Significance
Flowering plant reproduction involves two fertilization events: a sperm–egg fusion that forms the embryo, and a sperm–central cell fusion that forms the nutritive extraembryonic endosperm. Chromosomes in the embryo, endosperm, and other plant tissues are modified by methylation, a covalent addition to cytosine in DNA that regulates gene expression. Maternal endosperm chromosomes inherited from the central cell display a pattern of extensive demethylation, which is essential for seed development in Arabidopsis thaliana. Demethylation is presumed to occur in the central cell, but direct evidence for this is very limited. Here, we provide a genome-wide DNA methylation analysis of Arabidopsis and rice central cells, which demonstrates that the demethylation observed in the endosperm is indeed initiated in the central cell.
Keywords: DNA methylation, central cell, egg cell, epigenetics, gene imprinting
Abstract
Cytosine methylation is a DNA modification with important regulatory functions in eukaryotes. In flowering plants, sexual reproduction is accompanied by extensive DNA demethylation, which is required for proper gene expression in the endosperm, a nutritive extraembryonic seed tissue. Endosperm arises from a fusion of a sperm cell carried in the pollen and a female central cell. Endosperm DNA demethylation is observed specifically on the chromosomes inherited from the central cell in Arabidopsis thaliana, rice, and maize, and requires the DEMETER DNA demethylase in Arabidopsis. DEMETER is expressed in the central cell before fertilization, suggesting that endosperm demethylation patterns are inherited from the central cell. Down-regulation of the MET1 DNA methyltransferase has also been proposed to contribute to central cell demethylation. However, with the exception of three maize genes, central cell DNA methylation has not been directly measured, leaving the origin and mechanism of endosperm demethylation uncertain. Here, we report genome-wide analysis of DNA methylation in the central cells of Arabidopsis and rice—species that diverged 150 million years ago—as well as in rice egg cells. We find that DNA demethylation in both species is initiated in central cells, which requires DEMETER in Arabidopsis. However, we do not observe a global reduction of CG methylation that would be indicative of lowered MET1 activity; on the contrary, CG methylation efficiency is elevated in female gametes compared with nonsexual tissues. Our results demonstrate that locus-specific, active DNA demethylation in the central cell is the origin of maternal chromosome hypomethylation in the endosperm.
Flowering plant sexual reproduction is carried out by multicellular gametophytes that arise through mitosis from haploid meiotic spores (1–3). The male gametophyte, pollen, consists of two sperm cells and a companion vegetative cell. The vegetative cell forms the pollen tube that delivers the sperm into the female gametophyte, where one of the sperm cells fuses with an egg to form the zygote and the other sperm fuses with the homodiploid central cell to form the triploid endosperm (1–3). The endosperm is an extraembryonic, placenta-like tissue that nourishes the embryo and—particularly in monocots like rice and maize—the seedling after germination (4). Monocot endosperm also constitutes the bulk of human nutrition (4).
Proper gene expression in the endosperm requires extensive reprogramming of DNA methylation (5). Methylation is a covalent modification of cytosine that is accurately copied after DNA replication, thereby passing epigenetic information during cell division (6). Plant methylation is categorized into three contexts: CG, CHG (H = A, T, or G), and CHH (6). CG methylation, the most abundant and widespread of the three, is maintained by the MET1 DNA methyltransferase (6). All three types of methylation are found in transposable elements (TEs), which are transcriptionally repressed and heritably silenced by methylation (6). TE and TE-like methylated sequences that overlap gene regulatory regions influence gene expression—overlaps with transcriptional start sites and other regions that promote expression cause gene silencing, whereas overlaps with sequences that repress expression can enhance gene activity (5, 6). CG methylation is also common within the transcribed regions of genes, where its function is presently unclear (6).
In addition to methyltransferases, plants encode enzymes that can remove methylation from DNA (7). One of these, DEMETER (DME), is expressed in Arabidopsis thaliana male and female gametophytes, primarily in the vegetative and central cells, respectively (8, 9). In the vegetative cell, DME catalyzes demethylation of thousands of discrete loci, most of which are relatively euchromatic TEs (10, 11). The maternal endosperm chromosomes inherited from the central cell are also extensively demethylated at similar sequences in Arabidopsis (10) as well as in the distantly related monocots rice (12) and maize (13). As in the vegetative cell, demethylation of maternal endosperm chromosomes requires DME in Arabidopsis (10), and loss of DME function disrupts endosperm gene expression, gene imprinting, and causes seeds to abort (8, 14). Several lines of evidence strongly argue that the demethylation observed in the endosperm is inherited from the central cell: Only the central cell-derived chromosomes are demethylated (10, 12, 13), DME is rapidly down-regulated following sperm fusion (8), and genes activated by demethylation are expressed in the central cell (15, 16). Down-regulation of MET1 in the central cell has also been proposed to contribute to demethylation (2, 17). However, with the exception of three maize genes (16, 18), DNA methylation has not been analyzed in the central cell, leaving the origin of endosperm demethylation uncertain.
Here, we report genome-wide analysis of DNA methylation in the central cells of Arabidopsis and rice as well as in rice egg cells. We find that DNA demethylation in both species is initiated in central cells, which requires DME in Arabidopsis. However, we do not find evidence for a global reduction of CG methylation that would be indicative of reduced MET1 activity. Instead, we find that CG methylation is efficiently maintained in the central cells of both species and in rice eggs. Our results support locus-specific, active demethylation in the central cell as the cause of endosperm hypomethylation.
Results
Isolation of Arabidopsis Central Cells.
We used the INTACT (Isolation of Nuclei Tagged in specific Cell Types) approach (19, 20) to isolate central cell nuclei from wild-type Arabidopsis plants. A reporter protein (Nuclear Targeting Fusion, NTF) consisting of a nuclear envelope localization domain fused to GFP was expressed from the DD7 central cell-specific promoter (Fig. 1) (21). Nuclei were released from ovule tissue protoplasts and captured using anti-GFP antibodies; purity was calculated as the fraction of GFP+ nuclei in the obtained sample (Fig. S1). For DNA methylation analysis, we used two biological replicates, one with 85% purity (87 GFP+ nuclei; replicate 1) and one with 90% purity (75 GFP+ nuclei; replicate 2).
