Xu et al. show that LRCH1 interferes with the GEF activity of DOCK8 to inhibit Cdc42 activation. Upon chemokine stimulation, DOCK8 is phosphorylated and released from LRCH1 to drive cell migration. LRCH1 overexpression reduces CD4+ T cell migration to the CNS and ameliorates experimental autoimmune encephalomyelitis.
Abstract
Directional autoreactive CD4+ T cell migration into the central nervous system plays a critical role in multiple sclerosis. Recently, DOCK8 was identified as a guanine-nucleotide exchange factor (GEF) for Cdc42 activation and has been associated with human mental retardation. Little is known about whether DOCK8 is related to multiple sclerosis (MS) and how to restrict its GEF activity. Using two screening systems, we found that LRCH1 competes with Cdc42 for interaction with DOCK8 and restrains T cell migration. In response to chemokine stimulation, PKCα phosphorylates DOCK8 at its three serine sites, promoting DOCK8 separation from LRCH1 and translocation to the leading edge to guide T cell migration. Point mutations at the DOCK8 serine sites block chemokine- and PKCα-induced T cell migration. Importantly, Dock8 mutant mice or Lrch1 transgenic mice were protected from MOG (35–55) peptide–induced experimental autoimmune encephalomyelitis (EAE), whereas Lrch1-deficient mice displayed a more severe phenotype. Notably, DOCK8 expression was markedly increased in PBMCs from the acute phase of MS patients. Together, our study demonstrates LRCH1 as a novel effector to restrain PKCα–DOCK8–Cdc42 module–induced T cell migration and ameliorate EAE.
Introduction
Multiple sclerosis (MS) is considered a T cell–mediated inflammatory and demyelinating disease of the CNS with a complex genetic background (McFarland and Martin, 2007). Autoreactive CD4+ T lymphocyte extravasation and infiltration into the CNS is a finely regulated cascade of steps that is controlled by integrins, chemokines, or inflammatory cytokines (Sigal et al., 2000; Vajkoczy et al., 2001; Kerfoot and Kubes, 2002; Ransohoff et al., 2003). Chemokines such as SDF-1α (also termed CXCL12) and CCL5 are increased in MS lesions, which attract T lymphocytes across the endothelial cell monolayer (Sørensen et al., 1999; dos Santos et al., 2005; Krumbholz et al., 2006). Data from human MS genome–wide association studies and murine experimental autoimmune encephalomyelitis (EAE) models suggest that the dysregulation of T cell extravasation by key signaling proteins, such as S1P1 (sphingosine-1-phosphate receptor 1) and VCAM-1 (vascular cell adhesion molecule 1), might mediate MS development (De Jager et al., 2009; Sawcer et al., 2011; Damotte et al., 2014). Targeting VCAM-1or S1P1 by antibodies or small molecules represents a novel approach for treating MS (Steinman, 2005; Chun and Hartung, 2010). However, given their side effects, it is critical to explore new proteins that restrain T cell infiltration into the CNS and ameliorate MS.
Cdc42, a member of the Rho small GTPases, orchestrates the cell cytoskeleton for T cell migration. Cdc42 activity is precisely controlled temporally and spatially (Haddad et al., 2001; Etienne-Manneville, 2004). When responding to chemokines, Cdc42 switches from the GDP-bound inactive form to the GTP-bound active state in the presence of guanine-nucleotide exchange factors (GEFs; Randall et al., 2009). Several GEFs in T cells, including the VAV and DOCK (dedicator of cytokinesis) family proteins, ensure the proper activation of Cdc42 (Harada et al., 2012). Studies suggest that Vav1-deficient mice are prevented from EAE induction (Tybulewicz, 2005), and mice having Dock8 mutations or the loss of the Dock8 gene exhibit impaired immune responses to clear viral infection (Randall et al., 2009, 2011; Lambe et al., 2011; Jabara et al., 2012). Importantly, human DOCK8 mutations or SNPs are associated with immunodeficiency and mental retardation (Griggs et al., 2008; Zhang et al., 2009). Despite this, it is unknown whether DOCK8 is engaged in MS, and which negative regulators restrict DOCK8 GEF activity to prevent immune cell migration.
In this study, we identified LRCH1 as a novel binding partner to sequester DOCK8 from Cdc42. Upon chemokine stimulation, DOCK8 is phosphorylated by PKCα to separate from LRCH1 and relocate at the leading edge for T cell migration. By generation of Lrch1 transgenic, Lrch1 knockout and Dock8 mutant mice, we demonstrated their critical role in controlling the development of EAE in vivo.
Results
DOCK8 expression is enhanced in the acute phase of murine EAE
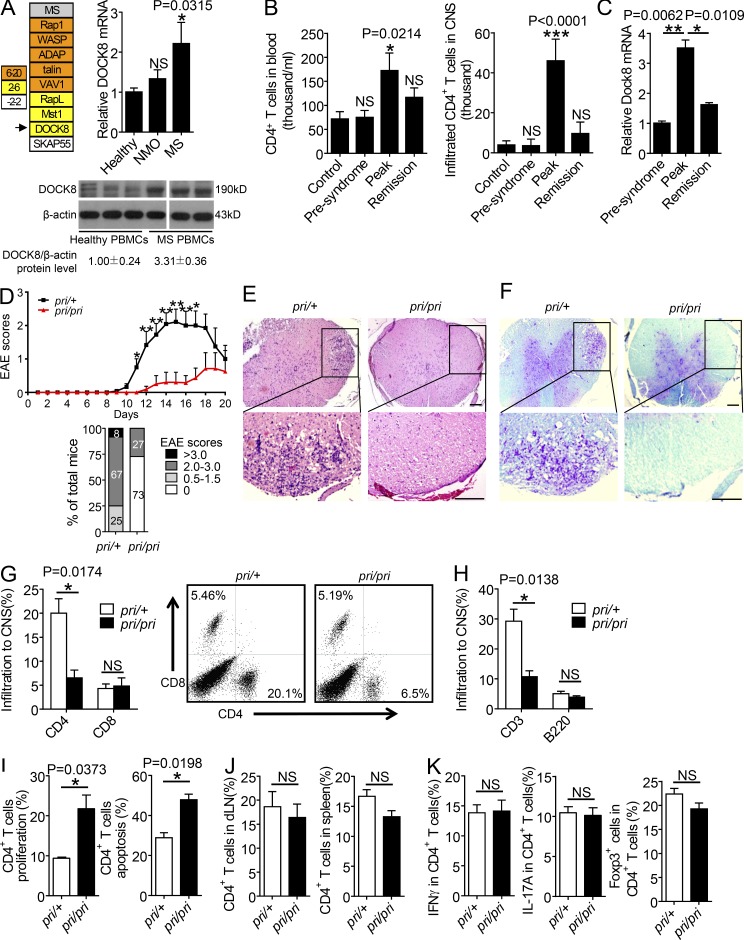
Great efforts have been made to identify critical signaling proteins involved in T lymphocyte adhesion and migration (Wang et al., 2010; Zhang and Wang, 2012; Yu et al., 2015). Some of these signaling proteins, including VAV1, ADAP, SKAP55, Rap1, RapL, Mst1, and DOCK8, also regulate T cell activation, apoptosis, or inflammation (Wang et al., 2003, 2004, 2007, 2009; Jo et al., 2005; Katagiri et al., 2006, 2011; Wang and Rudd, 2008; Li et al., 2015a,b,c). Considering the central role of myelin-specific CD4+ T cell activation and infiltration into the CNS in the pathogenesis of MS, we asked whether the expression levels of these molecules were associated with human MS patients. The mRNA levels of Rap1, WASP, VAV1, ADAP, talin, RapL, Mst1, or DOCK8 (but not SKAP55) were significantly enhanced in PBMCs from MS patients compared with age-matched healthy volunteers (Fig. 1 A, left). In agreement with our observation, previous studies suggest that a deficiency of VAV1 or ADAP ameliorates myelin oligodendrocyte glycoprotein peptide (MOG 35–55)–induced EAE, a mouse model that mimics human MS (Korn et al., 2003; Engelmann et al., 2013). Because Mst1 binds to the RapL–Rap1 complex, whereas DOCK8 is the key downstream effector of Mst1 (Mou et al., 2012), we asked whether DOCK8 influenced the pathogenesis of MS/EAE. First, we confirmed that the mRNA and protein levels of DOCK8 were significantly elevated in the PBMCs from MS patients, compared with those from healthy controls and neuromyelitis optica (NMO) patients who displayed similar symptoms to those of MS, but with a distinct etiology (Fig. 1 A, right). Furthermore, during the development of murine EAE model, we noticed that more CD4+ T cells circulated in the blood and infiltrated in the CNS at the peak stage than those at the presyndrome or remission stage (Fig. 1 B). Dock8 levels in the blood CD4+ T cells were significantly increased at the peak stage of EAE compared with at the presymptom or remission stage (Fig. 1 C). This suggests that DOCK8 expression levels are correlated with EAE severity.
Figure 1.
DOCK8 expression is positively associated with the peak phase of murine EAE. (A) The relative mRNA expression levels of the candidate genes in the PBMCs from MS patients and healthy volunteers (top left; n = 4). DOCK8 mRNA levels in the PBMCs (top right) from healthy volunteers (n = 42), NMO patients (n = 24), or MS patients (n = 38). DOCK8 expression in the PBMCs from healthy volunteers and MS patients by immunoblotting (bottom). (B) The total number of CD4+ T cells circulating in the blood (left) or infiltrating in the CNS (right) at different stages of murine EAE. n = 6. (C) Dock8 mRNA levels in CD4+ T cells from murine EAE at the presyndrome, peak, or remission stages. n = 3. (D) Clinical scores (top) and EAE incidence (bottom) of the Dock8pri/+ and Dock8pri/pri mice immunized with MOG (35–55). n = 10. (E and F) H&E and Luxol blue staining of the representative tissue sections of the spinal cords from the Dock8pri/+ and Dock8pri/pri mice on day 18 after EAE induction. Bars, 70 µm. (G and H) Frequency of CD4+ T, CD8+ T, and B220+ cells in the CNS by flow cytometry from the EAE-induced mice. n = 3. (I) The percentages of BrdU+CD4+ T cells and Annexin V+ CD4+ T cells in draining LNs from Dock8pri/+or Dock8pri/pri mice after EAE induction. n = 3. (J) The percentages of CD4+ T cells in the draining LNs and spleen. n = 3. (K) The percentages of IFN-γ+, IL-17A+, or Foxp3+ cells in CD4+ T cells from the draining LNs of Dock8pri/+ or Dock8pri/pri mice when EAE was induced at peak stage. n = 3. NS, not significant (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.005. Data are representative of three independent experiments (D, mean ± SEM; B, C, and G, mean ± SD) or two independent experiments (H, I, J, and K, mean ± SD). Statistical significance was determined using unpaired Student’s t test.
Next, we elucidated whether DOCK8 was a susceptible gene for the induction and development of EAE. As previously reported (Randall et al., 2009), Dock8pri/pri mice contain a serine-to-proline substitution in the DHR-2 (DOCK homology region 2) domain of Dock8, which abolishes the GEF activity for Cdc42 activation. In response to immunization with a MOG (35–55) peptide, all of the Dock8pri/+ mice developed EAE, whereas less than half of the Dock8pri/pri mice manifested EAE symptoms, which also reduced the disease severity (Fig. 1 D). Consistently, hematoxylin and eosin (H&E) or luxol fast blue staining revealed a decreased numbers of immune cells and a lower degree of demyelination in the spinal cord sections of the Dock8pri/pri mice compared with those of Dock8pri/+mice (Fig. 1, E and F). Additionally, the percentages of the CNS-infiltrated CD4+ T cells were remarkably decreased in the Dock8pri/pri mice when EAE developed at the peak phase (Fig. 1 G). In contrast, the percentages of CD8+ T cells or B220+ B cells were apparently not affected (Fig. 1, G and H).
Because Dock8 regulates naive T cell apoptosis (Harada et al., 2012), we injected BrdU into the MOG (35–55)–induced Dock8pri/+ and Dock8pri/pri EAE mice 1 d before the lymphocytes were collected to see whether Dock8pri/pri CD4+ T cells showed abnormal proliferation or apoptosis in vivo. Compared with the Dock8pri/+ CD4+ T cells, the Dock8pri/pri CD4+ T cells showed both increased proliferation and apoptosis in vivo (Fig. 1 I), resulting in the same percentages of CD4+ T cells in the draining LNs and spleen (Fig. 1 J). Moreover, IFN-γ and IL-17A production in the Dock8pri/pri CD4+ T cells, as well as the percentages of Foxp3+CD4+ regulatory T (T reg) cells in the Dock8pri/+mice, was not changed (Fig. 1 K). These results led us to speculate whether Dock8 regulated the induction of EAE mainly via affecting CD4+ T cell migration.
