Significance
Oxidative and inflammatory stresses (e.g., atheroprone flow and hyperlipidemia) induce innate immune responses in vascular endothelium, of which the assembly and activation of nucleotide oligomerization domain-like receptor family pyrin domain-containing protein 3 (NLRP3) inflammasome constitute a core mechanism. We show that TNF-α receptor-associated factor-interacting protein with a forkhead-associated domain (TIFA) is a mediator of NLRP3 inflammasome in endothelium. The involved regulations include that sterol regulatory element-binding protein 2 transcriptionally induces TIFA (signal 1) and that phosphorylated TIFA causes higher-order assembly of NLRP3 inflammasome (signal 2). Given that sustained innate immune response/activation in endothelium is common to a variety of vascular impairments, TIFA-mediated NLRP3 inflammasome constitutes a pathway by which sustained oxidative and inflammatory stresses lead to dysfunctional endothelium.
Keywords: endothelial cells, inflammasome, innate immunity, NLRP3, TIFA
Abstract
Toll-like receptor-mediated NF-κB activation is a major innate immune reaction of vascular endothelial cells (ECs) in response to prooxidative and proinflammatory stimuli. We identified that TNF-α receptor-associated factor-interacting protein with a forkhead-associated domain (TIFA) is a regulator of priming (signal 1) and activating (signal 2) signals of nucleotide oligomerization domain-like receptor family pyrin domain-containing protein 3 (NLRP3) inflammasome in ECs. Oxidative and inflammatory stresses such as atheroprone flow and hyperlipidemia induce and activate TIFA in vitro and in vivo. For the priming of signal 1, sterol regulatory element-binding protein 2 transactivates TIFA, which in turn induces NF-κB activation and augments the transcription of NLRP3 inflammasome components. For the activation of signal 2, Akt is involved in TIFA Thr9 phosphorylation, which is essential for TIFA–TIFA homophilic oligomerization. Thr9 phosphorylation-dependent TIFA oligomerization facilitates the higher-order assembly of NLRP3 inflammasome, as indicated by the interaction between TIFA and caspase-1 in the activated ECs. Our results suggest that TIFA is a crucial mediator in the endothelial innate immune response by potentiating and amplifying NLRP3 inflammasome via augmenting signals 1 and 2.
Pathogen-associated molecular patterns (PAMPs), such as LPS or viral DNA, activate NF-κB in macrophages to induce the expression of the inflammasome components and pro-IL-1β/pro-IL-18. Although these events are involved in the signal 1 priming step, a second signal (i.e., signal 2) triggered by danger-associated molecular patterns (DAMPs), is required for the assembly of inflammasome complex and the consequent activation (1–4). The induction of nucleotide oligomerization domain-like receptor family pyrin domain-containing protein 3 (NLRP3) inflammasome with the end-product of mature IL-1β or IL-18 causes aberrant lipid metabolism, unbalanced redox state, and enhanced innate immune response (2, 5). With DAMP-associated reactive oxygen species (ROS) triggering signal 2, an imbalanced redox state can enhance innate immunity in monocytes/macrophages (6). In contrast to the well-defined signals 1 and 2 in macrophages, little is known about the two-step mechanism of inflammasome activation in vascular endothelial cells (ECs).
TNF-α receptor-associated factor (TRAF)-interacting protein with a forkhead-associated (FHA) domain (TIFA) interacts with TRAF2 and TRAF6 through its FHA domain (7, 8). In vitro experiments showed that TIFA promotes oligomerization and ubiquitination of TRAF6, thereby activating IκB kinase (IKK) (9). Furthermore, ectopic expression of TIFA activates NF-κB and AP-1 in HEK293T cells (7, 10). TIFA exists as an intrinsic dimer, which becomes phosphorylated at Thr9 upon TNF-α treatment (10). TIFA phosphoThr9 (pT9) can bind to the FHA domain of an adjacent TIFA dimer. The crystal structure of TIFA indicates that the pT9–FHA interaction can occur only between different sets of dimers. Such homophilic TIFA interaction leads to the formation of the higher-order TIFA oligomer (11). This mode of TIFA oligomerization is required for TIFA interaction with TRAF6 and the ensuing activation of NF-κB (10, 11). By using a genome-wide RNA interference screening, Gaudet et al. recently showed that the pT9–FHA interaction is also essential for the innate immune response in the host induced by heptose-1,7-bisphosphate, a PAMP derived from Gram-negative bacteria (12). Although TIFA has emerged as a key player in innate immunity, the mechanism by which TIFA enhances inflammasome remains unclear.
Sterol regulatory element (SRE)-binding protein 2 (SREBP2) is a key transcription factor (TF) that regulates cholesterol biosynthesis (13). We and others have shown that a variety of stimuli that elicit cellular oxidative stress, such as oxidized phospholipids, angiotensin II, and oxidized LDL (oxLDL), promote the N-terminal cleavage and activation of SREBP2 [i.e., SREBP2(N)] in ECs (14, 15). As a result, SREBP2(N) transactivates NLRP3 (14). Regarding disease onset, EC-specific overexpression of SREBP2(N) in mice induces NLRP3 inflammasome to contribute to atherosclerosis (14). Interestingly, cross-talk between the Toll-like receptor (TLR) 4–MyD88–NF-κB pathway and SREBP2 can facilitate the formation of foam cells from macrophages (15). Collectively, our previous work and that from others suggest that SREBP2 plays a central role in the innate immune response in ECs and macrophages.
Oxidative stress (i.e., oxLDL and atheroprone flow) induces innate immune response as well as SREBP2 in ECs (14, 16). In the present study, we showed that SREBP2 transcriptionally regulates the TIFA–NF-κB axis, which contributes to signal 1 for priming NLRP3 inflammasome. Furthermore, we investigated whether TIFA is engaged in signal 2 for NLRP3 activation and found that the Akt-mediated phosphorylation of TIFA Thr9 promotes the assembly of NLRP3 inflammasome. The in vivo implications of the deduced mechanism were demonstrated by TIFA activation under several pathophysiological conditions in the vasculature.
Results
Oxidative Stress Induces TIFA in Vitro and in Vivo.
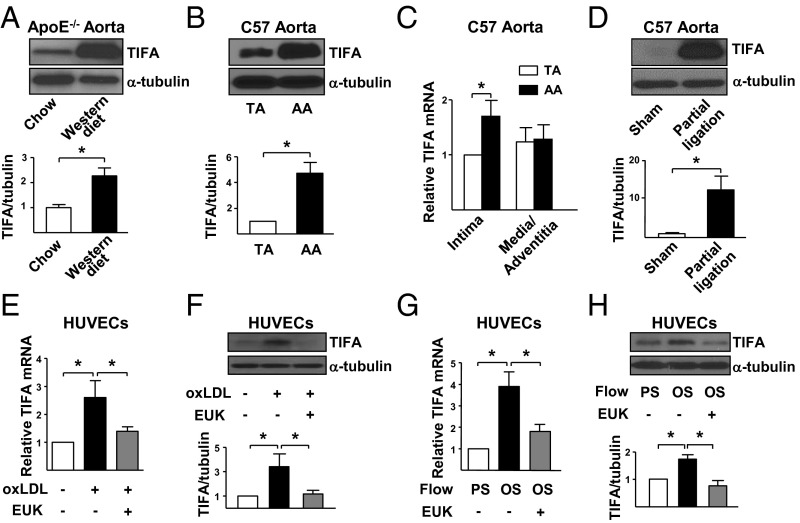
We first examined whether hyperlipidemia and atheroprone flow, both of which cause oxidative and/or inflammatory stresses, could induce TIFA in the vasculature in three mouse models. In the first set of experiments, TIFA was found to be markedly elevated in aortae from ApoE−/− mice receiving a Western diet compared with chow diet (Fig. 1A). In the second set of experiments, TIFA expression was significantly higher in atheroprone flow areas [i.e., aortic arch (AA)] than the atheroprotective thoracic aorta (TA) in C57BL/6 mice (Fig. 1B). Such differential expression of TIFA in AA vs. TA was mainly a result of the difference in intima, because the TIFA mRNA level was comparable in the media/adventitia (Fig. 1C). In the third set of experiments, TIFA expression in the partially ligated carotid artery, where the flow pattern is disturbed (17), was found to be elevated vs. the sham control (Fig. 1D). In line with the results from the hyperlipidemic ApoE−/− mice in vivo, oxLDL treatment increased mRNA and protein levels of TIFA in cultured human umbilical vein ECs (HUVECs; Fig. 1 E and F). Consistent with results from in vivo experiments involving AA and partially ligated carotid artery, in vitro studies with oscillatory shear stress (OS), which simulates atheroprone flow, caused a higher TIFA expression in cultured ECs than pulsatile stress (PS), which mimics atheroprotective flow (Fig. 1 G and H). Pretreating ECs with the antioxidant EUK-134 ablated the oxLDL or OS induction of TIFA (Fig. 1 E–H). Additionally, TNF-α treatment increased TIFA expression in ECs (Fig. S1). Taken together, Fig. 1 shows that oxidative and inflammatory stimuli induce TIFA in the vessel wall and in cultured ECs.
