Vaidyanathan et al. report that the environmental sensor aryl hydrocarbon receptor is inducibly expressed in B cells downstream of BCR signaling and that it controls B cell fates by negatively modulating class switching and plasma cell differentiation via aicda and prdm1, respectively.
Abstract
Generation of cellular heterogeneity is an essential feature of the adaptive immune system. This is best exemplified during humoral immune response when an expanding B cell clone assumes multiple cell fates, including class-switched B cells, antibody-secreting plasma cells, and memory B cells. Although each cell type is essential for immunity, their generation must be exquisitely controlled because a class-switched B cell cannot revert back to the parent isotype, and a terminally differentiated plasma cell cannot contribute to the memory pool. In this study, we show that an environmental sensor, the aryl hydrocarbon receptor (AhR) is highly induced upon B cell activation and serves a critical role in regulating activation-induced cell fate outcomes. We find that AhR negatively regulates class-switch recombination ex vivo by altering activation-induced cytidine deaminase expression. We further demonstrate that AhR suppresses class switching in vivo after influenza virus infection and immunization with model antigens. In addition, by regulating Blimp-1 expression via Bach2, AhR represses differentiation of B cells into plasmablasts ex vivo and antibody-secreting plasma cells in vivo. These experiments suggest that AhR serves as a molecular rheostat in B cells to brake the effector response, possibly to facilitate optimal recall responses. Thus, AhR might represent a novel molecular target for manipulation of B cell responses during vaccination.
Introduction
Antigen receptor gene assembly in the BM via V(D)J recombination endows B lymphocytes with a remarkable repertoire of specificities against a vast array of pathogens. In the wake of an infection, mature B cells in secondary lymphoid organs such as the spleen, LNs, and Peyer’s patches further tune this preformed repertoire through the secondary diversification reactions of somatic hypermutation and class-switch recombination (CSR) to generate cells with higher antigen affinity and distinct effector functions, respectively (Alt et al., 2013). Additionally, during the course of this response, B cells undergo plasma cell differentiation (PCD) to generate antibody-producing, terminally differentiated plasma cells. Finally, an imprint of the initial antigen challenge is engraved in the B cell pool via memory B cell differentiation (MBD; Kurosaki et al., 2015; Nutt et al., 2015). The mechanism by which a clonally expanding population of B cells in the germinal center commits to different cell fates including CSR, PCD, and MBD remains a major unsolved question. Recent studies have uncovered that instructive cues from neighboring T cells, intrinsic signals via asymmetric distribution of proteins, and stochastic cell-autonomous forces drive this heterogeneity (Tarlinton, 2012; Reiner and Adams, 2014). Although major positive effectors of CSR (e.g., activation-induced cytidine deaminase [AID]) and PCD (e.g., Blimp-1 and Irf4; Nutt et al., 2015) have been identified, little is known of the molecular mediators that serve as endogenous brakes to this effector B cell response, which may in turn balance the mutually exclusive branches of PCD and MBD (Gitlin et al., 2016; Shinnakasu et al., 2016; Weisel et al., 2016).
A factor that could regulate B cell fate decisions and bias a B cell to remember an antigenic challenge would likely be expressed and be functional in cognate B cells that receive instructive signals via the B cell antigen receptor (BCR), in essence, a sensory factor of the initial insult that might restrain master regulators of alternate cell fates. The aryl hydrocarbon receptor (AhR), a ligand-induced nuclear receptor transcription factor with a well characterized role as an environmental sensor for dioxins, was recently shown to regulate cell-fate decisions during T cell differentiation (Quintana et al., 2008; Gagliani et al., 2015). Here, we show that AhR is highly induced in B cells upon BCR cross-linking. Using pharmacological activation to engage the AhR pathway and genetic models of AhR deficiency, we demonstrate that AhR serves a unique role in negatively regulating the effector B cell response by dampening both CSR and PCD, a strategy that can potentially impact efficient MBD.
Results
B cell–extrinsic role of AhR during B cell development
Naive splenic B cells cross-linked with anti-IgD, a mimic for antigen encounter, induced a robust expression of AhR protein (Fig. S1 A) and its target gene cyp1a1 (Fig. S1 B; Mandal, 2005). This led us to explore the role of AhR in B cell biology. First, we characterized B cell development in AhR−/− mice. We observed modest yet statistically significant alterations in B cell development and maturation in AhR−/− mice (Schmidt et al., 1996). Compared with WT mice, AhR−/− mice showed increased frequency of pre–B cells (B220lo IgM− CD43−) in the BM (Fig. S1, C, D, and P) and decreased frequency of mature (IgDhi IgM+) B cells in the spleen but not in the LN (Fig. S1, F, G, and Q–S). There was a significant reduction in marginal zone (MZ) B cells and a trending increase in the frequency of transitional 1 (T1) and modest decrease in T2 B cells in the spleen of AhR−/− mice compared with WT mice (Fig. S1, I and J). The defects in BM B cell development were largely rescued in chimeras wherein congenically marked AhR−/− (CD45.2) or WT (CD45.1) BM precursor cells were transferred into irradiated RAG1-deficient recipients (hematopoietic KO and WT chimera, respectively; Fig. S1, E, H, K, and T–W). We additionally generated mixed BM chimeras in which BM cells from WT and AhR−/− mice were mixed 1:1 and adoptively transferred into irradiated RAG1-deficient recipients (Fig. S1, L and M) and observed a complete rescue of MZ, T1, and T2 phenotypes (Fig. S1, N and O). These results suggested a cell-extrinsic role of AhR in B cell development and homeostasis. Furthermore, restored B cell development allowed us to use the mixed BM chimeras as a convenient experimental system to compare the competence of WT and AhR−/− B cells during an immune response in vivo.
AhR negatively modulates CSR in a B cell–intrinsic manner
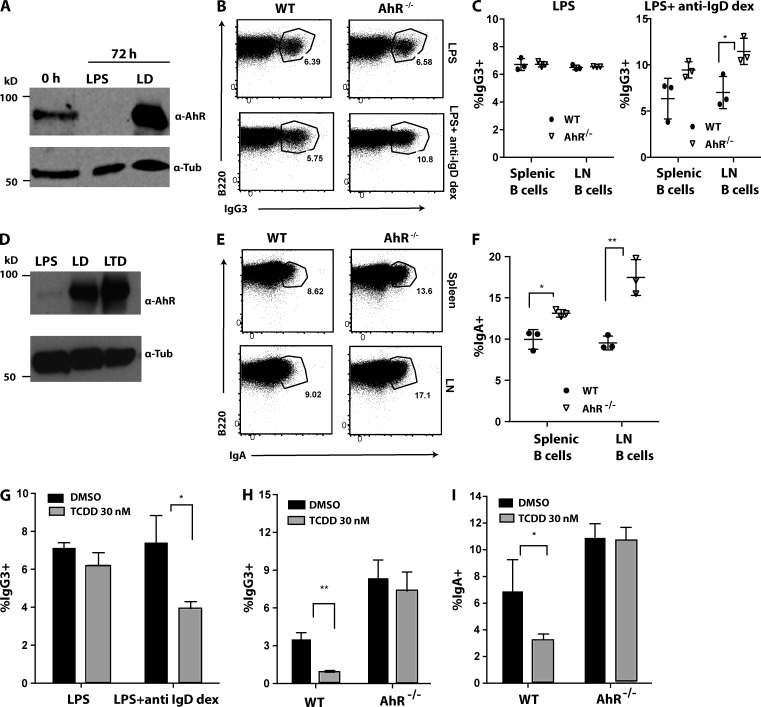
To test functionality of AhR in activated B cells, we queried whether AhR modulates CSR ex vivo. Purified naive B cells from spleen and LN of WT or AhR−/− mice were stimulated with either LPS or LPS + anti-IgD dextran (LD) to induce CSR to IgG3. LPS alone repressed basal AhR expression in resting B cells derived from WT mice (Fig. 1 A). In contrast, LPS stimulation in combination with BCR engagement (LD) induced significant AhR expression in WT B cells (Fig. 1 A), which was functional as measured by cyp1a1 induction (Fig. S2 A). Consistent with this expression pattern, there was no effect of AhR deficiency on CSR to IgG3 under LPS stimulation (Fig. 1, B and C). Strikingly, with stimulation with LD when AhR expression was induced, AhR−/− B cells exhibited a significant increase (about twofold) in CSR to IgG3 compared with their WT counterparts (Fig. 1, B and C).