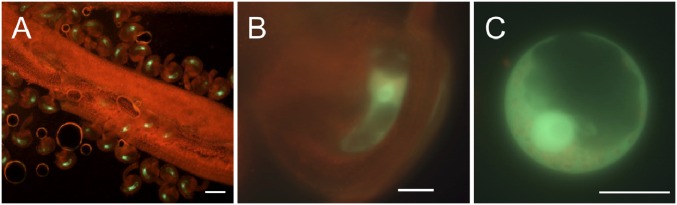
Fig. 1.
GFP expression in Arabidopsis central cells. Strong GFP fluorescence can be seen in ovules attached to a whole pistil (A), in the ovule (B), and in the nucleus of a central cell protoplast isolated from the ovule (C). GFP expression is primarily localized in the nucleus. [Scale bars, 100 μm (A) and 20 μm (B and C).]
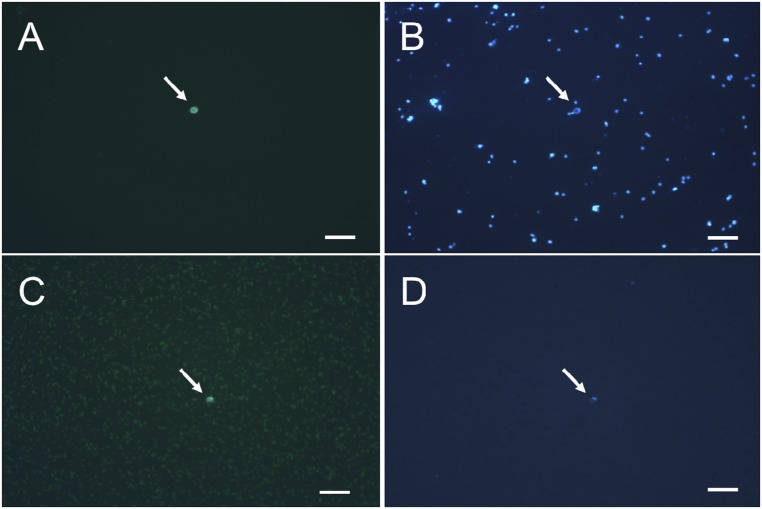
Fig. S1.
Purification of GFP-labeled central cell nuclei. Panels A and B show nuclei before binding to anti-GFP antibody beads. Panels C and D show cells after capture by anti-GFP antibody beads and purification. In panels A and C, fluorescence microscopy shows a GFP-labeled central cell nucleus. In panels B and D, light microscopy shows DAPI-stained nuclei and cells. White arrows indicate central cell nuclei. (Scale bars, 50 μm.)
DME-Dependent Demethylation Is Initiated in Arabidopsis Central Cells.
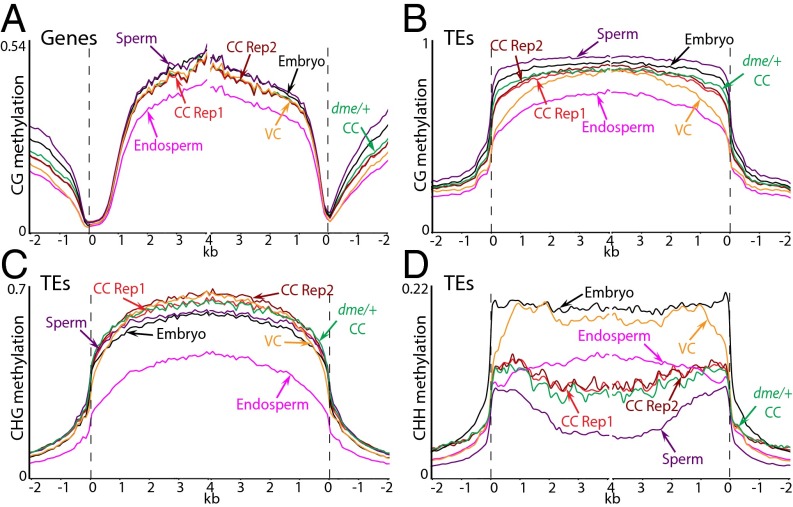
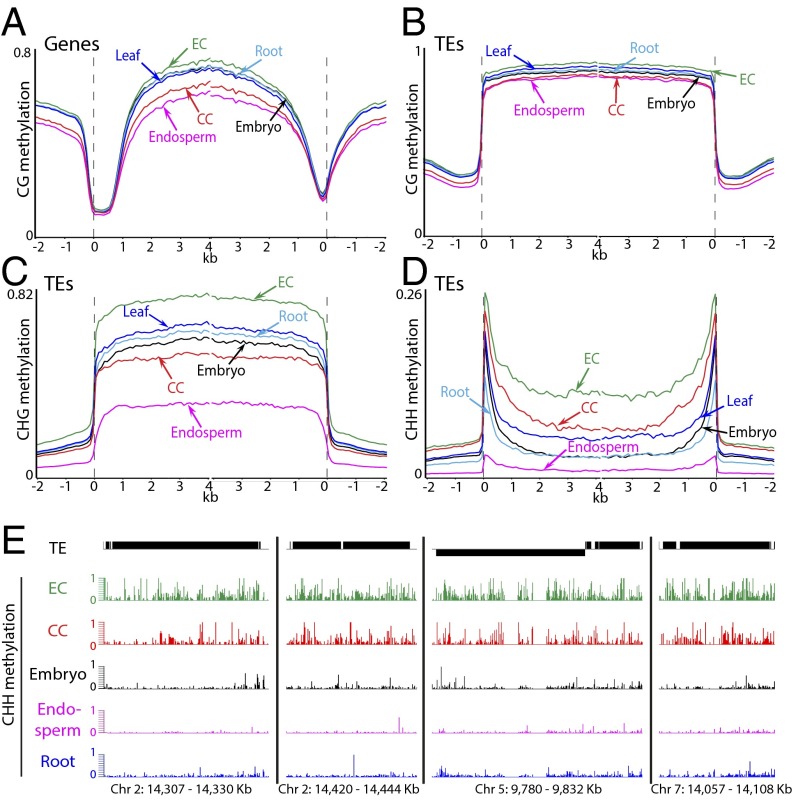
We obtained whole-genome DNA methylation data for both biological replicates (34-fold genome coverage for replicate 1 and 37-fold coverage for replicate 2; Table S1) using a modified version of a bisulfite sequencing protocol developed for small numbers of cells (22). The overall patterns of DNA methylation in genes and TEs are virtually identical between the two replicates and are similar between central cells and other cell types and tissues (Fig. 2) (10, 23–28). Genes and TEs are extensively methylated in the CG context, with CG methylation levels slightly lower than in sperm or embryos but higher than in endosperm (Fig. 2 A and B). CHG methylation of TEs is similar to embryos and male sex cells and substantially higher than in endosperm (Fig. 2C). TE CHH methylation patterns resemble sperm, with levels higher than those of sperm but lower than vegetative cells or embryos (Fig. 2D). The non-CG methylation patterns of neither sperm nor central cells closely resemble those of endosperm (Fig. 2 C and D), indicating that methylation is remodeled after gamete fusion.
