Significance
Structural analysis of microscale three-dimensional tissues (3D microtissues) in high-throughput is becoming increasingly important in drug discovery, regenerative medicine, and other biomedical areas because they recapitulate many in vivo biological features not present in 2D models. This can be done by using microfluidic technology to control and apply external forces to on-chip 3D microtissues, and imaging these organ-on-chip systems with confocal microscopy. However, the high cellular density of 3D microtissues scatters light and fundamentally limits the ability to characterize the entire tissue construct. We developed an on-chip strategy to rapidly clarify, image, and analyze whole intact microtissues without compromising internal structures. Our technique removes the imaging depth limit, allowing accurate analysis and characterization of entire tissues in microfluidic chips.
Keywords: microfluidic, CLARITY, fluorescence imaging, 3D imaging, computational analysis
Abstract
On-chip imaging of intact three-dimensional tissues within microfluidic devices is fundamentally hindered by intratissue optical scattering, which impedes their use as tissue models for high-throughput screening assays. Here, we engineered a microfluidic system that preserves and converts tissues into optically transparent structures in less than 1 d, which is 20× faster than current passive clearing approaches. Accelerated clearing was achieved because the microfluidic system enhanced the exchange of interstitial fluids by 567-fold, which increased the rate of removal of optically scattering lipid molecules from the cross-linked tissue. Our enhanced clearing process allowed us to fluorescently image and map the segregation and compartmentalization of different cells during the formation of tumor spheroids, and to track the degradation of vasculature over time within extracted murine pancreatic islets in static culture, which may have implications on the efficacy of beta-cell transplantation treatments for type 1 diabetes. We further developed an image analysis algorithm that automates the analysis of the vasculature connectivity, volume, and cellular spatial distribution of the intact tissue. Our technique allows whole tissue analysis in microfluidic systems, and has implications in the development of organ-on-a-chip systems, high-throughput drug screening devices, and in regenerative medicine.
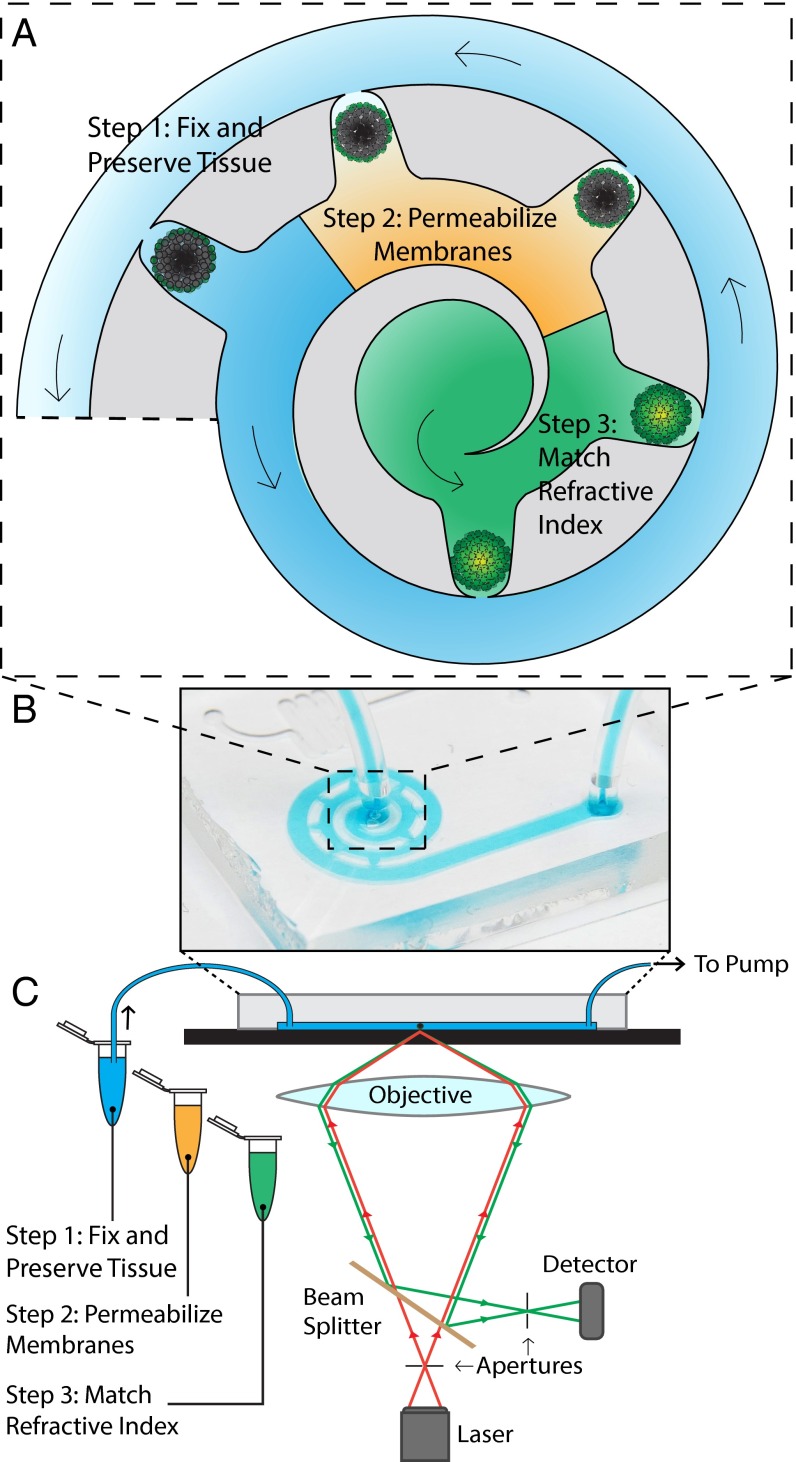
Developing cellular systems that mimic in vivo phenotype and function is important to drug screening, tissue engineering, and many other biomedical applications. Currently, high-throughput screening assays are based on culturing primary and immortalized cells in two dimensions (1, 2). However, tissue heterogeneity, coupled with the absence of cellular architecture, biological stresses, and dynamic changes in nutrient, waste, and oxygen gradients make these 2D cellular systems insufficient at recapitulating in vivo biological features and mechanisms (3–5). This has prompted researchers to use resected tissues or generate three-dimensional microscale tissue constructs (3D microtissues) such as spheroids and organoids as models for screening assays. These constructs have been combined with microfluidic technology to create organ-on-a-chip systems, where microfluidic technology is exploited to control flow conditions, throughput, and mechanical stressors in maintaining cellular function (6–9). The interior molecular expression and cellular organization of these microtissues are then analyzed using confocal microscopy (10), which optically sections them into a series of 2D images that are then computationally reconstructed into a 3D image (11–14). Despite these developments, the utility of these organ-on-a-chip systems is fundamentally limited by insufficient light penetration into these 3D microtissues due to their high cellular