CD4+ T cells from Dock8pri/pri mice ameliorate EAE with reduced CNS infiltration and migration
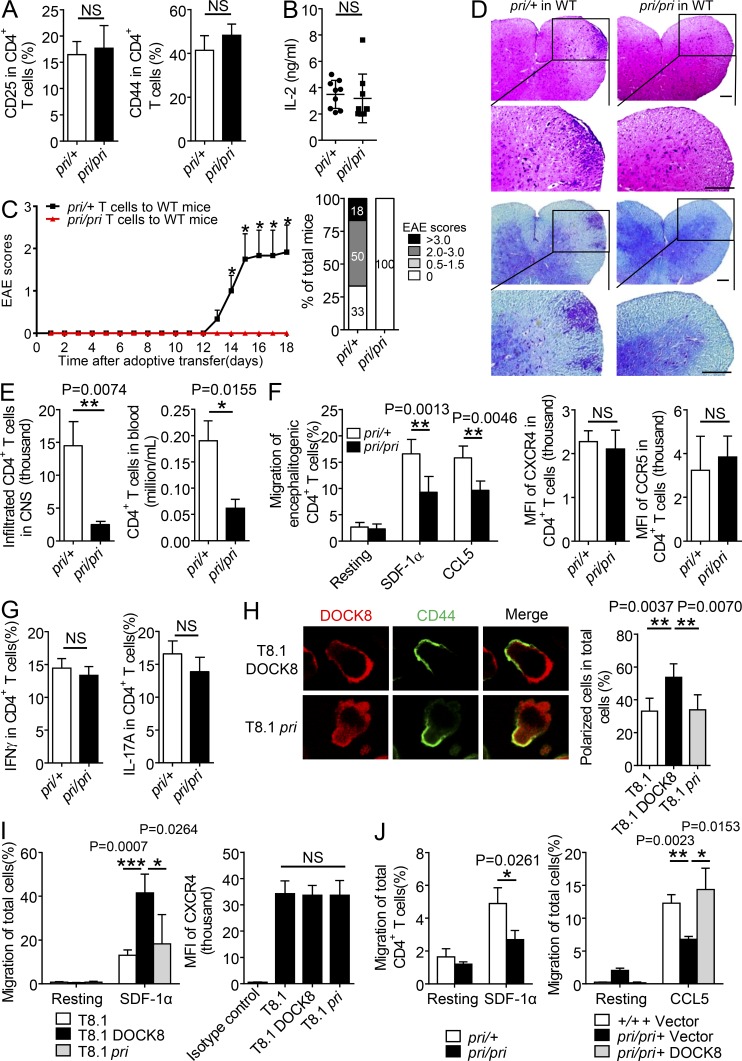
To answer this question, equal numbers of encephalitogenic CD4+ T cells were harvested from the Dock8pri/+ and Dock8pri/pri mice 8 d after MOG (35–55) immunization, and were transferred into the sublethally irradiated WT recipient mice. Before adoptive transfer, we confirmed that Dock8pri/+ and Dock8pri/pri encephalitogenic CD4+ T cells expressed the same levels of the T cell activation markers CD44 and CD25, as well as secreted similar amount of IL-2 (Fig. 2, A and B). More than 60% of the recipient mice developed EAE after being reconstituted with the Dock8pri/+ encephalitogenic CD4+ T cells. In contrast, no recipient mice showed the EAE phenotype after being injected with the Dock8pri/pri encephalitogenic CD4+ T cells (Fig. 2 C), which showed less immune cell infiltration (Fig. 2 D, top) and a lower degree of demyelination in the spinal cord (Fig. 2 D, bottom). Furthermore, we revealed a reduced numbers of the adoptively transferred Dock8pri/pri encephalitogenic CD4+ T cells in the CNS or blood (Fig. 2 E), which indeed showed impaired migration toward SDF-1α or CCL5 in a transwell assay ex vivo (Fig. 2 F, left). Nevertheless, the Dock8pri/pri encephalitogenic CD4+ T cells did not affect expression of the chemokine receptors CXCR4 and CCR5 (Fig. 2 F, right). In addition, Dock8pri/pri encephalitogenic CD4+ T cells secreted IFN-γ and IL-17A at levels that were similar to the control cells (Fig. 2 G).
Figure 2.
CD4+ T cells from Dock8pri/pri mice ameliorate EAE with reduced CNS infiltration and migration. (A and B) The encephalitogenic CD4+ T cells were purified from Dock8pri/+ or Dock8pri/pri mice to check surface expression of CD44 and CD25 by FACS (A). IL-2 concentrations were checked by ELISA in the supernatants of Dock8pri/pri or Dock8pri/+ encephalitogenic CD4+ T cells stimulation with MOG for 3 d (B). n = 6. (C–E) Adoptive transfer of encephalitogenic Dock8pri/+ and Dock8pri/priCD4+ T cells into the sublethally irradiated WT mice (n = 6) to assess clinical scores & EAE incidence (C). (D) H&E staining and Luxol blue staining of the representative tissue sections of the spinal cords. Bars, 70 μm. (E) Total number of CD4+ T cells in the CNS and in the blood by flow cytometry. n = 6. (F) The encephalitogenic CD4+ T cells were isolated from the Dock8pri/+ and Dock8pri/pri mice at day18 after EAE induction for SDF-1α– or CCL5-induced migration (left), or for a FACS assay to check the surface expression of CXCR4 and CCR5 (right). n = 4–6. (G) The percentages of IFN-γ+ cells and IL-17A+ cells in CD4+ T cells in spleen from the recipient mice presented in C. n = 5. (H–I) T8.1 cells were transfected with FLAG-tagged DOCK8 or the pri mutant and treated with or without SDF-1α, followed by immunostaining with anti-FLAG and anti-CD44 (H; bar, 5 µm; n = 50), or for a transwell assay (I; left; n = 3 per group), or for a FACS assay to check the surface expression of CXCR4 (I, right). n = 4. (J) The migration of CD4+ T cells from naive Dock8pri/+ or Dock8pri/pri mice was examined in response to SDF-1α (left). Or WT and Dock8pri/pri CD4+ T cells were reconstituted with the vector or DOCK8 to assess T cell migration in response to CCL5 (right). n = 3. NS, not significant (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.005. Data are representative of three independent experiments (C, mean ± SEM; E–F and H–I, mean ± SD), or two experiments (A, B, G, and J, mean ± SD). Statistical significance was determined using unpaired Student’s t test.
To accomplish directional migration to the sites of inflammation in the CNS, T cells need to polarize and transmigrate through the blood vessels. A polarized migrating T cell displays a typical hand-mirror–like morphology with key molecules being redistributed at the front region or the back tail (i.e., CD44 accumulation in the uropod; Ransohoff et al., 2003; Engelhardt and Ransohoff, 2005). We overexpressed wild-type DOCK8 or the mutant pri into the T cell line T8.1 cells. T8.1 cells expressed endogenous DOCK8, and the exogenous wild-type DOCK8 or the mutant pri did not affect surface CXCR4 expression levels (Fig. 2 I, right). In response to SDF-1α stimulation, >50% of the DOCK8-expressing T8.1 cells polarized with the DOCK8 translocation at the leading edge and CD44 accumulation in the uropod (Fig. 2 H); this was in agreement with the enhanced T cell migration in the transwell assay (Fig. 2 I, left). In contrast, only 30% of the mutant pri-expressing T8.1 cells showed CD44 accumulation in the uropod (Fig. 2 H), which failed to enhance cell migration (Fig. 2 I, left). In consistent, primary CD4+ T cells from the Dock8pri/pri mice reduced migration toward SDF-1α compared with those from the Dock8pri/+ mice (Fig. 2 J, left). Importantly, reconstitution of DOCK8 into Dock8pri/pri CD4+ T cells could restore CD4+ T cell migration (Fig. 2 J, right). Collectively, we have demonstrated that Dock8pri/pri CD4+ T cells are defective in chemokine-induced migration and protective against EAE.
LRCH1 is identified as a new binding partner of DOCK8
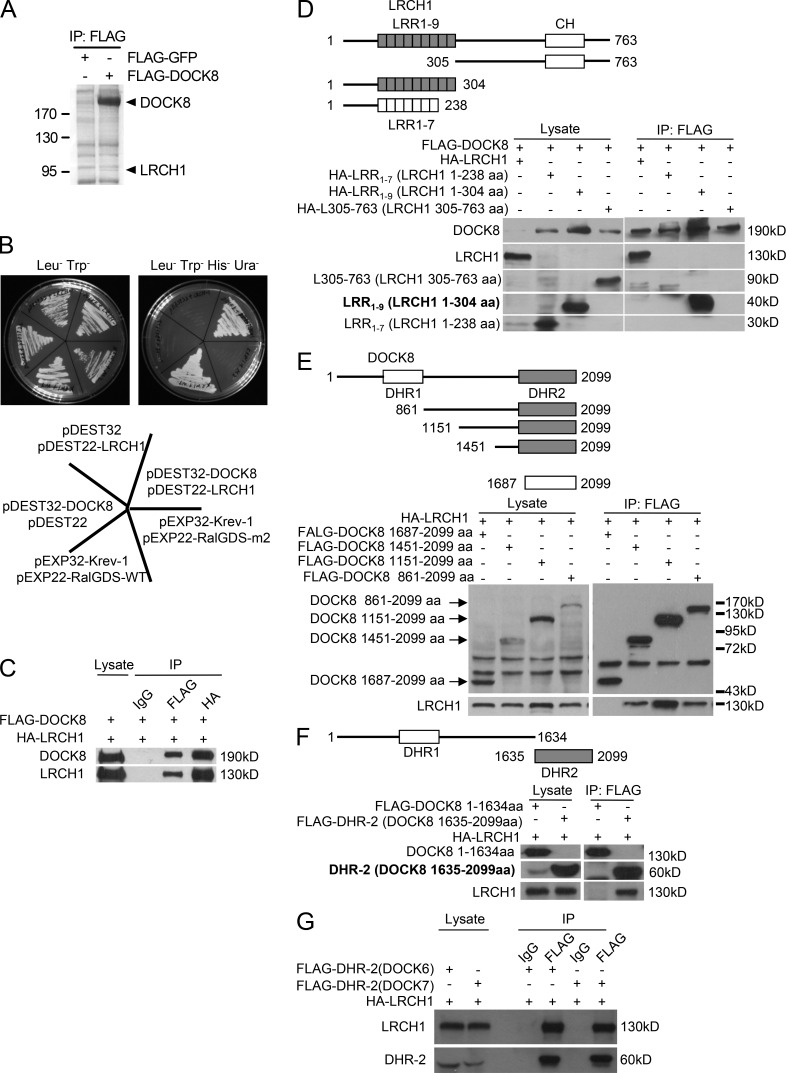
To understand the underline mechanism of DOCK8 function in T cell migration and EAE/MS, we searched DOCK8’s binding partners using mass spectrometry and yeast two-hybrid assays (see Materials and methods). Immunoprecipitation using an anti-FLAG antibody was performed and the pull-down proteins at 95 kD were identified by the mass spectrometry analysis from T8.1 cells, which was stably transfected to overexpress FLAG-tagged DOCK8. A specific band at a molecular weight 95 kD was identified as LRCH1 (Leucine-rich repeat [LRR] and calponin homology [CH] domain-containing protein 1; Fig. 3 A). In the yeast two-hybrid assay, DOCK8 was used as bait for screening a human cDNA library encoding >12,794 human genes (hORFeome V5.1). Positive clones were selected and LRCH1 was again identified in this method (Fig. 3 B). Currently, very limited studies have been reported regarding the function of LRCH1. The function of LRCH was initially reported in Drosophila, which has only one isoform named dLRCH. dLRCH colocalizes with F-actin and functions as a cytoskeleton regulator (Foussard et al., 2010). However, it is still a controversy about whether a C/T transition single-nucleotide polymorphism (SNP; rs912428) in LRCH1 is a risk factor for human osteoarthritis. We were therefore excited to explore whether and how LRCH1 functioned together with DOCK8 in the regulation of EAE.
