Significance
The p53 transcription factor is stabilized in response to cellular stress and regulates the expression of genes involved in numerous biological activities, thereby suppressing tumorigenesis. DNA damage and other stress signals upregulate p53, in part, by freeing p53 from negative regulation imposed by the Mdm2 and MdmX (Mdm4) oncoproteins. MDM proteins are subject to posttranslational modification, and accumulating evidence indicates that phosphorylation of Mdm2 by different stress-activated kinases such as ATM or c-Abl alters Mdm2-p53 signaling and profoundly affects p53 function. A better understanding of the in vivo effects of Mdm2 phosphorylation may facilitate the development of novel therapeutics capable of stimulating p53 antitumor activity or alleviating p53-dependent toxicities in nonmalignant tissues.
Keywords: c-Abl, Mdm2, DNA damage, p53, tumorigenesis
Abstract
The p53 tumor suppressor acts as a guardian of the genome by preventing the propagation of DNA damage-induced breaks and mutations to subsequent generations of cells. We have previously shown that phosphorylation of the Mdm2 oncoprotein at Ser394 by the ATM kinase is required for robust p53 stabilization and activation in cells treated with ionizing radiation, and that loss of Mdm2 Ser394 phosphorylation leads to spontaneous tumorigenesis and radioresistance in Mdm2S394A mice. Previous in vitro data indicate that the c-Abl kinase phosphorylates Mdm2 at the neighboring residue (Tyr393) in response to DNA damage to regulate p53-dependent apoptosis. In this present study, we have generated an Mdm2 mutant mouse (Mdm2Y393F) to determine whether c-Abl phosphorylation of Mdm2 regulates the p53-mediated DNA damage response or p53 tumor suppression in vivo. The Mdm2Y393F mice develop accelerated spontaneous and oncogene-induced tumors, yet display no defects in p53 stabilization and activity following acute genotoxic stress. Although apoptosis is unaltered in these mice, they recover more rapidly from radiation-induced bone marrow ablation and are more resistant to whole-body radiation-induced lethality. These data reveal an in vivo role for c-Abl phosphorylation of Mdm2 in regulation of p53 tumor suppression and bone marrow failure. However, c-Abl phosphorylation of Mdm2 Tyr393 appears to play a lesser role in governing Mdm2-p53 signaling than ATM phosphorylation of Mdm2 Ser394. Furthermore, the effects of these phosphorylation events on p53 regulation are not additive, as Mdm2Y393F/S394A mice and Mdm2S394A mice display similar phenotypes.
The significant role of p53 in human tumor suppression is evidenced by the fact that p53 is either mutated or functionally inactive in over 50% of human cancers (1). The tumor suppressive activity of p53 has been classically attributed to p53-dependent cellular responses of growth arrest and apoptosis in response to various stresses, although increasing evidence has implicated additional p53-target genes involved in regulating further cellular processes such as metabolic functions and DNA repair (2, 3). Stress-induced p53 responses are preceded by a profound increase in p53 protein levels and transcriptional activity. Accordingly, understanding the signaling events that lead to p53 stabilization and transcriptional activation has been the focus of extensive research. In order for p53 levels and activity to increase in the damaged cell, p53 must be relieved of the negative regulation imposed by the MDM oncoproteins, Mdm2 and MdmX.
Regulation of the DNA damage response (DDR) in mammals is governed by the PI3K-related ATM and ATR kinases. Activation of these transducer kinases depends on the type and amount of DNA damage and triggers the direct or indirect phosphorylation of numerous downstream proteins involved in the DDR (4, 5). ATM is activated primarily by double-strand breaks (DSBs), and its numerous target substrates include p53, Mdm2, and MdmX (6–10). We have previously reported the generation of a mouse model (Mdm2S394A) in which ATM phosphorylation of Mdm2 at serine residue 394 (Ser395 in human MDM2) was abolished (11). Cells and tissues of Mdm2S394A mice display profound defects in DNA damage-induced p53 protein stabilization and transcriptional activation. The diminished p53 response in these animals resulted in reduced p53-dependent apoptosis in hematopoietic tissues, radioresistance, and increased spontaneous tumorigenesis. These findings underscore that Mdm2 phosphorylation is a critical event in regulating Mdm2-p53 signaling and the induction of p53 activity during the DDR and in homeostatic tissues. However, Mdm2S394A mice display some p53 stabilization and activity following DNA damage and do not fully phenocopy p53−/− mice. This led us to examine whether the phosphorylation of additional Mdm2 residues contributes to p53 induction following DNA damage.
Intriguingly, the tyrosine residue immediately preceding Ser395 in human MDM2, Tyr394 (Tyr393 in mouse Mdm2), has been shown to be phosphorylated by the tyrosine kinase c-Abl (12, 13). Similar to ATM and ATR, c-Abl is activated by a variety of DNA damaging agents (14–16). Previous overexpression studies in cell lines indicate that c-Abl promotes growth arrest in a p53-dependent manner and apoptosis by both p53-dependent and independent mechanisms (17, 18). Furthermore, c-Abl can protect p53 from MDM2-mediated degradation, and c-Abl phosphorylation of MDM2 overcomes the inhibitory effect of MDM2 on p53 transcriptional activity and apoptosis (19). In addition, studies using c-abl−/− mouse embryonic fibroblasts (MEFs) indicate that c-Abl is required for maximal p53 accumulation in response to ionizing radiation (IR), doxorubicin, or mitomycin C treatment, and that coexpression of c-Abl overcomes MDM2-mediated ubiquitination and nuclear export of p53 (20). c-Abl phosphorylates MDM2 Tyr394 as well as Tyr276 and Tyr405 (12, 13), and c-Abl phosphorylation of MDM2 Tyr394 impairs the ability of MDM2 to inhibit p53 stabilization and transactivation and p53-mediated apoptosis (12). More recently, it was proposed that c-Abl phosphorylation of MDM2 increases MDM2–MDMX binding and promotes MDM2-directed MDMX ubiquitination, and that this ultimately destabilizes the MDM2–MDMX complex, promoting p53 stabilization (21).