Table S1.
Mean genomic coverage and DNA methylation for wild-type and mutant cell types
| Nuclear | Chloroplast | ||||
| Sample | Coverage | CG, % | CHG, % | CHH, % | Overall CHH, % |
| WT Arabidopsis central cell replicate 1 | 34.3 | 26.4 | 12.4 | 3.7 | 0.5 |
| WT Arabidopsis central cell replicate 2 | 37.3 | 26.7 | 12.7 | 3.7 | 0.6 |
| Arabidopsis central cells from dme/+ plants | 33.0 | 26.5 | 12.0 | 3.7 | 0.6 |
| WT rice central cell | 8.3 | 39.4 | 18.2 | 6.3 | 1.1 |
| WT rice egg cell | 6.4 | 41.9 | 26.2 | 7.4 | 1.0 |
| Arabidopsis root * | 5.2 | 25.3 | 8.3 | 2.3 | 0.2 |
| Arabidopsis seedling* | 53.6 | 22.6 | 7.7 | 2.4 | 0.5 |
| Arabidopsis root# | 18.2 | 26.0 | 8.7 | 2.5 | 0.1 |
| Arabidopsis cauline leaf# | 17.2 | 27.1 | 8.9 | 2.6 | 0.1 |
| Arabidopsis rosette leaf# | 13.2 | 26.5 | 8.2 | 2.4 | 0.1 |
| Arabidopsis rosette leaf replicate 1+ | 46.7 | 25.6 | 7.6 | 3.0 | 0.5 |
| Arabidopsis rosette leaf replicate 2$ | 34.2 | 27.4 | 10.4 | 5.6 | 1.0 |
| Arabidopsis rosette leaf replicate 3$ | 19.1 | 26.3 | 8.3 | 3.3 | 0.1 |
| Rice endosperm§ | 6.8 | 42.0 | 11.4 | 1.0 | 0.1 |
| Rice embryo§ | 8.8 | 44.6 | 20.9 | 4.5 | 0.1 |
| Rice root§ | 8.2 | 45.0 | 22.4 | 3.6 | 0.1 |
| Rice shoot§ | 11.6 | 46.6 | 23.2 | 3.6 | 0.1 |
| Rice leaf++ | 4.8 | 47.5 | 24.6 | 6.0 | 0.1 |
| WT Arabidopsis embryo, Col × Ler | 14.7 | 28.4 | 11.1 | 4.3 | 0.1 |
| dme Arabidopsis endosperm, dme2 × Ler | 50.1 | 26.5 | 4.5 | 0.5 | 0.2 |
| WT Arabidopsis endosperm, Col × Ler | 60.1 | 23.0 | 8.3 | 3.2 | 0.4 |
| WT Arabidopsis sperm | 119.2 | 30.2 | 11.9 | 2.0 | 0.5 |
| Arabidopsis vegetative cells from dme2/+ plants | 65.2 | 27.7 | 13.1 | 4.4 | 0.2 |
| WT Arabidopsis vegetative cell | 80.2 | 25.9 | 12.7 | 3.7 | 0.1 |
Chloroplast CHH methylation is an estimate of cytosine nonconversion rate and other errors. Mean methylation is calculated by averaging methylation of individual cytosines in each context. Published data are from the following: *, ref. 27; #, ref. 25; +, ref. 28; $, ref. 26; §, ref. 30; ++, ref. 35.
Fig. 2.
DNA methylation in Arabidopsis central cells in comparison with other cell types. Genes (A) and TEs (B–D) were aligned at the 5′ and 3′ ends. Methylation within each 100-bp interval was averaged and plotted from 2 kb away from the annotated gene or TE (negative numbers) to 4 kb into the annotated region (positive numbers). The dashed lines represent the points of alignment. Previously published data are from ref. 10.