Figure 3.
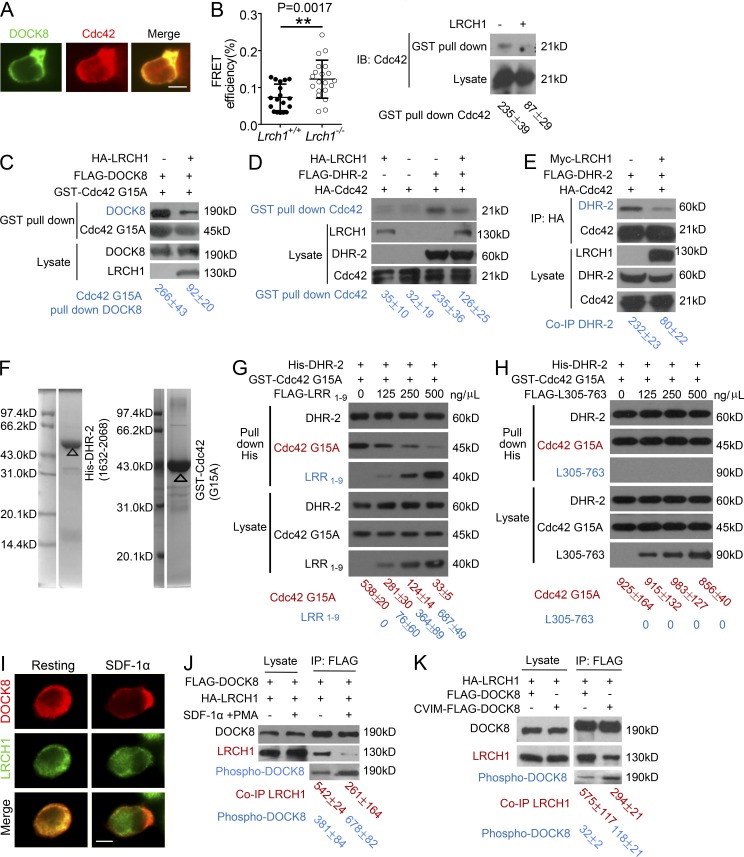
Identification of LRCH1 as a new binding partner of DOCK8. (A) An anti-FLAG IP was performed with FLAG-DOCK8–transfected T8.1 cells for the mass spectrometry assay, and the Coomassie blue staining is shown. (B) DOCK8 interaction with LRCH1 in the yeast two-hybrid system. DOCK8 and the pDEST22 vector, LRCH1 and the pDEST32 vector, and DOCK8 and LRCH1 were cotransfected into the yeast strain Mav 203-activated expression of β-glycosidase. Krev-1 and RalGDS-WT were cotransfected as a positive control. Krev-1 and RalGDS-m2 were cotransfected as a negative control. (C–F) 293T cells were cotransfected with FLAG-DOCK8 and HA-LRCH1 (C); FLAG-DOCK8 and HA-LRCH1 or its deletion mutants (HA-LRCH1 1–238 aa, 1–304 aa, or 305–763 aa; D); HA-LRCH1 and FLAG-DOCK8 or its mutants (DOCK8 861–2099 aa, 1151-2099 aa, 1451-2099 aa, and 1687–2099 aa, or 1–1634 aa, 1635–2099 aa; E and F) for immunoprecipitation followed by immunoblotting with the indicated antibodies. (G) 293T cells were cotransfected with HA-tagged LRCH1 and FLAG-tagged the DHR2 domain of DOCK6 or DOCK7. Cell lysates were immunoprecipitated with anti-FLAG antibody and followed by immunoblotting with the indicated antibodies. Data are representative of three experiments.
We first verified the interaction between DOCK8 and LRCH1 by immunoprecipitation using 293T cells that were transfected with a FLAG-tagged DOCK8 and an HA-tagged LRCH1. The FLAG-tagged DOCK8 could pull down the HA-tagged LRCH1 and vice versa (Fig. 3 C). Next, we truncated DOCK8 and LRCH1 to map their binding regions. LRCH1 contains nine leucine-rich repeats (LRRs) and a single CH domain. We found that the N-terminal fragment containing all nine LRRs (i.e., LRR1-9 and 1-304aa) bound to DOCK8. In contrast, after losing two LRRs, the first seven LRRs (i.e., LRR1-7 and 1-238aa) lost this interaction (Fig. 3 D), suggesting the importance of the whole nine LRRs for this interaction. Additionally, the fragment containing 305–763 aa of LRCH1 failed to interact with DOCK8 (Fig. 3 D). We further found that the DHR-2 domain (i.e., 1635–2099 aa) of DOCK8 interacted with LRCH1 (Fig. 3 F), whereas the deletion of the two α-helixes in the DHR-2 domain (i.e., 1687–2099 aa) or the N-terminal fragment (i.e., 1–1634 aa) lost this interaction (Fig. 3 E). The DHR-2 domain is conserved in the DOCK-C family, which contains DOCK6, DOCK7, and DOCK8. We found that LRCH1 also interacted with the DHR-2 domain of DOCK6 and DOCK7 (Fig. 3 G). Whether LRCH1 could affect DOCK6- or DOCK7-induced cell function is to be defined. We have elucidated that the DHR-2 domain of DOCK8 interacts with the nine LRRs of LRCH1.
Lrch1 transgenic mice are resistant to EAE with reduced T cell migration
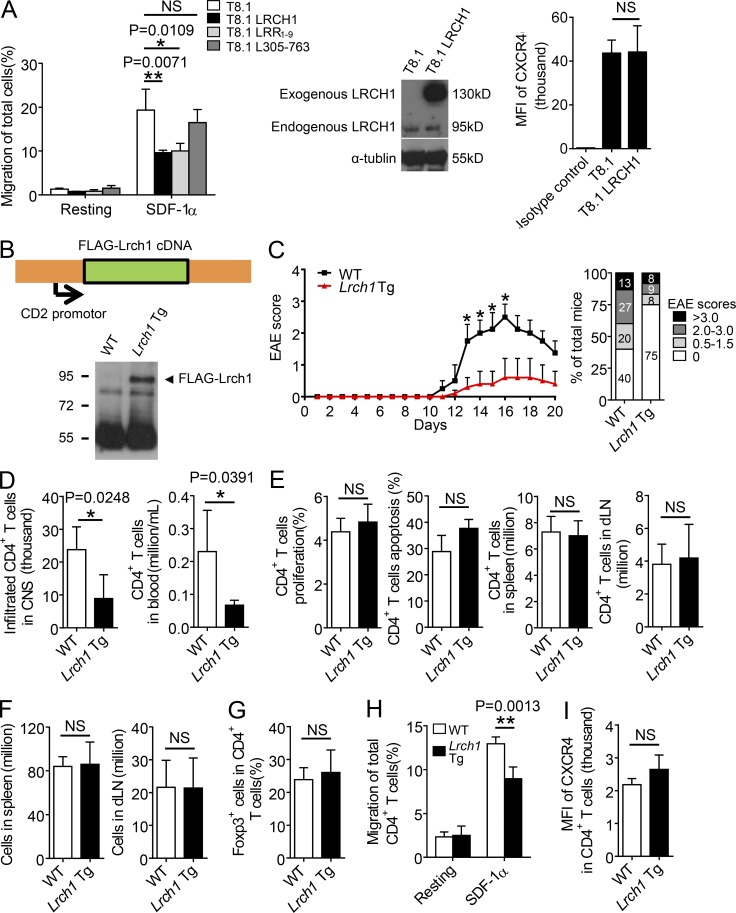
We next investigated the function of LRCH1 in T cells. T8.1 cells expressed endogenous murine LRCH1 (i.e., 90 kD), which were stably transduced to overexpress human wild-type LRCH1 (i.e., 130 kD; Fig. 4 A, middle) or its fragments LRR1-9 and L305-763 (305-763aa of LRCH1 that does not interact with DOCK8). To our surprise, T8.1 expressing the full length of LRCH1 decreased cell migration toward SDF-1α in a transwell assay (Fig. 4 A, left). The overexpression of LRR1-9, the fragment that binds DOCK8, impaired T cell migration toward SDF-1α, whereas the L305-763 fragment showed no interference (Fig. 4 A, left). Despite this, LRCH1 overexpression did not change the expression levels of the chemokine receptor CXCR4 (Fig. 4 A, right).
Figure 4.
Lrch1 transgenic mice are resistant to EAE with reduced T cell migration. (A) T8.1 cells were transfected with the vector control, LRCH1, or its fragments (LRR1-9, L305-763) for a transwell assay in response to SDF-1α (n = 3). FACS assay was performed to check the cell surface expression of CXCR4. The transfected exogenous human LRCH1 was detected at 130 kD and the endogenous murine LRCH1 in T8.1 cells was detected at 95 kD by Western blot. (B) The expression of FLAG-Lrch1 in thymus from Lrch1 transgenic mice was assessed by immunoblotting. (C) The clinical scores (left) and EAE incidence (right) of WT and Lrch1 transgenic mice induced by the MOG (35–55) peptide. n = 5. (D and E) The total numbers of CD4+ T cells in the CNS, blood (D), spleen, and draining LNs (E; middle right and right) of the WT or Lrch1 transgenic mice; percentages of BrdU+ or Annexin V+ CD4+ T cells in spleen were checked at the peak stage of EAE (E; left and middle left). n = 4–5. (F) The number of lymphocytes in spleen and draining LNs were counted from WT and Lrch1 Tg mice after EAE induction. n = 5. (G) The percentage of Foxp3+ CD4+ T reg cells in draining LNs from the WT or Lrch1 transgenic mice at the peak stage of EAE. n = 4. (H and I) The encephalitogenic CD4+ T cells were purified from the WT or Lrch1 transgenic mice for a transwell assay in response to SDF-1α (H), or for a FACS assay to check the surface expression of CXCR4 (I). n = 4–5. NS, not significant (P > 0.05); *, P < 0.05; **, P < 0.01. Data are representative of four experiments (A, mean ± SD), three experiments (C, mean ± SEM), or two experiments (D–I, mean ± SD). Statistical significance was determined using unpaired Student’s t test.
To examine the in vivo role of LRCH1, we generated Lrch1 transgenic mice (termed Lrch1 Tg) using a CD2 promoter–based vector that overexpressed FLAG-tagged Lrch1 only in T and B cells. Expression of FLAG-tagged Lrch1 was confirmed in primary T cells by Western blotting (Fig. 4 B). WT mice and Lrch1 transgenic mice were then immunized with the MOG (35–55) peptide to induce EAE. Compared with their WT littermates, Lrch1 transgenic mice were resistant to the development of EAE and showed reduced clinical scores (Fig. 4 C). This was in the agreement with a reduced numbers of the Lrch1 CD4+ Tg cells in the CNS or in the blood (Fig. 4 D). Importantly, we confirmed that this was not a result of the abnormal proliferation, apoptosis, or numbers of Lrch1 Tg CD4+cells in response to the MOG (35–55) peptide stimulation in vivo (Fig. 4 E). Additionally, the Lrch1 Tg mice showed normal numbers of lymphocytes in spleens and draining LNs (Fig. 4 F). The percentages of CD4+FoxP3+ T reg cells were also normal in Lrch1 transgenic mice after EAE induction (Fig. 4 G).
Next, encephalitogenic CD4+ T cells were purified from the Lrch1 transgenic mice when EAE was developed at the peak stage. Compared with the WT controls, encephalitogenic Lrch1 Tg CD4+ T cells impaired the ex vivo transmigration toward to SDF-1α (Fig. 4 H). Similar to our observation in the T8.1 cell line, encephalitogenic Lrch1 Tg CD4+ T cells did not affect the expression levels of CXCR4 (Fig. 4 I). These data together suggest that as a new binding partner of DOCK8, LRCH1, might play a distinctive role in T cell migration and the development of EAE.
