Significance
New discoveries have been continuously made in recent years on the roles of noncoding RNAs in regulating biological processes. Phased small-interfering RNAs (phasiRNAs) may be the newest member discovered in recent years. The photoperiod-sensitive male sterility (PSMS) rice is a very valuable germplasm that started the era of two-line hybrid rice. Here we show that phasiRNAs generated by a long-noncoding RNA PMS1T encoded by the Pms1 locus regulates PSMS in rice. This work provides a case associating the phasiRNAs with a biological trait, especially an agriculturally highly important trait, thus confirming that the phasiRNAs indeed have biological functions.
Keywords: photoperiod-sensitive male sterility, long-noncoding RNA, phasiRNA
Abstract
Phased small-interfering RNAs (phasiRNAs) are a special class of small RNAs, which are generated in 21- or 24-nt intervals from transcripts of precursor RNAs. Although phasiRNAs have been found in a range of organisms, their biological functions in plants have yet to be uncovered. Here we show that phasiRNAs generated by the photopheriod-sensetive genic male sterility 1 (Pms1) locus were associated with photoperiod-sensitive male sterility (PSMS) in rice, a germplasm that started the two-line hybrid rice breeding. The Pms1 locus encodes a long-noncoding RNA PMS1T that was preferentially expressed in young panicles. PMS1T was targeted by miR2118 to produce 21-nt phasiRNAs that preferentially accumulated in the PSMS line under long-day conditions. A single nucleotide polymorphism in PMS1T nearby the miR2118 recognition site was critical for fertility change, likely leading to differential accumulation of the phasiRNAs. This result suggested possible roles of phasiRNAs in reproductive development of rice, demonstrating the potential importance of this RNA class as regulators in biological processes.
Phased small-interfering RNAs (phasiRNAs) are a special class of small RNAs, which are generated in 21- or 24-nt intervals from transcripts of precursor RNAs (1, 2). PhasiRNAs have been found in a wide range of organisms (1–11). In animal, a class of phasiRNAs known as Zucchini-dependent piwi-interacting RNAs were required for spermatogenesis (3, 4). In some plant species, phasiRNA-generating loci (PHAS loci) are found to be distributed in genomic clusters and phasiRNAs preferentially accumulate in reproductive tissues (2, 10). In grass anthers, phasiRNAs are generated from precursor transcripts transcribed from PHAS genes or loci, triggered by cleavage of 22-nt miR2118 or miR2275 (1, 2). The 21-nt phasiRNAs are abundant in premeiotic anthers, and 24-nt phasiRNAs in meiotic anthers (10). However, the functions of these reproductive phasiRNAs are yet to be characterized.
The discovery of a photoperiod-sensitive male sterility (PSMS) rice (Oryza sativa L.) mutant started the development of two-line hybrid rice (12), which has made tremendous contributions to rice production, especially in China (13). Male fertility in PSMS rice is regulated by photoperiod: it is male-sterile under long-day conditions but fertile under short-day conditions. Studies have shown that PSMS segregates as two Mendelian loci, photopheriod-sensetive genic male sterility 1 (Pms1) and Pms3, in crosses between the original mutant Nongken 58S (abbreviated as 58S) and many rice varieties (14, 15). Recently, the Pms3 locus was shown to encode a long-noncoding RNA (lncRNA), LDMAR (16, 17), whose abundance is critical for male fertility in long days.
Here we show that Pms1 functions as a PHAS locus, which produces a transcript PMS1T targeted and triggered by miR2118 to generate 21-nt phasiRNAs. The abundance of the phasiRNAs was associated with the male fertility of 58S under long-day conditions.
Results
Map-Based Cloning of Pms1.
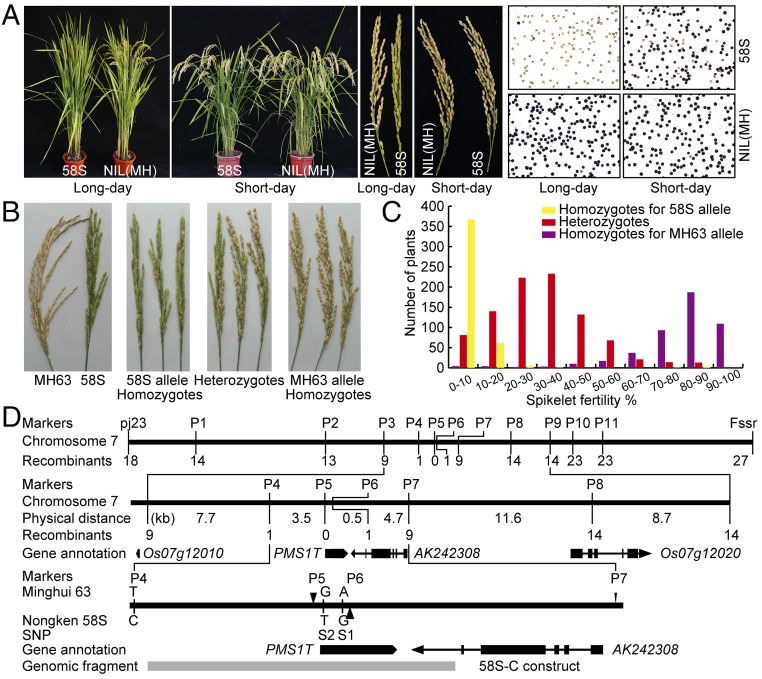
It was previously shown that fertility of PSMS rice under long-day conditions was governed by two independent segregating loci, Pms1 and Pms3, such that the pollen was completely sterile only when both loci were homozygous for the alleles from 58S (15). In this study, we developed an F2 population from a cross between 58S and NIL(MH) [near isogenic line developed by introgressing the Pms1 locus from Minghui 63 (MH63) to 58S background]. Plants homozygous for the 58S allele were highly sterile and plants homozygous for the MH63 allele were fertile under long-day conditions, whereas spikelet fertility of the heterozygotes varied from completely sterile to fully fertile (Fig. 1 A–C).
Fig. 1.
Genetic mapping of Pms1. (A) Fertility phenotypes of 58S and NIL(MH) under long- and short-day conditions. Whole plants are shown (Left), panicles (Center), and pollen (Right). (B and C) Spikelet fertility of plants (B) and their frequency distribution (C) in the F2 population from a cross between 58S and NIL(MH) under long-day conditions. The plants were genotyped using marker Fssr. (D) Mapping of Pms1 to a 4.0-kb region on chromosome 7 according to the MH63 genome sequence. The black blocks in the middle represent exons, and the lines between them represent introns, arrows indicate the directions of genes. Sequence polymorphisms between two parents MH63 and 58S are shown below, where vertical lines represent the single nucleotide polymorphisms, and triangles for insertions. The gray block at the bottom indicates the fragment amplified from 58S for preparing the construct 58S-C.
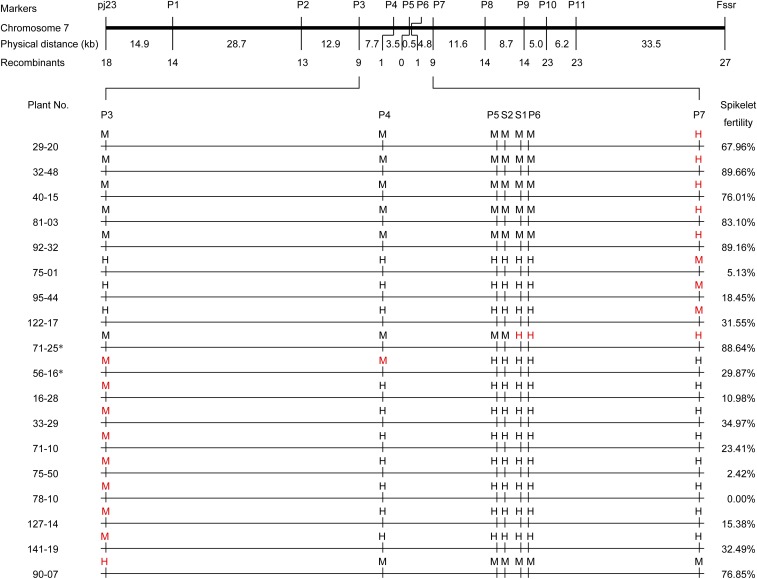
For fine-mapping Pms1, more molecular markers were developed surrounding the two markers Fssr and Rssr (now P5 in Fig. 1D) based on the previous work (18) to genotype 6,841 individuals from the F2-mapping population. Genotyping these plants identified 88 recombinants between two flanking markers, Fssr and pj23, covering a 138-kb genomic region in MH63 that corresponded to 300 kb in the Nipponbare reference genome (19). Because of extensive phenotypic overlap between heterozygotes and 58S homozygotes, only the recombinants between MH63 homozygotes and heterozygotes were used for further mapping. Genotyping the recombinants resolved the Pms1 locus to a 4.0-kb region flanked by markers P4 and P6, with one recombinant at each side (Fig. 1D), with the SSR marker P5 cosegregating with Pms1. A sequence comparison of the region bracketed by P4 and P7 between MH63 and 58S revealed two SNPs, S1 and S2, and a 65-bp InDel P6, in addition to P5 (Fig. 1D). One recombination event between SNPs S1 and S2 refined the Pms1 locus to a 3.8-kb region, whereas the S2 SNP with guanine (G) in MH63 and thymine (T) in 58S still cosegregated with Pms1 (Fig. 1D and Fig. S1).
Fig. S1.
Genotypes and fertility of the recombinants between markers P3 and P7. H, heterozygote; M, homozygous for MH63 genotype; red color indicates the recombination sites. Plant No. is the identification number for recombinants in the population; their spikelet fertility is shown on the right. An asterisk represents the most interesting recombinants.