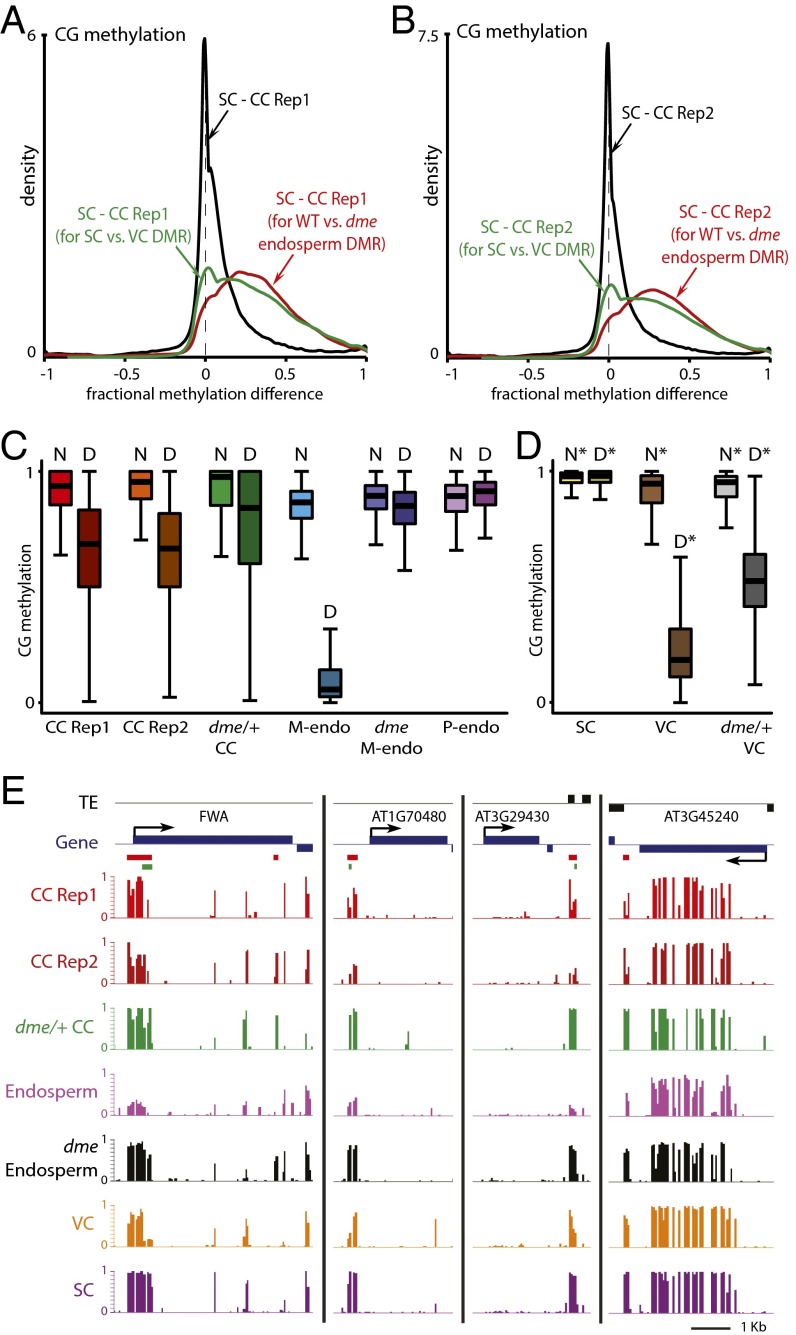
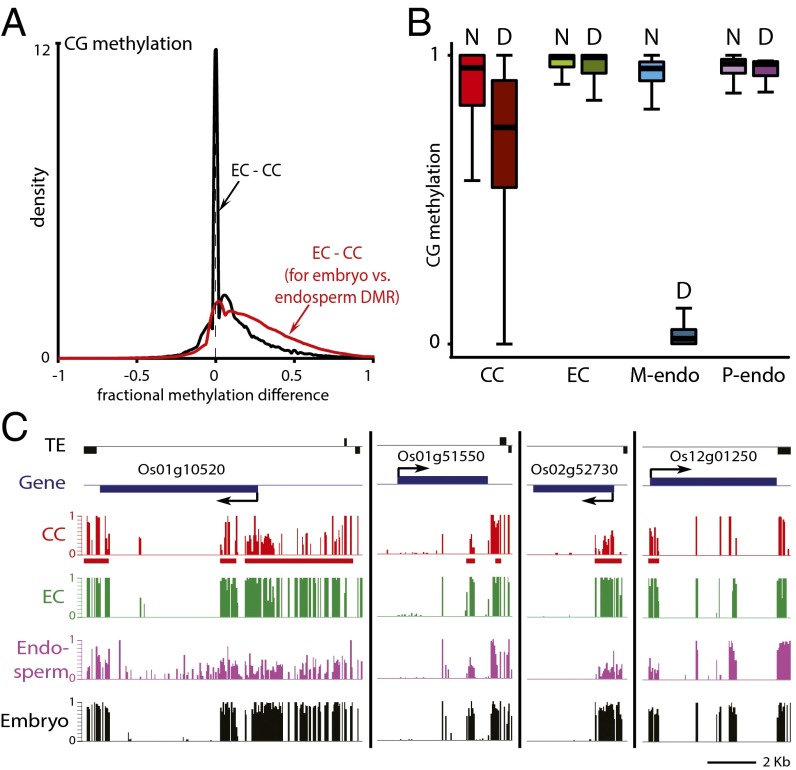
Comparison of CG methylation between sperm and either central cell replicate reveals that most loci are similarly methylated in the two cell types, as indicated by the peaks at zero in the density plots shown in Fig. 3 A and B. However, there are clear shoulders on the right sides of the distributions marking loci that are hypomethylated in central cells (Fig. 3 A and B). When analysis is confined to sequences that show demethylation of maternal chromosomes in the endosperm (10), nearly the entire distribution is on the right side of the graph for both replicates (red traces in Fig. 3 A and B), indicating that almost all of these loci are hypomethylated in the central cell. Analysis of loci demethylated in the vegetative cell (green traces in Fig. 3 A and B) yields a similar result, but with a small peak at zero, indicating that fewer of these sequences are hypomethylated in central cells—as would be expected from the partial overlap between endosperm and vegetative cell demethylation (10). Although central cells show demethylation of the vast majority of loci demethylated in the endosperm (Fig. 3 A and B), the extent of demethylation in the central cell is lower than that of the maternal endosperm chromosomes (Fig. 3C) or in the vegetative cell (Fig. 3D). This can be clearly seen at individual loci, which frequently show higher methylation in the central cell than in the endosperm (Fig. 3E), despite the averaging of methylation between demethylated maternal and fully methylated paternal chromosomes in Fig. 3E. Incomplete demethylation may be due to isolation of somewhat immature central cells, but we cannot rule out the possibility that demethylation continues during, or even after, fertilization. Because non-CG methylation shows substantial heterogeneity between Arabidopsis cell types (10, 11, 29–31) and non-CG methylation patterns of neither sperm nor central cells closely resemble those of endosperm (Fig. 2 C and D), we cannot unambiguously determine whether the loss of non-CG methylation observed at loci that show maternal CG demethylation in the endosperm (10) is initiated in the central cell.