Fig. 1.
Oxidative stress activates TIFA in vivo and in vitro. (A) TAs were isolated from 8-wk-old ApoE−/− mice fed regular chow or Western diet for 8 wk. The TIFA expression in isolated tissue extracts was examined by Western blot analysis. (B and C) TA and AA were isolated from C57BL/6 mice. (B) Protein level of TIFA was examined by Western blot analysis. (C) Isolated vessels were further dissected to intima and media/adventitia portions, and the mRNA level of TIFA was quantified by RT-qPCR with primers listed in Table S1. (D) Partial ligated or sham control carotid arteries were dissected from C57BL/6 mice. Tissue extracts from three animals were pooled, and TIFA level was assessed by Western blot analysis. (E–H) HUVECs were pretreated with or without antioxidant EUK-134 (1 μM) for 2 h and then oxLDL (100 μg/mL), PS (12 ± 4 dyn/cm2), or OS (0.5 ± 4 dyn/cm2) for 16 h before cell lysate collection. TIFA mRNA level (E and G) and protein level (F and H) in cells were assessed accordingly. Data are mean ± SEM from three independent experiments (*P < 0.05).
Fig. S1.
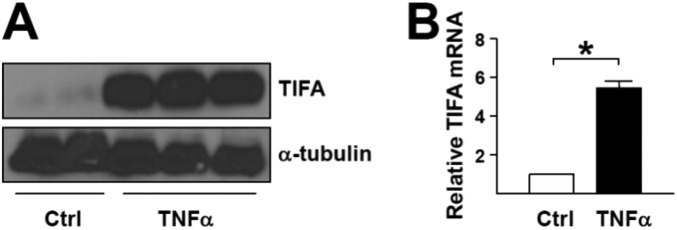
TNF-α induces TIFA expression. (A and B) HUVECs were treated with TNF-α (10 ng/mL) for 16 h. TIFA expression was measured by Western blot (A), and mRNA level was measured by RT-qPCR (B). Data are mean ± SEM from three independent experiments (*P < 0.05).
SREBP2 Transactivates TIFA.
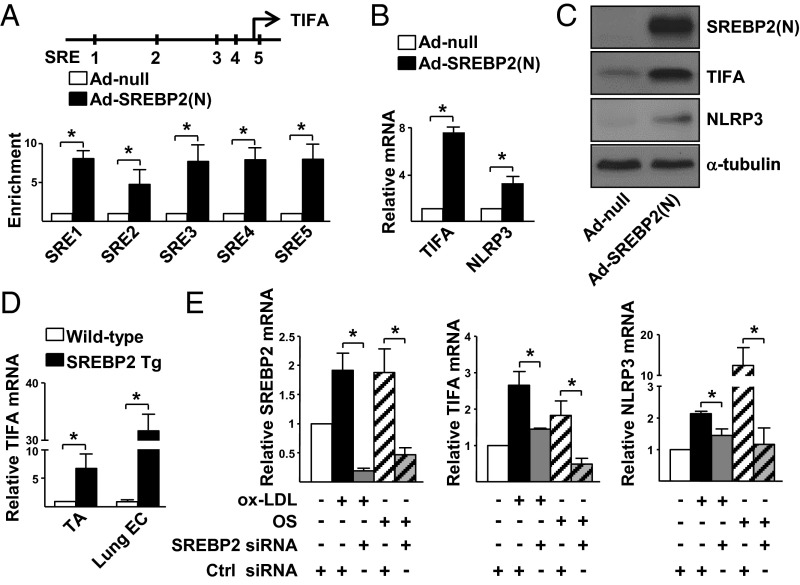
SREBP2 is an oxidative stress-activated TF in ECs, and its activation induces a panel of oxidative stress-sensitive genes, including those involved in the activation of NLRP3 inflammasome (14, 16). Having established the induction of TIFA by oxidative stress, and given the presence of five putative SREs in the upstream region of the human TIFA gene (−9,541, −7,671, −2,913, −2,318, and +282 bp from the TSS; Fig. 2A, Upper), we examined whether SREBP2 transactivates TIFA by overexpressing the active form of SREBP2, i.e., the N terminus [SREBP2(N)], in HUVECs. Overexpression of SREBP2(N) not only enhanced SREBP2(N) binding to the predicted SREs in ECs compared with mock control cells (Fig. 2A, Lower), but also raised the mRNA and protein levels of TIFA (Fig. 2 B and C). As a positive control, NLRP3 expression was increased, which we previously demonstrated to be an SREBP2 transcriptional target (14). In line with these in vitro results, TIFA expression was increased in ECs isolated from the aorta and lung of transgenic mice overexpressing SREBP2(N) in the endothelium [EC-SREBP2(N)-Tg; Fig. 2D] compared with those from the WT littermates. To examine whether SREBP2 is necessary for oxLDL- or OS-induced TIFA expression, we knocked down SREBP2 in ECs before oxLDL or OS stimulation, which led to an attenuation of the oxLDL- or OS-increased TIFA mRNA level (Fig. 2E).
Fig. 2.
SREBP2 transcriptionally up-regulates TIFA in ECs. (A) HUVECs were infected with Ad-SREBP2(N) for 48 h. ChIP assays were performed by using anti-SREBP2 or rabbit IgG as an isotype control. The enrichment of SREBP2(N) binding to the putative SREs in the promoter region of the TIFA gene (Upper) was quantified by RT-qPCR with primers listed in Table S2. (B and C) HUVECs were infected with Ad-SREBP2(N) or Ad-null. The mRNA and protein levels of TIFA were examined by RT-qPCR and Western blot, respectively. (D) TA and lung ECs were isolated from EC-SREBP2(N)-Tg mice and WT littermates. The levels of TIFA mRNA in the tissues were quantified by RT-qPCR. (E) HUVECs were transfected with SREBP2 or control siRNA (20 nM). The transfected cells were treated with or without oxLDL or OS. The levels of SREBP2, TIFA, and NLRP3 mRNA were examined by RT-qPCR. Data are mean ± SEM from three independent experiments (*P < 0.05 between compared groups).
TIFA Is Involved in Signal 1 of NLRP3 Inflammasome.
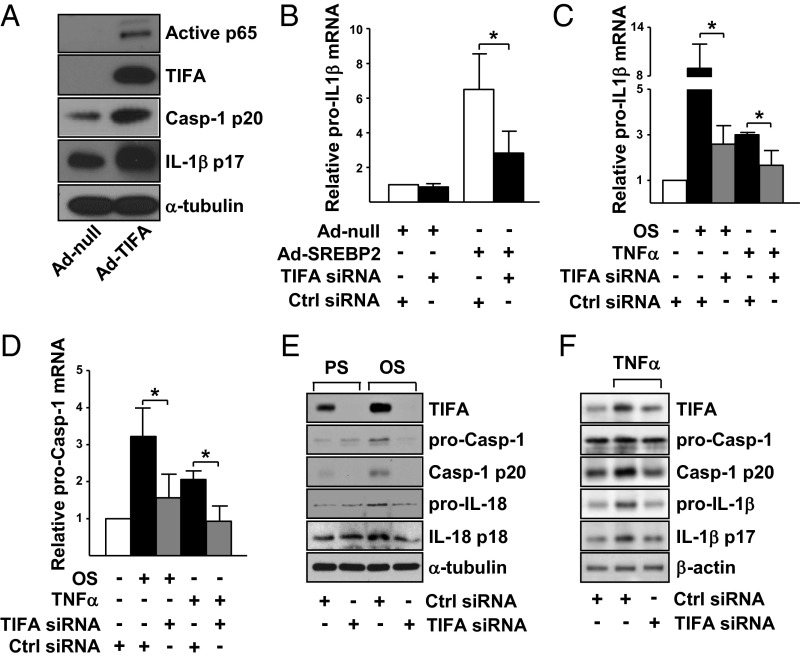
Because of the attendant induction of TIFA and NLRP3 by SREBP2 (14) (Fig. 2) and the activation of NF-κB by TIFA (10), the SREBP2–TIFA–NF-κB axis may be involved in signal 1 of inflammasome induction. To test this hypothesis, we first confirmed that NF-κB was indeed activated in ECs overexpressing SREBP2(N) (Fig. S2). Next, we overexpressed TIFA, which increased the expression of caspase-1 and IL-1β (Fig. 3A), indicative of inflammasome induction. Furthermore, SREBP2(N) induction of caspase-1 and IL-1β was inhibited following knockdown of TIFA (Fig. 3B). Because NF-κB–regulated signal 1 involves the transcriptional activation of NLRP3 inflammasome component proteins, we examined whether TIFA is necessary for the transcriptional induction of procaspase-1 and pro–IL-1β by various stimuli. TIFA knockdown attenuated the transcriptional induction of procaspase-1 and pro–IL-1β in ECs stimulated with OS or TNF-α (Fig. 3 C and D). TIFA knockdown also inhibited the protein expression of procaspase-1 and pro–IL-18/IL-1β with OS or TNF-α stimulation (Fig. 3 E and F). These results confirm the hierarchical order of the SREBP2–TIFA–NF-κB axis in signal 1 in ECs.