There was no predicted gene within this 3.8-kb region. The nearest predicted genes, LOC_07g12010 and LOC_07g12020, were ∼7.7- and 15.0-kb apart, respectively, from P5 according to the MH63 genomic sequence (Fig. 1D), and was even longer by the Nipponbare genome sequence. Both genes were annotated as a putative unclassified retrotransposon protein. A full-length cDNA AK242308 (cdna01.dna.affrc.go.jp/cDNA/) on the antisense chain was 1,139-bp apart from P6 (Fig. 1D). These data suggested the possibility that Pms1 encodes a previously unidentified genetic element.
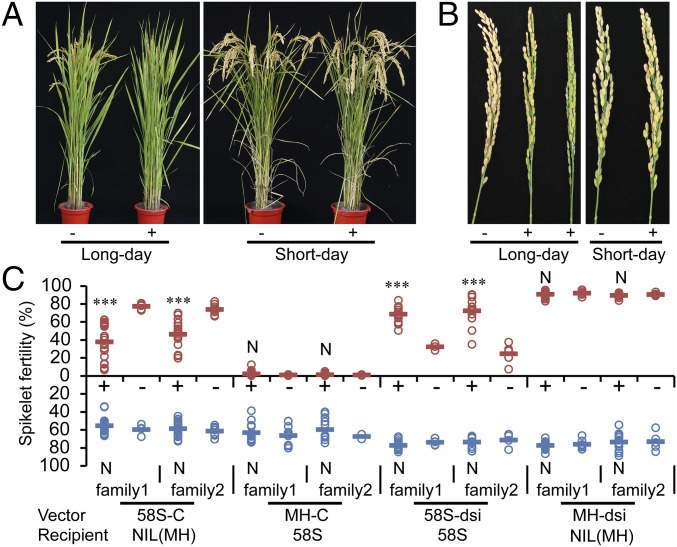
We transformed a construct (58S-C) containing a 5.6-kb genomic fragment from 58S into NIL(MH) (Fig. 1D) . Reciprocally, a construct (MH-C) carrying the homologous fragment from MH63 was introduced into 58S. Spikelet fertility of the transgene-positive plants in the 58S-C T1 families was significantly lower under long-day conditions than the negative segregants, but was not different under short-day conditions (Fig. 2). Analysis of the MH-C T1 families showed that the transgene had no effect on fertility under both long- and short-day conditions (Fig. 2C). These results confirmed that the transformed genomic fragment indeed contained the Pms1 locus, and also showed that long-day male sterility by Pms1 is semidominant rather than completely recessive, as previously assumed (14). We thus designated the allele from 58S as Pms1 and the one from MH63 as pms1.
Fig. 2.
Fertility of T1 transgenic plants under long- and short-day conditions. (A) Whole plants of transgenic segregants from the T1 family of 58S-C under long and short days. (B) Panicles of transgenic plants of 58S-C. +, positive plants; −, negative plants. (C) Spikelet fertility of T1 transgenic plants from various vectors under long- and short-day conditions. The bars indicate the means. ***P < 0.001 significant difference by t test between the positive and negative plants in each family; N indicates nonsignificant difference (P > 0.05).
The Candidate Transcript of Pms1.
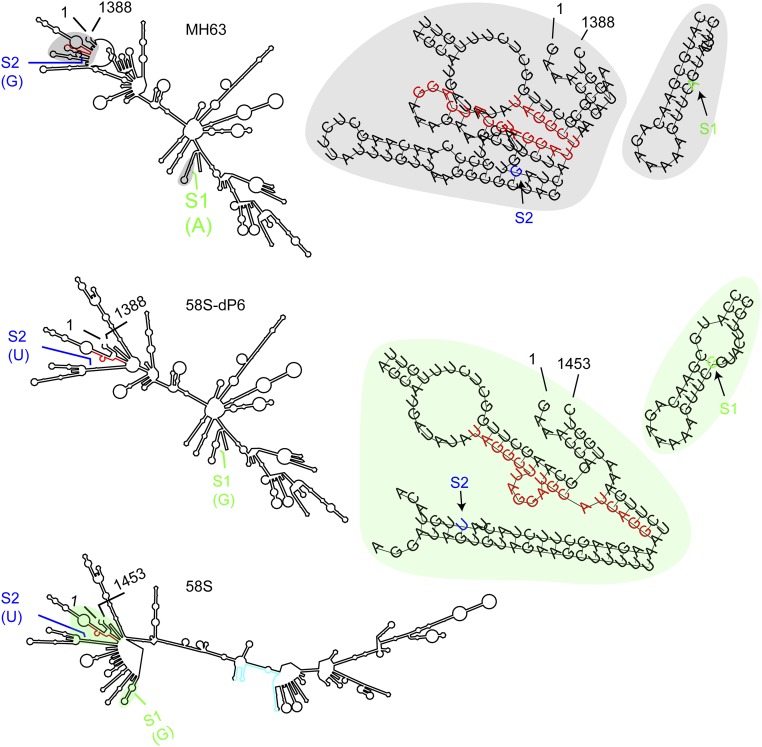
We identified from the sense strand within the transformed fragment a transcript (referred to as PMS1T) using 5′ RACE and 3′ RACE (Fig. 1D). This transcript had no intron, and the one from 58S was 1,453 nt, which was 65-nt longer than the one from MH63 (1,388 bp). This difference provided an InDel marker P6. The two SNPs, S1 and S2, were both located at the 5′ terminus of PMS1T (Fig. 1D and Fig. S2).
Fig. S2.
Comparison of the genomic sequences of MH63 and 58S that encode PMS1T. SNP S2 is in red and SNP S1 in blue. Vertical arrow indicates the cleavage site of miR2118. Predicted ORFs are boxed with colored lines: ORF1 is in red box, ORF2 in MH63 is in pink box, ORF2 in 58S is in yellow box, and ORF3 is in blue box overlapping with ORF2 in MH63. Sequences after 800 bp in MH63 are omitted. An asterisk indicates identical nucleotides between 58S and MH63.
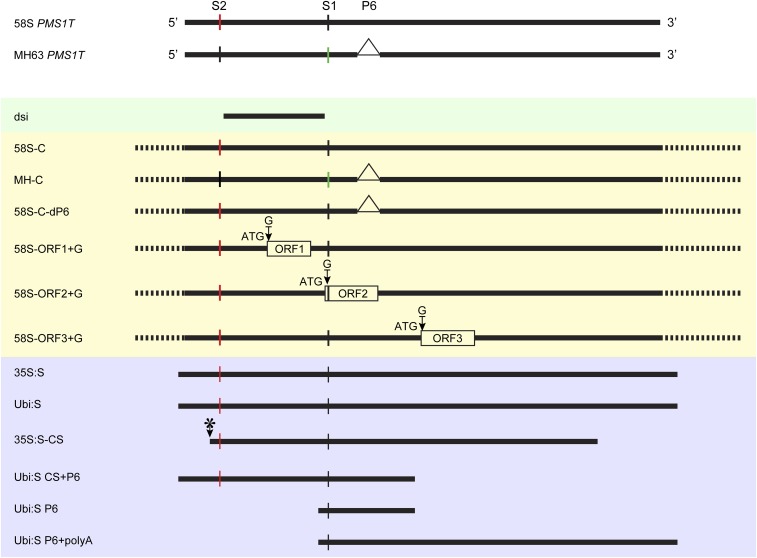
To investigate the effect of PMS1T on fertility, an RNAi construct (“dsi”) was transformed into 58S (58S-dsi) and NIL(MH) (MH-dsi), respectively, to knock down the expression of this transcript (see, for example, Fig. S4). Under long days, the spikelet fertility of 58S-dsi transgene-positive plants increased significantly versus negative segregants, but no difference was observed in short days (Fig. 2C and Fig. S3A). The spikelet fertility of MH-dsi transgenic plants did not change regardless of the day length (Fig. 2C). In contrast, overexpression of full-length PMS1T from 58S driven by the maize ubiquitin promoter (Ubi:S) in NIL(MH) greatly reduced fertility under long days (Table S1). This result indicates that PMS1T is the functional form of Pms1.
Fig. S4.
Schematic representation of constructs used for transformation. Constructs used for gene silencing by RNAi are in the green-shaded box, those for complementation tests are in the yellow-shaded box, and ones for overexpression are in the blue-shaded box. Short vertical lines represent the SNP: red, T; black, G; green, A. Triangle marks the 65-bp deletion at P6 marker in MH63. Dotted lines indicate omitted genome sequences. Arrow with asterisk indicates the cleavage site of miR2118.
Fig. S3.
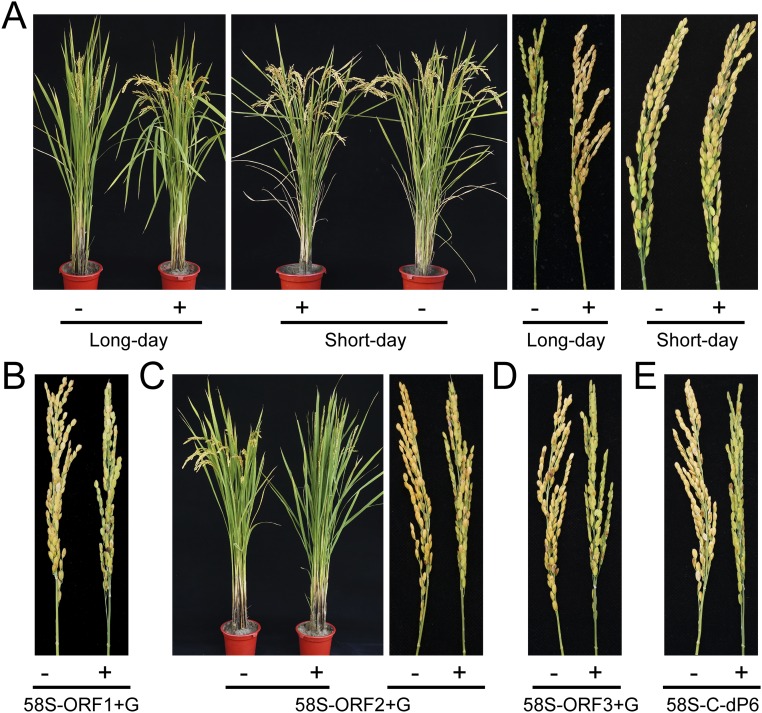
Phenotypes of T1 transgenic plants under long-day and short-day conditions. (A) T1 transgenic plants of 58S-dsi under long- and short-day conditions. +, positive plants; −, negative plants. (B–E) T1 transgenic plants of 58S-ORF1+G (B), 58S-ORF2+G (C), 58S-ORF3+G (D), and 58S-C-dP6 (E) under long-day conditions.
