Connor et al. show that transcriptomic profiling of DCs exposed to two different Th2 stimuli in vivo reveals large numbers of differentially expressed genes but few similarities between conditions.
Abstract
The dendritic cell signals required for the in vivo priming of IL-4–producing T cells are unknown. We used RNA sequencing to characterize DCs from skin LN of mice exposed to two different Th2 stimuli: the helminth parasite Nippostrongylus brasiliensis (Nb) and the contact sensitizer dibutyl phthalate (DBP)-FITC. Both Nb and DBP-FITC induced extensive transcriptional changes that involved multiple DC subsets. Surprisingly, these transcriptional changes were highly distinct in the two models, with only a small number of genes being similarly regulated in both conditions. Pathway analysis of expressed genes identified no shared pathways between Nb and DBP-FITC, but revealed a type-I IFN (IFN-I) signature unique to DCs from Nb-primed mice. Blocking the IFN-I receptor at the time of Nb treatment had little effect on DC migration and antigen transport to the LN, but inhibited the up-regulation of IFN-I–induced markers on DCs and effectively blunted Th2 development. In contrast, the response to DBP-FITC was not affected by IFN-I receptor blockade, a finding consistent with the known dependence of this response on the innate cytokine TSLP. Thus, the priming of Th2 responses is associated with distinct transcriptional signatures in DCs in vivo, reflecting the diverse environments in which Th2 immune responses are initiated.
Introduction
DCs are a specialized population of innate immune cells that play a pivotal role in the initiation of adaptive T cell immunity. In particular, CD11c+ DCs are essential for the priming of Th2 cells (Hammad et al., 2010; Phythian-Adams et al., 2010; Smith et al., 2011, 2012), which are associated with protective immunity against helminths and also responsible for the inappropriate response to innocuous antigens observed in allergic disease (Paul and Zhu, 2010; Pulendran and Artis, 2012). The mechanism by which DCs direct Th2 differentiation are not well understood, but are critical to developing effective treatment strategies against these diseases.
DCs that express the transcription factor IRF4 have been identified as essential for the development of Th2 immune responses in skin and airway (Gao et al., 2013; Kumamoto et al., 2013; Murakami et al., 2013; Williams et al., 2013). In the skin, IRF4-dependent DCs are Langerin−CD103−, and express the surface markers CD301b (Mgl2) and programmed cell death ligand 2 (PD-L2) (Gao et al., 2013; Kumamoto et al., 2013; Murakami et al., 2013). A key role of IRF4 is further supported by studies showing that the optimal induction of Th2 responses also requires DCs to express the transcriptional repressor Mbd2, which epigenetically regulates IRF4 expression (Cook et al., 2015). Similarly, the transcription factor KLF4 supports IRF4 expression during DC development and is necessary for the development of CD301b+ and CD301b− DC populations in LN, and the priming of Th2, but not Th17 responses (Tussiwand et al., 2015). Consistent with these observations, IRF4-independent DC subsets, which include the dermal CD103+ DCs and epidermal Langerhans cells in the skin, are either unnecessary or even inhibitory of Th2 responses (Everts et al., 2016). Studies from our laboratory also indicate that CD103−CD326− skin DC subsets are positively associated with the initiation of Th2 responses after skin immunization (Connor et al., 2014; Ochiai et al., 2014).
Innate cytokines also play an important role in DC activation during Th2 immune responses. In the skin, thymic stromal lymphopoietin (TSLP) is produced by epithelial cells subjected to barrier disruption (Oyoshi et al., 2010) or insults such as contact sensitizers (Larson et al., 2010), and directs DCs to express molecules such as OX40L (Ito et al., 2005), CCL17 and CCL22 (Soumelis et al., 2002) that promote Th2 priming. However, several Th2 immune responses, especially to parasite infections, appear to be TSLP independent (Massacand et al., 2009), suggesting that other cytokines or factors are involved. Expression of specific co-stimulatory molecules or cytokines and the ability to engage the TCR with altered avidity have both been proposed as possible mechanisms of Th2 induction (Bouchery et al., 2014; Hussaarts et al., 2014); however, conclusive evidence is not available.
In this study, we examined the transcriptional profiles of distinct DC populations isolated from skin LN after in vivo treatment with two different Th2 stimuli: intradermal transfer of nonviable Nippostrongylus brasiliensis larvae (Nb) as a model of skin immune response to a parasite (Camberis et al., 2013), or topical application of DBP-FITC as a model of contact hypersensitivity. These models were selected because they induce CD4 responses that are strongly dominated by IL-4 production with little or no IFN-γ or IL-17, and they require similar DC subsets for initiation. Therefore, comparison of DCs exposed to these two stimuli would presumably enable the identification of a shared Th2 transcriptional profile in DCs. We report that the CD11b+ and TN DC populations from Nb- or DBP-FITC–treated mice exhibited a noninflammatory profile and profound transcriptional changes compared with their respective controls. Unexpectedly, these transcriptional changes were specific to each Th2 condition such that only very few genes were similarly regulated in both Th2 models. Pathway analysis of expressed genes revealed a previously unreported type I IFN (IFN-I) signature that was unique to Nb, and necessary for the optimal induction of Th2 immune responses in this model. Our data thus reveal distinct patterns in the response of DCs to Th2 stimuli, despite the shared ability of these DCs to prime IL-4–producing T cells in vivo.
Results
The induction of Th2 responses by Nb and DBP-FITC involves similar subsets of DCs
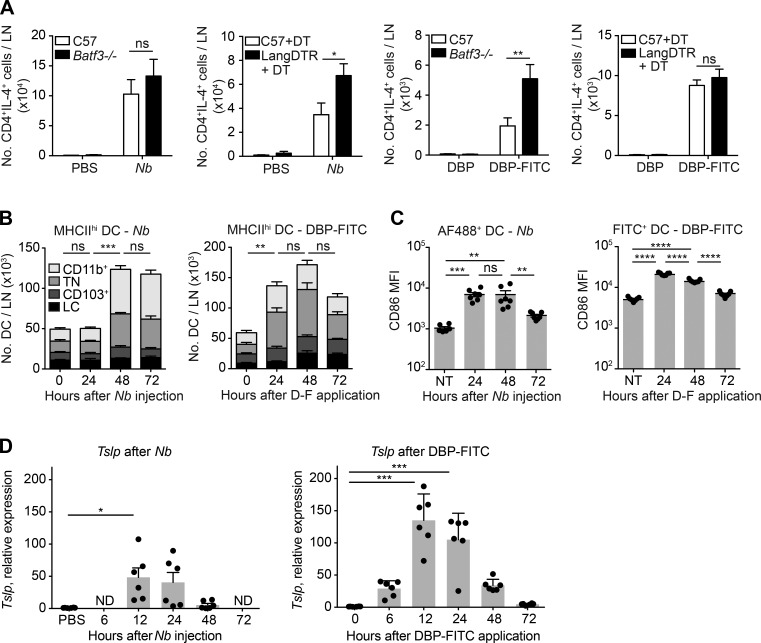
To assess whether induction of Th2 immunity by different stimuli involves similar skin DC subsets we used Batf3−/− mice, which constitutively lack dermal CD103+ DCs, and Langerin-diphtheria toxin receptor (DTR) mice that can be depleted of epidermal LCs and CD103+ DCs by DT treatment. The remaining CD326−CD103− MHCIIhi DC subsets (CD11b+ and CD103−CD11b−CD326−, or TN) are preserved in these mice (Kissenpfennig et al., 2005; Edelson et al., 2010). Mice were either injected with PBS or nonviable Nb i.d. into the ear, or were given one topical application of DBP ± FITC on ear skin. The number of IL-4–producing CD4+ T cells in draining LNs (dLNs) was determined on day 7 as a measure of Th2 induction. Langerin-DTR and Batf3−/− mice all exhibited robust IL-4 responses that were similar or higher than the response in C57BL/6 controls (Fig. 1 A).
Figure 1.
Characterization of DC subset contribution and Tslp induction during Th2 immune responses to Nb and DBP-FITC. LNs were collected from mice immunized with 600 Nb or DBP-FITC and respective controls at the indicated time points. (A) C57BL/6, Langerin-DTR, and Batf3−/− mice were immunized as indicated. DT was injected i.p. 1 d before and 1 d after immunization. On day 7 after immunization, LN cells were collected and processed for intracellular cytokine staining; in vitro restimulation was with PMA and ionomycin (Nb) or anti-CD3 and anti-CD28 (DBP-FITC). Bar graphs show mean + SEM for 4–8 mice/group. Data are from one of two experiments that gave similar results. P-values were determined using one-way ANOVA with the Bonferroni post-test. (B) C57BL/6 mice were immunized as indicated, and dLN were collected for flow cytometric analysis at the indicated times after treatment. DC subsets were defined as in Fig. S1. Bar graphs show the mean + SE of four (Nb) or seven (DBP-FITC) mice per time point. Data are from one of two experiments that gave similar results. Statistical significance values refer to the total number of DCs in LN, p-values were determined using one-way ANOVA with the Bonferroni post-test. (C) As in B, except that mice were immunized with AF488-Nb instead of Nb. CD86 expression in AF488+ or FITC+ DCs was determined on total MHCIIhi DC populations at the indicated time points. Each dot corresponds to one mouse. Data from AF488-Nb immunization show combination of two separate experiments with three to four mice/group, and data from DBP-FITC treatment are representative of two repeat experiments using six to seven mice/group. P-values were determined using one-way ANOVA with the Bonferroni post-test. (D) C57BL/6 mice were immunized on both ears as indicated. Epidermal layers were harvested for RNA preparation at the indicated times after treatment. Tslp data are expressed as relative to 18S and the respective control samples. Each dot represents one mouse (pool of two ears); mean and SEM for six mice/group are shown. Data are from one of two experiments that gave similar results. P-values were determined using the Kruskal-Wallis with the Dunn’s post-test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
To better assess the response of different DC subsets to Th2 stimuli, we performed a time course analysis of MHCIIhi DC subsets in the LN of C57BL/6 mice treated with Nb, DBP-FITC or the respective controls. After Nb injection, the number of MHCIIhi DCs in LN remained stable at 24 h, but was significantly increased at 48 and 72 h. Most of this increase was accounted for by higher numbers of CD11b+ and TN DCs (Fig. 1 B). DCs carrying AF488-labeled Nb (Nb-AF488) and expressing increased CD86 were detected in the LN by 24 h, but their number was highest at 48 h (Fig. 1 C; Connor et al., 2014). Treatment with DBP-FITC also led to increased numbers of MHCIIhi DCs in LN; however, this increase was already apparent at 24 h, with no further significant increases at 48 or 72 h (Fig. 1 C). Again, higher numbers of CD11b+ and TN DCs accounted for most of the increased cellularity. The proportion of FITC+ DCs, and their expression of CD86, was also highest at 24 h after DBP-FITC application (Fig. 1 C; Ochiai et al., 2014).
TSLP is essential for Th2 development after DBP-FITC, whereas Th2 response to Nb can develop in the absence of TSLPR (Larson et al., 2010; Connor et al., 2014). To determine whether Nb can trigger TSLP production in vivo, we performed quantitative RT-PCR on mRNA from the epidermis of mice exposed to Nb or DBP-FITC. Tslp transcripts were detected after both treatments and followed similar kinetics, although their levels were higher in DBP-FITC–treated mice compared with Nb (Fig. 1 D).
Together, these results are consistent with previous work identifying CD11b+ and TN DCs as key populations in the induction of TSLP-dependent and -independent Th2 responses (Gao et al., 2013; Kitajima and Ziegler, 2013; Kumamoto et al., 2013; Murakami et al., 2013; Williams et al., 2013; Connor et al., 2014; Ochiai et al., 2014; Tussiwand et al., 2015), whereas CD103+ DCs are not required or inhibitory (Everts et al., 2016).
