Significance
The voltage-gated sodium channel NaV1.7 is important for electrogenesis in sensory neurons. Insertion within the membrane is required for function of NaV1.7. However, the mechanisms determining how NaV1.7 is trafficked to neuronal cell membranes are poorly understood. Here, we elucidate a signaling program involving a complex and intriguing posttranslational modification regime of collapsin response mediator protein 2 (CRMP2), an NaV1.7-binding protein. NaV1.7 surface localization and currents are controlled by CRMP2 modifications. Activity of NaV1.7 is thought to modulate neuronal excitability that codes for several sensory modalities, including chronic pain, as inferred from human pain disorders caused by mutations in NaV1.7 channels. Understanding the role of cross-talk between CRMP2 modifications in modulation of NaV1.7 activity opens routes to exploit this system for pain.
Keywords: NaV1.7 sodium channel, trafficking, CRMP2, SUMOylation, phosphorylation
Abstract
Voltage-gated sodium channels are crucial determinants of neuronal excitability and signaling. Trafficking of the voltage-gated sodium channel NaV1.7 is dysregulated in neuropathic pain. We identify a trafficking program for NaV1.7 driven by hierarchical interactions with posttranslationally modified versions of the binding partner collapsin response mediator protein 2 (CRMP2). The binding described between CRMP2 and NaV1.7 was enhanced by conjugation of CRMP2 with small ubiquitin-like modifier (SUMO) and further controlled by the phosphorylation status of CRMP2. We determined that CRMP2 SUMOylation is enhanced by prior phosphorylation by cyclin-dependent kinase 5 and antagonized by Fyn phosphorylation. As a consequence of CRMP2 loss of SUMOylation and binding to NaV1.7, the channel displays decreased membrane localization and current density, and reduces neuronal excitability. Preventing CRMP2 SUMOylation with a SUMO-impaired CRMP2-K374A mutant triggered NaV1.7 internalization in a clathrin-dependent manner involving the E3 ubiquitin ligase Nedd4-2 (neural precursor cell expressed developmentally down-regulated protein 4) and endocytosis adaptor proteins Numb and epidermal growth factor receptor pathway substrate 15. Collectively, our work shows that diverse modifications of CRMP2 cross-talk to control NaV1.7 activity and illustrate a general principle for regulation of NaV1.7.
The tetrodotoxin-sensitive (TTX-S) voltage-gated sodium channel NaV1.7 generates thresholds for action potential firing in sensory neurons. Genetic and functional studies have established NaV1.7 as a major contributor to pain signaling in humans and have demonstrated that mutations in SCN9A, the gene encoding NaV1.7, produce distinct human pain syndromes (1–4). Pain also results from up-regulated NaV1.7 expression (5–7); however, dysregulation of NaV1.7 is poorly understood.
In search of NaV1.7 trafficking/regulatory events, we reported that surface expression and current density of NaV1.7 was controlled by SUMOylation of the cytosolic axonal collapsin response mediator protein 2 (CRMP2) (8). CRMP2 regulates multiple processes in neurons and was initially discovered to regulate mechanisms of neuronal polarity (9, 10). CRMP2 phosphorylation by cyclin-dependent kinase 5 (Cdk5) (11), glycogen synthase kinase 3β (10), Rho-associated protein kinase (12), or the Src-family kinases Fyn (13) and Yes (14) drives its diverse cellular functions, including neurite outgrowth, endocytosis, and ion-channel trafficking (8, 15–17). Studies of CRMP2 trafficking functions have revealed that CRMP2 facilitates endocytosis of L1-cell adhesion molecule by interacting with the endocytic protein Numb (18) that recruits epidermal growth factor receptor pathway substrate 15 (Eps15), an initiator of clathrin-mediated endocytosis (19).
In neuropathic pain, NaV1.7 surface localization is augmented by loss of Nedd4-2 (neural precursor cell expressed developmentally down-regulated protein 4), an E3 ubiquitin ligase that labels NaV1.7 for endocytosis (6). Inspired by these observations, we tested the hypothesis that a cross-talk between distinct CRMP2 posttranslational modifications is a key factor in determining NaV1.7 trafficking and localization. Here, we (i) demonstrate a pathway of NaV1.7 regulation dependent upon CRMP2 SUMOylation and phosphorylation states, (ii) identify interactions between CRMP2 and NaV1.7, and (iii) map molecular determinants of NaV1.7 endocytosis that result from altered CRMP2 interactions. We also report that the CRMP2-NaV1.7 signaling is conserved between rodent and human sensory neurons, supporting the exciting possibility that this pathway could be the target of future therapeutic agents aimed at controlling NaV1.7 in patients with neuropathic pain.
Results
CRMP2 Is Necessary for Maintaining TTX-S Sodium Currents in Sensory Neurons.
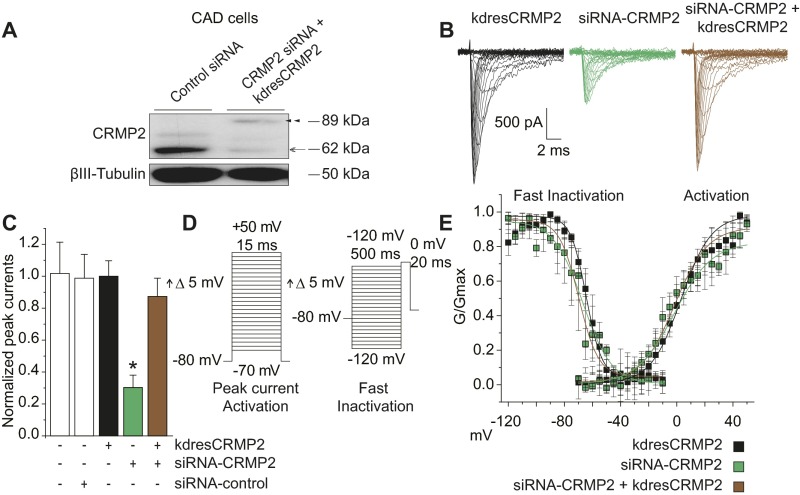
If CRMP2 loss of SUMOylation inhibits NaV1.7 activity, a similar reduction in NaV1.7 currents should be seen if CRMP2 expression is reduced, particularly if CRMP2 is necessary for maintaining NaV1.7 currents. NaV1.7 currents can be analyzed by whole-cell patch-clamp electrophysiology of mouse catecholamine A differentiated (CAD) cells, a central nervous system-derived catecholaminergic cell line that expresses high levels of NaV1.7 (20) and isolated dorsal root ganglia (DRG) sensory neurons responsible for transmission of noxious stimuli from glabrous skin through the sciatic nerve and to the spinal cord. A decrease in CRMP2 expression (Fig. S1A) caused a reduction in TTX-S sodium currents in rat DRG neurons (Fig. 1A, B) and in NaV1.7 currents in CAD cells without altering biophysical properties of the channels (Fig. S1 B–E and Table S1). Notably, sodium currents were normalized by “add-back” of a CRMP2 refractory to knockdown (Figs. 1 A and B and S1 A–C), confirming that CRMP2 is necessary for selectively maintaining TTX-S sodium currents.
Fig. S1.
Decreased TTX-S current densities in mouse CAD cell line. (A) Representative Western blot of CAD cells (n = 3) expressing control siRNA or CRMP2 siRNA and knockdown-resistant (kdres) CRMP2 plasmid. Open arrow indicates reduction of endogenous CRMP2 expression at 62 kDa by ∼83%, and closed arrow indicates de novo expression of exogenous CRMP2 at 90 kDa. (B) Representative traces of total sodium current from CAD cells expressing kdresCRMP2, siRNA-CRMP2, or both. (C) Quantitative analysis of normalized total sodium currents (pA/pF) from CAD cells expressing indicated combinations of siRNA and CRMP2 plasmids. Loss of CRMP2 expression produced a significant reduction to normalized peak current that was recovered by reintroduction of CRMP2 expression (compare green and brown bars). (D) Voltage protocols used to probe CAD cell peak current and activation (Left) and fast inactivation (Right). (E) Averaged Boltzmann fits from individual CAD cells expressing indicated combinations of siRNA and CRMP2 plasmids. No significant differences were detected in half-maximal voltage or slope properties of fast inactivation or activation. Data show means ± SEM (*P < 0.05 vs. siRNA control, Kruskal–Wallis test with Dunnett’s post hoc comparisons).
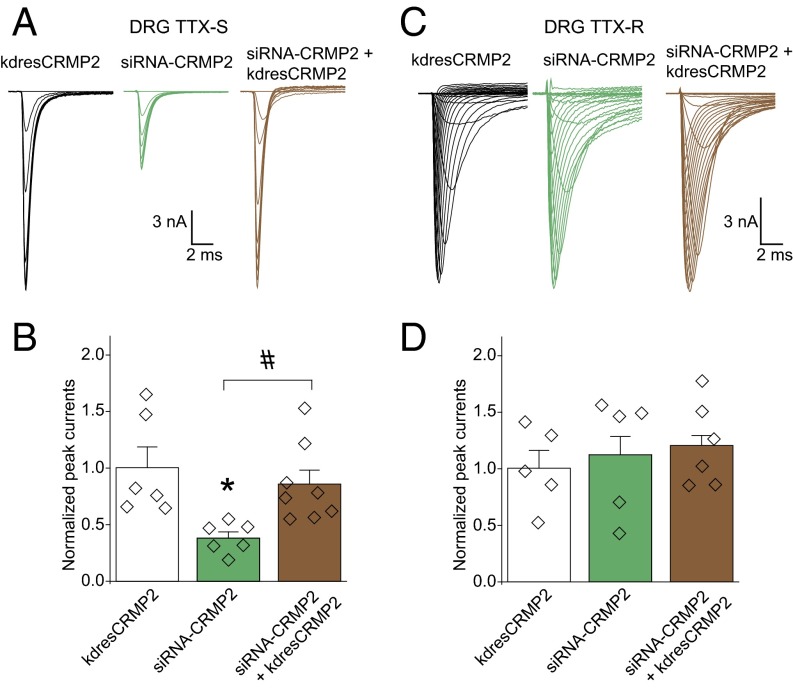
Fig. 1.