density. Large numbers of refractive index mismatches along the surfaces of the cell membranes all become light-scattering interfaces (15) that limit the penetration of light to less than 100 µm. This results in images that lack information from the tissue core, creating false negative data points at worse and incomplete data sets at best (10), and prevents the use of 3D microtissues for high-throughput screening applications. The rate of mass transport of staining molecules is also limited by diffusion in 3D microtissues, and labeling internal structures is especially difficult for intracellular targets of interest. Recent technologies, such as CLARITY by Chung et al. (16), were developed to address similar problems for imaging whole organs or animals, in which tissue proteins were cross-linked into a hydrogel construct, lipids were removed, and the refractive index of the remaining tissue components was homogenized to eliminate scattering interfaces and render the tissue transparent (16–20). However, applying these repeated solution changes to microtissues is labor-intensive, prone to tissue damage and sample loss, and limited by diffusion-dominated mass transport. Researchers have used electrophoresis to accelerate mass transport (21, 22), but this process does not affect nonelectromobile molecules which would still be limited to diffusional movement. These limitations greatly extend the processing time and impede the use of this tissue-clearing technique in high-throughput screening of microtissues. Here, we developed an on-chip strategy to rapidly label, clear, image, and analyze microtissues to enable high-throughput fluorescent imaging of 3D microtissues without compromising their internal structures. Our strategy overcomes diffusion limitations by inducing pressure-driven flow across the on-chip microtissues, thereby greatly enhancing interstitial fluid exchange and mass transport. As shown in Fig. 1, the first step in our on-chip tissue-clearing process is to infuse a monomer solution that preserves and protects tissue proteins and their spatial information within a tissue–gel hybrid network. The second step is to disrupt the cellular membranes with detergent to expose intracellular proteins for labeling, and is optional when searching for membrane-bound proteins. The last step is to match the refractive index of the resulting tissue–gel hybrid, which would eliminate the remaining scattering interfaces to render the tissue optically transparent. To match the refractive index, we infuse a refractive index matching solution [RIMS; 88% (wt/vol) Iohexol, 2.5% (wt/vol) 1,4-diazabicyclo[2.2.2]octane, 50 mM sodium borate, and 0.01% sodium azide]. Matching the refractive index will render the tissue transparent without any additional biological processes, but the increased viscosity of RIMS coupled with fluidic pressure differences within the chip will shrink and distort tissues that have not been fixed and gelled as described in the first step.
Fig. 1.
Schematic of on-chip clearing technique. Arrows show direction of fluid flow. In the first step, a monomer solution is infused into the device, which will preserve and fix the tissue after gelation. Following this, a solution containing SDS is infused to wash away the cellular membranes of the tissue, exposing the intracellular proteins for labeling. The final step is to infuse an RIMS to eliminate the scattering interfaces within the tissue, allowing the tissue core to be visualized and imaged. A shows the features of the microfluidic device that hold the microtissues in place. B shows the microfluidic device as seen by eye. (C) The microfluidic device is placed on an imaging stage of a confocal microscope, connected to the various solutions for steps 1–3, as well as a pump to draw the solutions through the device.
Results and Discussion
Optimization of Microfluidic-Based Tissue Clarification.