Table S1.
Spikelet fertility of T1 families from two representative T0 transgenic plants under long-day conditions
| Construct | Genotype | Spikelet fertility, % | |
| Family 1 | Family 2 | ||
| Ubi:S | Positive | 0.39 ± 0.13 (24) | 0.19 ± 0.07 (29) |
| Negative | 73.34 ± 1.67 (16) | 68.46 ± 2.54 (9) | |
| P value | 0.0000 | 0.0000 | |
| 35S:S | Positive | 14.72 ± 2.90 (27) | 35.51 ± 4.89 (15) |
| Negative | 66.82 ± 3.58 (8) | 66.18 ± 1.14 (3) | |
| P value | 0.0000 | 0.0146 | |
| 35S:S-CS | Positive | 68.15 ± 1.33 (18) | 71.52 ± 1.28 (20) |
| Negative | 65.51 ± 1.83 (8) | 71.77 ± 4.14 (5) | |
| P value | 0.2706 | 0.9396 | |
| Ubi:S CS+P6 | Positive | 8.94 ± 0.76 (33) | 0.18 ± 0.06 (20) |
| Negative | 74.41 ± 3.44 (13) | 70.57 ± 1.83 (16) | |
| P value | 0.0000 | 0.0000 | |
| Ubi:S P6 | Positive | 73.72 ± 1.58 (22) | 69.51 ± 2.66 (26) |
| Negative | 77.70 ± 3.98 (7) | 73.15 ± 1.84 (4) | |
| P value | 0.2736 | 0.6024 | |
| Ubi:S P6+polyA | Positive | 72.93 ± 1.40 (20) | 80.94 ± 1.31 (17) |
| Negative | 73.25 ± 1.40 (9) | 82.58 ± 1.44 (12) | |
| P value | 0.8890 | 0.4128 | |
| 58S-C-dP6 | Positive | 0.34 ± 0.10 (21) | 21.14 ± 4.31 (33) |
| Negative | 62.99 ± 6.05 (8) | 73.44 ± 1.28 (12) | |
| P value | 0.0000 | 0.0000 | |
| 58S-ORF1+G | Positive | 1.35 ± 0.30 (33) | 21.30 ± 5.38 (29) |
| Negative | 64.65 ± 5.70 (9) | 67.92 ± 2.42 (12) | |
| P value | 0.0000 | 0.0000 | |
| 58S-ORF2+G | Positive | 1.34 ± 0.22 (33) | 32.93 ± 4.06 (30) |
| Negative | 80.43 ± 1.04 (16) | 79.74 ± 1.05 (6) | |
| P value | 0.0000 | 0.0000 | |
| 58S-ORF3+G | Positive | 3.36 ± 2.40 (28) | 1.50 ± 0.49 (27) |
| Negative | 79.90 ± 3.75 (7) | 79.90 ± 2.59 (8) | |
| P value | 0.0000 | 0.0000 | |
All data presented are mean ± SEM. Numbers in parentheses are numbers of plants analyzed. The P value is obtained from the t test of positive and negative plants. All constructs were transformed into NIL(MH).
Pms1 Encodes a lncRNA.
PMS1T was predicted to have three short ORFs (Fig. S2). All had no homology to any known proteins. 58S and MH63 shared the same sequences for ORF1 and ORF3, predicted to encode 44 amino acids and 51 amino acids, respectively. ORF2 was predicted to encode 101 amino acids in MH63 but only 54 amino acids in 58S because of an early stop codon caused by the 65-bp insertion at the P6 marker. To inquire whether PMS1T encodes any small peptides or if it is a lncRNA, we modified the construct 58S-C by inserting a guanine residue immediately downstream the ATG start codons of putative ORFs to disrupt the reading frames. These constructs, named 58S-ORF1+G, 58S-ORF2+G, and 58S-ORF3+G, were independently transformed into NIL(MH) (Fig. S4). Under long-day conditions, all transgene-positive individuals showed decreased spikelet fertility, similar to 58S-C (Fig. S3 B–D and Table S1). Thus, altering the putative ORFs did not affect the function of 58S Pms1, suggesting that the PMS1T functions as an RNA rather than protein, regarded as a lncRNA.
The SNP S2 Is Crucial for PSMS.
There were three polymorphic sites between 58S and MH63 in the genomic sequences that transcribe PMS1T: the 65-bp InDel at P6 and the two SNPs, S2 and S1 (Fig. 1D). We compared sequences of 15 different rice lines, including eight PSMS lines bred from 58S, at these sites as well as the SSR marker P5 (Table S2). Lunhui 422 and 1514 were confirmed to contain the fertile allele at the Pms1 locus by genetic analysis (15), whereas Nongken 58 shared same genotype with 58S (20). Although the P5 marker cosegregated with Pms1 in the mapping population, it had varied numbers of AT repeats among these lines, thus is an unlikely candidate for PSMS (Table S2). A recombination event between SNP S1 and S2 ruled out the possibility for S1 to be the causal polymorphism (Fig. S1), narrowing the potential causal polymorphism to S2.
Table S2.
Comparative sequencing results of 15 rice lines in the Pms1 region
| Variety* | P5 | S2 | S1 | P6† (bp) | Sub species |
| Nongken 58S | (AT)14 | T | G | 65 | japonica |
| Nongken 58 | (AT)14 | T | G | 65 | japonica |
| 1514 | (AT)8 | G | A | — | japonica |
| Nipponbare | (AT)8 | G | A | — | japonica |
| Lunhui 422 | (AT)9 | G | A | — | japonica |
| Minghui 63 | (AT)40 | G | A | — | indica |
| 9311 | (AT)9 | G | A | — | indica |
| Peiai 64S | (AT)14 | T | G | 65 | indica |
| 32001S | (AT)14 | T | G | 65 | indica |
| HN5S | (AT)14 | T | G | 65 | indica |
| 95076S | (AT)14 | T | G | 65 | japonica |
| N5088S | (AT)14 | T | G | 65 | japonica |
| 7001S | (AT)14 | T | G | 65 | japonica |
| 29130S | (AT)14 | T | G | 65 | japonica |
| 31301S | (AT)14 | T | G | 65 | japonica |
All of the male sterile lines were bred from 58S.
Suffix letter “S” indicates PSMS line.
Dash represents the deletion of 65-bp at P6.
No similar sequence was found from other species, except rice using PMS1T as the query. The 65-bp inserted sequence, surrounded by sequences similar to those encoding transposon proteins, had high similarity to many loci throughout the genome. We suspected that the 65-bp insertion might be related to the transposon (21).
To investigate whether the 65-bp was responsible for fertility change, four constructs were independently transformed into NIL(MH): 58S-C-dP6 was obtained by deleting the 65-bp insertion of 58S-C; Ubi:S CS+P6, Ubi:S P6, and Ubi:S P6+polyA were designed to overexpress parts of PMS1T (Fig. S4). Ubi:S P6 contained a 288-bp fragment from 58S-C including the 65-bp sequence, whereas Ubi:S CS+P6 and Ubi:S P6+polyA included the same 288-bp fragment but extended to the 5′ terminus and 3′ terminus, respectively. Under long-day conditions, 58S-C-dP6 transgenic plants showed greatly reduced spikelet fertility (Fig. S3E), just like 58S-C. In contrast, there was no significant fertility difference between the transgene-negative and -positive plants of the constructs Ubi:S P6 and Ubi:S P6+polyA (Table S1). These results suggested that the 65-bp insertion in 58S might not be relevant to fertility difference. The T1-positive transgenic plants of Ubi:S CS+P6, producing a truncated transcript with a 696-bp sequence, showed as much fertility reduction as Ubi:S (Table S1), indicating that there was no important element for fertility change beyond this region, again supporting SNP S2 as the causal polymorphism.
Expression Pattern of PMS1T.
