Wang et al. show that antibody gene deamination by activation-induced cytidine deaminase (AID) is restricted to a short time window in early G1 as a result of AID’s transient nuclear localization and accessibility of the target sites.
Abstract
Activation-induced cytidine deaminase (AID) converts cytosine into uracil to initiate somatic hypermutation (SHM) and class switch recombination (CSR) of antibody genes. In addition, this enzyme produces DNA lesions at off-target sites that lead to mutations and chromosome translocations. However, AID is mostly cytoplasmic, and how and exactly when it accesses nuclear DNA remains enigmatic. Here, we show that AID is transiently in spatial contact with genomic DNA from the time the nuclear membrane breaks down in prometaphase until early G1, when it is actively exported into the cytoplasm. Consistent with this observation, the immunoglobulin (Igh) gene deamination as measured by uracil accumulation occurs primarily in early G1 after chromosomes decondense. Altering the timing of cell cycle–regulated AID nuclear residence increases DNA damage at off-target sites. Thus, the cell cycle–controlled breakdown and reassembly of the nuclear membrane and the restoration of transcription after mitosis constitute an essential time window for AID-induced deamination, and provide a novel DNA damage mechanism restricted to early G1.
B lymphocytes are rapidly dividing cells that suffer genome damage as a result of replication stress (Gaillard et al., 2015) and errors in chromosome segregation (Santaguida and Amon, 2015). In addition, activation-induced cytidine deaminase (AID), which is required for antibody gene diversification (Muramatsu et al., 2000; Di Noia and Neuberger, 2007; Alt et al., 2013), is also responsible for off-target DNA damage associated with the genesis of lymphoma (Robbiani and Nussenzweig, 2013; Qian et al., 2014; Casellas et al., 2016).
AID deaminates cytosine to uracil in single-stranded DNA (ssDNA; Chaudhuri et al., 2003; Dickerson et al., 2003) that is exposed by sequence-intrinsic mechanisms (Yeap et al., 2015; Zheng et al., 2015), R-loops (Yu et al., 2003), or simultaneous sense and antisense transcription (Meng et al., 2014; Pefanis et al., 2014). Once formed, AID-induced uracil is excised by UNG, producing an abasic site that is further processed by a variety of different mechanisms, some of which can lead to DNA breaks that are resolved by nonhomologous end joining (Di Noia and Neuberger, 2007; Robbiani and Nussenzweig, 2013). AID-mediated DNA damage is detectable primarily in G1 (Petersen et al., 2001; Faili et al., 2002), and it is limited by a variety of different mechanisms, including AID transcription, phosphorylation, ubiquitination, and Crm1-dependent extrusion from the nucleus (Casellas et al., 2016). However, precisely when AID induces deamination and how the cell cycle enables it to efficiently access the Igh locus and simultaneously limits its genotoxicity have not been determined.
Results and discussion
Transient nuclear localization of AID
To investigate how AID accesses genomic DNA, we analyzed its subcellular localization using primary B cells expressing a functional AID-EGFP fusion protein driven by the endogenous AID promoter (Crouch et al., 2007; Fig. S1 A). Consistent with previous studies (McBride et al., 2004), we found that AID-EGFP was cytoplasmic and absent from the nucleus in most cells (Fig. S1 B). However, cells occasionally displayed AID-EGFP in both nucleus and cytoplasm, and these cells were always observed in pairs (Fig. S1 C), suggesting that the cellular distribution of AID is associated with cell division.
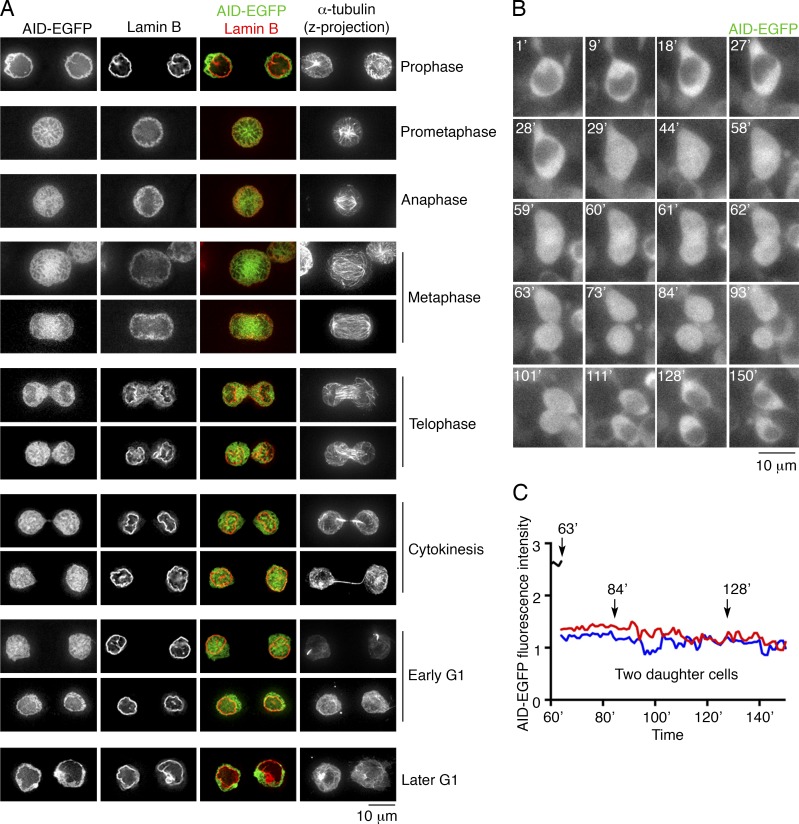
To test the possibility that AID gains access to the nucleus in a cell cycle–dependent manner, we examined specific stages of cell division (see Materials and methods). In agreement with previous observations (Lackey et al., 2012, 2013), in prometaphase, after the breakdown of the nuclear envelope, AID-EGFP was distributed throughout the cell body (Fig. 1 A). Upon nuclear envelope formation and during nuclear expansion (Anderson and Hetzer, 2007), AID-EGFP was present in the newly formed nucleus, and remained nuclear in early G1 cells (Fig. 1 A). Within 40–60 min of the beginning of cytokinesis, AID was restored to its cytoplasmic distribution (Fig. 1 B and Videos 1–5). The total fluorescence intensity in daughter cells did not change in the process, suggesting that nuclear AID-EGFP was exported into the cytoplasm rather than degraded in the nucleus (Fig. 1 C; and Fig. S1, D and E). Moreover, in some early G1 cells, the fluorescence of AID-EGFP was higher in the nucleus than in the cytoplasm (Fig. S1, F and G), which is consistent with previous findings that AID can also be actively imported into the nucleus (Patenaude et al., 2009). The data indicate that the breakdown of the nuclear membrane enables AID access to the genome during mitosis, cytokinesis and during a short time period in the early phase of G1, after which it is actively transported out.