Figure 1.
AhR negatively regulates CSR ex vivo. (A) Western blot showing expression of AhR and tubulin (Tub; loading control) under different stimulation conditions (LPS and LD) at 72 h compared with unstimulated (0 h) WT splenic B cells. A representative of two experiments is shown. (B) Representative flow cytometry plot of WT and AhR−/− purified LN B cells stimulated for 96 h with LPS (top) and LPS + anti-IgD dextran (dex; bottom) for CSR to IgG3. (C) Quantification of cell-surface IgG3 expression at 96 h in splenic and LN B cells of WT and AhR−/− mice stimulated with LPS (left) and LPS + anti-IgD dextran (right). n = 3. Mean ± SD is shown. (D) Western blot showing expression of AhR (loading control, tubulin; bottom) under different stimulation conditions in WT splenic B cells at 72 h after stimulation (LPS, LD [LPS + anti-IgD dextran], and LTD [LPS + TGFβ + anti-IgD dextran]). A representative of two experiments is shown. (E) Representative flow cytometry plot of purified splenic (top) and LN (bottom) B cells from WT and AhR−/− mice, stimulated for 96 h with LTD, showing cell-surface IgA expression as a measure of CSR. (F) Quantification of percent IgA+ splenic and LN B cells from WT and AhR−/− mice stimulated for 96 h with LTD. A representative of three experiments with three mice per group (mean ± SD) is shown. (G) Effect of TCDD treatment compared with vehicle control (DMSO) on CSR to IgG3 induced in WT splenic B cells treated with LPS or LD for 96 h. (H) IgG3 CSR in WT or AhR−/− splenic B cells treated with LD for 96 h in presence of TCDD (or DMSO). (I) IgA CSR in WT or AhR−/− splenic B cells treated with LTD for 96 h in the presence of TCDD (or DMSO). (G–I) n = 3. Mean ± SD is shown. *, P < 0.05; **, P < 0.005 (Student’s t test).
To investigate the role of AhR in CSR to IgA, we stimulated purified B cells with LPS + TGFβ + anti-IgD dextran (LTD). We observed a TGFβ-dependent increase in AhR expression in WT B cells stimulated with LPS, LD, and LTD (Fig. 1 D) and a concomitant AhR-dependent up-regulation of cyp1a1 mRNA (Fig. S2, B and C). Flow cytometric analysis of LTD-stimulated B cells revealed significantly increased CSR to IgA in AhR-deficient B cells compared with WT B cells (Fig. 1, E and F). It is to be noted that altered CSR was not caused by B cell developmental defects in AhR−/− mice, as similar results were also obtained in hematopoietic and mixed BM chimeras where B cell development was largely unaltered (Fig. S2, D–K).
In a complementary approach, we treated splenic B cells with a well characterized AhR agonist 2,3,7,8-tetrachlorodibenzo p-dioxin (TCDD; Murray et al., 2014). Consistent with the genetic data, pharmacological activation of AhR in WT B cells suppressed CSR to both IgG3 and IgA in the presence of LD and LTD, respectively (Fig. 1, G–I). These alterations were AhR dependent, as there was no significant effect of TCDD on CSR in AhR-deficient B cells (Fig. 1, H and I) or in WT cells stimulated with LPS (Fig. 1 G) that does not induce AhR expression. These data collectively strongly suggest that AhR negatively regulates CSR in a B cell–intrinsic manner.
AhR suppresses CSR by modulating AID expression
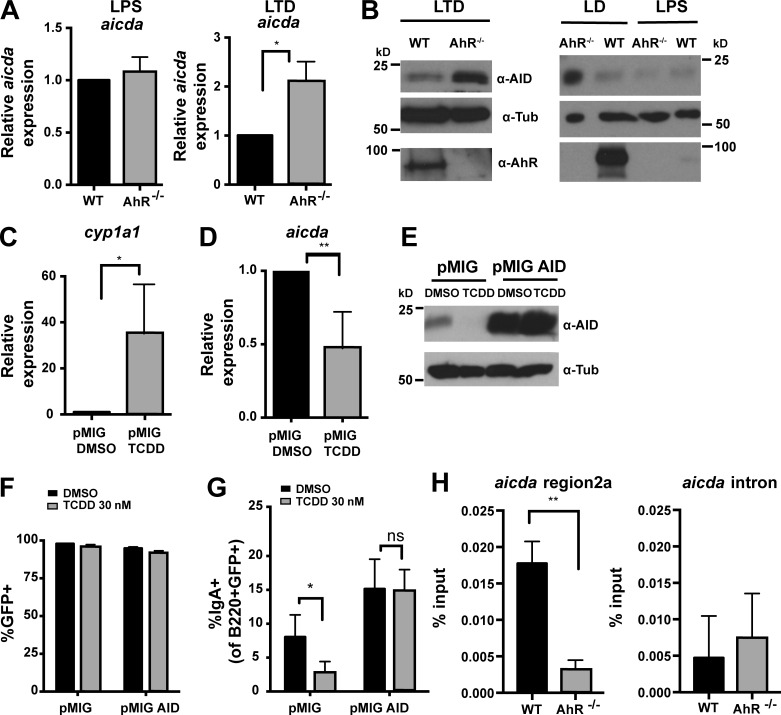
To explore the mechanism by which AhR regulates CSR, we examined known CSR parameters. Germline transcription (Fig. S3, A–F), B cell activation assessed by expression of activation markers CD69 and CD43 (Fig. S3 G), and proliferation measured by dilution of the cell surface–staining dye CellTrace violet (CTV; Fig. S3, H and I) were unaltered in AhR−/− B cells. However, expression of AID, a molecule essential for CSR (Muramatsu et al., 2000; Revy et al., 2000) was markedly altered. Relative to WT B cells, aicda mRNA (48 h) and AID protein levels (48–96 h) were significantly elevated in AhR−/− B cells under stimulations that induced AhR (LTD and LD but not LPS; Fig. 2, A and B). Similar observations were also made in chimeric mice (Fig. S3, J and K). These results suggested that AhR represses AID to restrict CSR.
Figure 2.
AhR represses AID expression. (A) Relative mRNA expression of aicda (normalized to β actin) under LPS and LTD stimulation at 48 h in WT and AhR−/− splenic B cells. n = 3. Mean ± SD is shown. (B) Western blot showing AID, AhR, and tubulin (Tub; loading control) expression under LTD 96-h stimulation of splenic B cells (left) and LD/LPS 48-h stimulation of LN B cells from WT and AhR−/− mice. A representative of two to three experiments is shown. (C) cyp1a1 expression in WT B cells stimulated with LTD for 48 h and infected with empty retroviral vector (pMIG) in the presence of DMSO or TCDD. n = 3. Mean ± SD is shown. (D) aicda expression in WT B cells stimulated with LTD for 48 h and infected with pMIG in the presence of DMSO or TCDD. n = 3. Mean ± SD is shown. (E) Western blot showing AID and tubulin (loading control) expression at 48 h LTD stimulated WT B cells infected with pMIG or pMIG-AID in the presence of DMSO or TCDD. A representative of two experiments is shown. (F) Percent GFP+ in pMIG- or pMIG-AID–infected WT B cells stimulated for 72 h with LTD in the presence of DMSO or TCDD. n = 4. Mean ± SD is shown. (G) Percent IgA+ cells of GFP-expressing WT B cells 72 h after pMIG or pMIG-AID infection in the presence of DMSO or TCDD. n = 4. Mean ± SD is shown. (H) ChIP using anti-AhR antibody. Binding of AhR to intronic silencer region 2a of aicda or to a region 2 kb downstream in intron 1 was examined. DNA in ChIP samples was quantified by quantitative PCR and normalized to input (5%). n = 3. Mean ± SD is shown. *, P < 0.05; **, P < 0.005 (Student’s t test).
To test whether AID repression was sufficient to explain the CSR-antagonizing effect of AhR, we overexpressed AID (Fig. 2 E) in WT B cells treated with AhR agonist TCDD (Fig. 2, C and D). Splenic B cells transduced with empty pMIG vector in the presence of TCDD induced cyp1a1 expression, indicating engagement of the AhR pathway (Fig. 2 C). In keeping with previous results, AhR activation reduced expression of both aicda mRNA (Fig. 2 D) and AID protein (Fig. 2 E) and suppressed CSR to IgA (Fig. 2 G). Remarkably, when AID was transduced into TCDD-treated B cells (Fig. 2, E and F), the defect in CSR was rescued (Fig. 2 G). Therefore, it appears that AID expression is a primary control node by which AhR modulates CSR.