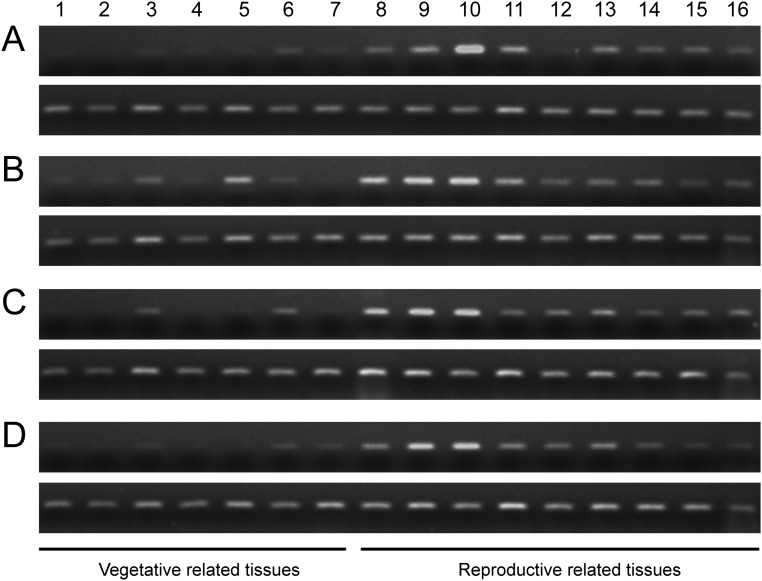
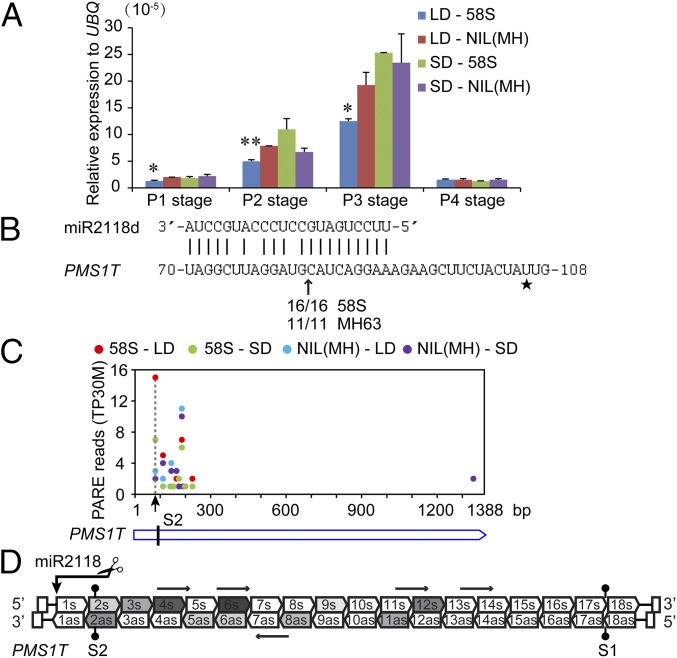
Sixteen different tissues from vegetative stage to reproductive stage in 58S and NIL(MH) grown under long- and short-day conditions were assayed for the expression pattern of PMS1T (Fig. S5). In general, PMS1T expression levels in both 58S and NIL(MH) were low in all tissues assayed, and especially so in green tissues, such as leaf and sheath (Fig. S5). It was expressed preferentially in young panicles and florets, with an expression peak when young panicles were ∼2 cm in length, corresponding to pollen mother cell formation stage (P3). This expression pattern corresponded well with previous results that the most critical period for PSMS occurred during the stages from P1 to P3 (22). Moreover, the transcript abundance was lower in 58S under long-day conditions than 58S in short days, and also than NIL(MH) under both long- and short-day conditions at these three stages (Fig. 3A).
Fig. S5.
Expression profiles of PMS1T. RT-PCR results of PMS1T in different tissues from 58S (A) and NIL(MH) (B) under long-day conditions, and 58S (C) and NIL(MH) (D) under short-day conditions. In each panel, the Upper row is the result from PMS1T, with 35 cycles of PCR amplification, and the Lower row is the reference gene Ubiquitin (LOC_Os03g13170) with 25 cycles. Tissues: 1, leaf at tillering stage; 2, leaf at young panicle stage (0.5–1.0 cm); 3, penultimate internode at young panicle stage (0.5–1.0 cm); 4, leaf sheath at young panicle stage (0.5–1.0 cm); 5, flag leaf; 6, penultimate internode at P7 stage (see description below); 7, sheath of flag leaf; 8, young panicle at secondary branch primordium differentiation stage (less than 0.5 cm, P1); 9, young panicle at pistil/stamen primordium differentiation stage (0.5–1.0 cm, P2); 10, young panicle at pollen mother cell formation stage (∼2.0 cm, P3); 11, floret at microsporocyte meiosis stage (3.0–4.0 mm in length, P4); 12, floret at tetrad stage of pollen mother cell meiosis (4.0–4.5 mm, P5); 13, floret at microspore stage (4.5–5.0 mm, P6); 14, floret at late microspore stage (5.0–6.0 mm, P7); 15, floret at bicellular pollen stage (6.0–7.0 mm, P8); 16, stamen at mature pollen stage.
Fig. 3.
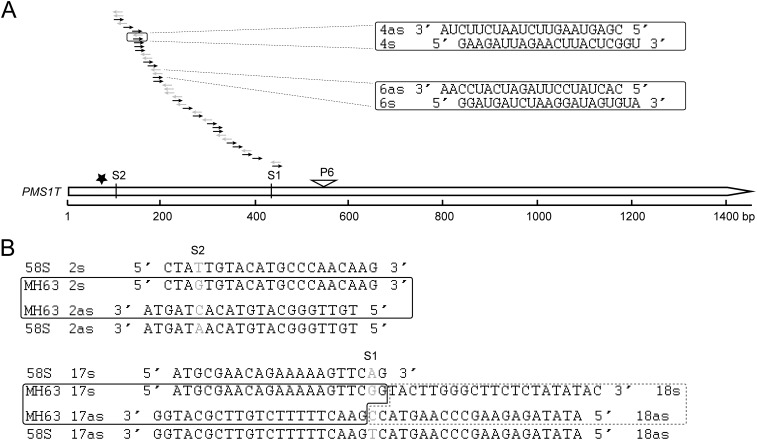
Targeting of PMS1T by miR2118 and production of phasiRNAs. (A) Relative abundance of PMS1T in 58S and NIL(MH) under long- and short-day conditions at the early stages of panicle development. The descriptions about different stages are as in the legend of Fig. S5. The expression levels are relative to the UBQ mRNA. Data are means ± SEM (n = 3). Statistically significant difference between LD-58S and LD-NIL(MH) by t tests at *P < 0.05 and **P < 0.01. (B) Validation by 5′ RLM-RACE that PMS1T is targeted by miR2118 using miR2118d as an example. The arrowhead indicates the cleavage site and sequence frequencies are shown below. Star marks the position of SNP S2. (C) PARE results from RNA degradome of PMS1T in 58S and NIL(MH) at P3 stage under long- and short-day conditions. Arrow indicates the cleavage sites directed by miR2118. The values of reads in each library are normalized to transcripts per 30 million (TP30M). (D) Schematic diagram of the 21-nt phasiRNAs generated from the PMS1T transcript in 58S at P3 stage under long-day conditions. The vertical arrow indicates the cleavage site directed by miR2118. Arrow-headed boxes show small RNAs in phase and horizontal arrows indicate ones that are not in phase. Intensity of gray color displays the abundance of 21-nt phasiRNA. as, phasiRNAs from antisense strand; s, phasiRNAs from sense strand.
PMS1T Is a Target of miR2118.
Many lncRNAs that are preferentially expressed in developing inflorescences in the grasses are typically targeted by either miR2118 or miR2275 (2, 23). PMS1T was predicted to be targeted by miR2118 (plantgrn.noble.org/psRNATarget/). We validated the cleavage site in PMS1T in both 58S and NIL(MH) by modified 5′ RLM-RACE (RNA ligase-mediated rapid amplification of cDNA ends), a site that mapped to the expected location between the 10th and 11th nucleotides from the 5′ end of miR2118, in the complementary region (24) (Fig. 3B), suggesting that PMS1T was a likely target of miR2118. This cleavage site was further confirmed using data of RNA degradome by parallel analysis of RNA ends (PARE) (Fig. 3C). The signature sequence of the highest abundance of PARE reads corresponded to the cleavage site, and the signal value from 58S under long days was higher than the others.
Mature miR2118 is 22 nt in length, and expressed exclusively in the immature inflorescence in rice (2, 25). Sequencing of small RNA libraries constructed using young panicle tissues from P2, P3, and P4 stages of 58S and NIL(MH) grown under long- and short-day conditions showed that miR2118 accumulated higher in P3 and P4 stages than in P2 (Fig. S6). The putative target sequence of PMS1T by miR2118 was identical between 58S and MH63. To learn whether cleavage triggered by miR2118 was essential for the function of PMS1T, we prepared construct 35S:S-CS containing the truncated PMS1T starting right from the miR2118 cleavage site, including only half of the target sequence (Fig. S4), and transformed it into NIL(MH). Another construct, 35S:S (having the same rice fragment as Ubi:S but driven by the 35S promoter), which would reduce the spikelet fertility of NIL(MH) under long-day conditions (Table S1), was used for comparison. Without the complete target sequence of miR2118, 35S:S-CS did not reduce spikelet fertility of NIL(MH), unlike the case of 35S:S or Ubi:S (Table S1). Thus, the 5′ region of PMS1T including the recognition site of miR2118 was necessary for causing male sterility by Pms1.
Fig. S6.
Abundance of miR2118 in panicles at three stages of young panicle development. Values are means of two biological replicates, and error bars represent SEM. Abundance of small RNAs in each library was normalized to TP10M based on the total count of genome-matched reads in that library. LD, long-day conditions; SD, short-day conditions.
PMS1T Produces 21-nt phasiRNAs Triggered by miR2118.
MiR2118 triggers 21-nt secondary siRNA biogenesis in plants from lncRNAs or protein coding genes (1, 2, 8, 9, 23, 26, 27). Sequencing of the small RNA libraries constructed from young panicles of 58S and NIL(MH) at P2, P3, and P4 stages under long- and short-day conditions revealed small RNAs of various lengths, from 18 nt to 30 nt within the PMS1T region, with 21-nt small RNAs being the most abundant, especially in 58S under long days (Table S3).
Table S3.