As we have shown that ATM phosphorylation of Mdm2 Ser394 profoundly impacts the p53 response to DNA damage in mice, we sought to determine whether c-Abl phosphorylation of Mdm2 Tyr393 similarly regulates p53 functions in vivo. To this end, we generated a knockin mouse model in which Mdm2 Tyr393 is substituted with phenylalanine (Mdm2Y393F), as well as a mouse in which both the c-Abl target residue Mdm2 Tyr393 and the adjacent ATM target residue Mdm2 Ser394 are mutated (Mdm2Y393F/S394A), enabling the study of whether phosphorylation of these residues has additive or redundant effects.
Results
Mdm2Y393F Mice Are Viable and Display Increased Spontaneous and Oncogene-Induced Tumorigenesis.
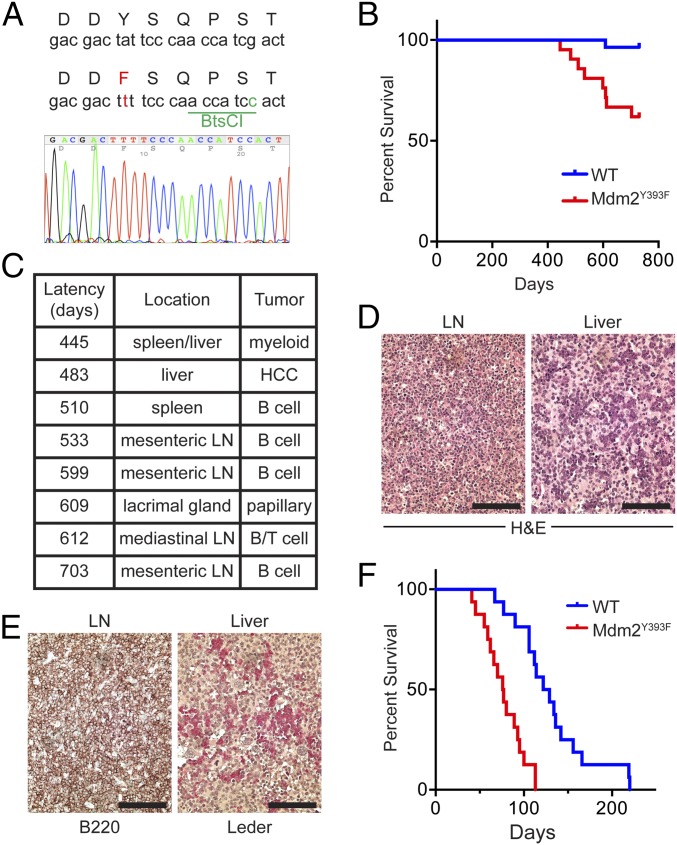
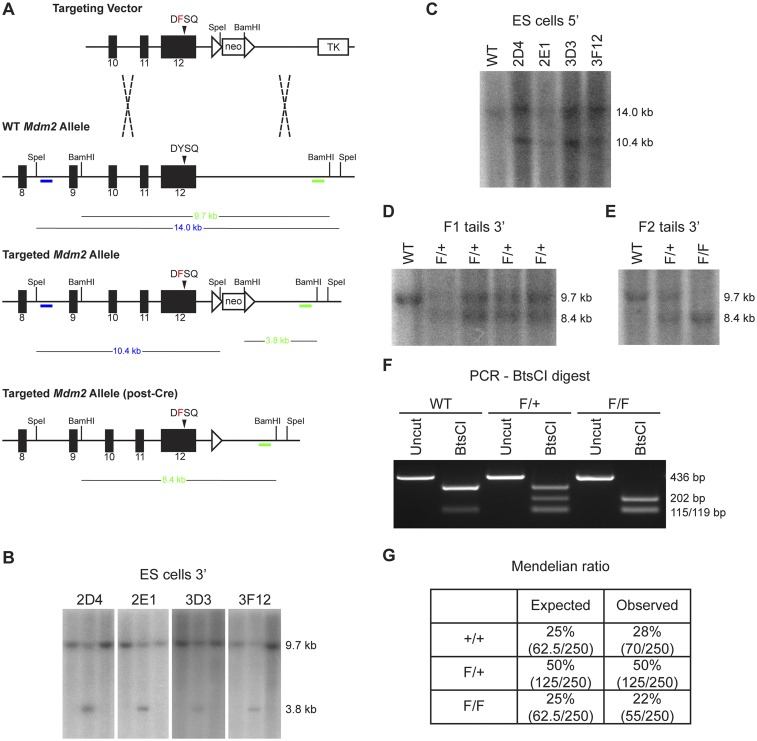
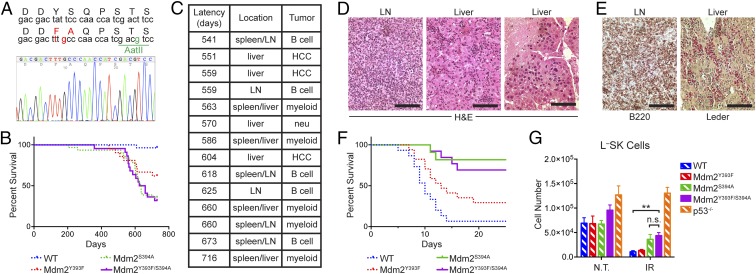
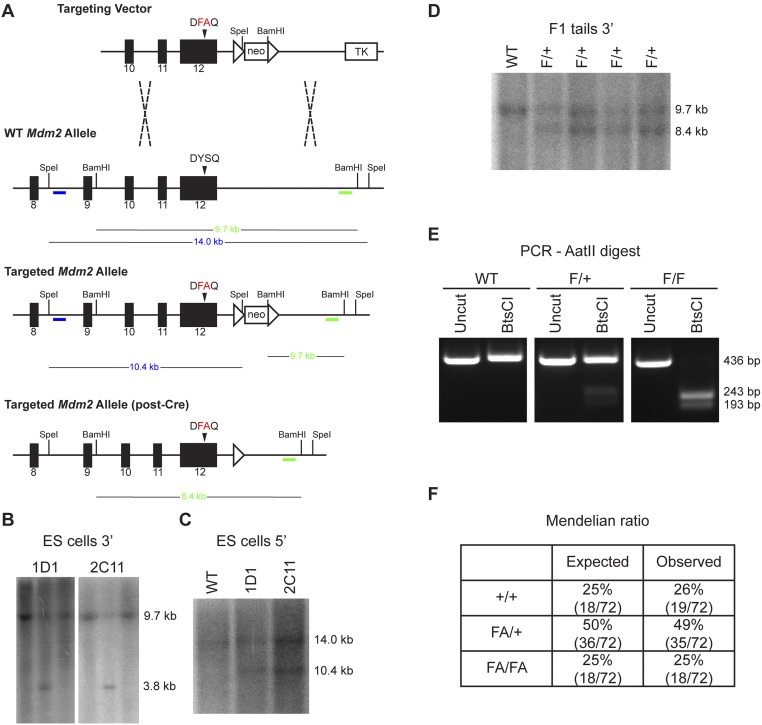
To investigate the role of Mdm2 Tyr393 phosphorylation under physiological conditions, we generated a mouse model in which this tyrosine residue is substituted with a phenylalanine residue (Y393F). Site-directed mutagenesis was performed to introduce an A-to-T missense mutation in the 393 codon, and a synonymous G-to-C mutation in the 397 codon of Mdm2 exon 12 (Fig. 1A). A gene-replacement vector was constructed to replace the endogenous Mdm2 exon 12 sequences with the mutated exon 12 (Fig. S1A). Gene targeting was performed in PC3 (129SV) embryonic stem (ES) cells (22), and homologous recombination was confirmed in G418-resistant clones by Southern blotting (Fig. S1 A–C). Blastocyst injection of targeted ES clones produced several high-degree male chimeras that passed the Mdm2 Y393F allele through their germ line. Southern blotting further confirmed proper targeting in F1 and F2 generation mice, along with protamine-Cre–directed deletion of the floxed neomycin cassette (Fig. S1 D and E). The presence of the additional synonymous G-to-C mutation within the 397 codon of the targeted allele introduced a novel BtsCI restriction digest site that allowed for identification of the Mdm2Y393F allele by PCR-digest strategy (Fig. 1A and Fig. S1F). Mdm2 transcripts from spleens of Mdm2Y393F mice were sequenced and confirmed as containing only the targeted mutations (Fig. 1A). Heterozygous intercrosses yielded homozygous Mdm2Y393F mice at Mendelian ratios, indicating that Mdm2Y393F is not compromised in its function during development (Fig. S1G). Additionally, no differences were observed in average litter size, body weights at 6 wk of age, or male-to-female sex distribution.
Fig. 1.
Mdm2Y393F mice are viable and display increased spontaneous and oncogene-induced tumorigenesis. (A) DNA sequence of WT and Mdm2Y393F allele surrounding codon 393 (Top). An A-to-T mutation changed codon 393 from Tyr to Phe. A silent G-to-C mutation in codon 397 introduced a BtsCI restriction site. Sequencing results of the corresponding region from Mdm2Y393F spleen cDNA are shown (Bottom). (B) Kaplan–Meier tumor-free survival curves of WT (n = 28) and Mdm2Y393F (n = 21) mice. Curves were compared by log-rank test: (P < 0.01). (C) Table displaying the latency, location, and tumor type for spontaneous tumors arising in Mdm2Y393F mice. LN, lymph node; HCC, hepatocellular carcinoma. (D) Representative H&E-stained tissue sections of a LN exhibiting a B-cell lymphoma (Left) and myeloid sarcoma present in the liver sinusoids (Right). (Scale bars, 100 μm.) (E) The B-cell lymphomas show expression of B220 (Left), and the myeloid neoplasms exhibit chloroacetate esterase (Leder) staining (Right). (Scale bars, 100 μm.) (F) Kaplan–Meier tumor-free survival curves of Eμ-Myc (n = 16) and Eμ-Myc;Mdm2Y393F (n = 16) mice. Median survival times were as follows: Eμ-Myc (125.5 d), Eμ-Myc;Mdm2Y393F (76.5 d). Curves were compared by log-rank test (P < 0.0001).
Fig. S1.
(A) Diagram of the targeting strategy used to generate the Mdm2Y393F allele. The targeting vector contains a floxed neomycin cassette (neo) and a 3′ thymidine kinase cassette (TK) for positive and negative drug selection, respectively. The targeting vector was linearized and electroporated into PC3 ES cells. ES cell targeting was confirmed by BamHI digest and Southern blot analysis with a 3′ external probe (green), as well as SpeI digest and Southern blot analysis with a 5′ external probe (blue). Protamine-Cre recombinase-directed excision of neo cassette was confirmed by BamHI digest of F1 and F2 generation tail DNA and Southern blot analysis with the same 3′ external probe as in ES cells (green). (B) BamHI digest and Southern blot analysis of ES cell genomic DNA. A 3′ external probe (green bar from A) detects a 9.7-kb WT allele and 3.8-kb targeted allele. Four clones with correct 3′ targeting are shown, each flanked by untargeted (WT) clones. (C) SpeI digest and Southern blot analysis of ES cell genomic DNA. A 5′ external probe (blue bar from A) detects a 14.0-kb WT allele and 10.4-kb targeted allele. (D) BamHI digest and Southern blot analysis of tail DNA from F1 generation mice. A 3′ external probe (green bar from A) detects a 9.7-kb WT allele and 8.4-kb targeted allele. (E) BamHI digest and Southern blot analysis of tail DNA from F2 generation mice. A 3′ external probe (green bar from A) detects a 9.7-kb WT allele and 8.4-kb targeted allele. (F) PCR-BtsCI digest analysis of F2 generation mice. Primers flanking Mdm2 codon 393 amplify a 436-bp fragment of DNA. The WT Mdm2 fragment contains a single BtsCI restriction site that generates 119- and 317-bp fragments. A silent mutation in the Mdm2Y393F allele introduces an additional BtsCI restriction site that results in 115-, 119-, and 202-bp fragments. (G) The expected and observed Mendelian ratios from Mdm2Y393F heterozygote breeding.
We have previously shown that mice deficient for phosphorylation of the neighboring residue (Mdm2 Ser394) by ATM are prone to spontaneous tumorigenesis (11). Therefore, we sought to examine whether c-Abl phosphorylation of Mdm2 Tyr393 impacted tumor suppression. Cohorts of WT and Mdm2Y393F mice were established and monitored for tumor formation. During the 24-mo tumor assay, 8 of 21 (38%) Mdm2Y393F mice developed spontaneous tumors (Fig. 1B), whereas only 1 of 28 (4%) WT mice presented with a tumor at 20 mo of age. Mdm2Y393F tumors arose between 14.5 and 24 mo of age, a similar latency as seen in Mdm2S394A mice (11). The majority of tumors arose in lymphatic tissues, and 5 of the 6 tumors (83%) were identified as B-cell lymphomas, with one of the B-cell lymphomas also containing atypical T cells. Other tumor types seen in the cohort included a myeloid sarcoma, a hepatocellular carcinoma, and a papillary tumor of lacrimal origin (Fig. 1 C–E).