Adoptive transfer of Lrch1 KO CD4+ T cells accelerates EAE with enhanced T cell migration
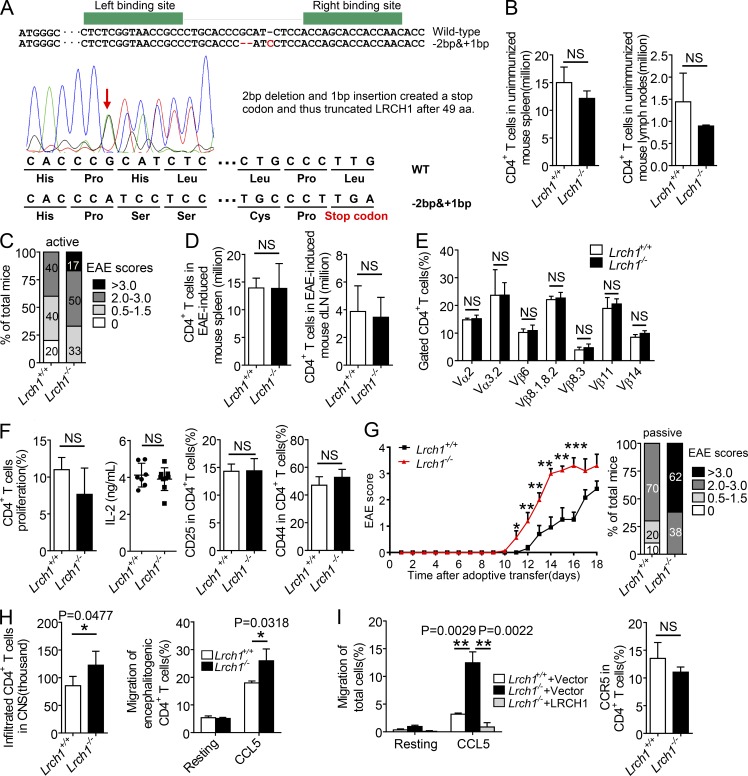
To further validate the opposite effect of LRCH1 from DOCK8, we generated Lrch1 KO mice by TALEN technology via targeting the first exon of Lrch1. One founder carried a 2-nt deletion and a 1-nt insertion in the ORF of the Lrch1 gene and created a stop code to KO Lrch1 (Fig. 5 A). The homozygous Lrch1-deficient mice (Lrch1−/−) were viable, fertile and showed normal populations of CD4+ cells in the spleen and LNs (Fig. 5 B). We induced active EAE using Lrch1−/−mice, which manifested a greater disease severity compared with WT mice (Fig. 5 C). During the induction of EAE, Lrch1 deficiency did not change the numbers of CD4+ T cells in spleen and draining LNs (Fig. 5 D). We confirmed that TCR usage in CD4+ T cells in response to MOG peptide stimulation was not changed in Lrch1−/− mice relative to that in Lrch1+/+ mice (Fig. 5 E). Moreover, Lrch1-deficient CD4+ T cells also proliferated and produced IL-2 at normal levels, and expressed normal levels of the activation markers CD44 and CD25 (Fig. 5 F).
Figure 5.
Adoptive transfer of Lrch1 KO CD4+T cells accelerates EAE with enhanced T cell migration. (A) Generation of Lrch1 KO mice. The exon 1 of the Lrch1 gene was specifically targeted by TALEN, and DNA sequencing confirmed the nucleotide mutation in the Lrch1 locus adjacent to the FOKI cleavage site (arrow). (B) Numbers of CD4+ T cells in spleen and LNs from unimmunized mice. n = 3. (C) EAE incidence of WT and Lrch1 KO mice in response to MOG (35–55) treatment. n = 5. (D–F) Numbers of CD4+ T cells in spleen and draining LNs (D); TCR usage analyzed by anti-TCR Vα and anti-TCR Vβ antibodies (E); and percentages of BrdU+ CD4+ T cells, IL-2 secretion, and the surface expression of CD25 and CD44 in CD4+ T cells from the draining LNs (F) of WT and Lrch1 KO mice after EAE induction. n = 5. (G) The sublethally irradiated WT recipient mice were reconstituted with WT or Lrch1−/− encephalitogenic CD4+ T cells to assess their clinical scores (left) and EAE incidence (right). n = 5. (H) The total numbers of WT or Lrch1−/− encephalitogenic CD4+ T cells in the CNS from the sublethally irradiated WT recipient mice presented in G. The encephalitogenic CD4+ T cells were purified from the WT or Lrch1−/− mice for a transwell assay in response to CCL5 (right). n = 5. (I) CD4+ T cells were reconstituted with LRCH1 or the vector control, for a transwell assay in response to CCL5 (left). The surface expression of CCR5 in the spleen CD4+ T cells from WT and Lrch1 KO mice after EAE induction (right). n = 3–4. NS, not significant (P > 0.05); *, P < 0.05; **, P < 0.01. Data are representative of three experiments (G, mean ± SEM), or two experiments (B–F and H–I, mean ± SD). Statistical significance was determined using unpaired Student’s t test.
To verify the role of Lrch1−/− CD4+ T cells in the development of EAE, we isolated equal numbers of encephalitogenic CD4+ T cells from the MOG (35–55) peptide-treated WT mice or Lrch1−/− mice, and transferred them into the sublethally irradiated WT recipient mice. The Lrch1−/− encephalitogenic CD4+ T cell–reconstituted mice showed an earlier disease onset with higher clinical scores (Fig. 5 G). Consistently, more Lrch1−/− encephalitogenic CD4+ T cells migrated into the CNS (Fig. 5 H, left), which also displayed an enhanced migration toward CCL5 in a transwell assay (Fig. 5 H, right). Reconstitution of LRCH1 expression in Lrch1−/− CD4+ T cells restored cell migration to normal levels as that in WT CD4+ T cells (Fig. 5 I, left). Despite of this, Lrch1−/− CD4+ T cells expressed normal levels of CCR5 (Fig. 5 I, right). After collecting our data from the Lrch1 Tg and Lrch1−/− mice, we suggest that LRCH1 protects mice against EAE as a result of the reduced CD4+ T cell migration.
LRCH1 attenuates DOCK8-mediated Cdc42 activation for T cell migration
Considering the opposite role of DOCK8 and LRCH1 in T cell migration without affecting the expression of chemokine receptors, we speculated how LRCH1 and DOCK8 cooperated together in response to chemokine stimulation. As a GEF protein, DOCK8 binds and activates Cdc42 (Harada et al., 2012). Using previously reported methods, we incubated T8.1 cells with the tetanus toxoid peptide (Ttox) peptide-pulsed APCs (i.e., APCs and L625.7) to form cell conjugates, and noticed that DOCK8 co-localized with Cdc42 at the immunological synapses (Fig. 6 A). Moreover, we examined Cdc42 activity in WT and Lrch1−/− CD4+ T cells by transfection of a fluorescent resonance energy transfer (FRET)–based biosensor Raichu-Cdc42, which consists of Cdc42, Cdc42-binding domain of PAK1, and a pair of green fluorescent protein mutants to monitor Cdc42 activation in vivo (Itoh et al., 2002; Shen et al., 2008). In response to SDF-1α treatment, FRET efficiency was increased in Lrch1−/− CD4+ T cells compared with that in WT cells, which indicates higher Cdc42 activation in Lrch1−/−CD4+ T cells (Fig. 6 B, left). Activated Cdc42-GTP interacts with the downstream effector PAK1, and we next determined how LRCH1 could affect the amount of activated Cdc42-GTP by a GST pull down assay using the GST-fused CRIB domain of PAK1 (Benard et al., 1999). Overexpression of LRCH1 in T8.1 cells reduced the amount of activated Cdc42-GTP by a GST pull-down assay compared with the control T8.1 cells (Fig. 6 B, right). Collectively, we suggest that LRCH1 functions as a negative regulator of Cdc42 activation. This led us to speculate whether LRCH1 interfered with Cdc42 activation by modulating the GEF activity of DOCK8.
Figure 6.
LRCH1 attenuates DOCK8-mediated Cdc42 activation for T cell migration. (A) T8.1 cells were transfected with FLAG-DOCK8, incubated with Ttox peptide-pulsed L625.7 cells to form cell conjugates, followed by immunostaining to visualize DOCK8 and Cdc42. Bar, 5 µm. (B) The FRET efficiency of biosensor Raichu-Cdc42 between WT and Lrch1−/−CD4+ T cells in response to SDF-1αtreatment. FRET efficiency was measured with donor dequenching approach, and was calculated as E = (Post −[Pre/Post]) × 100%, where Post and Pre represents the donor fluorescence before and after photo bleaching (left). n = 20. NS, not significant (P > 0.05); **, P < 0.01. Activated Cdc42 was precipitated by a GST-CRIB-PAK1 pull-down assay in T8.1 cells that overexpressed LRCH1 or the vector control (right). (C) 293T cells were transfected with FLAG-DOCK8 with or without HA-LRCH1, and the amount of DOCK8 binding to the GST-Cdc42G15A–coated beads was used to evaluate the GEF activity of DOCK8. (D) 293T cells were cotransfected with FLAG-DHR-2, HA-Cdc42 with or without HA-LRCH1. The cell lysates were subjected to a GST-CRIB-PAK1 pull-down assay to precipitate the active Cdc42. (E) 293T cells were transfected with HA-Cdc42, Myc-LRCH1, and FLAG-DHR-2, followed by immunoprecipitation with anti-HA and immunoblotting with anti-HA or anti-FLAG. (F) The purity of the purified His-DHR-2 (1632–2068 aa) and GST-Cdc42 G15A protein from E. coli was determined by Coomassie blue staining. (G and H) Increasing amounts of FLAG-LRR1-9 (G) or FLAG-L305-763 (H) were added into the solution containing the purified recombinant proteins His-DHR-2 and GST-Cdc42G15A, incubated and then subjected for precipitation using anti-His antibody. (I) T8.1 cells, which were transfected with FLAG-DOCK8 and HA-LRCH1, were treated with or without SDF-1α to assess localization of DOCK8 (red, top) and LRCH1 (green, middle). Bar, 5 µm. (J and K) T8.1 cells expressing FLAG-DOCK8 and HA-LRCH1 were treated or untreated with SDF-1α and PMA (J). 293T cells were transfected with HA-LRCH1, FLAG-DOCK8, or the membrane-localized CVIM-FLAG-DOCK8 (K), followed by immunoprecipitation with anti-FLAG to detect the phosphorylation levels of DOCK8 and the amount of LRCH1. Data presented are representative of two independent experiments (A–D, G, H, and J), or three independent experiments (E, I, and K). Intensity of the immunoblots was quantified and shown at the bottom (mean ± SD). Statistical significance was determined using unpaired Student’s t test.
Previous studies found that activated GEFs preferentially bind to guanine nucleotide–free forms of GTPases, suggesting that the Cdc42G15A mutant could be used to measure GEF activity (García-Mata et al., 2006). We therefore overexpressed DOCK8 with or without LRCH1 in the 293T cells and measured the amount of DOCK8 binding to the GST-fused-Cdc42G15A. Interestingly, fewer DOCK8 interacted with Cdc42G15A when LRCH1 was also present (Fig. 6 C). In addition, we transfected the FLAG-tagged catalytic DHR-2 domain of DOCK8 with or without LRCH1, followed by a GST pull-down assay to measure how LRCH1 affected Cdc42 activation. Although DOCK8 DHR-2 enhanced the amount of GTP-bound-Cdc42, the coexpression of LRCH1 substantially decreased this effect (Fig. 6 D). This led us to speculate whether LRCH1 and Cdc42 competed for binding to the DHR-2 domain of DOCK8. Indeed, in the presence of LRCH1, Cdc42 precipitated less DOCK8 DHR-2 (Fig. 6 E). To further confirm this phenotype, we purified the recombinant proteins His-DHR-2, GST-Cdc42G15A from Escherichia coli, and the recombinant proteins FLAG-LRR1-9 or FLAG-L305-763 from 293T cells. The purity and specificity of the purified His-DHR-2 and GST-Cdc42G15A protein from E. coli was shown in a Coomassie blue staining (Fig. 6 F). When GST-Cdc42G15A formed a complex with His-DHR-2 in vitro, addition of the increasing amount of FLAG-LRR1-9 could significantly decrease the amount of GST-Cdc42G15A binding to His-DHR-2 in a dose-dependent manner (Fig. 6 G). As an important negative control, addition of the purified recombinant protein FLAG-L305-763 did not affect the formation of the complex containing GST-Cdc42G15A and His-DHR-2 (Fig. 6 H). We propose the novel finding that LRCH1 attenuates DOCK8-mediated Cdc42 activation for T cell migration.