Abundance of small RNAs generated from the PMS1T region
| Length (nt) | Stage | 58S | NIL(MH) | OX-positive | OX–negative | dsi–positive | dsi–negative | ||
| LD | SD | LD | SD | LD | LD | LD | LD | ||
| 20 | P2 | 0 | 1 | 0 | 1 | ||||
| P3 | 5 | 4 | 3 | 0 | 16 | 3 | 280 | 1 | |
| P4 | 5 | 6 | 4 | 2 | |||||
| 21 | P2 | 19 | 162 | 47 | 33 | ||||
| P3 | 364 (357*) | 297 (293*) | 140 (137*) | 153 (153*) | 9,412 (678*) | 222 (218*) | 6,195 (189*) | 287 (281*) | |
| P4 | 169 | 125 | 68 | 56 | |||||
| 22 | P2 | 0 | 1 | 0 | 0 | ||||
| P3 | 0 | 3 | 2 | 0 | 300 | 2 | 7,658 | 8 | |
| P4 | 6 | 1 | 1 | 1 | |||||
| 23 | P2 | 0 | 1 | 0 | 0 | ||||
| P3 | 0 | 0 | 1 | 0 | 30 | 0 | 538 | 3 | |
| P4 | 1 | 0 | 0 | 1 | |||||
| 24 | P2 | 1 | 2 | 2 | 1 | ||||
| P3 | 2 | 3 | 2 | 1 | 81 | 4 | 2,682 | 13 | |
| P4 | 2 | 3 | 1 | 0 | |||||
All data are mean of two biological replicates. Abundances of small RNAs in each library were normalized to TP10M based on the total count of genome-matched reads in that library. dsi, 58S-dsi transgenic T1 plants; LD, long-day conditions; OX, Ubi:S transgenic T1 plants; SD, short-day conditions.
The numbers of 21-nt small RNAs in phase.
A cluster of 21-nt small RNAs was localized between the 82nd and 459th nucleotides of PMS1T, arranged in 18 phases or registers from both strands and beginning from the cleavage site of miR2118 (Fig. S7A). Additionally, a few nonphased small RNAs were also detected. Some of the sense and antisense phasiRNAs correspond to duplexes with 2-nt 3′-overhangs at both termini (Fig. 3D and Fig. S7A), consistent with processing by DICER-LIKE 4 (DCL4) and RNA-DEPENDENT RNA POLYMERASE 6 (RDR6) (23, 28).
Fig. S7.
The locations of 21-nt phasiRNAs generated from the PMS1T region. (A) Locations of 21-nt small RNAs produced from PMS1T. The data are generated from small RNA libraries of young panicles at P2, P3, and P4 stages from 58S and NIL(MH) grown under long- and short-day conditions. Examples of duplexes with 2-nt 3′ overhang formed between 21-nt sense and antisense phased small RNAs are illustrated in the boxes. (B) The location of SNP S2 and S1 at 21-nt phasiRNAs. Gray characters indicate the SNP S2 and S1.
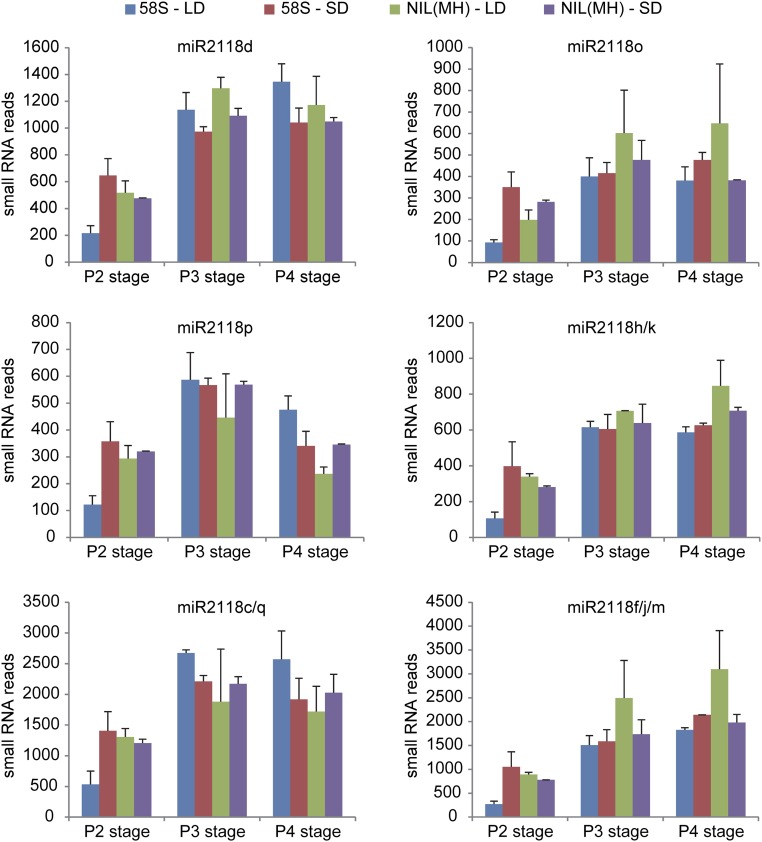
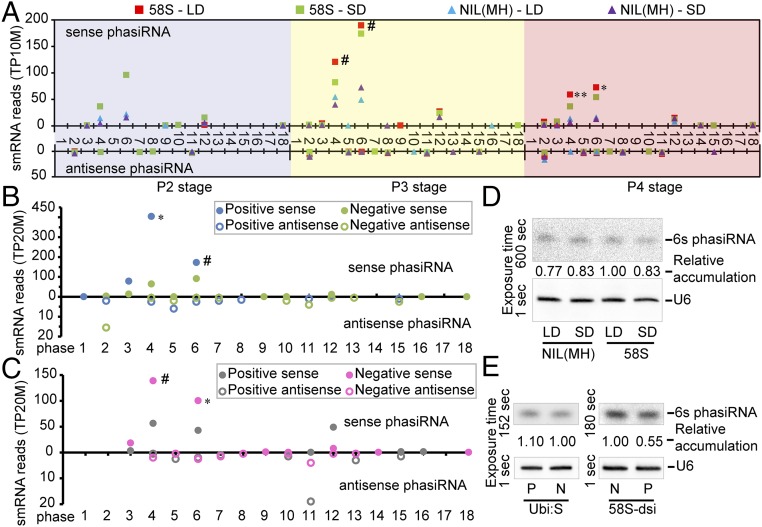
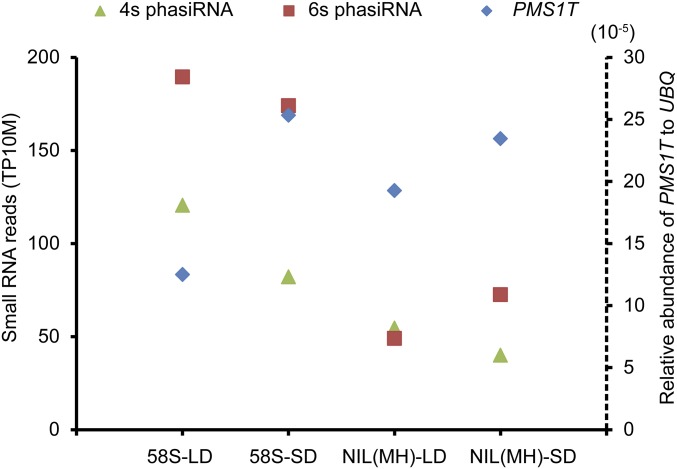
Comparative analysis revealed that the quantities of 21-nt PMS1T-phasiRNAs were higher in P3 and P4 stages than P2. Most phasiRNAs were generated from the sense strand and the 4s, 6s, and 12s phasiRNAs were more abundant than others (Fig. 4A). The phasiRNA reads in 58S under long days at P3 stage were the highest followed by 58S under short days, whereas the reads in NIL(MH) were much lower. We noticed a trend of negative relation between the abundance of phasiRNAs and the PMS1T transcript (Fig. S8), suggesting that more phasiRNAs and fewer intact PMS1T might be related. We also quantified small RNAs from P3 stage in the transgenic plants, which showed that the 21-nt phasiRNA reads of PMS1T were much higher in the Ubi:S transgenic-positive plants than in negative ones under long-day conditions (Fig. 4B). Conversely, their levels were lower in 58S-dsi transgenic-positive than in negative plants (Fig. 4C), despite the total number of 21-nt nonphased small RNA reads in 58S-dsi transgenic positive plants being much larger (Table S3). Examples for detecting the phasiRNAs by RNA blot are illustrated in Fig. 4 D and E, confirming the presence of the phasiRNAs.
Fig. 4.
Expression levels of PMS1T-phasiRNAs in various genotypes. (A) Abundance of 21-nt PMS1T-phasiRNAs from young panicles of 58S and NIL(MH) at P2, P3, and P4 stages under long- and short-day conditions. The smRNA reads are normalized to the whole library with transcripts per 10 million (TP10M) and presented as the mean from two biological replicates. Differences were detected between 58S-LD and NIL(MH)-LD by t test at **P < 0.01, *P < 0.05, or #P < 0.1. (B and C) Abundance of 21-nt phasiRNAs from young panicles of transgenic T1 plants from Ubi:S (B) and 58S-dsi (C) under long-day conditions at P3 stage. Differences were detected by t test at *P < 0.05 or #P < 0.1, respectively. The smRNA data collecting and handing were as in A. (D and E) RNA blot analysis of 21-nt 6s phasiRNA in transgenic plants. Ten-microgram small RNAs per sample from young panicles at P3 stage were loaded on the gel. The samples in E are collected under long-day conditions. The blots were stripped off and rehybridized with U6 probe. The exposure time of the membranes are listed on the left. LD, long days; N, negative plants; P, positive plants; SD, short days.
Fig. S8.
Comparison of the relative abundance of PMS1T-phasiRNAs and PMS1T in panicles at P3 stage under long- and short-day conditions. The small RNA reads and the relative abundance of PMS1T are collected from the data in Figs. 3A and 4A. The small RNA reads of 4s and 6s phasiRNAs are the average of two biological repeats, and the relative abundance of PMS1T are the average of qPCR results from three biological repeats.
Taken together, all of the results suggested an association between the abundance of the PMS1T-phasiRNAs and male sterility under long-day conditions, such that higher accumulation of phasiRNAs result in male sterility.
