dot represents mean particle size in one cell. Lines show total mean ± SEM. *, P < 0.05 (Mann-Whitney U test). (B) BMDCs from WT and Bg-J mice were preincubated with LysoTracker red and Lysosensor green. Fluorescence images were generated from live cells by confocal microscopy. Bars, 10 µm. DIC, differential interference contrast. (C) Phagocytic uptake of fluorescence-labeled E. coli by WT and Bg-J BMDCs was assessed by flow cytometric analysis at the indicated times. WT, shaded; Bg-J, red lines. (D) WT and Bg-J BMDMs were exposed to E. coli particles labeled with the pH-sensitive dye pHrodo. Phagosomal acidification was assessed by fluorescence microscopy after 40 min of incubation. pH sensitivity was controlled by analysis of pHrodo-labeled E. coli particles in buffers with different pH (bottom). Bars, 10 µm. (E and F) WT and Bg-J BMDMs were exposed to E. coli particles double labeled with the pH-sensitive dye pHrodo and with eFluor 670. Particle uptake and phagosomal acidification were assessed by flow cytometric analysis. (E) Flow cytometric profiles at 0 min (top) and 90 min (bottom) of incubation showing fluorescence staining for pHrodo versus eFluor 670. (F) Flow cytometric quantification of pHrodo-positive cells at the indicated time points. As specificity control, cells were incubated with 30 mM NH4Cl to quench lysosomal acidification, or cells were kept on ice. Data are mean ± SD from two independent cultures. All data are representative of at least two independent experiments.
Figure 6.
Dysregulated phagosomal maturation in Lyst-mutant Bg-J cells. (A and B) WT and Bg-J BMDCs were exposed to LPS beads (3–5 µm diameter). After 15 (A) and 60 (B) min of incubation, cells were fixed, permeabilized, and stained for EEA1 and Rab7. Nuclei were counterstained with DAPI. Arrowheads indicate beads. Insets show enlargements of bead-containing phagosomes. Bars, 5 µm. (Right) Line profiles from confocal microscopy images are shown. The fluorescence signal intensity (FL intensity) of EEA1 (red) and Rab7 (green) was quantified along the yellow line drawn across internalized LPS beads (shown in the overlay). Percentages of bead-containing phagosomes in A that were positive for EEA1 at 15 min: WT, 47.9% ± 27.4 (n = 18); Bg-J, 53.3% ± 13.9 (n = 6; P = 0.316; Mann-Whitney U test). Percentages of bead-containing phagosomes in B that were positive for Rab7 at 60 min: WT, 49.9% ± 27.9 (n = 21); Bg-J, 0% ± 0 (n = 12; P < 0.0001; Mann-Whitney U test). a.u., arbitrary units; DIC, differential interference contrast; rel., relative. (C) WT and Bg-J BMDMs were exposed to LPS beads (2 µm diameter). After 30 min of incubation, cells were fixed, permeabilized, and stained for EEA1 and Rab7. Nuclei were counterstained with DAPI. Bars, 5 µm. BF, bright field. (Right) Graphs show quantification of percentages of bead-containing phagosomes in WT and Bg-J cells that were positive for EEA1 (top) or positive for Rab7 (bottom). Number of evaluated bead-containing phagosomes: WT, n = 70; Bg-J, n = 127. **, P < 0.01 (Mann-Whitney U test). Error bars represent SD. All data are representative for at least three independent experiments.
These data reveal a specific defect in phagosomal maturation in Lyst-mutant Bg-J cells by indicating that Lyst is involved in the conversion from EEA1+ early phagosomal compartments to Rab7+ late phagosomes. Interestingly, despite defective Rab7 acquisition, phagosomes in Bg-J cells are not arrested in an early phagosomal state, as indicated by the loss of EEA1 at 60 min (Fig. 6 B) and progressive phagosomal acidification (Fig. 5 F). Thus, dysregulated endosomal/phagosomal maturation in Lyst-mutant Bg-J cells rather seems to result in missorting of endosomal vesicles and aberrant fusion with lysosomal compartments.
Rab7-positive endosomal compartments serve as a platform for TRIF-mediated signaling pathways
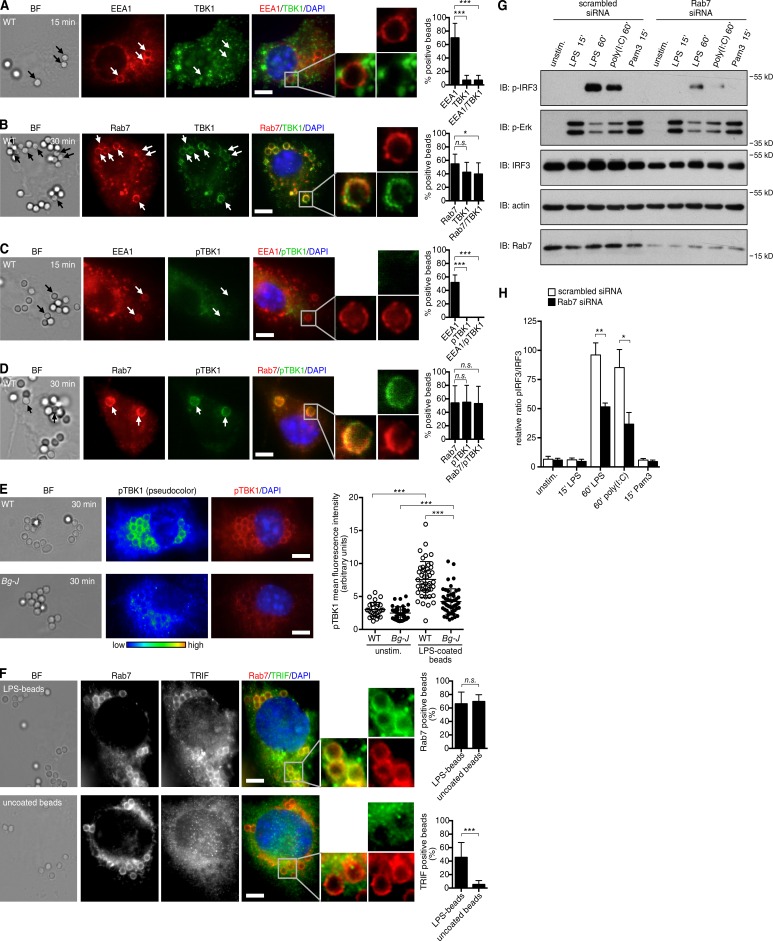
The correlation between defective Rab7 recruitment and impaired TRIF signaling in Bg-J cells suggested that Rab7+ subcellular compartments may provide a critical nidus for TRIF signaling. We addressed this hypothesis by performing a microscopy-based analysis of TRIF-mediated signaling at endosomal/phagosomal compartments in primary macrophages (BMDMs). To monitor the activation of the TRIF/IRF3 pathway, we investigated recruitment and activation of TBK1. This kinase is a key mediator of TRIF signaling. TBK1 interacts with TRIF and induces activation of its downstream target IRF3, finally leading to IFN-β induction (Fitzgerald et al., 2003). To achieve microscopic visualization of TBK1 recruitment and phosphorylation at endogenous protein levels, BMDMs were stimulated with relatively high numbers of LPS beads. Within 15 min of stimulation with LPS beads, EEA1 was found to localize to bead-containing phagosomes. At these early time points, no recruitment of TBK1 to bead-containing phagosomes was observed, and TBK1 was not significantly phosphorylated (Fig. 7, A and C). However, at later times of stimulation (30–60 min), when Rab7 recruitment to bead-containing phagosomes was readily detected, these Rab7+ phagosomal compartments also showed a clear recruitment of TBK1 (Fig. 7 B). Importantly, TBK1 at these Rab7+ sites was strongly phosphorylated at serine 172 (Fig. 7 D), indicative of TBK1 activation (McCoy et al., 2008). Lyst-mutant Bg-J cells, which exhibit defective Rab7 recruitment (Fig. 6, B and C), showed significantly impaired phosphorylation of TBK1 upon stimulation with LPS beads (Fig. 7 E). Moreover, the characteristic accumulation of phospho-TBK1 staining on LPS bead–containing phagosomes that was observed in WT cells was largely defective in Bg-J BMDMs (Fig. 7 E). Together, these data indicate that upon LPS stimulation, Rab7+ endosomal/phagosomal compartments recruit TBK1 and serve as major sites for active TBK1 signaling.