Upon SDF-1α stimulation, DOCK8 redistributed from the cytoplasm to the cell membrane. In contrast, LRCH1 was still ubiquitous in the cytoplasm (Fig. 6 I). We questioned whether chemokine signaling induced DOCK8 separation from LRCH1. Indeed, SDF-1α treatment could increase DOCK8 phosphorylation, and substantially reduced its interaction with LRCH1 (Fig. 6 J). To further confirm this, we generated a membrane-bound DOCK8 by fusing the CVIM motif to the C terminus of DOCK8. Critically, the membrane-localized CVIM-FLAG-DOCK8 recruited less LRCH1 and exhibited an enhanced phosphorylation (Fig. 6 K). This suggests that chemokine-induced DOCK8 phosphorylation might release DOCK8 from LRCH1, which is then able to interact with Cdc42 for further activation.
PKC phosphorylates DOCK8 for separation from LRCH1
The next critical question was which kinases could phosphorylate DOCK8. We searched the DOCK8 phosphorylation sites in the PhosphoSitePlus database, which provides the possible in vivo phosphorylation sites of proteins from published studies or from high-throughput phosphorylation site discovery programs. The database showed that the C-terminal motif of DOCK8 (i.e., 2077–2087 aa: SQKRDSFHRSS) is phosphorylated (Fig. 7 A), and kinases including PKCα and AKT are predicted to phosphorylate this motif by the program group base prediction 3.0. We therefore treated cells with either PKCα or AKT inhibitors, and then pulled down the membrane-bound CVIM-DOCK8 to assess DOCK8 phosphorylation. The PKCα inhibitor substantially reduced DOCK8 phosphorylation levels, whereas the AKT inhibitor showed little effect (Fig. 7 B). Moreover, the PKCα inhibitor abolished DOCK8-induced cell migration in response to SDF-1α stimulation (Fig. 7 C). This observation is in agreement with previous studies that PKCα is recruited to membrane for cytoskeleton rearrangement and chemokine-induced cell migration (Sun et al., 2014). Interestingly, PKCα was previously reported as a susceptible gene in MS patients (Barton et al., 2004).
Figure 7.
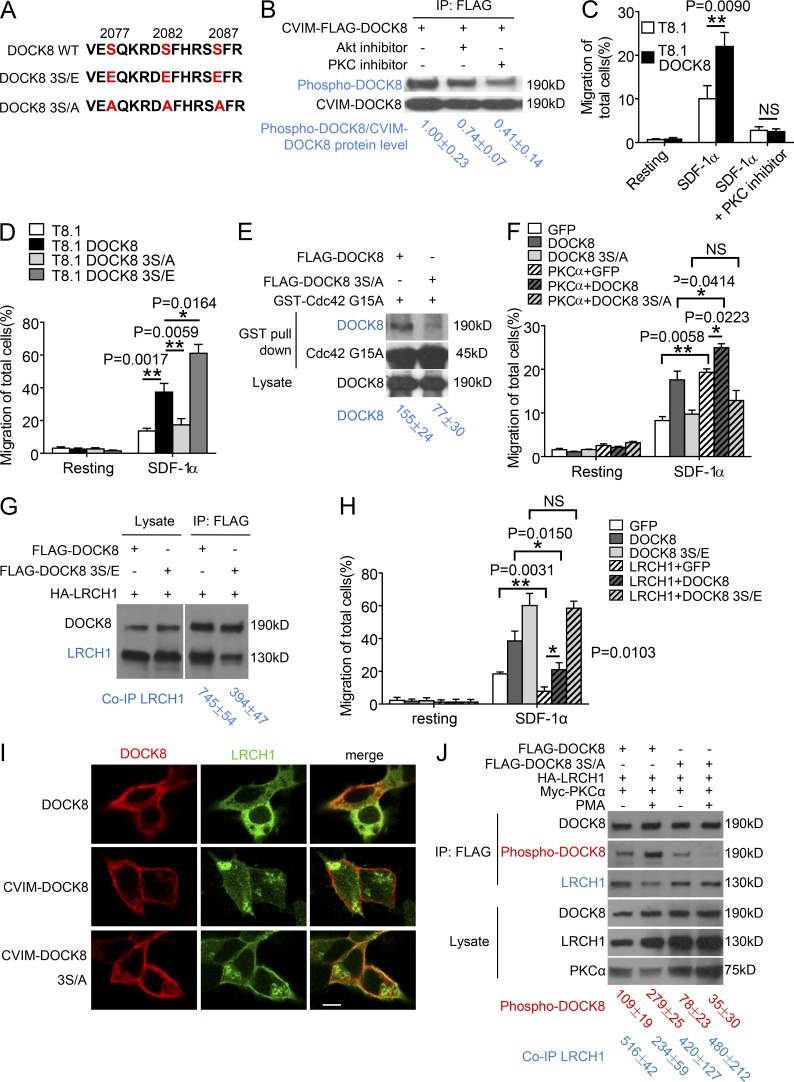
PKCα phosphorylates DOCK8 for separation from LRCH1. (A) The amino acid sequence (2075–2089) of DOCK8 and its mutants depict the three key serine residues. (B) 293T cells expressing CVIM-FLAG-DOCK8 were untreated or treated with the AKT and PKCα inhibitors, followed by immunoprecipitation with anti-FLAG to detect DOCK8 phosphorylation levels. (C and D) Migration of the T8.1 cells expressing the vector control, DOCK8, or the mutant 3S/E or 3S/A were examined by a transwell assay in response to SDF-1α in the presence or absence of the PKC inhibitor. n = 3. (E) 293T cells were transfected with FLAG-DOCK8 or the 3S/A mutant, followed by a GST-Cdc42G15A pull-down assay to measure their GEF activity. (F) The migration of T8.1 cells coexpressing PKCα with GFP, DOCK8, or the 3S/A mutant was assessed by a transwell assay. n = 3. (G) 293T cells were transfected with HA-LRCH1, FLAG-DOCK8, or 3S/E, followed by immunoprecipitation with anti-FLAG to analyze their binding to LRCH1. (H) The vector control, DOCK8, or the mutant 3S/E were coexpressed with or without LRCH1 into T8.1 cells and migration was examined by a transwell assay in response to SDF-1α. n = 3. (I) The localization of LRCH1 (green), CVIM-DOCK8, or CVIM-DOCK8 3S/A (red) was examined in 293T cells by immunostaining. Bar, 5 µm. (J) 293T cells were transfected with FLAG-DOCK8 or 3S/A together with HA-LRCH1 and Myc-PKCα, stimulated with or without PMA, followed by immunoprecipitation with anti-FLAG to analyze DOCK8 phosphorylation levels and binding to LRCH-1. NS, not significant (P > 0.05); *, P < 0.05; **, P < 0.01. Data are representative of three experiments (C, D, F, H, mean ± SD) or two experiments (B, E, G, and J). Intensity of the immunoblots was quantified and shown at the bottom (mean ± SD). Statistical significance was determined using unpaired Student’s t test.
Because PKCα phosphorylates serines (Parekh et al., 2000; Parker and Murray-Rust, 2004), we mutated the three key serines in the C-terminal motif of DOCK8 (Ser2077/2082/2087) to either glutamic acid or alanine, which, respectively, mimicked the phosphorylated DOCK8 (i.e., termed 3S/E) or abolished DOCK8 phosphorylation (i.e., termed 3S/A). Compared with the DOCK8-transfected T8.1 cells, the overexpression of 3S/E further enhanced cell migration in response to SDF-1α stimulation, whereas 3S/A markedly inhibited this effect (Fig. 7 D). Consistently, the 3S/A-transfected cells showed reduced GEF activity in the Cdc42G15A pull-down assay (Fig. 7 E). Moreover, when coexpression of PKCα with DOCK8 further promoted cell migration to SDF-1α, the 3S/A mutant failed to synergize with PKCα to increase migration (Fig. 7 F). Taken our data together, we suggest that PKCα phosphorylates DOCK8 at the Ser2077/2082/2087 sites to promote T cell migration.
Compared with the amount of LRCH1 binding to WT DOCK8, we found that the DOCK8 3S/E mutant (mimicking the phosphorylated DOCK8) recruited less LRCH1 (Fig. 7 G). Interestingly, coexpression of LRCH1 could block DOCK8-induced T cell migration, whereas LRCH1 could not inhibit the DOCK8 3S/E mutant-induced T cell migration (Fig. 7 H). In contrast, the membrane-bound CVIM-3S/A (abolishing DOCK8 phosphorylation) could still recruit more LRCH1 compared with the membrane-bound DOCK8 (Fig. 7 I). Furthermore, given that PMA stimulation triggers PKCα activation, DOCK8 was phosphorylated, which in turn reduced its interaction with LRCH1 (Fig. 7 J, lane 2 vs. lane 1). In contrast, the 3S/A mutation failed to be phosphorylated by PMA and still bound to LRCH1 at similar levels as that in resting T8.1 cells (Fig. 7 J, lane 4 vs. lane 3). Together, we have uncovered a novel mechanism that PKCα phosphorylates DOCK8 for separation from LRCH1, leading to Cdc42 activation and T cell migration.
Discussion
DOCK8 was recently identified as a key modulator for immune cell function, including B cell adhesion and integrin activation (Jabara et al., 2012). In agreement with this phenotype, Cdc42 was suggested to be DOCK8’s binding partner for cytoskeleton rearrangement and T cell migration. In this study, we identified LRCH1 as a novel DOCK8-interacting protein to restrain the GEF activity of DOCK8, resulting in the inhibition of Cdc42 activation and T cell migration. During the in vivo MOG (35–55) peptide-induced EAE, we observed the protective role of LRCH1 against EAE as a result of a blockage of CD4+ T cell migration into the CNS as demonstrated by Lrch1 transgenic or Lrch1 KO mice. Importantly, we also elucidated that LRCH1 deficiency did not affect CD4+ T cell proliferation, apoptosis, activation and production of IL-2. Next, we identified that LRCH1 competes with Cdc42 for binding to the catalytic DHR-2 domain of DOCK8 and restricts the GEF activity of DOCK8. Other members in the DOCK family contain an SH3 domain, which provides a self-inhibition mechanism to restrain their GEF activity (Lu et al., 2005). For example, the SH3 domain in DOCK1 interacts with its own DHR-2 domain and inhibits Rac binding (Meller et al., 2008). In contrast, DOCK8 has no SH3 domain. Our study suggests a possible mechanism that DOCK8 interacts with LRCH1 to lock DOCK8 at low GEF activity in resting T cells.
When T cells are stimulated with external signals such as chemokines, PI3K is activated and generates the product PI(3,4,5)P3 on the membrane, which recruits DOCK8 through its DHR-1 domain (Côté et al., 2005). We observed that DOCK8 was separated from LRCH1 in the cytoplasm, and relocated to the leading edge with Cdc42 after chemokine stimulation. Our findings are in agreement with previous reports that DOCK8 controls Cdc42 activity, specifically on the leading edge of the cell membrane rather than the globally distributed Cdc42 (Harada et al., 2012). Combining these observations, we propose a model that DOCK8 is located in the cytoplasm and interacts with LRCH1 to block Cdc42 binding in a resting T cell. In response to chemokine stimulation, DOCK8 is separated from LRCH1 and exposes its DHR-2 domain for Cdc42 activation, whereas its DHR-1 domain is recruited by PI(3,4,5)P3 to relocate DOCK8 at the leading edge of a migrating T cell.
In line with this scenario, we further demonstrated that in response to chemokine stimulation, PKCα phosphorylates DOCK8 at the Ser2077/2082/2087 sites to promote DOCK8 separation from LRCH1. The Ser2077/2082/2087 phosphoswitch in DOCK8 might alter the local electrostatic potential within the binding surface between DOCK8 and LRCH1, which perturbs the intermolecular interaction. Furthermore, we provided evidence that the DOCK8 3S/E mutant, but not the 3S/A mutant, could enhance T cell migration. Other studies have suggested that PKCα increases the integrin αIIbβ3 activation for platelet adhesion through the PKD–Rap1 pathway (Medeiros et al., 2005; Han et al., 2006; Abram and Lowell, 2009). We found that neither DOCK8 nor LRCH1 interacted with Rap1, and the activated Lrch1−/− platelets displayed normal levels of active Rap1 using a GST-RBD pull-down assay (unpublished data). Our study has therefore provided evidence to suggest DOCK8 as a new substrate of PKCα, which then activates Cdc42 to enhance T cell migration.