Discussion
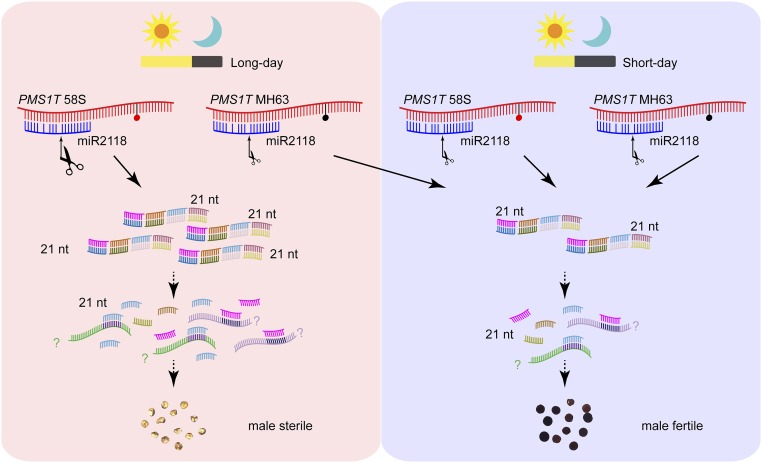
The results obtained in the study can be summarized as the following. The Pms1 locus encodes a lncRNA, PMS1T, and is a 21-PHAS gene. MiR2118 triggers the cleavage of PMS1T resulting in 21-nt phasiRNAs. These phasiRNAs differentially accumulated in the panicles of 58S compared with NIL(MH), especially under long-day conditions, and the elevated phasiRNAs eventually cause male sterility in rice through yet unknown pathways and genes (Fig. S9).
Fig. S9.
A hypothetic mechanistic model for Pms1–regulated PSMS in rice. The PMS1T transcript is targeted by miR2118, and more 21-nt phasiRNAs are produced in 58S than MH63 under long-day conditions because of the SNP S2 (red dot as base T, black dot as base G). The accumulation of phasiRNAs eventually causes male sterility in rice. The SNP S2 may influence the cleavage efficiency mediated by miR2118, resulting in more PMS1T-phasiRNAs in 58S under long days. On the other hand, PMS1T-phasiRNAs may function as secondary small RNA to target some unknown transcripts, which should play important roles in male sterility in rice.
It is somewhat surprising to find that male sterility conditioned by Pms1 to be a semidominant trait rather than completely recessive, as we previously expected based on the results of genetic analyses (15, 29). Such semidominance may explain the slow progress in the early days of two-line hybrid rice breeding in China. Knowledge gained from this study may help future two-line hybrid rice breeding using this PSMS system.
In some plant species, phasiRNAs have been found as a class of siRNAs preferentially accumulating in reproductive tissues, particularly anthers, suggesting a possible role in reproductive development (2, 10). The 21-nt phasiRNAs are abundant subepidermally in maize anthers, and 24-nt phasiRNAs in tapetum and meiocytes (10), but the functions of these reproductive phasiRNAs are not known. The rice Agronaute protein, MEIOSIS ARRESTED AT LEPTOTENE 1 (MEL1), binds to 21-nt phasiRNAs generated from over 700 long-intergenic noncoding RNAs (lincRNAs) dispersed on all of the chromosomes. These lincRNAs also contained one or multiple miR2118 recognition sites (1). MEL1 is required for homologous chromosome synapsis in the early meiosis stage and is expressed specifically in germ cells. The mel1 mutant is male-sterile with vacuolated pollen mother cells (30). Cytological studies by our group showed that the abnormalities in pollen development of 58S under long-day conditions occurred at the pollen mother cell-formation stage when pollen mother cells formed with lots of vacuoles in the large nucleus and the tapetum started to degenerate (16). Thus, there is a lot in common between MEL1 and PMS1T in their roles in regulating pollen development. Nonetheless, MEL1 encoding an AGO family protein impacts a key component of small RNA generation influencing many PHAS clusters, whereas PMS1T may impact only a single locus. Studies also showed that loss-of-function of OsDCL4, which is responsible for production of 21-nt phasiRNAs in rice (23), caused abnormal spikelet morphology and also sterility (31). These results also imply that 21-nt phasiRNAs play important roles in rice reproductive development. However, many questions remain to be answered to understand why and how the 21-nt PMS1T-phasiRNAs regulate male fertility.
A major question thus emerged concerning the targets of the PMS1T-phasiRNAs. The transcripts of TAS genes (tasiRNA-generating loci) are noncoding, and tasiRNAs are triggered and generated by one or two hits of miRNAs (7). TAS1- and TAS2-tasiRNAs target PPR (pentatricopeptide repeat) genes, as well as several genes of unknown function; TAS3-tasiRNAs target genes from ARF family; a group of MYB (myeloblastosis) genes are the targets of TAS4-tasiRNAs (32). A large number of NB-LRR (nucleotide-binding site and leucine-rich repeat) genes were discovered to be PHAS genes in Medicago that generated phasiRNAs targeting NB-LRR transcripts in cis or trans (8). In soybean, the phasiRNAs from a noncoding PHAS locus accumulating preferentially in anthers are predicted to target transposable elements (9). The PMS1T-phasiRNAs are also predicted to target many transcripts, but they did not belong to a special gene family or group. To identify whether one or more or the sum of the phasiRNAs regulate PSMS remains a challenge for future studies (Fig. S9).
Our study showed SNP S2 to be the causal polymorphism of PMS1T, and crucial for PSMS. It was located in the 4th nucleotide of the 2s phasiRNA and the 16th nucleotide of the 2as phasiRNA (Fig. S7B), 24 nt downstream of the cleavage site directed by miR2118 (Fig. 3C). SNPs outside of miRNA target sites could influence miRNA-mediated gene regulation (33–35). There are three speculated possibilities for the mechanism by which SNP S2 of PMS1T regulates PSMS. First, SNP S2 may influence the cleavage efficiency directed by miR2118. We analyzed the alteration of RNA secondary structure caused by PMS1T sequence variation (rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi), predicting a large difference between 58S and MH63 (Fig. S10). Because the 65-bp deletion did not affect male fertility, we used the sequence corresponding to construct 58S-C-dP6 for structure comparison. Contrastive analysis indicated that the S2 SNP brought about a large change in the RNA secondary structure, especially with respect to the miR2118 target region (Fig. S10). This different spatial structure may change the cleavage efficiency directed by miR2118, resulting in more phasiRNAs in 58S at long-day conditions (Fig. S9). Second, SNP S2 may cause differences in target selection of 2s and 2as phasiRNAs between 58S and MH63 under long days, such that one is more effective than the other in targeting the downstream genes for male sterility. Third, SNP S2 may influence the day-length–dependent preferential binding of some unknown RNA binding protein to the PMS1T, affecting the processing of phasiRNAs. However, more studies are needed to provide evidence to support these speculations.
Fig. S10.
Prediction of RNA secondary structure of PMS1T. The sequence of the miR2118 target is indicated in red, and the 65-bp region in cyan color. The positions and bases of SNP S2 and S1 are marked in blue and green colors, respectively. Magnification of the structures of the regions nearby the SNPs from MH63 and 58S are presented under the gray and green background, respectively.
Pms1 and Pms3 both confer PSMS in rice functioning in the same plant. The two share some common characters: they both encode lncRNAs that do not produce proteins; functional mutations are both SNPs; small RNAs can be detected in both transcripts (16, 36). However, there are important differences between them: sterility is of incomplete dominance for Pms1, whereas recessive for pms3; sufficient amount of the LDMAR transcript is required for fertility in long days, whereas abundance of PMS1T-phasiRNAs is the main cause for fertility reduction; data obtained at present suggest that RNA-directed DNA methylation is involved in pms3 (17), but phasiRNAs are produced in Pms1. How these two loci function in the same plant to cause male sterility remains to be characterized in future studies.
Nonetheless, the phasiRNAs of PMS1T provided an incidence associating the phasiRNAs with a biological trait, especially an agriculturally highly important trait that started the industry of two-line hybrid rice. Such an association confirmed that the phasiRNAs indeed have biological functions that expand our understanding of PSMS, and offer soundly based opportunities for artificially generating male-sterile lines for rice breeding, which may also be useful for other crops.
Materials and Methods
Plant Materials and Field Growth.
For natural field evaluation, all plants were grown and investigated under natural field conditions in the experimental farm of Huazhong Agricultural University (114.36°E, 30.48°N), Wuhan, China. For natural long-day conditions, seeds were sown in mid-April to mid-May to place the flowering time of 58S before September 3rd, ensuring the day-length at the young panicle-developing stage to be longer than 14 h. For a natural short day, seeds were sown in mid-to-late June to place the flowering time between September 8th and 20th, so that the day-length of the young panicle-developing stage to be shorter than 13.5 h (22). Plants with the heading time later than September 20th were not included because low temperature may cause sterility in the field.
Fine-Mapping of Pms1.
The mapping population was the BC5F2 generated from the cross between MH63 and 58S with five successive backcrosses using 58S as the female and recurrent parent. Further backcrosses were made to BC8 from which NIL(MH) (MH63 homozygote in near-isogenic lines) was obtained for subsequent molecular analysis. Molecular markers for mapping are listed in Table S4.
Table S4.