Figure 7.
Rab7+ endosomal compartments serve as a platform for TRIF signaling. (A–D) WT BMDMs were exposed to LPS beads (2 µm diameter). After 15 (A and C) and 30 (B and D) min of incubation, cells were fixed, permeabilized, and stained for EEA1 and TBK1 (A), Rab7 and TBK1 (B), EEA1 and phosphorylated TBK1 (pTBK1; C), and Rab7 and phosphorylated TBK1 (D). Nuclei were counterstained with DAPI. Arrows indicate EEA1+ or Rab7+ beads. Enlargements show EEA1+ or Rab7+ phagosomes. Bars, 5 µm. BF, bright field. (Right) Graphs show quantification of percentages of bead-containing phagosomes that were positive for the indicated markers. Number of evaluated bead-containing phagosomes: (A) n = 66; (B) n = 103; (C) n = 37; (D) n = 84. (E) WT and Bg-J BMDMs were exposed to LPS beads. After 30 min of incubation, cells were fixed and stained for phosphorylated TBK1. Nuclei were counterstained with DAPI. Phosphorylated TBK1 staining is shown in pseudocolor to highlight signal intensity. Bars, 5 µm. (Right) The graph on the right shows cellular pTBK1 staining intensity (mean fluorescence intensity in arbitrary units) in WT and Bg-J cells stimulated for 30 min with LPS beads and in unstimulated (unstim.) control cells. Each circle corresponds to an individual cell (n ≥ 65). (F) THP-1 cells were differentiated into macrophage-like cells and were exposed to LPS beads or uncoated control beads. After 60 min of incubation, cells were fixed, permeabilized, and stained for Rab7 and TRIF. Nuclei were counterstained with DAPI. Enlargements show bead-containing phagosomes. Bars, 5 µm. (Right) The graphs show quantification of percentages of bead-containing phagosomes (LPS beads and uncoated control beads, respectively) that were positive for Rab7 (top) or TRIF (bottom). Greater than 150 bead-containing phagosomes were evaluated for each condition. (A–F) Mann-Whitney U test was used. Error bars represent SD. (G and H) BMDMs transfected with siRNA against Rab7 or scrambled control siRNA were stimulated for the indicated times with 100 ng/ml LPS, 10 µg/ml poly(I:C), or 100 ng/ml Pam3CSK4 (Pam3). Whole cellular lysates were analyzed by immunoblotting (IB) for phosphorylated IRF3, phosphorylated Erk1/2, IRF3, actin, and Rab7. (H) Densitometric analysis of immunoblots showing the ratio of phosphorylated IRF3 band intensities to total IRF3 (arbitrary units; n ≥ 3). Data are mean ± SEM. Student’s t test was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001. All data are representative for at least three independent experiments.
As TBK1 has been reported to form a complex with TRIF to mediate IRF3 activation (Fitzgerald et al., 2003; Sato et al., 2003), we analyzed whether TRIF is also present on Rab7+ phagosomes. Immunofluorescence microscopy revealed TRIF protein localization on Rab7+ phagosomal compartments upon stimulation with LPS beads (Fig. 7 F, top). Interestingly, TRIF localization to Rab7+ phagosomes was not observed with uncoated control beads (Fig. 7 F, bottom), suggesting that TRIF recruitment to bead-containing phagosomes specifically depends on LPS-mediated signaling events. In summary, we conclude that Rab7+ compartments provide an important platform for TRIF-mediated signaling pathways.
We next investigated whether Rab7 is functionally involved in the regulation of TRIF signaling by applying RNA interference. Treatment of primary BMDMs with Rab7-specific siRNAs resulted in significantly reduced Rab7 protein expression levels (Fig. 7 G). Importantly, LPS- and poly(I:C)-induced IRF3 phosphorylation was significantly reduced in Rab7-siRNA–treated BMDMs compared with control treated cells (Fig. 7, G and H), indicating that Rab7 is a regulator of TRIF-mediated IRF3 signaling. Cell signaling was not generally affected by Rab7 knockdown, as TRIF-independent signaling events, such as LPS- and Pam3CSK4-induced Erk phosphorylation, were normal in Rab7-siRNA–treated cells (Fig. 7 G).
Together, our data provide a functional link between Rab7 and TRIF/IRF3 signaling and suggest that the active TRIF signalosome is formed or stabilized at Rab7+ phagosomal compartments.
