Significance
Cytosine methylation is a mechanism of epigenetic inheritance—the transmission across generations of information that does not reside in DNA sequence. This transmission is mediated by enzymes that copy methylation states following DNA replication. Only a small group of plant cells—gametes and their progenitors—participates in inheritance, yet methylation is usually studied in other cell types, in which cytosine methylation within CG dinucleotides appears to be too low for stable maintenance. Here, we examine methylation in the pollen grains of Arabidopsis thaliana plants with methyltransferase mutations and show that although methylation is maintained by similar mechanisms in pollen and somatic cells, maintenance of CG methylation is more efficient in pollen, explaining how methylation can be faithfully inherited across generations.
Keywords: DNA methylation, epigenetic inheritance, histone H1, pollen
Abstract
Cytosine DNA methylation regulates the expression of eukaryotic genes and transposons. Methylation is copied by methyltransferases after DNA replication, which results in faithful transmission of methylation patterns during cell division and, at least in flowering plants, across generations. Transgenerational inheritance is mediated by a small group of cells that includes gametes and their progenitors. However, methylation is usually analyzed in somatic tissues that do not contribute to the next generation, and the mechanisms of transgenerational inheritance are inferred from such studies. To gain a better understanding of how DNA methylation is inherited, we analyzed purified Arabidopsis thaliana sperm and vegetative cells—the cell types that comprise pollen—with mutations in the DRM, CMT2, and CMT3 methyltransferases. We find that DNA methylation dependency on these enzymes is similar in sperm, vegetative cells, and somatic tissues, although DRM activity extends into heterochromatin in vegetative cells, likely reflecting transcription of heterochromatic transposons in this cell type. We also show that lack of histone H1, which elevates heterochromatic DNA methylation in somatic tissues, does not have this effect in pollen. Instead, levels of CG methylation in wild-type sperm and vegetative cells, as well as in wild-type microspores from which both pollen cell types originate, are substantially higher than in wild-type somatic tissues and similar to those of H1-depleted roots. Our results demonstrate that the mechanisms of methylation maintenance are similar between pollen and somatic cells, but the efficiency of CG methylation is higher in pollen, allowing methylation patterns to be accurately inherited across generations.
Cytosine methylation is a covalent DNA modification that regulates transcription in eukaryotes (1). The highest levels of methylation in plant and animal genomes are typically located within symmetric CG dinucleotides (1). Methylation in this sequence context is virtually ubiquitous in plant transposable elements (TEs), which are transcriptionally silenced by methylation, but also occurs within many genes without disrupting their expression (1, 2). CG methylation is catalyzed by the Dnmt1 methyltransferase family, called MET1 in plants (1, 2). MET1 restores full methylation of hemimethylated CG dinucleotides generated by DNA replication, thereby perpetuating methylation patterns after cell division (1, 2). This maintenance activity is thought to allow DNA methylation to carry epigenetic information—and influence gene expression and phenotype—across generations (3, 4). The nature of this mechanism predicts that imperfect maintenance of CG methylation should lead to complete loss as methylation is diluted during each cell division, so that the only stable methylation states for a CG site in a population of cells should be fully methylated or fully unmethylated. However, the methylation levels measured at Arabidopsis thaliana CG sites appear to be too low for stable maintenance (5, 6). Therefore, exactly how CG methylation is so robustly inherited in flowering plants is not entirely clear.
In addition to MET1, plants possess the chromomethylase (CMT) and DRM methyltransferase families. In Arabidopsis, CMT3 catalyzes methylation of semisymmetric CNG sites, which is typically analyzed as CHG (H stands for A, T, or G) to avoid overlap with CG (1, 2). A related enzyme, CMT2, catalyzes asymmetric (CHH) methylation, primarily in heterochromatic TEs (7, 8). Both enzymes rely on dimethylation of lysine 9 of histone H3 (H3K9me2), a histone modification characteristic of plant heterochromatin (8, 9). DRM enzymes (DRM1 and DRM2 in Arabidopsis), which are guided by the small RNA-directed DNA methylation (RdDM) pathway (10), catalyze CHH methylation of more euchromatic TEs (7, 11, 12). Methylation mediated by CMT and DRM enzymes, collectively referred to as non-CG methylation, functions to repress TE expression and is almost completely absent from genes (1, 2). Non-CG methylation varies substantially between plant cell types and tissues (13–17) for reasons that remain largely unexplained.
Transgenerational inheritance of DNA methylation patterns is carried out by gametes and the cellular lineages from which they differentiate. The shoot apical meristem, a small group of stem cells that develops early during Arabidopsis embryogenesis, gives rise to all above-ground tissues, including the floral meristems that produce the sexual organs (18). In these, certain cells differentiate into meiocytes, which undergo meiosis to produce haploid spores (19, 20). The spores go on to divide by mitosis to create the multicellular male and female gametophytes. The male gametophyte, pollen, consists of two sperm cells and a vegetative cell, which forms the pollen tube that delivers the sperm into the female gametophyte (19, 20). As this developmental sequence illustrates, plants specify dedicated sexual lineages much later than animals, which set aside the germ line during embryogenesis (21). Nonetheless, only a very small fraction of plant cells can give rise to gametes. Despite their importance, these cells are rarely directly examined in studies of DNA methylation, so that most of our knowledge about the mechanisms of epigenetic methylation inheritance is inferred from analyses of differentiated tissues that do not contribute to the next generation.