Fig. 3.
DME-dependent demethylation is initiated in Arabidopsis central cells. (A and B) Density plots showing the frequency distribution of CG DNA methylation differences between sperm cell (SC) and central cell (CC) 50-bp windows with at least 10 informative sequenced cytosines in both samples and 70% methylation in one of the samples. The analysis marked by the red trace is confined to differentially methylated regions (DMRs) in the CG context between wild-type and dme mutant endosperm (10). The analysis marked by the green trace is confined to CG DMRs between sperm and vegetative cells (VCs) (10). (C and D) Box plots show CG methylation levels of 50-bp windows in the indicated cell type. Each box encloses the middle 50% of the distribution, with the horizontal line marking the median and vertical lines marking the minimum and maximum values that fall within 1.5 times the height of the box. (C) D, within CG DMRs between wild-type and dme mutant endosperm; M-endo, maternal endosperm; N, outside CG DMRs between wild-type and dme mutant endosperm; P-endo, paternal endosperm. Only windows with methylation greater than 70% in either wild-type or dme mutant endosperm, at least 20 informative sequenced cytosines in both tissues, and at least 20 sequenced cytosines in the graphed sample (10 for central cells) are included. (D) D*, within CG DMRs between sperm and vegetative cells; N*, outside such DMRs. Only windows with methylation greater than 70% in either wild-type sperm or vegetative cell, at least 20 informative sequenced cytosines in both cell types, and at least 20 sequenced cytosines in the graphed sample are included. (E) Snapshots of CG methylation in central cell (CC), endosperm, vegetative cell (VC), and sperm (SC). DMRs between wild-type and dme mutant endosperm are underlined in red; DMRs between sperm and vegetative cells are underlined in green. Previously published data are from ref. 10.
To determine whether central cell demethylation requires DME, we isolated central cells from plants heterozygous for the dme-2 loss-of-function mutation (homozygous dme-2 plants cannot be examined because seeds that inherit a maternal dme-2 allele abort) (8) using the INTACT system. We obtained 33-fold genome coverage (Table S1) from a 90% pure sample (67 GFP+ nuclei), with half the central cells expected to carry the dme-2 allele and half the wild-type allele. The overall methylation patterns are similar between central cells from wild-type and dme/+ plants (Fig. 2). Notably, overall CHG and CHH methylation does not appreciably change in central cells from dme/+ plants (Fig. 2 C and D), which contrasts with the greatly reduced non-CG methylation in dme mutant endosperm (29). This result supports a previous interpretation that the overall reduction of endosperm non-CG methylation outside DME target loci is not caused directly by lack of DME but rather by misregulation of the PRC2 complex and consequent abnormal endosperm development (10).
Although the overall patterns of CG methylation are similar between central cells from wild-type and dme/+ plants, loci demethylated on maternal endosperm chromosomes are more extensively methylated in central cells from dme/+ plants (Fig. 3 C and E), as they are in dme endosperm (Fig. 3 C and E) (10). The methylation levels at these loci in central cells from dme/+ plants are about half way between those of wild-type central cells and those of loci that are not demethylated in endosperm (marked by “N” in Fig. 3C), as would be expected if only half of the central cells harbor the dme mutation, and as is the case in vegetative cells from dme/+ plants (Fig. 3D) (10). Taken together, our results demonstrate that the DME-dependent demethylation observed on maternal endosperm chromosomes is initiated in the central cell.
DNA Demethylation Is Initiated in Rice Central Cells.
To complement our Arabidopsis experiments, we analyzed DNA methylation in central and egg cells of rice (Oryza sativa japonica cultivar Nipponbare), a species that shared a common ancestor with Arabidopsis about 150 million years ago (32). We isolated 20 rice central cells and 61 egg cells by microdissection (Fig. S2) (33, 34) and obtained whole-genome DNA methylation data for both cell types (eightfold genome coverage for central cells and sixfold coverage for eggs; Table S1). The methylation patterns of central and egg cells are similar to those of other tissues in genes (Fig. 4A) and TEs (Fig. 4 B–D) (30, 35). Central cell CG methylation levels are lower than those of embryos, roots, and leaves, resembling most closely the methylation of endosperm (Fig. 4 A and B), whereas CG methylation in the egg cell is slightly elevated compared with other cells (Fig. 4 A and B). CHG methylation in central cell TEs is a little lower than in embryos, roots, and leaves but much higher than in endosperm (Fig. 4C), whereas egg cell CHG methylation is much higher than in other tissues (Fig. 4C). Rice CHH methylation is concentrated in small, gene-adjacent TEs (30), a phenomenon dubbed CHH islands in maize (36). This manifests as dual peaks in the TE analysis (small TEs cluster near the points of alignment; Fig. 4D). CHH methylation is very low in endosperm compared with embryos, roots, and leaves (Fig. 4D) but comparatively very high in central and egg cells (Fig. 4D). Especially in egg cells, high CHH methylation levels are also found in long TEs (Fig. 4 D and E). As in Arabidopsis (Fig. 2 C and D), the levels of non-CG methylation in the central cell are not obviously predictive of those in endosperm (Fig. 4 C and D). The same holds for egg cells and embryos (Fig. 4 C and D). Overall, our results demonstrate that—as in Arabidopsis (10, 11, 29–31)—non-CG methylation shows large differences between rice cells and tissues, indicating that such fluctuations are a common feature of plant development.