The immunoregulatory role of Lyst on TLR3- and TLR4-mediated pathways is conserved in the human system
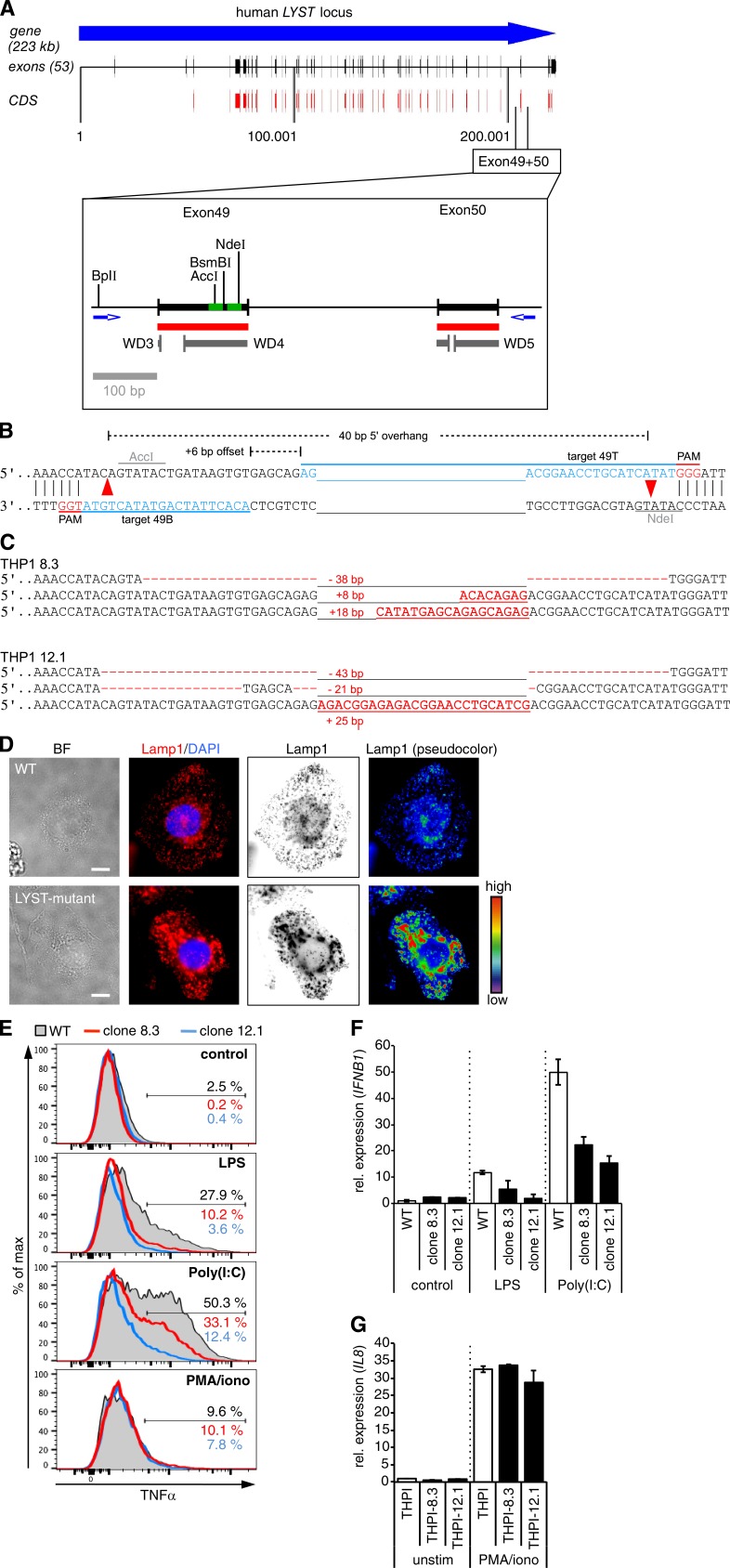
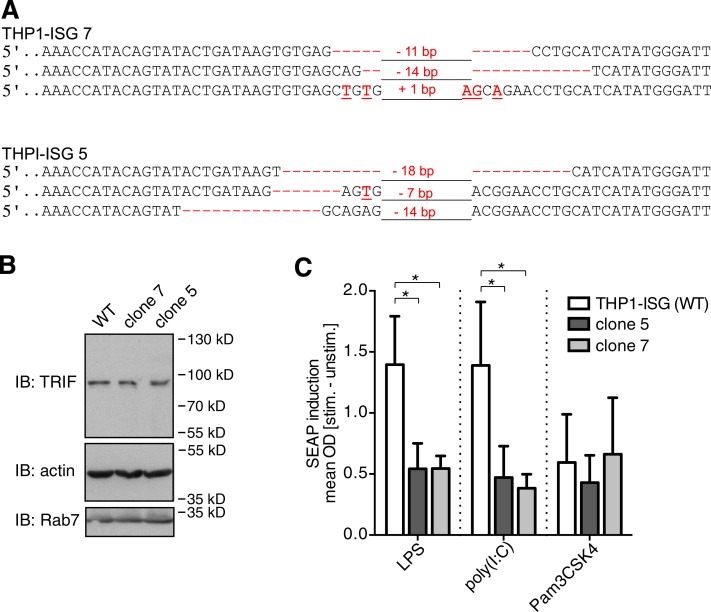
Enlarged lysosome-related organelles are a classical diagnostic feature of CHS and are associated with severe immunological defects in these patients (Kaplan et al., 2008). Thus, we next explored whether the immunoregulatory function of Lyst on selected TLR pathways is conserved in human cells. To this end, we generated LYST-mutant human monocytic THP-1 cells by using the recently developed CRISPR/Cas9 genome editing technology (Doudna and Charpentier, 2014; Hsu et al., 2014). In our mutational approach, we specifically targeted the WD40 domain of human LYST (Fig. 8 A), as mutations within this region have been described in patients with CHS (Lozano et al., 2014) and the beige mutation in Lyst-mutant mice (Lystbg-J) also affects the WD40 region (Trantow et al., 2009). To minimize potential off-target effects, we applied a double-nicking strategy (Mali et al., 2013; Ran et al., 2013a), in which a pair of single-guide RNA (sgRNA)–Cas9nickase complexes was used to generate two adjacent DNA single-strand breaks (nicks) on opposite DNA strands within exon 49 of human LYST (Fig. 8 B). Using this strategy, we obtained THP-1 cell clones with indel mutations within the WD40 domain in all three alleles of LYST (Fig. 8 C; the LYST locus [1q42.1–q42.2] is present in three copies in THP-1 cells, reflecting the special cytogenetic features of this cell line; Adati et al., 2009). For enhanced sensitivity to TLR ligands, monocytic THP-1 cells were differentiated into CD14-expressing macrophage-like cells (Park et al., 2007). Differentiated LYST-mutant THP-1 cells displayed enlarged lysosome-related structures (Fig. 8 D), a cellular phenotype reminiscent of that in cells from CHS patients. Flow cytometric analysis of intracellular cytokine production revealed that LYST-mutant THP-1–derived human macrophages were severely compromised in their ability to produce TNF in response to treatment with either LPS or poly(I:C) (Fig. 8 E). Consistent with a role of LYST in the control of intracellular TRIF signaling pathways, LYST-mutant THP-1–derived CD14+ macrophages also produced reduced levels of IFNB1 mRNA in response to stimulation with TLR4 ligand LPS or TLR3 ligand poly(I:C) (Fig. 8 F). However, cytokine responses were not generally impaired in LYST-mutant THP-1 clones, as these cells could still be activated by other stimuli (Fig. 8, E and G). To further determine the effect of LYST deficiency on signaling events downstream of the TRIF/IRF pathway, we also generated LYST-mutant THP-1 cells that carry an IRF-inducible reporter construct for monitoring IRF activity (Fig. 9 A). LYST mutation had no effect on TRIF expression (Fig. 9 B). However, in response to stimulation with LPS and poly(I:C), the activation of IFN-stimulated genes (ISGs) was significantly reduced in LYST-mutant THP-1 IRF reporter cells (Fig. 9 C). Pam3CSK4-induced responses were not affected by LYST mutation, indicating that cellular activation is not generally impaired in these cells (Fig. 9 C).
Figure 8.