DCs from the dLN of untreated, Nb-, or DBP-FITC–treated mice express a migratory DC gene signature and low levels of proinflammatory transcripts
To generate a full transcriptomic profile of the DCs involved in Th2 immune responses, CD11b+ and TN MHCIIhi DC populations were flow sorted from skin dLN of Nb- or DBP-FITC–treated mice according to the strategy in Fig. S1. CD11b+ and TN DCs were collected 48 h after Nb or PBS treatment, to correspond to the peak of DC cellularity in the dLN (Fig. 1 B). CD103+ DCs were also included in this analysis. Similarly, CD11b+ and TN DCs were flow sorted from the dLN of DBP-FITC–treated or untreated (UT) mice, except that dLN harvest was at 24 h to reflect earlier DC migration to the dLN in this model. The flow-sorting panel was also slightly modified for improved purity (Fig. S1, A–C).
To confirm that the desired populations of skin-derived migratory DCs had been purified from LN, we used previously published data (Miller et al., 2012) to generate a list of 23 migratory DC signature genes with high mean expression across all migratory DC populations compared with LN-resident DC populations, and vice versa for 14 resident DC signature genes. As shown in Fig. S2, individual DC subsets expressed high levels of most of the gene transcripts associated with migratory DCs, and low levels of the genes associated with resident DCs. We also assessed expression of genes that are expressed at a high levels in Ly6C+MHCII– monocytes or plasmacytoid DCs, and are low to medium in migratory DCs (Fig. S2). Both groups of transcripts were generally low in all populations.
Steady state migratory DCs are reported to express genes with immunoregulatory function (Miller et al., 2012; Manh et al., 2013; Dalod et al., 2014). Several of these genes: CD274 (PD-L1), Etv3, Sbno2, Spred1, Tnfaip3, Socs2, and Stat3, were highly expressed in all our DC populations and were often down-regulated in TN DCs from DBP-FITC–treated mice, but not in CD11b+ or TN DCs from Nb mice (Fig. 2). Interestingly, Stat4 was up-regulated in all DC subsets examined, except CD11b+ DCs in DBP-FITC–treated mice. TN DCs expressed low levels of the TGFβ-activating molecule Itgb8, reported to be necessary for Th17 and regulatory T cell (T reg) activation (Travis et al., 2007; Yoshida et al., 2014). All DC populations also expressed high levels of genes associated with antigen-presenting function (Cd74, H2-aa, Icam1, and Swap70), and variably up-regulated genes encoding co-stimulatory molecules after treatment with Nb or DBP-FITC (Fig. 2). Tnfsf9 and Tnfrsf9, or 4-1BB and 4-1BB ligand, were either down-regulated or not up-regulated. These transcripts are strongly induced in monocyte-derived DCs exposed to bacteria and, to a lesser extent, viruses (Huang et al., 2001).
Figure 2.
Expression of genes involved in DC function after immunization with Nb or DBP-FITC. LN were collected from C57BL/6 mice immunized with 600 Nb, DBP-FITC, or mock treated. DC subsets were flow sorted as in Fig. S1 for RNA preparation and sequencing. Heat maps on the left show the levels of expression of genes associated with different DC functions or with the induction of Th2 immune responses as indicated. Differential expression analysis in treated groups is on the right. DC expression data are the mean of three replicate samples each from an independent experiment for each Th2 condition. P-values were calculated by DESeq2; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Genes encoding inflammatory cytokines and chemokines were mostly expressed at low levels in all DC populations (Fig. 2). Nb immunization led to an up-regulation of these genes in CD103+ and TN DCs, but not in the CD11b+ subset. Similarly, no up-regulation was observed in CD11b+ or TN DCs from DBP-FITC–treated mice. The clear down-regulation of Il12b in TN and CD11b+ DCs after DBP-FITC was presumably caused by the effects of TSLP (Taylor et al., 2009), which is highly induced by DBP-FITC, but not Nb (Fig. 1 D). Transcripts for IL-12a and IL-23a were almost undetectable regardless of DC subset or condition (not depicted).
The expression of several genes associated with Th2 immunity was also examined (Fig. 2). Transcripts for the chemokine Ccl17 and the transcription factor Irf4 were up-regulated in both the CD11b+ and TN DC subsets from DBP-FITC and Nb-treated mice. Pdcd1lg2 (PD-L2) and Il4ra were also up-regulated, especially after Nb injection. Changes that were smaller or more variable across DC populations were also observed for Ccl22, Mbd2, Stat6, Il9r, Lsp1 (anti-Th1), and, surprisingly, Tnfsf4 (OX40L) transcripts. Cish, a TSLP-responsive negative regulator of cytokine signal transduction was especially up-regulated on TN DCs. Cxcr5 and the early innate cytokine Il33 were both variably up-regulated, but their baseline expression levels were very low. Dll4 and Vdr were strongly up-regulated by Nb treatment but not DBP-FITC. Dll4 is also up-regulated by DNFB (Tamoutounour et al., 2013) thus questioning its significance in Th2 immunity. The expression levels of Tyro3, a TAM family receptor tyrosine kinase recently reported to suppress Th2 responses (Chan et al., 2016), did not appear to be significantly regulated. Expression of Il1rl1 and Il17rb, the IL-33 and IL-25 receptors, respectively, was very low regardless of subset or condition (not depicted).
Th2 responses are associated with profound transcriptional changes involving multiple DC subsets
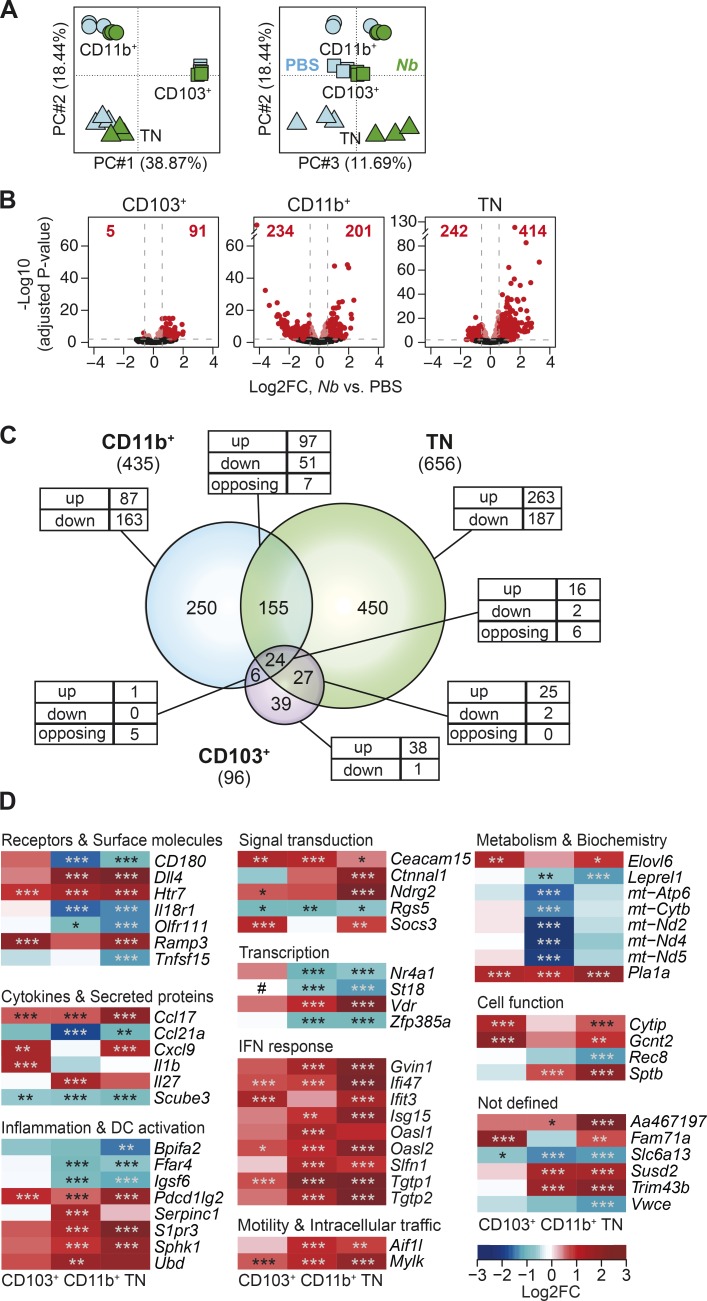
To investigate transcriptional changes that are associated with the immune response to Nb, we compared gene expression in CD103+, CD11b+, and TN DC populations from Nb and control mice in a principal component analysis (PCA; Fig. 3 A). We found good clustering of biological replicates for each DC subset and treatment. Individual DC subsets were segregated across the first two principal components, which together accounted for ∼57% of the total variability in the data. The second source of variation was associated with treatment conditions, with PC3 mostly separating the Nb-treated groups from the PBS controls.
Figure 3.
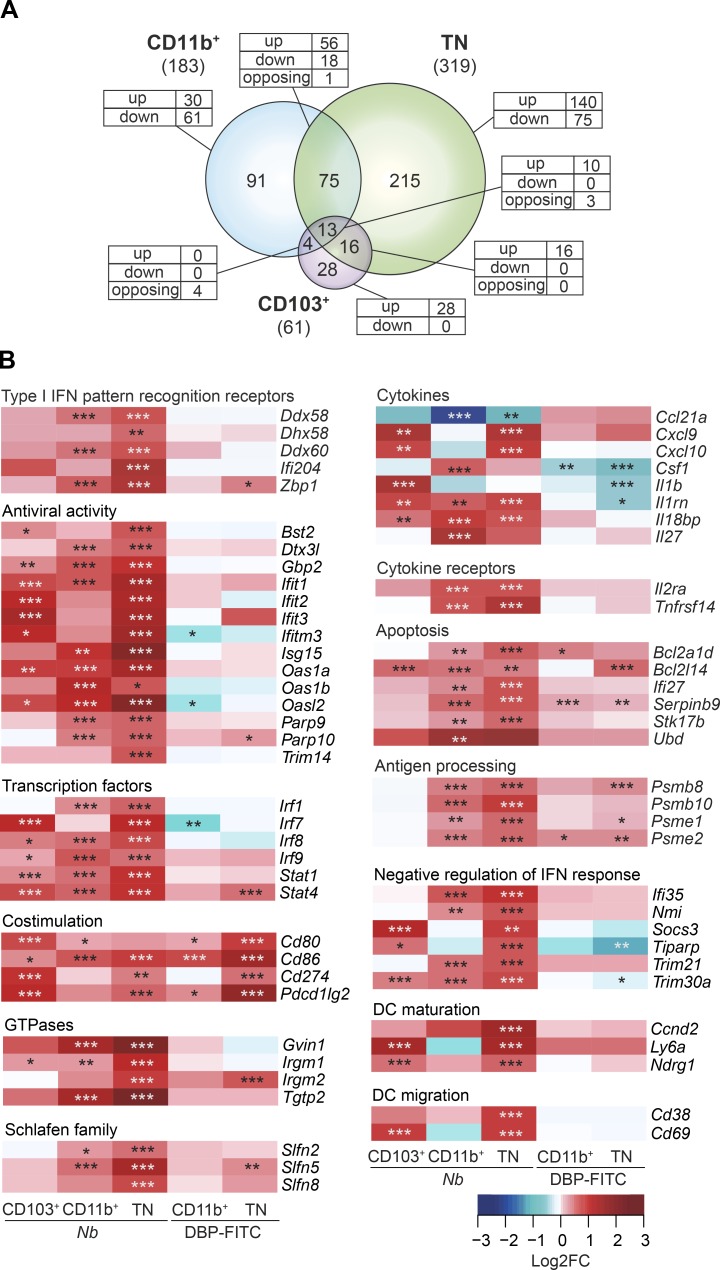
Treatment with Nb is associated with profound transcriptional changes affecting multiple DC subsets. (A) PCA of genes expressed in CD103+, CD11b+, and TN DCs from mice treated with 600 Nb or PBS. (B) Volcano plots comparing gene expression in DC subsets from Nb or PBS-treated mice. FC, fold change. Bright red dots and dashed lines identify the genes scored as differentially expressed (Log2FC > ±0.585 and adjusted P < 0.01). The number of up- or down-regulated genes for each comparison is shown in red. (C) Venn diagrams showing the relationship between the numbers of DEGs in different DC subsets after treatment with Nb versus PBS. The numbers of DEG in each subset is given in parentheses. The numbers of DEG unique to each subset or shared between populations are shown within the diagram; the numbers of genes that were up-regulated (up), down-regulated (down), or, for intersections, discordantly regulated (opposing) in different DC subsets compared to control are also shown. (D) Heat map showing a selection of top DEG in CD103+, CD11b+, and TN DCs. DC expression data are the mean of three replicate samples each from an independent experiment. P-values were calculated by DESeq2. *, P < 0.05; **, P < 0.01; ***, P < 0.001. #, expression below detection level.