CRMP2 is necessary for maintaining TTX-S sodium currents in sensory neurons. Sodium current traces representing TTX-S (A) and TTX-R (C) currents. Quantitative analysis of TTX-S (B) and TTX-R (D) DRG peak sodium current densities (picoamps/picofarad; pA/pF). DRG currents were 13.8 ± 4.2 nA and recorded from cells with conductances of 28.9 ± 9.1 pF. Unless otherwise indicated, for this and subsequent figures, all data represent mean ± SEM. Individual data points and n values are indicated by diamonds overlaid on the bar graphs [n = 5–7; *P < 0.05 vs. knockdown-resistant (kdres) CRMP2; #P < 0.05, Kruskal–Wallis test with Dunnett’s post hoc comparisons]. Voltage protocols and post hoc subtraction used to investigate DRG cell peak TTX-R and TTX-S currents are described in SI Materials and Methods.
Table S1.
Comparison of sodium channel currents in HEK and CAD cell lines
| Transfection condition | Peak current density, pA/pF | Voltage-dependence of activation | Voltage-dependence of fast-inactivation | ||
| V1/2, mV | Slope, mV/e-fold | V1/2, mV | Slope, mV/e-fold | ||
| HEK cells | |||||
| NaV1.1 | |||||
| WT | 100.0 ± 13.9 (5) | −11.8 ± 0.6 (5) | 6.3 ± 0.4 (5) | −55.7 ± 1.6 (5) | 5.6 ± 1.0 (5) |
| S522A | 87.3 ± 12.1 (5) | −9.7 ± 1.1 (5) | 7.4 ± 0.5 (5) | −54.7 ± 0.8 (5) | 5.3 ± 0.6 (5) |
| NaV1.3 | |||||
| WT | 100.0 ± 23.8 (5) | −10.7 ± 0.5 (4) | 5.3 ± 0.4 (4) | −55.8 ± 1.0 (4) | 7.2 ± 0.6 (4) |
| S522A | 89.6 ± 12.1 (5) | −13.6 ± 2.7 (4) | 4.5 ± 1.0 (4) | −54.8 ± 1.2 (4) | 7.6 ± 0.5 (4) |
| NaV1.5 | |||||
| WT | 100.0 ± 14.2 (5) | −32.5 ± 0.5 (4) | 5.5 ± 0.2 (4) | −76.9 ± 1.6 (5) | 4.4 ± 0.6 (5) |
| K374A | 106.3 ± 22.5 (5) | −32.6 ± 0.6 (4) | 5.1 ± 0.6 (4) | −75.1 ± 0.5 (4) | 5.1 ± 0.3 (4) |
| S522A | 97.2 ± 28.7 (5) | −32.8 ± 0.4 (5) | 5.8 ± 0.3 (5) | −75.0 ± 0.7 (4) | 4.4 ± 0.3 (4) |
| NaV1.7 | |||||
| WT | 100.0 ± 16.0 (5) | 0.9 ± 1.4 (5) | 7.2 ± 0.5 (5) | −53.5 ± 0.8 (5) | 7.5 ± 0.6 (5) |
| S522A | 42.5 ± 12.0 (5)* | −2.2 ± 0.5 (5) | 7.6 ± 0.5 (5) | −57.0 ± 2.3 (4) | 6.9 ± 1.2 (4) |
| CAD cells | |||||
| CRMP2 | 100.0 ± 9.7 (5) | 3.0 ± 1.3 (5) | 10.8 ± 1.0 (5) | −64.4 ± 1.9 (5) | 5.6 ± 0.7 (5) |
| Untransfected | 101.8 ± 17.9 (4) | 2.9 ± 2.8 (4) | 11.0 ± 1.2 (4) | −68.9 ± 1.1 (4) | 5.6 ± 0.6 (4) |
| siRNA-control | 98.8 ± 14.9 (6) | 3.4 ± 2.0 (5) | 14.1 ± 1.7 (5) | −68.0 ± 1.3 (4) | 6.2 ± 0.7 (4) |
| siRNA-CRMP2 | 30.2 ± 7.8 (7)* | −1.6 ± 3.2 (3) | 13.4 ± 2.1 (3) | −71.4 ± 2.6 (3) | 8.0 ± 1.4 (3) |
| siRNA-CRMP2+ kdresCRMP2 | 87.4 ± 11.5 (6) | −1.4 ± 1.8 (5) | 11.8 ± 1.2 (5) | −69.8 ± 1.3 (4) | 6.9 ± 0.6 (4) |
| kdresCRMP2-K374A | 42.9 ± 10.0 (5)* | 3.4 ± 1.4 (4) | 10.5 ± 0.9 (4) | −63.7 ± 3.1 (3) | 5.7 ± 0.9 (3) |
| siRNA-CRMP2+ kdresCRMP2-K374A | 34.1 ± 2.4 (5)* | 4.3 ± 1.1 (4) | 10.8 ± 0.7 (4) | −67.5 ± 0.9 (3) | 4.7 ± 1.1 (3) |
| SENP1 + SENP2 | 36.9 ± 9.0 (5)* | 3.5 ± 1.6 (4) | 10.4 ± 0.8 (4) | −65.5 ± 2.2 (3) | 6.1 ± 1.4 (3) |
| CRMP2-Y32F | 105.9 ± 25.0 (6) | 0.7 ± 1.2 (4) | 11.0 ± 1.0 (4) | −59.6 ± 2.1 (3) | 6.1 ± 1.3 (3) |
| CRMP2-Y32F/K374A | 107.0 ± 22.8 (6) | 1.3 ± 0.4 (4) | 8.3 ± 0.4 (4) | −63.8 ± 0.7 (5) | 5.0 ± 0.4 (5) |
| CRMP2-Y479F | 100.4 ± 9.1 (5) | 0.0 ± 1.1 (3) | 9.6 ± 0.7 (3) | −64.3 ± 1.3 (3) | 7.0 ± 0.7 (3) |
| CRMP2-K374A/Y479F | 38.0 ± 13.9 (5)* | 5.2 ± 1.8 (3) | 11.3 ± 0.8 (3) | −66.2 ± 1.6 (3) | 4.9 ± 1.0 (3) |
| CRMP2-T509A/T514A | 102.6 ± 15.1 (5) | 0.7 ± 1.1 (4) | 11.6 ± 0.8 (4) | −66.9 ± 1.0 (5) | 5.9 ± 0.7 (5) |
| CRMP2-K374A/T509A/T514A | 34.7 ± 12.5 (7)* | −1.4 ± 1.4 (3) | 10.3 ± 1.3 (3) | −66.6 ± 1.6 (3) | 5.7 ± 1.0 (3) |
| CRMP2-S522A | 44.7 ± 11.0 (5) * | 2.3 ± 2.0 (3) | 12.9 ± 1.4 (3) | −69.0 ± 1.6 (4) | 6.2 ± 0.8 (4) |
| CRMP2-K374A/S522A | 43.0 ± 11.3 (5)* | 0.9 ± 2.2 (3) | 14.0 ± 2.2 (3) | −64.2 ± 1.6 (3) | 3.5 ± 1.3 (3) |
| CRMP2-T555A | 97.7 ± 23.7 (5) | 1.0 ± 2.0 (4) | 12.9 ± 1.5 (4) | −67.6 ± 1.6 (3) | 6.7 ± 1.0 (3) |
| CRMP2-K374A/T555A | 50.6 ± 15.3 (4)* | 2.2 ± 2.7 (4) | 14.3 ± 2.2 (4) | −68.4 ± 1.1 (4) | 5.3 ± 1.0 (4) |
| CRMP2 + Fyn | 54.2 ± 9.6 (7) * | 1.6 ± 0.8 (5) | 12.1 ± 0.4 (5) | −67.1 ± 2.2 (3) | 4.7 ± 0.9 (3) |
n value provided in parentheses. Comparative sodium current statistics from HEK cells and CAD cells expressing indicated plasmids. HEK cells express single sodium channel isoforms and CAD cells predominantly express NaV1.7. Comparative normalized current densities and Boltzmann properties of half-maximal effect (V1/2) and slope (k) for activation and fast inactivation are presented. Only conditions designed to reduce CRMP2 SUMOylation state in Nav1.7 expressing cells demonstrate reduction of current density. CAD cell currents in this manuscript averaged 1.3 ± 0.1 nA and were recorded from cells 21.2 ± 1.7 pF. All HEK cells were of similar size, 17.5 ± 2.0 pF, but expressed variable current density depending on isoform; NaV1.1 1.4 ± 0.2 nA, NaV1.3 6.1 ± 1.1 nA, NaV1.5 9.8 ± 1.4 nA, NaV1.7 1.0 ± 0.2 nA. Data are presented as means ± SEM.
P < 0.05, Kruskal–Wallis test with pairwise comparisons.
Interplay Between CRMP2 Modifications Selectively Alter NaV1.7 Trafficking.