Figure 1.
AID accesses the genome during mitosis, cytokinesis, and a short time period in the early phase of G1. (A) Subcellular localization of AID-EGFP from mitosis to G1. AID-EGFP (green) and Lamin B (red, nuclear envelope) are represented by single optical slices (35–40 z-slices each cell). A projection of all slices of α-tubulin shows the microtubule network. The combination of nuclear envelope and microtubule network allows determination of the cell cycle stage. Bar, 10 µm. Three independent experiments. (B) Time-lapse imaging of AID-EGFP–expressing B cells during cell division (see Video 1). Single optical z-slices are shown for each time point (indicated in minutes in the top left corner). Bar, 10 µm. Two independent experiments were performed. (C) Relative fluorescence intensity of AID-EGFP after cell division. Total AID-EGFP fluorescence was measured over time, and no significant changes in total fluorescence are recorded in the daughter cells (red and blue lines).
AID deamination on the switch region is restricted to early G1
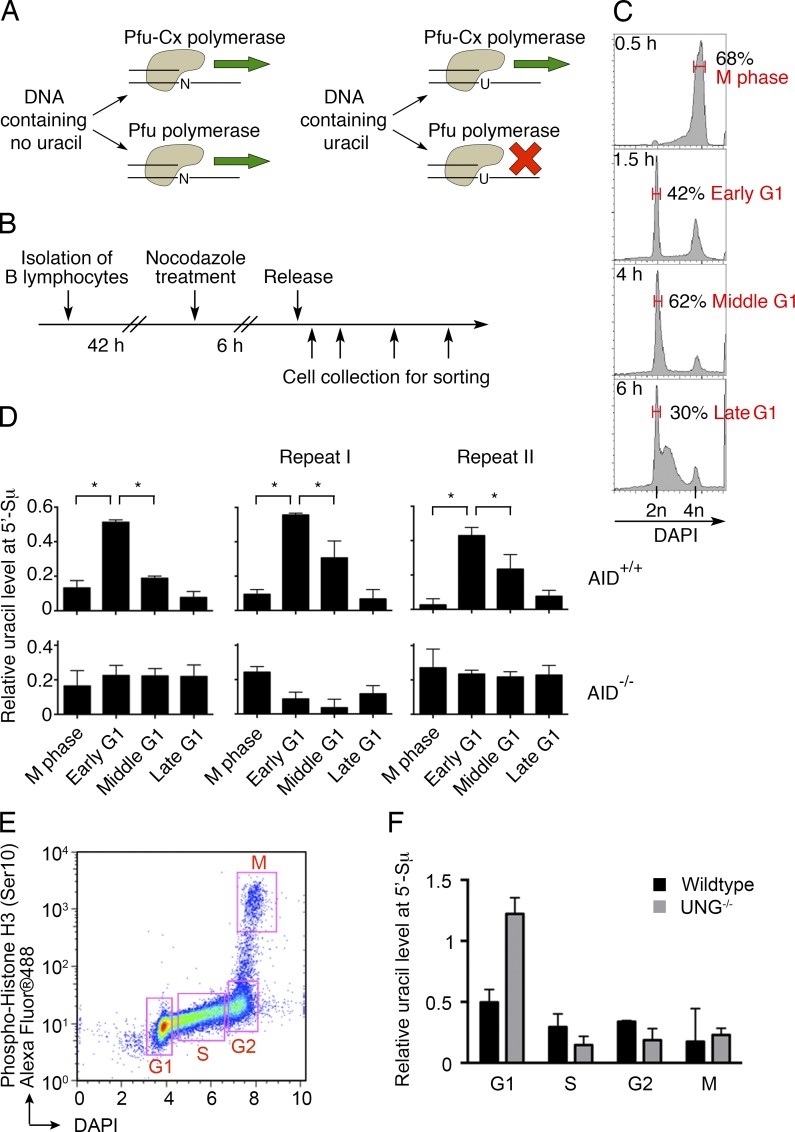
To directly determine when AID deaminates antibody genes, we used Pfu-Cx, a mutant polymerase that can amplify uracil-containing DNA (Horváth and Vértessy, 2010). Uracil levels are determined by comparing native and mutant Pfu polymerases on the same DNA samples (Fig. 2 A; Uracil-quantitative PCR [qPCR]; see Materials and methods). Activated B cells were blocked and thereby synchronized in M phase of the cell cycle with nocodazole, and then sorted and analyzed at different time points after release (Fig. 2, B and C; see Materials and methods). The uracil content of the Igh switch μ region (5′-Sμ), which is a physiological target of AID, was measured by Uracil-qPCR. Whereas control AID-deficient cells showed no change in uracil content, AID-sufficient early G1 cells showed significantly higher uracil levels than M phase cells, and uracil content dramatically decreased in cells in middle or late G1 (Fig. 2 D). Comparison of sorted cell fractions excluded the possibility that uracil accumulation at 5′-Sμ occurs in S, G2, or M (Fig. 2, E and F).
Figure 2.
Uracil-qPCR detects uracil accumulation at the antibody 5′-Sμ region. (A) Diagram of how Pfu polymerase and its mutant Pfu-Cx act on DNA. Uracil-free DNA can be amplified by both Pfu and Pfu-Cx polymerases (left). The lack of uracil recognition by Pfu-Cx polymerase allows it to replicate uracil-containing DNA, whereas Pfu polymerase stalls (right). This differential ability to amplify uracil-containing DNA is harnessed to measure the relative level of uracil (see Materials and methods; Horváth and Vértessy, 2010). (B) Experimental layout. In vitro–activated B lymphocytes were arrested with nocodazole 42 h after stimulation. 6 h later, nocodazole was removed and B cells were collected for analysis at different time points. (C) DAPI staining for DNA content at the indicated time points upon nocodazole release. Red, the sorted populations for experiments in Fig. 2 D and Fig. S2 (E and F). Upon release, B cells were still mainly in M phase at 0.5 h, whereas the released G1 population increased to ∼40% at 1.5 h (early G1) and 60% at 4 h (middle G1). At 6 h, some cells entered S phase, and 30% were still in G1 (late G1). (D) Relative uracil levels at the antibody 5′-Sμ region, as determined by Uracil-qPCR (see Materials and methods). AID-proficient and -deficient cells are compared. Error bars, SEM; *, P < 0.01, two-tailed Student t test. Three independent experiments are shown. (E) Diagram of the sorting strategy for the analysis of G1, S, G2, and M cells. In vitro–activated B cells at different cell cycle stages were sorted according to phospho-Histone H3 (Ser10) and DNA content (DAPI). (F) The relative uracil levels at 5′-Sμ at different cell cycle stages were determined by Uracil-qPCR in WT and UNG-deficient B cells. One experiment is shown.