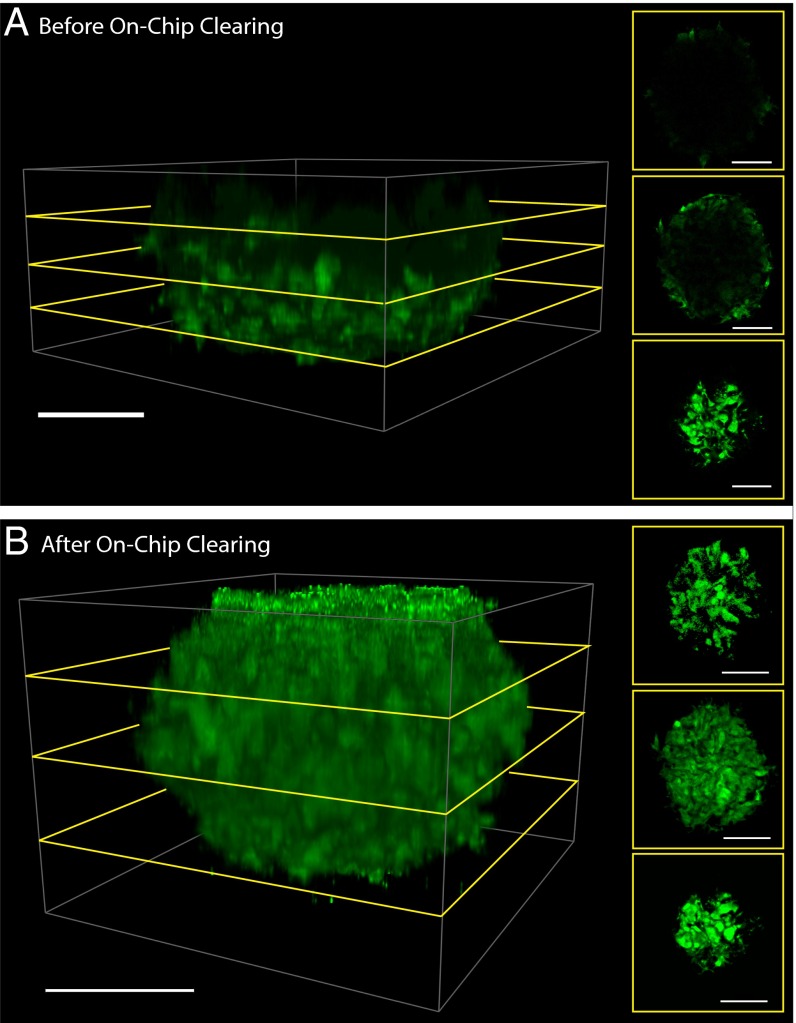
To develop our on-chip tissue-clearing system, we first determined the optimal monomer solution concentration that best preserves tissue proteins before rendering the tissue transparent. We generated 3D tumor spheroids via centrifugation (23) from the cancer cell lines MDA-MB-435, MCF7, and GFP-expressing U87MG to serve as tissue models for our optimization experiments. These 250-µm (MCF7) to 450-µm (U87MG-GFP) spheroids were loaded into our multichamber microfluidic chip using gravity-induced flow, as shown in Fig. 1A. This “spiraling” microfluidic chip was designed to ensure that each spheroid was subjected to equivalent shear stress, flow rates, and concentrations of fluids (24). Once loaded, a monomer solution of 2–8% (wt/vol) acrylamide, 4% (wt/vol) formaldehyde, and 2.5% (wt/vol) of the thermal radical initiator 2,2'-azobis[2-(2-imidazolin-2-yl)propane] dihydrochloride (Va-044) is infused at 800 µL/h through the spheroids in the microfluidic chip for 20 min (Fig. 1C). The combination of acrylamide and formaldehyde cross-links the tissue proteins and forms a hydrogel when the chip is placed at 37 °C for 2 h for acrylamide polymerization. Without disrupting their membranes, we then infused RIMS into the chip to render the microtissues transparent. The infusion of RIMS into the device will cause varying levels of microtissue deformation, which we quantified using a MATLAB script that measures the tissue volume shrinkage. We found that increasing acrylamide concentration decreases the extent of tissue shrinkage. We rationalized that a higher acrylamide concentration increases the level of protein cross-links and overall stiffness of the tissue–gel hybrid, and therefore its resistance to fluidic pressures exerted by the more viscous RIMS. As shown in SI Appendix, Fig. S1A and Movie S1, the tissues remained at 80–95% of their original volume after treatment with 8% (wt/vol) acrylamide. Without acrylamide treatment, the spheroid tissue deforms from fluidic forces within the chip during subsequent steps (SI Appendix, Fig. S1B and Movie S2). After RIMS infusion, the scattering interfaces within the tissue–gel hybrid were eliminated and the tissue became transparent. Using MATLAB and IMARIS image analysis software, we found that our process increased the imaging depth by 150% for MCF7 spheroids, and 250% for U87MG-GFP spheroids when compared against unprocessed spheroids, where only the periphery is visible (Fig. 2, SI Appendix, Fig. S3, and Movies S3 and S4). Both types of spheroids were completely transparent after our on-chip process, but the smaller size of the MCF7 spheroids artificially limits the ratio of imaging depth increase.
Fig. 2.
Increased imaging depth after on-chip clearing. Shown here is the same spheroid (A) before and (B) after the process. The application of our on-chip clearing technique on U87MG spheroids enhances the imaging depth by 250% compared with spheroids imaged through conventional confocal microscopy. Side panels show cross-sectional optical slices at different depths within the spheroid. Green, GFP signal, excited at 488 nm and emission captured from 493 to 598 nm. (Scale bar, 100 µm.) Movie S3 shows A, rotated in three dimensions, and Movie S4 shows B.
Assessment of Tissue Staining.