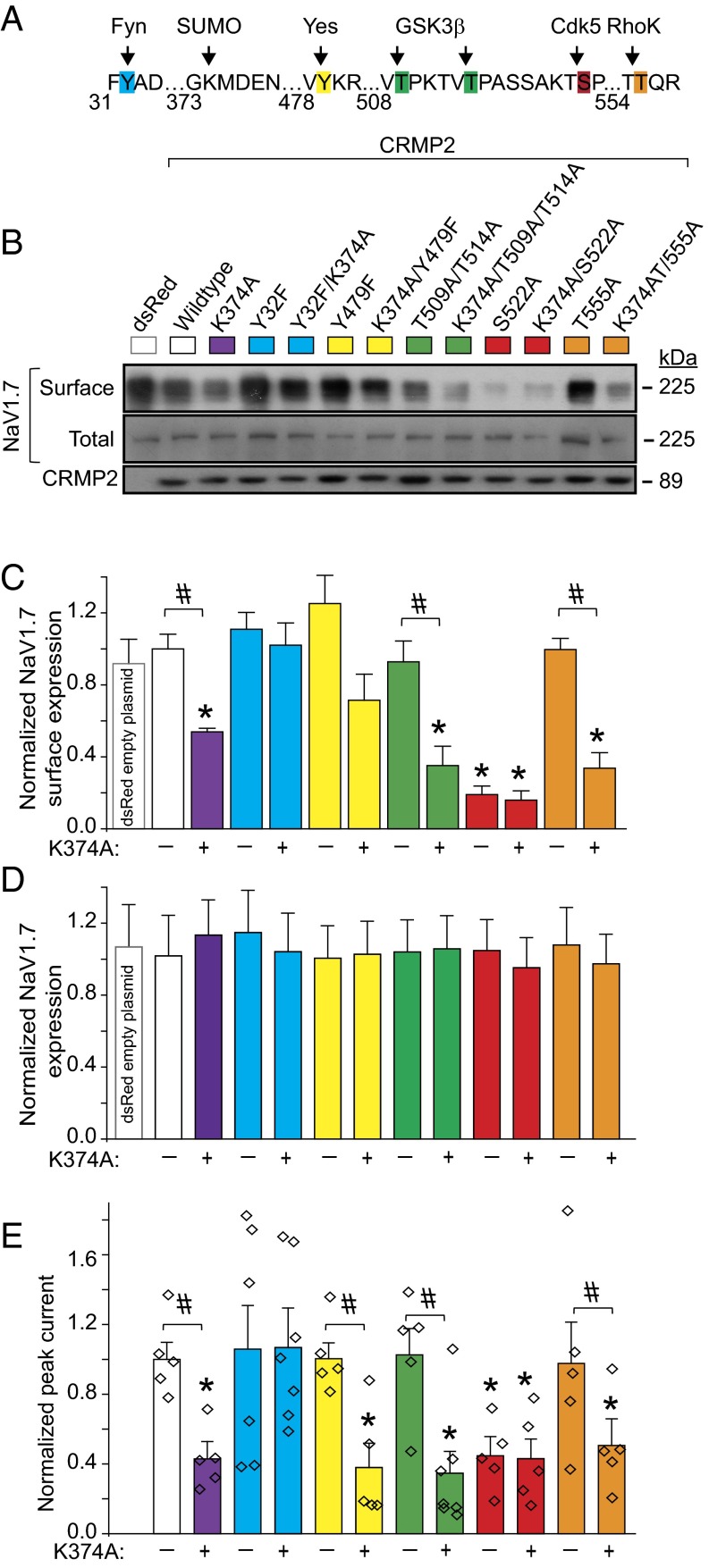
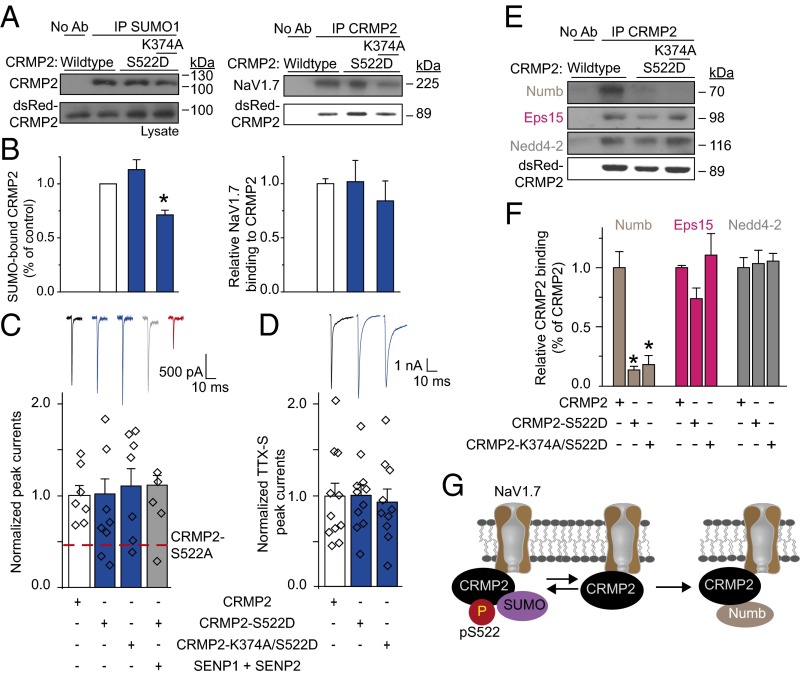
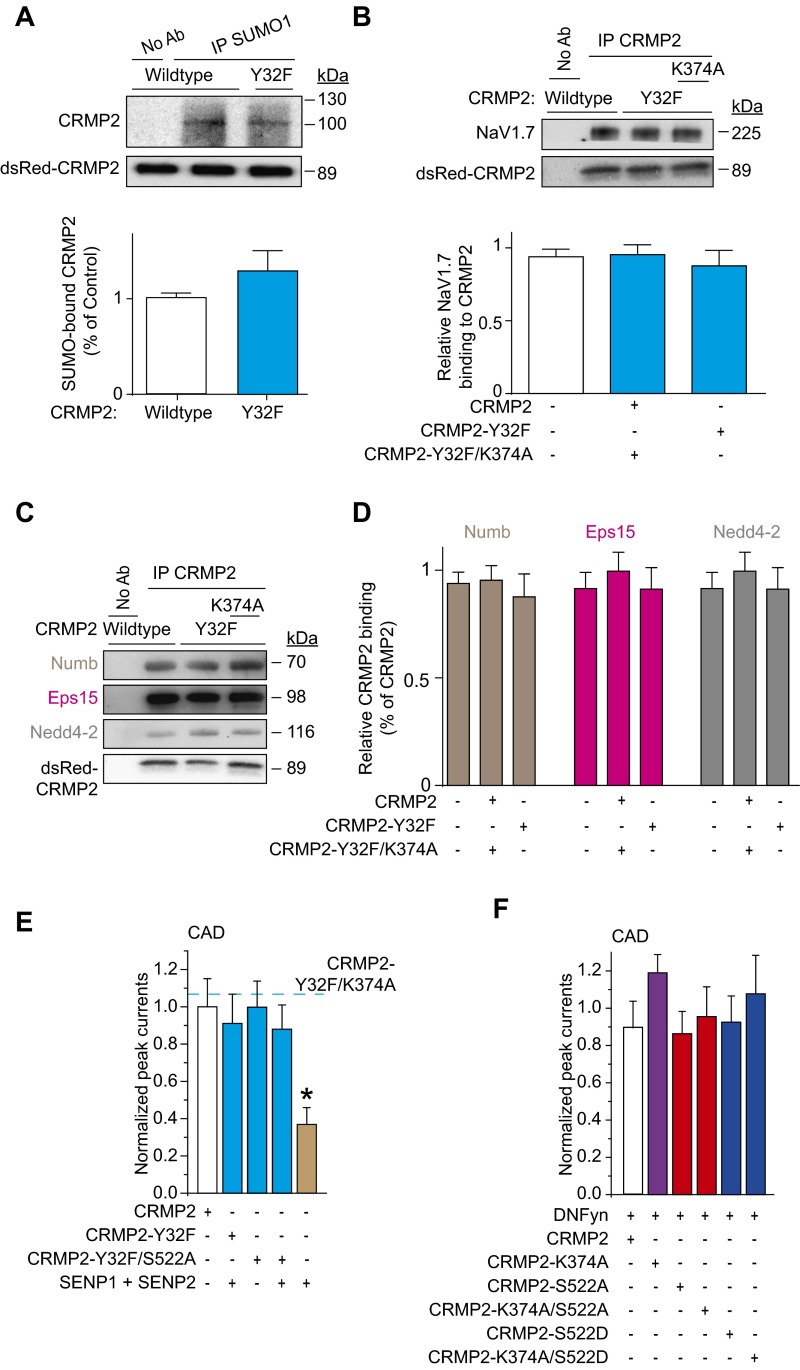
A loss of CRMP2 expression via knockdown (Fig. 1) or loss of CRMP2 SUMOylation via expression of SUMO-impaired mutant reduces sodium currents (8), but whether CRMP2 phosphorylation events also modulate sodium currents is unknown. To examine possible priming by CRMP2 modifications in modulation of NaV1.7 and to identify the relationships between CRMP2 modifications and their effects on NaV1.7, CRMP2 mutants with SUMO-site impaired, phospho-null sites, or both (Fig. 2A) were engineered. Consistent with previous findings (8), loss of CRMP2 SUMOylation reduced NaV1.7 surface fraction and currents (Fig. 2 B, C, and E). NaV1.7 surface localization and current density were reduced by mutations in CRMP2 at the site phosphorylated by Cdk5 (i.e., S522A) but not sites phosphorylated by other kinases [Fyn (Y32F), Yes (Y479F), GSK-3β (T509A/A514A), RhoK (T555A)]; Fig. 2 B, C, and E). Expression of WT CRMP2 or SUMO-impaired (i.e., K374A), or phospho-deficient mutants did not alter NaV1.7 protein expression (Fig. 2D). Concomitant elimination of CRMP2 SUMOylation and phosphorylation (Cdk5 site) did not further decrease NaV1.7 surface fraction or current compared with that observed with either mutation alone (Fig. 2 B, C, and E), suggesting a convergent mechanism of modulation. Concomitant elimination of CRMP2 SUMOylation and Fyn phosphorylation had no effect on NaV1.7 surface fraction or currents (Fig. 2 B, C, and E), suggesting that Fyn phosphorylation of CRMP2 is obligatory for eliciting the negative regulation induced by loss of CRMP2 SUMOylation. These results illustrate that at least three different posttranslational modifications of CRMP2, namely SUMOylation, Cdk5 phosphorylation, and Fyn phosphorylation, collaborate to control NaV1.7 trafficking and currents.