CRISPR/Cas9-mediated genome editing reveals impaired TLR3- and TLR4-induced cytokine production in LYST-mutant human cells. (A) Schematic representation of the human LYST locus. The enlargement shows the genomic region from exon 49 to 50 of human LYST. Red, coding sequence; gray, predicted WD40 repeats; green, target sequences for Cas9n-mediated double nicking in exon 49. CDS, coding sequence. (B) Strategy for Cas9n-mediated double nicking (red arrowheads) in exon 49 of human LYST. Target sequences (blue), protospacer-adjacent motifs (PAM; red), and the distance of Cas9n-induced nicks are indicated. (C) Sequence analysis shows indel mutations for all three alleles of LYST in THP-1 clone 8.3 and clone 12.1. Red dashes indicate deleted bases, red letters show inserted bases, and numerals indicate total number of deleted/inserted base pairs. (D) WT and LYST-mutant human THP-1 cells were differentiated into macrophage-like cells. After fixation and permeabilization, cells were stained with anti–human Lamp1 (Sigma-Aldrich) and DAPI. Representative individual and overlay fluorescence images were obtained on a microscope (IX81; Olympus). Subsequent image analysis and pseudocolor processing was performed by using iVision software (BioVision Technologies). Bars, 10 µm. BF, bright field. (E) Differentiated CD14+ WT and LYST-mutant THP-1 cells were stimulated for 4 h with the indicated reagents in the presence of brefeldin A. Flow cytometric analysis shows intracellular TNF expression on CD14+ cells. Shaded, WT; red lines, LYST-mutant clone 8.3; blue lines, LYST-mutant clone 12.1. (F) Differentiated CD14+ WT and LYST-mutant THP-1 cells (clones 8.3 and 12.1) were stimulated for 2 h with 20 ng/ml LPS, 10 µg/ml poly(I:C), or left untreated. IFNB1 mRNA levels were determined by quantitative real-time PCR of duplicate measurements. The expression level of nonstimulated WT THP-1 cells was set as 1. (G) Differentiated CD14+ WT and LYST-mutant THP-1 cells (clones 8.3 and 12.1) were stimulated for 2 h with PMA/ionomycin or left untreated. IL8 mRNA levels were determined by quantitative real-time PCR of duplicate measurements. (F and G) Data are mean ± SD. All data are representative for at least two independent experiments. iono, ionomycin; rel., relative.
Figure 9.
Impaired TLR3- and TLR4-mediated induction of IFN-responsive genes in LYST-mutant THP-1 cells. (A) Sequence analysis showing indel mutations for all three alleles of LYST in THP1-Blue ISG clones 7 and 5. Red dashes indicate deleted bases, red letters show inserted or substituted bases, and numerals indicate total number of deleted/inserted base pairs. (B) Whole cellular lysates of the indicated THP1-Blue ISG cell clones were immunoblotted (IB) with antibodies against TRIF, actin, and Rab7. Data are representative for three independent experiments. (C) Differentiated WT and LYST-mutant (clones 5 and 7) THP1-Blue–ISG cells that carry an IRF-inducible SEAP reporter construct, were stimulated (stim.) overnight with 1 µg/ml LPS, 20 µg/ml poly(I:C), or 100 ng/ml Pam3CSK4, and the IRF response was assessed by determining the SEAP activity from cell culture supernatants. Supernatants from Pam3CSK4-treated cells were diluted threefold. Data represent mean ± SEM. *, P < 0.05 (Student’s t test). Data are pooled from n = 4 independent experiments. unstim., unstimulated.
Together, these data indicate that the control of TLR3- and TLR4-mediated signaling pathways by lysosomal trafficking regulator Lyst is a more general phenomenon that is also conserved in the human system. Defects in these important innate immune pathways likely contribute to the severe immune deficits in patients with CHS.
Discussion
The concept of TLR signaling being functionally intertwined with the cellular membrane trafficking machinery has received much attention in recent years (Barton and Kagan, 2009; McGettrick and O’Neill, 2010). However, despite great progress in the identification of the molecular components of TLR signaling pathways, still little is known about whether and how regulators of the endosomal/phagosomal trafficking system affect TLR signaling and function, particularly under inflammatory in vivo conditions. Our discovery that lysosomal trafficking regulator Lyst selectively controls a specific subset of TLR signaling pathways provides novel insights into the complex relationship between TLR signaling and endolysosomal trafficking. Importantly, our data demonstrate the physiological relevance of this interrelation for inflammatory responses in vivo and suggest a mechanistic link to human immunopathology.
Lyst is known as an evolutionary conserved regulator of late endosome/lysosome-related organelle structure and trafficking (Kaplan et al., 2008). Although our results confirm a role of Lyst in the organization of endolysosomal networks, they also demonstrate that signaling via intracellular TLRs, which is thought to occur within endolysosomal compartments (Blasius and Beutler, 2010; Kawai and Akira, 2010), is not generally affected in Lyst-mutant Bg-J cells. Instead, the data revealed a regulatory function of Lyst that was restricted to only a specific subset of TLR pathways, namely endolysosomal TLR3- and TLR4-induced TRIF/IRF3 signaling pathways.
Early plasma membrane–initiated TLR4 signals, which are mediated via the MyD88 pathway (Barton and Kagan, 2009; Moresco et al., 2011), were not affected by Lyst, as indicated by normal activation of MAPKs in Lyst-mutant cells. Similarly, Lyst was not required for the initial phagocytic uptake of bacteria, which is in agreement with previous work in different species including humans, mice, Dictyostelium, and Drosophila (Mahoney et al., 1980; Stinchcombe et al., 2000; Harris et al., 2002; Rahman et al., 2012). However, the absence of functional Lyst resulted in defects later in the endosomal/phagosomal process, with particularly the conversion from EEA1+ early to Rab7+ late endosomes/phagosomes being affected. Loss of early endosomal markers and acquisition of the small GTPase Rab7 is a key feature of late endosomal/phagosomal maturation and cargo transport (Rink et al., 2005). Importantly, dysregulated late phagosomal maturation in Lyst-mutant Bg-J cells correlated with impaired TLR3- and TLR4-induced IRF3 activation, a key signaling event in the endosomal TRIF pathway. The adaptor molecule TRIF is an integral component of TLR3 and endosomal TLR4 signaling that is not used by any other TLR. The observation that the immunoregulatory function of Lyst was restricted to TLR3- and TLR4-induced responses thus further reinforces the view that the Lyst mutation specifically affects the TRIF pathway. Although the relevance of TLR-mediated TRIF pathways for protective immunity is well demonstrated for many infectious diseases (Hyun et al., 2013), the specific subcellular localization of endosomal TRIF signaling is still not very well defined. Recent data from overexpression studies have suggested that TLR4-mediated TRIF/IRF3 signaling occurs at early endosomes (Kagan et al., 2008; Palsson-McDermott et al., 2009), whereas our study’s use of macrophages and dendritic cells points to a later endosomal/lysosomal compartment. The relatively late timing of IRF3 activation, when TLR4-containing endosomal vesicles have already progressed to Rab7+ stages, further supports the idea of TLR-induced TRIF signaling originating from later endosomal/lysosomal compartments. Our data now directly demonstrate that TRIF localizes to Rab7+ phagosomes in an LPS-dependent manner. Moreover, our study also shows that activation of TBK1, a key mediator of the TRIF/IRF3 pathway, occurs at Rab7+ phagosomal compartments. These data suggest that Rab7+ endosomal/phagosomal compartments provide an important signaling platform for the assembly and activation of the TRIF signalosome, a multiprotein complex that mediates IRF3 phosphorylation. Furthermore, results from siRNA-mediated knockdown of Rab7 indicate that Rab7 function is actively involved in the regulation of TRIF signaling. Together, our study provides a functional link between Rab7 and TRIF/IRF3 signaling and suggests that efficient formation of the TRIF signalosome depends on Rab7+ endolysosomal compartments.