Fig. S2.
SREBP2 activates NF-κB. (A and B) HUVECs were infected with Ad-SREBP2 or Ad-null for 48 h, and NF-κB activity was detected through immunostaining (A) and Western blot (B).
Fig. 3.
TIFA is involved in signal 1 of NLRP3 inflammasome. (A) HUVECs were infected with Ad-null or Ad-TIFA. The protein level of phosphorylated p65 NF-κB, TIFA, caspase-1 p20, and IL-1β p17 was examined by Western blot. (B–F) HUVECs were transfected with TIFA siRNA (20 nM) or control siRNA (20 nM). (B) ECs were also infected with Ad-SREBP2. (C–F) ECs were stimulated with OS or TNF-α. The mRNA levels of pro–IL-1β mRNA and procaspase-1 were analyzed (B–D), along with the protein levels of TIFA, procaspase-1, caspase-1 p20, pro–IL-18 and IL-18 p18, or pro–IL-1β and IL-1β p17 (E and F). Data are mean ± SEM from three independent experiments (*P < 0.05).
Akt Promotes TIFA Oligomerization via TIFA Thr9 Phosphorylation.
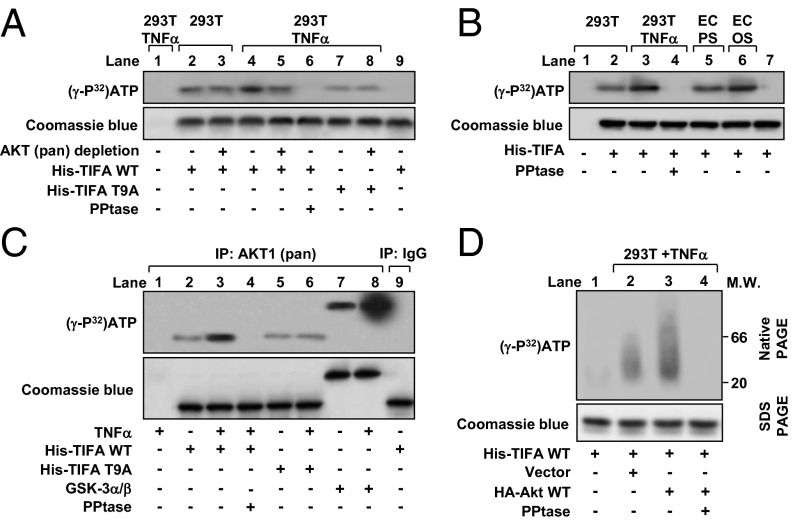
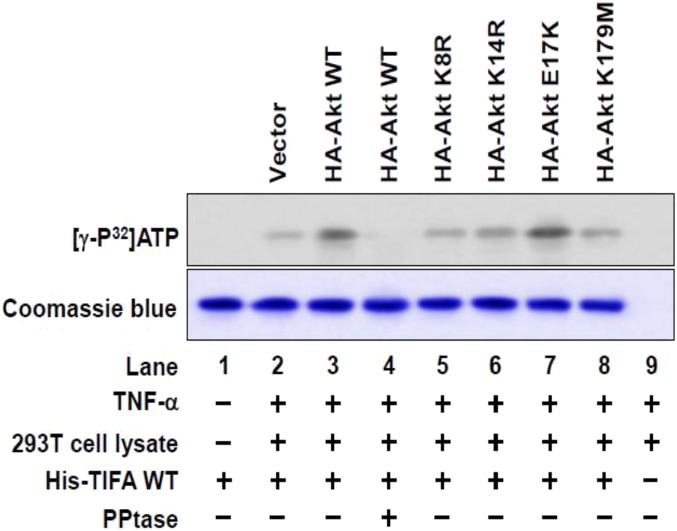
Given that oxidative and/or inflammatory stimuli, including OS, TNF-α, and oxLDL, all cause a sustained activation of Akt (18–20), and that TIFA Thr9 is phosphorylated under inflammatory conditions (10), we tested the hypothesis that Akt could increase the phosphorylation of TIFA Thr9, hence activating the NLRP3 inflammasome. We first found that incubation with lysates from TNF-α–treated HEK293T cells caused a greater phosphorylation of recombinant TIFA than those from the nontreated cells (Fig. 4A, lane 4 vs. lane 2). This effect was abolished following immunodepletion of Akt or addition of protein phosphatase to the kinase reaction mixture (Fig. 4A, lanes 5 and 6 vs. lane 4). As a negative control, recombinant TIFA-T9A (Thr9 replaced by an Ala) was not phosphorylated to the same level as the WT TIFA (Fig. 4A, lane 7 vs. lane 4). We have previously shown that OS caused sustained activation of Akt (18). Indeed, TIFA phosphorylation was greater when incubated with lysates from OS-treated ECs than with those from PS-treated cells (Fig. 4B, lane 6 vs. lane 5), and it was also greater when incubated with lysates from TNF-α–treated HEK293T cells than with those from untreated cells (Fig. 4B, lane 3 vs. lane 2). To test whether Akt could phosphorylate TIFA, we immunoprecipitated Akt from HEK293T cells, which was then incubated with recombinant TIFA WT, T9A, or GSK-3α/β (a canonical Akt substrate). Akt immunoprecipitated from TNF-α–treated cells increased the phosphorylation of coincubated TIFA-WT and GSK-3α/β, but not TIFA-T9A (Fig. 4C, lane 3 vs. lane 2, lane 6 vs. lane 5, lane 8 vs. lane 7). In contrast, the inclusion of protein phosphatase in the kinase reaction mixture abolished Akt phosphorylation of TIFA (Fig. 4C, lane 4 vs. lane 3). As a negative control, loss of function or kinase-dead mutants of Akt (i.e., K8R, K14R, or K179M) could not phosphorylate TIFA (Fig. S3). Because TIFA oligomerization has been shown to build on Thr9 phosphorylation (10), we tested whether the Akt-dependent TIFA Thr9 phosphorylation promotes TIFA oligomerization. Recombinant His-TIFA was incubated with lysates from HEK293T cells transfected with Akt and then treated with or without TNF-α. In vitro kinase assay followed by native gel electrophoresis revealed that Akt overexpression resulted in a TIFA band with increased molecular weight, indicative of enhanced TIFA oligomerization upon TNF-α treatment (Fig. 4D, lane 3 vs. lane 2). Such Akt-dependent TIFA oligomerization was abolished when protein phosphatase was included in the kinase reaction mixture (Fig. 4D, lane 4 vs. lane 3). Taken together, the results in Fig. 4 suggest that Akt kinase activity is required for the phosphorylation of TIFA Thr9 and the consequent TIFA oligomerization.
Fig. 4.