To explore the mechanism through which AhR represses AID expression, we examined AhR binding to the aicda locus, focusing on a silencer element in the first intron (region 2a) that has binding sites for ubiquitous repressive transcription factors like c-Myb and E2F (Tran et al., 2010). Chromatin immunoprecipitation (ChIP) with anti-AhR antibodies revealed a significant AhR-dependent enrichment of region 2a but not to a site 2-kb downstream in the intron (Fig. 2 H). Collectively, these results strongly suggest that AhR controls CSR by binding to aicda regulatory element and directly repressing aicda gene expression.
AhR dampens PCD via Blimp-1 repression
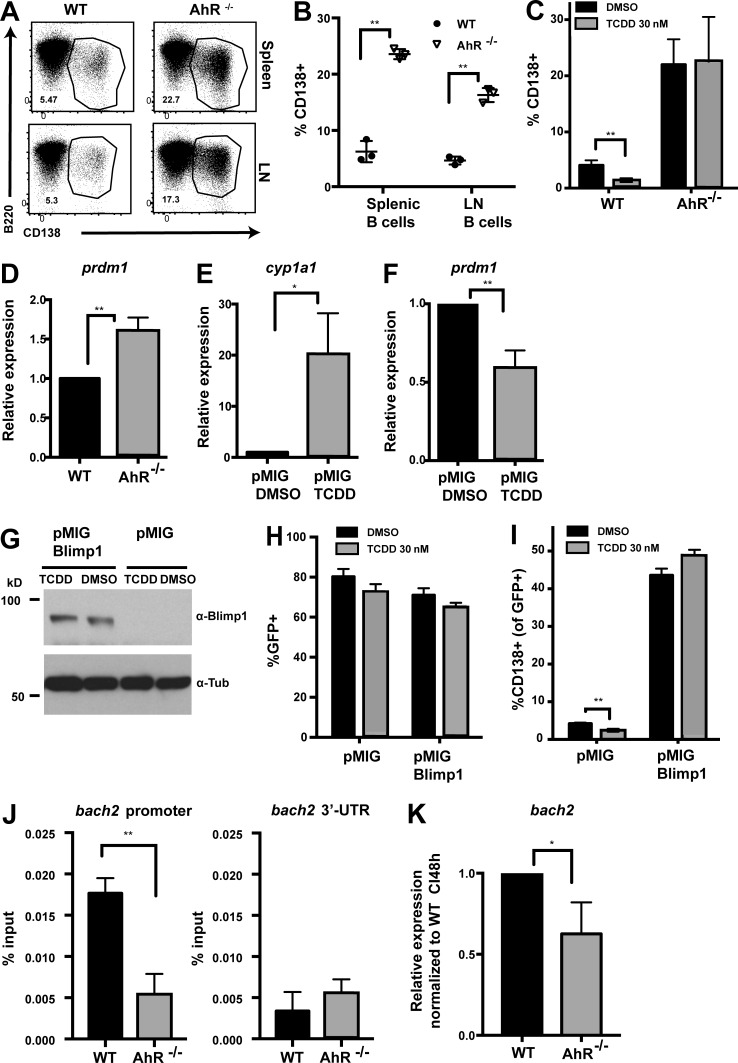
In addition to CSR, B cell activation also results in terminal differentiation into antibody-secreting plasma cells, the major effectors of humoral immunity (Nutt et al., 2015). Because these cells have a short life span and die within days of the primary response, it is important to modulate terminal differentiation to maintain the longevity of a responding B cell population and prevent exhaustion of an expanding B cell clone during an immune response. To test whether AhR can modulate PCD, naive splenic and LN B cells were stimulated with anti-CD40 + IL-4 (to mimic T cell–mediated activation in vivo) to induce plasmablast/plasma cell formation ex vivo. AhR−/− B cells differentiated at a significantly higher frequency to plasma cells (CD138+) compared with their WT counterparts (Fig. 3, A and B). Analysis of the single and mixed BM chimeras also revealed a similar increase in PCD in the absence of AhR (Fig. S3, L–N) buttressing the cell-intrinsic nature of the effect. In a complementary approach, using the pharmacologic agonist TCDD, we observed that AhR activation suppresses PCD, an effect that was completely AhR dependent (Fig. 3 C). Thus, AhR negatively regulates PCD in a B cell–intrinsic manner.
Figure 3.
AhR antagonizes PCD ex vivo via Blimp-1 repression. (A) Representative flow cytometry plot of WT and AhR−/− splenic (top) and LN (bottom) B cells stimulated with anti-CD40 + IL-4 (CI) for 96 h, showing CD138+ plasma cells. (B) Quantification summary of percent CD138+ splenic and LN B cells from WT and AhR−/− mice. A representative of three experiments with three mice per group is shown. (C) Percentage of CD138+ cells at 96 h after CI stimulation of WT and AhR−/− B cells treated with DMSO or TCDD. (D) Relative mRNA expression of prdm1 (normalized to β actin) at 48 h CI stimulation between WT and AhR−/− splenic B cells. (E) Relative cyp1a1 mRNA levels (normalized to β actin) as a measure of AhR activation in 48-h CI-treated WT B cells infected with pMIG in the presence of DMSO or TCDD. (F) Relative mRNA expression of prdm1 (normalized to β actin) in CI-stimulated WT B cells infected with pMIG (48 h) in the presence of DMSO or TCDD. (G) Western blot showing Blimp-1 and tubulin (Tub; loading control) expression in CI-stimulated WT B cells infected with empty vector (pMIG) or pMIG–Blimp-1 for 48 h in the presence of DMSO or TCDD. Endogenous Blimp-1 was not detectable using the Blimp-1 antibody. A representative of two experiments is shown. (H) Percent GFP+ in pMIG- or pMIG–Blimp-1–infected (96 h after infection) WT B cells stimulated with CI in the presence of DMSO or TCDD. (I) Percent CD138+ cells at 96 h after infection with empty vector or pMIG–Blimp-1 in WT B cells stimulated with CI in the presence of DMSO or TCDD. (J) ChIP using anti-AhR antibody. Binding of AhR to the bach2 promoter or region encoding the 3′ untranslated region (3′-UTR) was examined. DNA in ChIP samples was quantified by quantitative PCR and normalized to 5% of input. (K) Relative mRNA expression of bach2 (normalized to β actin) at 48 h CI stimulation between WT and AhR−/− splenic B cells. (C–F and H–K) n = 3. Mean ± SD is shown. *, P < 0.05; **, P < 0.005 (Student’s t test).
The increased potential of AhR−/− B cells to differentiate into plasma cells was not caused by altered activation (Fig. S3 O) and proliferation (Fig. S3 P). Rather, AhR antagonized expression of Blimp-1, the master regulator of PCD (Shaffer et al., 2001) as indicated by the significantly elevated prdm1 expression in AhR-deficient B cells relative to AhR-sufficient ones (Fig. 3 D and Fig. S3 Q). These observations suggest that AhR restricts PCD via Blimp-1 repression. To examine whether Blimp-1 repression by AhR was sufficient for dampened PCD, we overexpressed Blimp-1 (Fig. 3 G) in WT B cells treated with TCDD to activate AhR (Fig. 3 E) that decreases prdm1 expression (Fig. 3 F). Treatment of B cells with TCDD did not affect retroviral transduction of pMIG (empty vector) or pMIG–Blimp-1 (Fig. 3 H). Significantly, overexpression of Blimp-1 rescued the PCD defect in TCDD-treated B cells (Fig. 3, H and I). These results suggest that AhR regulates PCD through modulating expression of Blimp-1.
We subsequently explored the mechanism by which AhR could regulate expression of Blimp-1. Bach2 is a known repressor of Blimp-1 and has been implicated in TCDD-mediated down-regulation of Blimp-1 (De Abrew et al., 2011). We tested binding of AhR to the bach2 locus by ChIP. We found significant enrichment of AhR binding at the promoter and not the 3′ untranslated region only in WT and not in AhR-deficient B cells (Fig. 3 J). This correlated with a small but significant decrease in bach2 expression at 48 h of stimulation in AhR−/− B cells relative to WT B cells (Fig. 3 K), suggesting that AhR promotes bach2 expression to curb Blimp-1, the master regulator of PCD.