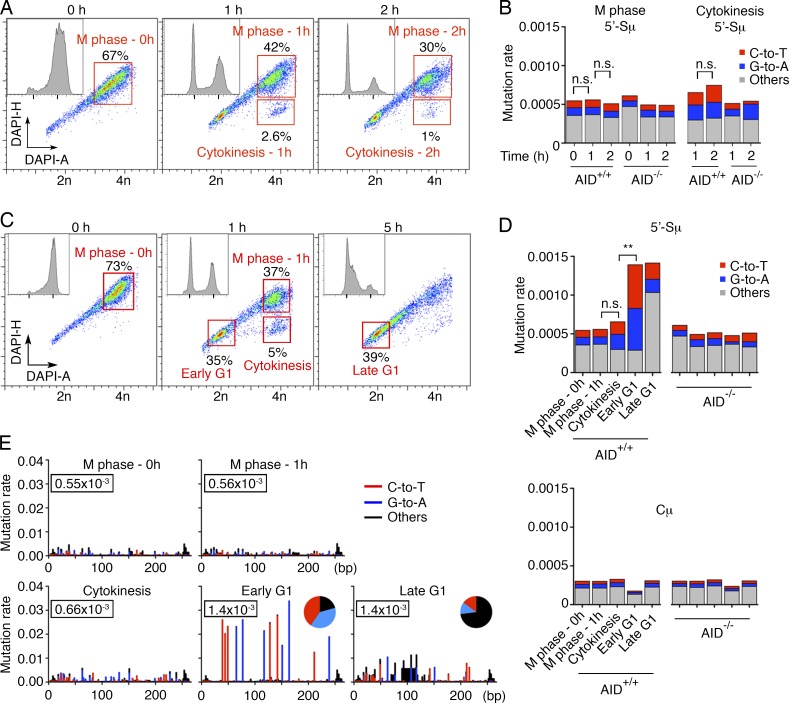
Pfu-Cx polymerase copies uracils to produce A:U base pairs. By combining Pfu-Cx polymerase and next generation sequencing (MutPE-Seq; Fig. S2 A), we were able to capture the AID-induced uracils as mutations in Igh DNA (C-to-T and G-to-A; Fig. S2 B). Purified M phase cells showed background rates of mutation at the 5′-Sμ region irrespective of the amount of time they remained in M (Fig. 3, A and B). Similarly, cells in cytokinesis displayed a low level of mutation despite the fact that AID has access to the DNA during this cell cycle stage, possibly because chromosomes are still partially condensed (Fig. 1 A; Fig. 3, C–E; and Fig. S2, C and D). In contrast, C-to-T and G-to-A mutations were significantly increased in early G1 cells (Fig. 3, C–E; and Fig. S2, C and D). No further increase was observed in late G1 cells, but a substantial fraction of C-to-T and G-to-A mutations were converted into other types of mutations in this phase of the cell cycle (Fig. 3, D and E; and Fig. S2 D). Negative controls included AID-deficient cells and the Cμ region, which is not targeted by AID (Fig. 3 D and Fig. S2 D). To rule out the possibility that the increase of uracil in early G1 is caused by reduced UNG activity (Haug et al., 1998; Hagen et al., 2008), we measured uracil accumulation in UNG-deficient cells. Similar to WT B cells, C-to-T and G-to-A mutations increased exclusively in early G1 in UNG−/−AID+/+ B cells, with no significant further increase in middle or late G1 (Fig. S2, E and F). This indicates that even in the absence of UNG, deamination occurs in early G1, and does not extend throughout the whole G1. We conclude that AID deaminates cytosine after cytokinesis in early G1, and that processing of these lesions continues throughout the G1 phase of the cell cycle.
Figure 3.
AID-induced deamination at antibody switch region is restricted to early G1. (A and B) Prolonged staying in M phase upon nocodazole release does not significantly increase the level of AID-induced mutations. (A) Cell cycle distribution of In vitro–cultured B lymphocytes at different time points upon nocodazole release. Cells in M, cytokinesis, and G1 are resolved by combining DNA fluorescence peak (DAPI-H) and intensity (DAPI-A; Gasnereau et al., 2007). Insets show DAPI-A profiles only. Nocodazole-treated cells were collected at 0, 1, and 2 h after release. In red are the sorted cell populations analyzed in Fig. 3 B. (B) Mutation rates at 5′-Sμ, as determined by MutPE-Seq. Red, blue, and gray represent C-to-T, G-to-A, and all other mutations, respectively. n.s., nonsignificant, one-tailed bootstrap test. One experiment is shown. (C) Nocodazole-treated cells were collected at 0, 1, and 5 h after release for mutational analysis with Pfu-Cx. Red, the sorted cell populations analyzed in D and E. (D and E) Mutation frequencies at 5′-Sμ are shown as a histogram (D) or at single-nucleotide resolution (E). The numbers in the rectangles indicate the overall mutation rate. Red, blue, and black represent C-to-T, G-to-A, and all other mutations, respectively. n.s., nonsignificant; **, P < 0.01, one-tailed bootstrap test. Two independent experiments were performed.