To help address this deficiency, we analyzed DNA methylation in purified Arabidopsis sperm and vegetative cells with mutations in CMT3, CMT2, and both DRM1 and DRM2, respectively. We also analyzed sperm and vegetative cells with mutation of both genes encoding canonical histone H1, a chromatin protein that globally reduces heterochromatic DNA methylation in all sequence contexts (7). Despite the reported absence of H3K9me2 from the vegetative cell nucleus (22), we find that methyltransferase dependencies of non-CG methylation in the sperm and vegetative cells are similar to those of leaves and other examined tissues, although RdDM partially extends into vegetative cell heterochromatin. Unlike in somatic tissues, mutation of H1 does not substantially increase heterochromatic methylation in either sperm or vegetative cell. Instead, methylation of CG sites is elevated in wild-type (WT) pollen, resembling h1 mutant somatic tissues. The higher CG methylation levels in pollen are easier to reconcile with stable transgenerational maintenance, indicating that CG methylation efficiency is reduced in somatic cells with limited division potential. This in turn suggests that small DNA methylation differences between somatic cells or tissues may be caused by variance in maintenance efficiency instead of active developmental reprogramming.
Results
Mechanisms of DNA Methylation Maintenance Are Similar Between Pollen and Leaves.
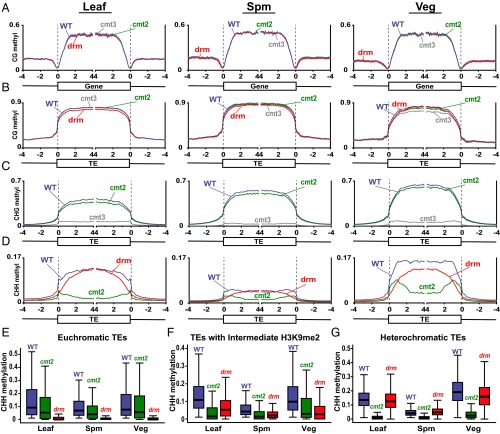
We isolated sperm and vegetative cell nuclei from cmt3, cmt2, drm1drm2 (drm), and control WT plants by fluorescence-activated cell sorting (FACS) (15). Genome-wide analysis of DNA methylation by bisulfite sequencing (Table S1) revealed that, as expected, none of the mutations have a major effect on CG DNA methylation in either genes or TEs (Fig. 1 A and B), just as in leaves (Fig. 1 A and B) and other somatic tissues (5–8, 23). The cmt3 mutation has a similarly strong effect on TE CHG methylation in sperm, vegetative cells, and leaves (Fig. 1C), indicating that CHG methylation in both pollen cell types is maintained primarily by CMT3, despite the reported lack of H3K9me2 in the vegetative nucleus (22). Likewise, mutation of either cmt2 or drm affects the patterns of CHH methylation similarly in sperm, vegetative cells, and leaves (Fig. 1D), even though CHH methylation is about threefold higher in vegetative cells compared with sperm (Fig. 1D) (15, 16). As in leaves and other tissues (7, 8), sperm and vegetative cell CHH methylation in euchromatic TEs—here defined as those with a low level of H3K9me2 in leaves (Fig. S1) (7)—is primarily dependent on drm (Fig. 1E and Fig. S2A). CHH methylation of TEs with intermediate H3K9me2 is dependent on drm and cmt2 (Fig. 1F), with cmt2 causing a stronger reduction in TE bodies and drm in TE edges (Fig. S2B) (7), and methylation of the most heterochromatic TEs is performed primarily by CMT2 in sperm, vegetative cells, and leaves (Fig. 1G and Fig. S2C). An interesting feature of the data is that vegetative cells have only modestly higher CHH methylation of euchromatic TEs compared with sperm (Fig. 1E), whereas heterochromatic TEs are far more methylated in the vegetative cell (Fig. 1G). The elevated CHH methylation in the vegetative cell is thus largely caused by increased activity of H3K9me2-dependent CMT2 in heterochromatin (Fig. 1G). Overall, our results demonstrate that the roles of the major Arabidopsis DNA methyltransferases in pollen are remarkably similar to those in somatic tissues.
Table S1.
Mean genomic coverage and DNA methylation for wild type (WT) and mutants sequenced in this paper
| Nuclear | Chloroplast | ||||
| Sample | Coverage | CG, % | CHG, % | CHH, % | Overall CHH, % |
| WT sperm | 115.46 | 30.34 | 11.25 | 1.46 | 0.55 |
| cmt2 sperm | 30.08 | 29.35 | 10.06 | 0.68 | 0.22 |
| cmt3 sperm | 16.56 | 28.64 | 0.99 | 0.69 | 0.20 |
| drm sperm | 94.81 | 28.79 | 10.37 | 1.13 | 0.62 |
| h1 sperm | 34.90 | 30.34 | 11.05 | 1.23 | 0.43 |
| WT vegetative nuclei | 40.66 | 26.68 | 12.35 | 3.40 | 0.56 |
| cmt2 vegetative nuclei | 28.83 | 24.77 | 11.30 | 1.38 | 0.22 |
| cmt3 vegetative nuclei | 10.54 | 22.76 | 1.48 | 1.86 | 0.34 |
| drm vegetative nuclei | 23.04 | 25.78 | 11.38 | 2.25 | 0.67 |
| h1 vegetative nuclei | 38.47 | 26.69 | 12.45 | 3.14 | 0.51 |
Chloroplast CHH methylation is an estimate of cytosine nonconversion rate and other errors (contaminating chloroplast DNA is present in the purified vegetative nuclei samples). Mean methylation is calculated by averaging methylation of individual cytosines in each context.
Fig. 1.
DNA methylation in Arabidopsis leaf, sperm (Spm), and vegetative cell (Veg). Genes (A) and TEs (B–D) were aligned at the 5′ and 3′ ends. Methylation within each 100-bp interval was averaged and plotted from 4 kb away from the annotated gene or TE (negative numbers) to 4 kb into the annotated region (positive numbers). The dashed lines represent the points of alignment. (E–G) Box plots show CHH methylation levels within 50-bp windows in WT, cmt2, and drm mutants in TEs with low H3K9me2 in leaves (euchromatic TEs; E), TEs with intermediate H3K9me2 (F), and TEs with high H3K9me2 (heterochromatic TEs; G). Each box encloses the middle 50% of the distribution, with the horizontal line marking the median and vertical lines marking the minimum and maximum values that fall within 1.5 times the height of the box. Only windows with methylation greater than 1% in WT tissues and with at least 20 informative sequenced cytosines across all samples are included.