To investigate the genes and signaling pathways that are associated with DC responses to Th2 stimuli, we performed a differential expression analysis between PBS and Nb for individual DC subsets. Treatment with Nb resulted in transcriptional changes in all DC subsets examined, albeit to differing degrees (Fig. 3, B and C; and Table S1). CD103+ DCs, which are not required for the response to Nb, exhibited the least number of DEG, and few with log2 fold-changes (FCs) outside the –1 to +1 range. CD11b+ DCs, which transport Nb antigen into the LN (Connor et al., 2014), showed an intermediate number of DEG, with comparable numbers of down-regulated and up-regulated genes. Finally, TN DCs, which are also involved in Nb antigen transport, had the most DEG, and these were mostly up-regulated by Nb. Interestingly, 24 DEG were shared between the three subsets and only 18 were similarly regulated, indicating that Nb immunization induced a unique although related gene signature in each DC population.
Heat maps illustrating a selection of the top DEG from each DC subset also revealed that the response to Nb varied considerably among DC populations (Fig. 3 D). This variability was most evident for genes encoding cytokines and secreted proteins, but was also observed in genes involved in cell metabolism, where a clear reduction in mitochondrial DNA transcription was detected selectively in CD11b+ DCs. Genes sharing similar expression patterns across DC subsets included those encoding molecules involved in DC activation and motility. Ubd, which regulates NF-κB activity via IkB ubiquitination, and Serpinc1 or antithrombin, which may moderate DC activation and inflammation during sepsis (Niessen et al., 2008), were both selectively up-regulated in the CD11b+ subset. Message for the IL-27 p28 subunit Il27 was also up-regulated in CD11b+ DCs, whereas mRNA for the second subunit of this heterodimeric cytokine, IL-27B or Ebi3, was expressed at stable but detectable levels in all DC subsets (not depicted). Notably, IFN-I response genes were also up-regulated in all three DC subsets.
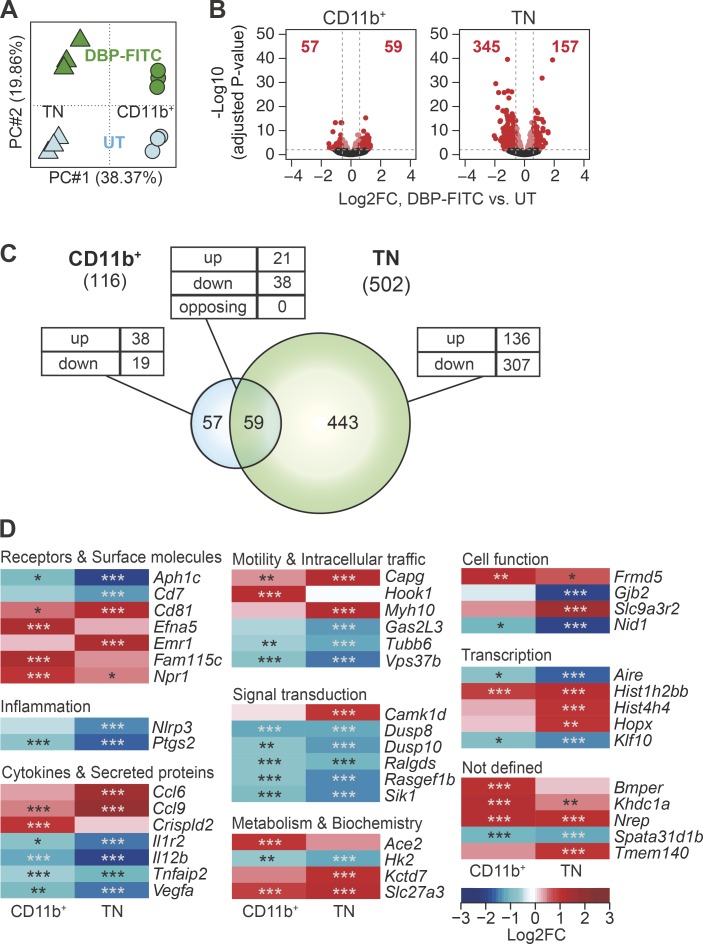
Similar analyses were performed on DCs after DBP-FITC treatment. Again, a PCA indicated that subset differences accounted for the largest proportion of the variability among samples (PC1; 38.37%), whereas transcriptional changes associated with DBP-FITC treatment were revealed on PC2 and represented ∼20% of the variability (Fig. 4 A). Comparison of DEG numbers in the CD11b+ and TN DC subsets showed that DBP-FITC treatment induced transcriptional changes in both populations, but DEG were much more abundant in the TN DC subset and more frequently down-regulated (Fig. 4, B and C; and Table S1). A substantial proportion of the DEG were shared between CD11b and TN DCs, with all 59 shared DEG concordantly regulated. As shown in the heat maps in Fig. 4 D, several genes associated with inflammation, cytokine secretion, and signal transduction were down-regulated in both DC subsets. However, genes associated with chemokine secretion, motility, and metabolism were up-regulated compared with untreated DCs, suggesting that DCs were functional and maintained their ability to interact with T cells.
Figure 4.
Treatment with DBP-FITC is associated with profound transcriptional changes preferentially affecting TN DCs. (A) PCA of genes expressed in CD11b+ and TN DCs from mice treated with DBP-FITC or mock-treated (UT). (B) Volcano plots comparing gene expression in DC subsets from DBP-FITC–treated or UT mice. FC, fold change. Bright red dots and dashed lines identify the genes scored as differentially expressed in each DC subset (Log2FC > ±0.585 and adjusted P < 0.01). The number of up- or down-regulated genes for each comparison is shown in red. (C) Venn diagram showing the relationship between numbers of DEG after treatment with DBP-FITC versus UT in the CD11b+ and TN subsets. The total number of DEG in each subset is given in parentheses. The numbers of DEG unique to each DC subset or shared between populations are shown within the diagram; the numbers of genes that were up-regulated (up), down-regulated (down), or, for intersections, discordantly regulated (opposing) in different DC subsets compared to control are also shown. (D) Heat map showing a selection of the top DEG in CD11b+ and TN DCs. DC expression data are the mean of three replicate samples each from an independent experiment. P-values were calculated by DESeq2. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The DCs that prime Th2 responses express diverse transcriptional profiles
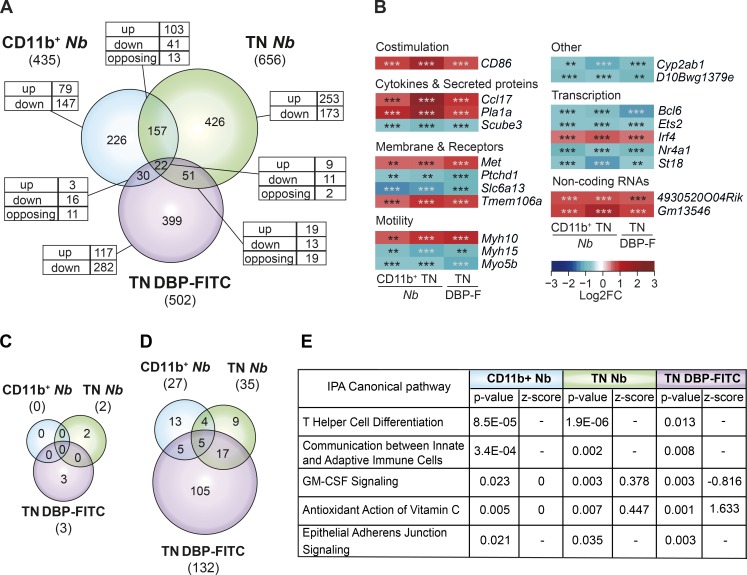
Nb and DBP-FITC induce CD4 responses that are dominated by IL-4 production. To identify potential transcriptional similarities between the DCs that drive these responses, we compared genes that are differentially expressed in CD11b+ or TN DCs exposed to Nb, and TN DCs exposed to DBP-FITC. CD11b+ DCs from DBP-FITC–treated mice were excluded from the comparison because of the low number of DEG in this population. Comparing DEG in these three DC populations revealed 20 genes that were similarly regulated in all subsets, corresponding to only 3–5% of the DEG in each population (Fig. 5 A). The most substantial similarity was between CD11b+ and TN DCs from Nb-treated mice, with 144 of 157 genes being similarly regulated in both populations. In contrast, the DEG shared between TN DCs from DBP-FITC–treated mice, and either CD11b+ or TN DCs from Nb-treated mice, were much fewer and more likely to be discordantly regulated, suggesting that the transcriptional programs initiated by Nb or DBP-FITC were considerably different from each other.
Figure 5.
The transcriptional profiles of DCs that prime Th2 responses are heterogeneous. (A) Venn diagram showing the relationship between the numbers of DEG in CD11b+ and TN DCs from mice treated with 600 Nb versus PBS, and TN DCs from mice treated with DBP-FITC versus UT. The total number of DEG in each subset is given in parentheses. The numbers of DEG unique to each DC subset or shared between populations are shown within the diagram; the numbers of genes that were up-regulated (up), down-regulated (down), or, for the intersection, discordantly regulated (opposing) are also shown. (B) Heat map of the 20 genes that were concordantly regulated in CD11b+ and TN DCs from Nb mice, and TN DCs from DBP-FITC. DC expression data are the mean of three replicate samples each from an independent experiment for each Th2 condition. P-values were calculated by DESeq2; **, P < 0.01; ***, P < 0.001. (C) Venn diagram showing the number of canonical signaling pathways identified as differentially regulated in each DC subset using IPA. Pathways were scored as differentially regulated when the z-score was >+2 or <−2, and P < 0.05. (D) As in C, except that differentially regulated pathways were selected only on the basis of P < 0.05, and no z-score threshold was applied. (E) Description, p-value, and z-score of differentially regulated pathways shared between CD11b+ and TN DCs from Nb mice, and TN DCs from DBP-FITC as identified in D. Where z-score = 0, the direction of a pathway could not be predicted; −, z-score was not assigned.
Heat maps of the 20 shared DEG showed a predictable regulation of genes involved in DC motility and co-stimulation (Fig. 5 B). The up-regulation of the gene encoding the Th2-associated chemokine CCL17 and the down-regulation of Scube3 that encodes for a TGFβRII ligand are also notable. Interestingly, several transcription factors were also similarly regulated in each DC population. Irf4, a key regulator of the differentiation and function of Th2-inducing DCs, was up-regulated, whereas Bcl6, Ets2, Nr4a1 (Nur77), and St18 were down-regulated.
To evaluate more broadly the relationship among DC subsets, we used the Canonical Pathway function of Ingenuity Pathway Analysis (IPA) to search for common signaling pathways in Nb and DBP-FITC DCs. Initial analyses using the recommended IPA thresholds of z-scores >+2 or <–2, and P < 0.05, identified very few potential pathways, which were specific to either Nb or DBP-FITC (Fig. 5 C). Similar results were obtained when using conditions that were made less stringent by either lowering the IPA z-score threshold to ±1.5, or by increasing the number of DEG by removing the cut-off of a log2FC of at least ±0.585. In either case, no shared pathways between Nb and DBP-FITC conditions became apparent, despite a substantial increase in potential pathways specific to either condition (unpublished data). When the requirement for a minimum IPA z-score was removed altogether, a small number of shared pathways could be identified (Fig. 5 D and Table S2), these included generic pathways such as communication between immune cells and GM-CSF signaling. Interestingly, similarities with signaling in Th cells were also detected.