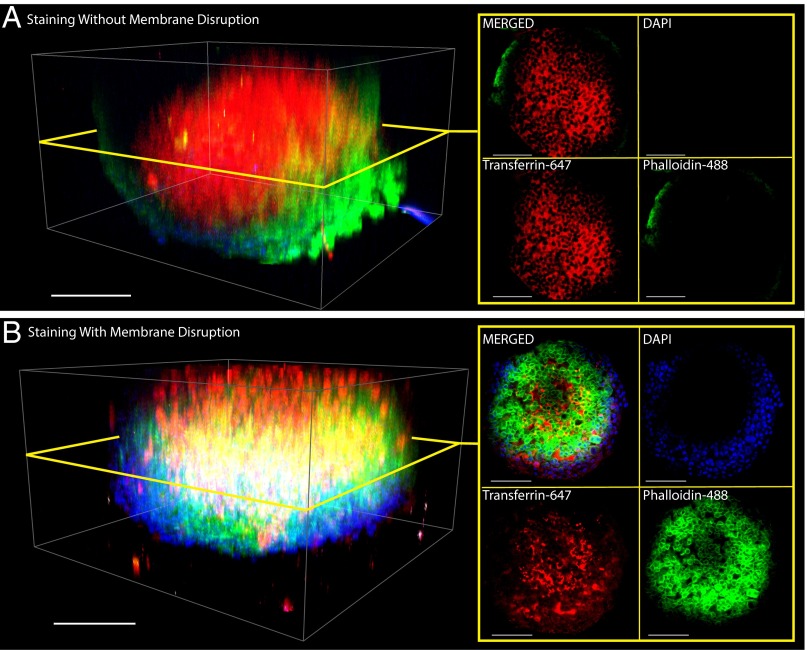
With tissue preservation optimized, we proceeded to further reduce the time needed to stain protein targets of interest, before rendering tissues transparent. Although lipid membrane disruption is not necessary for rendering microtissues transparent, minimizing the staining time is needed to make this method amenable for high-throughput screening applications. We used MDA-MB-231 spheroids and the lipophilic dye DiO to optimize the membrane disruption protocol. The membranes were found to be disrupted and washed out of the tissue within 5–10 min (SI Appendix, Fig. S2 and Movie S5) when 8% wt/vol SDS was infused at 800 µL/h. To determine if membrane disruption is critical for staining and identifying intracellular targets, we generated MDA-MB-435 spheroids, preserved them with 4% monomer solution, and disrupted their membranes. We then infused transferrin-647, phalloidin-488, and DAPI at 400 µL/h for 15 min to stain the membrane-bound transferrin receptors, intracellular actin, and nuclear DNA, respectively. After staining and RIMS infusion (seen in Movie S6), we observed in Fig. 3 that phalloidin-488 was able to easily stain actin only within spheroids that were membrane-disrupted, whereas the cell-surface transferrin receptors were labeled in both sample groups.
Fig. 3.
Membrane disruption enables intracellular staining. The success of labeling intracellular proteins depends heavily on whether or not the cellular membranes within the tissue is disrupted. MDA-MB-435 spheroids were fixed and cross-linked, then in (A) cellular membranes within MDA-MB-435 spheroids were left intact, and in (B) the membranes were disrupted with 4% SDS. Spheroids were then stained on-chip for 15 min under 400 µL/h by Transferrin-647 [red, excited (ex.) 633 nm, emission (em.) 640–735 nm], Phalloidin-488 (green, ex. 488 nm, em. 511–597 nm), and DAPI (blue, 2-photon ex. 810 nm, em. 416–483 nm) to label the extracellular transferrin receptor, intracellular actin, and intracellular nucleus, respectively. (Scale bar, 100 µm.) Spheroid of A is rotated in Movie S7, and spheroid of B is rotated in Movie S8.
Characterizing Flow Rate Effects on Interstitial Fluid Exchange.
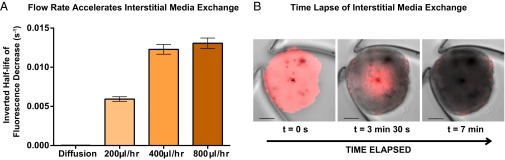
The entire on-chip process of preserving, staining, and rendering microscale tissues optically transparent was found to take as little as 5 h, and varied depending on the user’s staining protocol and targets of interest. In contrast, off-chip techniques for 3D interrogation found in the literature require longer processing times, ranging from several days to a week for similarly sized tissues (25). We hypothesized that this difference in processing time was due to an enhanced rate of interstitial fluid exchange within the tissues, granted by fluidic forces within microfluidic devices (26, 27). To verify this hypothesis, we devised an experiment to estimate the relative rates of on-chip and off-chip interstitial fluid exchange. We infused RIMS into B16F10 melanoma spheroids (expressing tdTomato) to maximize its detectable fluorescent signal, then replaced the RIMS with PBS while imaging the same area within an internal z slice (seen in Movie S9). Because PBS does not match the interstitial refractive indices, the fluorescence signals will decrease as the PBS replaces the RIMS, and the half-life of this decrease gives an estimate of the rate of interstitial fluid replacement. The PBS infusion flow rates were then increased because faster flow rates would feasibly allow faster tissue-clearing rates and therefore decrease the total processing time. We plotted the decrease of fluorescence intensity in SI Appendix, Fig. S4B, and the inverted half-life of fluorescence decrease in Fig. 4 under increasing PBS flow rates of 200, 400, or 800 µL/h. We found that fluid exchange occurs 567-fold faster (SI Appendix, Table 1) than diffusion under the highest flow rate of 800 µL/h. We rationalized this with the convection–diffusion equation, assuming a constant diffusion coefficient: , where represents the porosity of the spheroid, c represents the concentration of the RIMS inside the spheroid, and represents the fluid velocity. When flow is absent, the second term is eliminated, and the movement of media is dominated by diffusion. With flow, the advection term enhances media exchange, and this can be accelerated by increasing the flow velocity within the device. This decrease in tissue-clearing time within a microfluidic system will enable researchers to develop high-throughput screening assays on 3D tissue models.
Fig. 4.
Complete tissue transparency is accelerated by microfluidic technology. Plotted here is the inverted half-life of fluorescence decay within B16F10 melanoma tumor spheroids. The fluorescence signal depends on the interstitial fluids, with high fluorescence occurring when the refractive index is matched by RIMS within the tissue. The decay of fluorescence signal was measured as RIMS was purged by infused PBS. (A) As the flow rate of PBS increases, the fluid exchange within the tissues is accelerated, thus decreasing the half-life of fluorescence decay as RIMS is purged by PBS. Flow assistance shrinks the clearing process from a week to an afternoon. (B) Fluorescence signal from B16F10 cells decreases rapidly over 7 min as RIMS is replaced by PBS. Red: tdTomato fluorescence signal (excited 543 nm, emission 552–691 nm). (Scale bar, 100 µm.) See SI Appendix, Fig. S4B for fluorescence decay curves. See Movie S9 to watch the fluorescence decay.