As Mdm2Y393F mice were more prone to develop spontaneous tumors of lymphoid origin, we further examined the effects of Mdm2-Y393 phosphorylation on tumor suppression using the Eμ-Myc mouse model (23). Cohorts of Eμ-Myc and Eμ-Myc;Mdm2Y393F mice were generated (C57BL/6 background) and assayed for tumor formation. Eμ-Myc mice develop pre-B/B-cell lymphomas within 3–6 mo of age, and we observed a median time to tumorigenesis of 125.5 d in the Eμ-Myc model cohort (Fig. 1F), a duration in close agreement with previous studies (23, 24). However, Eμ-Myc;Mdm2Y393F mice displayed significantly accelerated tumorigenesis, with a median time to tumor formation of only 76.5 d. Thus, whereas dispensable for development, c-Abl phosphorylation of Mdm2-Y393 significantly impacts tumorigenesis by preventing the formation of both spontaneous and oncogene-induced tumors.
c-Abl Phosphorylation of Mdm2 Tyr393 Does Not Influence DNA Damage-Induced p53 Stabilization and Activation in Spleen and Thymus.
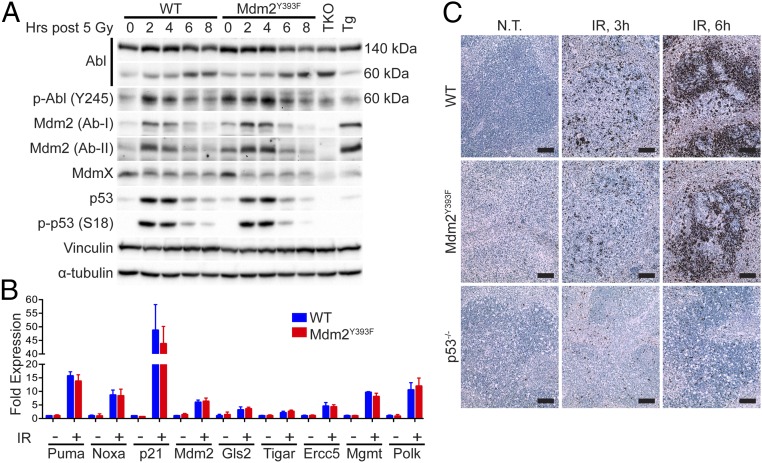
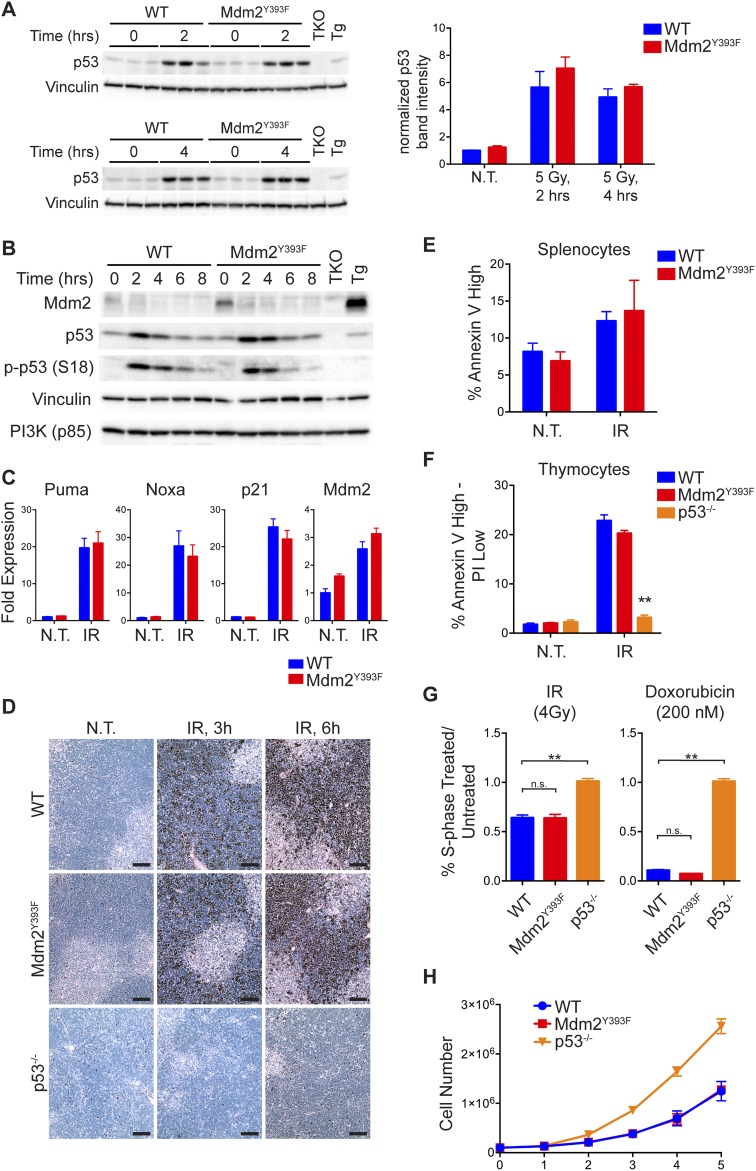
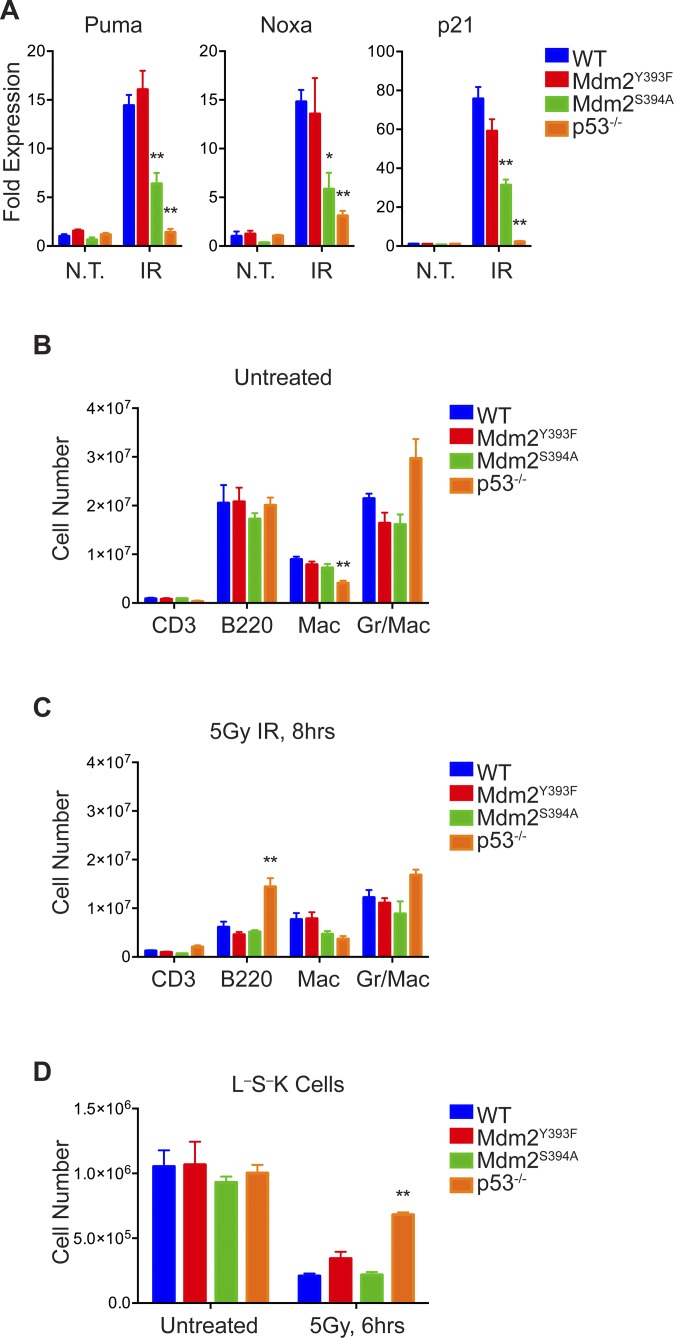
Having observed increased lymphomagenesis in Mdm2Y393F mice, and as c-Abl is activated by a variety of DNA damaging agents (14–16), we next examined whether c-Abl phosphorylation of Mdm2 Tyr393 impacts DNA damage-induced p53 stabilization and activation in lymphatic tissues of Mdm2Y393F mice. WT and Mdm2Y393F mice were exposed to 5 Gy IR and spleens were analyzed for p53 and Mdm2 levels by immunoblotting (Fig. 2A). In both genotypes, c-Abl cleavage was observed in response to IR, similar to previous observations in cells undergoing stress-induced apoptosis (25, 26). c-Abl phosphorylation at Tyr245 confirmed kinase activation. No qualitative differences were detected in basal or IR-induced p53 protein levels. Similarly, no obvious differences were observed in p53 activation as indicated by levels of p53-Ser18 phosphorylation. No differences were observed in the levels of Mdm2 protein, which increased at 2–4 h following IR, before decreasing as p53 activity diminished, similar to what has previously been reported in this tissue (11, 27). The lack of a difference in p53 stabilization was confirmed quantitatively in biological triplicates at 0, 2, and 4 h (Fig. S2A). Similar results were observed in thymi from these same animals, with no qualitative differences detected in the levels of total p53, phosphorylated p53 (S18), or Mdm2, save for slightly higher basal Mdm2 levels in Mdm2Y393F mice (Fig. S2B). Consistent with equal levels of basal and IR-induced p53 protein stabilization and phosphorylation, real-time quantitative PCR (RT-qPCR) performed on cDNA from untreated and irradiated spleens of WT and Mdm2Y393F mice detected no differences in the basal or DNA damage-induced expression levels of a selection of p53-target genes involved in growth arrest, apoptosis, DNA repair, or metabolic regulation (Fig. 2B). Similar results were observed in thymi (Fig. S2C).