Altogether, these analyses identified few similarities between the transcriptomic profiles of DCs from Nb- or DBP-FITC–treated mice.
DCs isolated from Nb-treated mice express a pronounced IFN-I signature
Data in Fig. 3 show that several IFN-I signaling pathway transcripts were up-regulated in Nb-treated DCs. Therefore, we used the Interferome database (v2.01; Rusinova et al., 2013) to identify all potential IFN-I–regulated genes in our DC populations. As shown in Fig. 6 A and Table S3, as many as 30% of the DEG in DCs from Nb-immunized mice could be linked to IFN-I signaling, including transcripts for molecules involved in sensing nucleic acids, antiviral defense mechanisms, IFN-α/β receptor (IFNAR) signaling, apoptosis, and immune functions such as antigen presentation and cytokine production (Fig. 6 B). No similar changes could be detected in DBP-FITC DCs, suggesting that IFN-I signaling was not activated by this treatment. Interestingly, IFN-I cytokine transcripts were undetectable in every DC subset (not depicted). Thus, DCs from Nb-treated mice had been recently exposed to IFN-I, but whether the source of IFN-I was cell-autonomous or from other cells could not be established from the single time point measured.
Figure 6.
Expression of genes associated with IFN-I signaling in DCs from Nb or DBP-FITC–treated mice. (A) Venn diagram showing the relationship between numbers of IFN-I-regulated DEG after treatment with Nb versus PBS in CD11b+, TN, and CD103+ DC subsets. The total numbers of IFN-I–regulated DEG in each subset is given in parentheses. The numbers of IFN-I–regulated DEG unique to each DC subset or shared between populations are shown within the diagram; the numbers of genes that are up-regulated (up), down-regulated (down), or, for intersections, discordantly regulated (opposing) in different DC subsets compared with control are also shown. (B) Heat maps showing a selection of IFN-I–regulated DEG in DC subsets from Nb-treated and DBP-FITC–treated mice. DC expression data are the mean of three replicate samples each from an independent experiment for each Th2 condition. P-values were calculated by DESeq2; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
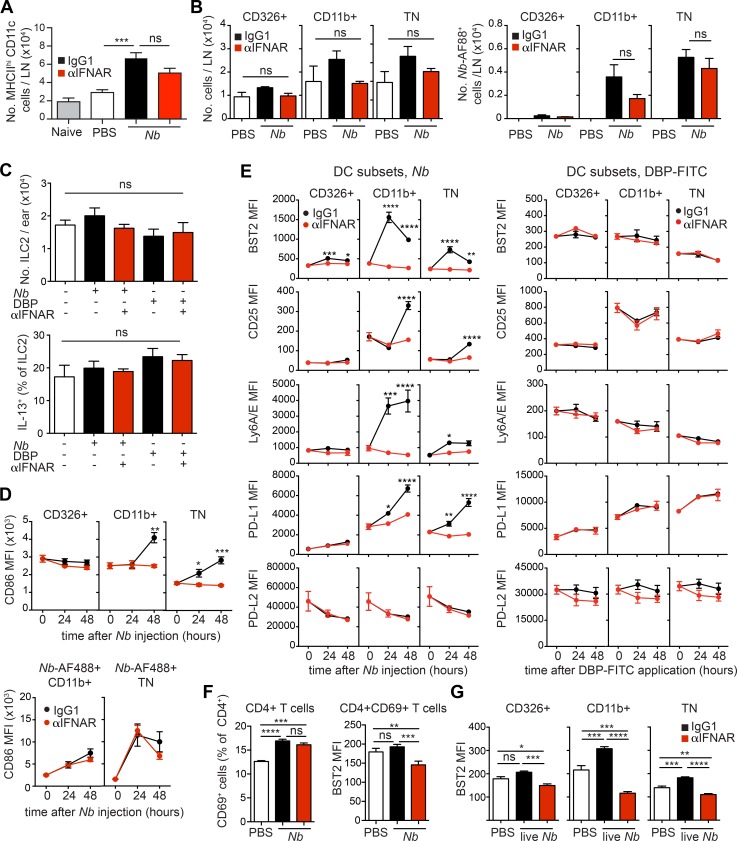
To validate the observation that IFN-I signaling is activated in DCs exposed to Nb, we used an anti-IFNAR (αIFNAR) antibody to block IFN-I signaling at the time of Nb immunization. Mice were injected i.d. with Nb-AF488 together with αIFNAR or isotype control, and dLNs were examined 2 d later. Treatment with αIFNAR had a modest effect on DC migration to the dLN. The number of total MHCIIhi DCs in LN (Fig. 7 A), and the number of DCs in each subset (Fig. 7 B), were decreased by αIFNAR, but this was not statistically significant. Similarly, comparable numbers of Nb-AF488+ CD11b+ and TN DCs could be demonstrated in the dLN of IgG1-treated and αIFNAR-treated mice (Fig. 7 B). The number of type 2 innate lymphocytes (iLC2) in the skin, and their expression of IL-13 (Fig. 7 C), were also not affected by immunization or by αIFNAR treatment, suggesting that differences in iLC2 activity were unlikely to explain differences in dLN DC numbers, phenotype, or function.
Figure 7.
DCs from Nb-treated mice express a pronounced IFN-I signature. C57BL/6 and (C57BL/6 × 4C13R)F1 IL-4/IL-13 double reporter mice were treated with PBS, 300 Nb, or DBP-FITC coadministered with αIFNAR or IgG1 isotype control as indicated. Ear skin or dLN were harvested at the indicated time points for flow cytometric analysis. (A) Numbers of total MHCIIhiCD11c+ DCs in the dLN of C57BL/6 mice treated 48 h earlier with Nb and αIFNAR, as indicated. Bar graphs show mean + SEM for six separate experiments each with four to five mice/group, except for the Naive group, which refers to one experiment with four mice. (B) As in A, except that mice were injected with Nb-AF88 48 h earlier. Numbers of total CD326+ (which includes the CD103+ DC and LC subsets), CD11b+, and TN DCs, and of Nb-AF488+ DCs, are shown. Bar graphs show mean + SEM for three separate experiments each with three to five mice/group. (C) Numbers of iLC2 per ear and percentage of IL-13-dsRed+ iLC2 as measured in 4C13R double reporter mice 24 h after the indicated treatments. Bar graphs show mean + SEM for three to four mice/group. Data refer to one of two repeat experiments that gave similar results. (D) Flow cytometric analysis of DC subsets from C57BL/6 mice immunized with Nb or Nb-AF488 and treated with αIFNAR or IgG1 isotype control as indicated. Mean MFI values ± SEM for five mice/group are shown for each time point. Data refer to one of two repeat experiments that gave similar results. (E) Flow cytometric analysis of DC subsets from C57BL/6 mice immunized with Nb or DBP-FITC and treated with αIFNAR or IgG1 isotype control as indicated. Mean MFI values ± SEM for five mice/group are shown for each time point. Analyses of Nb or DBP-FITC responses were performed in different experiments, thus MFI values are not directly comparable across treatments. Data refer to one of two repeat experiments that gave similar results. (F) C57BL/6 mice were treated as in B. Bar graphs show expression of CD69 and BST2 on LN CD4+ T cells at 48 h after Nb immunization. Mean + SEM for five mice/group; data refer to one of two repeat experiments that gave similar results. (G) As in B, except that C57BL/6 mice were injected with 300 live N. brasiliensis L3 larvae; dLN were collected for flow cytometric analysis at 48 h after immunization. Bar graphs show mean + SEM for five mice/group. Data refer to one of two repeat experiments that gave similar results. P-values were determined using one-way ANOVA with the Bonferroni post-test, except for panels in E, which were determined using a two-way ANOVA with the Bonferroni post-test; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
We then assessed expression of IFN-I–induced markers and co-stimulatory molecules on DC subsets over time. CD86 was up-regulated on total and Nb-AF488+ CD11b+ and TN DCs (Fig. 7 D). Treatment with αIFNAR reversed CD86 up-regulation as assessed on the total DC subset, but had little effect on Nb-AF488+ DCs, suggesting that IFN-I was mainly driving bystander CD86 expression. Expression of BST2 (CD317 or PDCA-1; Blasius et al., 2006), CD25, Ly6A/E (Sca-1), and PD-L1 on CD11b+ and TN DC was up-regulated after Nb immunization (Fig. 7 E). Treatment with αIFNAR reversed this up-regulation, confirming that increased expression of these markers was driven by IFN-I. In contrast, expression of PD-L2 remained stable after Nb, and was not affected by αIFNAR. Expression of all these markers on DCs from DBP-FITC mice was not affected by αIFNAR (Fig. 7 E).
To determine whether other LN populations were also exposed to IFN-I, we assessed expression of IFN-I–induced markers on CD4+ T cells 2 d after Nb. As shown in Fig. 7 F, CD69 was up-regulated on CD4+ T cells after immunization, but this up-regulation was not affected by αIFNAR, suggesting that IFN-I was not involved. Expression of BST2 on total CD4+ T cells (not depicted) or CD4+CD69+ T cells (Fig. 7 F) was not increased after Nb immunization. As also observed in DCs (Fig. 7 E), treatment with αIFNAR reduced BST2 expression to below the levels in PBS-treated mice, suggesting the presence of homeostatic IFN-I signaling in vivo. Thus, IFN-I induced through Nb immunization appeared to have little or no impact on CD4+ T cells in LN.
Finally, we also assessed expression of BST2 on skin LN DCs 48 h after injection with live N. brasiliensis. As shown in Fig. 7 G, BST2 expression on CD11b+ and TN DCs was increased in an IFN-I–dependent fashion, suggesting that induction of IFN-I was not limited to immunization with nonviable Nb.
Thus, αIFNAR treatment could prevent IFN-I signaling in DCs with little impact on DC migration to the dLN, or their transport of AF488-labeled Nb material.