Convergent transcription in early G1 during AID targeting
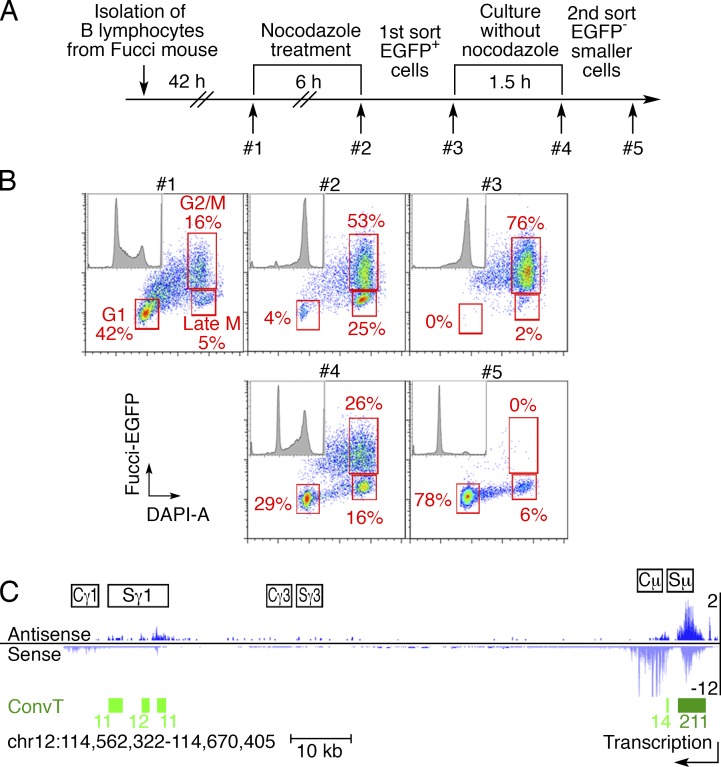
SHMs are associated with convergent transcription, which exposes the ssDNA targets of AID (Meng et al., 2014). To determine whether there is convergent transcription at AID target sites in early G1, we established a strategy to purify the live early G1 B cells from Fucci-EGFP+ mice (Sakaue-Sawano et al., 2008; Fig. 4, A and B; see Materials and methods). As measured by GRO-Seq, genome-wide transcription was similar in early G1 and nonsynchronized B cells (Fig. S3 A). Convergent transcription was evident over the Igh Sμ and Sγ1 regions, but not at constant regions (Fig. 4 C and Fig. S3 B). We conclude that the switch regions are convergently transcribed in early G1.
Figure 4.
Convergent transcription is detected at Igh switch regions in early G1 cells. (A) Experimental layout of the double-sorting strategy to isolate live early G1 cells. After 6-h nocodazole treatment, the Fucci-EGFP+ B cells (mostly M phase) were sorted and replated in nocodazole-free medium for 1.5 h. After the second sort, EGFP− small cells (mostly early G1) were purified (see Materials and methods). (B) FACS analysis of the double-sorting strategy. At each step, a small aliquot of cells was fixed and stained with DAPI (quality control, #1–#5). G1, G2/M, and late M populations (including cells in cytokinesis) are indicated on the representative plots. (C) GRO-Seq and convergent transcription (ConvT) profiles at the Igh locus in early G1 cells. Sense (light blue) and antisense transcription (dark blue) are displayed. ConvT regions are shown as green bars, and the numbers indicate the levels of ConvT, which were calculated by the geometric means of sense and antisense transcription reads as described previously (Meng et al., 2014). The arrow shows the direction of sense transcription. Two independent experiments were performed.
Extended AID nuclear residence leads to more genomic damage
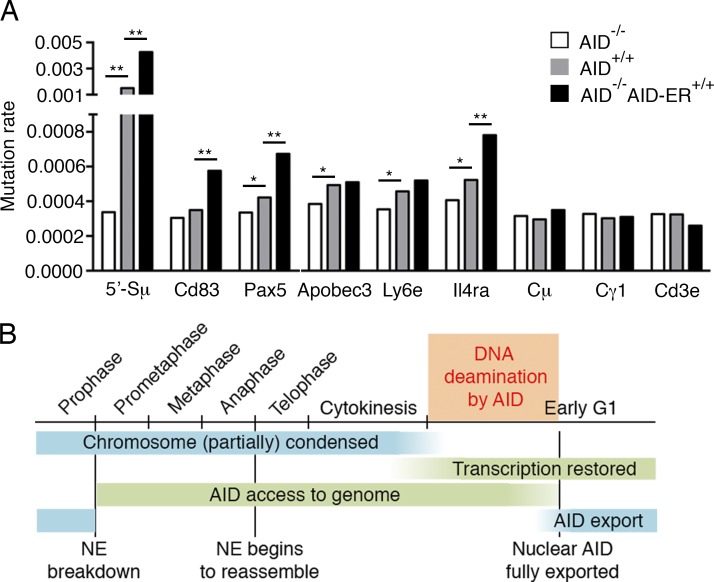
To determine whether extended AID residence in the nucleus produces additional off-target mutations, we analyzed transgenic B cells expressing AID fused to the estrogen receptor ligand-binding domain (AID-ER; Wang et al., 2014). Upon tamoxifen administration, AID-ER is actively transported into the nucleus, extending its presence in this compartment compared with the WT protein (Doi et al., 2003). Although AID-ER is expressed at fivefold lower levels than WT AID (Fig. S3 C), it induces higher levels of antibody CSR (Doi et al., 2003), and 5′-Sμ and off-target mutation (Cd83, Pax5, and Il4ra; Fig. 5 A and Fig. S3 D). These findings are consistent with previous observations (Le and Maizels, 2015; Methot et al., 2015). Thus, extending AID residence in the nucleus enhances its off-target activity.
Figure 5.
Extended presence of AID in the nucleus causes more mutations. (A) Relative mutation rates at 5′-Sμ, AID off-target sites (Cd83, Pax5, Apobec3, Ly6e, and Il4ra), and non-AID targets (Cμ, Cγ1, and Cd3e), revealed by MutPE-Seq. *, P < 0.05; **, P < 0.01, one-tailed bootstrap test. Two independent experiments were performed. (B) Proposed model. AID induces deamination at the Igh gene only during a short time window in early G1. Before then, although AID has spatial contact with chromosomes after breakdown of the nuclear envelope (NE), chromosomal DNA is condensed and not available for deamination. Shortly thereafter, AID is quickly exported into the cytoplasm by Crm1-mediated nuclear export. During the rest of the cell cycle, AID is cytoplasmic and no significant AID activity is detected. Therefore, this limited time window for AID to induce deamination is the combined consequence of transient nuclear residence of AID and the recovered transcription in the early G1 phase. The AID-induced uracil in this short time period is processed in G1 to enable SHM and CSR.
Concluding remarks
AID activity has long been linked to cell division and to a specific stage in the cell cycle, but how this might be regulated has remained unclear. Our data indicate that proliferation is necessary for AID activity because the associated breakdown of the nuclear membrane provides access to DNA (Fig. 5 B). This observation explains why SHM and CSR are dependent on, and accumulate with, each cell division cycle. Although the breakdown of the nuclear membrane exposes nuclear DNA to AID in mitosis and cytokinesis, chromosome condensation protects the genome during these cell cycle stages. Chromosome decondensation and reinitiation of transcription in the early phase of G1 expose the ssDNA substrate to AID. DNA damage is then limited by reassembly of the nuclear envelope and restoration of Crm1-mediated AID nuclear export in B lymphocytes undergoing rapid cell division in the dark zone of the germinal center (McBride et al., 2004; Victora et al., 2010).