Primers used in this study
| Name | Sequence (5′ to 3′) | Remark |
| Markers for map-based cloning | ||
| pj23-F | AGAGAGAACTCTGCCAATATC | SSR |
| pj23-R | AAAGTAACCCCTGCATAGACT | |
| P1-F | TCCTGTGGTCCAATCAAACG | CAPS, MunI |
| P1-R | CAGATCGTCTGATGATGTTGC | |
| P2-F | GCAATGGCTGTTGTCCGAAT | CAPS, Cfr10I |
| P2-R | CTATGACGTTTGCTTTGTGCC | |
| P3-F | CTCCTTCGCAGACTCTACAC | CAPS, Eco52I |
| P3-R | ATGTCAATCTCTGATATCGGC | |
| P4-F | GCGAGCACATAAGTCCTCTA | dCAPS, AluI |
| P4-R | ATAATTGTTTAAAGGTGTCAG | |
| P5-F | AGGCGCAGTAAAAACACCTG | SSR |
| P5-R | CTTGCGTGGTTTCAAGGG | |
| P6-F | CATTAGGCGGAGATGGCAAT | InDel |
| P6-R | ATAGCCAACACTCATCACTGTCG | |
| P7-F | AAGGAGATGCAGAGAGGGTTTAC | InDel |
| P7-R | AGGAAGGATAGGTGTTTGATGAAC | |
| P8-F | TCAGATGACTTAGCAACACAAGC | CAPS, DraI |
| P8-R | CTTGCTTATCCCAAATGTCGT | |
| P9-F | AGGAAACGATAGGCGAACAAAC | CAPS, EcoRV |
| P9-R | ACAAGGCTTCCAAACTTCCAA | |
| P10-F | CACGACCTTACTTATGAGCATG | SSR |
| P10-R | AAAAGAACACAGCAGGGGAG | |
| P11-F | GTCCCTCATCTTTAGGCTGTG | SSR |
| P11-R | GCCTAAAGAGGAGGGACGAA | |
| Fssr-F | TTAGTGCGTGTGGCATAGATG | SSR |
| Fssr-R | GGCCTTTTTGTGAATTGGG | |
| Vector construction | ||
| 58S-C-F | ATGAAGGACCGAGAAGAAGC | 58S-C |
| 58S-C-R | GTTCTCAGATGATATGGAACTGTG | |
| dsi-F | TCGACTAGTTGGCAGGGTACCTGTACATGCCCAACAAGCTCT | dsi |
| dsi-R | GGCGAGCTCGGATCCTGTTCGCATGGACAATTGGTG | |
| OX-1F | TATGGTACCGACTACATGGGCACCCCTTGAA | 35S:S, Ubi:S |
| OX-1R | TATGGATCCCGTGATTCAGCAGGTGGAGTTAA | |
| OX-2F | TCAGAGCTCGCATCAGGAAAGAAGCTTCTACTA | 35S:S-CS |
| OX-2R | TGACCTGCAGTACTTCAGTACCATCAGCGTGAC | |
| OX-3F | TATGGTACCAATTGTCCATGCGAACAGAAAA | Ubi:S P6 |
| OX-3R | TATGGATCCCCGTAAACAATTATCGGTGATA | |
| ORF1G-R | CATTAGGCGGAGATGGCAAT | 58S-ORF1+G |
| ORF1G-muF | CAAAATCACAATAAAGATATATGGATCAAAACCTTAATAAT | |
| ORF1G-muR | ATTATTAAGGTTTTGATCCATATATCTTTATTGTGATTTTG | |
| ORF1G-F | AGGTGTCGGTTCTAATGGTAGAAC | |
| ORF2G-F | CCATTTATAGGCTACTCCTTTCC | 58S-ORF2+G |
| ORF2G-muR | AACTAGTATATAGTATATAGAGAAGCCCAAGTACCGAACTTTTTCTGTTCGCCAT | |
| ORF3G-R | ATTGTTGCAGGTGAAGGTGAGC | 58S-ORF3+G |
| ORF3G-muF | CCCATTGCCATCTCCGCCTAATGGAACGCCTTGCATGTTGG | |
| ORF3G-muR | CCAACATGCAAGGCGTTCCATTAGGCGGAGATGGCAATGGG | |
| ORF3G-F | CAATTTTGCCTGGTATCACCAA | |
| Positive test | ||
| PMCGF | GGCTCACCAAACCTTAAACAA | |
| PMCGR | CTGAGCTACACATGCTCAGGTT | |
| GUS1.6F | CCAGGCAGTTTTAACGATCAGTTCGC | |
| GUS1.6R | GAGTGAAGATCCCTTTCTTGTTACCG | |
| P6-F2 | AATTGTCCATGCGAACAGAAAA | |
| P6-R2 | CCGTAAACAATTATCGGTGATA | |
| RACE | ||
| RACE5-1 | CTGATGACTGTGTTCCAGTATTTG | 5′-RACE |
| RACE5-2 | ACGCAAGCTTGGCTCTTTGTT | 5′-RACE |
| RACE5-3 | ACCTGGCATAGACCGATAGTTAC | 5′ RLM-RACE |
| RACE5-4 | CTGATGACTGTGTTCCAGTATTTG | 5′ RLM-RACE |
| RACE3-1 | CAATTTTGCCTGGTATCACCAA | 3′-RACE |
| RACE3-2 | CAACATCATTAGGTTGCTGTGAT | 3′-RACE |
| qPCR | ||
| RT-F | AAAGTTCGGTACTTGGGCTTCTCT | |
| RT-R | ACTCCCATTCGATATTGTTGCAAGGGC | |
| UBQ-F | AACCAGCTGAGGCCCAAGA | |
| UBQ-R | ACGATTGATTTAACCAGTCCATGA | |
CAPS, cleaved amplified polymorphic sequences marker; dCAPS, derived cleaved amplified polymorphic sequences marker; InDel, insertion/deletion marker; SSR, simple sequence repeat marker.
Full-Length cDNA Determination and Modified 5′ RLM-RACE.
Young panicles were collected from NIL(MH) and 58S plants growing in the field and stored in liquid nitrogen. Total RNA was isolated following the instruction of RNA extraction kit (TRIzol reagent, Invitrogen).
To synthesize the first-strand cDNA, 1 μg total RNA was reverse-transcribed using the protocol provided by the SMART RACE cDNA Amplification Kit (Clontech), with a final volume of 100 μL. For 5′-RACE, two rounds of PCR amplification were conducted according to the protocol provided with the Advantage 2 PCR kit (Clontech), using two nested adapter primers (UPM and NUPM) in the kit and gene-specific primers (RACE5-1 paired with UPM, RACE5-2 paired with NUPM). The 3′-RACE was essentially the same except that primer RACE3-1 was used to pair with UPM in the first round of reaction, and RACE3-2 with NUPM in the second round of reaction.
The modified 5′ RLM-RACE, which was adapted to validate the cleavage site of miRNA on mRNA, was performed using the FirstChoice RLM RACE Kit (Ambion) by modifying the manufacturer’s instructions without enzymatic pretreatment, as described previously (24). During the nested PCR, primer RACE5-3 and RACE5-4 were paired with adapter primer 5′ RACE outer primer and 5′ RACE inner primer, respectively, as supplied in the kit.
All above PCR products were gel-purified, cloned (pGEM-T Easy, Promega), and sequenced. The sequences of the primers are listed in Table S4.
Small RNA-seq and PARE Analysis.
Following the TruSeq Small RNA Sample Preparation Guide supplied in the TruSeq Small RNA Sample Prep Kit (Illumina), the libraries for Small RNA-seq were constructed and immediately sequenced on an Illumina HiSEq. 2000 instrument. The small RNA data were analyzed as previously described (8). PARE library construction and sequencing followed previously described methods (37).
Small RNA Page-Northern Blotting Analysis.
Total RNA extracted from the young panicles at P3 stage with use of TRIzol reagent (Invitrogen) was mixed with one-third volume 25% (wt/vol) PEG8000 and one-third volume 2.5M NaCl to isolate low molecular weight RNAs. Ten micrograms of low molecular weight RNAs was separated by 15% (wt/vol) denaturing TBE-urea polyacrylamide gels (Invitrogen) and transferred to Hybond-N+ membranes (GE Healthcare) by use of a transblot semidry transfer cell (Bio-Rad). Antisense DNA oligonucleotides of 21-nt complementary to 6s phasiRNA were used as probes (5′-TACACTATCCTTAGATCATCC-3′). The probe was modified with biotin on the 5′ terminus. The membrane was hybridized with hybridization buffer containing 50-pmol/mL–labeled probes. The hybridization was carried out with previously described methods (38). After probe detection, the blots were stripped and reanalyzed with U6 biotin-labeled probe (5′-TGTATCGTTCCAATTTTATCGGATGT-3′), and 1-pmol/mL–labeled probe was added to the hybridization buffer. The biotin-labeled probes were detected using Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Scientific). Hybridized membranes were placed in Molecular imager ChemiDOC XRS+ (Bio-Rad) for exposure. The exposed images were captured and analyzed with Image Lab software to quantify the relative abundance of small RNAs.
Details of the other materials and methods are provided in SI Materials and Methods.
SI Materials and Methods
Construct Preparation and Transformation.
The rice genomic fragment for complementation construct MH-C was obtained from the BAC clone 21O9 (GenBank accession no. DQ989628) of the Minghui 63 BAC library (39), which contained the putative Pms1 locus, by digesting with restriction enzymes PstI and SacI. A fragment of 5,682 bp was recovered and ligated into the binary vector pCAMBIA1301. A genomic fragment amplified from 58S was used to generate the construct 58S-C. The constructs 58S-ORF1+G, 58S-ORF2+G, and 58S-ORF3+G were modified from 58S-C, and the residue G inserted at downstream of ATG start codon was generated by overlap-extension PCR. The 65-bp deletion in 58S-C (58S-C-dP6) was realized by amplifying a fragment from MH63 genomic sequence and replacing the original 58S sequence by double digestion (SpeI and SacI) and ligation. The DNA fragment for the RNAi construct dsi was cloned into the vector pDS1301 (40) by two rounds of double digestion and ligation with SpeI and SacI, and KpnI and BamHI, respectively.
For preparation of the overexpression construct with 35S promoter, the PCR product amplified from the cDNA of 58S was digested with both PstI and SacI and then ligated into the pCAMBIA1301S vector (41), which contained the CaMV 35S promoter. Those with the ubiquitin promoter were generated by cleaving PCR product with both KpnI and BamHI, and then inserted into the vector pU1301 (42), where the gene was under the control of a maize ubiquitin gene promoter.