Akt is involved in the phosphorylation of TIFA Thr9. (A) HEK293T cells were treated with or without TNF-α for 30 min. Cell lysates were extracted, and Akt was depleted by immunoprecipitation (lanes 3, 5, and 8) before incubation with recombinant WT TIFA (His-TIFA-WT; lanes 2–6) or TIFA-T9A mutant (lanes 7 and 8) and [γ-32P]ATP. Alkaline phosphatase (PPtase) was added as indicated (lane 6). The reaction mixtures were separated by SDS-PAGE, and the bands were revealed by autoradiography. Lane 9 was a reaction mixture without cell lysates. (B) HUVECs were exposed to PS (lane 5) or OS (lane 6) for 30 min, and the collected cell lysates were incubated with recombinant WT TIFA and [γ-32P]ATP for in vitro kinase assay. Lysates from HEK293T cells treated with or without TNF-α (lanes 1–4) were used as positive controls, and lane 7 was reaction mixture without cell lysates. (C) Akt was immunoprecipitated from HEK293T cells treated with or without TNF-α for 30 min. The cell lysates were incubated with recombinant WT TIFA (lanes 2–4), TIFA-T9A mutant (lanes 5 and 6), or GSK-3α/β (lanes 7 and 8) together with [γ-32P]ATP for in vitro kinase assay. Rabbit IgG was an isotype control (lane 9). (D) HEK293T cells were transfected with or without the WT Akt and then treated with TNF-α for 30 min as indicated. His-TIFA-WT was incubated with cell lysates for in vitro kinase reaction. The polymerization of His-TIFA was analyzed by native gel electrophoresis. Coomassie blue staining was used for visualization and quantitation of TIFA protein in SDS-PAGE.
Fig. S3.
Akt is involved in TIFA phosphorylation. HEK293T cells were transiently transfected with Akt WT or mutants for 48 h followed by treatment with TNF-α (10 ng/mL). Recombinant TIFA protein was incubated with cell extracts in the presence of [γ-32P]ATP. Phosphorylated recombinant proteins were then immunoprecipitated by using anti-His mAb and eluted with His peptide. Eluted recombinant proteins were separated by SDS-PAGE and revealed by autoradiography or Coomassie blue stain as the loading control.
TIFA Induces the Assembly and Activation of NLRP3 Inflammasome via Signal 2.
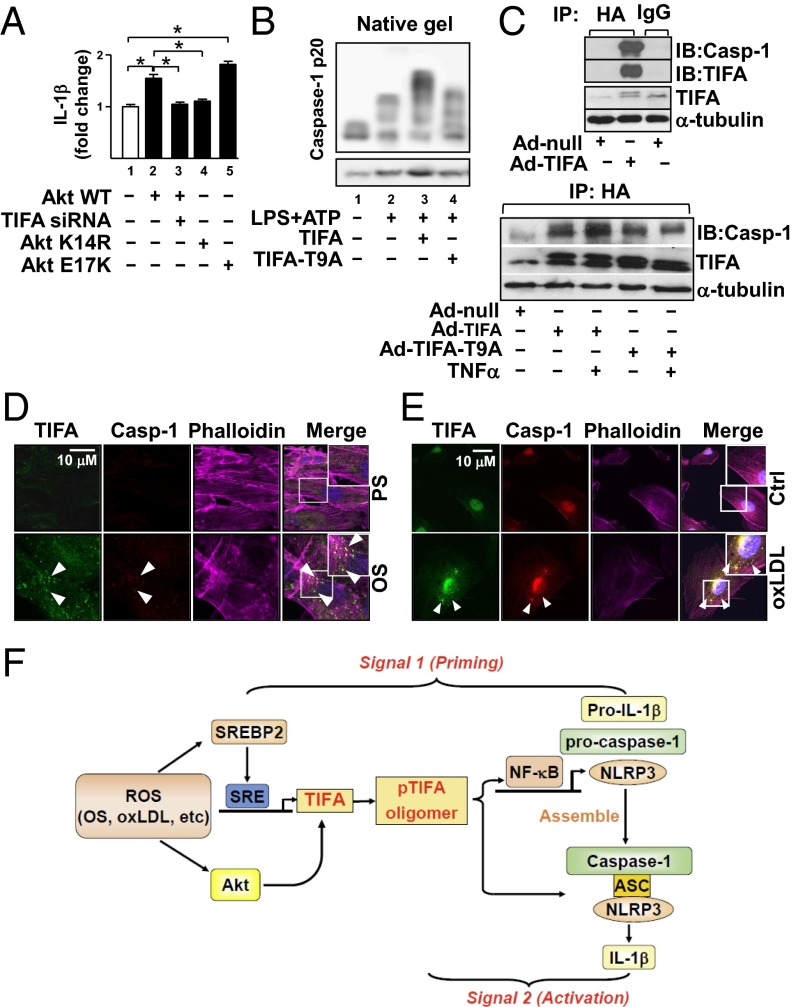
Higher-order assemblies of innate immune receptors such as those in the TNFR superfamily and the TLR/IL-1 receptor superfamily appear to be common in innate immune signaling (21, 22). Because TIFA activates TRAF and NLRP3 inflammasome (9) (Fig. 3), we examined whether TIFA phosphorylation and oligomerization can induce the assembly and activation of NLRP3 inflammasome. To functionally link Akt phosphorylation of TIFA with inflammasome activation, we performed an NLRP3 inflammasome reconstitution assay in HEK293T cells lacking the endogenous inflammasome core proteins (23, 24). HEK293T cells were cotransfected with NLRP3, ASC, procaspase-1, and pro–IL-1β together with Akt WT, TIFA siRNA, Akt K14R (i.e., loss-of-function mutant), or Akt E17K (i.e., gain-of-function mutant). As shown in Fig. 5A, Akt WT and Akt E17K enhanced IL-1β production (Fig. 5A, lane 2 vs. lane 1 and lane 5 vs. lane 1). The increased IL-1β production was attenuated when endogenous TIFA was knocked down (Fig. 5A, lane 3 vs. lane 2) or Akt K14R mutant was coexpressed (Fig. 5A, lane 4 vs. lane 2). These results suggest that the Akt-activated TIFA is involved in the NLRP3 inflammasome activation.
Fig. 5.
TIFA pT9 is involved in signal 2 of NLRP3 inflammasome in ECs. (A) HEK293T cells were transfected with pcDNA3 (lane 1), DNA plasmid encoding WT Akt (lane 2), TIFA siRNA (lane 3), Akt K14R (lane 4), or a constitutively active Akt E17K (lane 5) together with ASC, NLRP3, caspase-1, or IL-1β for 48 h. The level of IL-1β in the supernatant was measured by ELISA. The data are fold change with the value in lane 1 set to 1. Data are mean ± SEM from three independent experiments (*P < 0.05). (B) HUVECs were transfected with WT TIFA (lane 3) or TIFA-T9A (lane 4) and then treated with LPS (100 ng/mL, 16 h) and ATP (5 mM, 30 min; lanes 2–4). Cell lysates were separated by native gel electrophoresis (Top) or SDS-PAGE (Bottom). Caspase-1 oligomerization was detected by Western blot. (C) HUVECs were infected with various adenovirus as indicated; (Lower) cells were also stimulated with or without TNF-α for 4 h as shown. HA-TIFA was immunoprecipitated by anti-HA. Western blots showed the coimmunoprecipitated caspase-1 or TIFA. (D and E) HUVECs were stimulated with PS, OS, or oxLDL for 16 h and then immunostained with anti-TIFA or anti–caspase-1 antibodies. The merged images reveal the colocalized TIFA and caspase-1 in OS- or oxLDL-stimulated cells. (F) Graphic illustration of TIFA involvement in signal 1 and signal 2 for NLRP3 inflammasome activation in ECs.
To examine the causality of TIFA activation with that of NLRP3 inflammasome, we investigated whether TIFA overexpression combined with inflammasome stimuli (e.g., LPS/ATP) results in oligomerization of inflammasome component proteins (e.g., caspase-1). LPS and ATP cotreatment indeed induced caspase-1 oligomerization (Fig. 5B, lane 2 vs. lane 1). Overexpression of TIFA further increased the level of oligomerized caspase-1 (Fig. 5B, lane 3 vs. lane 2). However, such augmented caspase-1 oligomerization was not seen in cells overexpressing TIFA-T9A (Fig. 5B, lane 4 vs. lane 3). Besides oligomerization, caspase-1 expression would also contribute to the increased band intensity of caspase-1. This is evident by the increased high molecular weight of caspase-1 in lane 3 in the native gel compared with lanes 2 and 4 in Fig. 5B.