Fig. 2.
c-Abl phosphorylation of Mdm2 Tyr393 does not influence DNA damage-induced p53 stabilization and activation in spleen and thymus. (A) WT and Mdm2Y393F mice were left untreated or exposed to 5 Gy IR and spleens were harvested at 2-h intervals. Protein levels were analyzed by Western blotting. TKO indicates Mdm2−/−, MdmX−/−, p53−/− triple knockout control; Tg indicates Mdm2Tg/+ Mdm2 overexpressing control. (B) Mice were left untreated or exposed to 5 Gy IR and spleens were harvested at 4 h. Fold expression of p53-target genes was determined by RT-qPCR, relative to untreated WT samples using Rplp0 as internal reference (n = 3–4, ± SEM). Significance was determined by Student’s t test of ΔΔCt values. (C) TUNEL staining of spleens of mice treated as in A and harvested at 3 and 6 h.
Fig. S2.
(A) Splenic protein levels in biological triplicates of WT and Mdm2Y393F mice left untreated or exposed to 5 Gy IR and harvested at 2 and 4 h were analyzed by Western blotting (Left). Band intensities were determined by densitometry. p53 levels were normalized for Vinculin levels and average values plotted (±SEM) (Right). (B) WT and Mdm2Y393F mice were left untreated or exposed to 5 Gy IR and thymi were harvested at 2-h intervals. Protein levels were analyzed by Western blotting. TKO indicates Mdm2−/−, MdmX−/−, p53−/− control; Tg indicates Mdm2Tg/+ Mdm2 overexpressing control. (C) WT and Mdm2Y393F mice were left untreated or exposed to 5 Gy IR and thymi were harvested at 4 h. Fold expression of p53-target genes was determined by RT-qPCR, relative to untreated WT samples using Rplp0 as internal reference (n = 3–4, ±SEM). Significance was determined by Student’s t test of ΔΔCt values. (D) TUNEL staining of thymi from WT, Mdm2Y393F, and p53−/− mice treated as in A and harvested at 3 and 6 h. (E) Quantification of apoptotic splenocytes by flow cytometry. WT and Mdm2Y393F mice were left untreated or irradiated with 5 Gy IR. Spleens were harvested 5 h later, and single cell suspensions of splenocytes were generated and stained with Annexin V-FITC for FACS analysis. The percentage of Annexin VHigh cells was quantified (n = 3, ±SEM). (F) Quantification of apoptotic thymocytes by flow cytometry. WT, Mdm2Y393F, and p53−/− mice were left untreated or irradiated with 5 Gy IR. Thymi were harvested 5 h later, and single cell suspensions of thymocytes were generated and stained with AnnexinV-FITC and propidium iodide (PI) for FACS analysis. The percentage of Annexin VHigh–PILow cells was quantified (n = 3, ±SEM). Significance was determined relative to similarly treated WT samples, **P < 0.01 (Student’s t tests). (G) Cell-cycle arrest analysis. MEFs were either left untreated or exposed to 4 Gy IR (Left) or 200 nM doxorubicin (Right) for 18 h. Cells were pulse labeled with 50 µM BrdU for the final 3 h before being harvested and fixed for FACS analysis. Fixed cells were stained with a FITC-conjugated anti-BrdU antibody and PI and analyzed by flow cytometry. Data are presented as the percentage of cells in S phase in the treated samples relative to untreated samples (n = 3, ±SEM). **P < 0.01 (Student’s t tests). (H) Primary MEF proliferation. The 106 cells were seeded in six-well plates and cell number per well was counted each day (n = 3 cell lines per genotype, ±SD).
Hematopoietic organs are highly radiosensitive, and p53 activation in these tissues triggers widespread apoptosis following IR (28). In keeping with the absence of observable differences in p53 stabilization and activation in Mdm2Y393F spleens and thymi, we detected no differences in splenic or thymic apoptosis in untreated and irradiated Mdm2Y393F mice by TUNEL (Fig. 2C and Fig. S2D). This observation was confirmed quantitatively by flow cytometry of Annexin V-FITC–stained splenocytes and thymocytes (Fig. S2 E and F). Thus, the IR-induced stabilization and activation of p53 and the resultant apoptotic response is unaltered in Mdm2Y393F spleens and thymi.
We also examined whether Mdm2 Tyr393 phosphorylation affected p53-dependent growth arrest. MEFs from WT, Mdm2Y393F, and p53−/− mice were untreated or exposed to either 5 Gy IR or 150 nM doxorubicin for 18 h. Flow cytometric cell-cycle analysis revealed no differences in growth arrest in Mdm2Y393F MEFs following either irradiation or treatment with doxorubicin (Fig. S2G). Furthermore, WT and Mdm2Y393F MEFs exhibited similar growth rates in standard cell proliferation assays (Fig. S2H).
Mdm2Y393F Mice Are Radioresistant and Display Enhanced Bone Marrow Repopulating Abilities Following IR Exposure.
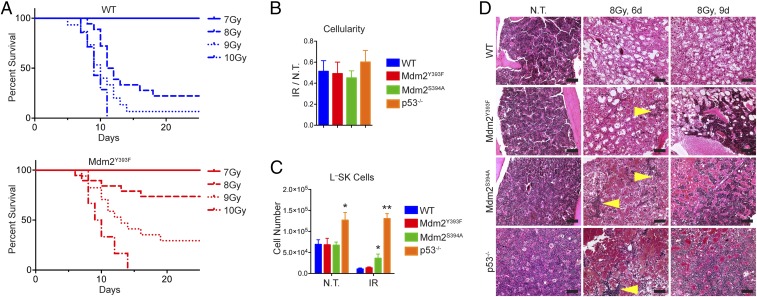
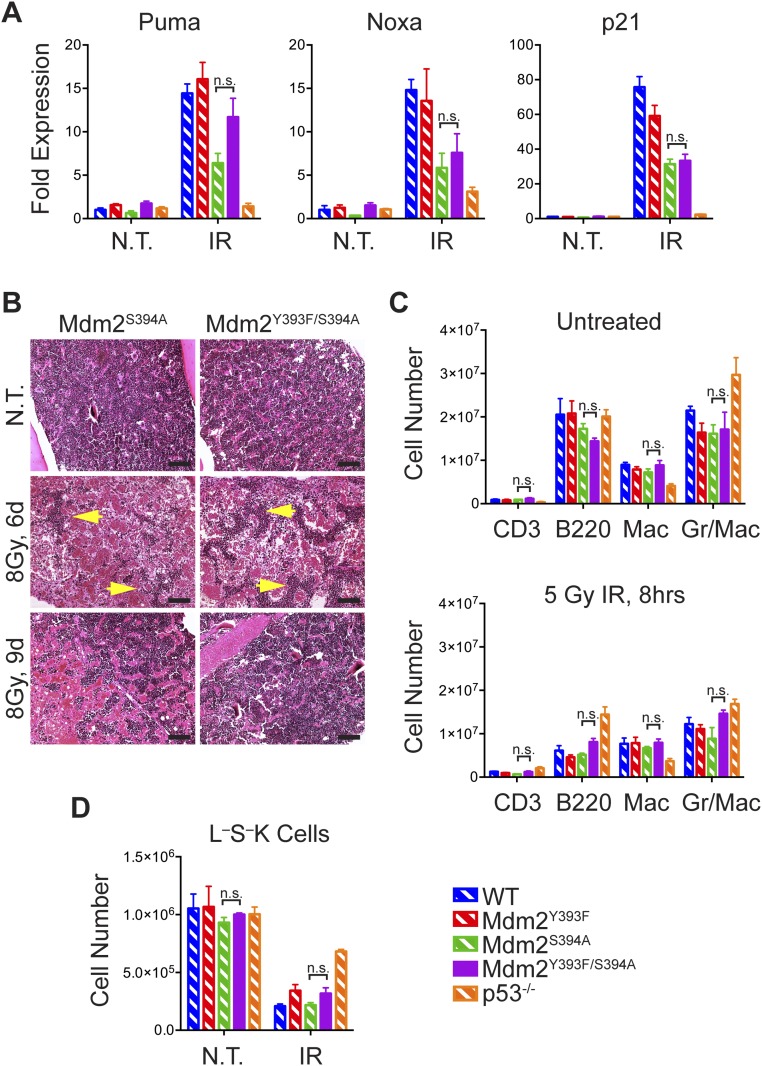
The proposed effects of c-Abl phosphorylation of Mdm2 on p53 are similar to those of ATM phosphorylation of Mdm2, namely p53 stabilization and activation. Conversely, our findings that DNA damage-induced p53 stabilization and activation in spleen and thymus, and p53-dependent apoptosis and growth arrest in Mdm2Y393F tissues and cells, are unaltered contrasts with what we have reported with Mdm2S394A mice. Thus, c-Abl’s phosphorylation of Mdm2-Y393 may either have limited effect on the p53-dependent DDR or may be adequately compensated for by ATM phosphorylation of Mdm2 Ser394. However, when we challenged cohorts of WT and Mdm2Y393F mice to a series of IR doses spanning the threshold-lethal range, we observed a significant resistance to whole-body IR-induced lethality in Mdm2Y393F mice (Fig. 3A). Following treatment with 8 Gy IR, 74% of Mdm2Y393F mice survived to 4 wk compared with 22% survival of WT mice. Similarly, 29% of Mdm2Y393F mice survived to 4 wk after exposure to 9 Gy IR compared with 7% of WT mice. All mice survived 7 Gy IR for both genotypes, whereas no mice of either genotype survived past 2 wk following 10 Gy IR. The majority of mice succumbed between 1 and 3 wk post-IR. Lethality in mice treated with this range of IR doses is routinely attributed to p53-dependent bone marrow failure, referred to as “hematopoietic syndrome” (29). As we have described a similar resistance to whole-body IR in Mdm2S394A mice (11), we next compared the effects of IR on bone marrow from WT, Mdm2Y393F, Mdm2S394A, and p53−/− mice. Expression of p53-target genes Puma and p21 are unaltered in Mdm2Y393F bone marrow, in agreement with what was observed in irradiated spleen and thymus (Fig. S3A). Conversely, expression levels of those same target genes are significantly reduced in Mdm2S394A bone marrow, in keeping with the previously described reduced IR-induced p53 responses in these animals (11). However, similar initial reductions in bone marrow cellularity were observed in both mutants, as well as in WT mice and in p53−/− mice following treatment with 5 Gy IR (Fig. 3B). Thus, the initial reduction in gross cellularity appears, to some extent, to be independent of p53. Flow cytometry analysis of untreated bone marrow from WT, Mdm2Y393F, Mdm2S394A, and p53−/− mice revealed no differences in the populations of lineage-defined, mature hematopoietic cells, save for statistically fewer Cd11b+ cells in p53−/− mice (Fig. S3B). In agreement with the observed reduction in bone marrow cellularity in all genotypes, IR treatment induced a cumulative decline in populations of mature cells in bone marrow from each genotype (Fig. S3C). Only p53−/− mice displayed significantly more B220+ cells. Again, no differences were observed in the absence of treatment, in the more primitive Lin−Sca1−cKit+ (L−S−K) progenitor populations of WT, Mdm2Y393F, Mdm2S394A, or p53−/− mice (Fig. S3D). However, whereas L−S−K cell numbers decreased 