AhR restricts primary B cell responses to model antigen-induced T cell–dependent and T cell–independent responses
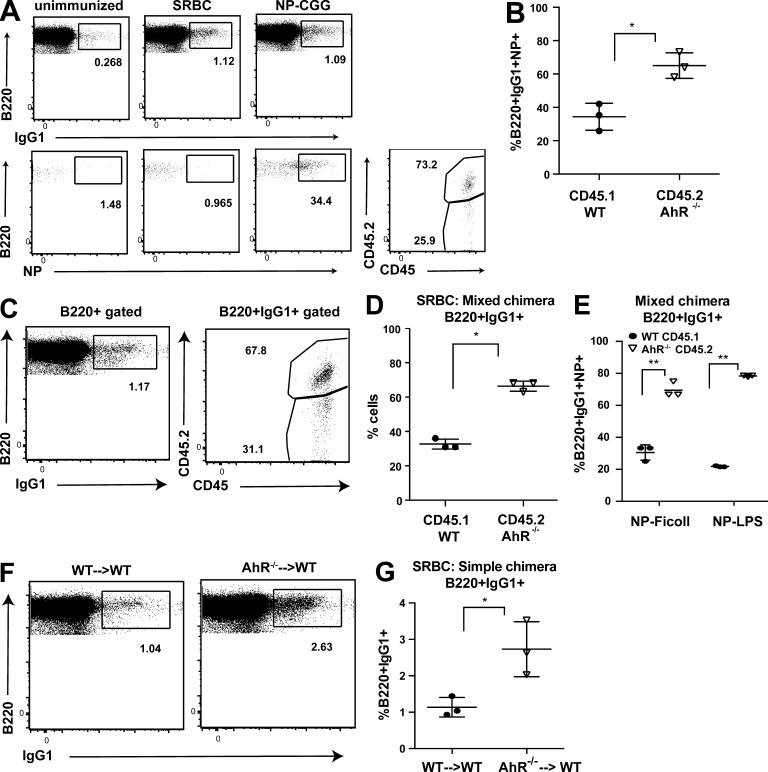
Our ex vivo findings led us to test the impact of AhR deficiency on primary B cell responses in vivo. To this end, we used a model system of T cell–dependent soluble antigen (NP-CGG)–driven B cell response that allows tracking of the primary response against the hapten (4-hydroxy-3-nitrophenylacetyl [NP]). We immunized AhR mixed BM chimeric mice with NP-CGG in alum and analyzed the primary response 2 wk later. Mice injected with PBS alone (unimmunized) or immunized with sheep RBCs (SRBCs), a T cell–dependent particulate antigen, served as controls for switching and NP specificity, respectively (Fig. 4 A). AhR-deficient B cells contributed significantly more to the class-switched (IgG1+) antigen (NP)-specific compartment (Fig. 4, A and B). Similar skewing toward AhR−/− B cells was also observed upon SRBC immunization (Fig. 4, C and D). Given the role of AhR in T cells, we also analyzed B cell responses to T cell–independent type I (NP-LPS) and TI type II (NP-Ficoll) model antigens and found that antigen-specific switched B cells come predominantly from the AhR−/−, suggesting that the phenotype is B cell intrinsic and not caused by extrinsic T cell cues governed by AhR (Fig. 4 E). Additionally, AhR−/− B cells in KO hematopoietic chimeras underwent significantly increased CSR to IgG1 in response to SRBCs, compared with that in WT chimeras (WT→WT; Fig. 4, F and G). These findings suggested that AhR provides a robust cell-intrinsic endogenous braking mechanism imparted to the primary B cell responses.
Figure 4.
AhR dampens both T cell–dependent and T cell–independent B cell responses. (A, left) Representative flow cytometry plot showing live singlet B220+ cells that are IgG1+ (top)- and B220+IgG1+NP-specific (bottom) in unimmunized (left), SRBC-immunized (middle), and NP-CGG–immunized (right) AhR−/− mix chimeras at day 14. (Bottom right) Contribution of WT (CD45.1) and AhR−/− (CD45.2) cells to the antigen-specific class-switched pool in NP-CGG–immunized AhR−/− mix chimeras. (B) Quantification summary of percent contribution of WT (CD45.1) and AhR−/− (CD45.2) cells to the splenic B220+IgG1+NP-specific pool on day 14 after NP-CGG immunization. (C) Representative flow cytometry plot showing IgG1+ B cells in the spleen and the percent contribution of WT (CD45.1) and AhR−/− (CD45.2) to this pool in SRBC-immunized AhR mixed chimeras at day 14. (D) Quantification summary of percent contribution of WT (CD45.1) and AhR−/− (CD45.2) cells to the B220+IgG1+ pool. (E) Quantification summary of percent contribution of WT (CD45.1) and AhR−/− (CD45.2) cells to the B220+IgG1+NP-specific pool in AhR mixed chimeras immunized with T cell–independent type I (NP-Ficoll) or type II (NP-LPS) antigens at day 14. (F) Representative flow cytometry plot showing IgG1+ B cells in the spleen of SRBC-immunized AhR WT only chimera (WT→WT) and AhR KO only (hematopoietic KO; AhR−/−→WT) mice. (G) Quantification summary of percent of B220+IgG1+ cells at day 14 after SRBC immunization of AhR WT only chimera (WT→WT) and AhR KO only (hematopoietic KO; AhR−/− →WT) mice. (B, D, E, and G) n = 3. Mean ± SD is shown. *, P < 0.05; **, P < 0.005 (Student’s t test).
AhR dampens the primary B cell response to influenza virus
We next examined whether AhR could influence primary immune response in vivo during influenza virus infection. We intranasally infected AhR mixed BM chimeras with the influenza strain H1NI (PR8) to ascertain B cell–intrinsic functionality of AhR in a competitive setting. At baseline, the chimerism of B cells, T cells, and myeloid (CD11b+) cells in peripheral blood was ∼50:50 (Fig. S4 A). This ratio mirrored the chimerism in control mix chimeras where WT (CD45.1) and WT (CD45.2) BM cells were injected into RAG-1−/− recipients (Fig. S4 B), demonstrating that congenic differences do not per se skew B cell development. Splenic B cell chimerism was also identical at homeostasis (Fig. S4 C).
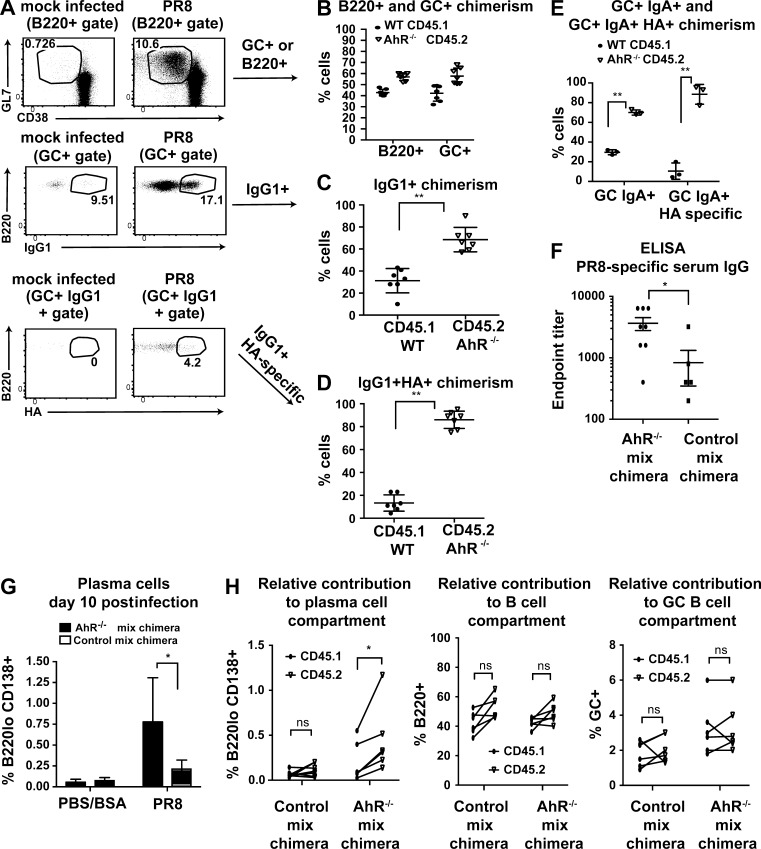
2 wk after PR8 infection, lung-draining mediastinal LNs were analyzed for the relative abundance of WT (CD45.1) versus AhR−/− (CD45.2) B cells. There were no overt perturbations of the WT/AhR−/− ratio either in the total B cell pool or in the germinal center population, and this ratio was indistinguishable from that observed in control (WT [CD45.1]/WT [CD45.2]) mix chimeras (Fig. 5, A and B; and Fig. S4 E). Thus, there was no gross competitive advantage of AhR-deficient B cells in vivo in seeding or persisting in the germinal center compartment. However, within the germinal centers, we observed that AhR−/− B cells contributed markedly more than WT B cells to the class-switched IgG1 and IgA population (Fig. 5, A, C, and E). This overrepresentation of AhR−/− B cells was even more striking when antibody response to influenza-specific hemagglutinin (HA) protein was analyzed. Almost 90% of the HA-specific IgG1+ or IgA+ B cells in the germinal center were AhR deficient (Fig. 5, A, D, and E). It is to be noted that to ensure the validity of the data, we verified the specificity of HA staining, IgG1, and IgA antibodies used in the experiments with relevant controls (Fig. S4 D). Importantly, the skew toward CD45.2 in the class-switched antigen-specific B cells was absent in control mix chimeras, ruling out congenic marker differences (Fig. S4 E).