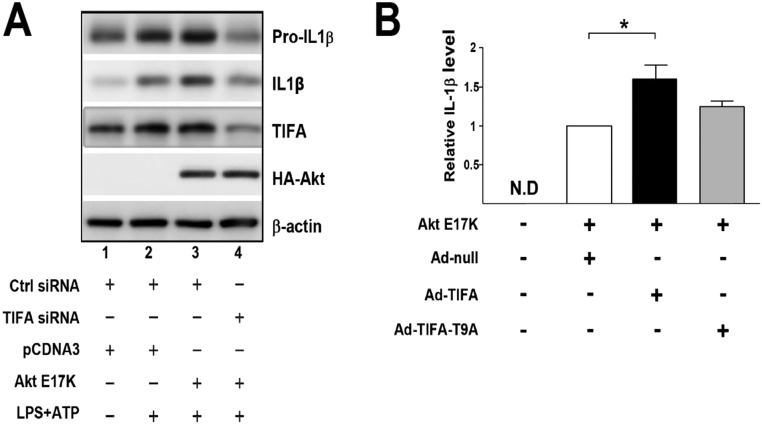
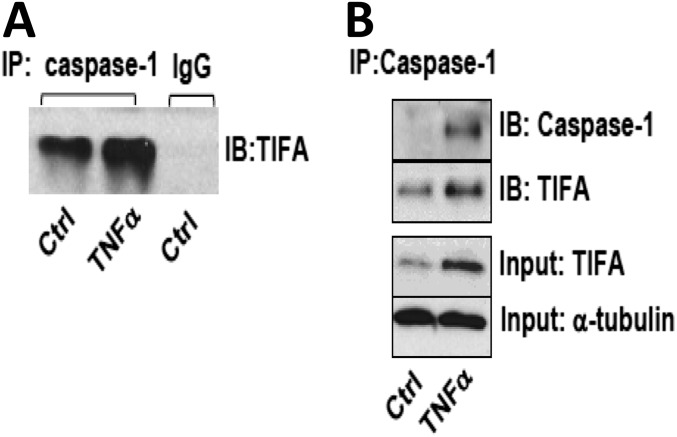
To this end, the Akt–TIFA–NLRP3 inflammasome axis may constitute an innate immune response in ECs. To test this hypothesis, we overexpressed Akt E17K in HUVECs in which TIFA siRNA or control siRNA was cotransfected. As shown in Fig. S4A, Akt E17K overexpression together with LPS/ATP stimulation potentiated pro–IL-1β expression and its cleavage (Fig. S4A, lane 3 vs. lane 2). TIFA knockdown abolished such activation of IL-1β (Fig. S4A, lane 4 vs. lane 3). Moreover, overexpression of TIFA, but not TIFA-T9A, increased the Akt E17K-increased IL-1β (Fig. S4B). To study the molecular basis underlying TIFA oligomerization of caspase-1, we overexpressed TIFA in HUVECs. Coimmunoprecipitation (co-IP) experiments show the interaction of TIFA with caspase-1 (Fig. 5C, Upper). This interaction was enhanced in ECs treated with TNF-α (Fig. S5). To further elucidate the role of TIFA Thr9 phosphorylation in NLRP3 inflammasome activation, we infected HUVECs with Ad-TIFA WT or Ad-TIFA-T9A and then examined TIFA-caspase-1 association under TNF-α treatment. Results in Fig. 5C, Lower, showed that TIFA WT, but not T9A mutant, increased the interaction between TIFA and caspase-1.
Fig. S4.
Akt activates the TIFA-NLRP3 inflammasome axis in ECs. (A) HUVECs were transfected with control siRNA, TIFA siRNA, pCDNA3, or Akt E17K as indicated. The cells were then stimulated with or without LPS and ATP. Cells were lysed, and IL-1β cleavage was measured by Western blot. (B) HUVECs were transfected with Akt E17K and infected with Ad-null, Ad-TIFA WT, or Ad-TIFA T9A as indicated. Cells in the various experimental groups were lysed, and IL-1β contents were measured by ELISA (*P < 0.05).
Fig. S5.
TNF-α increased TIFA interaction with caspase-1. HUVECs were treated with TNF-α for 4 h. (A) IgG control. (B) Caspase-1 was immunoprecipitated, and immunoblotting was then performed to show the increased interaction between TIFA and caspase-1.
In macrophages, NLRP3 inflammasome activation can be observed microscopically by the newly formed speckles, an indication of aggregated inflammasome components (25). We also assessed the role of TIFA in OS- and oxLDL-activated NLRP3 inflammasome by examining whether OS or oxLDL induces the inflammasome speckles, and, if so, whether such aggregates colocalize with TIFA and/or caspase-1. ECs under OS (compared with PS) or treated with oxLDL (compared with untreated control) showed a marked increase in the amount of speckles coimmunostained with anti-TIFA and anti–caspase-1 (Fig. 5 D and E). Together, the results in Fig. 5 suggest that the Akt-enhanced phosphorylation of TIFA Thr9 induces TIFA interaction with caspase-1, which would constitute “signal 2” to promote the assembly of NLRP3 inflammasome, a necessary step in inflammasome activation.
Discussion
In this study, we investigated the mechanism by which TIFA activates NLRP3 inflammasome in the context of atheroprone flow and other prooxidative and proinflammatory stimuli known to induce innate immune response in the endothelium. Our results indicate that TIFA is engaged in signal 1 (i.e., priming) and signal 2 (i.e., activation) required for NLRP3 inflammasome activation. In particular, SREBP2 induction of TIFA is required for the transcriptional activation of proinflammatory cytokines (i.e., pro–IL-1β) and inflammasome components (i.e., procaspase-1), which would be considered signal 1. Additionally, Akt-enhanced phosphorylation of TIFA Thr9 promotes the assembly of NLRP3 inflammasome, which is considered as signal 2 of inflammasome activation (Fig. 5F).
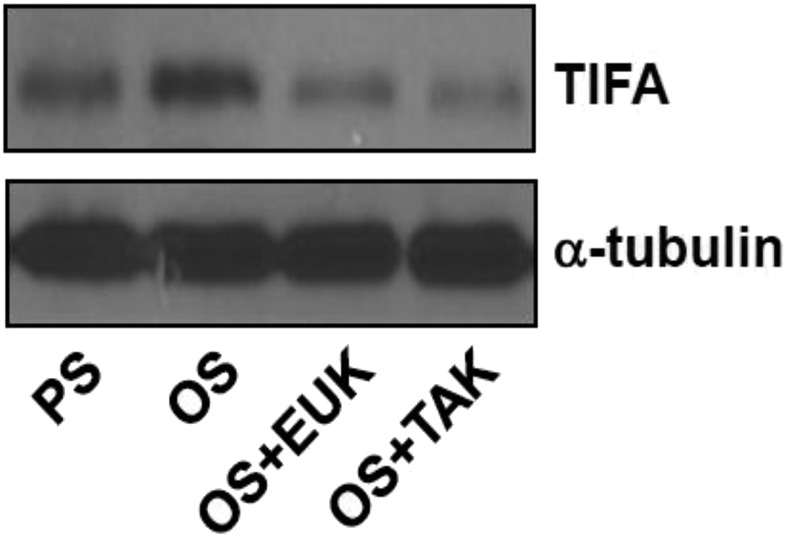
SREBP2, with its canonical role in regulating cholesterol homeostasis in the cell, is implicated in innate immunity, in part through its activation of NLRP3 inflammasome (26). Activated by atheroprone flow and a number of proinflammatory stimuli, the SREBP2 transactivation of TIFA, illustrated in Fig. 2, would be a new mechanism of signal 1 in the endothelial innate immune response. Given that TIFA activates NF-κB (10) and that NF-κB–mediated transcription primes the signal 1 of inflammasome (27), the SREBP2–TIFA–NF-κB axis would be a nexus linking proinflammatory and prooxidative stresses with the innate immune response in the endothelium. In monocytes/macrophages, oxLDL activates NF-κB via a complex encompassing CD36, TLR4, and TLR6 (28). Therefore, TLR activation may be an upstream molecular event leading to signal 1 activation. In ECs, OS and oxLDL induce TIFA (Fig. 1) and SREBP2 (14, 17). LPS stimulation also activates SREBP2 (29, 30), presumably via TLRs and their downstream effector TRAF6 (10). Collectively, TLRs could be common innate immune receptors in response to oxidative stress, including OS and oxLDL that induce signal 1. In support of this proposition, blocking the TLR pathway inhibited TIFA induction (Fig. S6).
Fig. S6.
OS-induced TIFA is attenuated by EUK-134 and TAK-242. HUVECs were pretreated with EUK-134 (1 mM) or TAK-242 (1 mM) for 2 h before OS for 16 h. TIFA protein levels were analyzed by Western blot.