70–80% in WT, Mdm2Y393F, and Mdm2S394A bone marrows, bone marrow from p53−/− mice retained significantly more L−S−K cells following IR. This finding suggests that increased survival of hematopoietic progenitors underlies the resistance to IR-induced bone marrow failure in mice with compromised p53. Indeed, when Lin−Sca1+cKit+ (L−SK) hematopoietic stem and progenitor cells (HSPCs) were quantified in these same animals, we observed no decrease following irradiation in p53−/− mice (Fig. 3C). Whereas greater numbers of HSPCs were observed in untreated p53−/− mice, no differences were observed in HSPC number in untreated WT, Mdm2Y393F, and Mdm2S394A mice. However, there were significantly more HSPCs in Mdm2S394A bone marrow following IR, and HSPC levels also appeared slightly (although not statistically) elevated in irradiated Mdm2Y393F bone marrow. We examined whether the resistance to acute whole-body IR-associated lethality observed in Mdm2Y393F and Mdm2S394A mice was due to an increased capacity to repopulate irradiated marrow by performing H&E stains on bone marrow from mice either untreated or 6 and 9 d following 8 Gy IR (Fig. 3D). At 6 d post-IR, whereas mice of all three genotypes displayed evidence of a significant decrease in cellularity, both p53−/−and Mdm2S394A bone marrow, and to a lesser extent Mdm2Y393F bone marrow, contained multiple colonies of hematopoietic cells that were not apparent in WT bone marrow, as well as visibly more erythrocytes. By 9 d post-IR, coinciding with the period of observed morbidity in threshold-lethally irradiated animals, an even larger discrepancy in cellularity was observable between WT and mutant bone marrows. Few hematopoietic colonies were visible in WT marrow, whereas colonies present in p53−/−and Mdm2S394A mice had expanded significantly and largely replenished the medullary cavity. Whereas there was greater visible repopulation in Mdm2Y393F bone marrow compared with WT, this repopulation appeared intermediate to that observed in Mdm2S394A bone marrow, trending with the observed improved HSPC survival following IR.
Fig. 3.
Mdm2Y393F mice are radioresistant and display improved bone marrow repopulation following IR exposure. (A) Kaplan–Meier survival curves of WT (n = 7–18) and Mdm2Y393F (n = 6–19) mice exposed to 7, 8, 9, and 10 Gy whole-body IR. WT and Mdm2Y393F curves were compared by log-rank test: 7 Gy (n.s.), 8 Gy (P = 0.003), 9 Gy (P = 0.030), and 10 Gy (n.s.). (B) Quantification of numbers of nucleated cells in bone marrow from both hindlimbs of WT, Mdm2Y393F, Mdm2S394A, and p53−/− mice either untreated or 8 h after exposure to 5 Gy IR. (n = 3–6, ± SEM). *P < 0.05, **P < 0.01 (Student’s t tests). (C) Quantification of L–SK HSPCs in bone marrow of WT, Mdm2Y393F, Mdm2S394A, and p53−/− mice treated as described in B. *P < 0.05, **P < 0.01 (Student’s t tests). (D) H&E stained bone marrow from WT, Mdm2Y393F, Mdm2S394A, and p53−/− mice exposed to 8 Gy IR. Yellow arrows indicate nascent hematopoietic cell colonies. (Scale bars, 100 μm.)
Fig. S3.
(A) Fold expression of p53-target genes Puma, Noxa, and p21 in bone marrow of WT, Mdm2Y393F, Mdm2S394A, and p53−/− mice, untreated and 6 h after 5 Gy IR, was determined by RT-qPCR. Fold expression was calculated relative to untreated WT samples using Rplp0 as internal reference (n = 3–4 mice, ±SEM). *P < 0.05, **P < 0.01 (Student’s t tests of ΔΔCt values). (B) Quantification of lineage-defined hematopoietic cells in untreated bone marrow of WT, Mdm2Y393F, Mdm2S394A, and p53−/− mice (n = 3–4, ±SEM). **P < 0.01 (Student’s t tests). (C) Quantification of lineage-defined hematopoietic cells in bone marrow of WT, Mdm2Y393F, Mdm2S394A, and p53−/− mice 8 h after 5 Gy IR (n = 3–4, ±SEM). **P < 0.01 (Student’s t tests). (D) Quantification of L−S−K hematopoietic progenitor cells in bone marrow of WT, Mdm2Y393F, Mdm2S394A, and p53−/− mice, either untreated or 8 h after 5 Gy IR (n = 3–4, ±SEM). **P < 0.01 (Student’s t tests).
Mdm2 Tyr393 and Ser394 Phosphorylations Are Not Additive in Their Impact on Tumor Suppression and Radioresistance.
To determine whether the shared effects of Mdm2 Tyr393 and Ser394 phosphorylation were redundant or additive, we generated a knockin mouse in which both the Tyr393 residue is substituted with a phenylalanine residue (Y393F) and the Ser394 residue is substituted with an alanine (S394A) (Fig. 4A). Gene targeting was carried out following a similar strategy as used for Mdm2Y393F mice (Fig. S4 A–E). The targeting construct coded for an A-to-T missense mutation in the 393 codon, a T-to-G missense mutation in the 394 codon, and a synonymous T-to-G mutation in the 398 codon of Mdm2 exon 12, which also facilitated PCR-digest genotyping of cells and mice. As observed for Mdm2S394A mice (11), Mdm2Y393F/S394A mice were recovered from heterozygous intercrosses at Mendelian ratios (Fig. S4F). A cohort of biallelic Mdm2Y393F/S394A mice was monitored for spontaneous tumor presentation, and by 24 mo, 14 of 21 (67%) of these mice presented with tumors (Fig. 4B). This result mirrors the 65% of Mdm2S394A mice that developed spontaneous tumors in our previous study (11), as does the observed latency of 18–24 mo for the majority of tumors. Consequently, the tumor suppressive effects of Mdm2 Tyr393 phosphorylation by c-Abl and Mdm2 Ser394 phosphorylation by ATM do not appear to be additive or synergistic. The predominant tumor type in Mdm2Y393F/S394A mice was B-cell lymphoma (6/15, 40%), along with myeloid sarcomas (5/15, 33%), hepatocellular carcinomas (3/15, 20%), and one tumor that appeared to be a liver metastasis of neuroendocrine cell origin (Fig. 4 C–E).
Fig. 4.
Mdm2 Tyr393 and Ser394 phosphorylations are not additive in their impact on tumor suppression or radioresistance. (A) DNA sequence of WT and Mdm2Y393F/S394A allele surrounding codons 393 and 394 (Top). An A-to-T mutation changed codon 393 from Tyr to Phe. A T-to-C mutation changed codon 394 from Ser to Ala. A silent T-to-G mutation in codon 398 introduced a AatII restriction site. Sequencing results of the corresponding region from Mdm2Y393F/S394A spleen cDNA are shown (Bottom). (B) Kaplan–Meier tumor-free survival curve of Mdm2Y393F/S394A mice (n = 21). Included are tumor-free survival curves of WT and Mdm2Y393F mice described in Fig. 1B and Mdm2S394A mice described previously (11). Curves were compared by log-rank test: Mdm2Y393F/S394A to WT (P < 0.0001) and Mdm2Y393F/S394A to Mdm2Y393F (P = 0.080). (C) Table displaying the latency, location, and tumor type for spontaneous tumors arising in Mdm2Y393F/S394A mice. LN, lymph node; HCC, hepatocellular carcinoma; Neu, neuroendocrine. (D) Representative H&E-stained tissue sections of a B-cell lymphoma (Left), myeloid sarcoma (Middle), and hepatoma (Right) that developed in Mdm2Y393F/S394A mice. (Scale bars, 100 μm.) (E) B-cell lymphoma shows expression of B220 (Left), and a myeloid neoplasm exhibits chloroacetate esterase (Leder) activity (Right). (Scale bars, 100 μm.) (F) Kaplan–Meier survival curves of WT (n = 15), Mdm2Y393F (n = 17), Mdm2S394A (n = 11), and Mdm2Y393F/S394A (n = 13) mice exposed to 9 Gy whole-body IR. Curves were compared by log-rank test: WT to Mdm2Y393F (P = 0.030), WT to Mdm2S394A (P < 0.0001), WT to Mdm2Y393F/S394A (P < 0.0001), Mdm2Y393F to Mdm2S394A (P = 0.010), Mdm2Y393F to Mdm2Y393F/S394A (P = 0.020), and Mdm2S394A to Mdm2Y393F/S394A (n.s.). (G) Quantification of L–SK HSPCs in bone marrow of WT, Mdm2Y393F, Mdm2S394A, and Mdm2Y393F/S394A mice untreated or 8 h following 5 Gy IR (n = 3–6, ± SEM). *P < 0.05, **P < 0.01 (Student’s t tests).