Fig. S1.
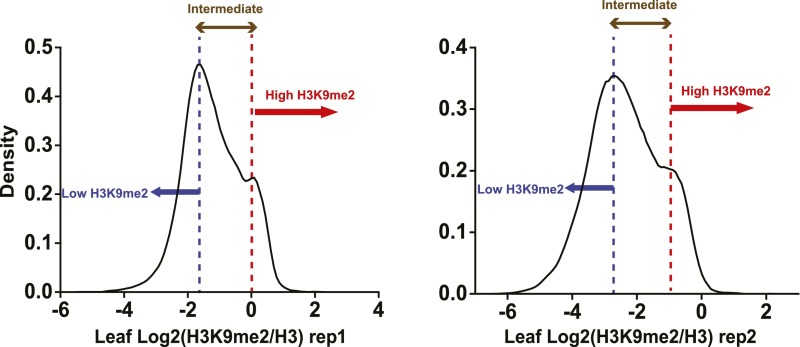
Grouping of TEs based on the level of H3K9me2. TEs were classified into three groups based on density plots: TEs with log2 (H3K9me2 IP/H3 IP) below the lower peak in both replicates were classified as euchromatic; TEs with log2 (H3K9me2 IP/H3 IP) between the peaks in both replicates were classified as intermediate; TEs with log2 (H3K9me2 IP/H3 IP) above the higher peak in both replicates were classified as heterochromatic.
Fig. S2.
DNA methylation in Arabidopsis leaf, sperm (Spm), and vegetative cell (Veg) TEs. (A–C) Euchromatic TEs, TEs with intermediate H3K9me2, and heterochromatic TEs were aligned at the 5′ and 3′ ends. Methylation within each 100-bp interval was averaged and plotted from 4 kb away from the annotated gene or TE (negative numbers) to 4 kb into the annotated region (positive numbers). The dashed lines represent the points of alignment. (D) Snapshots of CHH methylation of TEs with intermediate H3K9me2 in leaf, Spm, and Veg of WT, cmt2, and drm mutants.
New Heterochromatic RdDM Targets in the Vegetative Cell.
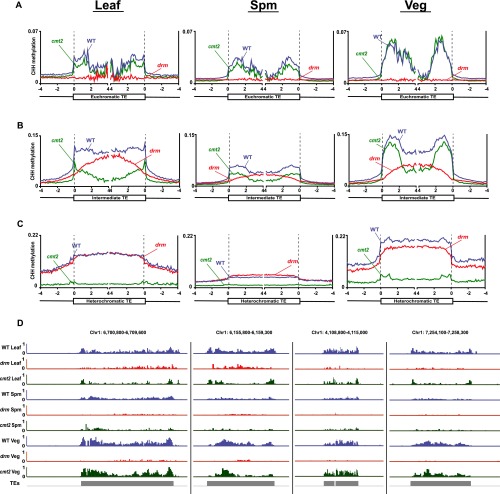
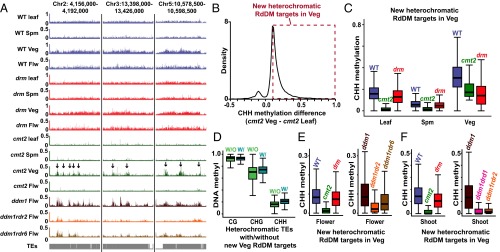
Although pollen and somatic heterochromatic CHH methylation is largely dependent on CMT2, the effect of the cmt2 mutation is weaker in the vegetative cell compared with leaf and sperm (Fig. 1G and Fig. S2C). The cmt2 mutation also does not reduce CHH methylation in TEs with intermediate H3K9me2 as much in the vegetative cell as it does in leaf (Fig. 1F and Fig. S2B). This is easily noticeable in the methylation data as heterochromatic TEs that retain substantial CHH methylation in cmt2 vegetative cells (Fig. 2A) and as intermediate TEs that retain essentially WT methylation levels in cmt2 vegetative cells (Fig. S2D). To systematically identify heterochromatic loci with CMT2-independent CHH methylation, we compared methylation levels between all 50-bp windows in heterochromatic TEs that retain CHH methylation in either cmt2 vegetative cell or cmt2 leaf (Fig. 2B). The vast majority of windows have more methylation in the vegetative cell (Fig. 2B). As expected, CHH methylation of these windows is dependent on CMT2 in leaf and sperm (Fig. 2C). WT vegetative cells have higher CHH methylation of these loci than leaf or sperm, which is dependent on both CMT2 and DRM (Fig. 2C), resembling the pattern observed in TEs with intermediate H3K9me2 (Fig. 1F). TEs containing such loci also exhibit higher CHH and CHG methylation than other heterochromatic TEs in the vegetative cell (Fig. 2D). Thus, in addition to the usual maintenance of heterochromatic CHH methylation by CMT2, some parts of vegetative cell heterochromatin are also targeted by DRM via RdDM, suggesting that these loci are less heterochromatic in this cell type. However, the 466,450 bp covered by such loci in our analysis (Dataset S1) represent only 7.9% of CMT2-dependent heterochromatic loci in the vegetative cell (5,905,450 bp), so that the vast majority of vegetative cell heterochromatic CHH methylation requires CMT2 (Fig. 1G).
Fig. 2.