There is ample evidence that the stimulation of TLRs activates PI3K/Akt (31). However, PI3K/Akt can also negatively regulate TLR signaling in a feedback manner (32, 33). We demonstrated that Akt is a kinase involved in TIFA Thr9 phosphorylation, which leads to TIFA oligomerization in ECs (Fig. 4). The aberrant oxidative stress in ECs may elicit sequential events that encompass Akt phosphorylation of TIFA Thr9 and the consequent binding of TIFA’s FHA domain to pT9, thus causing the TIFA–TIFA homophilic association. In the context of atherosclerosis susceptibility, we previously showed that atheroprone flow persistently activates Akt (18). Hence, Akt phosphorylation of TIFA with the ensuing TIFA oligomerization might contribute to the unresolved innate immune response in atheroprone areas (34). Interestingly, TRAF6 can function as an E3 ligase to ubiquitinate and activate Akt and IKK via K63 linkage (35, 36). Moreover, NLRP3 inflammasome activity can be enhanced by the TRAF3-dependent K63 polyubiquitination of ASC (37). Thus, a comprehensive network, rather than a simple hierarchical axis, may exist among Akt, TIFA, TRAF6, and TRAF3 to regulate the innate immune response.
In macrophages, signal 2-dependent stimuli, including exogenous ATP, K+ efflux, and ROS, initiate the assembly and activation of NLRP3 inflammasome (27, 38–40). Our results in Fig. 5 suggest that TIFA pT9 guides the higher-order assembly of inflammasome core proteins (i.e., NLRP3, ASC, and caspase-1), which would constitute signal 2 in activating NLRP3 inflammasome in ECs. Significantly, TIFA oligomerization seems to predominantly depend on pT9 rather than FHA or TRAF6 binding sites (i.e., Ser66 and Glu178, respectively) (10). The recently reported TIFA crystal structure supports such higher-order assembly of TIFA (11). Because TIFA activates signal 1 via TRAF6 oligomerization and signal 2 via the assembly of the inflammasome component proteins, phosphorylation of TIFA T9 would be essential for the innate immune response.
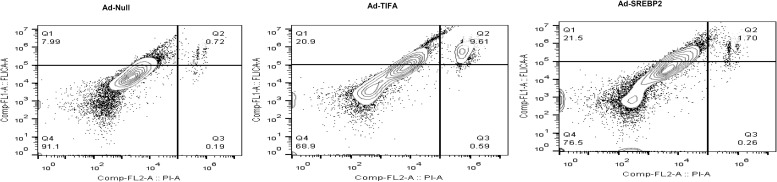
OS, oxLDL, and TNF-α increased the interaction between TIFA and procaspase-1, as shown by the co-IP and coimmunostaining experiments (Fig. 5 C–E). The caspase-1–dependent pyroptosis is a form of cell death triggered by the inflammasome (41). Thus, TIFA activation of caspase-1 in ECs is also supported by the increased pyroptosis in ECs overexpressing SREBP2 or TIFA (Fig. S7). ASC provides a scaffold between NLRP3 and procaspase-1 to form supramolecular structures of inflammasome, which relies on the interactions of the pyrin domain (PYD) in NLRP3 and ASC, as well as the caspase activation and recruitment domain (CARD) in ASC and procaspase-1 (1, 23, 42). TIFA, which does not contain PYD- or CARD-like domains, may nucleate the NLRP3 inflammasome by directly binding to NLRP3, ASC, and/or procaspase-1. Alternatively, TIFA might act as a prion-like “proteinaceous nucleating particle” (43) to enhance the higher-order assembly of the NLRP3–ASC–procasepase-1 complex.
Fig. S7.
TIFA or SREBP2 overexpression induces EC pyroptosis. HUVECs were infected with Ad-TIFA or Ad-SREBP2 for 48 h. Caspase-1 activity was determined by flow cytometry with the use of a FAM FLICA Caspase-1 Assay kit. Membrane integrity was analyzed by PI staining.
In summary, we have demonstrated that TIFA plays a crucial role in the innate immune response in ECs induced by prooxidative and proinflammatory stimuli, including atheroprone flow, oxLDL, and TNF-α. The mechanisms involve the transcriptional up-regulation of TIFA by SREBP2 (signal 1) and the oligomerization of TIFA by TIFA pT9 to induce the higher-order assembly of NLRP3 inflammasome (signal 2; Fig. 5F). Because unresolved innate immunity in the endothelium plays a pathophysiological role in vascular impairments, targeting TIFA may provide a new therapeutic strategy for vascular diseases. In light of this, Aurora A kinase has recently been shown to phosphorylate Thr9 of TIFA as well, and targeting TIFA can enhance therapeutic efficacies in the treatment of acute myeloid leukemia (44).
Materials and Methods
The sources of antibodies and reagents and detailed methods for cell culture, shear stress experiments, RNAi inhibition, adenovirus infection, bioinformatics methods, ChIP assay, immunoblotting, real-time quantitative PCR (RT-qPCR), and caspase-1 activity/pyroptosis are described in SI Materials and Methods.
Animal Experiments.
Animal experimental protocols were approved by the institutional animal care and use committee of the University of California, San Diego. ApoE−/− or EC-SREBP2(N)-Tg-ApoE−/− genotypes were generated by crossing EC-SREBP2(N)-Tg mice (14) and ApoE−/− mice. All mice were housed in colony cages with a 12-h light/dark cycle and fed ad libitum unless otherwise indicated. Mouse aortae and lung ECs were isolated for RNA or protein extraction. In animals receiving Western diet, it was started at 8 wk of age. Male EC-SREBP2(N)+/+ApoE−/− and their EC-SREBP2(N)−/−ApoE−/− littermates were fed a Western diet containing 15% (wt/wt) fat, 1.25% (wt/wt) cholesterol, and 0.5% sodium cholate (Harlan Teklad) ad libitum. Mice were killed 8 wk later, and aortae were isolated for RNA or protein extraction. The isolation of primary mouse lung ECs was described previously (14). The partial carotid ligation model was as described by Nam et al. (17).
In Vitro Kinase Assay.
HEK293T cells under the indicated conditions were lysed with CHAPS lysis buffer containing 20 mM Pipes, 1 mM Na3VO4, 1 mM EGTA, 50 mM Tris⋅HCl, 150 mM NaCl, 50 mM NaF, 1% CHAPS, and 10% (vol/vol) glycerol supplemented with protease inhibitor mixture (Roche) and phosphatase inhibitor (Sigma). The cell extracts were precleaned with sheep anti-mouse IgG M-280 Dynabeads (Invitrogen) at room temperature for 30 min. The resulting cell extracts were incubated with the recombinant His-tagged WT or T9A mutant TIFA in the reaction buffer (40 mM Hepes, pH 7.5, 20 mM MgCl2, and 100 μM ATP) in the presence of 10 μM [γ-32P]ATP (Perkin-Elmer) at 37 °C for 30–90 min. His-tagged proteins were pulled down with M-280 Dynabeads coated with anti-His mAb. The reaction was terminated by the addition of SDS sample buffer and heating at 95 °C for 10 min before gel electrophoresis.
Inflammasome Reconstitution Assay.
HEK293T cells at 70% confluence were seeded onto six-well plates overnight and then transfected with plasmids encoding inflammasome component proteins, including 200 ng pcDNA4–pro-IL-1β, 25 ng pcDNA4-NLRP3-Myc, 20 ng pcDNA-ASC, and 10 ng pcDNA4–procaspase-1-Myc. Cell pellets were collected 48 h after transfection and lysed in cell lysis buffer for immunoblotting analysis. Cell supernatants were collected for ELISA to detect IL-1β secretion (R&D Systems).
Immunoprecipitation.
Cells were washed with PBS solution, UV cross-linked, and then lysed. After centrifugation at 13,000 × g at 4 °C for 15 min, Dynabead Protein G (DynaG) was added and washed with PBS solution with 0.1% BSA before incubation with indicated antibodies at 4 °C for 45 min. The supernatants were precleared with anti-mouse IgG and then incubated with antibody-coated DynaG at 4 °C for 12 h. The beads were washed with PBS solution and reconstituted in 50 μL lysis buffer before immunoblotting.
Statistical Analysis.
Data are expressed as mean ± SEM (n = 3 unless otherwise noted). For parametric data, Student’s t test or ANOVA was used to analyze differences among groups if data were normally distributed. For nonparametric data, Mann–Whitney U test with the exact method was used to analyze differences between two groups. P < 0.05 was considered statistically significant.