Materials and methods
Mice
AID−/−, AID−/−ROSAAID-ER/AID-ER, UNG−/−, AID-EGFP, and Fucci-EGFP+ mice have been reported previously (Muramatsu et al., 2000; Endres et al., 2004; Crouch et al., 2007; Sakaue-Sawano et al., 2008; Robbiani et al., 2015). With the exception of UNG−/−, which are in a mixed background, all other mice were generated or backcrossed to C57BL/6 for at least 11 generations. All experiments were performed in accordance with protocols approved by the Rockefeller University Animal Care and Use Committee.
B cell culture, synchronization, and sorting
The isolation of B cells and in vitro activation were performed as previously described (Wang et al., 2014). In brief, resting B cells were purified from mouse spleens by CD43-negative selection and cultured in supplemented RPMI medium (RPMI-1640, 10% FBS, 2 mM l-glutamate, 1x Antibiotic-Antimycotic, 10 mM Hepes, 10 mM sodium pyruvate, and 55 µM β-mercaptoethanol) with 500 ng/ml RP105 (BD), 25 µg/ml LPS (Sigma-Aldrich), and 5 ng/ml mouse recombinant IL-4 (Sigma-Aldrich). Where indicated, nocodazole (100 ng/ml; Sigma-Aldrich) was added at 42 h after isolation for synchronization. After 6 h of nocodazole treatment, cells were washed and released in nocodazole-free supplemented RPMI for 0–6 h. Cells were then collected and fixed with 4.2% formaldehyde at room temperature for 20 min. After DAPI staining (0.5 µg/ml) at room temperature for 5 min, cells were sorted using a FACSAria (BD). The cytokinesis population was determined by combining DNA fluorescence peak (DAPI-H) and intensity (DAPI-A) according to a previous study (Gasnereau et al., 2007). For mutational analysis (Fig. 5 A), B cells of all genotypes were treated with 1 µg/ml 4-hydroxytamoxifen (Sigma-Aldrich) on day 2 and collected on day 6 to sort live cells. The sorted cell pellets were stored at –80°C. For sorting cells at different cell cycle stages (Fig. 2, E and F), in vitro–activated B cells (48 h after isolation) were stained with Alexa Fluor 488–conjugated anti–phospho-Histone H3, Ser10 antibody (Cell Signaling Technology), and DAPI (0.5 µg/ml), before sorting with FACSAria (BD).
Immunofluorescence and time-lapse imaging
In vitro–cultured AID-EGFP–expressing B cells (nontreated, nocodazole-arrested, or nocodazole-released) were fixed with 4% paraformaldehyde in PBS, and then subjected to DAPI staining (0.5 µg/ml in PBS at room temperature for 5 min) before microscopic analysis. For immunofluorescence, fixed cells were incubated with antibodies after permeabilization (0.5% Triton X-100 in PBS) and blocking (3% BSA in PBS). The following primary antibodies were used (all at 1:200 dilution): anti-Lamin B1 rabbit polyclonal antibody (Abcam) and anti–α-tubulin mouse monoclonal antibody (DM1A; Abcam). The following secondary antibodies were used (at 1:250 dilution): Alexa Fluor 568/647–conjugated goat anti–mouse/rabbit IgG antibodies (Thermo Fisher Scientific). Image capture and analyses were performed using a spectral confocal microscope Leica TCS SP (Leica) or DeltaVision Image Restoration Microscope system (Applied Precision). For live cell imaging, in vitro–cultured cells were maintained at 37°C and observed under DeltaVision Image Restoration Microscope system (1 min interval, 1.5 µm/layer × 12 layers). The relative fluorescent intensity was measured by ImageJ software (National Institutes of Health).
Genomic DNA isolation
The sorted cell pellets were resuspended in 400 µl proteinase K buffer (100 mM Tris-HCl 8.5, 0.2% SDS, 200 mM NaCl, and 5 mM EDTA) in the presence of 0.25 mg/ml proteinase K. Incubation at 55°C overnight was followed by phenol-chloroform extraction and ethanol precipitation. The DNA pellet was finally dissolved in distilled water.
Uracil-qPCR
Uracil-qPCR was performed as previously described (Horváth and Vértessy, 2010), with minor modifications. The reaction mix for quantitative real-time PCR was in a final volume of 20 µl, and contained 1x Cloned PFU reaction buffer (Agilent Technologies), 1x EvaGreen (Biotium), 30 nM passive reference dye ROX (Thermo Fisher Scientific), 0.25 mM dNTP, and 0.5 µM of each primer (F-qPCR-Sμ, TCTCTGAGTGCTTCTAAAATGCG; and R-qPCR-Sμ, TCACCCCAACACAGCGTAGC). 50 ng of template genomic DNA was added to each reaction, with either 0.5 µl PfuTurbo Hotstart or PfuTurbo Cx Hotstart DNA polymerase (Agilent Technologies). The PfuTurbo Cx Hotstart DNA polymerase was engineered to bear a point mutation that overcomes uracil stalling during PCR amplification, allowing the polymerase to read through uracil on the template strand. Real-time PCR reactions were performed in QuantStudio 6 Flex system (Thermo Fisher Scientific) according to the manufacturer's instructions. Reaction conditions were as follows: 95°C for 2 min, 40 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 1 min. Dissociation curve analysis, gel electrophoresis, and sequencing of the PCR products confirmed the specificity of amplification and the absence of primer dimers. Non-template control was measured in all cases. For data analysis, Ct values obtained with PfuTurbo Hotstart DNA polymerase were used as reference, and the 2-ΔΔCt method was used to calculate the relative uracil level. The calculated relative uracil levels were represented as mean ± SEM from three to four parallel reactions.