Proof-of-Concept Biological Applications of Microfluidic Clearing System.
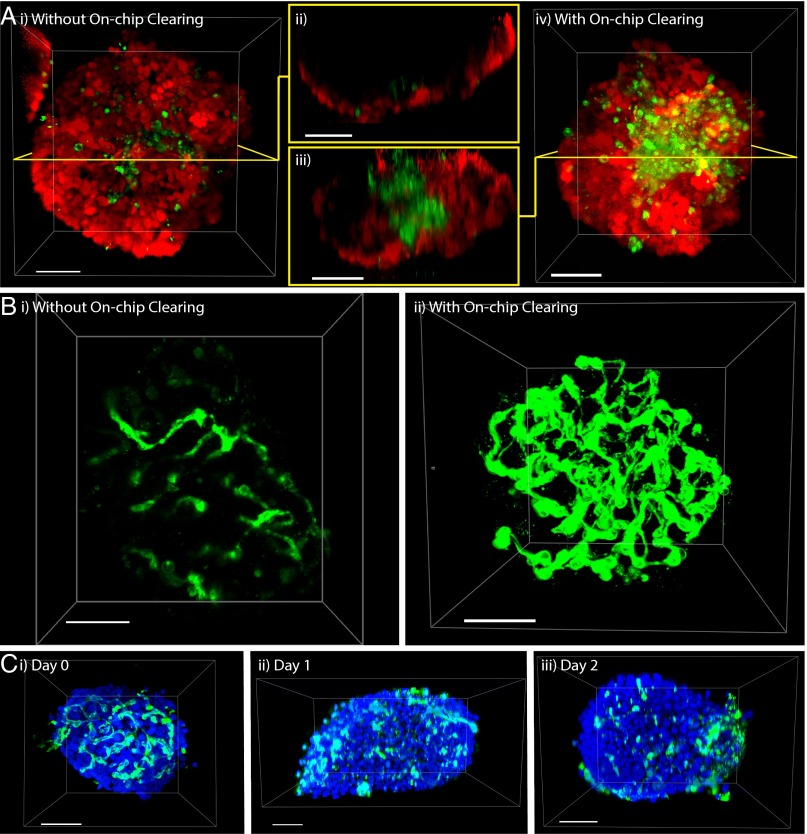
Our microfluidic system empowers researchers with the ability to collect complete optical data from 3D microtissues in their intact form. This is especially important for heterogeneous tissues which may have interior multicellular structures that are only revealed after our microfluidic technique is applied. We demonstrate this concept by creating a hybrid tumor spheroid from a mixture of B16F10 melanoma cancer cells and human umbilical vein endothelial cells (HUVECs, expressing GFP). After our microfluidic clearing technique was applied, we discovered that the HUVECs gathered in the core of the spheroid, whereas the B16F10 cells remained along the periphery (Fig. 5A). This heterogeneity cannot be observed without our microfluidic process (Fig. 5 A, i and ii and Movie S10), and was revealed after our on-chip clearing process was applied to the spheroids (Fig. 5 A, iii and iv and Movie S11). This segregation behavior was unexpected because the cells were initially homogeneously mixed before spheroid generation (SI Appendix, Fig. S5A). We found that the formation of the HUVEC core is not universal and varies across other cancer cell lines (SI Appendix, Fig. S5 B and C). A possible explanation for this behavior is that HUVECs are attracted by hypoxia-induced factors that may be secreted by internal cancer cells. Alternatively, this could be evidence to support the differential adhesion hypothesis (28), which posits that cells with similar adhesive strength will preferentially adhere together to reduce surface tension. In any case, our ability to probe this heterogeneity will become more important as tissue engineers develop more complex models to mirror patient tumor variability.
Fig. 5.
Visualization and monitoring of tissue heterogeneity and vascularity. Hybrid spheroids generated from a mixture of HUVECs [green, expressing GFP, excited (ex.) 488 nm, emission (em.) 493–536 nm] and B16F10 melanoma cancer cells (red, expressing tdtomato, ex. 543 nm, em. 565–702 nm), form a core–shell structure that is (A, i and ii) undetectable before on-chip clearing (see Movie S10) and appears in A, iii and iv after on-chip clearing (see Movie S11). A, ii and iii show cross-sectional views, before and after on-chip clearing. The same spheroid is shown here, before and after on-chip clearing, and all images were taken with the same objective lens (Objective Plan-Apochromat 20x/0.8 M27 - 420650–9901 from Zeiss). (Scale bar, 100 µm.) (B) Islets of Langerhans were extracted from C57BL/6 mice, stained on-chip with Lectin-555 (ex. 543 nm, em. 549–697 nm), and rendered transparent. Shown here is the same islet: (B, i) before treatment (see Movie S12); (B, ii) after treatment (see Movie S13). In C, the islet vasculature (stained with Lectin-647, shown in green – ex. 633 nm, em. 663–755 nm) is shown to degrade over the course of 2 d, a conclusion that cannot be drawn from conventional imaging: (C, i) day 0; (C, ii) day 1; (C, iii) day 2. Day 0 islet is shown in Movie S14; day 1 is shown in Movie S15; day 2 is shown in Movie S16. Blue, SytoxGreen staining of nucleus, ex. 488 nm, em. 500–551 nm. (Scale bar, 50 µm.)