Different modes of phagosome maturation have been observed in macrophages and dendritic cells, constitutive and TLR signal–dependent inducible (Blander and Medzhitov, 2004). Thus, it is also conceivable that, upon pathogen uptake, innate immune cells use a specialized so far uncharacterized intracellular signaling compartment for TRIF activation that differs from classical endosomal trafficking compartments found under steady-state conditions. Given that the localization of TLR-sorting adaptors is controlled by interaction with phosphoinositides (Kagan et al., 2008; Bonham et al., 2014), it is tempting to speculate that such a specialized signaling compartment might have a particular membrane composition, e.g., specific phospholipids, that mediate recruitment of appropriate signaling adaptors. Together, our results suggest that Lyst orchestrates the formation of a Rab7+ endosomal/phagosomal compartment, which allows for optimal TRIF signalosome assembly and activity.
The underlying molecular mechanism by which Lyst controls Rab7 recruitment and phagosomal maturation/trafficking in innate immune cells is currently unclear. As a BEACH (beige and Chediak-Higashi) domain–containing protein, Lyst has been proposed to act as an anchoring scaffold for molecules controlling membrane trafficking processes (Cullinane et al., 2013). Potentially, this involves direct interaction with Rab7, as LysB, the Dictyostelium homologue of Lyst, has been reported to colocalize with Rab7 on a subset of endolysosomal vesicles (Kypri et al., 2007). Alternatively, Lyst might exhibit regulatory function via control of other Rab GTPases, such as Rab14 (Kypri et al., 2013), or interaction with soluble N-ethylmaleimide–sensitive fusion attachment protein receptor (SNARE) complexes (Tchernev et al., 2002).
Evidence for endosomal TLR signaling comes mainly from cell biological in vitro studies, as a lack of appropriate genetic animal models has so far largely hampered the analysis of its physiological relevance in intact organisms. Our study shows that in innate immune cells, such as macrophages and dendritic cells, lysosomal-trafficking regulator Lyst selectively controls TLR3- and TLR4-mediated pathways for the induction of proinflammatory cytokines. Importantly, by using Lyst-mutant mice, we could demonstrate the relevance of these cellular defects for in vivo immunity in experimental models of acute inflammation. Generally, loss of functional Lyst resulted in impaired inflammatory responses and protected from endotoxin-induced excessive inflammation during septic shock. These results are in line with the reported proinflammatory function of TRIF which, as an adaptor protein in TLR3- and TLR4-mediated inflammatory responses, is involved in both antiviral and antibacterial defense mechanisms but can also trigger immunopathology and organ damage (Hyun et al., 2013).
The finding that the immunoregulatory function of Lyst is conserved in human cells suggests that defects in TLR signaling pathways (caused by mutations in the human LYST gene) contribute to the frequent and severe infections observed in patients with CHS. Although speculative, impaired TLR3/TRIF-mediated antiviral immune responses may even facilitate the development of the life-threatening lymphoma-resembling accelerated phase of CHS, which seems to be triggered by Epstein-Barr virus infection (Rubin et al., 1985; Certain et al., 2000).
Collectively, the identification of Lyst as a specific regulator of TLR3- and TLR4-mediated TRIF signaling pathways reveals how the regulation of the intracellular membrane trafficking network is functionally linked to specific TLR signaling pathways. Importantly, our results highlight the significance of this interrelation for inflammatory reactions in vivo. The findings not only have implications for our understanding of normal and pathophysiological processes during inflammatory reactions, but may also open up prospects for future therapeutic interventions by specifically targeting intracellular TLR trafficking routes.
Materials and methods
Mice and cell culture
Lyst-mutant beige mice (C57BL/6J-Lystbg-J) and WT control C57BL/6J mice were obtained from the Jackson Laboratory. Mice were housed under specific pathogen–free conditions. Animal experiments were performed in accordance with institutional guidelines and were approved by the local authorities (Ministry of Agriculture, the Environment, and Rural Areas, Schleswig-Holstein and the Lower Saxony State Office for Consumer Protection and Food Safety).
Total bone marrow cells were isolated from 7–10-wk-old mice and were cultured in the presence of mouse M-CSF (Promokine) with a final concentration of 20 ng/ml for 7 d to differentiate BMDMs. Alternatively, bone marrow cells were cultured in the presence mouse GM-CSF (Promokine) with a final concentration of 20 ng/ml for 9 d to differentiate BMDCs.
Human monocytic THP-1 cells were maintained in RPMI 1640 supplemented with 10% heat-inactivated FCS, 2 mM l-glutamine, 10 mM Hepes, nonessential amino acids, and antibiotics. Differentiation into macrophage-like cells was induced by treating THP-1 cells overnight with 100 ng/ml PMA followed by 5–7 d resting in normal culture medium. Differentiated CD14+ cells were then purified via magnetic cell sorting with human CD14 Microbeads (Miltenyi Biotec) and were used subsequently for experiments.
THP-1–Blue ISG cells (InvivoGen), which express a SEAP reporter gene under the control of an ISG54 promoter in conjunction with five IFN-stimulated response elements, were cultured in THP-1 growth medium containing 100 µg/ml Normocin and 100 µg/ml Zeocin. The cell response was monitored by measuring SEAP activity in cell culture supernatants with QUANTI-Blue medium (InvivoGen) according to the supplier’s instructions.
Purification of LPS from S. Typhimurium
S. Typhimurium cells were extracted with aq 45% phenol at 68°C, and the aqueous layer was successively dialyzed against tap and distilled water and then lyophilized. This smooth LPS was purified by ultracentrifugation at 105,000 g, freeze dried, and then digested with RNase and DNase (37°C for 4 h) and with proteinase K (1 h at 56°C). The suspension was dialyzed against distilled water (48 h at 4°C) and freeze dried (yield: 79% of crude LPS).
Measurement of cytokines
BMDMs or BMDCs were stimulated with different ligands for TLRs: 100 ng/ml Pam3CSK4 (InvivoGen), 10 µg/ml poly(I:C) RNA (Sigma-Aldrich), 100 ng/ml LPS, 10 µg/ml R848 (InvivoGen), or 5 µM CpG-C DNA (ODN2395; InvivoGen) for 6 h. TNF, IL-12, or IFN-β protein levels in cell culture supernatants were then determined by specific ELISA kits (eBioscience) according to the manufacturer’s protocol. For the detection of intracellular cytokines, cells were stimulated with various TLR ligands in the presence of brefeldin A (eBioscience) for blockade of Golgi function. Cells were fixed with 4% paraformaldehyde in PBS, permeabilized with 0.1% saponin (eBioscience), and subjected to intracellular FACS staining using antibodies against TNF (BioLegend).
After stimulation of differentiated THP-1 cells with TLR ligands in the presence of brefeldin A, cells were surface stained with anti–human CD14 (BioLegend) followed by intracellular staining with anti–human TNF (BioLegend).
Real-time quantitative RT-PCR analysis
Total RNA was purified from cells using a PureLink RNA Mini kit (Invitrogen) or the RNeasy Micro kit (QIAGEN) according to the manufacturer’s instructions and reverse transcribed using the Superscript II first-strand synthesis system (Invitrogen). For real-time quantitative RT-PCR, Lyst, Tnf, Ifnb1, Tlr3, and Tlr4 gene expression was measured relative to Hprt using the Universal Probe Library (Roche) TaqMan-based system (Applied Biosystems). Amplification was performed in a fluorescence temperature cycler (LightCycler 2.0 or LightCycler 480; Roche). Primers and the Universal Probe Library ID numbers are listed in Table S1.