SI Materials and Methods
Anti-TIFA monoclonal antibody was raised by the antibody core facility in Academia Sinica; anti-SREBP2 antibodies were obtained from the BD Transduction Laboratory and Abcam; anti-Flag, anti–IL-1β, anti–caspase-1, anti–α-tubulin, anti–β-actin, and HRP-conjugated anti-rabbit and anti-mouse antibodies were from Cell Signaling Technology; anti-NLRP3 was from Abcam; anti-ASC antibody was from Enzo Life Science; antibodies for NF-κB p65, phospho-p65 NF-κB, and anti–IL-18 were from Santa Cruz Biotechnology and that for active-p65 NF-κB was from Millipore; oxLDL was from Athens Research & Technology; LPS and ATP were from Invivogen; and TNF-α was from R&D Systems. Human IL-1b ELISA kit was from BD Biosciences, and FAM FLICATM Caspase 1 assay kit was from ImmunoChemistry.
Cell Culture and Shear Stress Experiments.
HUVECs were cultured in medium M199 (Gibco) supplemented with 15% (vol/vol) FBS (Fisher), 3 ng/mL β-EC growth factor, 4 U/mL heparin, 100 mg/L sodium pyruvate, and 100 U/mL penicillin/streptomycin. HEK293T cells were maintained in DMEM-high glucose (Gibco) supplemented with 10% (vol/vol) FBS, 6 mM l-glutamine, and 100 U/mL penicillin/streptomycin. All cells were incubated in a humidified atmosphere at 37 °C with 5% CO2. When necessary, cells were starved in serum-free condition for 8 h before the addition of 10 ng/mL of TNF-α or 100 μg/mL oxLDL at the indicated times. Shear stress experiments were conducted in a parallel plate flow system. The atheroprotective PS was 12 ± 4 dyn/cm2, and the atheroprone OS was 0.5 ± 4 dyn/cm2.
RNAi Inhibition and Adenovirus Infection.
Cells were seeded on culture dishes overnight at 70% confluence. Cells were then transfected with SREBP2 siRNA (Qiagen), TIFA siRNA (Invitrogen), or control siRNA at 20 nM by using Lipofectamine RNAi Max (Invitrogen). Culture medium was changed after 4 h incubation with transfection complexes, and cells were collected after 48 h incubation. To overexpress TIFA or SREBP2 by adenovirus in HUVECs, 70% confluent cultured HUVECs were treated with the adenoviruses and incubated for 4 h before medium change. The infected cells were then incubated for 48 h before harvest and detection of mRNA or protein level of interest.
Binding Site Prediction and ChIP Assay.
The position weight matrix algorithm from TRANSFAC 15 was used to scan the promoter region of the human TIFA gene for the putative SREBP2 binding sites. The promoter region used for prediction was defined as approximately −10,000 to +1,000 from the transcriptional start site of TIFA mRNA. ChIP assays were performed with anti-SREBP2 (Abcam) and rabbit IgG as an isotype control (Cell Signaling). In brief, cells were treated with formaldehyde to cross-link protein and DNA. Cells were collected in a lysis buffer (50 Mm Hepes⋅KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, pH 8, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, protease inhibitors) and sonicated to shear the DNA to approximately 500–1,000 bp, and then cells were centrifuged and supernatants were immunoprecipitated with anti-SREBP2 antibody (Abcam) overnight. The bound DNA was eluted and underwent quantitative PCR with primers targeting the predicted binding sites in the TIFA promoter region. The sequences of these primers are listed in Table S2.
Table S2.
ChIP primer sequences
| SRE | Location | Sequence | |
| 1 | −9541 to TSS | Forward | CAAAGTGCTGGGATTACAG |
| Reverse | AGGCTGAGTGAGCTGTGAT | ||
| 2 | −7671 to TSS | Forward | GTGGGTGGATTAGGAGGCAA |
| Reverse | TCCTGTACATTCCCCTTTGGT | ||
| 3 | −2913 to TSS | Forward | ACACATGCCTACTGTTTTCGTC |
| Reverse | CTCATGGGCTGCCAACATCT | ||
| 4 | −2318 to TSS | Forward | AAATGTCTAGTACCTTTCATTGC |
| Reverse | GGGACATTTGCAAAAGAAGCA | ||
| 5 | 282 to TSS | Forward | AGAGAGTTCACTGACTCCCCA |
| Reverse | TTTTGCAGGCTTTCCTTAGCA | ||
Immunoblotting.
Cell or tissue samples were lysed in Nonidet P-40 lysis buffer containing 10 mM Tris, pH 7.4, 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10% (vol/vol) glycerol, 1 mM PMSF, and protease inhibitor mixture (Roche). The extracted proteins were resolved by SDS-PAGE and then transferred to PVDF membrane (Millipore) for immunoblotting with antibodies as indicated. Protein bands were revealed by the ECL system (Millipore).
RT-qPCR.
Total RNA from cells or tissues was isolated by TRIzol reagent (Invitrogen). cDNAs were synthesized with use of the PrimeScript RT kit (Takara). Quantitative PCR involved use of the iCycler real-time PCR Detection System (Bio-Rad) with iQ SYBR Green Supermix (Bio-Rad). Quantification of mRNA levels involved use of CFX Manager software (Bio-Rad). Primers used in the PCR are listed in the Table S1.
Table S1.
Primer sequences for real-time PCR
| Gene | Species | Sequence | |
| β-Actin | Human | Forward | CATGTACGTTGCTATCCAGGC |
| Reverse | CTCCTTAATGTCACGCACGAT | ||
| β-Actin | Mouse | Forward | GGCTGTATTCCCCTCCATCG |
| Reverse | CCAGTTGGTAACAATGCCATGT | ||
| SREBP2 | Human | Forward | CCCTGGGAGACATCGACGA |
| Reverse | CGTTGCACTGAAGGGTCCA | ||
| Srebp2 | Mouse | Forward | GCAGCAACGGGACCATTCT |
| Reverse | CCCCATGACTAAGTCCTTCAACT | ||
| TIFA | Human | Forward | CAAACAGGTTTCCCGAGTTCA |
| Reverse | TGTCCACGATCAGATTGGTCTT | ||
| Tifa | Mouse | Forward | ACGCAATTCCAACATGTGCC |
| Reverse | CCGAGCTCCTGGTTGTCTAC | ||
| IL-1β | Human | Forward | AGGCACAAGGCACAACAGGCTG |
| Reverse | GTCCTGGAAGCACTTCATCTGT | ||
| Caspase-1 | Human | Forward | CTTCCTTTCCAGCTCCTCAGGCA |
| Reverse | CGTGTGCGGCTTGACTTGTCC | ||
| NLRP3 | Human | Forward | TGCCGGGGCCTCTTTTCAGT |
| Reverse | CCACAGCGCCCCAACCACAA | ||
Caspase-1 Activity and Pyroptosis.
Caspase-1 activity and cell pyroptosis were measured with use of a FAM-FLICA Caspase-1 Assay Kit. The fluorescent inhibitor probe FAM-YVAD-FMK labels active caspase-1 enzyme in living cells. In brief, cell were trypsinized and incubated FLICA reagent for 1 h. Cells were then washed with PBS solution three times, counterstained with propidium iodide (PI), and analyzed with a flow cytometer.
Acknowledgments
The authors thank Drs. Feng-Mao Lin, Hsi-Yuan Huang, and Hsien-Da Huang at National Chao-Tung University, Taiwan, for their bioinformatics analysis; Dr. Pei-Yu Gabriel Wu at the Institute of Biological Chemistry (IBC) of Academia Sinica for technical advice; Drs. Jui-Hung Weng and Chia-Chi Flora Huang at IBC for providing recombinant TIFA protein; and Dr. Ming-Zong Lai at the Institute of Molecular Biology of Academia Sinica for providing the plasmids encoding inflammasome components. This work was supported by National Institutes of Health Grants HL108735 (to J.Y.-J.S. and S.C.) and K99HL122368 (to Z.C.), Taiwan National Health Research Institute Grant NHRI-EX100-10002NI (to M.-D.T.), and Taiwan Protein Project of Academia Sinica funded by Ministry of Science and Technology Grant MOST105-0210-01-12-01 (to M.-D.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618773114/-/DCSupplemental.