RdDM extends into heterochromatin in the vegetative cell. (A) Snapshots of CHH methylation in leaf, sperm (Spm), vegetative cell (Veg), and flower (Flw) of WT, cmt2, and drm mutants (the arrows point to loci that retain CHH methylation in cmt2 mutant vegetative cell, but not in leaf, sperm, and flower). (B) A density plot showing the frequency distribution of CHH DNA methylation differences between 50-bp windows in heterochromatic TEs that retain at least 10% CHH methylation in either cmt2 vegetative cell or cmt2 leaf. Only windows with at least 20 informative sequenced cytosines in both samples are included. The dashed box indicates windows in which CHH methylation is at least 10% greater in cmt2 vegetative cell than in cmt2 leaf. These windows are defined as new heterochromatic RdDM targets in the vegetative cell. (C) Box plots show CHH methylation levels of 50-bp windows marked by the dashed box in B (heterochromatic RdDM targets in the vegetative cell) in WT, cmt2, and drm mutants. Only windows with at least 20 informative sequenced cytosines in each sample are included. (D) Box plots show vegetative cell methylation levels of heterochromatic TEs with RdDM targets in the vegetative cell (W/) and other heterochromatic TEs (W/O). (E and F) Box plots show CHH methylation levels of 50-bp windows marked by the dashed box in B in the indicated genotypes for flowers (E) and shoots (F).
RdDM-Targeted Heterochromatic TEs Are Likely Transcribed in the Vegetative Cell.
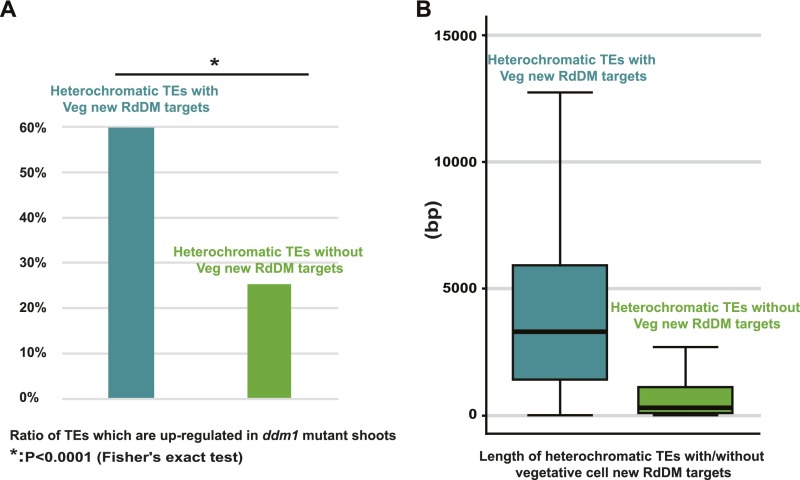
Arabidopsis TEs are generally transcriptionally silenced, but activation of heterochromatic TEs has been reported in the vegetative cell (24). Furthermore, transcriptional activation of such TEs in the ddm1 mutant background has been associated with RdDM targeting (25), suggesting that DRM-targeted heterochromatic TEs (Fig. 2 A–C) may be transcribed in the vegetative cell. To test this hypothesis, we examined how TEs targeted by RdDM in the vegetative cell are methylated in ddm1 mutant flowers (25) and shoots (7). As expected, CHH methylation of these loci is maintained by CMT2 in WT flowers and shoots (Fig. 2 A, E, and F). However, CHH methylation becomes RdDM-dependent in both tissues in ddm1 plants, as it is greatly reduced by additional mutation of either RDR2 or DRD1 (Fig. 2 E and F), genes that are required for RdDM (10). CHH methylation in ddm1 flowers is also reduced by mutation of RDR6, which contributes to RdDM of transcriptionally active loci (Fig. 2E) (25). This can be seen at individual TEs, where the RDR2- and RDR6-dependent methylation patterns of ddm1 flowers resemble those of cmt2 vegetative cells (Fig. 2A). Heterochromatic TEs targeted by RdDM in the vegetative cell are also more likely to be transcribed in ddm1 mutants than other heterochromatic TEs (Fig. S3A and Dataset S2), and they tend to be much longer (Fig. S3B), suggesting they are mostly full-length, transcriptionally competent elements (25). Overall, our results support the interpretation that RdDM targets transcriptionally activated heterochromatic TEs in the vegetative cell.
Fig. S3.
Heterochromatic TEs targeted by RdDM in the vegetative cell are transcriptionally competent. (A) RdDM-targeted TEs are more likely to be transcribed in ddm1 mutant shoots (7). Eight hundred and four out of the 1,349 heterochromatic TEs targeted by RdDM in the vegetative cell are up-regulated in ddm1 mutant shoots, whereas only 375 out of the 1,479 heterochromatic TEs not targeted by RdDM in the vegetative cell are up-regulated. (B) Heterochromatic TEs targeted by RdDM in the vegetative cell (1,349) are much longer than heterochromatic TEs not targeted by RdDM (1,479).
Histone H1 Is Present in Sperm but Absent from the Vegetative Cell.