Flow cytometric analysis
Single-cell suspensions of BMDMs or BMDCs were blocked with FcBlock (anti-CD16/CD32; BioLegend) and subsequently stained with fluorescence-labeled mAbs for CD11b, CD11c, Gr-1, Ly6C, TLR4/MD2 (clone MTS510), and TLR4 (clone SA15-21; all BioLegend). Intracellular staining of human and mouse TNF (both BioLegend) and mouse TLR3 (Imgenex) was performed in permeabilized cells according to standard procedures. Flow cytometric measurements were performed on a FACSCantoII cell analyzer and an LSRII flow cytometer (both BD). Data were analyzed with FlowJo software (version 9.8.3, Mac; version 10.0.7, PC; Tree Star).
TLR4 internalization
Cells were stimulated with 100 ng/ml LPS at 37°C for the indicated times. TLR4 internalization was terminated by adding ice-cold 0.1% azide–containing medium and stored on ice. Surface-remaining TLR4 was detected by staining with an anti–mouse TLR4 antibody (SA15-21; BioLegend). TLR4 binding by clone SA15-21 is not interrupted by LPS stimulation. Cells were then fixed with 2% paraformaldehyde before analysis by flow cytometry.
In vivo Salmonella infection
Age- and gender-matched mice at an age of 10–12 wk were treated with 20 mg streptomycin per mouse by oral gavage and infected with 3 × 106 S. Typhimurium SL1344 by oral gavage 24 h later. Mice were sacrificed 1 or 4 d after infection, and tissue samples were homogenized in 1 ml of sterile PBS using a Tissue Lyser II (QIAGEN). Serial dilutions were plated on Luria broth agar plates containing 100 µg/ml streptomycin to determine bacterial colonization.
In vivo models of acute inflammation
To induce in vivo cytokine secretion, 10-wk-old female mice were injected i.p. with 40 ng/g body weight LPS from S. Typhimurium, 2 µg/g body weight poly(I:C), or 3.5 µg/g body weight Pam3CSK4. At the indicated times after injection, cytokine levels in the sera were measured by specific ELISA. To induce septic shock, mice were injected i.p. with 8 µg/g body weight LPS from S. Typhimurium, and their health status was monitored at regular intervals, every 12 h for 7 d. Acute pulmonary inflammation in mice was induced by intranasal instillation of 1 µg LPS. Infiltrated immune cells in the airway were collected after 24 h by BAL. Recovered BAL cells were counted and further analyzed by flow cytometry.
Western blotting
After stimulation of cells with 100 ng/ml LPS or 10 µg/ml poly(I:C) for the indicated times, cells were lysed in whole-cell lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 1% NP-40, 1 mM Na3VO4, 10 mM NaF, and the complete Mini EDTA-free Protease Inhibitor cocktail; Roche) for 20 min on ice. For the nuclear fractionation assay, nuclear protein extraction was performed by using a Nuclear Protein Extraction kit (NE-PER; Thermo Fisher Scientific) according to the manufacturer’s protocol. Whole cellular lysates or nuclear fractions were resolved by SDS-PAGE, and proteins were detected by Western blotting using antibodies against phospho-Erk1/2 (Thr202/204), phospho-JNK (Thr183/185), phospho-p38 MAPK (Thr180/182), phospho-IRF3 (Ser396), phospho-TBK1 (Ser172), phospho-IκBα, Erk2, p38 MAPK, IRF3, TBK1, TRAF3, IκBα, Rab7, lamin A/C (all from Cell Signaling Technology), TRIF-related adaptor molecule (TRAM), TRIF (Acris), actin, and GAPDH (Santa Cruz Biotechnology, Inc.).
Immunofluorescence microscopy
For confocal microscopy, BMDCs, BMDMs, and differentiated THP-1 cells were grown on flow chamber microslides (Ibidi). After fixation in 4% paraformaldehyde, cells were permeabilized in 0.2% saponin and blocked with 5% goat serum. Then, cells were stained with the appropriate antibodies in staining buffer containing 0.1% saponin. Antibodies used in the microscopy studies included: anti-EEA1 (rabbit antibody, Cell Signaling Technology; mouse antibody, MBL), anti-Rab4 and anti-Rab5 (BD), anti-Rab9, anti-Rab11, anti–phsopho-TBK1 (Cell Signaling Technology), anti-Rab7 (rabbit antibody, Cell Signaling Technology; mouse antibody, Acris), biotinylated anti–mouse Lamp1 (eBioscience), anti-TBK1 (Acris), and anti-TRIF (BioLegend). Secondary reagents, Alexa Fluor 647–conjugated streptavidin, Alexa Fluor 546– or Alexa Fluor 488–conjugated goat anti–rabbit IgG, and goat anti–mouse IgG, were obtained from Jackson ImmunoResearch Laboratories, Inc. DNA was visualized by staining with DAPI (Invitrogen).
To assess LPS bead phagocytosis, streptavidin beads (PolySciences) were conjugated with biotinylated LPS (1,000 ng LPS/5 × 105 beads/1 ml) in glass tubes and sonicated afterward. LPS-conjugated beads were added to the cells at a ratio of five beads (3–5 µm diameter) per cell for phagosomal maturations studies or ∼50 beads (2 µm diameter) per cell for signaling visualization, and cultures were incubated at 37°C for the indicated times. Phagocytosis was stopped with ice-cold PBS and subsequent fixation with 4% paraformaldehyde in PBS. Images were acquired on an SP5 confocal microscope (Leica Biosystems) or an IX81 microscope (Olympus). Subsequent analysis of individual and overlay fluorescence images was performed using LAS AF (Leica Biosystems), ImageJ (1.50c; National Institutes of Health), and iVision (BioVision Technologies) software. Quantification of particle size and fluorescence intensities and the generation of line profile data were performed using ImageJ software (1.50c).
Phagocytosis assay, measurement of acidification, and live-cell imaging
The quantitative analysis of phagocytosis was performed by measuring the uptake of CFSE-labeled E. coli particles by flow cytometry. The fluorescence of extracellular bacterial particles was quenched by adding 0.4% trypan blue to measure only internal fluorescence of ingested bacteria. The acidification of phagosomal compartments was assessed through the uptake of E. coli particles labeled with a low pH–sensitive dye (pHrodo Green; Invitrogen). To control phagocytosis, bacteria were co-labeled with eFluor 670 (eBioscience) in some experiments. Analysis of fluorescence from ingested E. coli was monitored by flow cytometry or fluorescence microscopy. To detect acidic lysosomal compartments, cells were stained with 75 nm LysoTracker red and 1 mM Lysosensor green (all from Invitrogen). Images of live cells containing ingested E. coli were captured at 37°C using a confocal microscope (SP5; Leica Biosystems) with a heated stage.
siRNA-mediated gene knockdown
BMDMs were transfected overnight with a mix of four siRNAs against mouse Rab7 (FlexiTube siRNA; QIAGEN) or scrambled control siRNA (QIAGEN) by using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. At day 2 after transfection cells, were stimulated with LPS or poly(I:C) for the indicated time, and whole cellular lysates were prepared.