Fig. 2.
Expression of SUMOylation- and phosphorylation-deficient CRMP2 plasmids regulates NaV1.7 surface expression and current density in mouse CAD cells. (A) Amino acid sequence of CRMP2 highlighting sites of phosphorylation and SUMOylation. Numbers refer to amino acids of rat CRMP2. (B) Representative immunoblots of streptavidin-enriched surface fraction probed with a NaV1.7 antibody (Top) and total fraction probed with a pan-NaV antibody (Middle). (Bottom) Blot of lysates probed with CRMP2 antibody to verify expression of exogenous ∼89-kDa CRMP2 (n = 5). (C) Bar graphs summarizing mean surface NaV1.7 in CAD cells transfected with the indicated plasmids. Data are normalized to total NaV1.7 protein and then to the CRMP2 in each lane and plotted as ratio of the WT CRMP2 condition. (D) Bar graphs summarizing mean total NaV1.7 in CAD cells transfected with the indicated plasmids. Data are normalized to total NaV1.7 protein and then to β-tubulin in each lane and plotted as ratio of the WT CRMP2 condition. (E) Quantitative analysis of CAD cell peak current density following expression of indicated CRMP2 plasmids (n = 5–7). CAD cell currents throughout this study averaged 1.3 ± 0.1 nA and were recorded from cells with conductances of 21.2 ± 1.7 pF.
The reduction in DRG sodium currents imposed by loss of CRMP2 SUMOylation and loss of Cdk5 phosphorylation was limited to NaV1.7, as no changes were observed in TTX-resistant (TTX-R) currents (predominantly NaV1.8; Fig. 3A) or in non-NaV1.7 TTX-S (predominantly NaV1.1 and NaV1.6) currents upon their isolation with electrophysiological and pharmacological protocols (Fig. 3A). Additionally, hNaV1.1, rNaV1.3, or the cardiac hNaV1.5 expressed in heterologous cells were also unaffected by loss of CRMP2 SUMOylation (8) or phosphorylation (Table S1).
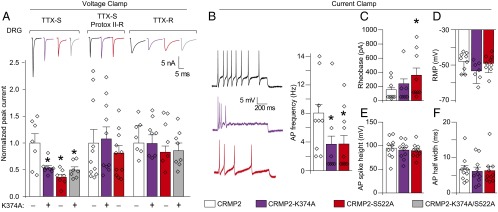
Fig. 3.
CRMP2 modifications control NaV1.7 currents and excitability (A). Normalized peak sodium current density (picoamps/picofarad; pA/pF) from DRG neurons transfected with WT (CRMP2), SUMO-impaired (CRMP2-K374A), phospho-null (CRMP2-S522A), or dual SUMO-impaired/phospho-null (CRMP2-K374A/S522A) CRMP2 (n = 7–11). Representative traces are displayed above the bar graphs. Total sodium current was separated electrically and pharmacologically into TTX-S (NaV1.1, NaV1.6, NaV1.7), TTX-S/Protox-II–resistant (NaV1.1, NaV1.6; 5 nM Protox-II), and TTX-R (NaV1.8; 500 nM TTX). DRG currents were 13.9 ± 2.7 nA and recorded from cells with conductances of 33.3 ± 6.6 pF (n = 7–11). Summary of current-clamp properties: action potentials (AP) and representative traces (B), rheobase (C), resting membrane potential (RMP) (D), action potential spike height (E), and action potential half width (F) (n = 7–10 per condition; *P < 0.05 vs. WT CRMP2; Kruskal–Wallis test with Dunnett’s post hoc comparisons).
CRMP2 Modifications Control Excitability of DRG Neurons.
NaV1.7 contributes to DRG excitability as evident in human gain-of-function mutations that increase action potential firing (21), a phenomenon directly correlated to and underlying transmission of painful stimuli. Consequently, we tested excitability properties of DRGs expressing SUMO-impaired or Cdk5 phospho-null CRMP2. Loss of these CRMP2 modifications reduced evoked action potential frequency by half (Fig. 3B). Rheobase, the current required to initiate an action potential, was increased in cells expressing the Cdk5 phospho-null CRMP2; however, interpretation of this result as dependent on CRMP2 SUMOylation status or NaV1.7 trafficking is made difficult by previously described relationships between CRMP2 phosphorylation by Cdk5 and modulation of N-type voltage-gated calcium (CaV2.2) channels that may also modulate rheobase (22) (Fig. 3C). The resting membrane potential, action potential spike height, and action potential half width were equivalent among all conditions (Fig. 3 D–F). These findings strengthen the hypothesis that CRMP2 modifications are linked to NaV1.7 activity.
Biochemical Mapping of CRMP2 Modifications and Binding to NaV1.7.
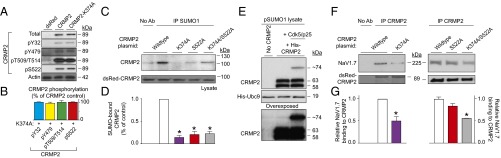
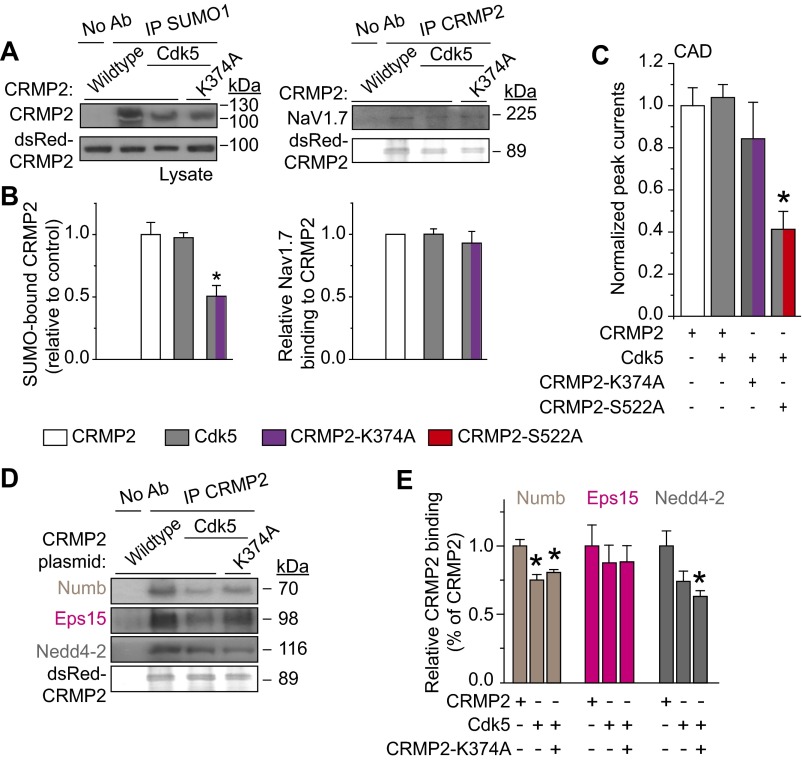
Next, we asked if CRMP2 modifications influence each other to choreograph CRMP2’s activity toward NaV1.7. Eliminating CRMP2 SUMOylation did not affect CRMP2 phosphorylation (Fig. 4 A and B). In contrast, elimination of the CRMP2 Cdk5 phosphorylation site prevented CRMP2 SUMOylation in CAD cells (Fig. 4 C and D). Additionally, Cdk5, together with its cofactor p25, facilitated in vitro SUMOylation of recombinant CRMP2 (Fig. 4E). Finally, loss of CRMP2 SUMOylation alone or together with loss of Cdk5 phosphorylation of CRMP2 reduced NaV1.7–CRMP2 binding (Fig. 4 F and G). Together, these observations place Cdk5-mediated phosphorylation of CRMP2 upstream of SUMOylation to control NaV1.7 surface expression, current density, and DRG neuron excitability.
Fig. 4.
CRMP2 SUMOylation status is affected by its phosphorylation, and both modifications contribute to NaV1.7 binding. (A) Representative Western blot of CRMP2 and CRMP2-K374A expressing CAD cells indicating CRMP2 phosphorylation levels at indicated kinase target sites (n = 3). No signal was detected with a CRMP2 antibody against pThreonine-555 (n = 3). (B) Quantitative analysis of CRMP2-K374A phosphorylation compared with total CRMP2. Representative immunoblot (C) and summary (D) to detect SUMOylated CRMP2 from CAD cells transfected with indicated plasmids (n = 3). (E) Representative immunoblot of in vitro SUMOylation of CRMP2. A SUMOylated CRMP2 band (∼74 kDa) was detected only when Cdk5 and its cofactor p25 were added to the reaction. The E2 SUMO-conjugating enzyme Ubc9, the key component of protein SUMOylation, is unchanged by the kinase. The overexposed blot shows no SUMOylated CRMP2 when the Cdk5/p25 complex is not present. Representative immunoblot (F) and summary (G) of immunoprecipitation (IP) with CRMP2 to detect NaV1.7 from CAD cells transfected with indicated plasmids (n = 6). Asterisks indicate statistically significant differences compared with WT CRMP2-expressing control cells (P < 0.05, one-way ANOVA with Tukey’s post hoc test; n = 3).