MutPE-Seq
To determine the frequency of AID-induced mutations at specific loci, we performed mutational analysis by paired-end sequencing, as previously described (Robbiani et al., 2015). 50 ng of genomic DNA from sorted in vitro–cultured B cells were amplified by two rounds of PCR and prepared for deep sequencing. To capture uracils and mutations simultaneously, the first round of PCR was performed using PfuTurbo Cx Hotstart DNA polymerase, in the presence of 1x PfuTurbo Cx reaction buffer and primers. F-first-Sμ and R-first-Sμ for 5′-Sμ region (269 base pairs), and F-first-Cμ and R-first-Cμ for Cμ region (244 bp). Reaction conditions were as follows: 95°C for 2 min, 20 cycles of 95°C for 15 s, 55°C for 30 s, and 72°C for 1 min. For mutational analysis in Fig. 5 A, phusion polymerase (Thermo Fisher Scientific) was used in the first round of PCR, instead of PfuTurbo Cx Hotstart DNA polymerase. The first round PCR primers used to amplify the 5′-Sμ, Cμ, and Cγ1 regions were: F-first-Sμ, 5′-TCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTCTCTGAGTGCTTCTAAAATGCG-3′; R-first-Sμ, 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTCACCCCAACACAGCGTAGC-3′; F-first-Cμ, 5′-TCTACACTCTTTCCCTACACGACGCTCTTCCGATCTCGGTTTTGGAGTGAAGTTCGTG-3′; R-first-Cμ, 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTGTGCCCATTCCAGGTAAGAAC-3′; F-first-Cγ1, 5′-TCTACACTCTTTCCCTACACGACGCTCTTCCGATCTAACAAGGGGCAGCATGGGTTGGAA-3′; and R-first-Cγ1, 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTAGCGCTGCTGTCACACTCTTCA-3′. Those used to amplify regions of Cd83, Pax5, Apobec3, Ly6e, Il4ra, and Cd3e were as follows: F-first-Cd83, 5′-TCTACACTCTTTCCCTACACGACGCTCTTCCGATCTATGCAGTGTCCTGGGCCAAGGTAA-3′; R-first-Cd83, 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTCACGCCAAGCCGCTTTTGA-3′; F-first-Pax5, 5′-TCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTGCAGGCCTTTGGATTTCTGCCA-3′; R-first-Pax5, 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTAGCCCCAACGATTCCTCCCCTTTA-3′; F-first-Apobec3, 5′-TCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTCAAAGGTTCAGGGAGTTGGGGGT-3′; R-first-Apobec3, 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTGGTGAAGGGAGACGGGTGAAT-3′; F-first-Ly6e, 5′-TCTACACTCTTTCCCTACACGACGCTCTTCCGATCTCGACTCCACTTGTCTGCTTGG-3′; R-first-Ly6e, 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGGTTGTTCAGGTTACCCCCG; F-first-Il4ra, 5′-TCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTGTGGGAGAACACTTTGGCAGG-3′; R-first-Il4ra, 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTCAAAGCAAGGAATTCAGGGGC-3′; F-first-Cd3e, 5′-TCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTCATGCTTCTGAGGCAGCTCTTGG-3′; and R-first-Cd3e, 5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTACCAAGCAAACCTCGACACCCA-3′. DNA was extracted and purified from the first-round PCR reaction by NucleoSpin Gel and PCR Clean-up kit (Takara Bio Inc.), and half of the samples were used as template for the second round of PCR. For the second round of PCR, phusion polymerase was used and reaction conditions were as follows: 98°C for 2 min, 15 cycles of 98°C for 15 s, 65°C for 30 s, and 72°C for 30 s. Second round primers were as follows: F-second-Common, 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGAC-3′; R-second-Index, 5′-CAAGCAGAAGACGGCATACGAGATNNNNNNGTGACTGGAGTTCAGACGTGTG-3′. Different index sequences (6 nt) on the primers were used to multiplex different samples. Finally, gel-extracted bands were pooled together and sequenced from both ends by Mi-Seq 300 × 2 (Illumina). Only mutations present on both paired reads were considered authentic mutations. After removing all undetermined nucleotides, the sequencing coverage for 5′-Sμ and Cμ regions is always >104 for each nucleotide position (Fig. S3 E).
Retroviral infection and Ig switching
Primers for cloning AID-EGFP into the pMX retroviral vector: F-BamHI-kozak-AID, 5′-GGGGGATCCGCCACCATGGACAGCCTTCTGATGAAGCA-3′; and R-NotI-TAA-EGFP, 5′-TTTGCGGCCGCTCATTACTTGTACAGCTCGTCCATGCCG-3′. Retroviral supernatants were produced as reported (Wang et al., 2014) and harvested 72 h after co-transfection of BOSC23 cells with pCL-Eco and pMX-EGFP-based plasmids. For the B cell infection, retroviral supernatants were filtered, supplemented with 5 µg/ml polybrene (Sigma-Aldrich), and then used to replace the culture medium at 20 and 44 h after B cell isolation. B cells were then spinoculated at 1,150 g for 1.5 h, followed by 4 h recovery at 37°C. Supernatants were then replaced by the original supplemented RPMI medium. At 96 h, B cells were collected for flow cytometry analysis. To measure the rate of Ig switching, live B cells were stained with APC-conjugated rat anti–mouse IgG1 antibody (BD) on ice for 20 min. The percentage of IgG1-switched (APC+) cells was determined by flow cytometry on an LSRFortessa (BD).
Quantitative real-time PCR
B cells from the various genotypes were collected at days 2, 3, and 4 from the beginning of the culture. Total RNA was extracted using RNeasy plus mini kit (QIAGEN). Subsequently, 1 µg of total RNA was used to synthesize the cDNA by Superscript III Reverse transcription (Thermo Fisher Scientific) according to the manufacturer’s instruction. Quantitative real-time PCR was performed using SYBR Green qPCR Master Mix (Agilent Technologies) on QuantStudio 6 Flex system (Thermo Fisher Scientific). The relative mRNA levels of AID were normalized to actin and calculated by the 2–ΔΔCt method. The sequence of the primers used for this analysis: F-qPCR-Aicda, 5′-CCTCCTGCTCACTGGACTTC-3′; R-qPCR-Aicda, 5′-GAAATGCATCTCGCAAGTCA-3′; F-qPCR-Actb, 5′-CTTTGCAGCTCCTTCGTTGC-3′; and R-qPCR-Actb, 5′-ACGATGGAGGGGAATACAGC-3′. The AID-ER fusion protein was undetectable by Western blot using three different anti-AID antibodies (Fig. S3 F). The antibodies were mAID-2 mouse monoclonal (eBioscience), and two rabbit polyclonal antibodies (generated and purified in house).