We further demonstrate another application of our technique by examining the cytotoxic effects of small-molecule drugs on 3D tissue structures. We statically incubated B16F10 spheroids for 3 h in complete media supplemented with 180 µM of anticancer drugs doxorubicin, imatinib, or sunitinib. Spheroids were removed to fresh media and then loaded onto the microfluidic device. To quantify the extent of cellular death, a near-IR live/dead stain (Life Technologies) was infused into the device at 400 µL/h for 40 min. Spheroids were then imaged with confocal microscopy, and the extent of cellular death was compared against untreated spheroids. SI Appendix, Fig. S6 shows that the anticancer activity is greatest for sunitinib, followed by imatinib, then by doxorubicin. Whereas sunitinib showed complete cell death in the entire spheroid, spheroids treated with imatinib showed cellular death mainly in the periphery of the microtissue, and doxorubicin-treated spheroids showed no difference compared with the untreated spheroid controls. This proof-of-concept study provides evidence that our approach can characterize and discriminate between the cytotoxic effects of different small-molecule drugs on biological structures. Further work will be required to explore the limits of our technique for drug screening on 3D tissues, including dose–response studies, and refinements to tissue structure analysis methods.
Finally, we demonstrate a third application of our system to visualize the vasculature of pancreatic islets that were extracted from C57BL/6 mice, because these biological features are finer, more organized, and vary over a longer range than the features seen in B16F10/HUVEC hybrid spheroids. In addition, the health of the islet vasculature may have implications on the efficiency (29) and overall success rate of transplantation, a proposed method for treating type 1 diabetes (30–32). We infused AlexaFluor 555-labeled GSL-1 lectin to stain the vasculature before our on-chip clearing process and found that the majority of the islet vasculature cannot be optically captured if on-chip clearing is omitted (Fig. 5 B, i and Movie S12). With a complete picture of the islet (seen in Fig. 5 B, ii and Movie S13), it is now possible to accurately determine 3D characteristics such as vessel connectivity, spatial distribution, and total vasculature volume that may be critical for tissue engineering and regenerative medicine.
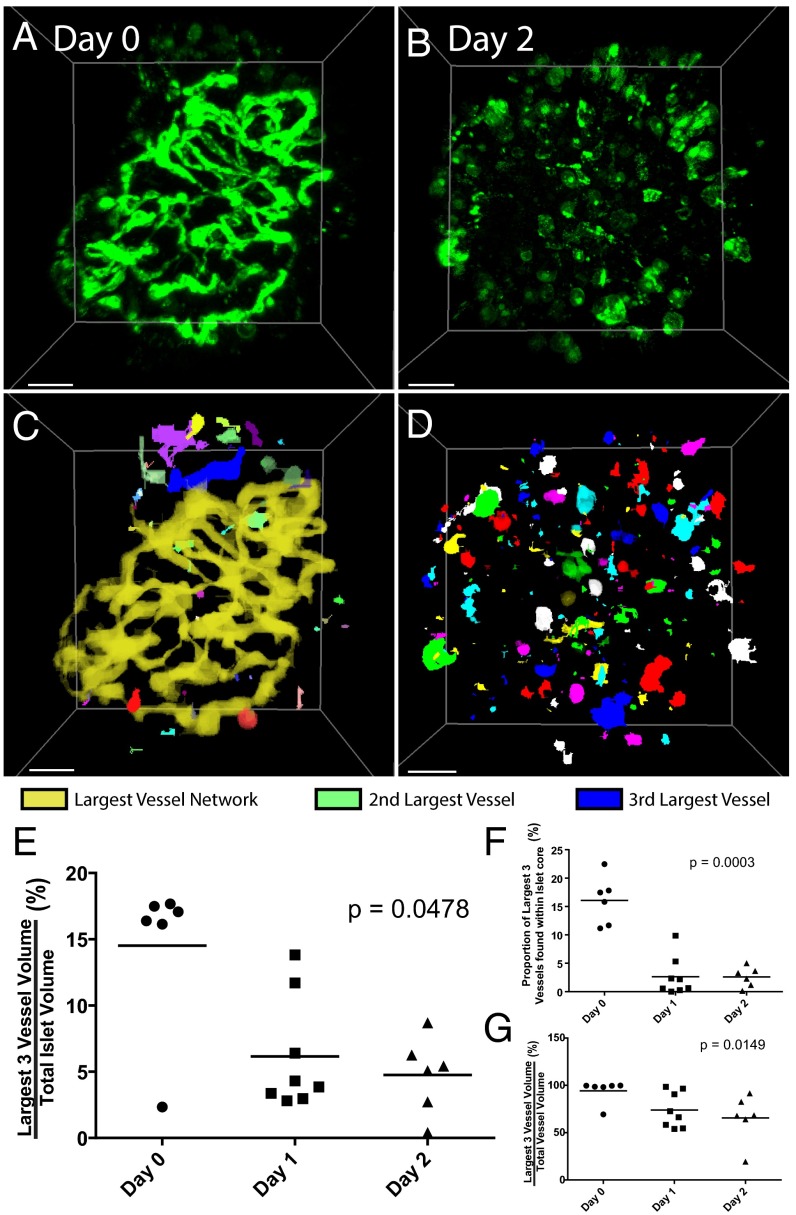
To use these images to draw conclusions from high-throughput screening assays, we still require an unbiased method to quantify the signals that we obtain after our on-chip process. Therefore, we wrote a MATLAB script to analyze the pancreatic islet vasculature, which has been previously shown to degrade rapidly upon extraction from the donor (33). To track this degradation (shown in Fig. 5C), islets were extracted and incubated in static culture within a Petri dish over a total period of 2 d. Islets were taken from the Petri dish on day 0 (extraction day), day 1, or day 2 of static incubation, and subjected to our on-chip staining and clearing process. Islets on these three days are rotated in Movie S14, Movie S15, and Movie S16, respectively. After the islets are imaged with confocal microscopy, our MATLAB script applies a local threshold to the image, converts it to a binary format, and analyzes the existence of a joined neighbor to each pixel to automatically determine the vessel networks within islets (as shown in Fig. 6 A–D). Each network is assigned a label, and its properties, including volume, surface area, or mean fluorescent intensity, are measured using the original image. We found that vessel volume was 17% of total islet volume on day 0, and dramatically decreased to below 6% and 5% within 24 and 48 h, respectively (Fig. 6E). Through our quantification, we determined that the proportion of the three largest vessel networks within the islet core is 16% on Day 0, and drops to 3% on subsequent days (Fig. 6F), suggesting that the majority of the blood vessels that persist after 24 h are not located within the core. From this, we can conclude that degradation begins in the core of the islet. The overall decrease in vessel connectivity shown in Fig. 6G indicates that the vasculature is fragmenting before the endothelial cells die completely. Our on-chip clearing technique and the subsequent image analysis can both be automated, making it possible to develop high-throughput screening assays to probe pro- or antiangiogenic drugs and draw accurate conclusions from a complete data set.