References
- 1.Martinon F, Burns K, Tschopp J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10(2):417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 2.Tschopp J, Schroder K. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat Rev Immunol. 2010;10(3):210–215. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 3.Agostini L, et al. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20(3):319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 4.Mariathasan S, et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430(6996):213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 5.Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481(7381):278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 6.Henao-Mejia J, Elinav E, Strowig T, Flavell RA. Inflammasomes: Far beyond inflammation. Nat Immunol. 2012;13(4):321–324. doi: 10.1038/ni.2257. [DOI] [PubMed] [Google Scholar]
- 7.Kanamori M, Suzuki H, Saito R, Muramatsu M, Hayashizaki Y. T2BP, a novel TRAF2 binding protein, can activate NF-kappaB and AP-1 without TNF stimulation. Biochem Biophys Res Commun. 2002;290(3):1108–1113. doi: 10.1006/bbrc.2001.6315. [DOI] [PubMed] [Google Scholar]
- 8.Takatsuna H, et al. Identification of TIFA as an adapter protein that links tumor necrosis factor receptor-associated factor 6 (TRAF6) to interleukin-1 (IL-1) receptor-associated kinase-1 (IRAK-1) in IL-1 receptor signaling. J Biol Chem. 2003;278(14):12144–12150. doi: 10.1074/jbc.M300720200. [DOI] [PubMed] [Google Scholar]
- 9.Ea CK, Sun L, Inoue J, Chen ZJ. TIFA activates IkappaB kinase (IKK) by promoting oligomerization and ubiquitination of TRAF6. Proc Natl Acad Sci USA. 2004;101(43):15318–15323. doi: 10.1073/pnas.0404132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang CC, et al. Intermolecular binding between TIFA-FHA and TIFA-pT mediates tumor necrosis factor alpha stimulation and NF-κB activation. Mol Cell Biol. 2012;32(14):2664–2673. doi: 10.1128/MCB.00438-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weng JH, et al. Uncovering the mechanism of Forkhead-Associated Domain-mediated TIFA oligomerization that plays a central role in immune responses. Biochemistry. 2015;54(40):6219–6229. doi: 10.1021/acs.biochem.5b00500. [DOI] [PubMed] [Google Scholar]
- 12.Gaudet RG, et al. INNATE IMMUNITY. Cytosolic detection of the bacterial metabolite HBP activates TIFA-dependent innate immunity. Science. 2015;348(6240):1251–1255. doi: 10.1126/science.aaa4921. [DOI] [PubMed] [Google Scholar]
- 13.Horton JD, Goldstein JL, Brown MS. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109(9):1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao H, et al. Sterol regulatory element binding protein 2 activation of NLRP3 inflammasome in endothelium mediates hemodynamic-induced atherosclerosis susceptibility. Circulation. 2013;128(6):632–642. doi: 10.1161/CIRCULATIONAHA.113.002714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li LC, et al. Cross-talk between TLR4-MyD88-NF-κB and SCAP-SREBP2 pathways mediates macrophage foam cell formation. Am J Physiol Heart Circ Physiol. 2013;304(6):H874–H884. doi: 10.1152/ajpheart.00096.2012. [DOI] [PubMed] [Google Scholar]
- 16.Chen Z, et al. Oxidative stress activates endothelial innate immunity via sterol regulatory element binding protein 2 (SREBP2) transactivation of microRNA-92a. Circulation. 2015;131(9):805–814. doi: 10.1161/CIRCULATIONAHA.114.013675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nam D, et al. Partial carotid ligation is a model of acutely induced disturbed flow, leading to rapid endothelial dysfunction and atherosclerosis. Am J Physiol Heart Circ Physiol. 2009;297(4):H1535–H1543. doi: 10.1152/ajpheart.00510.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo D, Chien S, Shyy JY. Regulation of endothelial cell cycle by laminar versus oscillatory flow: Distinct modes of interactions of AMP-activated protein kinase and Akt pathways. Circ Res. 2007;100(4):564–571. doi: 10.1161/01.RES.0000259561.23876.c5. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Himi T, Morita I, Murota S. Inhibition of phosphatidylinositol-3 kinase/Akt or mitogen-activated protein kinase signaling sensitizes endothelial cells to TNF-alpha cytotoxicity. Cell Death Differ. 2001;8(5):528–536. doi: 10.1038/sj.cdd.4400838. [DOI] [PubMed] [Google Scholar]
- 20.Chavakis E, et al. Oxidized LDL inhibits vascular endothelial growth factor-induced endothelial cell migration by an inhibitory effect on the Akt/endothelial nitric oxide synthase pathway. Circulation. 2001;103(16):2102–2107. doi: 10.1161/01.cir.103.16.2102. [DOI] [PubMed] [Google Scholar]
- 21.Netea MG, et al. Nucleotide-binding oligomerization domain-2 modulates specific TLR pathways for the induction of cytokine release. J Immunol. 2005;174(10):6518–6523. doi: 10.4049/jimmunol.174.10.6518. [DOI] [PubMed] [Google Scholar]
- 22.Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153(2):287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu JW, et al. Cryopyrin and pyrin activate caspase-1, but not NF-kappaB, via ASC oligomerization. Cell Death Differ. 2006;13(2):236–249. doi: 10.1038/sj.cdd.4401734. [DOI] [PubMed] [Google Scholar]
- 24.Chuang YT, et al. Tumor suppressor death-associated protein kinase is required for full IL-1β production. Blood. 2011;117(3):960–970. doi: 10.1182/blood-2010-08-303115. [DOI] [PubMed] [Google Scholar]
- 25.Richards N, et al. Interaction between pyrin and the apoptotic speck protein (ASC) modulates ASC-induced apoptosis. J Biol Chem. 2001;276(42):39320–39329. doi: 10.1074/jbc.M104730200. [DOI] [PubMed] [Google Scholar]
- 26.Chen Z, Martin M, Li Z, Shyy JY. Endothelial dysfunction: The role of sterol regulatory element-binding protein-induced NOD-like receptor family pyrin domain-containing protein 3 inflammasome in atherosclerosis. Curr Opin Lipidol. 2014;25(5):339–349. doi: 10.1097/MOL.0000000000000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13(6):397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart CR, et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat Immunol. 2010;11(2):155–161. doi: 10.1038/ni.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Costales P, et al. Lipopolysaccharide downregulates CD91/low-density lipoprotein receptor-related protein 1 expression through SREBP-1 overexpression in human macrophages. Atherosclerosis. 2013;227(1):79–88. doi: 10.1016/j.atherosclerosis.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 30.Im SS, et al. Linking lipid metabolism to the innate immune response in macrophages through sterol regulatory element binding protein-1a. Cell Metab. 2011;13(5):540–549. doi: 10.1016/j.cmet.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arbibe L, et al. Toll-like receptor 2-mediated NF-kappa B activation requires a Rac1-dependent pathway. Nat Immunol. 2000;1(6):533–540. doi: 10.1038/82797. [DOI] [PubMed] [Google Scholar]
- 32.Fukao T, et al. PI3K-mediated negative feedback regulation of IL-12 production in DCs. Nat Immunol. 2002;3(9):875–881. doi: 10.1038/ni825. [DOI] [PubMed] [Google Scholar]
- 33.Guha M, Mackman N. The phosphatidylinositol 3-kinase-Akt pathway limits lipopolysaccharide activation of signaling pathways and expression of inflammatory mediators in human monocytic cells. J Biol Chem. 2002;277(35):32124–32132. doi: 10.1074/jbc.M203298200. [DOI] [PubMed] [Google Scholar]
- 34.Hajra L, et al. The NF-kappa B signal transduction pathway in aortic endothelial cells is primed for activation in regions predisposed to atherosclerotic lesion formation. Proc Natl Acad Sci USA. 2000;97(16):9052–9057. doi: 10.1073/pnas.97.16.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang WL, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325(5944):1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng L, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103(2):351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 37.Guan K, et al. MAVS promotes inflammasome activation by targeting ASC for K63-linked ubiquitination via the E3 ligase TRAF3. J Immunol. 2015;194(10):4880–4890. doi: 10.4049/jimmunol.1402851. [DOI] [PubMed] [Google Scholar]
- 38.Duncan JA, et al. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc Natl Acad Sci USA. 2007;104(19):8041–8046. doi: 10.1073/pnas.0611496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pétrilli V, et al. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 2007;14(9):1583–1589. doi: 10.1038/sj.cdd.4402195. [DOI] [PubMed] [Google Scholar]
- 40.Dostert C, et al. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320(5876):674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: Host cell death and inflammation. Nat Rev Microbiol. 2009;7(2):99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandes-Alnemri T, et al. The pyroptosome: A supramolecular assembly of ASC dimers mediating inflammatory cell death via caspase-1 activation. Cell Death Differ. 2007;14(9):1590–1604. doi: 10.1038/sj.cdd.4402194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai X, et al. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 2014;156(6):1207–1222. doi: 10.1016/j.cell.2014.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei TYW, et al. Aurora A and the NF-κB survival pathway drive chemoresistance in acute myeloid leukemia via TRAF-interacting protein TIFA. Cancer Res. November 10, 2016 doi: 10.1158/0008-5472.CAN-16-1004. [DOI] [PubMed] [Google Scholar]