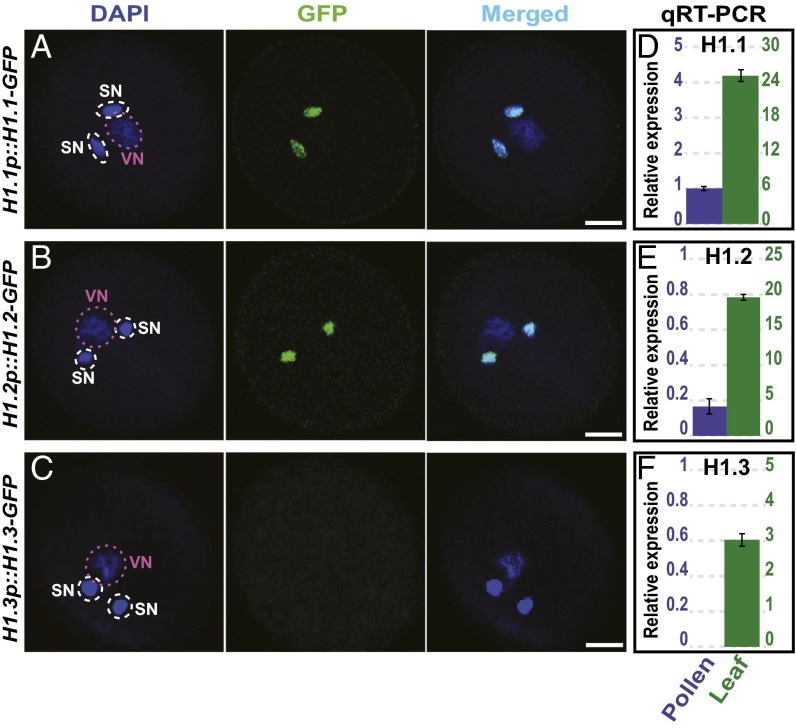
The targeting of heterochromatic loci by RdDM in the vegetative cell (Fig. 2) as well as much higher levels of heterochromatic CHH methylation compared with sperm (Fig. 1G) suggest that heterochromatin may be more accessible to DNA methyltransferases in the vegetative nucleus. This is consistent with the decondensation of chromatin and lack of heterochromatic foci reported in the vegetative cell (22). Histone H1 is a chromatin protein that reduces the efficiency of heterochromatic DNA methylation (7). We therefore wondered if increased heterochromatic methylation may be caused by reduced abundance of H1 in the vegetative cell. The Arabidopsis genome encodes two canonical, widely expressed H1 genes, H1.1 and H1.2, as well as a truncated, stress-induced H1.3 gene generally expressed at a much lower level (26). We find that H1.1-GFP and H1.2-GFP reporter proteins (27) are present in sperm but undetectable in vegetative nuclei (Fig. 3 A and B). H1.3-GFP is not detectable in pollen (Fig. 3C). Quantitative RT-PCR experiments show that H1.1 and H1.2 transcript levels are much lower in pollen than in leaf (Fig. 3 D and E), whereas H1.3 is undetectable in pollen (Fig. 3F). Vegetative cell chromatin thus appears to be depleted of histone H1, and sperm chromatin may contain less H1 than leaves and other tissues.
Fig. 3.
H1 expression in pollen. (A–C) H1.1-GFP (A) and H1.2-GFP (B) reporter constructs (27) are expressed in the sperm cells but not in the vegetative cell of pollen, whereas H1.3-GFP is not expressed in pollen (C). DAPI stains DNA in the pollen nuclei. SN, sperm nucleus; VN, vegetative nucleus. (Scale bar, 5 μm.) (D–F) Quantitative RT-PCR of H1.1 (D), H1.2 (E), and H1.3 (F) in pollen and leaf. The y axis scales show relative expression in pollen (left) and leaf (right).
Lack of H1 Does Not Increase Heterochromatic Methylation in Pollen.
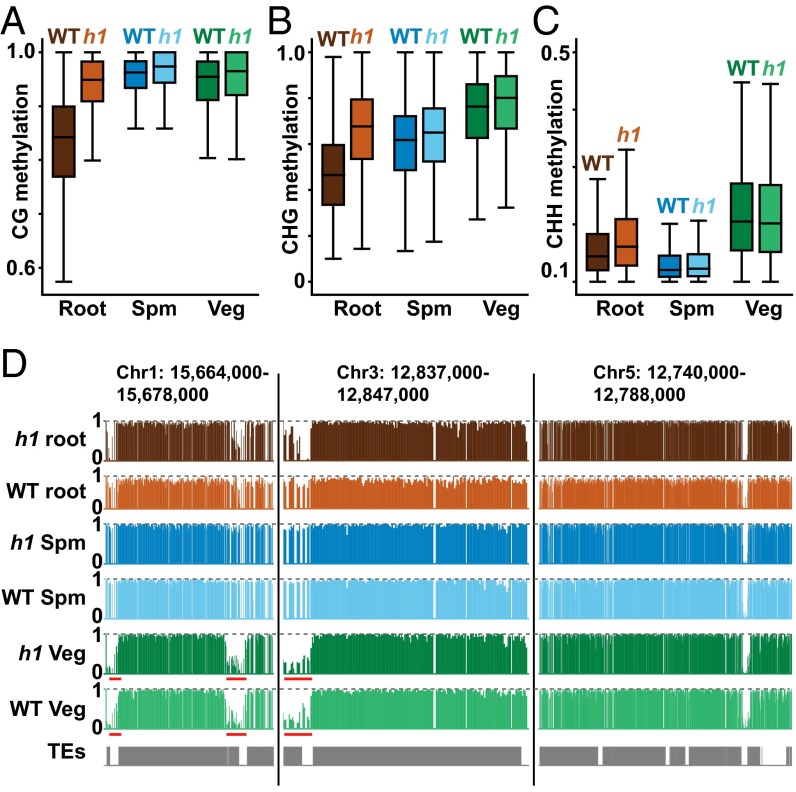
To determine how histone H1 influences DNA methylation in pollen, we analyzed methylation of FACS-purified sperm and vegetative cells with mutations in H1.1 and H1.2 (h1 mutants) (7) (Table S1). Unlike somatic tissues (7), vegetative cells do not show substantially increased heterochromatic methylation in any sequence context (Fig. 4 A–C), consistent with undetectable expression of H1 in the vegetative cell (Fig. 3). Heterochromatic methylation is also largely unaffected in sperm cells (Fig. 4 A–C), perhaps due to lower levels of H1 in this cell type (Fig. 3). Overall, heterochromatic CG methylation in WT sperm and vegetative cells is similar to h1 roots and substantially higher than in WT roots (Fig. 4A), which is readily apparent even at individual loci (Fig. 4D). Pollen heterochromatic CG methylation is therefore substantially higher than in somatic tissues, potentially due to reduced levels of H1.
Fig. 4.