Interfering with SUMOylation and Cdk5-Mediated Phosphorylation of CRMP2 Promotes Clathrin-Mediated Endocytosis of NaV1.7.
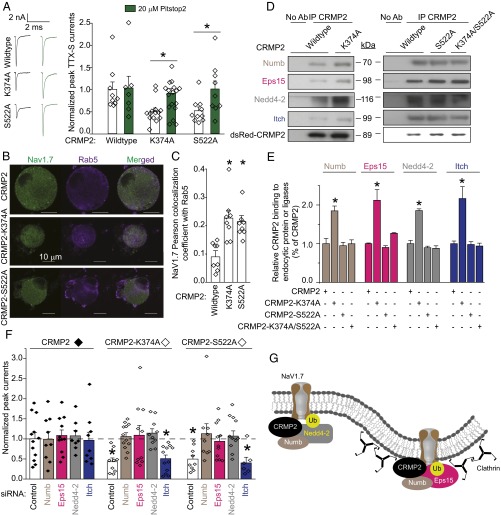
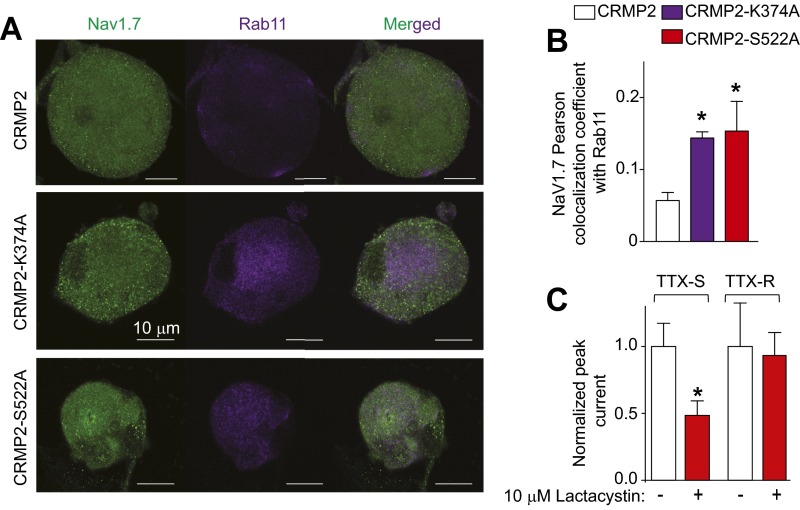
CRMP2 SUMOylation controls surface, but not total, NaV1.7 expression (Fig. 2 B–D), suggesting that CRMP2 can relocalize the channel away from the membrane to produce reduced current densities. Because CRMP2 interacts with the endocytic adaptor Numb (18) to regulate L1-cell adhesion molecule internalization via clathrin mediated endocytosis (19), we asked if loss of CRMP2 SUMOylation prevents NaV1.7 membrane localization by inducing its endocytosis. Pitstop2, a clathrin assembly inhibitor (23), prevented current density reductions imposed by loss of CRMP2 SUMOylation or Cdk5 phosphorylation (Fig. 5A), arguing for an involvement of clathrin-mediated endocytosis in NaV1.7 regulation by CRMP2. The internalized NaV1.7 relocalized to early (Fig. 5 B and C) and recycling (Fig. S2 A and B) endosomal compartments when these CRMP2 modifications were lost. The proteasome inhibitor Lactacystin did not rescue the reduction in NaV1.7 current imposed by loss of CRMP2 modifications (Fig. S2C), thus ruling out a degradation of the channel. These results identify a trafficking route followed by NaV1.7 under control of CRMP2 modifications and demonstrate that CRMP2’s activity toward NaV1.7 involves clathrin-mediated endocytosis.
Fig. 5.
CRMP2 modifications control its interactions with endocytosis-related proteins, NaV1.7 trafficking, and endocytosis via clathrin-mediated endocytosis. (A) Normalized peak TTX-S sodium current density (picoamps/picofarad; pA/pF) from DRG neurons transfected with CRMP2, CRMP2-K374A, or CRMP2-S522A before and 30 min after application of 20 µM of Pitstop2, a clathrin-mediated endocytosis inhibitor. Representative traces are displayed adjacent to the bar graphs (n = 7–17). (B) Representative confocal images of DRG neurons costained for NaV1.7 and the endosome marker Rab5. (Scale bar, 10 μm.) (C) Summary of Pearson correlation coefficient of NaV1.7 and Rab5 costaining (n = 8). Representative immunoblots (D) and summary (E) of CRMP2 IP from CAD cells transfected with the indicated plasmids and probed with antibodies for endocytosis proteins Numb and Eps15 and E3 ubiquitin ligases Nedd4-2 and Itch (n = 3). (F) Normalized peak TTX-S sodium current density (pA/pF) from DRG neurons transfected with indicated plasmids and siRNAs to knock down Numb, Eps15, Nedd4-2, or Itch. DRG currents were 11.3 ± 0.8 nA and recorded from cells 36.0 ± 2.6 pF (n = 6–12; *P < 0.05 vs. CRMP2 control, Kruskal–Wallis test with Dunnett’s post hoc comparisons). (G) Schematic of proposed mechanism of CRMP2-mediated endocytosis of NaV1.7. Non-SUMOylated CRMP2 binds to the endocytic protein Numb to recruit the E3 ligase Nedd4-2, which monoubiquitinates NaV1.7 (Left). Recruitment of Eps15 to this complex elicits clathrin-mediated endocytosis of NaV1.7 (Right).
Fig. S2.
CRMP2 modification relocalizes NaV1.7 to recycling endosomes. (A) Representative confocal images of NaV1.7 colocalization with Rab11 in CRMP2, CRMP2-K374A, and CRMP2-S522A expressing DRG neurons (n = 8). (B) Quantitative analysis of NaV1.7 colocalization with Rab11, a specific marker or recycling endosomes. (C) Quantitative analysis of TTX-S and TTX-R DRG sodium currents in cells expressing CRMP2-K374A/S522A treated with 10 µM Lactacystin for 24 h. Lactacystin did not alter any current properties of CRMP2-K374A/S522A–expressing cells. Data show means ± SEM (*P < 0.05, Kruskal–Wallis test with Dunnett’s post hoc comparisons in B and t test in C).
CRMP2 Recruits an Endocytic Complex to Down-Regulate NaV1.7.
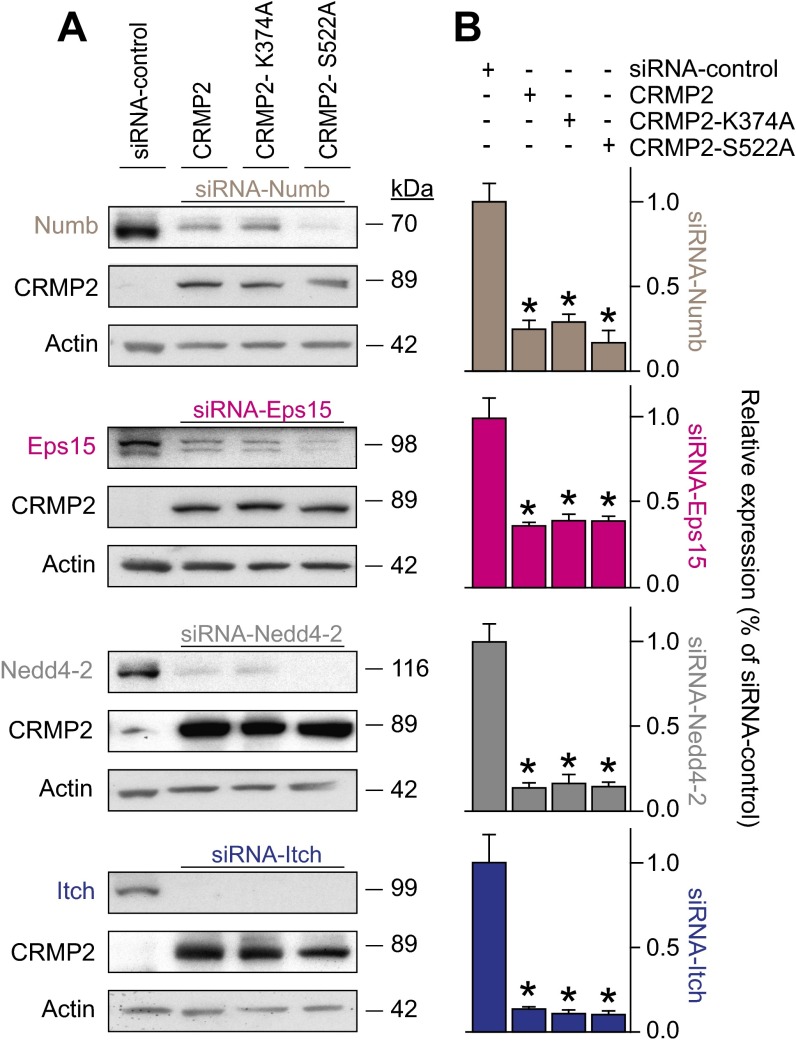
Having demonstrated that CRMP2 regulates endocytosis of NaV1.7, we next investigated the molecular mechanism involved. CRMP2 reportedly controls trafficking of membrane proteins and vesicles (8, 15, 18). As NaV1.7 endocytosis requires monoubiquitination by the ubiquitin ligase Nedd4-2 (24) and Numb—a CRMP2 protein partner acts as a scaffold between Nedd4 (25) and the endocytic machinery (19)—we hypothesized that recruitment of these proteins by CRMP2 may promote NaV1.7 internalization. Additionally, Numb can recruit Eps15 (19, 26, 27). Together, these proteins compose the machinery required to mark a protein for endocytosis (28) and initiate clathrin-mediated endocytosis by enabling interactions between Eps15 and a monoubiquitinated cargo. To test this hypothesis, we examined CRMP2 interactions with endocytic proteins Numb and Eps15 and Numb-interacting E3 ubiquitin ligases Nedd4-2 and Itch (29). Loss of CRMP2 SUMOylation, but not Cdk5-mediated phosphorylation or simultaneous loss of both modifications, increased CRMP2’s association with each of these proteins (Fig. 5 D and E). If these CRMP2-interacting proteins participate in NaV1.7 internalization, eliminating their expression should prevent CRMP2-mediated reductions of current density. Indeed, following specific reduction of these CRMP2-interacting proteins (Fig. S3 A and B), the inhibition in NaV1.7 currents forced by loss of CRMP2 modifications was rescued (Fig. 5F). Taken together, our data support the conclusion that CRMP2 association with proteins Numb, Eps15, and Nedd4-2 triggers endocytosis of NaV1.7 (Fig. 5G).