Figure 5.
AhR restrains primary B cell response to influenza virus. (A) Representative flow cytometry plot showing B220+ cells that are within germinal centers (GC; GL7+; top), B220+GL7+IgG1+ (middle), and B220+GL7+IgG1+ HA specific (bottom) in mock (PBS + 0.1% BSA)-infected or PR8-infected AhR mixed chimeras at day 16 after intranasal challenge. (B) Contribution of WT (CD45.1) and AhR−/− (CD45.2) cells to the B220+ and B220+GL7+ pool. (C) Contribution of WT (CD45.1) and AhR−/− (CD45.2) cells to the B220+GL7+IgG1+ pool. (D) Contribution of WT (CD45.1) and AhR−/− (CD45.2) cells to the B220+GL7+IgG1+ HA-specific pool. (B–D) n = 7. Mean ± SD is shown. (E) Contribution of WT (CD45.1) and AhR−/− (CD45.2) cells to the B220+GL7+IgA+ and B220+GL7+IgA+ HA-specific pool. n = 3. Mean ± SD is shown. (F) ELISA data showing endpoint titer (reciprocal of lowest serum dilution above background of day 0 sera) of PR8-specific IgG at day 10 after infection in AhR−/− mix chimera and control mix chimera. n = 6–8. Mean ± SEM is shown. (G) Percentage of plasma cells (B220lo CD138+) at day 10 after PR8 infection (or mock infected) in AhR−/− mix chimera and control mix chimera. n = 2 for mock and n = 3–5 for PR8 infected. Mean ± SD is shown. (H) Percentage of plasma cells (left), total B220+ (middle), and germinal center+ (right) cells that come from CD45.1 versus CD45.2 in PR8-infected AhR−/− mix chimera and control mix chimera. n = 5–6. Mean ± SD is shown. *, P < 0.05; **, P < 0.005 (Student’s t test).
We next compared the serum titers of virus-specific IgG at day 10 after infection in AhR mix chimeras versus control mix chimeras and observed a fourfold increase in the endpoint titer (Fig. 5 F). To investigate whether this enhanced serum antibody was caused by increased PCD, we detected B220lo CD138+ plasma cells by flow cytometry and found that the AhR mix chimeras had a higher frequency of plasma cells compared with control mix chimeras (Fig. 5 G). Importantly, AhR−/− B cells contributed significantly more than WT cells to the total plasma cell pool in AhR mix chimeras, a skewing not observed in control mix chimeras and in chimerism of total B cells or the germinal center B cell pool (Fig. 5 H). This competitive advantage suggests that AhR deficiency might lead to a marked increase in the primary immune response–associated fates (class switching and PCD) during influenza virus infection. Overall, these results reveal the existence of a negative regulatory circuitry during effector B cell responses involving the environmental sensor AhR, which serves to fetter CSR and PCD, a possible function that might promote memory responses.
Discussion
Activation of a naive B cell upon antigen receptor engagement prompts its clonal expansion, antibody isotype switching, and differentiation into antibody-secreting plasma cells to effectuate a robust primary immune response (Taylor et al., 2015). Significant work over the past decades has revealed effectors that promote class switching and PCD. However, regulators that negatively impact these processes, a warranted moderation given the risks posed to genomic integrity and exhaustion, are not well characterized. Our mechanistic studies using ex vivo activated B cells combined with assessment of the immune response after antigen exposure in mixed BM chimeras lead us to propose a model wherein AhR is induced in B cells upon antigen receptor engagement and plays a key role in negatively regulating CSR and PCD both ex vivo and in vivo. Although the effect of AhR on plasmablasts ex vivo was very robust, it did not translate into an equally strong in vivo effect on plasma cells, suggesting that there might also be AhR-dependent extrinsic restraining influences in vivo.
CSR proceeds through deliberate generation of AID-instigated DNA double-strand breaks, one of the most toxic lesions that can occur in a cell (Nussenzweig and Nussenzweig, 2010). Thus, temporal fine-tuning of AID expression is extremely important not only during CSR, but also after the process to preserve genomic integrity and switched memory B cell identity. Additionally, the existence of a robust pool of IgM memory B cells (Capolunghi et al., 2013) suggests that CSR should be actively constrained during a primary response to prevent depletion of antigen-experienced unswitched B cells that would be best fit to undergo all possible fates during a recall response. We identify AhR as a novel repressor of AID expression and, thus, CSR. Our results suggest that this repressor activity is elicited via binding to a well characterized silencer element in the aicda locus and likely through cooperation of other factors that bind this region.
Terminal differentiation of B cells into antibody-secreting plasma cells is indeed mandatory for humoral responses, but during a primary response, this fate is mutually exclusive to MBD, and uncontrolled PCD might also exhaust the pool of B cells that have first responded to the antigen (McHeyzer-Williams et al., 2012; Weisel et al., 2016). Both CSR and PCD are irreversible processes in that the original B cell clone is effectively lost from the population. However, the essence of adaptive immunity is that an infection leaves a footprint that serves as a recognition module during a reinfection. For B cells, this footprint is a memory B cell clone generated during the primary response that was somehow stalled from undergoing the irreversible processes of CSR and PCD. Additional studies will be required to delineate whether AhR directly participates in the generation and/or maintenance of the memory B cell pool.
Memory B cells are not only instrumental in the physiological setting for immunity during reinfection with pathogens, but they are also key to effective preventative vaccination. However, unlike, memory T cell differentiation, MBD is understudied, and key transcription factors and master regulators are elusive (Rankin et al., 2011; McHeyzer-Williams et al., 2012). A recent study points toward Bach2 being a key player in B cell memory (Shinnakasu et al., 2016), and our finding that AhR positively regulates Bach2 suggests that they might be part of the same circuitry that balances primary and secondary responses. Understanding the molecular framework of MBD is integral to rational vaccine design. Factors regulating B cell memory can be pharmacologically targeted in vaccine adjuvant formulations to effectuate better recall responses. AhR is a well studied pharmacologically targeted molecule, with potent agonists and antagonists readily available (Zhao et al., 2010; Murray et al., 2014). Our results establishing the existence of a negative modulatory circuit during primary response involving AhR opens the field toward modules that might promote secondary responses. Follow up studies on whether AhR promotes B cell memory will potentially open a therapeutic window of opportunity to use AhR agonists in vaccination regimens (e.g., flu vaccine) against pathogens (e.g., influenza virus) where B cell responses are crucial for protection (Chen et al., 2014; Ellebedy et al., 2014).