Because DOCK8 expression in CD4+ T cells was significantly enhanced in the acute phase of EAE, it is interesting to further explore whether DOCK8 in autoreactive CD4+ T cells might be used as valuable biomarkers for the assessment of prognosis or drug responses in MS patients. Considering that drugs targeting cell trafficking to the CNS are now applied to treat MS patients, for example IFN-β–derived products (De Jager and Hafler, 2007), targeting DOCK8 may offer a new strategy to ameliorate MS. DOCK8 belongs to the DOCK-C family, which contains DOCK6 and DOCK7. The DOCK-C family also contains the DHR-1 domain for membrane binding to PIP3, and the DHR-2 domain for activation of the Rho small GTPases. We found LRCH1 also interacted with the DHR-2 domain of DOCK6 and DOCK7. And overexpression of LRCH1 reduced the amount of activated Cdc42 in T8.1 cells by a GST pull-down assay. Considering that the global Cdc42 activation was intact in Dock8-deficient dendritic cells examined by the same method, this might be caused by the different cell types (T cells versus dendritic cells) used to test Cdc42 activation. Also, it is possible that LRCH1 could restrain Cdc42 activation via binding to all the DOCK-C family members, including DOCK6, DOCK7, and DOCK8. This possibility is to be further investigated. However, whether DOCK6 and DOCK7 are involved in the development of MS is not clear. In addition, little data is available about the function of LRCH1 in mammalian cells. There are conflicting reports about whether a C/T transition SNP (rs912428) in LRCH1 is a risk factor for human osteoarthritis (Snelling et al., 2007; Jiang et al., 2008). Considering that SNP polymorphisms of PKCα have been identified in MS patients, and PKCα-deficient mice are resistant to EAE (Barton et al., 2004; Meisel et al., 2013; Paraboschi et al., 2014), this makes us even more interesting to further investigate whether SNP polymorphisms in DOCK8 or LRCH1 are associated with MS using a large population of MS samples.
Material and methods
Mice
The Dock8pri/pri mice were kindly provided by C.C. Goodnow (John Curtin School of Medical Research, The Australian Nation University, Canberra, Australia). To specifically overexpress Lrch1 in T or B lymphocytes, cDNA encoding murine Lrch1 was tagged with HA and inserted to the human CD2 plasmid (provided by Paul Love, Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD). The Lrch1 transgenic mice in the C57BL/6 mouse background were generated by X. Liu's group at the Shanghai Institute of Biochemistry and CellBiology, Chinese Academy of Sciences (CAS); the mice specifically overexpress FLAG-tagged Lrch1 only in T and B cells using a CD2-promoter based vector. The Lrch1 transgenic positive mice were identified by PCR with the following primers: forward, 5′-CACTCGGGACTTATGAACT-3′; reverse, 5′-GATCGTAAACTGTGGGTCT-3′. The Lrch1 KO mice were generated by transcription activator-like effector nuclease (TALEN) technology specifically targeting the exon 1 of Lrch1 (SIDANSAI Biotechnology Co.). PCR and DNA sequencing were used to identify the Lrch1 KO founder mice, which were selected and used for further breeding and characterization. All mice were bred under specific pathogenic–free conditions at the Animal Care Facility of Shanghai Institute of Biochemistry and Cell Biology (SIBCB, CAS, Shanghai, China). The animal experiments were performed in compliance with the guidance for the care and use of laboratory animals and were approved by the institutional biomedical research ethics committee of SIBCB.
Enrollment of patients and healthy subjects
Patients clinically diagnosed with MS or NMO and healthy volunteers from the outpatient clinic were enrolled from Huashan Hospital, Shanghai, China. After informed consent, blood samples from the subjects were collected and prepared in accordance with the guidelines of the Review Boards of Huashan Hospital (Shanghai, China).
Cell lines
T8.1 cells, a murine cell line, were stably transfected with an antigen receptor specific for Ttox, and the murine fibroblast cell line L625.7 cells were used as APCs that express HLA-DR*1102, CD80, ICAM-2 and ICAM-1 (Michel and Acuto, 1996). T8.1 cells expressed endogenous Dock8 and Lrch1. 293T is a Human Embryonic Kidney Cell line. T8.1 and 293T cells were cultured in DMEM supplemented with 10% (vol/vol) fetal calf serum and 50 U/ml penicillin/streptomycin.
Reagents
The following antibodies were purchased from eBioscience and used for FACS assay: anti–mouse CD3e (145-2C11), anti–mouse CD4 (GK1.5), anti–mouse CD8a (53–6.7), anti–mouse CD45R (B220; RA3-6B2), anti–mouse IL-17A (eBio17B7), anti–mouse IFN-γ (XMG1.2), anti–mouse CD11b (M1/70), anti–mouse CD29 (eBioHMb1-1), and anti–mouse CXCR4 (2B11). The following antibodies were purchased from BD: anti–mouse-Vα2 (B20.1), anti–mouse-Vα3.2 (553219), anti–mouse-Vβ6 (553194), anti–mouse-Vβ8.1/8.2 (118405), anti–mouse-Vβ8.3 (118603), anti–mouse-Vβ11 (125907), and anti–mouse-Vβ14 (553258). The following antibodies were purchased from to be added: anti–mouse CCR5 (HM-CCR5; BD), anti–mouse CD44 (IM7; eBioscience), and anti–mouse Foxp3 (FJK-16S; eBioscience). Anti–human CD4 (S5.3; Invitrogen), anti–human DOCK8 (sc-104911; Santa Cruz Biotechnology, Inc.), and anti–human LRCH1 (sc-84195; Santa Cruz Biotechnology, Inc.) were used for FACS or immunoblotting assay. Anti-Phosphoserine/threonine/tyrosine antibody (ab15556; Abcam), anti-FLAG (F3165; Sigma-Aldrich), anti-HA (H3663; Sigma-Aldrich), anti-c-Myc (C3956; Sigma-Aldrich), and anti-Cdc42 (sc-87; Santa Cruz Biotechnology, Inc.) were used for immunoprecipitation or immunoblot assay.
The induction of EAE
The encephalitogenic peptide MOG (35–55; GL Biochem) used to induce EAE had a purity of 95%. For EAE induction, 8–10-wk-old C57BL/6 mice were immunized s.c. with 200 µg MOG (35–55) in complete Freund’s adjuvant containing heat-killed Mycobacterium tuberculosis (H37Ra strain; 5 mg/ml; Sigma-Aldrich). Pertussis toxin (200 ng per mouse; EMD Millipore) in PBS was administered i.p. on days 0 and 2. Mice were examined daily for disease signs by researchers blinded to experimental conditions and were assigned scores on a scale of 0–5 as follows: 0, no clinical signs; 1, paralyzed tail; 2, paresis (weakness, incomplete paralysis of one or two hindlimbs); 3, paraplegia (complete paralysis of both hindlimbs); 4, paraplegia with forelimb weakness or paralysis; and 5, moribund state or death (Jin et al., 2009). For analysis of CNS infiltrates, brain and spinal cord tissues were collected from perfused mice and mononuclear cells were prepared by Percoll gradient centrifugation. For histological analysis, the same tissue samples were immediately fixed in 4% (wt/vol) paraformaldehyde. Paraffin-embedded sections of spinal cord were stained with H&E or with Luxol fast blue for analysis of inflammation or demyelination, respectively. For adoptive transfer of encephalitogenic CD4+ T cells experiments, splenocytes were isolated from mice 8 d after active MOG immunization and cultured for 3 d in the growth media containing 20 µg/ml MOG; 2 × 106 encephalitogenic CD4+ T cells were i.v. injected into the sublethally irradiated recipient mice.
Intracellular staining and flow cytometry
For intracellular cytokine staining, cells obtained from DLNs of mice with EAE were incubated in a tissue culture incubator for 5 h at 37°C with phorbol 12-myristate 13-acetate (50 ng/ml; Sigma-Aldrich), ionomycin (1 µg/ml; Sigma-Aldrich), and Brefeldin A (10 µg/ml; Sigma-Aldrich). Surface staining was performed with the corresponding fluorescence-labeled surface antibodies in PBS buffer for 30 min . After surface staining, cells were resuspended in Fixation/Permeabilization solution (Cytofix/Cytoperm kit; eBioscience), and intracellular cytokine staining was done according to the manufacturer’s protocol. For Foxp3 staining, cells were isolated from the EAE mice and prepared for intracellular staining using the Foxp3 Staining Buffer set as suggested (eBioscience). FACSCalibur (BD), FlowJo software, or Accuri C6 was used for flow cytometry.
Plasmids and qRT-PCR
Flag-tagged human DOCK8 (WT or pri mutation) were subcloned into the retroviral vector pMX-IRES-GFP; HA-tagged human LRCH1 was subcloned into the MIGR-IRES-GFP vector. These plasmids were transfected to 293T cells using the reagent pCL-10A, and the retroviral supernatants were collected to infect T8.1 cells, followed by sorting of GFP+ cells. Total RNA was isolated from cells or tissues using TRIzol Reagent (TIANGEN), and converted to cDNA using M-MLV reverse transcription (Takara Bio Inc.). Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed on a CFX-96 machine (Bio-Rad Laboratories) using SYBR Green master mix (DBI Bioscience).
CD4+ T cells transfection
A 293T platinum-E cell line (Plat-E; a gift from C. Xu, SIBCB, CAS, Shanghai, China) was cultured in DMEM supplemented with 10% (vol/vol) fetal calf serum and antibiotics, including puromycin and blasticidin. Plat-E cells were transfected with pMX-FLAG-DOCK8 or MIGR-HA-LRCH1 to produce retrovirus supernatants. CD4+ T cells were isolated from C57BL/6 mice and activated with 2 µg/ml anti-CD3 and 2 µg/ml anti-CD28. 24 h after activation, the cells were infected with retrovirus supernatants in the presence of 12 µg/ml polybrene by spinning at 2,500 rpm for 120 min, 30°C. The second infection was repeated in the next day, transduction efficiency was determined by the percentages of GFP+ cells by FACS. Overexpression levels of DOCK8 and LRCH1 were examined by intracellular staining with anti-FLAG or anti-HA. Chemotaxis assay was performed using these transduced CD4+ T cells and the percentages of GFP+CD4+ T cells before and after a transwell assay were analyzed by FACS. The migration efficiency was calculated by a formula Efficiency = (% GFP+ cells × total cells in the bottom well)/(% GFP+ cells × total cells before transwell).
Chemotaxis assay
The chemotaxis assay was performed using a transwell chamber (5 µm; Corning). 200,000 cells suspended in 100 µl medium were placed into the top chamber, and 600 µl medium containing 50 ng/ml human SDF-1α (PeproTech) was added to the bottom well. After 4 h of incubation, cells in the bottom well were collected and the cell number was counted using an Accuri C6 (BD).
Polarization and immunofluorescence
T8.1 cells were placed on ICAM-1–coated plates that were centrifuged at 100 g for 1 min. 100 ng/ml SDF-1α was added into the plates. For T-APC conjugate formation, T8.1 cells were transfected with FLAG-DOCK8 and HA-Cdc42, and incubated with Ttox-pulsed murine fibroblast L625.7 cells to form cell conjugates. After 10-min incubation at 37°C, cells were fixed with 4% PFA in PBS and stained with anti-CD44. For intracellular staining, fixed cells were permeabilized with 0.1% Triton X-100 in PBS and stained with anti-FLAG and anti-HA. Images were captured with Olympus BX51 microscope and polarized cells were counted.
Immunoprecipitation and immunoblotting assay
293T cells overexpressed indicated proteins for 36 h were immediately washed twice with ice-cold PBS before harvested in ice-cold lysis buffer (PBS containing 1% Triton X-100, 2 mM EDTA, and protease and phosphatase inhibitors). Whole-cell lysate was incubated with anti-FLAG or anti-HA beads and at 4°C for 2 h. The beads were washed three times with lysis buffer and then resuspended in an appropriate amount of SDS-PAGE loading buffer. Proteins were loaded onto SDS-PAGE gel and analyzed via immunoblotting. To measure DOCK8 phosphorylation, the membrane-bound DOCK8 (FLAG-CVIM-DOCK8) was transfected to 293T cells, and treated with the indicated inhibitors for 2 h. Cell lysates were prepared for immunoprecipitation with anti-FLAG antibody to pull-down FLAG-DOCK8, and the phosphorylation levels of DOCK8 were then assessed by immunoblotting with anti-phosphoserine/threonine antibody.