Fig. S3.
Numb, Eps15, Nedd4-2, and Itch expression in CAD cells can be reduced by siRNA under coexpression with various CRMP2 plasmids. (A) Representative Western blots of CAD cell endocytosis protein expression in CAD cells expressing siRNA control, CRMP2, CRMP2-K374A, or CRMP2-S522A (n = 3). (B) Quantitative analysis of Numb, Eps15, Nedd4-2, and Itch expression following knockdown in CRMP2 plasmid expressing CAD cells. Data show means ± SEM (*P < 0.05, Kruskal–Wallis test with Dunnett’s post hoc comparisons).
Hierarchical Interactions Between CRMP2 Modifications Modulate NaV1.7 Endocytosis.
To further investigate the underlying mechanism by which CRMP2 Cdk5 phosphorylation and SUMOylation control NaV1.7 endocytosis and current density, Cdk5 phosphorylation levels were enhanced with a Cdk5 site phospho-mimetic CRMP2 (i.e., pseudophosphorylated) mutant or overexpression of Cdk5 itself. CRMP2 SUMOylation was unaffected by mimicking constitutive phosphorylation by Cdk5 (Figs. 6 A and B and S4 A and B). Whereas CRMP2 loss of SUMOylation decreased NaV1.7–CRMP2 interaction (Fig. 4 F and G), mimicking constitutive phosphorylation by Cdk5 prevented this effect (Figs. 6 A and B and S4 A and B). Supporting these results, mimicking constitutive Cdk5 phosphorylation in CAD cells and in DRG neurons prevented loss of NaV1.7 currents imposed by non-SUMOylated CRMP2 (Figs. 6 C and D and S4C). Finally, inhibition of NaV1.7 currents by a Cdk5 phosphorylation-deficient CRMP2 mutant (CRMP2-S522A; Figs. 2E and 3A) was refractory to rescue by simultaneous Cdk5 overexpression (Fig. S4C). These outcomes demonstrate dominance of CRMP2 Cdk5 phosphorylation over SUMOylation in control of NaV1.7 currents.
Fig. 6.
Constitutive Cdk5 phosphorylation of CRMP2 overrides CRMP2 deSUMOylation-mediated loss of NaV1.7 current density via changes in interactions with endocytic machinery. Representative immunoblots (A) and summary (B) of IP with SUMO1 or CRMP2 antibodies from CAD cells transfected with the indicated plasmids and probed with antibodies against CRMP2 (Left) or NaV1.7 (Right; n = 3). (C) Representative traces and normalized peak sodium current density (picoamps/picofarad; pA/pF) from CAD cells (n = 5–8) or TTX-S current density from DRGs (n = 10–11) (D) transfected with the indicated plasmids. DRG currents were 9.9 ± 1.8 nA in amplitude and recorded from cells with average conductances of 30.0 ± 5.3 pF. For comparison, the red dashed line, representing data from Fig. 2E, is illustrated in C. Representative immunoblots (E) and summary (F) of IP with CRMP2 antibody from CAD cells transfected with the indicated plasmids and probed with antibodies against Numb, Eps15, Nedd4-2, and CRMP2 (n = 3; *P < 0.05 vs. CRMP2 WT, Kruskal–Wallis test with Dunnett’s post hoc comparisons). (G) Model proposing that elimination of Cdk5-mediated CRMP2 phosphorylation promotes CRMP2’s interaction with Numb.
Fig. S4.
Cdk5 expression in CAD cells prevents CRMP2-mediated effects on NaV1.7. (A) Representative Western blots depicting SUMO1 IP to identify CRMP2 SUMOylation and CRMP2 IP to identify interaction with NaV1.7. Experiments were performed on CAD cells expressing Cdk5 and CRMP2 or CRMP2-K374A (n = 3). (B) Quantitative analysis of CRMP2 SUMOylation and NaV1.7 interaction. (C) Quantitative analysis of CAD cell total sodium current density following expression of the indicated CRMP2 and Cdk5 plasmids. (D) Representative Western blot of CRMP2 immunoprecipitation to detect interaction with Numb, Eps15, and Nedd4-2 in cells coexpressing Cdk5 and CRMP2 or CRMP2-K374A (n = 3). (E) Quantitative analysis of CRMP2 interaction with indicated endocytosis-related proteins. Data show means ± SEM (*P < 0.05, Kruskal–Wallis test with Dunnett’s post hoc comparisons).
Cdk5 phosphorylation of CRMP2 could regulate NaV1.7 trafficking by preventing assembly of a CRMP2-endocytic complex. To test this hypothesis, interactions between CRMP2 and Numb, Eps15, or Nedd4-2 were examined. Mimicking constitutive phosphorylation of CRMP2 by Cdk5 (Fig. 6 E and F) and overexpression of Cdk5 inhibited the CRMP2–Numb interaction (Fig. S4 D and E). Furthermore, enhanced interactions between non-SUMOylated CRMP2 and Numb, Eps15, or Nedd4-2 (Fig. 5 D and E) were prevented by constitutive phosphorylation of CRMP2 by Cdk5 (Fig. 6 E and F and Fig. S4 D and E). These findings identify that a non-SUMOylated, nonphosphorylated (by Cdk5) CRMP2 recruits Numb to trigger NaV1.7 endocytosis (Fig. 6G) and illustrate that CRMP2’s activity on NaV1.7 is negatively regulated by Cdk5 phosphorylation of CRMP2.
Antagonism Between CRMP2 Fyn Phosphorylation and SUMOylation Contributes to NaV1.7 Endocytosis.
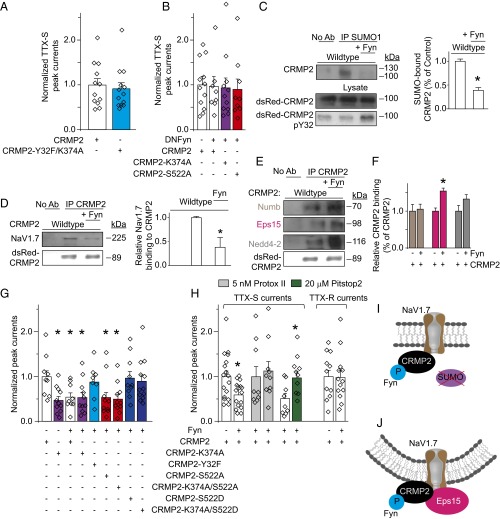
Simultaneous loss of Fyn phosphorylation and SUMOylation of CRMP2 overcame the NaV1.7 current reduction imposed by loss of SUMOylation alone (Figs. 2E and 7A). Although loss of Fyn phosphorylation did not change CRMP2’s SUMOylation state (Fig. S5A), it prevented the loss of binding to NaV1.7 imposed by loss of SUMOylation (Fig. S5B). Concomitant loss of Fyn phosphorylation and SUMOylation of CRMP2 suppressed the internalization of NaV1.7 imposed by loss of SUMOylation alone (Fig. 2 B and C). Loss of Fyn phosphorylation prevented the increased binding to Numb, Eps15, and Nedd4-2 imposed by loss of CRMP2 SUMOylation (Fig. S5 C and D). Thus, loss of Fyn phosphorylation is dominant over CRMP2 SUMOylation in the control of NaV1.7 internalization.
Fig. 7.
Gain of CRMP2 phosphorylation by Fyn overrides the reduction in NaV1.7 current density imposed by loss of CRMP2 SUMOylation or CRMP2 phosphorylation by Cdk5. Normalized peak TTX-S current density (picoamps/picofarad; pA/pF) (A and B) from DRGs transfected with WT or the double Fyn-null/SUMO-impaired (CRMP2-Y32F/K374A) CRMP2 (n = 12–13) (A) or the inactive dominant-negative Fyn kinase (n = 8–12) in addition to the indicated CRMP2 plasmids (B). (C) Representative immunoblots (Left) and summary (Right) of denaturing IP with SUMO1 antibody from CAD cells transfected with the indicated plasmids and probed with CRMP2 antibody. Lysates were also probed for expression of dsRed-CRMP2 (Middle) and pY32 CRMP2 (Bottom; n = 3). (D) Representative immunoblots (Left) and summary (Right) of IP with CRMP2 antibody from CAD cells transfected with the indicated plasmids and probed with a NaV1.7 antibody (n = 3). Representative immunoblots (E) and summary (F) of IP with CRMP2 antibody from CAD cells transfected with the indicated plasmids and probed with antibodies for Numb, Eps15, Nedd4-2, and CRMP2 (n = 3). Normalized peak sodium current density (pA/pF) from transfected CAD cells (n = 8–12) (G) or TTX-S, non–NaV1.7-TTX-S-fraction, TTX-S fraction following Pitstop2 application (20 µM, 30 min), or TTX-R sodium current density from DRGs (n = 9–17) (H). DRG currents were 10.3 ± 1.5 nA in amplitude and recorded from cells with average conductances of 34.6 ± 5.6 pF (*P < 0.05 vs. CRMP2 WT, Kruskal–Wallis test with Dunnett’s post hoc comparisons). (I) Model proposing that Fyn-mediated CRMP2 phosphorylation impairs CRMP2 SUMOylation, but enhances its interaction with Eps15 (J).