AhR has been extensively studied as an environmental sensor for dioxins. The recent upsurge in research on the functions of AhR beyond toxicity, especially in the immune system, has unraveled the importance of this xenobiotic sensor in modulating physiological aspects of both innate and adaptive immunity (Stevens et al., 2009; Nguyen et al., 2013; Stockinger et al., 2014; Cella and Colonna, 2015; Zhou, 2016). AhR has been shown to regulate the T regulatory–T helper 17 cell balance in a ligand-specific manner and also to control the transdifferentiation of pathogenic T helper 17 cells into immunosuppressive Tr1 cells (Quintana et al., 2008; Gagliani et al., 2015). Additionally, AhR has been implicated in the biology of innate immune cells including macrophages, NK cells, and innate lymphoid 3 cells, especially in the context of infection with intestinal bacteria and gut homeostasis (Qiu et al., 2012; Qiu and Zhou, 2013; Moura-Alves et al., 2014; Wagage et al., 2014). Our findings further AhR functions to the B lymphocyte lineage, where it serves a unique role in dictating cell-fate decisions and exemplifies the versatility of this prototypical environmental sensor.
Materials and methods
Mice
AhR−/− mice (C57BL/6J; generation 19; Schmidt et al., 1996) bred as heterozygous and littermates (6–10 wk) were used for experiments, or AhR−/− males were bred to AhR+/− females, and age/sex-matched WT C57BL6/J (purchased from The Jackson Laboratory) mice were used as controls. For BM chimeras, WT CD45.1 congenic and AhR−/− mice (or WT CD45.2 C57BL6/J; 6–8-wk-old females) were used as a source of BM cells. Single suspensions of BM extracted from femurs and tibias were depleted of T cells with Thy1 antibody–coated magnetic beads. 3–5 × 106 T cell–depleted BM cells were transferred (intravenous) individually or at a 1:1 ratio into irradiated (650 cGy) 8–10-wk-old Rag1−/− female recipients (The Jackson Laboratory). BM engraftment was examined by flow cytometric analysis of peripheral blood lymphocytes 8–10 wk after BM transfer. Three independent sets of chimera were generated. Mice were cheek bled at 7–8 wk after reconstitution to determine chimerism of B, T, and myeloid cells by flow cytometry. All animals were maintained and euthanized as per the Memorial Sloan Kettering Cancer Center Research and animal resource center guidelines.
B cell harvest and stimulations
For ex vivo assays, splenic and LN B cells were purified by CD43 negative selection (no. 130-049-801; Miltenyi Biotec) or CD19 positive selection (no. 130-052-201) following the manufacturer’s protocol, cultured at a density of 1 million/ml, re-fed with fresh media (with cytokines) at 48 and 72 h, and analyzed by flow cytometry at 96 h.
Cytokines, reagents, and antibodies
LPS (no. L4130; Sigma-Aldrich) at 10 µg/ml or 30 µg/ml, TGFβ (recombinant human TGFβ1; no. 240-B; R&D Systems) at 2 ng/ml, anti-IgD dextran conjugates (no. 0001; Fina Biosolution) at 300 ng/ml, anti-CD40 (FGK45; Memorial Sloan Kettering Cancer Center core facility) at 2 µg/ml, and mouse IL-4 (no. 404-ML; R&D Systems) at 12.5 ng/ml were used to stimulate B cells ex vivo. Cells were treated with vehicle control 0.1% DMSO (no. D8418-100ML; Sigma-Aldrich) or 30 nM TCDD in DMSO (custom made through AccuStandard Inc.). For Western blotting, ∼25–100 µg protein extracts (radioimmunoprecipitation assay buffer [Zheng et al., 2015] lysates of three to five million B cells) were resolved on 10% SDS-polyacrylamide gels (Bio-Rad Laboratories) and, after semidry transfer, probed with anti-AID (Chaudhuri et al., 2003), anti–α tubulin (no. T9026; Sigma-Aldrich), anti-AhR (pAb; no. BML-SA210-0100; Enzo Life Sciences), anti–Blimp-1 (clone 6D3; no. 14-5963-82; eBioscience), anti-rabbit HRP (no. 711-035-152; Jackson ImmunoResearch Laboratories, Inc.), anti–rat HRP (no. 712-035-153; Jackson ImmunoResearch Laboratories, Inc.), and anti–mouse HRP (no. 115-035-003; Jackson ImmunoResearch Laboratories, Inc.). For flow cytometry, anti–mouse B220 (clone RA3-6B2), anti-CD21 (PE and efluor450; clone eBio4E3), anti-CD23 (clone B3B4), anti-CD38 (AF700; clone eBio 90), anti-CD43 (FITC, biotin, PE; clone eBio R2/60), anti-CD45 (clone 30-F11), anti-CD45.1 (clone A20), anti-CD45.2 (clone 104), anti-CD138 (clone 281-2), anti-Fas (clone Jo2), anti-GL7 (clone GL7), anti-IgA (FITC; clone C10-3), anti-IgA (PE; clone mA-6E1), anti-IgD (clone 11.26.2c), anti-IgG1 (clones X56 and A85-1), anti-IgG3 (clone R40-82), anti-IgM (clone II/41), anti-Ki67 (eBio clone SolA15), anti-NP (PE; no. 5070; LGC Biosearch Technologies), streptavidin-APC efluor780/PeCy7, CD69 (clone H1.2F3), DAPI (no. D1306; Invitrogen), Ghost dye violet 510 (Tonbo Bioscience), and Zombie red fixable viability dye (BioLegend) were purchased from eBioscience, BD, and BioLegend unless specified otherwise. Flow cytometric analyses of single-cell suspension of cells from BM, spleen, LNs, and purified B cells was done on an LSRII flow cytometer (BD), and data analysis was performed using FlowJo software (version 9.9; Tree Star).
Retroviral transduction
HEK293T cells were transfected with 10 µg pCL-Eco packaging construct and 15 µg retroviral pMIG vectors (either empty or AID; Blimp-1 expressing) to generate virus. B cells were harvested and stimulated for 1 d with anti-IgD alone (for AID rescue) or anti-CD40 + IL-4 (for Blimp-1 rescue). Viral supernatants were added to B cells in the presence of 10 µg/ml polybrene, and spinfection was performed on days 2 and 3 as described previously (Vuong et al., 2009). CSR was assessed 72 h and PCD at 96 h after infection. Blimp-1–expressing retroviral construct was provided by S. Crotty (La Jolla Institute, San Diego, CA).
RNA extraction and quantitative reverse transcription PCR
RNA was extracted using TRIzol (15596-018; Invitrogen) following the manufacturer’s protocol. cDNA was made using a qscript cDNA synthesis kit (no. 95047; Quanta bioscience), and quantitative PCR was performed using gene-specific primers (as listed in Table 1) using iQ SYBR green supermix (Bio-Rad Laboratories). Relative gene expression was calculated according to the Δ Ct method.
Table 1. Quantitative PCR primer list.
| Primer name | Primer sequence (5′–3′) |
|---|---|
| Imu FP | CTCTGGCCCTGCTTATTGTTG |
| Cmu RP | GAAGACATTTGGGAAGGACTGACT |
| AID FP | GCCACCTTCGCAACAAGTCT |
| AID RP | CCGGGCACAGTCATAGCAC |
| Cyp1a1 FP | AAGTGCAGATGCGGTCTTCT |
| Cyp1a1 RP | AAAGTAGGAGGCAGGCACAA |
| Prdm1 FP | TTCTCTTGGAAAAACGTGTGGG |
| Prdm1 RP | GGAGCCGGAGCTAGACTTG |
| β actin FP | TGCGTGACATCAAAGAGAAG |
| β actin RP | CGGATGTCAACGTCACACTT |
| Bach2 FP | AGGGCTAGAGGCCAATGGTA |
| Bach2 RP | CTTCCCCATTAAGCAGCCCA |
| Iα FP | GACATGATCACAGGCACAGG |
| Cα RP | TTCCCCAGGTCACATTCATCGT |
| Iy3 FP | AACTACTGCTACCACCACCACCAG |
| Cy3 RP | ACCAAGGGATAGACAGATGGGG |
Abbreviations used: FP, forward primer; RP, reverse primer.
ChIP
In brief, cells were stimulated with anti-CD40 + IL-4 or LPS + TGFβ + anti-IgD dextran for 42–48 h and cross-linked with 1% formaldehyde. ChIP was performed as previously described (Vuong et al., 2009) with 2 µg anti-AhR (pAb; no. BML-SA210-0100; Enzo Life Sciences) antibody using a ChIP assay kit (EMD Millipore). Quantitative PCR using iQ SYBR green supermix (Bio-Rad Laboratories) was performed with the primers in Table 2. Percent input for immunoprecipitation was calculated by normalizing the raw Ct values to the input (5%). The primers used have been previously described (Tran et al., 2010; De Abrew et al., 2011).