CRISPR/Cas9-mediated genome editing in human THP-1 cells
A pair of annealed sgRNA-specifying oligos for each targeting site (Table S2) was cloned into the bicistronic expression vector pX461 (plasmid no. 48140; Addgene; gift from Feng Zhang, Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, MA,; Ran et al., 2013b), which contains the D10A nickase mutant of Cas9 (Cas9n)-2A-GFP and a cloning backbone for sgRNA. sgRNAs were chosen to target exon 49 of human LYST with minimal potential off-target effects by bioinformatics analysis. The plasmids were cotransfected into THP-1 cells by using the Nucleofector transfection system according to the manufacturer’s instructions (program T20; Lonza), and after 24–48 h, GFPhigh cells were single cell sorted (XDPs; Beckman Coulter) into 96-well plates. After 4–6 wk, genomic DNA was extracted from individual colonies, the targeted locus was PCR amplified with Phusion High Fidelity DNA polymerase (New England Biolabs, Inc.), and mutant clones were identified by sequencing (oligonucleotides used for PCR amplification and sequencing are listed in Table S2). In addition, PCR products were cloned into pcDNA3.1+, and plasmid DNA from multiple bacterial colonies corresponding to each PCR product was analyzed by sequencing.
Online supplemental material
Table S1 describes primers and Universal Probe Library ID numbers used for quantitative real-time PCR analysis. Table S2 lists oligonucleotides related to CRISPR/Cas9-mediated genome editing.
Supplementary Material
Acknowledgments
We greatly appreciate the excellent technical support of Katrin Westphal, P. Prilla, L. Nath, and M. Hein.
This work was supported in part by grants from the European Union FP7 Marie Curie International Reintegration Grants (202287), the Deutsche Forschungsgemeinschaft (DFG) cluster of excellence Inflammation at Interfaces (306/1 and 306/2), and DFG (1254/3-1).
The authors declare no competing financial interests.
Author contributions: conceptualization, K.-H. Lee and N. Föger; methodology, A. Westphal, W. Cheng, and G. Grassl; investigation, A. Westphal, W. Cheng, J. Yu, M. Krautkrämer, and G. Grassl; writing (original draft), N. Föger and K.-H. Lee; writing (review and editing), N. Föger and K.-H. Lee; resources, G. Grassl and O. Holst; supervision, N. Föger and K.-H. Lee; and funding acquisition, K.-H. Lee.
Footnotes
Abbreviations used:
- BAL
- bronchoalveolar lavage
- CHS
- Chédiak-Higashi syndrome
- IRF
- IFN regulatory factor
- ISG
- IFN-stimulated gene
- poly(I:C)
- polyinosinic-polycytidylic acid
- sgRNA
- single-guide RNA
- TRIF
- TIR domain–containing adapter-inducing IFN-β
References
- Adati N., Huang M.C., Suzuki T., Suzuki H., and Kojima T.. 2009. High-resolution analysis of aberrant regions in autosomal chromosomes in human leukemia THP-1 cell line. BMC Res. Notes. 2:153 10.1186/1756-0500-2-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S., Uematsu S., and Takeuchi O.. 2006. Pathogen recognition and innate immunity. Cell. 124:783–801. 10.1016/j.cell.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Arpaia N., Godec J., Lau L., Sivick K.E., McLaughlin L.M., Jones M.B., Dracheva T., Peterson S.N., Monack D.M., and Barton G.M.. 2011. TLR signaling is required for Salmonella typhimurium virulence. Cell. 144:675–688. 10.1016/j.cell.2011.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa M.D., Nguyen Q.A., Tchernev V.T., Ashley J.A., Detter J.C., Blaydes S.M., Brandt S.J., Chotai D., Hodgman C., Solari R.C., et al. 1996. Identification of the homologous beige and Chediak-Higashi syndrome genes. Nature. 382:262–265. 10.1038/382262a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton G.M., and Kagan J.C.. 2009. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat. Rev. Immunol. 9:535–542. 10.1038/nri2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander J.M., and Medzhitov R.. 2004. Regulation of phagosome maturation by signals from toll-like receptors. Science. 304:1014–1018. 10.1126/science.1096158 [DOI] [PubMed] [Google Scholar]
- Blasius A.L., and Beutler B.. 2010. Intracellular toll-like receptors. Immunity. 32:305–315. 10.1016/j.immuni.2010.03.012 [DOI] [PubMed] [Google Scholar]
- Bonham K.S., Orzalli M.H., Hayashi K., Wolf A.I., Glanemann C., Weninger W., Iwasaki A., Knipe D.M., and Kagan J.C.. 2014. A promiscuous lipid-binding protein diversifies the subcellular sites of toll-like receptor signal transduction. Cell. 156:705–716. 10.1016/j.cell.2014.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt J.K., Wiebel F.A., Hester S., and Argon Y.. 1993. The giant organelles in beige and Chediak-Higashi fibroblasts are derived from late endosomes and mature lysosomes. J. Exp. Med. 178:1845–1856. 10.1084/jem.178.6.1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certain S., Barrat F., Pastural E., Le Deist F., Goyo-Rivas J., Jabado N., Benkerrou M., Seger R., Vilmer E., Beullier G., et al. 2000. Protein truncation test of LYST reveals heterogenous mutations in patients with Chediak-Higashi syndrome. Blood. 95:979–983. [PubMed] [Google Scholar]
- Cullinane A.R., Schäffer A.A., and Huizing M.. 2013. The BEACH is hot: a LYST of emerging roles for BEACH-domain containing proteins in human disease. Traffic. 14:749–766. 10.1111/tra.12069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J.A., and Charpentier E.. 2014. The new frontier of genome engineering with CRISPR-Cas9. Science. 346:1258096 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Doyle S., Vaidya S., O’Connell R., Dadgostar H., Dempsey P., Wu T., Rao G., Sun R., Haberland M., Modlin R., and Cheng G.. 2002. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 17:251–263. 10.1016/S1074-7613(02)00390-4 [DOI] [PubMed] [Google Scholar]
- Fitzgerald K.A., McWhirter S.M., Faia K.L., Rowe D.C., Latz E., Golenbock D.T., Coyle A.J., Liao S.M., and Maniatis T.. 2003. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491–496. 10.1038/ni921 [DOI] [PubMed] [Google Scholar]
- Harris E., Wang N., Wu Wl W.L., Weatherford A., De Lozanne A., and Cardelli J.. 2002. Dictyostelium LvsB mutants model the lysosomal defects associated with Chediak-Higashi syndrome. Mol. Biol. Cell. 13:656–669. 10.1091/mbc.01-09-0454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebe K., Du X., Georgel P., Janssen E., Tabeta K., Kim S.O., Goode J., Lin P., Mann N., Mudd S., et al. 2003. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 424:743–748. 10.1038/nature01889 [DOI] [PubMed] [Google Scholar]