Fig. S5.
CRMP2 loss of Fyn phosphorylation prevents CRMP2-mediated reduction of sodium current. Representative Western blots of SUMO1 IP to identify CRMP2 SUMOylation (A) or CRMP2 IP to detect binding to NaV1.7 (B) for the indicated constructs expressed in CAD cells (n = 3). Bar graphs below the blots denote a summary of the mean data. (C) Representative Western blots of CRMP2 immunoprecipitation to detect interaction with Numb, Eps15, and Nedd4-2 for the indicated conditions (n = 3). (D) Quantitative analysis of CRMP2 interaction with the indicated endocytosis-related proteins. Quantitative analysis of sodium current densities following loss of CRMP2 Fyn phosphorylation by phospho-null constructs (E) or expression of a dominant-negative Fyn (DNFyn) (F). Peak sodium currents from CAD cells expressing CRMP2-Y32F/K374A (dashed blue line), is replotted from Fig. 2D for comparison. Data show means ± SEM (*P < 0.05, Kruskal–Wallis test with Dunnett’s post hoc comparisons).
To further resolve the hierarchy of CRMP2 posttranslational modifications in control of NaV1.7, we asked if loss of Fyn phosphorylation could be dominant over loss of Cdk5 phosphorylation of CRMP2. NaV1.7 current reductions induced by (i) SUMO-impaired CRMP2 mutant; (ii) SUMO proteases SENP1/2, which remove SUMO from conjugated proteins; or (iii) loss of Cdk5-mediated phosphorylation were prevented when Fyn phosphorylation was also eliminated (Fig. S5E). Additionally, a dominant-negative Fyn kinase (DNFyn-K299M) (30) normalized the current density reductions observed with loss of CRMP2 SUMOylation or phosphorylation (Cdk5 site; Figs. 7B and S5F). Therefore, loss of Fyn phosphorylation of CRMP2 dominates all CRMP2-dependent posttranslational modifications that reduce NaV1.7 currents.
To determine the mechanism by which Fyn phosphorylation of CRMP2 regulates NaV1.7, we next investigated CRMP2 SUMOylation and CRMP2’s interactions with NaV1.7 and endocytosis proteins. Overexpression of a competent Fyn kinase increased CRMP2 phosphorylation (at Y32), decreased CRMP2 SUMOylation (Fig. 7C), decreased CRMP2’s interaction with NaV1.7 (Fig. 7D), and increased CRMP2’s interaction with the endocytic scaffolding protein Eps15 (Fig. 7 E and F). Because loss of CRMP2 SUMOylation translates into a reduction in NaV1.7 current density (Fig. 2E), likely as a result of a decrease in binding to NaV1.7 (Fig. 4 F and G), we hypothesized that Fyn-phosphorylated CRMP2 would lead to a suppression of NaV1.7 currents in CAD cells and in DRG neurons. As expected, forcing Fyn phosphorylation of CRMP2 led to a specific reduction in NaV1.7 currents compared with cells expressing WT CRMP2 (Fig. 7 G and H). Forcing Fyn phosphorylation of a SUMO-incompetent CRMP2 did not result in any additional reduction in NaV1.7 currents (Fig. 7G). Conversely, coexpression of competent Fyn kinase and a Fyn phosphorylation-incompetent CRMP2 rescued baseline NaV1.7 current densities (Fig. 7G), directly implicating Fyn phosphorylation of CRMP2 in control of NaV1.7 activity.
Next, we manipulated Cdk5 phosphorylation of CRMP2 to further dissect the interplay between CRMP2 posttranslational modifications in control of NaV1.7. Loss of Cdk5 phosphorylation of CRMP2 together with forcing Fyn phosphorylation did not result in a further decrease of NaV1.7 currents, even when the SUMOylation site was deleted (Fig. 7G). On the contrary, gain of Cdk5 phosphorylation of CRMP2 was dominant over the gain of Fyn phosphorylation of CRMP2 (Fig. 7G), and this effect occurred regardless of CRMP2’s SUMOylation status. Loss of NaV1.7 currents imposed by forcing Fyn phosphorylation of WT CRMP2 in DRG neurons was normalized by inhibition of clathrin mediated endocytosis with Pitstop2 (Fig. 7H). Under no conditions were the biophysical properties of NaV1.7 channels changed (Table S1). Thus, CRMP2-dependent internalization of NaV1.7 requires the simultaneous gain of Fyn phosphorylation coupled with loss of Cdk5 phosphorylation of CRMP2, which result in reduced SUMOylation of CRMP2 (Fig. 7I) and NaV1.7 endocytosis (Fig. 7J). These three posttranslational modifications coordinate CRMP2’s activity toward NaV1.7.
CRMP2 Modulation of NaV1.7 Currents Is Maintained in Human DRGs.
The relationship between CRMP2 posttranslational modifications and NaV1.7 currents in human DRGs has not been defined. Validating the relationships between CRMP2 posttranslational modifications and NaV1.7 trafficking in humans is critical to understanding the mechanistic underpinnings of channel dysregulation in human disease and the eventual translation of cell signaling studies to preclinical models. Therefore, human DRGs were obtained and transfected with SUMO-impaired or phospho-null CRMP2 mutants (Fig. 8A), and sodium currents were recorded. Appropriately compensated recordings did not reveal any differences in biophysical channel properties (Fig. S6 A–C), and use-dependence was unaffected (Fig. S6 D and E). Importantly, both the total (Fig. 8 B and C) and the TTX-S fraction (Fig. 8 D and E) of sodium currents were decreased by loss of these CRMP2 modifications in human DRGs. The congruence between CRMP2 modulation of sodium currents in rat and human DRGs validates the CRMP2-NaV1.7 signaling model (Fig. 9 A and B) and provides a strong mechanistic understanding from which future studies can target CRMP2-NaV1.7 interactions in preclinical animal models of human pain syndromes.
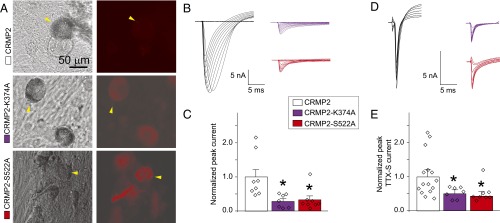
Fig. 8.
CRMP2 modifications control NaV1.7 currents in human DRGs. (A) Representative differential interference contrast (Left) and fluorescence (dsRed; Right) images showing transfection of human DRGs (yellow arrowheads). Representative families of total sodium current traces (B) or TTX-S sodium current traces (D) from human DRGs expressing WT, SUMO-impaired (CRMP2-K374A), or phospho-null (CRMP2-S522A) CRMP2. Normalized peak total sodium current density (n = 7–8) (C) or TTX-S sodium current density [picoamps/picofarad (pA/pF); n = 7–15] (E) from human DRGs expressing the indicated plasmids. Human DRG currents were 23.1 ± 4.0 nA and recorded from cells with average conductances of 84.3 ± 16.5 pF (*P < 0.05 vs. CRMP2 WT, Kruskal–Wallis test with Dunnett’s post hoc comparisons).
Fig. S6.
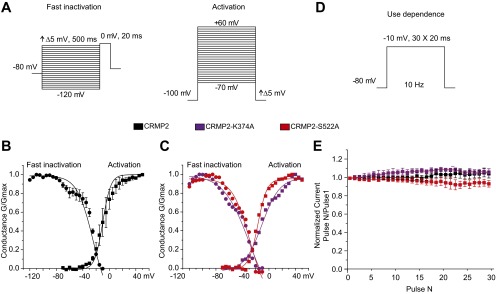
Properties of human DRG sodium currents following expression of CRMP2 plasmids. Voltage protocols used to elicit fast inactivation and activation (A) and use dependence (D). Graphs of human DRG sodium channel fast inactivation and activation for CRMP2-expressing cells (n = 3) or (B) CRMP2-K374A– or CRMP2-S522A–expressing cells (n = 1). (C) Complex arborization of mature hDRG and glial mixed cultures made series resistance compensation difficult and prevented collection of channel kinetic data from most recordings. (E) Quantitative analysis of human DRG use dependence in cells expressing indicated CRMP2 plasmids (n = 4). Data show means ± SEM when more than one cell was recorded.
Fig. 9.