Sorting early G1 cells for GRO-Seq
Collection of live B cells in early G1. Splenic B cells were purified from Fucci-EGFP+ mouse spleens and activated in vitro. Nocodazole (100 ng/ml; Sigma-Aldrich) was added at 42 h after isolation. After 6 h, synchronized cells were collected and sorted at 4°C with a FACSAria (BD). Sorted EGFP+ cells (mostly G2/M) were put back into culture in prewarmed supplemented RPMI medium (no nocodazole). After 1.5 h at 37°C, cells were collected for sorting of the small EGFP− population (mostly early G1). At each step, an aliquot of cells was collected, fixed with 4.2% formaldehyde at room temperature for 20 min, and stained with DAPI (0.5 µg/ml) for quality control analysis. GRO-Seq. Global run-on and, after library preparation for sequencing, were done as described with minor modifications (Kaikkonen et al., 2013). GRO-Seq libraries were prepared with two biological replicates for each condition. In brief, nuclei were extracted from ∼20 million sorted live B cells in early G1 or nonsynchronized cells. After run-on reaction in the presence of BrUTP, the RNA was extracted with TRIzol LS Reagent (Thermo Fisher Scientific). RNA was treated with TURBO DNase (Ambion), fragmented using RNA Fragmentation Reagents (Ambion), and purified by running through a P-30 column (Bio-Rad Laboratories). Fragmented RNA was dephosphorylated with PNK (New England Biolabs), followed by heat inactivation. Dephosphorylation reactions were purified and precipitated using anti-BrUTP beads (Santa Cruz Biotechnology, Inc.). Poly(A)-tailing and cDNA synthesis was performed. The cDNA fragments were purified on a denaturing Novex 10% polyacrylamide TBE-urea gel (Thermo Fisher Scientific). The recovered cDNA was circularized, linearized, and amplified for 10–14 cycles. The final product was run on a Novex 10% TBE gel, purified, and cleaned-up using ChIP DNA Clean & Concentrator kit (Zymo Research Corporation). Finally, the libraries were sequenced with Next-Seq Sequencing System (Illumina).
Data analysis
Sequence alignment and convergent transcription analysis were conducted as previously described (Meng et al., 2014). We aligned the short-reads from GRO-Seq libraries to the mouse genome build mm9/NCBI37 using Bowtie2 software. We applied Homer software to de novo identify transcripts by detecting regions of continuous GRO-Seq signal along each strand of each chromosome. The minimum length of predicted transcripts was 150 bp. Antisense/sense-overlapping regions >100 base pairs were classified as convergent transcription. GRO-Seq profiles were displayed with IGV software.
Accession no.
GRO-Seq data were deposited to the Gene Expression Omnibus (accession no. GSE89464).
Online supplemental material
Fig. S1 shows that the localization of AID-EGFP shifts from the nucleus to the cytoplasm shortly after cell division, and that AID is slightly enriched in the nucleus of early G1 B cells. Fig. S2 shows that the increase of AID-induced uracils detected by MutPE-Seq is restricted to early G1, regardless of UNG. Fig. S3 provides additional information on GRO-Seq and AID-ER. Videos 1–5 show the time-lapse imaging of AID-EGFP during cell division.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the laboratory for discussions and suggestions, Dr. Xiaolei Su for comments on the manuscript, Dr. Yige Guo for advice with nocodazole experiments, Dr. Weiran Feng for discussion, Dr. Alexander Gitlin for help with Fucci-GFP+ mice and Dr. Zhou Du and Dr. Fred Alt for providing the algorithm for calculating convergent transcription. A particular thank you goes to Neena Thomas and Klara Velinzon for FACS sorting and to Tom Eisenreich for help managing the mouse colony.
This work was supported in part by National Institutes of Health grants AI112602 (to D.F. Robbiani), and AI037526 and AI072529 (to M.C. Nussenzweig). M.C. Nussenzweig is an HHMI Investigator.
The authors declare no competing financial interests.
Author contributions: Q. Wang planned and performed experiments and wrote the manuscript. K.-R. Kieffer-Kwon and R. Casellas planned and performed GRO-Seq experiments. T.Y. Oliveira, J. Pai, and M. Dose performed bioinformatic analysis. K. Yao, C.T. Mayer, and Z. Cao performed experiments. D.F. Robbiani, M. Jankovic, and M.C. Nussenzweig planned experiments and wrote the manuscript.
Footnotes
Abbreviations used:
- AID
- activation-induced cytidine deaminase
- Cμ
- constant μ region
- CSR
- class switch recombination
- ER
- estrogen receptor ligand-binding domain
- GRO-Seq
- global run-on sequencing
- MutPE-Seq
- mutational analysis by paired-end deep sequencing
- qPCR
- quantitative PCR
- SHM
- somatic hypermutation
- Sμ
- switch μ region
- ssDNA
- single-stranded DNA
References
- Alt F.W., Zhang Y., Meng F.L., Guo C., and Schwer B.. 2013. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 152:417–429. 10.1016/j.cell.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D.J., and Hetzer M.W.. 2007. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat. Cell Biol. 9:1160–1166. 10.1038/ncb1636 [DOI] [PubMed] [Google Scholar]
- Casellas R., Basu U., Yewdell W.T., Chaudhuri J., Robbiani D.F., and Di Noia J.M.. 2016. Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity. Nat. Rev. Immunol. 16:164–176. 10.1038/nri.2016.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J., Tian M., Khuong C., Chua K., Pinaud E., and Alt F.W.. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 422:726–730. 10.1038/nature01574 [DOI] [PubMed] [Google Scholar]
- Crouch E.E., Li Z., Takizawa M., Fichtner-Feigl S., Gourzi P., Montaño C., Feigenbaum L., Wilson P., Janz S., Papavasiliou F.N., and Casellas R.. 2007. Regulation of AID expression in the immune response. J. Exp. Med. 204:1145–1156. 10.1084/jem.20061952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson S.K., Market E., Besmer E., and Papavasiliou F.N.. 2003. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197:1291–1296. 10.1084/jem.20030481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J.M., and Neuberger M.S.. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76:1–22. 10.1146/annurev.biochem.76.061705.090740 [DOI] [PubMed] [Google Scholar]
- Doi T., Kinoshita K., Ikegawa M., Muramatsu M., and Honjo T.. 2003. De novo protein synthesis is required for the activation-induced cytidine deaminase function in class-switch recombination. Proc. Natl. Acad. Sci. USA. 100:2634–2638. 10.1073/pnas.0437710100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres M., Biniszkiewicz D., Sobol R.W., Harms C., Ahmadi M., Lipski A., Katchanov J., Mergenthaler P., Dirnagl U., Wilson S.H., et al. 2004. Increased postischemic brain injury in mice deficient in uracil-DNA glycosylase. J. Clin. Invest. 113:1711–1721. 10.1172/JCI200420926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faili A., Aoufouchi S., Guéranger Q., Zober C., Léon A., Bertocci B., Weill J.C., and Reynaud C.A.. 2002. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat. Immunol. 3:815–821. 10.1038/ni826 [DOI] [PubMed] [Google Scholar]