Lack of H1 does not increase heterochromatic methylation in pollen. (A–C) Box plots show DNA methylation levels in 50-bp windows within heterochromatic TEs in WT and h1 mutant root, sperm (Spm), and vegetative cell (Veg). Only windows with at least 10 informative sequenced cytosines and methylation of at least 30% for CG and 10% for CHG and CHH are included. (D) Snapshots of CG methylation in root, sperm, and vegetative cell of WT and h1 mutants. Note how DNA methylation levels compare with the horizontal dashed line that denotes 100% methylation. Underlined regions bordering heterochromatic TEs are subject to active DNA demethylation in the vegetative cell (15).
CG Methylation Is More Robustly Maintained in Pollen than in Leaf or Root.
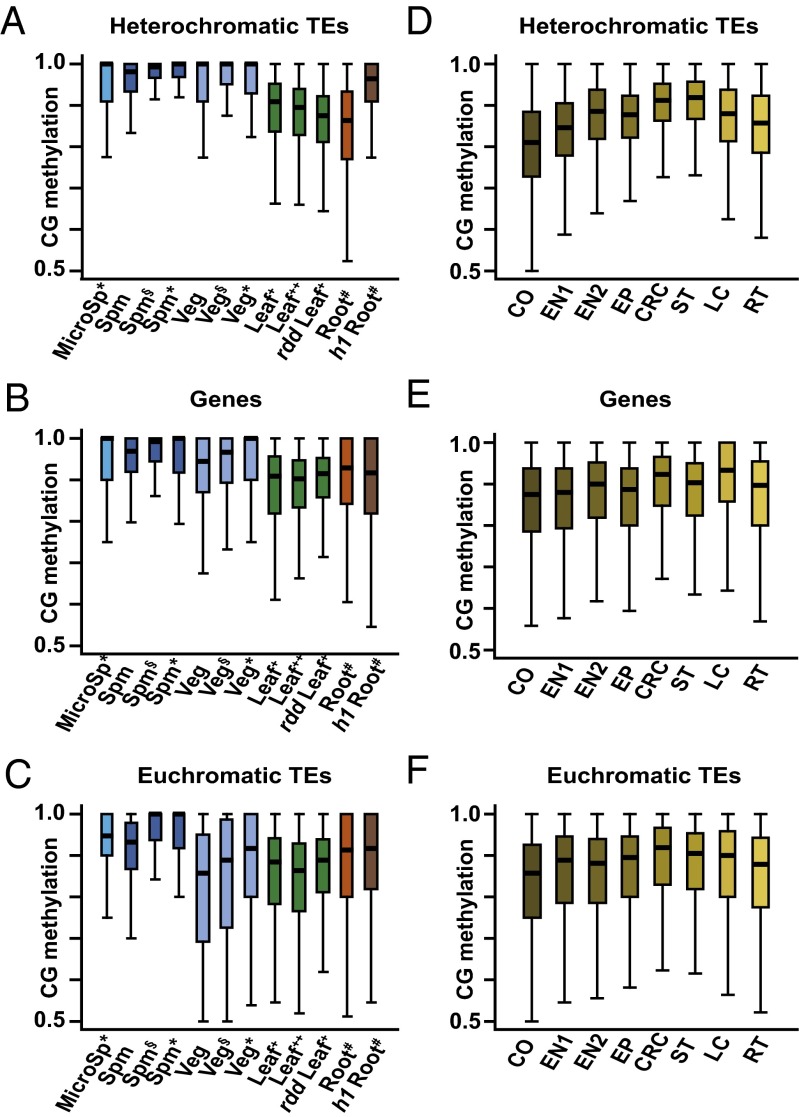
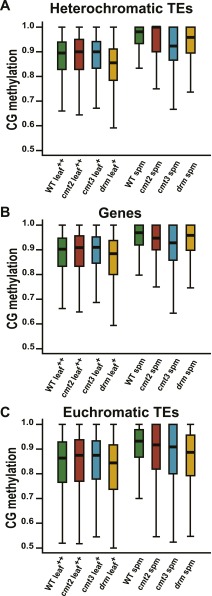
We were intrigued by the higher levels of heterochromatic CG methylation in sperm and vegetative cells (Fig. 4 A and D) because they suggested an explanation for a longstanding mystery. The semiconservative model of CG methylation maintenance (1) implies that even a modest reduction in maintenance efficiency below 100% should lead to dilution of methylation with each cell division that eventually causes complete loss (28). However, CG methylation levels measured in somatic cells (Fig. 4A) (5, 6) appear to be below what would be required for stable maintenance (28). Somatic tissues do not contribute to the next generation, so higher methylation efficiency in cells that do, including gametes, would solve the problem. Indeed, methylation of individual CG sites in sperm and vegetative cells, as well as in the microspores from which they arise, is much higher than in leaves and roots, both in heterochromatic TEs and genes (Fig. 5 A and B). CG methylation of euchromatic TEs is also higher in sperm and microspores than in somatic tissues (Fig. 5C). The low methylation of euchromatic TEs in vegetative cells (Fig. 5C) is attributable to extensive active DNA demethylation of such sequences in this cell type (Fig. 4D) (15, 16), which may also explain the somewhat lower CG methylation of genes compared with sperm and microspores (Fig. 5A).
Fig. 5.
More robust maintenance of CG methylation in pollen. (A–F) Box plots show CG methylation for individual CG sites with methylation greater than 50% and at least 10 informative sequenced cytosines. Published data in A–C are from the following: *, ref. 16; §, ref. 15; +, ref. 23; ++, ref. 8; #, ref. 7. Published data in D–F are from ref. 17. CO, cortex; CRC, columella root cap; EN, endodermis; EP, epidermis; LC, lower columella; MicroSp, microspore; RT, root tip; Spm, sperm; ST, stele; Veg, vegetative cell.
A potential concern regarding our analysis is that we are comparing methylation between pure male reproductive cells and complex somatic tissues. Methylation heterogeneity between cell types within leaves and roots could, when averaged, create the impression of an overall lower methylation efficiency even though methylation within each cell type is as robust as in pollen. To circumvent this issue, we analyzed published methylation data from multiple purified root cell types (17). These cells show CG methylation levels comparable to those of whole roots and well below those of male reproductive cells (Fig. 5 D–F). Therefore, the lower CG methylation levels we observe in somatic cells and tissues are caused by reduced methylation efficiency rather than tissue heterogeneity.