Fig. S4.
(A) Diagram of the targeting strategy used to generate the Mdm2Y393F/S394A allele. The targeting vector contains a floxed neomycin cassette (neo) and a 3′ thymidine kinase cassette (TK) for positive and negative drug selection, respectively. The targeting vector was linearized and electroporated into PC3 ES cells. ES cell targeting was confirmed by BamHI digest and Southern blot analysis with a 3′ external probe (green), as well as SpeI digest and Southern blot analysis with a 5′ external probe (blue). Protamine-cre recombinase-directed excision of neo cassette was confirmed by BamHI digest of F1 generation tail DNA and Southern blot analysis with the same 3′ external probe as in ES cells (green). (B) BamHI digest and Southern blot analysis of ES cell genomic DNA. A 3′ external probe (green bar from A) detects a 9.7-kb WT allele and 3.8-kb targeted allele. Two clones with correct 3′ targeting are shown, each flanked by untargeted (WT) clones. (C) SpeI digest and Southern blot analysis of ES cell genomic DNA. A 5′ external probe (blue bar from A) detects a 14.0-kb WT allele and 10.4-kb targeted allele. (D) BamHI digest and Southern blot analysis of tail DNA from F1 generation mice. A 3′ external probe (green bar from A) detects a 9.7-kb WT allele and 8.4-kb targeted allele. (E) PCR-BtsCI digest analysis of F2 generation mice. Primers flanking Mdm2 codons 393 and 394 amplify a 436-bp fragment of DNA. A silent mutation in the Mdm2Y393F/S394A allele introduces a novel AatII restriction site that results in 193- and 243-bp fragments. (F) The expected and observed Mendelian ratios from Mdm2Y393F/S394A heterozygote breeding.
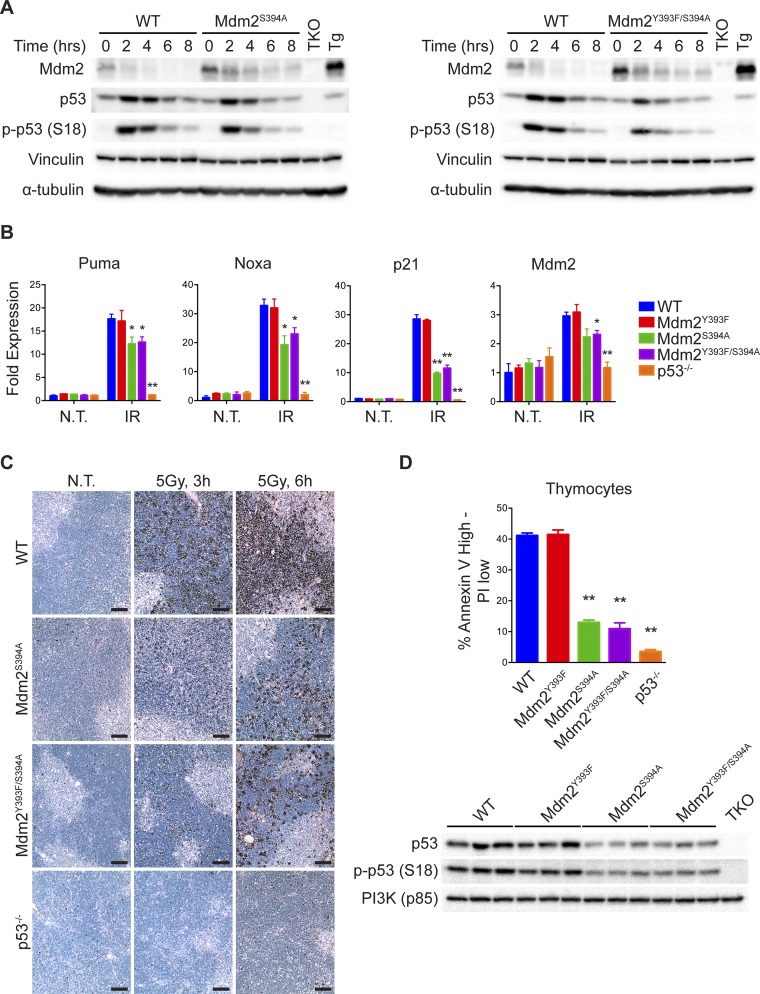
Irradiated thymi from Mdm2Y393F/S394A mice were examined by immunoblotting, as well as by qPCR of p53-target gene transcripts, and showed a similar reduction of p53 stabilization and activity as observed in Mdm2S394A mice (Fig. S5 A and B). This finding is in keeping with the absence of observable defects in the p53 response to DNA damage in Mdm2Y393F mice described in Fig. 2 and Fig. S2. Accordingly, whereas reduced in comparison with WT, no additional deficits in apoptosis were observed by TUNEL staining or Annexin V staining followed by flow cytometry of Mdm2Y393F/S394A thymi relative to those seen in Mdm2S394A thymi (Fig. S5 C and D).
Fig. S5.
(A) WT and Mdm2S394A mice (Left) or Mdm2Y393F/S394A mice (Right) were left untreated or exposed to 5 Gy IR and thymi were harvested at 2-h intervals. Protein levels were analyzed by Western blotting. TKO indicates Mdm2−/−, MdmX−/−, p53−/− control; Tg indicates Mdm2Tg/+ Mdm2 overexpressing control. (B) WT, Mdm2Y393F, Mdm2S394A, Mdm2Y393F/S394A, and p53−/− mice were left untreated or exposed to 5 Gy IR and thymi were harvested at 4 h. Fold expression of p53-target genes was determined by RT-qPCR, relative to untreated WT samples using Rplp0 as internal reference (n = 3–4, ±SEM). *P < 0.05, **P < 0.01 (Student’s t tests of ΔΔCt values). (C) TUNEL staining of thymi from WT, Mdm2S394A, Mdm2Y393F/S394A, and p53−/− mice treated as in A and harvested at 3 and 6 h. (D) Quantification of apoptotic thymocytes by flow cytometry (Top) in WT, Mdm2Y393F, Mdm2S394A, Mdm2Y393F/S394A, and p53−/− mice. Thymi were harvested 5 h after treatment with 5 Gy IR and single cell suspensions of thymocytes were generated and stained with AnnexinV-FITC and PI for FACS analysis. The percentage of Annexin VHigh–PILow cells was quantified (n = 3, ±SEM). **P < 0.01 (Student’s t tests). Protein lysates generated from fractions of WT, Mdm2Y393F, Mdm2S394A, and Mdm2Y393F/S394A cell suspensions were analyzed for p53 and p-p53 (S18) protein levels (Bottom).
Finally, we examined whether the radioresistant phenotypes observed in Mdm2Y393F and Mdm2S394A mice were exacerbated in Mdm2Y393F/S394A mice. Mdm2S394A and Mdm2Y393F/S394A mice were exposed to 9 Gy whole-body IR (as used for Mdm2Y393F mice) and monitored for signs of morbidity (Fig. 4F). No significant difference in survival was observed between Mdm2S394A and Mdm2Y393F/S394A mice at this dose, with 82% of Mdm2S394A mice and 69% of Mdm2Y393F/S394A mice surviving at 4 wk, respectively. However, both genotypes are significantly more radioresistant than Mdm2Y393F mice, which are themselves significantly more radioresistant than WT mice at this dose. Thus, radioresistance resulting from the loss of either Mdm2 Tyr393 phosphorylation by c-Abl or Mdm2 Ser394 phosphorylation by ATM is not increased by mutation of both phospho-target residues. Accordingly, IR-induced expression levels of p53-target genes in irradiated bone marrow of Mdm2S394A and Mdm2Y393F/S394A mice are similarly reduced (Fig. S6A). The equivalent deficiency in the p53 response in Mdm2S394A and Mdm2Y393F/S394A bone marrow follows with comparably increased numbers of L−SK HSPCs surviving in Mdm2Y393F/S394A bone marrow following IR (Fig. 4G). Furthermore, bone marrow cells from Mdm2Y393F/S394A or Mdm2S394A mice exhibit similar hematopoietic repopulating abilities in vivo (Fig. S6B). As was observed with Mdm2Y393F and Mdm2S394A mice, no differences were seen in the number of lineage-defined or L−S−K hematopoietic cells (Fig. S6 C and D).
Fig. S6.
(A) Fold expression of p53-target genes in bone marrow of WT, Mdm2Y393F, Mdm2S394A, and p53−/− mice, as shown in Fig. S3A, untreated and 6 h after 5 Gy IR, including expression data from Mdm2Y393F/S394A bone marrow (n = 3–4 mice, ±SEM). No significant difference is observed between Mdm2S394A and Mdm2Y393F/S394A expression levels (Student’s t tests of ΔΔCt values). (B) H&E-stained bone marrow from Mdm2S394A and Mdm2Y393F/S394A mice exposed to 8 Gy IR. Yellow arrows indicate nascent hematopoietic cell colonies. (Scale bars, 100 μm.) (C) Quantification of lineage-defined hematopoietic cells in untreated bone marrow (Top) or 8 h after 5 Gy IR (Bottom) of WT, Mdm2Y393F, Mdm2S394A, and p53−/− mice, as shown in Fig. S3 B and C, including data from Mdm2Y393F/S394A bone marrow (n = 3–4, ±SEM). No significant difference is observed between Mdm2S394A and Mdm2Y393F/S394A lineage-defined cell numbers (Student’s t tests). (D) Quantification of L−S−K hematopoietic progenitor cells in bone marrow of WT, Mdm2Y393F, Mdm2S394A, and p53−/− mice, as shown in Fig. S3D, either untreated or 8 h after 5 Gy IR, including data from Mdm2Y393F/S394A bone marrow (n = 3–4, ±SEM). No significant difference is observed between Mdm2S394A and Mdm2Y393F/S394A L−S−K progenitor cell numbers (Student’s t tests).