- Gaillard H., García-Muse T., and Aguilera A.. 2015. Replication stress and cancer. Nat. Rev. Cancer. 15:276–289. 10.1038/nrc3916 [DOI] [PubMed] [Google Scholar]
- Gasnereau I., Ganier O., Bourgain F., de Gramont A., Gendron M.C., and Sobczak-Thépot J.. 2007. Flow cytometry to sort mammalian cells in cytokinesis. Cytometry A. 71:1–7. 10.1002/cyto.a.20352 [DOI] [PubMed] [Google Scholar]
- Hagen L., Kavli B., Sousa M.M., Torseth K., Liabakk N.B., Sundheim O., Pena-Diaz J., Otterlei M., Hørning O., Jensen O.N., et al. 2008. Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J. 27:51–61. 10.1038/sj.emboj.7601958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haug T., Skorpen F., Aas P.A., Malm V., Skjelbred C., and Krokan H.E.. 1998. Regulation of expression of nuclear and mitochondrial forms of human uracil-DNA glycosylase. Nucleic Acids Res. 26:1449–1457. 10.1093/nar/26.6.1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth A., and Vértessy B.G.. 2010. A one-step method for quantitative determination of uracil in DNA by real-time PCR. Nucleic Acids Res. 38:e196 10.1093/nar/gkq815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen M.U., Spann N.J., Heinz S., Romanoski C.E., Allison K.A., Stender J.D., Chun H.B., Tough D.F., Prinjha R.K., Benner C., and Glass C.K.. 2013. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell. 51:310–325. 10.1016/j.molcel.2013.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey L., Demorest Z.L., Land A.M., Hultquist J.F., Brown W.L., and Harris R.S.. 2012. APOBEC3B and AID have similar nuclear import mechanisms. J. Mol. Biol. 419:301–314. 10.1016/j.jmb.2012.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey L., Law E.K., Brown W.L., and Harris R.S.. 2013. Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle. 12:762–772. 10.4161/cc.23713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Q., and Maizels N.. 2015. Cell Cycle Regulates Nuclear Stability of AID and Determines the Cellular Response to AID. PLoS Genet. 11:e1005411 10.1371/journal.pgen.1005411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride K.M., Barreto V., Ramiro A.R., Stavropoulos P., and Nussenzweig M.C.. 2004. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J. Exp. Med. 199:1235–1244. 10.1084/jem.20040373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F.L., Du Z., Federation A., Hu J., Wang Q., Kieffer-Kwon K.R., Meyers R.M., Amor C., Wasserman C.R., Neuberg D., et al. 2014. Convergent transcription at intragenic super-enhancers targets AID-initiated genomic instability. Cell. 159:1538–1548. 10.1016/j.cell.2014.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot S.P., Litzler L.C., Trajtenberg F., Zahn A., Robert F., Pelletier J., Buschiazzo A., Magor B.G., and Di Noia J.M.. 2015. Consecutive interactions with HSP90 and eEF1A underlie a functional maturation and storage pathway of AID in the cytoplasm. J. Exp. Med. 212:581–596. 10.1084/jem.20141157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., and Honjo T.. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. 10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Patenaude A.M., Orthwein A., Hu Y., Campo V.A., Kavli B., Buschiazzo A., and Di Noia J.M.. 2009. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat. Struct. Mol. Biol. 16:517–527. 10.1038/nsmb.1598 [DOI] [PubMed] [Google Scholar]
- Pefanis E., Wang J., Rothschild G., Lim J., Chao J., Rabadan R., Economides A.N., and Basu U.. 2014. Noncoding RNA transcription targets AID to divergently transcribed loci in B cells. Nature. 514:389–393. 10.1038/nature13580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen S., Casellas R., Reina-San-Martin B., Chen H.T., Difilippantonio M.J., Wilson P.C., Hanitsch L., Celeste A., Muramatsu M., Pilch D.R., et al. 2001. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 414:660–665. 10.1038/414660a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J., Wang Q., Dose M., Pruett N., Kieffer-Kwon K.-R., Resch W., Liang G., Tang Z., Mathé E., Benner C., et al. 2014. B cell super-enhancers recruit AID tumorigenic activity. Cell. 159:1524–1537. 10.1016/j.cell.2014.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., and Nussenzweig M.C.. 2013. Chromosome translocation, B cell lymphoma, and activation-induced cytidine deaminase. Annu. Rev. Pathol. 8:79–103. 10.1146/annurev-pathol-020712-164004 [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., Deroubaix S., Feldhahn N., Oliveira T.Y., Callen E., Wang Q., Jankovic M., Silva I.T., Rommel P.C., Bosque D., et al. 2015. Plasmodium infection promotes genomic instability and AID-dependent B cell lymphoma. Cell. 162:727–737. 10.1016/j.cell.2015.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H., et al. 2008. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 132:487–498. 10.1016/j.cell.2007.12.033 [DOI] [PubMed] [Google Scholar]
- Santaguida S., and Amon A.. 2015. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 16:473–485. 10.1038/nrm4025 [DOI] [PubMed] [Google Scholar]
- Victora G.D., Schwickert T.A., Fooksman D.R., Kamphorst A.O., Meyer-Hermann M., Dustin M.L., and Nussenzweig M.C.. 2010. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 143:592–605. 10.1016/j.cell.2010.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Oliveira T., Jankovic M., Silva I.T., Hakim O., Yao K., Gazumyan A., Mayer C.T., Pavri R., Casellas R., et al. 2014. Epigenetic targeting of activation-induced cytidine deaminase. Proc. Natl. Acad. Sci. USA. 111:18667–18672. 10.1073/pnas.1420575111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeap L.S., Hwang J.K., Du Z., Meyers R.M., Meng F.L., Jakubauskaitė A., Liu M., Mani V., Neuberg D., Kepler T.B., et al. 2015. Sequence-Intrinsic Mechanisms that Target AID Mutational Outcomes on Antibody Genes. Cell. 163:1124–1137. 10.1016/j.cell.2015.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Chedin F., Hsieh C.L., Wilson T.E., and Lieber M.R.. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4:442–451. 10.1038/ni919 [DOI] [PubMed] [Google Scholar]
- Zheng S., Vuong B.Q., Vaidyanathan B., Lin J.Y., Huang F.T., and Chaudhuri J.. 2015. Non-coding RNA Generated following Lariat Debranching Mediates Targeting of AID to DNA. Cell. 161:762–773. 10.1016/j.cell.2015.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.