To understand the mechanism of increased CG methylation efficiency in pollen, we tested several key known pathways. First, we asked if active DNA demethylation contributes to reduced efficiency in somatic tissues by analyzing methylation of leaves lacking ROS1, DML2, and DML3, the Arabidopsis DNA demethylases expressed in somatic tissues (29). The ros1dml2dml3 (rdd) triple mutant did not substantially affect CG methylation in leaves compared with sperm and vegetative cells (Fig. 5 A–C), consistent with the limited genomic hypermethylation observed in this mutant (6). We also found that CG methylation of drm, cmt2, and cmt3 mutant sperm remained higher than in leaves in TEs and genes (Fig. S4). Our data indicate that—with the exception of MET1—no single known pathway can explain the increased CG methylation efficiency in pollen.
Fig. S4.
CG methylation of drm, cmt2, and cmt3 mutant sperm remains higher than in leaves in TEs and genes. (A–C) Box plots show CG methylation for individual CG sites with methylation greater than 50% and at least 10 informative sequenced cytosines.
Discussion
The nuclei of sperm and vegetative cells are drastically different. The sperm nucleus is small, with densely packed chromatin (Fig. 3) and obvious H3K9me2-containing heterochromatic foci (22). The vegetative nucleus is larger (Fig. 3), lacks heterochromatic foci and cytologically detectable H3K9me2 (22), and has much higher levels of CHH methylation (Fig. 1D) (15, 16). CMTs are dependent on H3K9me2, so it is reasonable to hypothesize that CHH and even CHG methylation in the vegetative nucleus may be largely dependent on RdDM. The columella cells in the Arabidopsis root cap also have much higher CHH methylation than neighboring cells, with elevated RdDM proposed as the cause (17). Our data do show that RdDM extends somewhat into heterochromatic TEs in vegetative cells (Fig. 2), which is consistent with published results (22) and may reflect TE activation in the vegetative nucleus (24). However, CHG methylation is still dependent on CMT3 (Fig. 1C) and heterochromatic CHH methylation primarily on CMT2 (Fig. 1G). In general, maintenance of non-CG methylation is remarkably similar between vegetative, sperm, and leaf cells (Fig. 1 C–G). It is likely that the active removal of H3K9me2 in the vegetative nucleus (30) occurs after DNA methylation is deposited, allowing CMTs to work. Overall, our results demonstrate that large, global CHH methylation changes can occur with only minor alterations of pathway specificity.
Although DNA methylation pathways function similarly in pollen and somatic tissues, the efficiency of CG methylation is substantially higher in pollen (Fig. 5). The semiconservative model of CG methylation inheritance (1), and the considerable genetic evidence in support of this model (31–33), is more compatible with the higher efficiency of CG methylation in the male germ line (microspores and sperm), and with the similarly high efficiency observed in female plant gametes (34), than with that observed in somatic tissues (28). In heterochromatic TEs, much or all of the increased efficiency might be accounted by reduced levels of histone H1 (Fig. 3), but the mechanism must be different in genes and euchromatic TEs, where loss of H1 does not facilitate CG methylation (Fig. 5 B and C). Instead of a unifying mechanism, the similar pollen–soma methylation efficiency differences in genes and TEs (Fig. 5) may be an unavoidable consequence of distinct selection pressures. Gametes—and cells that might give rise to gametes—have the potential to undergo an essentially unlimited number of divisions that span generations and should therefore be under strong selection for very efficient methylation maintenance. In comparison, somatic cells will divide very few times and need a methylation maintenance activity that is just sufficient to keep TE silencing and other methylation functions from degenerating. Therefore, mutations that reduce methylation efficiency in somatic cells but keep it above this threshold would not be counterselected. Under these conditions, somatic methylation efficiency would be expected to settle at this equilibrium threshold. An important consequence of less efficient somatic methylation is that small methylation differences between somatic tissues or cell types (17) may be caused by maintenance fluctuations rather than developmental reprogramming.
Materials and Methods
Isolation of A. thaliana Sperm and Vegetative Cell Nuclei.
A. thaliana plants were grown under 16 h light/8 h dark in a growth chamber (20 °C, 80% humidity). Sperm and vegetative cell nuclei were isolated by FACS based on SYBR Green staining, as previously described (15).
Whole-Genome Bisulfite Sequencing.
Bisulfite sequencing libraries for sperm and vegetative cells were constructed using the Ovation Ultralow Methyl-Seq Library Systems (Nugen, 0336) and EpiTect Fast Bisulfite Conversion (Qiagen, 59802) kits according to the kit protocols, except the incorporation of two rounds of bisulfite conversion. Illumina sequencing was performed at the UC Berkeley Vincent J. Coates Genomics Sequencing Laboratory, the DNA sequencing facility of the University of Cambridge Department of Biochemistry, Novogene Ltd., and the Bauer Core Facility at Harvard University. Sequenced reads (75 or 100 base single end) were mapped to the TAIR10 reference genome, and DNA methylation of each cytosine was ascertained as previously described (15).
Published Genomic Data.
DNA methylation data for WT, cmt3, cmt2, drm1drm2 (drm), and ros1dml2dml3 (rdd) leaf tissue are from refs. 8 and 23. DNA methylation data for WT and h1 root are from ref. 7. DNA methylation data for sperm, vegetative cell, and microspore are from refs. 15 and 16. DNA methylation data for WT, cmt2, drm2, ddm1, ddm1rdr2, ddm1rdr6, and ddm1drd1 flower and shoot tissues are from refs. 25 and 7. Leaf H3K9me2 and histone H3 ChIP-seq data are from ref. 8.