GST pull-down and Cdc42 activity assay
GST-PAK1-PBD (amino acids 69–150 of human PAK1) was kindly provided by Z. Chen (SIBCB, CAS). 293T cells transfected with indicated proteins were washed twice with the ice-cold PBS and lysed in Mg2+lysis buffer (MLB) as described by the manual (EMD Millipore). The supernatants were incubated with GST-PBD-agarose beads at 4°C for 2 h. The beads were washed three times by MLB and resuspended in the loading buffer for the immunoblotting assay.
Fluorescence resonance energy transfer measurement
FRET efficiency was measured with donor dequenching approach and the filter sets (458 nm for CFP, 514 nm for YFP) were used as previously described (Xu et al., 2008). Images were captured by Leica TCS SP2 AOBS microscope. The FRET efficiency was calculated as E = (Post −[Pre/Post]) × 100%, where Post and Pre represents the donor fluorescence before and after photo bleaching.
Cloning and purification of recombinant protein
The DHR-2 domain of DOCK8 (residues 1632–2086) was cloned into the pET-28a plasmid with a N-terminal His6tag (Novagen), and Cdc42G15A was inserted into the pGEX4T1 plasmid (GE Healthcare). Recombinant proteins were expressed in E. coli BL21 (DE3) Codon-Plus strain (Novagen). The transformed cells were grown at 37°C in LB medium containing 0.05 mg/ml ampicillin until OD600 reached 0.8, and then induced with 0.25 mM IPTG at 16°C for 24 h His-DHR-2 was purified by Ni-NTA affinity chromatography (QIAGEN) and GST-Cdc42G15A was purified by glutathione Sepharose beads as previously reported (Yang et al., 2013; Zhang et al., 2014). FLAG-LRR1-9 and FLAG-L305-763 were transfected into 293T cells and purified with anti-FLAG antibody. The increasing amount of the purified FLAG-LRR1-9 or FLAG-L305-763 was added to a solution containing His-DHR-2 and GST-Cdc42G15A. The mixture were incubated at 4°C for 60 min and then subjected to anti-His antibody precipitation.
MS and yeast two-hybrid assays
Immunoprecipitation using an anti-FLAG antibody was performed from T8.1 cells overexpressing FLAG-tagged DOCK8. The pull-down proteins at 90 kD were identified by the mass spectrometry analysis (below panel). DOCK8 was used as bait for screening a human cDNA library encoding over 12,794 human genes (hORFeome V5.1) by a yeast two-hybrid assay, and LRCH1 was found as a positive clone.
Statistics
All statistical analyses were performed with Prism6 software (GraphPad Software). Student's t test was used for comparisons between two groups. P < 0.05 was considered statistically significant.
Study approval
All procedures of animal experiments were conducted in accordance with the institutional guidelines and were approved by the Institutional Animal Care and Use Committee of Shanghai Institute of Biochemistry and Cell Biology (Protocol No. IBCB0057). For collecting human MS patients’ blood samples, the study was approved by the Ethical Committee for Clinical Research of Huashan Hospital, Fudan University.
Acknowledgments
We thank Dr. Rosemary Boyton (Imperial College London, UK) for her helpful discussion; Dr. Christopher C. Goodnow (The Australian Nation University, Canberra, Australia) for providing the DOCK8pri mice; and Drs. Xueliang Zhu, Jianfen Chen and Zhengjun Chen for providing the plasmids (SIBCB, CAS, China).
This work was supported by grants from the Ministry of Science and Technology of China (2016YFD0500207, 2016YFC0902200, and 2016YFD0500407), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000), National Natural Science Foundation of China (31422018, 81630043, 81571617, 81571552, 31300723, 81671572, and 81301027), and the State Key Laboratory of Cell Biology, SIBCB, CAS (SKLCBKF2013003). B. Wei is funded by the Hundred Talents Plan in HuBei Province and H. Wang is supported by the Hundred Talents Program of the Chinese Academy of Sciences.
The authors declare no competing financial interests.
Author contributions: X. Xu, L. Han, G. Zhao, S. Xue, J. Xiao, Y. Gao, S. Zhang, and P.Chen performed experiments; X. Xu and L. Han analyzed data; H. Wang, X. Xu, L. Han, B. Wei, Z. Wu, R. Hu, and J. Ding designed experiments; X. Xu, H. Wang, L. Han, and B.W. wrote the paper.
Footnotes
Abbreviations used:
- CH
- calponin homology
- DHR-2
- DOCK homology region 2
- EAE
- experimental autoimmune encephalomyelitis
- GEF
- guanine-nucleotide exchange factor
- LRR
- leucine-rich repeat
- MOG
- myelin oligodendrocyte glycoprotein peptide
- MS
- multiple sclerosis
- NMO
- neuromyelitis optica
- SNP
- single-nucleotide polymorphism
- Ttox
- tetanus toxoid peptide
References
- Abram C.L., and Lowell C.A.. 2009. The ins and outs of leukocyte integrin signaling. Annu. Rev. Immunol. 27:339–362. 10.1146/annurev.immunol.021908.132554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton A., Woolmore J.A., Ward D., Eyre S., Hinks A., Ollier W.E., Strange R.C., Fryer A.A., John S., Hawkins C.P., and Worthington J.. 2004. Association of protein kinase C alpha (PRKCA) gene with multiple sclerosis in a UK population. Brain. 127:1717–1722. 10.1093/brain/awh193 [DOI] [PubMed] [Google Scholar]
- Benard V., Bohl B.P., and Bokoch G.M.. 1999. Characterization of rac and cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J. Biol. Chem. 274:13198–13204. 10.1074/jbc.274.19.13198 [DOI] [PubMed] [Google Scholar]
- Chun J., and Hartung H.P.. 2010. Mechanism of action of oral fingolimod (FTY720) in multiple sclerosis. Clin. Neuropharmacol. 33:91–101. 10.1097/WNF.0b013e3181cbf825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté J.F., Motoyama A.B., Bush J.A., and Vuori K.. 2005. A novel and evolutionarily conserved PtdIns(3,4,5)P3-binding domain is necessary for DOCK180 signalling. Nat. Cell Biol. 7:797–807. 10.1038/ncb1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damotte V., Guillot-Noel L., Patsopoulos N.A., Madireddy L., El Behi M., De Jager P.L., Baranzini S.E., Cournu-Rebeix I., Fontaine B. International Multiple Sclerosis Genetics Consortium. Wellcome Trust Case Control Consortium 2 . 2014. A gene pathway analysis highlights the role of cellular adhesion molecules in multiple sclerosis susceptibility. Genes Immun. 15:126–132. 10.1038/gene.2013.70 [DOI] [PubMed] [Google Scholar]
- De Jager P.L., and Hafler D.A.. 2007. New therapeutic approaches for multiple sclerosis. Annu. Rev. Med. 58:417–432. 10.1146/annurev.med.58.071105.111552 [DOI] [PubMed] [Google Scholar]
- De Jager P.L., Jia X., Wang J., de Bakker P.I., Ottoboni L., Aggarwal N.T., Piccio L., Raychaudhuri S., Tran D., Aubin C., et al. International MS Genetics Consortium . 2009. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat. Genet. 41:776–782. 10.1038/ng.401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos A.C., Barsante M.M., Arantes R.M., Bernard C.C., Teixeira M.M., and Carvalho-Tavares J.. 2005. CCL2 and CCL5 mediate leukocyte adhesion in experimental autoimmune encephalomyelitis--an intravital microscopy study. J. Neuroimmunol. 162:122–129. 10.1016/j.jneuroim.2005.01.020 [DOI] [PubMed] [Google Scholar]
- Engelhardt B., and Ransohoff R.M.. 2005. The ins and outs of T-lymphocyte trafficking to the CNS: anatomical sites and molecular mechanisms. Trends Immunol. 26:485–495. 10.1016/j.it.2005.07.004 [DOI] [PubMed] [Google Scholar]
- Engelmann S., Togni M., Thielitz A., Reichardt P., Kliche S., Reinhold D., Schraven B., and Reinhold A.. 2013. T cell-independent modulation of experimental autoimmune encephalomyelitis in ADAP-deficient mice. J. Immunol. 191:4950–4959. 10.4049/jimmunol.1203340 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. 2004. Cdc42--the centre of polarity. J. Cell Sci. 117:1291–1300. 10.1242/jcs.01115 [DOI] [PubMed] [Google Scholar]
- Foussard H., Ferrer P., Valenti P., Polesello C., Carreno S., and Payre F.. 2010. LRCH proteins: a novel family of cytoskeletal regulators. PLoS One. 5:e12257 10.1371/journal.pone.0012257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata R., Wennerberg K., Arthur W.T., Noren N.K., Ellerbroek S.M., and Burridge K.. 2006. Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol. 406:425–437. 10.1016/S0076-6879(06)06031-9 [DOI] [PubMed] [Google Scholar]
- Griggs B.L., Ladd S., Saul R.A., DuPont B.R., and Srivastava A.K.. 2008. Dedicator of cytokinesis 8 is disrupted in two patients with mental retardation and developmental disabilities. Genomics. 91:195–202. 10.1016/j.ygeno.2007.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad E., Zugaza J.L., Louache F., Debili N., Crouin C., Schwarz K., Fischer A., Vainchenker W., and Bertoglio J.. 2001. The interaction between Cdc42 and WASP is required for SDF-1-induced T-lymphocyte chemotaxis. Blood. 97:33–38. 10.1182/blood.V97.1.33 [DOI] [PubMed] [Google Scholar]
- Han J., Lim C.J., Watanabe N., Soriani A., Ratnikov B., Calderwood D.A., Puzon-McLaughlin W., Lafuente E.M., Boussiotis V.A., Shattil S.J., and Ginsberg M.H.. 2006. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr. Biol. 16:1796–1806. 10.1016/j.cub.2006.08.035 [DOI] [PubMed] [Google Scholar]
- Harada Y., Tanaka Y., Terasawa M., Pieczyk M., Habiro K., Katakai T., Hanawa-Suetsugu K., Kukimoto-Niino M., Nishizaki T., Shirouzu M., et al. 2012. DOCK8 is a Cdc42 activator critical for interstitial dendritic cell migration during immune responses. Blood. 119:4451–4461. 10.1182/blood-2012-01-407098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh R.E., Kurokawa K., Ohba Y., Yoshizaki H., Mochizuki N., and Matsuda M.. 2002. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell. Biol. 22:6582–6591. 10.1128/MCB.22.18.6582-6591.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabara H.H., McDonald D.R., Janssen E., Massaad M.J., Ramesh N., Borzutzky A., Rauter I., Benson H., Schneider L., Baxi S., et al. 2012. DOCK8 functions as an adaptor that links TLR-MyD88 signaling to B cell activation. Nat. Immunol. 13:612–620. 10.1038/ni.2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q., Shi D., Nakajima M., Dai J., Wei J., Malizos K.N., Qin J., Miyamoto Y., Kamatani N., Liu B., et al. 2008. Lack of association of single nucleotide polymorphism in LRCH1 with knee osteoarthritis susceptibility. J. Hum. Genet. 53:42–47. 10.1007/s10038-007-0216-4 [DOI] [PubMed] [Google Scholar]
- Jin W., Zhou X.F., Yu J., Cheng X., and Sun S.C.. 2009. Regulation of Th17 cell differentiation and EAE induction by MAP3K NIK. Blood. 113:6603–6610. 10.1182/blood-2008-12-192914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo E.K., Wang H., and Rudd C.E.. 2005. An essential role for SKAP-55 in LFA-1 clustering on T cells that cannot be substituted by SKAP-55R. J. Exp. Med. 201:1733–1739. 10.1084/jem.20042577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri K., Imamura M., and Kinashi T.. 2006. Spatiotemporal regulation of the kinase Mst1 by binding protein RAPL is critical for lymphocyte polarity and adhesion. Nat. Immunol. 7:919–928. 10.1038/ni1374 [DOI] [PubMed] [Google Scholar]
- Katagiri K., Ueda Y., Tomiyama T., Yasuda K., Toda Y., Ikehara S., Nakayama K.I., and Kinashi T.. 2011. Deficiency of Rap1-binding protein RAPL causes lymphoproliferative disorders through mislocalization of p27kip1. Immunity. 34:24–38. 10.1016/j.immuni.2010.12.010 [DOI] [PubMed] [Google Scholar]