Discussion
These findings provide further evidence of the significance of phosphorylation of Mdm2 by DNA damage-activated kinases in regulating p53-dependent organismal responses. Mdm2Y393F mice are viable and display no developmental defects, yet they are significantly more prone to developing spontaneous tumors over their lifespan (Fig. 1B). The spontaneous tumors arising in Mdm2Y393F mice are primarily hematopoietic in nature and led us to examine whether Mdm2 Tyr393 phosphorylation impacted oncogene-induced tumorigenesis using the Eμ-Myc allele. Indeed, Eμ-Myc;Mdm2Y393F mice developed tumors at a significantly accelerated rate (Fig. 1F).
The increased spontaneous tumorigenesis rates and accelerated B-cell lymphomagenesis are a likely result of reduced p53 activity in Mdm2Y393F and Eμ-Myc;Mdm2Y393F mice, as p53-dependent apoptosis and senescence have been shown to inhibit spontaneous tumorigenesis and B-cell tumors induced by aberrant Myc activity (24, 30–32). However, we observed no defects in p53-dependent apoptosis in spleens or thymi of Mdm2Y393F mice following IR, or p53-dependent growth arrest in MEFs treated with IR or doxorubicin (Fig. 2 and Fig. S2). It is conceivable that Mdm2 phosphorylation by c-Abl can regulate p53 tumor suppressing effects other than apoptosis or growth arrest, and/or that subtle differences in these p53 functions in Mdm2Y393F mice are not detectable by examining the effects of acute damage on p53 tumor suppression. Furthermore, it is possible that additional c-Abl target residues on Mdm2 can compensate for the loss of Mdm2 Tyr393 phosphorylation (12, 13).
Seemingly incongruously, Mdm2Y393F mice are resistant to threshold-lethal doses of radiation (Fig. 3A). This radioresistance parallels our previous observations with Mdm2S394A mice, which display profound defects in p53-dependent apoptosis and tissues. HSPC or bone marrow cells from both mutants display improved repopulating functions following IR exposure, albeit to a greater extent in Mdm2S394A mice (Fig. 3D). In keeping with this difference, HSPCs in Mdm2S394A bone marrow display a significant survival advantage following IR, whereas Mdm2Y393F HSPCs display only a marginal increase in survival after DNA damage. However, the slight increase in the survival of Mdm2Y393F HSPCs manifests increased radioresistance in Mdm2Y393F mice relative to WT mice.
The generation of Mdm2Y393F/S394A mice allowed us to examine whether the common effects of Mdm2 Tyr393 phosphorylation by c-Abl and Mdm2 Ser394 phosphorylation by ATM are additive or redundant. We observed no additive or synergistic effects of the loss of both phosphorylation events on the incidence of spontaneous tumorigenesis, with Mdm2Y393F/S394A mice developing spontaneous tumors at a frequency and latency that nearly overlaps what we have reported with Mdm2S394A mice (Fig. 4B). Similarly, we observed no additive effects on radioresistance, with Mdm2Y393F/S394A and Mdm2S394A mice displaying comparable survival, HSPC numbers, and bone marrow reconstitution following whole-body IR (Fig. 4 F and G and Fig. S6). Hence, there is an apparent redundancy of the shared phenotypes between Mdm2Y393F and Mdm2S394A mice, with tumorigenesis and radioresistance in Mdm2Y393F/S394A mice never exceeding that observed in Mdm2S394A mice. This may reflect the proposed interdependence of c-Abl and ATM for their respective activities (16, 33, 34). However, ATM phosphorylation of Mdm2 Ser394 clearly has a predominant effect on Mdm2-p53 signaling and p53 functions, relative to the effects induced by c-Abl phosphorylation of Mdm2 Tyr393.
Methods
Mice and Animal Studies.
All animals described in this study were on a C57BL/6 background. Mice and cells were irradiated with a cesium-137 source (Gammacell 40). The generation of Mdm2S394A mice has been previously described (11). Eμ-Myc mice were a gift from Christine Eischen, Vanderbilt University, Nashville. TN. A detailed description of the generation and genotyping of Mdm2Y393F and Mdm2Y393F/S394A mice is provided in SI Methods. All animals used in this study were maintained and assayed in accordance with federal guidelines and those established by the Institutional Animal Care and Use Committee at the University of Massachusetts Medical School (UMMS).
Immunoblotting.
Tissues and cells were lysed in Nonidet P-40 lysis buffer supplemented with protease and phosphatase inhibitors. A detailed description of the methods used, including antibodies and clones, is provided in SI Methods.
Gene Expression Analysis and Sequencing.
Total RNA was isolated from tissues by RNeasy Mini Kit (Qiagen) and cDNA synthesized by the SuperScript III First Strand Synthesis System (Invitrogen). qPCR was performed using SYBR Select Master Mix (Applied Biosystems) in conjunction with a 7300 Real-Time PCR System (Applied Biosystems). A detailed description of the methods used for qPCR and sequencing is provided in SI Methods.
Histopathology.
Tissue samples were fixed in 10% (vol/vol) formalin for 24 h. The UMMS Morphology Core Laboratory performed embedding, sectioning, and staining. TUNEL staining was performed using the In Situ Cell Death Detection Kit, POD (Roche) according to manufacturer’s instructions. Immunohistochemistry was performed with antibodies specific for B220 (550286; BD Pharmingen) and CD3 (A0452; Dako). Naphthol chloroacetate esterase staining was performed to detect cells with myeloid differentiation. Stained tissue was analyzed using an Olympus CX41 microscope fitted with a PixeLINK camera and software.
Bone Marrow Analysis.
Total bone marrow from both hind limbs was harvested, RBCs were lysed, and single-cell suspensions were stained with cell-surface antibodies for Gr-1, CD11B, CD3, and B220. For LSK analysis, bone marrow cells were stained with a biotin lineage mixture and antibodies for Sca-1, c-Kit, CD34, and Flk2. All samples were run on a BD LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo software (Tree Star). A complete list of antibodies including clone numbers is given in Table S1.
Table S1.
Flow cytometry antibodies
| Antibody | Clone | Fluorophore | Source |
| CD3 | 145–2C11 | Biotin, APC | BD Biosciences |
| B220 | RA3-6B2 | Biotin, PE | BD Biosciences |
| Ter119 | TER-119 | Biotin | Biolegend |
| Gr-1 | RB6-8C5 | Biotin, APC, PE | Biolegend |
| Mac-1 | M1/70 | Biotin, FITC | Biolegend |
| Sca-1 | D7 | APC/Cy7 | Biolegend |
| CD117 | 2B8 | APC | BD Biosciences |
| CD34 | RAM34 | FITC | BD Biosciences |
| Flk2 | A2F10 | PE | Biolegend |
| CD45.2 | 104 | FITC, PE/Cy7 | Biolegend |
| CD45.1 | A20 | PercP/Cy5.5 | Biolegend |
Statistical Analysis.
Statistical analyses were performed using GraphPad Prism software, version 6.0d. Kaplan–Meier survival curves were analyzed by log-rank test. A P value of <0.05 was considered statistically significant for Student t tests.
SI Methods
Generation and Genotyping of Mdm2Y393F and Mdm2Y393F/S394A Mice.
Targeting vectors were constructed from subcloned fragments of the Mdm2 genomic sequence (129Sv strain). Site-directed mutagenesis was performed according to manufacturer instructions using a QuikChange II Site-Directed Mutagenesis Kit (Stratagene). Targeting vectors were sequenced to ensure only the desired mutations were introduced. Linearized targeting vectors were introduced into PC3 ES cells by electroporation. Targeted homologous recombination was detected by Southern blotting using 5′ and 3′ external probes after SpeI and BamHI restriction digests, respectively. Targeted cells were microinjected into E3.5 blastocysts (C57BL/6 strain), and embryos were surgically implanted into pseudopregnant foster mice by standard procedures. Transmission of the knockin allele and excision of the neo cassette in F1 and F2 offspring of male chimeric mice was confirmed by Southern blotting using the same 3′ external probe and BamHI restriction digest strategy as in ES cells. Mdm2Y393F and Mdm2Y393F/S394A alleles were followed by genomic PCR using the primer pair: forward 5′ AAAGATGCTGGACCCTTCGTGAGA3′ and reverse 5′ GCACACGTGAAACATGACATGAGG3′, followed by BtsCI digest to identify the Mdm2Y393F allele or AatII digest to identify the Mdm2Y393F/S394A allele.
Immunoblotting.
Tissues and cells were lysed in Nonidet P-40 lysis buffer [50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 0.5% Nonidet P-40] supplemented with protease and phosphatase inhibitor mixture tablets (Roche). Protein extracts (50 µg) were analyzed by direct Western blotting with antibodies specific for Mdm2 [NBP1-02158; Novus (Ab-I) and Ab-5; Calbiochem (Ab-II)], p53 (CM5; Novocastra), phospho-p53 (S15) (9284; Cell Signaling), α-tubulin (B-5–1-2; Sigma), Vinculin (hVin-1; Sigma), PI3K p85 (06-496; Millipore), MdmX (MDMX-82; Sigma), c-Abl (2862; Cell Signaling), and phospho–c-Abl (Y245) (2861; Cell Signaling). Mdm2 was detected using NBP1-02158; Novus (Ab-I) unless otherwise specified. Blots were imaged on a Chemidoc MP (Bio-Rad) and relative band intensities determined by densitometry using Image Lab software (v4.1, Bio-Rad). All Western blots shown in the manuscript are of samples run together on 15-well gels. To generate figure panels displaying multiple proteins, multiple gels were run using the same sets of samples, and blots were never stripped and reprobed more than once.