Fig. 6.
Automated image analysis of islet vasculature. (A) Raw images of pancreatic islets imaged on day 0 and (B) day 2. (Scale bar, 30 µm.) Lectin-647, shown in green (excited 633 nm, emission 663–755 nm). (C and D) Images generated from the MATLAB script, where each interconnected vessel network is assigned a specific color. In D, vasculature was so fragmented that colors were reused to assign vessel networks. (E) Graph depicting the amount of volume that the three largest vessel networks occupy within each islet, which decreases as total vasculature dies over time. (F) Graph shows the proportion of the three largest surviving vessel networks that are in the islet core. The decrease indicates vasculature death starts in the islet core. (G) Graph shows how much of the remaining vasculature comprises the three largest networks, an indication of the degree of connectivity of the three largest networks. The decrease over time indicates that vessels are fragmenting as endothelial cells die.
Conclusion
High-throughput screening with 3D microtissues will allow researchers to screen drugs for biological outcomes in features that are not present in 2D assays. Many cellular responses are dependent on their immediate surroundings, and these characteristics are lost in 2D monolayer cultures. However, high-throughput screening with 3D tissues is currently impossible, because conventional techniques to assess 3D structures are either off-chip or fall victim to scattering interfaces present within the tissue. We have developed a method that enables high-throughput, deep-tissue imaging with high numbers of statistical replicates without the need to aggregate and average the data collected from individual tissues. With access to signals from the entire microtissue, it is now possible to develop high-throughput assays that screen drugs for their effects on multicellular biological features, such as vasculature, necrosis, hypoxia, or region-dependent protein expression. Internal proteins of interest can be labeled and identified, and their spatial location within tissues may hint at their function within the larger 3D system. Other assays will enable tissue engineers to test the efficacy of their scaffolds, or chemists to screen the potency of their drugs in a model that more closely resembles the in vivo environment. Our microfluidic process has the potential to be automated and integrated into a wide variety of other applications outside of high-throughput screening or drug discovery.
Materials and Methods
Microtissues were loaded via gravity flow, and confocal images were taken with a Zeiss LSM 710 microscope. The monomer solution was infused at 600–800 µL/h (200 µL/h for islets) for 20 min, then incubated at 37 °C for 2 h to gel the monomer. A brief PBS rinse (iBMHH for islets) was gently flushed through the device to restart flow. After monomer infusion and gelation, spheroids in the microfluidic device were infused with 8% clearing solution for 7–10 min at 600–800 µL/h, followed by a 20-min wash with PBS. This step is optional, and is only needed for staining intracellular targets of interest. Microtissues are rendered transparent when infused with RIMS at 800 µL/h (for tumor spheroids), or at 400 µL/h (for murine islets) for 10–15 min. Confocal images were then taken and then fed through the MATLAB script for analysis. See SI Appendix for more details regarding materials and methods.
Supplementary Material
Acknowledgments
We thank Dr. Steven Proulx and Dr. Michael Detmar for donating the tdTomato-expressing B16F10 melanoma cell line, Dr. Gang Zheng for donating the GFP-expressing U87MG cell line, and Dr. Kathleen Kelly for donating the tdTomato-expressing MDA-MB-231 cell line. We acknowledge the Canadian Institute of Health Research (CIHR) (Grants MOP 130143 and GCS105653-1) and Natural Sciences and Engineering Research Council (NSERC) (Grant RGPIN288231) for funding support for the project. Y.Y.C., P.N.S., and A.M.S. acknowledge NSERC for student fellowships. S.S. acknowledges CIHR for student fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609569114/-/DCSupplemental.