Treatment with IFNAR-blocking antibodies impairs Th2 induction after Nb, but not DBP-FITC, immunization
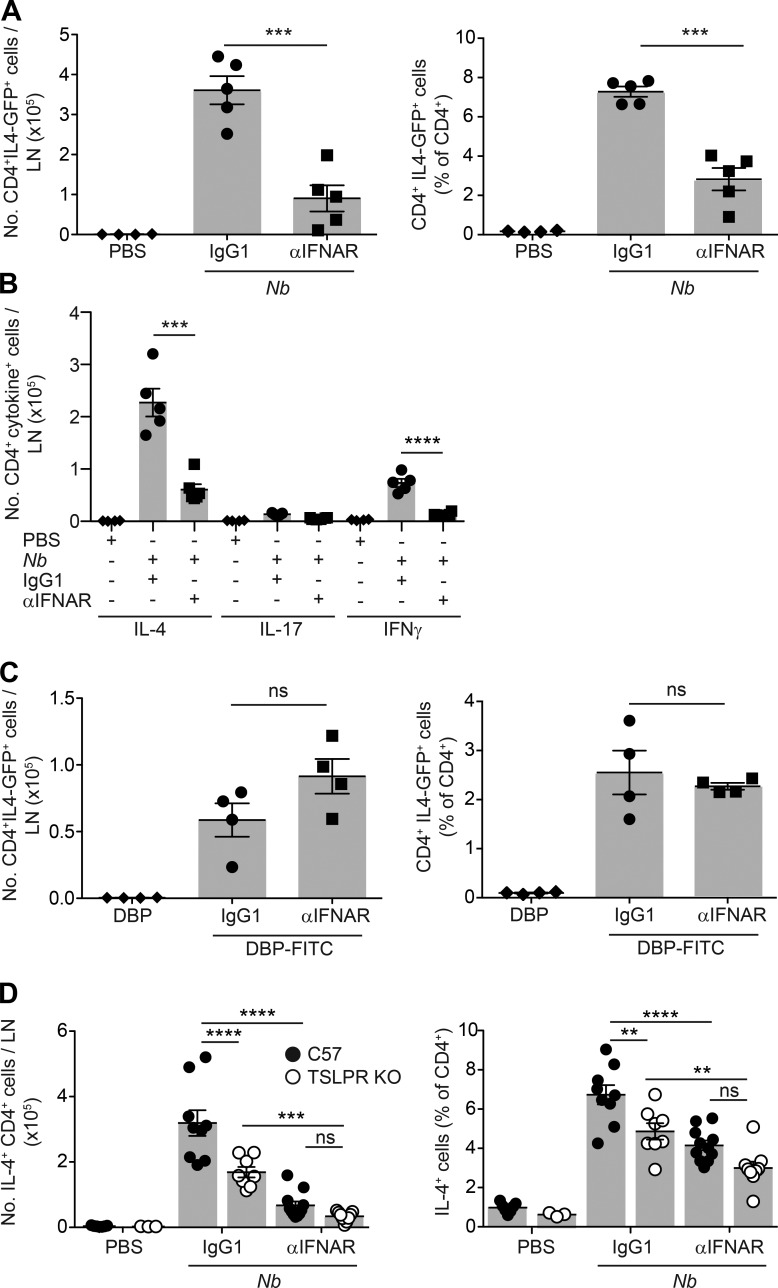
To assess the effect of αIFNAR on the priming of IL-4–producing T cells, we used IL-4-GFP reporter mice treated with Nb. Compared with isotype controls, treatment with αIFNAR led to a marked reduction in the number and proportion of CD4+ IL-4-GFP+ T cells in dLN (Fig. 8 A). Reduced numbers of IL-4+ CD4+ cells were also observed when cytokine production was assessed by in vitro restimulation and intracellular cytokine staining. In addition, the IFN-γ response observed after Nb immunization was not increased by αIFNAR treatment, but reduced, and the IL-17 response remained very low (Fig. 8 B). In contrast, treatment with αIFNAR did not affect the number or percentage of IL-4-GFP+ T cells after DBP-FITC treatment (Fig. 8 C).
Figure 8.
IFN-I signaling is required for optimal priming of IL-4–producing T cells after Nb immunization and cooperates with TSLP in promoting Th2 responses. IL-4-GFP+/−, C57BL/6, or TSLPR KO mice were treated with 300 Nb, DBP-FITC, or the respective controls coadministered with αIFNAR or IgG1 isotype as indicated. 1 wk later, dLN were harvested for cytokine production assessment. (A) Numbers and percentages of IL-4-GFP+ CD4+ T cells in dLN as measured in IL-4-GFP+/− mice on day 7 after treatment with Nb and αIFNAR or IgG1 isotype control. (B) As in A, except that the number of cytokine+ T cells was measured in C57BL/6 mice by intracellular staining after in vitro restimulation with PMA and ionomycin. (C) As in A, except that IL-4-GFP mice were treated with DBP-FITC or DBP only. (D) As in A, except that the number of cytokine+ T cells was measured in C57BL/6 or TSLPR KO mice by intracellular staining after in vitro restimulation with PMA and ionomycin. Bar graphs show mean ± SEM for 3–11 mice/group; each symbol corresponds to one mouse. A–C show data from one of at least two repeat experiments that gave similar results. D shows combined data from two repeat experiments. P-values were determined using one-way (A–C) or two-way (D) ANOVA with the Bonferroni post-test; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
Treatment with αIFNAR reduced but did not block Th2 development after Nb immunization. To establish whether IFN-I may act in concert with other Th2-inducing cytokines, we assessed the effect of αIFNAR in TSLPR KO mice, as Nb injection induced low but detectable levels of Tslp transcripts. Compared with C57BL/6 mice, TSLPR KO mice generated lower numbers and lower percentages of IL-4–producing T cells after Nb immunization. Treatment with αIFNAR further reduced the response in TSLPR KO mice to very low levels, suggesting that IFN-I and TSLP promote Th2 immune responses via independent pathways (Fig. 8 D).
Discussion
In this study, we show that the transcriptional profiles of DC subsets from skin dLN of mice treated with two different Th2-inducing agents, Nb and DBP-FITC, are diverse and may represent differential abilities and modes to instruct Th2 immune responses. Among the four main migratory DC populations in the skin dLN, the IRF4-dependent PD-L2+ CD11b+ subset and the TN DC subset have both been shown to be necessary for Th2 immunity (Gao et al., 2013; Kumamoto et al., 2013; Murakami et al., 2013; Tussiwand et al., 2015). Here, we show that both of these subsets undergo substantial transcriptional changes after exposure to Th2-inducing agents, and that a large proportion of those changes are both DC subset–specific and treatment-specific, suggesting a considerable degree of specialization within DC subsets.
The divergence of the DC transcriptomes associated with Th2 responses to Nb and DBP-FITC is consistent with the observation in this paper that these agents elicit different cytokines. Previous work has demonstrated that TSLPR signaling in DCs is essential for Th2 responses to DBP-FITC (Bell et al., 2013), whereas the immune response to Nb can develop in the absence of TSLPR (Connor et al., 2014). Accordingly, we find that DCs from DBP-FITC-treated mice up-regulate TSLP-regulated genes such as Cd86, Ccl17, and Cish (Zhong et al., 2014) and down-regulate IL12b (Taylor et al., 2009). This signature was only partly observed after Nb immunization, suggesting that additional factors other than TSLP were influencing expression of those markers. We show in this paper that DCs from Nb-treated mice, and especially the CD11b+ and TN subsets, express a clear IFN-I signature. Flow cytometry experiments confirmed that the IFN-I–responsive markers BST2, Ly6A/E, CD25, and PD-L1 were up-regulated on CD11b+ and TN DCs after Nb, but not after DBP-FITC, and that this up-regulation could be blocked by αIFNAR treatment. We also show that IFNAR signaling is functionally important, as blocking IFNAR signaling led to impaired IL-4 responses after Nb immunization without affecting the response to DBP-FITC. Thus, the effects of the powerful innate cytokines TSLP and IFN-I can explain the transcriptional heterogeneity of DCs in the Th2 immune response models used in this study.
Our observation that IFN-I is involved in the priming of Th2 cells was surprising because this cytokine has been shown to play important roles in the induction of type I CD4+ and CD8+ T cell responses to viruses and tumors (Trinchieri, 2010), and immune responses to fungal pathogens, such as Candida albicans or vaccine adjuvants like chitosan (del Fresno et al., 2013; Carroll et al., 2016). Interestingly, immune responses to these agents are reported to require DC subsets, such as the CD103+ DCs for antiviral and antitumor immune responses (Hildner et al., 2008), and Langerhans cells for the immune response to Candida (del Fresno et al., 2013; Smeekens et al., 2013), that are distinct from the DC subsets that induce Th2 responses. These results may suggest that other DC subset-specific and/or antigen-specific signals, cytokines, or cell populations differentially induced by these very different agents may also contribute to the diversity of the resulting response.
Experiments in TSLPR KO mice suggested that IFN-I and TSLP both contributed to the Th2 response to Nb, with IFNAR signaling making the larger contribution to the number, but not the percentage, of IL-4–producing T cells in dLN. However, simultaneously blocking signaling by both cytokines was still insufficient to fully ablate Th2 priming after Nb injection, suggesting that additional cytokines or cell-mediated signals were probably involved. This observation may reflect the complexity of multicellular organisms such as parasites, which are likely to trigger multiple innate response mechanisms. The production of IFN-I by DCs exposed in vitro to parasite products, such as SEA has been reported in previous studies (Trottein et al., 2004), and we show in this paper that live N. brasiliensis infection could also induce expression of IFN-I responsive markers in DCs. In addition, experiments in our laboratory showed that IFN-I signaling was also required for immune responses to nonparasitic allergens, such as the common household allergen house dust mite (unpublished data). Experiments to identify the cellular sources and signaling pathways that drive IFN-I production after Nb injection will be essential in understanding the basis of these responses.
The IFNAR is widely expressed on most cells in the body, and our IFNAR-blocking experiments could not distinguish between IFN-I effects on DCs versus other cell types. Therefore, we used BST2 expression as a readout of IFN-I exposure in vivo as this marker is sensitively and specifically induced by IFN-I (Blasius et al., 2006). We found that after Nb immunization BST2 was only marginally up-regulated on CD4+ T cells, and was highest on MHCIIhi CD11b+ and TN DCs, suggesting that these DC populations were preferentially exposed to IFN-I. We also examined iLC2 in skin, as IFN-I can affect the function of these cells (Duerr et al., 2016). We found that iLC2 numbers and their production of IL-13, which can support DC migration and function (Halim et al., 2014, 2016), were also not affected by IFN-I. Thus, our data are consistent with IFN-I primarily affecting the function of the DC subsets that transport Nb-AF488 to the LN.
IFN-I–dependent activation of DCs is critical for the immunostimulatory function of adjuvants such as polyI:C, through the up-regulation of the co-stimulatory molecules CD86 and CD70 (Longhi et al., 2009). In our model, blocking IFN-I signaling did not significantly affect the number of Nb-AF488+ DCs in the LN or their up-regulation of CD86, suggesting that DC migration and activation were not exclusively IFN-I dependent. Nonetheless, adaptive T cell immunity to Nb was impaired by anti-IFNAR treatment, perhaps suggesting that other DC functions were affected. In this context, it is interesting to note that IFN-I is reported to inhibit inflammasome activation (Guarda et al., 2011) and down-regulate TLR-dependent production of inflammatory cytokines during late phases of the immune response (Rothlin et al., 2015). Consistent with this possibility, we show that genes that inhibit cytokine production, such as Cish, Socs2, and CD200, were up-regulated in DCs conditioned by either Nb or DBP-FITC.
The absence of proinflammatory cytokine production by DCs has previously been linked to Th2 development. Mice lacking MyD88, a critical component of the TLR signaling pathway and the initiation of proinflammatory cytokine production, develop a Th2 immune profile when exposed to Th1 microbial stimuli (Jankovic et al., 2002). Similarly, DCs activated in vitro under suboptimal conditions fail to produce proinflammatory cytokines and preferentially prime T cells to Th2 (Pletinckx et al., 2011). We show that DCs exposed to Th2 conditions in vivo up-regulate transcripts for some proinflammatory molecules; however, up-regulation was variable with respect to Th2 condition, DC subset, and the proinflammatory molecule involved. Comparison to published data shows that CD11b+ DCs from DNFB-treated mice (Tamoutounour et al., 2013) and human monocyte-derived DCs exposed to E. coli or influenza virus in culture (Huang et al., 2001) up-regulate transcripts for Tnfsf9 (CD137L), Tnfrsf9 (CD137) and Icam1. These molecules, which are reported to favor Th1 differentiation by supporting IL-12 production by DCs and strengthening DC–T cell interaction (Futagawa et al., 2002; Smits et al., 2002; Wilcox et al., 2002), were either stably expressed or down-regulated in our Th2 conditions. In addition, transcripts for the transcription factors Ets2 and St18 were also down-regulated; these molecules are reported to promote inflammatory cytokine production and apoptosis in different models (Wolvetang et al., 2003; Yang et al., 2008; van der Pouw Kraan et al., 2009). Thus, the simple lack of proinflammatory cytokine production by DCs, also known as the default hypothesis, may be one element involved in DC instruction of Th2 responses.