Fig. S2.
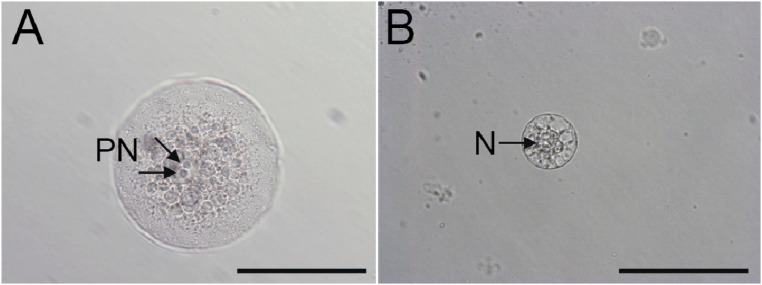
Manually dissected female rice gametes. (A) The central cell was isolated from an ovule by enzyme digestion. Two polar nuclei (PN, arrows) are observed inside the cell. (B) The egg cell was isolated nonenzymatically by making an incision at the basal part of the ovary. N, nucleus. (Scale bars, 100 μm.)
Fig. 4.
DNA methylation in rice central and egg cells in comparison with other tissues. (A–D) Genes (A) and TEs (B–D) were aligned at the 5′ and 3′ ends, as in Fig. 2. (E) Snapshots of TE CHH methylation in rice egg cell (EC), central cell (CC), embryo, endosperm, and root. Previously published data are from refs. 30, 35.
Comparison of CG methylation between central and egg cells shows that most loci have the same methylation levels in these cell types, as indicated by the sharp, narrow peak at zero in Fig. 5A. However, there is a substantial group of loci hypomethylated in central cells, as evidenced by the shoulder on the right side of the distribution (Fig. 5A). As in Arabidopsis (Fig. 3 A and B), analysis of the loci that show demethylation of maternal chromosomes in the endosperm (12) demonstrates that almost all of these are hypomethylated in the central cell (red trace in Fig. 5A). Also as in Arabidopsis (Fig. 3C), the extent of demethylation in the central cell is lower than in endosperm (Fig. 5B), which is evident at individual loci (Fig. 5C). Taken together, our data indicate that the DNA demethylation of maternal endosperm chromosomes observed in flowering plants is initiated in the central cell.
Fig. 5.
DNA demethylation is initiated in rice central cells. (A) Density plot showing the frequency distribution of CG DNA methylation differences between egg cell (EC) and central cell (CC) 50-bp windows with at least 10 informative sequenced cytosines in both samples and 70% methylation in one of the samples. The analysis marked by the red trace is confined to DMRs in the CG context between embryo and endosperm (12). (B) Box plots show CG methylation levels of 50-bp windows in the indicated cell type. Only windows with methylation greater than 70% in either embryo or endosperm, at least 20 informative sequenced cytosines in both tissues, and at least 20 sequenced cytosines in the graphed sample (10 for central and egg cells) are included. D, within CG DMRs between embryo and endosperm; M-endo, maternal endosperm; N, outside CG DMRs between embryo and endosperm; P-endo, paternal endosperm. (C) Snapshots of CG methylation in central cell (CC), egg cell (EC), endosperm, and embryo. DMRs between embryo and endosperm are underlined in red.
CG Methylation Is Efficiently Maintained in Female Sex Cells.
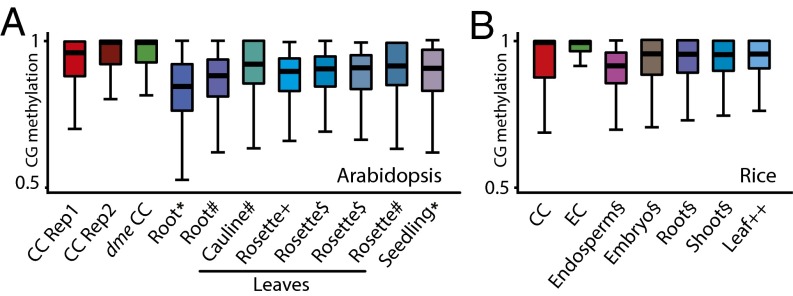
In addition to active DNA demethylation by DME, passive hypomethylation caused by down-regulation of the MET1 methyltransferase has been proposed to contribute to central cell demethylation (2, 17). Such passive demethylation should manifest itself as reduced CG methylation outside the loci targeted by DME. Therefore, we tested the passive demethylation hypothesis by analyzing methylation of individual CG sites within loci that do not show maternal endosperm demethylation (10). However, instead of the expected reduction compared with other tissues, we found that CG methylation levels in the central cell are substantially higher (Fig. 6A), indicating that CG methylation is maintained more, not less, efficiently. Analysis of rice central and egg cells revealed a similarly increased efficiency of CG methylation (Fig. 6B). Therefore, our data indicate that central cell demethylation occurs by an active process unaided by passive demethylation.
Fig. 6.
CG methylation is efficiently maintained in female sex cells. (A and B) Box plots show CG methylation for individual CG sites with methylation greater than 50% and located within TEs. (A) Only sites outside DMRs in the CG context between wild-type and dme mutant Arabidopsis endosperm (10) and with at least 10 informative sequenced cytosines are included. Published data are from the following: *, ref. 27; #, ref. 25; +, ref. 28; $, ref. 26. (B) Only sites outside rice embryo–endosperm CG DMRs (12) and with at least 20 informative sequenced cytosines are included. Published data are from the following: §, ref. 30; ++, ref. 35.