Definition of Genomic Features.
Only genes with CG methylation between 20% and 60% were used for analysis in this paper. Overlapping TE annotations were merged.
Confocal Microscopy.
Pollen grains were isolated by vortexing open flowers in PBS with 1% of Triton-X-100 and 0.1 µg/mL of DAPI and spread on slides for confocal microscopy under DAPI and GFP channels, respectively.
Quantitative RT-PCR.
Total RNA was extracted from mature pollen and rosette leaves, respectively, and treated with DNase I. Equal amounts of total RNA from the two samples were used for reverse transcription and quantitative PCR. The ACT8 gene was used as an internal control.
Supplementary Material
Acknowledgments
We thank Andrzej Jerzmanowski for kind sharing of the H1 reporter lines, Célia Baroux for discussion of H1 expression in the male sexual lineage, the John Innes Centre Computing Infrastructure for Science Facility and Bioimaging facility and staff, and the Norwich BioScience Institute Partnership Computing Infrastructure for Science group for High Performance Computing Resources. This work was partially funded by National Science Foundation Grant IOS-1025890 and NIH Grant R01-GM069415 (to R.L.F. and D.Z.), and Biotechnology and Biological Sciences Research Council (BBSRC) David Phillips Fellowship BB/L025043/1 (to X.F.). This work was also partially supported by the Centre of Excellence for Plant and Microbial Sciences (CEPAMS), established between the John Innes Centre and the Chinese Academy of Sciences (X.F. and S.H.). This work used the Vincent J. Coates Genomics Sequencing Laboratory at University of California, Berkeley, supported by NIH Instrumentation Grants S10RR029668 and S10RR027303.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE87170).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619074114/-/DCSupplemental.
References
- 1.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11(3):204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim MY, Zilberman D. DNA methylation as a system of plant genomic immunity. Trends Plant Sci. 2014;19(5):320–326. doi: 10.1016/j.tplants.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Weigel D, Colot V. Epialleles in plant evolution. Genome Biol. 2012;13(10):249. doi: 10.1186/gb-2012-13-10-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heard E, Martienssen RA. Transgenerational epigenetic inheritance: Myths and mechanisms. Cell. 2014;157(1):95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133(3):523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zemach A, et al. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell. 2013;153(1):193–205. doi: 10.1016/j.cell.2013.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stroud H, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21(1):64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du J, et al. Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell. 2012;151(1):167–180. doi: 10.1016/j.cell.2012.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matzke MA, Mosher RA. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15(6):394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- 11.Zhong X, et al. DDR complex facilitates global association of RNA polymerase V to promoters and evolutionarily young transposons. Nat Struct Mol Biol. 2012;19(9):870–875. doi: 10.1038/nsmb.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huettel B, et al. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25(12):2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsieh T-F, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324(5933):1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zemach A, et al. Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA. 2010;107(43):18729–18734. doi: 10.1073/pnas.1009695107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibarra CA, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337(6100):1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calarco JP, et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151(1):194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawakatsu T, et al. Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat Plants. 2016;2(5):16058. doi: 10.1038/nplants.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irish VF. The flowering of Arabidopsis flower development. Plant J. 2010;61(6):1014–1028. doi: 10.1111/j.1365-313X.2009.04065.x. [DOI] [PubMed] [Google Scholar]
- 19.Feng X, Zilberman D, Dickinson H. A conversation across generations: Soma-germ cell crosstalk in plants. Dev Cell. 2013;24(3):215–225. doi: 10.1016/j.devcel.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt A, Schmid MW, Grossniklaus U. Plant germline formation: Common concepts and developmental flexibility in sexual and asexual reproduction. Development. 2015;142(2):229–241. doi: 10.1242/dev.102103. [DOI] [PubMed] [Google Scholar]
- 21.Saffman EE, Lasko P. Germline development in vertebrates and invertebrates. Cell Mol Life Sci. 1999;55(8-9):1141–1163. doi: 10.1007/s000180050363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoft VK, et al. Induction of RNA-directed DNA methylation upon decondensation of constitutive heterochromatin. EMBO Rep. 2009;10(9):1015–1021. doi: 10.1038/embor.2009.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152(1-2):352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136(3):461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panda K, et al. Full-length autonomous transposable elements are preferentially targeted by expression-dependent forms of RNA-directed DNA methylation. Genome Biol. 2016;17(1):170. doi: 10.1186/s13059-016-1032-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jerzmanowski A, Przewloka M, Grasser KD. Linker histones and HMG1 proteins of higher plants. Plant Biol. 2000;2:586–597. [Google Scholar]
- 27.She W, et al. Chromatin reprogramming during the somatic-to-reproductive cell fate transition in plants. Development. 2013;140(19):4008–4019. doi: 10.1242/dev.095034. [DOI] [PubMed] [Google Scholar]
- 28.Haerter JO, Lövkvist C, Dodd IB, Sneppen K. Collaboration between CpG sites is needed for stable somatic inheritance of DNA methylation states. Nucleic Acids Res. 2014;42(4):2235–2244. doi: 10.1093/nar/gkt1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penterman J, et al. DNA demethylation in the Arabidopsis genome. Proc Natl Acad Sci USA. 2007;104(16):6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mérai Z, et al. The AAA-ATPase molecular chaperone Cdc48/p97 disassembles sumoylated centromeres, decondenses heterochromatin, and activates ribosomal RNA genes. Proc Natl Acad Sci USA. 2014;111(45):16166–16171. doi: 10.1073/pnas.1418564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330(6004):622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker C, et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature. 2011;480(7376):245–249. doi: 10.1038/nature10555. [DOI] [PubMed] [Google Scholar]
- 33.Schmitz RJ, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334(6054):369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park K, et al. DNA demethylation is initiated in the central cells of Arabidopsis and rice. Proc Natl Acad Sci USA. 2016;113:15138–15143. doi: 10.1073/pnas.1619047114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.