Our analysis of DC subsets isolated from Th2-inducing conditions revealed few genes that were concordantly regulated by Nb and DBP-FITC, a finding also reflected in the very low number of shared signaling pathways as identified by IPA. However, among the shared DEG were several that have been previously associated with DCs and Th2 immunity, including Ccl17 (Alferink et al., 2003; Gros et al., 2009; Fujita et al., 2011; Kitajima and Ziegler, 2013) and Irf4 (Gao et al., 2013; Williams et al., 2013). The transcription factor IRF4 has a well-recognized role in the development and function of Th2-inducing DCs, but was initially identified for its role in modulating IL-4 responsiveness and production in lymphocytes (Gupta et al., 1999; Rengarajan et al., 2002). Interestingly, analysis of shared DEG in DCs identified additional transcripts that can be linked to Irf4 through their known interactions in lymphocytes. BCL6 and NR4A1, whose transcripts are down-regulated in Th2 DCs, are reported to suppress IRF4 activity in B cells and CD8+ T cells, respectively (Gupta et al., 1999; Nowyhed et al., 2015). In addition, BCL6 KO mice develop exaggerated Th2 inflammation caused by dysregulated STAT6 activity (Dent et al., 1997). Although the precise role of BCL6 in DC function is only starting to be characterized (Zhang et al., 2014), the observation that multiple cell types similarly regulate overlapping transcriptional networks during Th2 responses may suggest the activation of common genetic programs after exposure to Th2-inducing agents.
In conclusion, we report an unanticipated diversity in the transcriptional profile of CD11b+ and TN DC subsets priming different Th2 responses in the skin. This diversity reflects differences in the cytokines, IFN-I and TSLP, driving the Th2 responses under investigation, and might in turn underlie different properties of the Th2 immune responses being generated (Bouchery et al., 2014). Studies to examine additional allergens might reveal further heterogeneity. Altogether, our findings pose the question of how the observed diversity may translate into a common instructive DC signal that drives Th2 differentiation.
Materials and methods
Mice
Age- and sex-matched specific pathogen–free C57BL/6J, Langerin-DTR (Kissenpfennig et al., 2005), TSLPR KO (Al-Shami et al., 2004), C57BL/6IL4-GFP/wt (Hu-Li et al., 2001), and (C57BL/6 × 4C13R)F1 (Roediger et al., 2013) mice, all on a C57BL/6 background, were bred and housed at the Malaghan Institute of Medical Research Animal Facility. Batf3−/− (Hildner et al., 2008) were backcrossed to C57BL/6J in our facility for 10 generations. In all experiments, C57BL/6 mice were used as controls. To deplete Langerin+ DCs, Langerin-DTR mice were given i.p injections of 0.35–1 µg DT (Sigma-Aldrich) 1 d before and 1 d after Nb or DBP-FITC treatment, respectively. All experimental protocols were approved by the Victoria University Animal Ethics Committee and performed according to institutional guidelines.
Immunizations and in vivo treatments
N. brasiliensis infective L3 larvae were prepared, washed in sterile PBS, killed by three freeze–thaw cycles, and injected into the ear pinna of anaesthetized mice, as previously described (Camberis et al., 2013). In some experiments live L3 larvae were injected using the same protocol. For fluorescent labeling, nonviable L3 were incubated in 0.05 M NaHCO3 buffer and 0.1 mg of Alexa Fluor 488 Microscale Protein labeling dye (Molecular Probes), and then washed with 0.1 M Tris buffer. For DBP-FITC application, hair was removed from recipient mice 1 wk before sensitization and 20 µl of a solution of 0.5% FITC (Sigma-Aldrich) in 50% DBP (BDH Laboratory Supplies)/50% acetone (Pacific Sphere Limited) were applied epicutaneously on the back, abdomen, and ear as previously described (Ochiai et al., 2014). To block IFN-I cytokine signaling in vivo, recipient mice were injected on day 0 with 200 µg anti–mouse IFNAR1 (MAR1-5A3) or mouse IgG1 (MOPC-21) antibodies (Bio X Cell) given i.d. with the Th2 immunizations. A second injection of 200 µg antibody was given i.p. on day 2.
Quantitative real-time RT-PCR
Ears were collected at indicated times after injection with 600 Nb, or topical application of DBP-FITC. Epidermal layers were separated from the dermis by digestion with Dispase II (Roche), and total RNA was extracted from the tissue using an RNeasy mini kit (QIAGEN). cDNA was synthesized by using a High-Capacity RNA-to-cDNA kit (Applied Biosystems), and quantitative PCR was performed using TaqMan gene expression master mix and primers (Applied Biosystems). TSLP data are normalized to 18S RNA and expressed as relative to control samples.
DC sorting
For Nb-treated mice, auricular LN were collected from 15 mice at 48 h after treatment and made into single-cell suspensions by digestion with DNase I and Liberase TL (Roche). CD11c+ cells were enriched by negative magnetic selection (Dynabead Mouse DC Enrichment Kit; Invitrogen). Enriched cells were labeled with a cocktail of fluorescent antibodies specific for: CD11c (HL3) and CD11b (M1/70; both from BD); and MHCII (M5-114), CD326 (G8.8), and CD103 (2E7; all from BioLegend). Anti-Ly6C (AL-21; BD) was also added when sorting CD11b+ DCs from Nb-treated mice. For DBP-FITC treatment, brachial, axillary, inguinal, and auricular LN were collected from seven donor mice per group at 24 h after treatment, and processed as for Nb DC preparation. Enriched cells were labeled with fluorescent antibodies specific for CD11c (HL3), MHCII (M5-114), CD11b (M1/70), CD326 (G8.8), CD24 (M1/69), and anti-Ly6C (AL-21; eBioscience). Dead cells were identified and excluded from analysis using DAPI labeling (Molecular Probes). DC populations were sorted and isolated using a FACSVantage Diva (BD) with BD FACSDiva software, version 5.0.3 (BD). Cell pellets of sorted DC populations were frozen at –80°C until RNA extraction. Three biological replicates for each treatment were prepared in three separate DC sorting experiments.
RNA isolation and sequencing
Total RNA was prepared from frozen cell pellets using QIAzol Lysis Reagent and the RNeasy Micro Kit (all from QIAGEN). RNA was quantified using a Qubit Fluorometer (Invitrogen) and RNA integrity was checked using an Agilent Bioanalyzer and RNA 6000 Nano Kit (Agilent Technologies).
Library preparation and RNA sequencing were contracted out to New Zealand Genomics Limited. Samples were spiked with External RNA Controls Consortium mix (Ambion; Life Technologies) and ribosomal RNA was depleted using RiboZero (Illumina) before library preparation. Paired-end stranded RNA sequencing was performed on an Illumina HiSeq system using Illumina TruSeq kits. Between 10 and 30 million read pairs were generated per sample.
Read mapping and differential expression analysis
Paired-end raw read FASTq files were preprocessed using Trimmomatic (v0.32) and quality checked using the qrqc R package (v1.20.0). Reads were mapped to the C57BL/6J mouse reference genome, version GRCm38.p3, using TopHat2 (v2.0.10). Read hits to annotated genes were counted using HtSeq-count (HT-Seq, v0.6.0) excluding Y chromosome genes. Statistical analysis of gene expression counts was performed in R (v3.1.3) using the R package DESeq2 (v1.6.3; Anders and Huber, 2010). A variance-stabilizing transformation (VST) was applied to count data using the DESeq2 function varianceStabilizingTransformation, producing gene expression values that are adjusted for variance across sample conditions. To compare different genes to each other, this matrix was further transformed by dividing by transcript length in kbp to generate VSTPk values. For each desired comparison, gene differential expression was calculated from regularized log-fold-changes and multiple-testing-corrected P-values. Where mouse sex was a covariate, it was included in the experimental model supplied to DESeq2. Genes were considered to be differentially expressed when P < 0.01 and log2FC was >0.585 in either direction (which corresponds to FC > 1.5 in either direction).
Volcano plots, PCA plots (using R package FactoMineR), and Venn diagrams were generated from genes that were initially filtered to exclude those with low overall expression in all samples. VSTPk values were compared across all samples and genes with a maximum VSTPk of −2 or less were excluded, leading to a quasi-normal VSTPk distribution. Heat maps were generated excluding noncoding genes, genes where transcript mapping was unclear, and genes that are known not to be expressed in cDCs.
For comparisons to published data (Fig. S2; Miller et al., 2012), a generalized linear model was fitted to the relationship between log-transformed microarray and VSTPk expression levels obtained from the ImmGen Project database, and was used to transform the microarray data into values comparable to our VSTPks.
Pathway analysis
Datasets obtained after DESeq2 analysis were analyzed through the use of QIAGEN’s Ingenuity Pathway Analysis (IPA). Canonical pathway and upstream regulator analyses were performed on genes that were up- and down-regulated, using a P-value cut-off of 0.01 and a log2FC of 0.585. In IPA, the pathways were compared using a P-value cut-off of 0.05 with and without an activation z-score threshold of 2.
Cell preparations
For DC preparation, auricular LN were collected and digested with DNase I and Liberase TL (Roche) for 25 min at 37°C. For skin ILC2 preparation, ears were split into dorsal and ventral layers and digested in collagenase IV (Sigma-Aldrich) and DNase I (Roche) for 30 min at 37°C. Cells were passed through a 70-µm nylon cell strainer and washed in flow cytometry buffer in preparation for staining. ILC2 cells were identified as CD45+CD90.2+CD2−CD3−NK1.1−. To assess IL-13 production by iLC2 cells, 4C13R mice were used and IL-13-DSred reporter expression was detected by flow cytometry.
Flow cytometry
For staining of cell surface molecules, cells were incubated with anti–mouse CD16/32 (clone 2.4G2; affinity purified from hybridoma culture supernatant) before labeling with cocktails of fluorescent antibodies specific for: CD11c (HL3), CD86 (GL1), MHCII (M5/114), CD326 (G8.8), CD4 (RM4-5), CD3 (145-2C11), and CD44 (IM7; all from BD); CD11b (M1/70), CD25 (7D4), Ly6A/E (D7), CD45 (30-F11), CD90.2 (53.2.1), CD2 (RM2-5), NK1.1 (PK136; BD); Ly6C (HK1.4), CD317 (BST2, PDCA-1; clone 927), CD273 (PDL2, Ty25), CD69 (H1.2F3; all from BioLegend); CD274 (PDL1, MIH5) and B220 (RA3-6B2; both from eBioscience). For intracellular cytokine staining, cells were surface stained, and then fixed and permeabilized using the Cytofix/Cytoperm kit (BD), and labeled with anti–IL-4 (11B11), anti–IFN-γ (XMG1.2), or respective IgG1 isotype controls (all from BD). Anti–IL-17A (eBio17B7) was obtained from eBioscience. Dead cells and doublets were identified and excluded from analysis using DAPI labeling or LIVE/DEAD Fixable Blue dead cell stain kit (both from Molecular Probes). Compensation was set in each experiment using CompBeads (BD). All samples were collected on a LSRII SORP flow cytometer (BD) and analyzed using FlowJo version 9.6.2 (Tree Star).
Assessment of in vivo T cell responses
7 d after i.d. challenge, mice were sacrificed and auricular LN were collected for analysis. To assess IL-4 production using IL-4-GFP reporter expression, live CD3+ CD4+ T cells were identified by flow cytometry, and the percentage of IL-4-GFP+ cells was determined by comparison to an IL-4-GFPneg C57BL/6 control. To assess IL-4, IFN-γ, and IL-17 production by intracellular cytokine staining LN cells were cultured in fetal bovine serum–supplemented Iscove’s modified Dulbecco medium in the presence of 1 µg/ml ionomycin, 50 ng/ml PMA, and GolgiStop (BD) for 5 h, or cultured on plate-coated anti-CD3 with soluble anti-CD28 and IL-2 for 4 h, with GolgiStop added during the final 2 h. Cells were then fixed and permeabilized as described (Connor et al., 2014). Statistical analyses were performed using Prism 5.0 GraphPad Software. Mean ± SEM is shown in all graphs. Data were analyzed using one-way or two-way ANOVA with Bonferroni post-test; p-values < 0.05 were considered significant.
Data access
RNA-seq data are deposited in Gene Expression Omnibus, available under accession no. GSE88998.