Table 2. ChIP quantitative PCR primer list.
| Name of primer | Primer sequence (5′–3′) |
|---|---|
| aicda region 2a FP | CCACTTAATTACATCCTGAGCCC |
| aicda region 2a RP | CTATAAACCCAGAAGCAGCTCA |
| aicda intron FP | CTGACCCAGCAAAATCCAAA |
| aicda intron RP | CGAGGGATCAAACTCAAGAC |
| Bach2 FP promoter | AACCCTGCTCGTACATGACA |
| Bach2 RP promoter | TTCCAGGTAGCAAAGGCTAGA |
| Bach2 FP Untr6 | TCAGGCATGAACCACCATAC |
| Bach2 RP Untr6 | AACATCCACACGTCCAGTGA |
Abbreviations used: FP, forward primer; RP, reverse primer.
Cell proliferation
CD19+ B naive splenic B cells from WT and AhR−/− mice were labeled with 1.5 µM CTV (no. C34557; Thermo Fisher Scientific) following the manufacturer’s protocol. Equal labeling between genotypes was ensured by flow cytometric analysis at 0 h immediately after labeling. Proliferation of B cells was tracked by dye dilution at 96 h after stimulation. Switching and plasmablast formation as a function of dye dilution (proliferation) was analyzed using the proliferation platform on FlowJo.
Immunizations
Mice were intraperitoneally injected with 50 µg NP-CGG (in alum; range 30–39; no. N5055D; LGC Biosearch Technologies), NP-Ficoll (no. F-1420; LGC Biosearch Technologies), NP-LPS (no. N5065; LGC Biosearch Technologies), and 100 million SRBCs (no. IC10-0210; Innovative Research) on day 0, followed by a boost on day 10 and analysis of immune response on day 14. For the memory experiment, after the initial scheme as the primary response immunization, the mice were again challenged with 50 µg NP-CGG in PBS at 8 wk and analyzed 1 wk after the rechallenge.
Influenza infection
Isoflurane-anesthetized mice were infected intranasally with the influenza virus A H1N1 A/Puerto Rico/8/34 (PR8) with 50 TCID50 (50% tissue culture infective dose) in PBS + 0.1% BSA or mock (PBS + 0.1% BSA) infected (Frank et al., 2015). Mediastinal LNs were harvested at ∼2 wk after infection, and single-cell suspensions were analyzed by flow cytometry. Biotinylated HA with a mutation in the receptor-binding site (rHAPR8Y98F) was used to detect B cells expressing antibodies specific for influenza (Whittle et al., 2014; Frank et al., 2015).
ELISA
Serum from PBS/BSA-infected and PR8-infected mixed chimeras was taken by cheek bleeding mice (Goldenrod animal lancet 5 mm; MEDIpoint Inc.) on days 0 and 10 using Microtainer serum collection tubes (BD). Immulon high-binding ELISA plates (Thermo Fisher Scientific) were coated overnight at 4°C with PR8 virus in allantoic fluid. Plates were blocked with PBS/4% milk for 1 h at room temperature. After three washes with PBS + 0.05% Tween 20, plates were incubated with 50 µl of sera, which was twofold serially diluted starting from 1:100 in PBS + 0.05% Tween 20 for 1 h at room temperature. After three washes, plates were incubated with biotinylated anti–mouse IgG (Vector Laboratories) diluted 1:1,000 for 45 min at room temperature. After three washes, plates were incubated with avidin D–HRP (Vector Laboratories) diluted 1:2,500 for 45 min at room temperature. After three washes, plates were developed for 5 min using TMB substrate (KPL biomedical) and halted with 0.1 N HCl. Plates were read at 450 nm. Endpoint titer was determined as the reciprocal of lowest dilution above background of day 0 sera. Uninfected mice were seronegative.
Statistical analyses
Prism 6 (GraphPad Software) was used for data representations and statistical calculations. Student’s t test (paired and unpaired) was used for determining statistical significance. *, P < 0.05; **, P < 0.005.
Online supplemental material
Fig. S1 shows AhR induction in B cells and the role of AhR in B cell development. Fig. S2 shows cyp1a1 expression and CSR in AhR single and mixed chimeras. Fig. S3 shows germline transcription, proliferation in AhR-deficient B cells, and AID and Blimp-1 expression in AhR single chimeras. Fig. S4 shows reconstitution of AhR mixed and control chimeras and HA stain specificity.
Supplementary Material
Acknowledgments
The authors wish to thank members of the Chaudhuri and Rudensky laboratory for technical assistance and discussion, especially Joseph N. Pucella and Montserrat Cols Vidal for help with mouse organ harvests and A. Bravo for help with maintenance of the mouse colony. The authors appreciate the kind provision of retroviral vector for Blimp-1 overexpression by Shane Crotty.
This work was supported by grants from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH; 1RO1AI072194 and 1RO1AI124186) and the Starr Cancer Research Foundation (I7-A767) to J. Chaudhuri, grants from the Ludwig Center at Memorial Sloan Kettering Cancer Center, the Hilton-Ludwig Cancer Prevention Initiative (Conrad N. Hilton Foundation and Ludgwig Cancer Research), and the Howard Hughes Medical Institute to A.Y. Rudensky, an NIH/National Cancer Institute Cancer Center Support Grant (P30 CA008748) to A.Y. Rudensky and J. Chaudhuri, a grant from the Division of Intramural Research, NIAID, NIH to J.W. Yewdell and A.B. McDermott, a grant from the Intramural Research Program of the Vaccine Research Center, NIAID, NIH to A.B. McDermott, and a grant from the NIH (R37-ES005703) to C.A. Bradfield.
The authors declare no competing financial interests.
Author contributions: B. Vaidyanathan, A. Chaudhry, A.Y. Rudensky, and J. Chaudhuri developed ideas and designed experiments. B. Vaidyanathan and A. Chaudhry performed most experiments. W.T. Yewdell and D. Angeletti helped with influenza infection, and D. Angeletti performed ELISA experiments. J.W. Yewdell, A.K. Wheatley, and A.B. McDermott provided reagents and helped set up influenza infection experiments. W.T. Yewdell and W.F. Yen helped with BM chimera generation. C.A. Bradfield provided mice and reagents. B. Vaidyanathan and J. Chaudhuri wrote the manuscript.