Quantification of Apoptosis.
Animals were treated as described. Thymi and spleens were harvested and single cell suspensions generated by mechanical disruption and screening through 70-µm mesh. RBCs were lysed in splenocyte samples by suspension in ammonium-chloride-potassium (ACK) buffer. Samples were stained using an Annexin V-FITC Apoptosis Detection Kit I (BD Pharmingen 556547) according to manufacturer protocol. Flow cytometry was performed by the UMMS Flow Cytometry Core Lab. Early apoptotic cells (Annexin Vhigh PIlow) were quantified using FlowJo software (Tree Star).
Cell-Cycle Analysis.
Treated MEFs were pulse labeled with 50 μM bromodeoxyuridine (BrdU) for 3 h before being harvested by trypsinization and fixed in 70% ethanol overnight. Fixed cells were stained with FITC-conjugated anti-BrdU antibody (Becton Dickinson 347583) and propidium iodide (PI), and analyzed by flow cytometry by the UMMS Flow Cytometry Core Lab. Cell-cycle populations were quantified using FlowJo software (Tree Star).
Gene Expression Analysis and Sequencing.
Total RNA was isolated from tissues by RNeasy Mini Kit (Qiagen) and cDNA synthesized by SuperScript III First Strand Synthesis System (oligo-dT priming) (Invitrogen). qPCR was performed using SYBR Select Master Mix (Applied Biosystems) in conjunction with a 7300 Real-Time PCR System (Applied Biosystems). Thymus cDNA input was 10 ng and bone marrow cDNA input was 100 ng. Fold expression was calculated using the ΔΔCt method relative to untreated WT samples using Rplp0 as internal reference. Primers used were as follows: Puma, 5′ ACGACCTCAACGCGCAGTACG3′ and 5′ GAGGAGTCCCATGAAGAGATTG3′; Noxa, 5′CTCAGGAAGATCGGAGACAAAG3′ and 5′ GCACACTCGTCCTTCAAGT3′; p21, 5′ CTGAGCGGCCTGAAGATT3′ and 5′ ATCTGCGCTTGGAGTGATAG3′; Mdm2, 5′ AGTCTCTGGACTCGGAAGATTA3′ and 5′ CTGTATCGCTTTCTCCTGTCTG3′; Rplp0, 5′ CTGAGTACACCTTCCCACTTAC3′ and 5′ CTCTTCCTTTGCTTCAGCTTTG3′; Gls2, 5′ GAACAAGATGGCTGGGAACGA3′ and 5′ CGGAGCCGATGGCGTAATTCCG3′; Tigar, 5′ AAAGACATGGCGGTGAAGTACG3′ and 5′ CGGATTAGCGGCTTGCCTTCCG3′; Ercc5, 5′ GGACACAGAGAAGCTGACATA3′ and 5′ GTTACTTTCCAGGGCATCCA3′; Mgmt, 5′ AAGCCTATTTCCGTGAACCC3′ and 5′ CACCTGTCTGGTGAATGAATCT3′; Polk, 5′ CCCAAAGAAAGCTCGAGAAGTA3′ and 5′ AATCCATCAAGGGTGTGTCTG3′. Mdm2 message was amplified from spleen cDNA in overlapping fragments using two separate primer pairs: Mdm2seq1, 5′ CCAGGCCAATGTGCAATACCAACA3′ and 5′ TCCTCAGCACATGGCTCTTTAGCA3′; Mdm2seq2, 5′ TCTTGACGATGGCGTAAGTGAGCA3′ and 5′ TCCAAAGTCCTTCCAAGCGGAGAT3′. Amplified fragments were gel extracted (Qiagen) and sequenced by Sequegen.
Acknowledgments
This research was supported by NIH Grants R01-CA077735 (to S.N.J.) and R01-CA096899 (to M.A.K.). J.E.R. was supported by Postdoctoral Fellowship 125087-PF-13-247-01-LIB from the American Cancer Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611798114/-/DCSupplemental.
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Li T, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149(6):1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valente LJ, et al. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Reports. 2013;3(5):1339–1345. doi: 10.1016/j.celrep.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciccia A, Elledge SJ. The DNA damage response: Making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shieh S-Y, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91(3):325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 7.Shieh S-Y, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14(3):289–300. [PMC free article] [PubMed] [Google Scholar]
- 8.Maya R, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: Role in p53 activation by DNA damage. Genes Dev. 2001;15(9):1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pereg Y, et al. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci USA. 2005;102(14):5056–5061. doi: 10.1073/pnas.0408595102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Gilkes DM, Pan Y, Lane WS, Chen J. ATM and Chk2-dependent phosphorylation of MDMX contribute to p53 activation after DNA damage. EMBO J. 2005;24(19):3411–3422. doi: 10.1038/sj.emboj.7600812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gannon HS, Woda BA, Jones SN. ATM phosphorylation of Mdm2 Ser394 regulates the amplitude and duration of the DNA damage response in mice. Cancer Cell. 2012;21(5):668–679. doi: 10.1016/j.ccr.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg Z, et al. Tyrosine phosphorylation of Mdm2 by c-Abl: Implications for p53 regulation. EMBO J. 2002;21(14):3715–3727. doi: 10.1093/emboj/cdf384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dias SS, Milne DM, Meek DW. c-Abl phosphorylates Hdm2 at tyrosine 276 in response to DNA damage and regulates interaction with ARF. Oncogene. 2006;25(50):6666–6671. doi: 10.1038/sj.onc.1209671. [DOI] [PubMed] [Google Scholar]
- 14.Kharbanda S, et al. Activation of the c-Abl tyrosine kinase in the stress response to DNA-damaging agents. Nature. 1995;376(6543):785–788. doi: 10.1038/376785a0. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z-G, et al. Three distinct signalling responses by murine fibroblasts to genotoxic stress. Nature. 1996;384(6606):273–276. doi: 10.1038/384273a0. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, et al. A positive role for c-Abl in Atm and Atr activation in DNA damage response. Cell Death Differ. 2011;18(1):5–15. doi: 10.1038/cdd.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen ST, Jackson PK, Van Etten RA. The cytostatic function of c-Abl is controlled by multiple nuclear localization signals and requires the p53 and Rb tumor suppressor gene products. EMBO J. 1996;15(7):1583–1595. [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan Z-M, et al. Regulation of DNA damage-induced apoptosis by the c-Abl tyrosine kinase. Proc Natl Acad Sci USA. 1997;94(4):1437–1440. doi: 10.1073/pnas.94.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sionov RV, et al. c-Abl neutralizes the inhibitory effect of Mdm2 on p53. J Biol Chem. 1999;274(13):8371–8374. doi: 10.1074/jbc.274.13.8371. [DOI] [PubMed] [Google Scholar]
- 20.Sionov RV, et al. c-Abl regulates p53 levels under normal and stress conditions by preventing its nuclear export and ubiquitination. Mol Cell Biol. 2001;21(17):5869–5878. doi: 10.1128/MCB.21.17.5869-5878.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waning DL, Lehman JA, Batuello CN, Mayo LD. c-Abl phosphorylation of Mdm2 facilitates Mdm2-Mdmx complex formation. J Biol Chem. 2011;286(1):216–222. doi: 10.1074/jbc.M110.183012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci USA. 1997;94(26):14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams JM, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 24.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13(20):2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barilà D, et al. Caspase-dependent cleavage of c-Abl contributes to apoptosis. Mol Cell Biol. 2003;23(8):2790–2799. doi: 10.1128/MCB.23.8.2790-2799.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machuy N, Rajalingam K, Rudel T. Requirement of caspase-mediated cleavage of c-Abl during stress-induced apoptosis. Cell Death Differ. 2004;11(3):290–300. doi: 10.1038/sj.cdd.4401336. [DOI] [PubMed] [Google Scholar]
- 27.Pant V, et al. The p53-Mdm2 feedback loop protects against DNA damage by inhibiting p53 activity but is dispensable for p53 stability, development, and longevity. Genes Dev. 2013;27(17):1857–1867. doi: 10.1101/gad.227249.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gudkov AV, Komarova EA. The role of p53 in determining sensitivity to radiotherapy. Nat Rev Cancer. 2003;3(2):117–129. doi: 10.1038/nrc992. [DOI] [PubMed] [Google Scholar]
- 29.Komarova EA, et al. Dual effect of p53 on radiation sensitivity in vivo: p53 promotes hematopoietic injury, but protects from gastro-intestinal syndrome in mice. Oncogene. 2004;23(19):3265–3271. doi: 10.1038/sj.onc.1207494. [DOI] [PubMed] [Google Scholar]
- 30.Eischen CM, Roussel MF, Korsmeyer SJ, Cleveland JL. Bax loss impairs Myc-induced apoptosis and circumvents the selection of p53 mutations during Myc-mediated lymphomagenesis. Mol Cell Biol. 2001;21(22):7653–7662. doi: 10.1128/MCB.21.22.7653-7662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt CA, et al. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1(3):289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 32.Post SM, et al. p53-dependent senescence delays Emu-myc-induced B-cell lymphomagenesis. Oncogene. 2010;29(9):1260–1269. doi: 10.1038/onc.2009.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baskaran R, et al. Ataxia telangiectasia mutant protein activates c-Abl tyrosine kinase in response to ionizing radiation. Nature. 1997;387(6632):516–519. doi: 10.1038/387516a0. [DOI] [PubMed] [Google Scholar]
- 34.Shafman T, et al. Interaction between ATM protein and c-Abl in response to DNA damage. Nature. 1997;387(6632):520–523. doi: 10.1038/387520a0. [DOI] [PubMed] [Google Scholar]