Online supplemental material
Fig. S1 shows the gating strategy used for DC flow-sorting and back-gating analysis of individual DC subsets. Fig. S2 shows the expression of genes associated with LN migratory DCs, LN resident DCs, LN monocytes, and LN pDCs in individual DC subsets in this study. Table S1 shows a list of all DEG in DC subsets, by DC subset and Th2 condition. Table S2 shows a list of canonical pathways in DC subsets as detected by IPA. Table S3 shows the list of IFN-I regulated genes in each DC subset as determined using the Interferome database. Tables S1–S3 are available as Excel files.
Supplementary Material
Acknowledgments
The authors wish to thank Andrew MacDonald, University of Manchester, for discussion; Aaron Jeffs and Dan Jones, New Zealand Genomics Limited, for invaluable advice on RNA sequencing experiments; and A. Kissenpfennig, G. Le Gros, W. Leonard, K. Murphy, and the late W.E. Paul for providing mouse strains. The expert support of the MIMR Flow Cytometry, Information Technologies, and Animal Facility staff are also gratefully acknowledged. This work benefitted from data assembled by the ImmGen consortium.
This work was funded by research grants from the Health Research Council of New Zealand to F. Ronchese. S. Ochiai and K.L. Hilligan were supported by PhD Scholarships from the University of Otago, New Zealand.
L.M. Connor and F. Ronchese are listed as inventors on a provisional patent application concerned with the subject matter of this paper. The authors declare no additional competing financial interests.
Footnotes
Abbreviations used:
- DBP
- dibutyl phthalate
- DEG
- differentially expressed gene
- dLN
- draining LN
- DTR
- diphtheria toxin receptor
- FC
- fold change
- IFNAR
- IFN α/β receptor
- IFN-I
- type I IFN
- IPA
- Ingenuity Pathway Analysis
- Nb
- N. brasiliensis
- PCA
- principal component analysis
- TN
- triple negative
- TSLP
- thymic stromal lymphopoietin
- UT
- untreated
References
- Alferink J., Lieberam I., Reindl W., Behrens A., Weiss S., Hüser N., Gerauer K., Ross R., Reske-Kunz A.B., Ahmad-Nejad P., et al. 2003. Compartmentalized production of CCL17 in vivo: strong inducibility in peripheral dendritic cells contrasts selective absence from the spleen. J. Exp. Med. 197:585–599. 10.1084/jem.20021859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shami A., Spolski R., Kelly J., Fry T., Schwartzberg P.L., Pandey A., Mackall C.L., and Leonard W.J.. 2004. A role for thymic stromal lymphopoietin in CD4+ T cell development. J. Exp. Med. 200:159–168. 10.1084/jem.20031975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., and Huber W.. 2010. Differential expression analysis for sequence count data. Genome Biol. 11:R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell B.D., Kitajima M., Larson R.P., Stoklasek T.A., Dang K., Sakamoto K., Wagner K.U., Kaplan D.H., Reizis B., Hennighausen L., and Ziegler S.F.. 2013. The transcription factor STAT5 is critical in dendritic cells for the development of TH2 but not TH1 responses. Nat. Immunol. 14:364–371. 10.1038/ni.2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius A.L., Giurisato E., Cella M., Schreiber R.D., Shaw A.S., and Colonna M.. 2006. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J. Immunol. 177:3260–3265. 10.4049/jimmunol.177.5.3260 [DOI] [PubMed] [Google Scholar]
- Bouchery T., Kyle R., Ronchese F., and Le Gros G.. 2014. The differentiation of CD4+ T-helper cell subsets in the context of helminth parasiteinfection. Front. Immunol. 5:487 10.3389/fimmu.2014.00487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camberis M., Prout M., Tang S.C., Forbes-Blom E., Robinson M., Kyle R., Belkaid Y., Paul W., and Le Gros G.. 2013. Evaluating the in vivo Th2 priming potential among common allergens. J. Immunol. Methods. 394:62–72. 10.1016/j.jim.2013.05.004 [DOI] [PubMed] [Google Scholar]
- Carroll E.C., Jin L., Mori A., Muñoz-Wolf N., Oleszycka E., Moran H.B., Mansouri S., McEntee C.P., Lambe E., Agger E.M., et al. 2016. The vaccine adjuvant chitosan promotes cellular immunity via DNA sensor cGAS-STING-dependent induction of type I interferons. Immunity. 44:597–608. 10.1016/j.immuni.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P.Y., Carrera Silva E.A., De Kouchkovsky D., Joannas L.D., Hao L., Hu D., Huntsman S., Eng C., Licona-Limón P., Weinstein J.S., et al. 2016. The TAM family receptor tyrosine kinase TYRO3 is a negative regulator of type 2 immunity. Science. 352:99–103. 10.1126/science.aaf1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor L.M., Tang S.C., Camberis M., Le Gros G., and Ronchese F.. 2014. Helminth-conditioned dendritic cells prime CD4+ T cells to IL-4 production in vivo. J. Immunol. 193:2709–2717. 10.4049/jimmunol.1400374 [DOI] [PubMed] [Google Scholar]
- Cook P.C., Owen H., Deaton A.M., Borger J.G., Brown S.L., Clouaire T., Jones G.R., Jones L.H., Lundie R.J., Marley A.K., et al. 2015. A dominant role for the methyl-CpG-binding protein Mbd2 in controlling Th2 induction by dendritic cells. Nat. Commun. 6:6920 10.1038/ncomms7920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalod M., Chelbi R., Malissen B., and Lawrence T.. 2014. Dendritic cell maturation: functional specialization through signaling specificity and transcriptional programming. EMBO J. 33:1104–1116. 10.1002/embj.201488027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Fresno C., Soulat D., Roth S., Blazek K., Udalova I., Sancho D., Ruland J., and Ardavín C.. 2013. Interferon-β production via Dectin-1-Syk-IRF5 signaling in dendritic cells is crucial for immunity to C. albicans. Immunity. 38:1176–1186. 10.1016/j.immuni.2013.05.010 [DOI] [PubMed] [Google Scholar]
- Dent A.L., Shaffer A.L., Yu X., Allman D., and Staudt L.M.. 1997. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 276:589–592. 10.1126/science.276.5312.589 [DOI] [PubMed] [Google Scholar]
- Duerr C.U., McCarthy C.D., Mindt B.C., Rubio M., Meli A.P., Pothlichet J., Eva M.M., Gauchat J.F., Qureshi S.T., Mazer B.D., et al. 2016. Type I interferon restricts type 2 immunopathology through the regulation of group 2 innate lymphoid cells. Nat. Immunol. 17:65–75. 10.1038/ni.3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelson B.T., Kc W., Juang R., Kohyama M., Benoit L.A., Klekotka P.A., Moon C., Albring J.C., Ise W., Michael D.G., et al. 2010. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J. Exp. Med. 207:823–836. 10.1084/jem.20091627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B., Tussiwand R., Dreesen L., Fairfax K.C., Huang S.C., Smith A.M., O’Neill C.M., Lam W.Y., Edelson B.T., Urban J.F. Jr., et al. 2016. Migratory CD103+ dendritic cells suppress helminth-driven type 2 immunity through constitutive expression of IL-12. J. Exp. Med. 213:35–51. 10.1084/jem.20150235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H., Shemer A., Suárez-Fariñas M., Johnson-Huang L.M., Tintle S., Cardinale I., Fuentes-Duculan J., Novitskaya I., Carucci J.A., Krueger J.G., and Guttman-Yassky E.. 2011. Lesional dendritic cells in patients with chronic atopic dermatitis and psoriasis exhibit parallel ability to activate T-cell subsets. J. Allergy Clin. Immunol. 128:574–82.e1: 12. 10.1016/j.jaci.2011.05.016 [DOI] [PubMed] [Google Scholar]
- Futagawa T., Akiba H., Kodama T., Takeda K., Hosoda Y., Yagita H., and Okumura K.. 2002. Expression and function of 4-1BB and 4-1BB ligand on murine dendritic cells. Int. Immunol. 14:275–286. 10.1093/intimm/14.3.275 [DOI] [PubMed] [Google Scholar]
- Gao Y., Nish S.A., Jiang R., Hou L., Licona-Limón P., Weinstein J.S., Zhao H., and Medzhitov R.. 2013. Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity. 39:722–732. 10.1016/j.immuni.2013.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros E., Bussmann C., Bieber T., Förster I., and Novak N.. 2009. Expression of chemokines and chemokine receptors in lesional and nonlesional upper skin of patients with atopic dermatitis. J. Allergy Clin. Immunol. 124:753–60.e1. 10.1016/j.jaci.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Guarda G., Braun M., Staehli F., Tardivel A., Mattmann C., Förster I., Farlik M., Decker T., Du Pasquier R.A., Romero P., and Tschopp J.. 2011. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 34:213–223. 10.1016/j.immuni.2011.02.006 [DOI] [PubMed] [Google Scholar]
- Gupta S., Jiang M., Anthony A., and Pernis A.B.. 1999. Lineage-specific modulation of interleukin 4 signaling by interferon regulatory factor 4. J. Exp. Med. 190:1837–1848. 10.1084/jem.190.12.1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim T.Y., Steer C.A., Mathä L., Gold M.J., Martinez-Gonzalez I., McNagny K.M., McKenzie A.N., and Takei F.. 2014. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 40:425–435. 10.1016/j.immuni.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim T.Y., Hwang Y.Y., Scanlon S.T., Zaghouani H., Garbi N., Fallon P.G., and McKenzie A.N.. 2016. Group 2 innate lymphoid cells license dendritic cells to potentiate memory TH2 cell responses. Nat. Immunol. 17:57–64. 10.1038/ni.3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad H., Plantinga M., Deswarte K., Pouliot P., Willart M.A.M., Kool M., Muskens F., and Lambrecht B.N.. 2010. Inflammatory dendritic cells--not basophils--are necessary and sufficient for induction of Th2 immunity to inhaled house dust mite allergen. J. Exp. Med. 207:2097–2111. 10.1084/jem.20101563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100. 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Liu D., Majewski P., Schulte L.C., Korn J.M., Young R.A., Lander E.S., and Hacohen N.. 2001. The plasticity of dendritic cell responses to pathogens and their components. Science. 294:870–875. 10.1126/science.294.5543.870 [DOI] [PubMed] [Google Scholar]
- Hu-Li J., Pannetier C., Guo L., Löhning M., Gu H., Watson C., Assenmacher M., Radbruch A., and Paul W.E.. 2001. Regulation of expression of IL-4 alleles: analysis using a chimeric GFP/IL-4 gene. Immunity. 14:1–11. 10.1016/S1074-7613(01)00084-X [DOI] [PubMed] [Google Scholar]
- Hussaarts L., Yazdanbakhsh M., and Guigas B.. 2014. Priming dendritic cells for th2 polarization: lessons learned from helminths and implications for metabolic disorders. Front. Immunol. 5:499 10.3389/fimmu.2014.00499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Wang Y.H., Duramad O., Hori T., Delespesse G.J., Watanabe N., Qin F.X., Yao Z., Cao W., and Liu Y.J.. 2005. TSLP-activated dendritic cells induce an inflammatory T helper type 2 cell response through OX40 ligand. J. Exp. Med. 202:1213–1223. 10.1084/jem.20051135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic D., Kullberg M.C., Hieny S., Caspar P., Collazo C.M., and Sher A.. 2002. In the absence of IL-12, CD4+ T cell responses to intracellular pathogens fail to default to a Th2 pattern and are host protective in an IL-10(-/-) setting. Immunity. 16:429–439. 10.1016/S1074-7613(02)00278-9 [DOI] [PubMed] [Google Scholar]