- Hsu P.D., Lander E.S., and Zhang F.. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 157:1262–1278. 10.1016/j.cell.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M., Helip-Wooley A., Westbroek W., Gunay-Aygun M., and Gahl W.A.. 2008. Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu. Rev. Genomics Hum. Genet. 9:359–386. 10.1146/annurev.genom.9.081307.164303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye H., Halaas Ø., Stenmark H., Tunheim G., Sandanger Ø., Bogen B., Brech A., Latz E., and Espevik T.. 2006. Endocytic pathways regulate Toll-like receptor 4 signaling and link innate and adaptive immunity. EMBO J. 25:683–692. 10.1038/sj.emboj.7600991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun J., Kanagavelu S., and Fukata M.. 2013. A unique host defense pathway: TRIF mediates both antiviral and antibacterial immune responses. Microbes Infect. 15:1–10. 10.1016/j.micinf.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Introne W., Boissy R.E., and Gahl W.A.. 1999. Clinical, molecular, and cell biological aspects of Chediak-Higashi syndrome. Mol. Genet. Metab. 68:283–303. 10.1006/mgme.1999.2927 [DOI] [PubMed] [Google Scholar]
- Kagan J.C. 2012. Signaling organelles of the innate immune system. Cell. 151:1168–1178. 10.1016/j.cell.2012.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan J.C., Su T., Horng T., Chow A., Akira S., and Medzhitov R.. 2008. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nat. Immunol. 9:361–368. 10.1038/ni1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J., De Domenico I., and Ward D.M.. 2008. Chediak-Higashi syndrome. Curr. Opin. Hematol. 15:22–29. 10.1097/MOH.0b013e3282f2bcce [DOI] [PubMed] [Google Scholar]
- Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 441:101–105. 10.1038/nature04734 [DOI] [PubMed] [Google Scholar]
- Kawai T., and Akira S.. 2006. TLR signaling. Cell Death Differ. 13:816–825. 10.1038/sj.cdd.4401850 [DOI] [PubMed] [Google Scholar]
- Kawai T., and Akira S.. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11:373–384. 10.1038/ni.1863 [DOI] [PubMed] [Google Scholar]
- Krzewski K., and Cullinane A.R.. 2013. Evidence for defective Rab GTPase-dependent cargo traffic in immune disorders. Exp. Cell Res. 319:2360–2367. 10.1016/j.yexcr.2013.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kypri E., Schmauch C., Maniak M., and De Lozanne A.. 2007. The BEACH protein LvsB is localized on lysosomes and postlysosomes and limits their fusion with early endosomes. Traffic. 8:774–783. 10.1111/j.1600-0854.2007.00567.x [DOI] [PubMed] [Google Scholar]
- Kypri E., Falkenstein K., and De Lozanne A.. 2013. Antagonistic control of lysosomal fusion by Rab14 and the Lyst-related protein LvsB. Traffic. 14:599–609. 10.1111/tra.12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano M.L., Rivera J., Sánchez-Guiu I., and Vicente V.. 2014. Towards the targeted management of Chediak-Higashi syndrome. Orphanet J. Rare Dis. 9:132 10.1186/s13023-014-0132-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney K.H., Morse S.S., and Morahan P.S.. 1980. Macrophage functions in beige (Chédiak-Higashi syndrome) mice. Cancer Res. 40:3934–3939. [PubMed] [Google Scholar]
- Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., and Church G.M.. 2013. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 31:833–838. 10.1038/nbt.2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur R., Oh H., Zhang D., Park S.-G., Seo J., Koblansky A., Hayden M.S., and Ghosh S.. 2012. A mouse model of Salmonella typhi infection. Cell. 151:590–602. 10.1016/j.cell.2012.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy C.E., Carpenter S., Pålsson-McDermott E.M., Gearing L.J., and O’Neill L.A.. 2008. Glucocorticoids inhibit IRF3 phosphorylation in response to Toll-like receptor-3 and -4 by targeting TBK1 activation. J. Biol. Chem. 283:14277–14285. 10.1074/jbc.M709731200 [DOI] [PubMed] [Google Scholar]
- McGettrick A.F., and O’Neill L.A.J.. 2010. Localisation and trafficking of Toll-like receptors: an important mode of regulation. Curr. Opin. Immunol. 22:20–27. 10.1016/j.coi.2009.12.002 [DOI] [PubMed] [Google Scholar]
- Moresco E.M.Y., LaVine D., and Beutler B.. 2011. Toll-like receptors. Curr. Biol. 21:R488–R493. 10.1016/j.cub.2011.05.039 [DOI] [PubMed] [Google Scholar]
- Palsson-McDermott E.M., Doyle S.L., McGettrick A.F., Hardy M., Husebye H., Banahan K., Gong M., Golenbock D., Espevik T., and O’Neill L.A.J.. 2009. TAG, a splice variant of the adaptor TRAM, negatively regulates the adaptor MyD88-independent TLR4 pathway. Nat. Immunol. 10:579–586. 10.1038/ni.1727 [DOI] [PubMed] [Google Scholar]
- Park E.K., Jung H.S., Yang H.I., Yoo M.C., Kim C., and Kim K.S.. 2007. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm. Res. 56:45–50. 10.1007/s00011-007-6115-5 [DOI] [PubMed] [Google Scholar]
- Rahman M., Haberman A., Tracy C., Ray S., and Krämer H.. 2012. Drosophila mauve mutants reveal a role of LYST homologs late in the maturation of phagosomes and autophagosomes. Traffic. 13:1680–1692. 10.1111/tra.12005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., and Zhang F.. 2013a Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 154:1380–1389. 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013b Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y., and Zerial M.. 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 122:735–749. 10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Rubin C.M., Burke B.A., McKenna R.W., McClain K.L., White J.G., Nesbit M.E. Jr., and Filipovich A.H.. 1985. The accelerated phase of Chediak-Higashi syndrome. An expression of the virus-associated hemophagocytic syndrome? Cancer. 56:524–530. [DOI] [PubMed] [Google Scholar]
- Sato S., Sugiyama M., Yamamoto M., Watanabe Y., Kawai T., Takeda K., and Akira S.. 2003. Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-κB and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 171:4304–4310. 10.4049/jimmunol.171.8.4304 [DOI] [PubMed] [Google Scholar]
- Sorkin A., and von Zastrow M.. 2009. Endocytosis and signalling: intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 10:609–622. 10.1038/nrm2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe J.C., Page L.J., and Griffiths G.M.. 2000. Secretory lysosome biogenesis in cytotoxic T lymphocytes from normal and Chediak Higashi syndrome patients. Traffic. 1:435–444. 10.1034/j.1600-0854.2000.010508.x [DOI] [PubMed] [Google Scholar]
- Tchernev V.T., Mansfield T.A., Giot L., Kumar A.M., Nandabalan K., Li Y., Mishra V.S., Detter J.C., Rothberg J.M., Wallace M.R., et al. 2002. The Chediak-Higashi protein interacts with SNARE complex and signal transduction proteins. Mol. Med. 8:56–64. [PMC free article] [PubMed] [Google Scholar]
- Trantow C.M., Mao M., Petersen G.E., Alward E.M., Alward W.L., Fingert J.H., and Anderson M.G.. 2009. Lyst mutation in mice recapitulates iris defects of human exfoliation syndrome. Invest. Ophthalmol. Vis. Sci. 50:1205–1214. 10.1167/iovs.08-2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., and Akira S.. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 301:640–643. 10.1126/science.1087262 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.