Discussion
DNA demethylation of maternal endosperm chromosomes at thousands of loci regulates gene expression and gene imprinting, and is required for seed viability (5). Despite extensive indirect evidence that DNA demethylation occurs in the central cell and is inherited by the endosperm, central cell methylation has only been directly measured at three maize genes: fie1, fie2, and mee1 (16, 18). Here, we present genome-wide methylation data for Arabidopsis and rice central cells, which demonstrate that endosperm DNA demethylation is indeed initiated in the central cell. We also confirm that this process is dependent on the DME DNA demethylase in Arabidopsis. The similar features of DNA demethylation in rice (12) strongly suggest the involvement of a DME-related enzyme or enzymes, such as ROS1a (37). Arabidopsis and rice belong to two major branches of flowering plants (dicots and monocots, respectively) that separated around 150 million years ago (32), indicating that DNA demethylation in the central cell is an ancient and conserved feature of plant sexual development.
Although our data indicate that demethylation is generally initiated in the central cell (Figs. 3 A and B and 5A), central cell demethylation is less extensive than that of maternal endosperm chromosomes (Figs. 3C and 5B). This difference could be attributed to several causes. The most trivial explanation is contamination with other cells. This is certainly an issue with Arabidopsis samples, which are 85–90% pure. However, this level of contamination is not sufficient to explain the methylation differences between central cells and endosperm. Furthermore, rice central cells were individually isolated by microdissection (Fig. S2A) and should have little, if any, contamination with other genetic material. An alternative explanation is that the central cells we analyzed were not fully mature. DME expression is highest in mature central cells (8), suggesting that demethylation occurs near the end of central cell development. Arabidopsis central cell nuclei were isolated at the time fertilization normally occurs—24 hours after flowers reached stage 12 (38)—but may not have developed fully nonetheless. Similarly, we isolated rice central cells from flowers that are within 1 day of pollination and fertilization (33), which may be a relatively early stage for DNA demethylation. We also cannot rule out the possibility that demethylation continues during or even after the fusion of central and sperm cells in one or both species.
In addition to active demethylation by DME, reduced MET1 expression has been proposed to passively contribute to central cell demethylation (2, 17). A similar hypothesis has been proposed to explain demethylation in the pollen vegetative cell (2). However, our data show that CG methylation is maintained more efficiently in Arabidopsis central cells than in leaves and roots (Fig. 6A) and that the same applies to rice central and egg cells (Fig. 6B). We also recently obtained similar results for male Arabidopsis sex cells, including the vegetative cell (39). Therefore, CG methylation maintenance is apparently generally more efficient in sex cells, which are responsible for faithful propagation of methylation patterns across generations. Overall, our rice and Arabidopsis data indicate that DNA demethylation in central and vegetative cells is an active process that does not involve overall genomic hypomethylation.
Materials and Methods
Isolation of A. thaliana Central Cell Nuclei.
A. thaliana plants homozygous for the DD7:NTF transgene were grown on soil in either a greenhouse or in an environment-controlled room with a long-day photoperiod (16 h light, 8 h dark). We emasculated stage 12 flowers (38), which have yellow anthers that have not dehisced. The emasculated plants were then incubated in an environment-controlled room or in a growth chamber with a long-day photoperiod for 24 h. Pistils were dissected, opened, and ovules exposed to a protoplasting enzyme solution in a vacuum based on methods described in ref. 40. Protoplasts were gently pelleted by centrifugation, the supernatant was removed, and the pellet was resuspended in protoplast lysis buffer adapted from ref. 41. Modified procedures based on the INTACT method (19) were used to purify central cell nuclei. To remove cell debris, samples were preincubated with Dynal Protein-G magnetic beads (Invitrogen: 100–03D), which were pelleted by exposure to a magnetic field. The supernatant was removed and incubated with anti-GFP antibodies (Invitrogen: G10362), which were precipitated with Protein-G magnetic beads. Central cell nuclei were dissociated from the anti-GFP beads and stored at –80 °C. Please refer to ref. 20 for a more detailed description.
GFP Fluorescence Microscopy.
GFP fluorescence in central cell nuclei and DAPI staining were observed under an Axio Imager A1 (Carl Zeiss) microscope as described in ref. 42.
Isolation of Rice Central and Egg Cells.
O. sativa cv. Nipponbare was grown in environmental chambers at 26 °C in a 13/11 h light/dark cycle. Rice central cells were isolated according to ref. 33 with a small modification. Central cells were collected from rice ovules with an incision in the peripheral region. Ovules were incubated in 650 mOsmol/kg H2O mannitol solution containing enzymes (33) for 30 min to 1 h at room temperature until the central cell was released from the embryo sac. Central cells with visible polar nuclei and large vacuoles were collected. Rice egg cells were isolated according to ref. 34. Egg cells released from ovaries with an incision in the basal portion were collected and washed in 370 mOsmol/kg H2O mannitol solution without enzymes. The isolated cells were stored at –80 °C.
Whole-Genome Bisulfite Sequencing.
Bisulfite sequencing libraries were constructed as described in ref. 22. We followed the single-cell library preparation protocol using Klenow exo− from Enzymatics. Library amplification was performed with 13 PCR cycles using the EPIK Amplification Kit (Bioline GmbH). Illumina sequencing was performed using the NextSeq 500 platform (75 nt single-end reads) at the DNA sequencing facility of the University of Cambridge Department of Biochemistry and the Bauer Core Facility at Harvard University and the HiSeq 4000 platform (100 nt single-end reads) at Novogene Ltd. Sequenced reads from Arabidopsis and rice cells were mapped to the TAIR10 and MSU7 reference genomes, respectively. Alignment and DNA methylation analyses were performed as previously described (22), except the Trim Galore and Bismark software settings were adjusted to the single-end mode.
Acknowledgments
We thank Jen Sheen and Jennifer Frost for helpful discussions about optimizing yield and purity of Arabidopsis central cell nuclei, Hongbo Gao for assistance with figure preparation, and the Norwich BioScience Institute Partnership Computing infrastructure for Science Group for High Performance Computing Resources. This work was partially funded by Next-Generation BioGreen 21 Program Grant PJ011018 and National Research Foundation of Korea Grant 2014R1A2A2A01004887 (to Y.C.), NSF Grant IOS-1025890 and NIH Grant R01-GM069415 (to R.L.F. and D.Z.), and Biotechnology and Biological Sciences Research Council (BBSRC) David Phillips Fellowship BB/L025043/1 (to X.F.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE89789).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619047114/-/DCSupplemental.