- Kissenpfennig A., Henri S., Dubois B., Laplace-Builhé C., Perrin P., Romani N., Tripp C.H., Douillard P., Leserman L., Kaiserlian D., et al. 2005. Dynamics and function of Langerhans cells in vivo: dermal dendritic cells colonize lymph node areas distinct from slower migrating Langerhans cells. Immunity. 22:643–654. 10.1016/j.immuni.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Kitajima M., and Ziegler S.F.. 2013. Cutting edge: identification of the thymic stromal lymphopoietin-responsive dendritic cell subset critical for initiation of type 2 contact hypersensitivity. J. Immunol. 191:4903–4907. 10.4049/jimmunol.1302175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto Y., Linehan M., Weinstein J.S., Laidlaw B.J., Craft J.E., and Iwasaki A.. 2013. CD301b+ dermal dendritic cells drive T helper 2 cell-mediated immunity. Immunity. 39:733–743. 10.1016/j.immuni.2013.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson R.P., Zimmerli S.C., Comeau M.R., Itano A., Omori M., Iseki M., Hauser C., and Ziegler S.F.. 2010. Dibutyl phthalate-induced thymic stromal lymphopoietin is required for Th2 contact hypersensitivity responses. J. Immunol. 184:2974–2984. 10.4049/jimmunol.0803478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhi M.P., Trumpfheller C., Idoyaga J., Caskey M., Matos I., Kluger C., Salazar A.M., Colonna M., and Steinman R.M.. 2009. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J. Exp. Med. 206:1589–1602. 10.1084/jem.20090247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manh T.P., Alexandre Y., Baranek T., Crozat K., and Dalod M.. 2013. Plasmacytoid, conventional, and monocyte-derived dendritic cells undergo a profound and convergent genetic reprogramming during their maturation. Eur. J. Immunol. 43:1706–1715. 10.1002/eji.201243106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massacand J.C., Stettler R.C., Meier R., Humphreys N.E., Grencis R.K., Marsland B.J., and Harris N.L.. 2009. Helminth products bypass the need for TSLP in Th2 immune responses by directly modulating dendritic cell function. Proc. Natl. Acad. Sci. USA. 106:13968–13973. 10.1073/pnas.0906367106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.C., Brown B.D., Shay T., Gautier E.L., Jojic V., Cohain A., Pandey G., Leboeuf M., Elpek K.G., Helft J., et al. Immunological Genome Consortium . 2012. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13:888–899. 10.1038/ni.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami R., Denda-Nagai K., Hashimoto S., Nagai S., Hattori M., and Irimura T.. 2013. A unique dermal dendritic cell subset that skews the immune response toward Th2. PLoS One. 8:e73270 10.1371/journal.pone.0073270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessen F., Schaffner F., Furlan-Freguia C., Pawlinski R., Bhattacharjee G., Chun J., Derian C.K., Andrade-Gordon P., Rosen H., and Ruf W.. 2008. Dendritic cell PAR1-S1P3 signalling couples coagulation and inflammation. Nature. 452:654–658. 10.1038/nature06663 [DOI] [PubMed] [Google Scholar]
- Nowyhed H.N., Huynh T.R., Thomas G.D., Blatchley A., and Hedrick C.C.. 2015. Cutting Edge: The Orphan Nuclear Receptor Nr4a1 Regulates CD8+ T Cell Expansion and Effector Function through Direct Repression of Irf4. J. Immunol. 195:3515–3519. 10.4049/jimmunol.1403027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai S., Roediger B., Abtin A., Shklovskaya E., Fazekas de St Groth B., Yamane H., Weninger W., Le Gros G., and Ronchese F.. 2014. CD326lo CD103lo CD11blo dermal dendritic cells are activated by thymic stromal lymphopoietin during contact sensitization in mice. J. Immunol. 193:2504–2511. 10.4049/jimmunol.1400536 [DOI] [PubMed] [Google Scholar]
- Oyoshi M.K., Larson R.P., Ziegler S.F., and Geha R.S.. 2010. Mechanical injury polarizes skin dendritic cells to elicit a T(H)2 response by inducing cutaneous thymic stromal lymphopoietin expression. J. Allergy Clin. Immunol. 126:976–984: 984.e1–984.e5. 10.1016/j.jaci.2010.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul W.E., and Zhu J.. 2010. How are T(H)2-type immune responses initiated and amplified? Nat. Rev. Immunol. 10:225–235. 10.1038/nri2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phythian-Adams A.T., Cook P.C., Lundie R.J., Jones L.H., Smith K.A., Barr T.A., Hochweller K., Anderton S.M., Hämmerling G.J., Maizels R.M., and MacDonald A.S.. 2010. CD11c depletion severely disrupts Th2 induction and development in vivo. J. Exp. Med. 207:2089–2096. 10.1084/jem.20100734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pletinckx K., Stijlemans B., Pavlovic V., Laube R., Brandl C., Kneitz S., Beschin A., De Baetselier P., and Lutz M.B.. 2011. Similar inflammatory DC maturation signatures induced by TNF or Trypanosoma brucei antigens instruct default Th2-cell responses. Eur. J. Immunol. 41:3479–3494. 10.1002/eji.201141631 [DOI] [PubMed] [Google Scholar]
- Pulendran B., and Artis D.. 2012. New paradigms in type 2 immunity. Science. 337:431–435. 10.1126/science.1221064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan J., Mowen K.A., McBride K.D., Smith E.D., Singh H., and Glimcher L.H.. 2002. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J. Exp. Med. 195:1003–1012. 10.1084/jem.20011128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roediger B., Kyle R., Yip K.H., Sumaria N., Guy T.V., Kim B.S., Mitchell A.J., Tay S.S., Jain R., Forbes-Blom E., et al. 2013. Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat. Immunol. 14:564–573. 10.1038/ni.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothlin C.V., Carrera-Silva E.A., Bosurgi L., and Ghosh S.. 2015. TAM receptor signaling in immune homeostasis. Annu. Rev. Immunol. 33:355–391. 10.1146/annurev-immunol-032414-112103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H., Chapman R., and Hertzog P.J.. 2013. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 41(D1):D1040–D1046. 10.1093/nar/gks1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens S.P., Ng A., Kumar V., Johnson M.D., Plantinga T.S., van Diemen C., Arts P., Verwiel E.T., Gresnigt M.S., Fransen K., et al. 2013. Functional genomics identifies type I interferon pathway as central for host defense against Candida albicans. Nat. Commun. 4:1342 10.1038/ncomms2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.A., Hochweller K., Hämmerling G.J., Boon L., MacDonald A.S., and Maizels R.M.. 2011. Chronic helminth infection promotes immune regulation in vivo through dominance of CD11cloCD103− dendritic cells. J. Immunol. 186:7098–7109. 10.4049/jimmunol.1003636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.A., Harcus Y., Garbi N., Hämmerling G.J., MacDonald A.S., and Maizels R.M.. 2012. Type 2 innate immunity in helminth infection is induced redundantly and acts autonomously following CD11c+ cell depletion. Infect. Immun. 80:3481–3489. 10.1128/IAI.00436-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits H.H., de Jong E.C., Schuitemaker J.H., Geijtenbeek T.B., van Kooyk Y., Kapsenberg M.L., and Wierenga E.A.. 2002. Intercellular adhesion molecule-1/LFA-1 ligation favors human Th1 development. J. Immunol. 168:1710–1716. 10.4049/jimmunol.168.4.1710 [DOI] [PubMed] [Google Scholar]
- Soumelis V., Reche P.A., Kanzler H., Yuan W., Edward G., Homey B., Gilliet M., Ho S., Antonenko S., Lauerma A., et al. 2002. Human epithelial cells trigger dendritic cell mediated allergic inflammation by producing TSLP. Nat. Immunol. 3:673–680. [DOI] [PubMed] [Google Scholar]
- Tamoutounour S., Guilliams M., Montanana Sanchis F., Liu H., Terhorst D., Malosse C., Pollet E., Ardouin L., Luche H., Sanchez C., et al. 2013. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity. 39:925–938. 10.1016/j.immuni.2013.10.004 [DOI] [PubMed] [Google Scholar]
- Taylor B.C., Zaph C., Troy A.E., Du Y., Guild K.J., Comeau M.R., and Artis D.. 2009. TSLP regulates intestinal immunity and inflammation in mouse models of helminth infection and colitis. J. Exp. Med. 206:655–667. 10.1084/jem.20081499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis M.A., Reizis B., Melton A.C., Masteller E., Tang Q., Proctor J.M., Wang Y., Bernstein X., Huang X., Reichardt L.F., et al. 2007. Loss of integrin alpha(v)beta8 on dendritic cells causes autoimmunity and colitis in mice. Nature. 449:361–365. 10.1038/nature06110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri G. 2010. Type I interferon: friend or foe? J. Exp. Med. 207:2053–2063. 10.1084/jem.20101664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottein F., Pavelka N., Vizzardelli C., Angeli V., Zouain C.S., Pelizzola M., Capozzoli M., Urbano M., Capron M., Belardelli F., et al. 2004. A type I IFN-dependent pathway induced by Schistosoma mansoni eggs in mouse myeloid dendritic cells generates an inflammatory signature. J. Immunol. 172:3011–3017. 10.4049/jimmunol.172.5.3011 [DOI] [PubMed] [Google Scholar]
- Tussiwand R., Everts B., Grajales-Reyes G.E., Kretzer N.M., Iwata A., Bagaitkar J., Wu X., Wong R., Anderson D.A., Murphy T.L., et al. 2015. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity. 42:916–928. 10.1016/j.immuni.2015.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Pouw Kraan T.C., Zwiers A., Mulder C.J., Kraal G., and Bouma G.. 2009. Acute experimental colitis and human chronic inflammatory diseases share expression of inflammation-related genes with conserved Ets2 binding sites. Inflamm. Bowel Dis. 15:224–235. 10.1002/ibd.20747 [DOI] [PubMed] [Google Scholar]
- Wilcox R.A., Chapoval A.I., Gorski K.S., Otsuji M., Shin T., Flies D.B., Tamada K., Mittler R.S., Tsuchiya H., Pardoll D.M., and Chen L.. 2002. Cutting edge: Expression of functional CD137 receptor by dendritic cells. J. Immunol. 168:4262–4267. 10.4049/jimmunol.168.9.4262 [DOI] [PubMed] [Google Scholar]
- Williams J.W., Tjota M.Y., Clay B.S., Vander Lugt B., Bandukwala H.S., Hrusch C.L., Decker D.C., Blaine K.M., Fixsen B.R., Singh H., et al. 2013. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat. Commun. 4:2990 10.1038/ncomms3990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolvetang E.J., Wilson T.J., Sanij E., Busciglio J., Hatzistavrou T., Seth A., Hertzog P.J., and Kola I.. 2003. ETS2 overexpression in transgenic models and in Down syndrome predisposes to apoptosis via the p53 pathway. Hum. Mol. Genet. 12:247–255. 10.1093/hmg/ddg015 [DOI] [PubMed] [Google Scholar]
- Yang J., Siqueira M.F., Behl Y., Alikhani M., and Graves D.T.. 2008. The transcription factor ST18 regulates proapoptotic and proinflammatory gene expression in fibroblasts. FASEB J. 22:3956–3967. 10.1096/fj.08-111013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., Yoshimi R., Yoshii H., Kim D., Dey A., Xiong H., Munasinghe J., Yazawa I., O’Donovan M.J., Maximova O.A., et al. 2014. The transcription factor IRF8 activates integrin-mediated TGF-β signaling and promotes neuroinflammation. Immunity. 40:187–198. 10.1016/j.immuni.2013.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.T., Liu D., Calabro S., Eisenbarth S.C., Cattoretti G., and Haberman A.M.. 2014. Dynamic expression of BCL6 in murine conventional dendritic cells during in vivo development and activation. PLoS One. 9:e101208 10.1371/journal.pone.0101208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Sharma J., Raju R., Palapetta S.M., Prasad T.S., Huang T.C., Yoda A., Tyner J.W., van Bodegom D., Weinstock D.M., et al. 2014. TSLP signaling pathway map: a platform for analysis of TSLP-mediated signaling. Database (Oxford). 2014:bau007 10.1093/database/bau007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.