- Kerfoot S.M., and Kubes P.. 2002. Overlapping roles of P-selectin and alpha 4 integrin to recruit leukocytes to the central nervous system in experimental autoimmune encephalomyelitis. J. Immunol. 169:1000–1006. 10.4049/jimmunol.169.2.1000 [DOI] [PubMed] [Google Scholar]
- Korn T., Fischer K.D., Girkontaite I., Köllner G., Toyka K., and Jung S.. 2003. Vav1-deficient mice are resistant to MOG-induced experimental autoimmune encephalomyelitis due to impaired antigen priming. J. Neuroimmunol. 139:17–26. 10.1016/S0165-5728(03)00128-0 [DOI] [PubMed] [Google Scholar]
- Krumbholz M., Theil D., Cepok S., Hemmer B., Kivisäkk P., Ransohoff R.M., Hofbauer M., Farina C., Derfuss T., Hartle C., et al. 2006. Chemokines in multiple sclerosis: CXCL12 and CXCL13 up-regulation is differentially linked to CNS immune cell recruitment. Brain. 129:200–211. 10.1093/brain/awh680 [DOI] [PubMed] [Google Scholar]
- Lambe T., Crawford G., Johnson A.L., Crockford T.L., Bouriez-Jones T., Smyth A.M., Pham T.H., Zhang Q., Freeman A.F., Cyster J.G., et al. 2011. DOCK8 is essential for T-cell survival and the maintenance of CD8+ T-cell memory. Eur. J. Immunol. 41:3423–3435. 10.1002/eji.201141759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Jiao S., Wang G., Gao Y., Liu C., He X., Zhang C., Xiao J., Li W., Zhang G., et al. 2015a The immune adaptor ADAP regulates reciprocal TGF-β1-integrin crosstalk to protect from influenza virus infection. PLoS Pathog. 11:e1004824 10.1371/journal.ppat.1004824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Li W., Xiao J., Jiao S., Teng F., Xue S., Zhang C., Sheng C., Leng Q., Rudd C.E., et al. 2015b ADAP and SKAP55 deficiency suppresses PD-1 expression in CD8+ cytotoxic T lymphocytes for enhanced anti-tumor immunotherapy. EMBO Mol. Med. 7:754–769. 10.15252/emmm.201404578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Xiao J., Zhou X., Xu M., Hu C., Xu X., Lu Y., Liu C., Xue S., Nie L., et al. 2015c STK4 regulates TLR pathways and protects against chronic inflammation-related hepatocellular carcinoma. J. Clin. Invest. 125:4239–4254. 10.1172/JCI81203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Kinchen J.M., Rossman K.L., Grimsley C., Hall M., Sondek J., Hengartner M.O., Yajnik V., and Ravichandran K.S.. 2005. A Steric-inhibition model for regulation of nucleotide exchange via the Dock180 family of GEFs. Curr. Biol. 15:371–377. 10.1016/j.cub.2005.01.050 [DOI] [PubMed] [Google Scholar]
- McFarland H.F., and Martin R.. 2007. Multiple sclerosis: a complicated picture of autoimmunity. Nat. Immunol. 8:913–919. 10.1038/ni1507 [DOI] [PubMed] [Google Scholar]
- Medeiros R.B., Dickey D.M., Chung H., Quale A.C., Nagarajan L.R., Billadeau D.D., and Shimizu Y.. 2005. Protein kinase D1 and the β1 integrin cytoplasmic domain control β1 integrin function via regulation of Rap1 activation. Immunity. 23:213–226. 10.1016/j.immuni.2005.07.006 [DOI] [PubMed] [Google Scholar]
- Meisel M., Hermann-Kleiter N., Hinterleitner R., Gruber T., Wachowicz K., Pfeifhofer-Obermair C., Fresser F., Leitges M., Soldani C., Viola A., et al. 2013. The kinase PKCα selectively upregulates interleukin-17A during Th17 cell immune responses. Immunity. 38:41–52. 10.1016/j.immuni.2012.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller N., Westbrook M.J., Shannon J.D., Guda C., and Schwartz M.A.. 2008. Function of the N-terminus of zizimin1: autoinhibition and membrane targeting. Biochem. J. 409:525–533. 10.1042/BJ20071263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., and Acuto O.. 1996. Induction of T cell adhesion by antigen stimulation and modulation by the coreceptor CD4. Cell. Immunol. 173:165–175. 10.1006/cimm.1996.0264 [DOI] [PubMed] [Google Scholar]
- Mou F., Praskova M., Xia F., Van Buren D., Hock H., Avruch J., and Zhou D.. 2012. The Mst1 and Mst2 kinases control activation of rho family GTPases and thymic egress of mature thymocytes. J. Exp. Med. 209:741–759. 10.1084/jem.20111692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraboschi E.M., Rimoldi V., Soldà G., Tabaglio T., Dall’Osso C., Saba E., Vigliano M., Salviati A., Leone M., Benedetti M.D., et al. 2014. Functional variations modulating PRKCA expression and alternative splicing predispose to multiple sclerosis. Hum. Mol. Genet. 23:6746–6761. 10.1093/hmg/ddu392 [DOI] [PubMed] [Google Scholar]
- Parekh D.B., Ziegler W., and Parker P.J.. 2000. Multiple pathways control protein kinase C phosphorylation. EMBO J. 19:496–503. 10.1093/emboj/19.4.496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker P.J., and Murray-Rust J.. 2004. PKC at a glance. J. Cell Sci. 117:131–132. 10.1242/jcs.00982 [DOI] [PubMed] [Google Scholar]
- Randall K.L., Lambe T., Johnson A.L., Treanor B., Kucharska E., Domaschenz H., Whittle B., Tze L.E., Enders A., Crockford T.L., et al. 2009. Dock8 mutations cripple B cell immunological synapses, germinal centers and long-lived antibody production. Nat. Immunol. 10:1283–1291. 10.1038/ni.1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall K.L., Chan S.S., Ma C.S., Fung I., Mei Y., Yabas M., Tan A., Arkwright P.D., Al Suwairi W., Lugo Reyes S.O., et al. 2011. DOCK8 deficiency impairs CD8 T cell survival and function in humans and mice. J. Exp. Med. 208:2305–2320. 10.1084/jem.20110345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransohoff R.M., Kivisäkk P., and Kidd G.. 2003. Three or more routes for leukocyte migration into the central nervous system. Nat. Rev. Immunol. 3:569–581. 10.1038/nri1130 [DOI] [PubMed] [Google Scholar]
- Sawcer S., Hellenthal G., Pirinen M., Spencer C.C., Patsopoulos N.A., Moutsianas L., Dilthey A., Su Z., Freeman C., Hunt S.E., et al. Wellcome Trust Case Control Consortium 2 . 2011. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 476:214–219. 10.1038/nature10251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Li N., Wu S., Zhou Y., Shan Y., Zhang Q., Ding C., Yuan Q., Zhao F., Zeng R., and Zhu X.. 2008. Nudel binds Cdc42GAP to modulate Cdc42 activity at the leading edge of migrating cells. Dev. Cell. 14:342–353. 10.1016/j.devcel.2008.01.001 [DOI] [PubMed] [Google Scholar]
- Sigal A., Bleijs D.A., Grabovsky V., van Vliet S.J., Dwir O., Figdor C.G., van Kooyk Y., and Alon R.. 2000. The LFA-1 integrin supports rolling adhesions on ICAM-1 under physiological shear flow in a permissive cellular environment. J. Immunol. 165:442–452. 10.4049/jimmunol.165.1.442 [DOI] [PubMed] [Google Scholar]
- Snelling S., Sinsheimer J.S., Carr A., and Loughlin J.. 2007. Genetic association analysis of LRCH1 as an osteoarthritis susceptibility locus. Rheumatology (Oxford). 46:250–252. 10.1093/rheumatology/kel265 [DOI] [PubMed] [Google Scholar]
- Sørensen T.L., Tani M., Jensen J., Pierce V., Lucchinetti C., Folcik V.A., Qin S., Rottman J., Sellebjerg F., Strieter R.M., et al. 1999. Expression of specific chemokines and chemokine receptors in the central nervous system of multiple sclerosis patients. J. Clin. Invest. 103:807–815. 10.1172/JCI5150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman L. 2005. Blocking adhesion molecules as therapy for multiple sclerosis: natalizumab. Nat. Rev. Drug Discov. 4:510–518. 10.1038/nrd1752 [DOI] [PubMed] [Google Scholar]
- Sun H., Liu J., Zheng Y., Pan Y., Zhang K., and Chen J.. 2014. Distinct chemokine signaling regulates integrin ligand specificity to dictate tissue-specific lymphocyte homing. Dev. Cell. 30:61–70. 10.1016/j.devcel.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Tybulewicz V.L. 2005. Vav-family proteins in T-cell signalling. Curr. Opin. Immunol. 17:267–274. 10.1016/j.coi.2005.04.003 [DOI] [PubMed] [Google Scholar]
- Vajkoczy P., Laschinger M., and Engelhardt B.. 2001. Alpha4-integrin-VCAM-1 binding mediates G protein-independent capture of encephalitogenic T cell blasts to CNS white matter microvessels. J. Clin. Invest. 108:557–565. 10.1172/JCI12440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., and Rudd C.E.. 2008. SKAP-55, SKAP-55-related and ADAP adaptors modulate integrin-mediated immune-cell adhesion. Trends Cell Biol. 18:486–493. 10.1016/j.tcb.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Moon E.Y., Azouz A., Wu X., Smith A., Schneider H., Hogg N., and Rudd C.E.. 2003. SKAP-55 regulates integrin adhesion and formation of T cell-APC conjugates. Nat. Immunol. 4:366–374. 10.1038/ni913 [DOI] [PubMed] [Google Scholar]
- Wang H., McCann F.E., Gordan J.D., Wu X., Raab M., Malik T.H., Davis D.M., and Rudd C.E.. 2004. ADAP-SLP-76 binding differentially regulates supramolecular activation cluster (SMAC) formation relative to T cell-APC conjugation. J. Exp. Med. 200:1063–1074. 10.1084/jem.20040780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liu H., Lu Y., Lovatt M., Wei B., and Rudd C.E.. 2007. Functional defects of SKAP-55-deficient T cells identify a regulatory role for the adaptor in LFA-1 adhesion. Mol. Cell. Biol. 27:6863–6875. 10.1128/MCB.00556-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wei B., Bismuth G., and Rudd C.E.. 2009. SLP-76-ADAP adaptor module regulates LFA-1 mediated costimulation and T cell motility. Proc. Natl. Acad. Sci. USA. 106:12436–12441. 10.1073/pnas.0900510106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Lim D., and Rudd C.E.. 2010. Immunopathologies linked to integrin signalling. Semin. Immunopathol. 32:173–182. 10.1007/s00281-010-0202-3 [DOI] [PubMed] [Google Scholar]
- Xu C., Gagnon E., Call M.E., Schnell J.R., Schwieters C.D., Carman C.V., Chou J.J., and Wucherpfennig K.W.. 2008. Regulation of T cell receptor activation by dynamic membrane binding of the CD3epsilon cytoplasmic tyrosine-based motif. Cell. 135:702–713. 10.1016/j.cell.2008.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Zhang T., Tao Y., Wang F., Tong L., and Ding J.. 2013. Structural insights into the functions of the FANCM-FAAP24 complex in DNA repair. Nucleic Acids Res. 41:10573–10583. 10.1093/nar/gkt788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Zhou X., Chang M., Nakaya M., Chang J.H., Xiao Y., Lindsey J.W., Dorta-Estremera S., Cao W., Zal A., et al. 2015. Regulation of T-cell activation and migration by the kinase TBK1 during neuroinflammation. Nat. Commun. 6:6074 10.1038/ncomms7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., and Wang H.. 2012. Integrin signalling and function in immune cells. Immunology. 135:268–275. 10.1111/j.1365-2567.2011.03549.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Davis J.C., Lamborn I.T., Freeman A.F., Jing H., Favreau A.J., Matthews H.F., Davis J., Turner M.L., Uzel G., et al. 2009. Combined immunodeficiency associated with DOCK8 mutations. N. Engl. J. Med. 361:2046–2055. 10.1056/NEJMoa0905506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Zhang T., Wang S., Gong Z., Tang C., Chen J., and Ding J.. 2014. Molecular mechanism for Rabex-5 GEF activation by Rabaptin-5. eLife. 3:3 10.7554/eLife.02687 [DOI] [PMC free article] [PubMed] [Google Scholar]