References
- 1.Mordwinkin NM, Burridge PW, Wu JC. A review of human pluripotent stem cell-derived cardiomyocytes for high-throughput drug discovery, cardiotoxicity screening, and publication standards. J Cardiovasc Transl Res. 2013;6(1):22–30. doi: 10.1007/s12265-012-9423-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Held MA, et al. Genotype-selective combination therapies for melanoma identified by high-throughput drug screening. Cancer Discov. 2013;3(1):52–67. doi: 10.1158/2159-8290.CD-12-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Astashkina A, Mann B, Grainger DW. A critical evaluation of in vitro cell culture models for high-throughput drug screening and toxicity. Pharmacol Ther. 2012;134(1):82–106. doi: 10.1016/j.pharmthera.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Hirschhaeuser F, et al. Multicellular tumor spheroids: An underestimated tool is catching up again. J Biotechnol. 2010;148(1):3–15. doi: 10.1016/j.jbiotec.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Goodman TT, Ng CP, Pun SH. 3-D tissue culture systems for the evaluation and optimization of nanoparticle-based drug carriers. Bioconjug Chem. 2008;19(10):1951–1959. doi: 10.1021/bc800233a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu LY, Di Carlo D, Lee LP. Microfluidic self-assembly of tumor spheroids for anticancer drug discovery. Biomed Microdevices. 2008;10(2):197–202. doi: 10.1007/s10544-007-9125-8. [DOI] [PubMed] [Google Scholar]
- 7.Hsiao AY, et al. Microfluidic system for formation of PC-3 prostate cancer co-culture spheroids. Biomaterials. 2009;30(16):3020–3027. doi: 10.1016/j.biomaterials.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wlodkowic D, Cooper JM. Tumors on chips: Oncology meets microfluidics. Curr Opin Chem Biol. 2010;14(5):556–567. doi: 10.1016/j.cbpa.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Albanese A, Lam AK, Sykes EA, Rocheleau JV, Chan WC. Tumour-on-a-chip provides an optical window into nanoparticle tissue transport. Nat Commun. 2013;4:2718. doi: 10.1038/ncomms3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pampaloni F, Ansari N, Stelzer EHK. High-resolution deep imaging of live cellular spheroids with light-sheet-based fluorescence microscopy. Cell Tissue Res. 2013;352(1):161–177. doi: 10.1007/s00441-013-1589-7. [DOI] [PubMed] [Google Scholar]
- 11.Bersini S, et al. A microfluidic 3D in vitro model for specificity of breast cancer metastasis to bone. Biomaterials. 2014;35(8):2454–2461. doi: 10.1016/j.biomaterials.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajadhyaksha M, Grossman M, Esterowitz D, Webb RH, Anderson RR. In vivo confocal scanning laser microscopy of human skin: Melanin provides strong contrast. J Invest Dermatol. 1995;104(6):946–952. doi: 10.1111/1523-1747.ep12606215. [DOI] [PubMed] [Google Scholar]
- 13.Shotton DM. Confocal scanning optical microscopy and its applications for biological specimens. J Cell Sci. 1989;94(2):175–206. [Google Scholar]
- 14.Cavanagh HD, et al. Clinical and diagnostic use of in vivo confocal microscopy in patients with corneal disease. Ophthalmology. 1993;100(10):1444–1454. doi: 10.1016/s0161-6420(93)31457-0. [DOI] [PubMed] [Google Scholar]
- 15.Hell S, Reiner G, Cremer C, Stelzer EHK. Aberrations in confocal fluorescence microscopy induced by mismatches in refractive index. J Microsc. 1993;169(3):391–405. [Google Scholar]
- 16.Chung K, Deisseroth K. CLARITY for mapping the nervous system. Nat Methods. 2013;10(6):508–513. doi: 10.1038/nmeth.2481. [DOI] [PubMed] [Google Scholar]
- 17.Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc. 2014;9(7):1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung K, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497(7449):332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang B, et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158(4):945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sindhwani S, et al. Three-dimensional optical mapping of nanoparticle distribution in intact tissues. ACS Nano. 2016;10(5):5468–5478. doi: 10.1021/acsnano.6b01879. [DOI] [PubMed] [Google Scholar]
- 21.Kim S-Y, et al. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc Natl Acad Sci USA. 2015;112(46):E6274–E6283. doi: 10.1073/pnas.1510133112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Epp JR, et al. Optimization of CLARITY for clearing whole-brain and other intact organs. eNeuro. 2015;2(3):ENEURO.0022–15.2015. doi: 10.1523/ENEURO.0022-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivascu A, Kubbies M. Diversity of cell-mediated adhesions in breast cancer spheroids. Int J Oncol. 2007;31(6):1403–1413. [PubMed] [Google Scholar]
- 24.Silva PN, et al. Highly efficient adenoviral transduction of pancreatic islets using a microfluidic device. Lab Chip. 2016;16(15):2921–2934. doi: 10.1039/c6lc00345a. [DOI] [PubMed] [Google Scholar]
- 25.El-Gohary Y, et al. Three-dimensional analysis of the islet vasculature. Anat Rec (Hoboken) 2012;295(9):1473–1481. doi: 10.1002/ar.22530. [DOI] [PubMed] [Google Scholar]
- 26.Choi NW, et al. Microfluidic scaffolds for tissue engineering. Nat Mater. 2007;6(11):908–915. doi: 10.1038/nmat2022. [DOI] [PubMed] [Google Scholar]
- 27.Hsu Y-H, et al. Full range physiological mass transport control in 3D tissue cultures. Lab Chip. 2013;13(1):81–89. doi: 10.1039/c2lc40787f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foty RA, Steinberg MS. The differential adhesion hypothesis: A direct evaluation. Dev Biol. 2005;278(1):255–263. doi: 10.1016/j.ydbio.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Brissova M, Powers AC. Revascularization of transplanted islets: Can it be improved? Diabetes. 2008;57(9):2269–2271. doi: 10.2337/db08-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phelps EA, Headen DM, Taylor WR, Thulé PM, García AJ. Vasculogenic bio-synthetic hydrogel for enhancement of pancreatic islet engraftment and function in type 1 diabetes. Biomaterials. 2013;34(19):4602–4611. doi: 10.1016/j.biomaterials.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruni A, Gala-Lopez B, Pepper AR, Abualhassan NS, Shapiro AJ. Islet cell transplantation for the treatment of type 1 diabetes: Recent advances and future challenges. Diabetes Metab Syndr Obes. 2014;7:211–223. doi: 10.2147/DMSO.S50789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapiro AMJ. Islet transplantation in type 1 diabetes: Ongoing challenges, refined procedures, and long-term outcome. Rev Diabet Stud. 2012;9(4):385–406. doi: 10.1900/RDS.2012.9.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nyqvist D, Köhler M, Wahlstedt H, Berggren PO. Donor islet endothelial cells participate in formation of functional vessels within pancreatic islet grafts. Diabetes. 2005;54(8):2287–2293. doi: 10.2337/diabetes.54.8.2287. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.