Primers used for construct preparation are listed in Table S4. All constructs were transformed into the Agrobacterium strain EHA105, and rice transformation was done according to a previously described method (43).
Genotype Detection and Pollen Fertility Investigation.
The genotypes of complementation and overexpression transgenic plants were determined by either the P6 marker or a primer pair P6-F2 and P6-R2 based on the 65-bp insertion in 58S genome. Cosegregation analysis of RNAi plants was carried out using primer pair PMCGF and PMCGR. Other transgenic plants were analyzed by amplifying the GUS (β-glucuronidase) gene with primer pair GUS1.6F and GUS1.6R (Table S4).
For investigation of pollen fertility, mature pollen grains sampled from six flowers of a plant were stained with 1% I2–KI solution and observed with a Leica DM4000B microscope. Spikelet fertility was measured as the proportion of well-developed fertile spikelets in total number of spikelets using three panicles per plant.
RT-PCR and Quantitative Real-Time PCR.
For examine the expression profile of PMS1T, total RNA was isolated from various tissues mentioned in Fig. S5 using TRIzol (Invitrogen). Approximately 2 μg DNaseI-treated total RNA was reverse-transcribed using the M-MLV reverse transcriptase (Invitrogen) in a volume of 80 μL to obtain cDNA. We performed the PCR using equal amounts of RT products to amplify the transcript with primer RT-F and RT-R. PCR products were separated in 2% (wt/vol) agarose gels and normalized against the rice ubiquitin gene (UBQ, LOC_Os03g13170).
Quantitative real-time PCR was carried out in a total volume of 25 μL containing 2 μL of the reverse-transcribed product, 0.25 mM gene-specific primers, and 12.5 μL SYBR Green Master Mix (Applied Biosystems) on an Applied Biosystems 7500 Real-Time PCR System following the manufacturer’s instructions. The expression levels of genes were calculated using the relative quantification method (44). The primer sequences of different genes for quantitative PCR are listed in Table S4.
Acknowledgments
We thank Qili Fei, Jixian Zhai, and Mayumi Nakano for assistance with data handling and helpful discussions. This work was supported by National Natural Science Foundation Grant 91540101, the National Key Research and Development Program Grant 2016YFD0100903 of China, and a grant from the Bill & Melinda Gates Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo [accession nos. GSE84885 (small RNA and PARE) and KX578835 and KX578836 (PMS1T in 58S and MH63)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619159114/-/DCSupplemental.
References
- 1.Komiya R, et al. Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 2014;78(3):385–397. doi: 10.1111/tpj.12483. [DOI] [PubMed] [Google Scholar]
- 2.Johnson C, et al. Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res. 2009;19(8):1429–1440. doi: 10.1101/gr.089854.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han BW, Wang W, Li C, Weng Z, Zamore PD. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science. 2015;348(6236):817–821. doi: 10.1126/science.aaa1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohn F, Handler D, Brennecke J. Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science. 2015;348(6236):812–817. doi: 10.1126/science.aaa1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121(2):207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127(3):565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 7.Chen HM, Li YH, Wu SH. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc Natl Acad Sci USA. 2007;104(9):3318–3323. doi: 10.1073/pnas.0611119104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhai J, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25(23):2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arikit S, et al. An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant Cell. 2014;26(12):4584–4601. doi: 10.1105/tpc.114.131847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhai J, et al. Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc Natl Acad Sci USA. 2015;112(10):3146–3151. doi: 10.1073/pnas.1418918112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia R, Ye S, Liu Z, Meyers B, Liu Z. Novel and recently evolved miRNA clusters regulate expansive F-box gene networks through phasiRNAs in wild diploid strawberry. Plant Physiol. 2015;169(1):594–610. doi: 10.1104/pp.15.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi M. The discovery and preliminary studies of the photoperiod-sensitive recessive male sterile rice (Oryza sativa L. subsp. japonica) Zhongguo Nong Ye Ke Xue. 1985;2:44–48. [Google Scholar]
- 13.Si H, Liu W, Fu Y, Sun Z, Hu G. Current situation and suggestions for development of two-line hybrid rice in China. Chin J Rice Sci. 2011;25(5):544–552. [Google Scholar]
- 14.Zhang Q, et al. Using bulked extremes and recessive class to map genes for photoperiod-sensitive genic male sterility in rice. Proc Natl Acad Sci USA. 1994;91(18):8675–8679. doi: 10.1073/pnas.91.18.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mei M, Dai X, Xu C, Zhang Q. Mapping and genetic analysis of the genes for photoperiod-sensitive genic male sterility in rice using the original mutant Nongken 58S. Crop Sci. 1999;39(6):1711–1715. [Google Scholar]
- 16.Ding J, et al. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci USA. 2012;109(7):2654–2659. doi: 10.1073/pnas.1121374109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding J, et al. RNA-directed DNA methylation is involved in regulating photoperiod-sensitive male sterility in rice. Mol Plant. 2012;5(6):1210–1216. doi: 10.1093/mp/sss095. [DOI] [PubMed] [Google Scholar]
- 18.Liu N, et al. Identification of an 85-kb DNA fragment containing pms1, a locus for photoperiod-sensitive genic male sterility in rice. Mol Genet Genomics. 2001;266(2):271–275. doi: 10.1007/s004380100553. [DOI] [PubMed] [Google Scholar]
- 19.Yu J, et al. Rapid genome evolution in Pms1 region of rice revealed by comparative sequence analysis. Chin Sci Bull. 2007;52(7):912–921. [Google Scholar]
- 20.Wang F, Mei M, Xu C, Zhang Q. pms1 genomic region does not cause fertility difference between the photoperiod-sensitive male sterile rice Nongken 58S and normal Nongken 58. Acta Bot Sin. 1997;39(10):922–925. [Google Scholar]
- 21.Kelley D, Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012;13(11):R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan S, Zhang Z, Xu C. Studies on the critical stage of fertility change induced by light and its phase development in HPGMR. Acta Agron Sin. 1988;14(1):7–13. [Google Scholar]
- 23.Song X, et al. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 2012;69(3):462–474. doi: 10.1111/j.1365-313X.2011.04805.x. [DOI] [PubMed] [Google Scholar]
- 24.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297(5589):2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 25.Song X, et al. Rice RNA-dependent RNA polymerase 6 acts in small RNA biogenesis and spikelet development. Plant J. 2012;71(3):378–389. doi: 10.1111/j.1365-313X.2012.05001.x. [DOI] [PubMed] [Google Scholar]
- 26.Chen HM, et al. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA. 2010;107(34):15269–15274. doi: 10.1073/pnas.1001738107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shivaprasad PV, et al. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell. 2012;24(3):859–874. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajeswaran R, Pooggin MM. RDR6-mediated synthesis of complementary RNA is terminated by miRNA stably bound to template RNA. Nucleic Acids Res. 2012;40(2):594–599. doi: 10.1093/nar/gkr760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mei M, et al. pms3 is the locus causing the original photoperiod-sensitive male sterility mutation of ‘Nongken 58S’. Sci China C Life Sci. 1999;42(3):316–322. doi: 10.1007/BF03183609. [DOI] [PubMed] [Google Scholar]
- 30.Nonomura K, et al. A germ cell specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell. 2007;19(8):2583–2594. doi: 10.1105/tpc.107.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu B, et al. Oryza sativa dicer-like4 reveals a key role for small interfering RNA silencing in plant development. Plant Cell. 2007;19(9):2705–2718. doi: 10.1105/tpc.107.052209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howell MD, et al. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19(3):926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishra PJ, et al. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci USA. 2007;104(33):13513–13518. doi: 10.1073/pnas.0706217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen AX, et al. Germline genetic variants disturbing the Let-7/LIN28 double-negative feedback loop alter breast cancer susceptibility. PLoS Genet. 2011;7(9):e1002259. doi: 10.1371/journal.pgen.1002259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Reichel M, Millar AA. Determinants beyond both complementarity and cleavage govern microR159 efficacy in Arabidopsis. PLoS Genet. 2014;10(3):e1004232. doi: 10.1371/journal.pgen.1004232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou H, et al. Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 2012;22(4):649–660. doi: 10.1038/cr.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhai J, Arikit S, Simon SA, Kingham BF, Meyers BC. Rapid construction of parallel analysis of RNA end (PARE) libraries for Illumina sequencing. Methods. 2014;67(1):84–90. doi: 10.1016/j.ymeth.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 38.Huang Q, Mao Z, Li S, Hu J, Zhu Y. A non-radioactive method for small RNA detection by northern blotting. Rice (N Y) 2014;7(1):26. doi: 10.1186/s12284-014-0026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng K, Zhang H, Zhang Q. BAC library constructed to the rice cultivar “Minghui 63” for cloning gene of agronomic importance. Acta Bot Sin. 1998;40(12):1108–1114. [Google Scholar]
- 40.Yuan B, Shen X, Li X, Xu C, Wang S. Mitogen-activated protein kinase OsMPK6 negatively regulates rice disease resistance to bacterial pathogens. Planta. 2007;226(4):953–960. doi: 10.1007/s00425-007-0541-z. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Y, et al. Over-expression of aspartate aminotransferase genes in rice resulted in altered nitrogen metabolism and increased amino acid content in seeds. Theor Appl Genet. 2009;118(7):1381–1390. doi: 10.1007/s00122-009-0988-3. [DOI] [PubMed] [Google Scholar]
- 42.Qiu D, et al. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol Plant Microbe Interact. 2007;20(5):492–499. doi: 10.1094/MPMI-20-5-0492. [DOI] [PubMed] [Google Scholar]
- 43.Lin YJ, Zhang Q. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Rep. 2005;23(8):540–547. doi: 10.1007/s00299-004-0843-6. [DOI] [PubMed] [Google Scholar]
- 44.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]