References
- 1.Feng X, Zilberman D, Dickinson H. A conversation across generations: Soma-germ cell crosstalk in plants. Dev Cell. 2013;24(3):215–225. doi: 10.1016/j.devcel.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Kawashima T, Berger F. Epigenetic reprogramming in plant sexual reproduction. Nat Rev Genet. 2014;15(9):613–624. doi: 10.1038/nrg3685. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt A, Schmid MW, Grossniklaus U. Plant germline formation: Common concepts and developmental flexibility in sexual and asexual reproduction. Development. 2015;142(2):229–241. doi: 10.1242/dev.102103. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Berger F. Endosperm: Food for humankind and fodder for scientific discoveries. New Phytol. 2012;195(2):290–305. doi: 10.1111/j.1469-8137.2012.04182.x. [DOI] [PubMed] [Google Scholar]
- 5.Rodrigues JA, Zilberman D. Evolution and function of genomic imprinting in plants. Genes Dev. 2015;29(24):2517–2531. doi: 10.1101/gad.269902.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim MY, Zilberman D. DNA methylation as a system of plant genomic immunity. Trends Plant Sci. 2014;19(5):320–326. doi: 10.1016/j.tplants.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Zhu JK. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi Y, et al. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell. 2002;110(1):33–42. doi: 10.1016/s0092-8674(02)00807-3. [DOI] [PubMed] [Google Scholar]
- 9.Schoft VK, et al. Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc Natl Acad Sci USA. 2011;108(19):8042–8047. doi: 10.1073/pnas.1105117108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibarra CA, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337(6100):1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calarco JP, et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151(1):194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodrigues JA, et al. Imprinted expression of genes and small RNA is associated with localized hypomethylation of the maternal genome in rice endosperm. Proc Natl Acad Sci USA. 2013;110(19):7934–7939. doi: 10.1073/pnas.1306164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang M, et al. Genome-wide high resolution parental-specific DNA and histone methylation maps uncover patterns of imprinting regulation in maize. Genome Res. 2014;24(1):167–176. doi: 10.1101/gr.155879.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh T-F, et al. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci USA. 2011;108(5):1755–1762. doi: 10.1073/pnas.1019273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kinoshita T, et al. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science. 2004;303(5657):521–523. doi: 10.1126/science.1089835. [DOI] [PubMed] [Google Scholar]
- 16.Jahnke S, Scholten S. Epigenetic resetting of a gene imprinted in plant embryos. Curr Biol. 2009;19(19):1677–1681. doi: 10.1016/j.cub.2009.08.053. [DOI] [PubMed] [Google Scholar]
- 17.Jullien PE, et al. Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 2008;6(8):e194. doi: 10.1371/journal.pbio.0060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutiérrez-Marcos JF, et al. Epigenetic asymmetry of imprinted genes in plant gametes. Nat Genet. 2006;38(8):876–878. doi: 10.1038/ng1828. [DOI] [PubMed] [Google Scholar]
- 19.Deal RB, Henikoff S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc. 2011;6(1):56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park K, et al. Optimized methods for the isolation of Arabidopsis female central cells and their nuclei. Mol Cells. 2016;39(10):768–775. doi: 10.14348/molcells.2016.0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steffen JG, Kang I-H, Macfarlane J, Drews GN. Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 2007;51(2):281–292. doi: 10.1111/j.1365-313X.2007.03137.x. [DOI] [PubMed] [Google Scholar]
- 22.Smallwood SA, et al. Single-cell genome-wide bisulfite sequencing for assessing epigenetic heterogeneity. Nat Methods. 2014;11(8):817–820. doi: 10.1038/nmeth.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133(3):523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coleman-Derr D, Zilberman D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 2012;8(10):e1002988. doi: 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152(1-2):352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153(1):193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stroud H, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21(1):64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsieh T-F, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324(5933):1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zemach A, et al. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA. 2010;107(43):18729–18734. doi: 10.1073/pnas.1009695107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawakatsu T, et al. Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat Plants. 2016;2(5):16058. doi: 10.1038/nplants.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaw SM, Chang CC, Chen HL, Li WH. Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol. 2004;58(4):424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- 33.Uchiumi T, Komatsu S, Koshiba T, Okamoto T. Isolation of gametes and central cells from Oryza sativa L. Sex Plant Reprod. 2006;19:37–45. [Google Scholar]
- 34.Abiko M, Maeda H, Tamura K, Hara-Nishimura I, Okamoto T. Gene expression profiles in rice gametes and zygotes: Identification of gamete-enriched genes and up- or down-regulated genes in zygotes after fertilization. J Exp Bot. 2013;64(7):1927–1940. doi: 10.1093/jxb/ert054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 36.Gent JI, et al. CHH islands: De novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 2013;23(4):628–637. doi: 10.1101/gr.146985.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ono A, et al. A null mutation of ROS1a for DNA demethylation in rice is not transmittable to progeny. Plant J. 2012;71(4):564–574. doi: 10.1111/j.1365-313X.2012.05009.x. [DOI] [PubMed] [Google Scholar]
- 38.Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2(8):755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsieh P-H, et al. The Arabidopsis male sexual lineage exhibits more robust maintenance of CG methylation than somatic tissues. Proc Natl Acad Sci USA. 113:15132–15137. doi: 10.1073/pnas.1619074114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2(7):1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 41.Sheen J. Protein phosphatase activity is required for light-inducible gene expression in maize. EMBO J. 1993;12(9):3497–3505. doi: 10.1002/j.1460-2075.1993.tb06024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yadegari R, et al. Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell. 2000;12(12):2367–2382. doi: 10.1105/tpc.12.12.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]