Footnotes
Abbreviations used:
- AhR
- aryl hydrocarbon receptor
- AID
- activation-induced cytidine deaminase
- BCR
- B cell antigen receptor
- ChIP
- chromatin immunoprecipitation
- CSR
- class-switch recombination
- CTV
- CellTrace violet
- HA
- hemagglutinin
- MBD
- memory B cell differentiation
- MZ
- marginal zone
- PCD
- plasma cell differentiation
- SRBC
- sheep RBC
- TCDD
- 2,3,7,8-tetrachlorodibenzo p-dioxin
References
- Alt F.W., Zhang Y., Meng F.L., Guo C., and Schwer B.. 2013. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 152:417–429. 10.1016/j.cell.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capolunghi F., Rosado M.M., Sinibaldi M., Aranburu A., and Carsetti R.. 2013. Why do we need IgM memory B cells? Immunol. Lett. 152:114–120. 10.1016/j.imlet.2013.04.007 [DOI] [PubMed] [Google Scholar]
- Cella M., and Colonna M.. 2015. Aryl hydrocarbon receptor: Linking environment to immunity. Semin. Immunol. 27:310–314. 10.1016/j.smim.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., and Alt F.W.. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 422:726–730. 10.1038/nature01574 [DOI] [PubMed] [Google Scholar]
- Chen M., Hong M.J., Sun H., Wang L., Shi X., Gilbert B.E., Corry D.B., Kheradmand F., and Wang J.. 2014. Essential role for autophagy in the maintenance of immunological memory against influenza infection. Nat. Med. 20:503–510. 10.1038/nm.3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Abrew K.N., Phadnis A.S., Crawford R.B., Kaminski N.E., and Thomas R.S.. 2011. Regulation of Bach2 by the aryl hydrocarbon receptor as a mechanism for suppression of B-cell differentiation by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol. Appl. Pharmacol. 252:150–158. 10.1016/j.taap.2011.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellebedy A.H., Krammer F., Li G.M., Miller M.S., Chiu C., Wrammert J., Chang C.Y., Davis C.W., McCausland M., Elbein R., et al. 2014. Induction of broadly cross-reactive antibody responses to the influenza HA stem region following H5N1 vaccination in humans. Proc. Natl. Acad. Sci. USA. 111:13133–13138. 10.1073/pnas.1414070111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank G.M., Angeletti D., Ince W.L., Gibbs J.S., Khurana S., Wheatley A.K., Max E.E., McDermott A.B., Golding H., Stevens J., et al. 2015. A simple flow-cytometric method measuring B cell surface immunoglobulin avidity enables characterization of affinity maturation to influenza A virus. MBio. 6:e01156-15 10.1128/mBio.01156-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliani N., Amezcua Vesely M.C., Iseppon A., Brockmann L., Xu H., Palm N.W., de Zoete M.R., Licona-Limón P., Paiva R.S., Ching T., et al. 2015. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature. 523:221–225. 10.1038/nature14452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin A.D., von Boehmer L., Gazumyan A., Shulman Z., Oliveira T.Y., and Nussenzweig M.C.. 2016. Independent roles of switching and hypermutation in the development and persistence of B lymphocyte memory. Immunity. 44:769–781. 10.1016/j.immuni.2016.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T., Kometani K., and Ise W.. 2015. Memory B cells. Nat. Rev. Immunol. 15:149–159. 10.1038/nri3802 [DOI] [PubMed] [Google Scholar]
- Mandal P.K. 2005. Dioxin: a review of its environmental effects and its aryl hydrocarbon receptor biology. J. Comp. Physiol. B. 175:221–230. 10.1007/s00360-005-0483-3 [DOI] [PubMed] [Google Scholar]
- McHeyzer-Williams M., Okitsu S., Wang N., and McHeyzer-Williams L.. 2012. Molecular programming of B cell memory. Nat. Rev. Immunol. 12:24–34. 10.1038/nri3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura-Alves P., Faé K., Houthuys E., Dorhoi A., Kreuchwig A., Furkert J., Barison N., Diehl A., Munder A., Constant P., et al. 2014. AhR sensing of bacterial pigments regulates antibacterial defence. Nature. 512:387–392. 10.1038/nature13684 [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., and Honjo T.. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. 10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Murray I.A., Patterson A.D., and Perdew G.H.. 2014. Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat. Rev. Cancer. 14:801–814. 10.1038/nrc3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen N.T., Hanieh H., Nakahama T., and Kishimoto T.. 2013. The roles of aryl hydrocarbon receptor in immune responses. Int. Immunol. 25:335–343. 10.1093/intimm/dxt011 [DOI] [PubMed] [Google Scholar]
- Nussenzweig A., and Nussenzweig M.C.. 2010. Origin of chromosomal translocations in lymphoid cancer. Cell. 141:27–38. 10.1016/j.cell.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt S.L., Hodgkin P.D., Tarlinton D.M., and Corcoran L.M.. 2015. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 15:160–171. 10.1038/nri3795 [DOI] [PubMed] [Google Scholar]
- Qiu J., and Zhou L.. 2013. Aryl hydrocarbon receptor promotes RORγt+ group 3 ILCs and controls intestinal immunity and inflammation. Semin. Immunopathol. 35:657–670. 10.1007/s00281-013-0393-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., Heller J.J., Guo X., Chen Z.M., Fish K., Fu Y.X., and Zhou L.. 2012. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 36:92–104. 10.1016/j.immuni.2011.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana F.J., Basso A.S., Iglesias A.H., Korn T., Farez M.F., Bettelli E., Caccamo M., Oukka M., and Weiner H.L.. 2008. Control of Treg and TH17 cell differentiation by the aryl hydrocarbon receptor. Nature. 453:65–71. 10.1038/nature06880 [DOI] [PubMed] [Google Scholar]
- Rankin A.L., MacLeod H., Keegan S., Andreyeva T., Lowe L., Bloom L., Collins M., Nickerson-Nutter C., Young D., and Guay H.. 2011. IL-21 receptor is critical for the development of memory B cell responses. J. Immunol. 186:667–674. 10.4049/jimmunol.0903207 [DOI] [PubMed] [Google Scholar]
- Reiner S.L., and Adams W.C.. 2014. Lymphocyte fate specification as a deterministic but highly plastic process. Nat. Rev. Immunol. 14:699–704. 10.1038/nri3734 [DOI] [PubMed] [Google Scholar]
- Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A., et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575. 10.1016/S0092-8674(00)00079-9 [DOI] [PubMed] [Google Scholar]
- Schmidt J.V., Su G.H., Reddy J.K., Simon M.C., and Bradfield C.A.. 1996. Characterization of a murine Ahr null allele: involvement of the Ah receptor in hepatic growth and development. Proc. Natl. Acad. Sci. USA. 93:6731–6736. 10.1073/pnas.93.13.6731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer A.L., Rosenwald A., Hurt E.M., Giltnane J.M., Lam L.T., Pickeral O.K., and Staudt L.M.. 2001. Signatures of the immune response. Immunity. 15:375–385. 10.1016/S1074-7613(01)00194-7 [DOI] [PubMed] [Google Scholar]
- Shinnakasu R., Inoue T., Kometani K., Moriyama S., Adachi Y., Nakayama M., Takahashi Y., Fukuyama H., Okada T., and Kurosaki T.. 2016. Regulated selection of germinal-center cells into the memory B cell compartment. Nat. Immunol. 17:861–869. 10.1038/ni.3460 [DOI] [PubMed] [Google Scholar]
- Stevens E.A., Mezrich J.D., and Bradfield C.A.. 2009. The aryl hydrocarbon receptor: a perspective on potential roles in the immune system. Immunology. 127:299–311. 10.1111/j.1365-2567.2009.03054.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger B., Di Meglio P., Gialitakis M., and Duarte J.H.. 2014. The aryl hydrocarbon receptor: multitasking in the immune system. Annu. Rev. Immunol. 32:403–432. 10.1146/annurev-immunol-032713-120245 [DOI] [PubMed] [Google Scholar]
- Tarlinton D. 2012. B-cell differentiation: instructive one day, stochastic the next. Curr. Biol. 22:R235–R237. 10.1016/j.cub.2012.02.045 [DOI] [PubMed] [Google Scholar]
- Taylor J.J., Pape K.A., Steach H.R., and Jenkins M.K.. 2015. Apoptosis and antigen affinity limit effector cell differentiation of a single naïve B cell. Science. 347:784–787. 10.1126/science.aaa1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran T.H., Nakata M., Suzuki K., Begum N.A., Shinkura R., Fagarasan S., Honjo T., and Nagaoka H.. 2010. B cell-specific and stimulation-responsive enhancers derepress Aicda by overcoming the effects of silencers. Nat. Immunol. 11:148–154. 10.1038/ni.1829 [DOI] [PubMed] [Google Scholar]
- Vuong B.Q., Lee M., Kabir S., Irimia C., Macchiarulo S., McKnight G.S., and Chaudhuri J.. 2009. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nat. Immunol. 10:420–426. 10.1038/ni.1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagage S., John B., Krock B.L., Hall A.O., Randall L.M., Karp C.L., Simon M.C., and Hunter C.A.. 2014. The aryl hydrocarbon receptor promotes IL-10 production by NK cells. J. Immunol. 192:1661–1670. 10.4049/jimmunol.1300497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisel F.J., Zuccarino-Catania G.V., Chikina M., and Shlomchik M.J.. 2016. A temporal switch in the germinal center determines differential output of memory B and plasma cells. Immunity. 44:116–130. 10.1016/j.immuni.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle J.R., Wheatley A.K., Wu L., Lingwood D., Kanekiyo M., Ma S.S., Narpala S.R., Yassine H.M., Frank G.M., Yewdell J.W., et al. 2014. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J. Virol. 88:4047–4057. 10.1128/JVI.03422-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Degroot D.E., Hayashi A., He G., and Denison M.S.. 2010. CH223191 is a ligand-selective antagonist of the Ah (Dioxin) receptor. Toxicol. Sci. 117:393–403. 10.1093/toxsci/kfq217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Vuong B.Q., Vaidyanathan B., Lin J.Y., Huang F.T., and Chaudhuri J.. 2015. Non-coding RNA generated following lariat debranching mediates targeting of AID to DNA. Cell. 161:762–773. 10.1016/j.cell.2015.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L. 2016. AHR function in lymphocytes: Emerging concepts. Trends Immunol. 37:17–31. 10.1016/j.it.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.