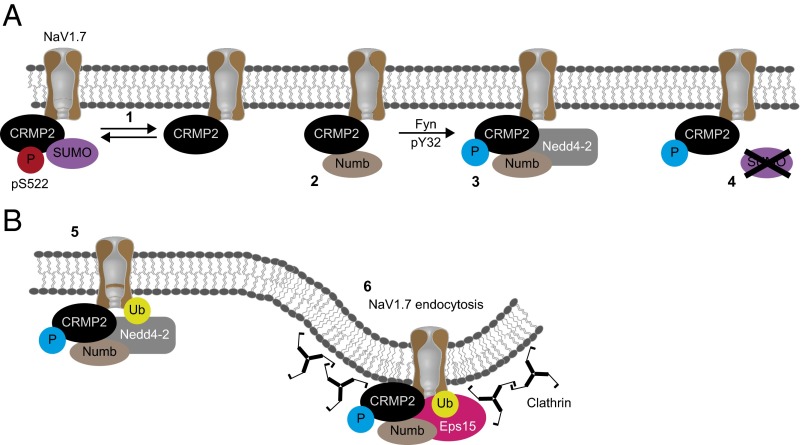
Schematic of CRMP2 modifications and their contribution to the regulation of NaV1.7 trafficking. (A) CRMP2 SUMOylation (1) is prevented when CRMP2 phosphorylation by Cdk5 at S522 is also prevented. Consequently, nonSUMOylated CRMP2 has an enhanced interaction with Numb (2). The enhanced interaction with Numb and subsequent phosphorylation of CRMP2 by Fyn kinase (3) can facilitate further recruitment of the E3 ubiquitin ligase Nedd4-2. Furthermore, when phosphorylated by Fyn, CRMP2 SUMOylation is prevented (4). (B) Within the CRMP2/NaV1.7/endocytic machinery complex, Nedd4-2 monoubiquitinates NaV1.7 (5), which, in turn, permits Eps15/clathrin-mediated endocytosis of the channel (6).
Discussion
The present findings make a compelling case for targeting CRMP2 posttranslational modifications, loss of which selectively attenuates NaV1.7 currents and dampens nociceptor excitability, as a pathway for development of therapeutic agents for the treatment of neuropathic pain. Although CRMP2 posttranslational modifications serve to direct protein activity to a variety of functional cellular processes, CRMP2 SUMOylation does not feed back onto CRMP2 phosphorylation as evident by unchanged CRMP2 phosphorylation between WT and K374A mutant CRMP2 (Fig. 4 A and B). Therefore, although posttranslational modifications codify a molecular language that instruct CRMP2 to positively or negatively influence NaV1.7 surface expression, a direct intervention of CRMP2 SUMOylation has potential to attenuate pain states without modifying CRMP2 phosphorylation or other functions of CRMP2. It may also be possible that the CRMP2-NaV1.7 trafficking pathway dissected in our work (Fig. 9), which is congruent between rodent and human DRGs, is dysfunctional within human disease pain states and contributes to nociceptor excitability. Future studies will aim to dissect if subtle tuning of the CRMP2 modifications that alter NaV1.7 currents can result in significant reduction of pain in animal models.
From a mechanistic perspective, the NaV1.7 trafficking pathway identified here greatly extends the findings of Laedermann et al., who reported that the Nedd4-2 ubiquitin ligase monoubiquitinates NaV1.7 to signal the channel’s endocytosis (6). Our results suggest that induction of membrane curvature by Eps15 (26, 27) to trigger clathrin-mediated endocytosis is required for CRMP2-mediated endocytosis of NaV1.7 via the formation of a CRMP2/Numb/Eps15/Nedd4-2 complex. If CRMP2 is not SUMOylated, these interactions are strengthened, culminating in accumulation of the channel into early and recycling endosomes. This study also extends our own previous work that demonstrated a link between NaV1.7 function and CRMP2 SUMOylation (8) to now illustrate that the trafficking program of NaV1.7 is dependent on multiple modifications of CRMP2 and details the mechanism of endocytosis of NaV1.7.
Akin to the relationship described here between CRMP2 and NaV1.7, several studies have been successful in identifying links between protein SUMOylation and trafficking of an accessory protein. For instance, the immediate early gene Activity-regulated cytoskeleton-associated protein/Activity-regulated gene 3.1 (Arc), which couples changes in neuronal activity to synaptic plasticity events (31), is SUMOylated (32). Arc SUMOylation impacts the trafficking of AMPARs, which are heterotetrameric glutamate-gated ion channels that underpin the vast majority of fast excitatory glutamate neurotransmission in the central nervous system (33). Preventing Arc SUMOylation may control AMPARs. Rab3-Interacting Molecule 1α (RIM1α) is a presynaptic protein implicated in the docking/priming of synaptic vesicles and in the development of short- and long-term synaptic plasticity (34, 35). RIM1α SUMOylation is required for presynaptic targeting and clustering of Cav2.1 calcium channels (36). Our study adds to the growing evidence that protein SUMOylation is critical for trafficking processes in neurons.
A salient finding of our work is the selectivity in CRMP2 modulation of NaV1.7, but not other channels. How CRMP2 regulates NaV1.7 specifically is a critical question. One possibility may be differential recruitment of the endocytic machinery or CRMP2 itself. Nedd4 proteins regulate trafficking of multiple sodium channels (6, 37) whereas Numb and Eps15 proteins are general players in clathrin-mediated endocytosis (18, 19, 27), so specificity is unlikely endowed by these proteins. Likely, the specificity is conferred by CRMP2, as its expression and modifications causally influence NaV1.7. CRMP2 may have differential binding affinities for NaV1.x channels with the highest affinity for NaV1.7. Although this binding has not been mapped for each of the channels, a hypothesis to explain selective regulation of NaV1.7 may be differential binding of CRMP2 to intracellular loops of sodium channels, especially those that are not conserved between NaV1.x isoforms (e.g., the N terminus and loop 2 connecting the second and third transmembrane domain modules). Because CRMPs are heterotetrameric, we cannot rule out contributions to channel specificity by other CRMP isoforms. For instance, CRMP4 expression increases following sciatic nerve axotomy (38), and a CRMP2–CRMP4 tetramer may permit binding to NaV1.7, but not other NaV1.x channels. CRMP4-KO mice exhibit impaired olfactory ability (39), and loss-of-function mutations in NaV1.7 cause anosmia (40), thus providing further evidence of a possible link between CRMP4 and NaV1.7. However, another possibility is that there may be a still unknown protein that is required for the CRMP2-NaV1.7 specificity.
Many proteins bear multiple, distinct modifications, and the ability of one modification to antagonize or synergize the addition of another can have significant biological consequences. Of the multiple kinases that target CRMP2, it is interesting that phosphorylation by Cdk5 and Fyn confers regulatory checkpoints upon CRMP2 SUMOylation that steer CRMP2/NaV1.7 signaling in opposing directions (Fig. 9). Notably, both kinases have been linked to pain. Increased Cdk5 activity has been reported in inflammatory (41) and neuropathic (42) pain models, and roscovitine, a Cdk5-inhibiting compound, blunted neuropathic pain after chronic compression injury of DRGs (42). Constitutive action of Fyn produced mechanical allodynia and thermal hyperalgesia via enhanced expression of ionotropic glutamate receptors (43). Despite the breadth of pain research and continuous identification of novel targets of protein SUMOylation, no previous reports have linked this modification to pain as far as we are aware. A weak link can be surmised from data demonstrating an enhanced susceptibility to symptomatic knee osteoarthritis and multiple regional pain in patients who show expression of the NaV1.7 R1150W mutation (44) and an increase in global SUMOylation rate in rheumatoid arthritis (45).
NaV1.7 is a high-value target for development of new pain therapeutic agents. Tarantula peptide toxins are selective for NaV1.7 in vitro but fail to provide analgesia in pain models (46), likely because of a lack of penetration past the perineural barrier (47). A NaV1.7-selective centipede-venom peptide elicited antinociception in acute pain models but has not yet been tested in more complex rodent pain models (48). A monoclonal antibody against the voltage sensor of NaV channels exhibited selectivity for NaV1.7, with partial effects on NaV1.6 (49), and several small-molecule blockers of human NaV1.7 have been described (50). Thus, strategies targeting NaV1.7 channels are ongoing and show promise in pain therapy. Disrupting CRMP2 SUMOylation is an approach to controlling NaV1.7 channels that overcomes the selectivity hurdle of targeting NaV1.7 directly. When posttranslationally modified, CRMP2 reduces NaV1.7 currents and excitability. Thus, modification of CRMP2 by SUMOylation is a mechanism to selectively regulate NaV1.7, and interacting with this pathway to curb NaV1.7 activity could normalize nociceptor excitability. From a translational perspective, the concordance in NaV1.7 regulation by CRMP2 in rodent and human sensory neurons will likely mean better success when this strategy is pursued in future clinical trials.
Materials and Methods
Detailed descriptions of methods used and any associated references are available in SI Materials and Methods. Briefly, all electrophysiology and biochemistry experiments were performed according to established protocols (8, 15). All animal protocols were approved by the institutional animal care and use committee of the College of Medicine at the University of Arizona and conducted in accordance with the Guide for Care and Use of Laboratory Animals published by the National Institutes of Health.
SI Materials and Methods
Animals.
Adult male Sprague–Dawley rats (Harlan) were kept in a temperature-controlled environment with lights on from 0700 h to 1900 h with food and water available ad libitum. All animal procedures were performed in accordance with the policies and recommendations of the International Association for the Study of Pain and the National Institutes of Health, and with approval from the animal care and use committee of the University of Arizona for the handling and use of laboratory animals.
Plasmids, Antibodies, and Other Materials.
Biochemical analysis of protein content by Western blot and immunocytochemical analysis used the following antibodies: NaV1.7 (cat. no. 75–103, RRID:AB_2184355; University of California, Davis/National Institutes of Health NeuroMab Facility), pan-NaV (cat. no. ASC-003 RRID:AB_2040204; Alomone Laboratories), βIII-Tubulin (cat. no. G7121 RRID:AB_430874; Promega), CRMP2 (cat. no. C2993 RRID:AB_1078573; Sigma-Aldrich), CRMP2 pTyr32 (provided by Yoshio Goshima, Yokohama City University Graduate School of Medicine, Yokohama, Japan), CRMP2 pTyr479 (provided by Pascale Giraudon, Lyon Neuroscience Research Center, Lyon, France), CRMP2 pThr509/Thr514 (cat. no. PB-043, RRID:AB_2620175; MRC), CRMP2 pSer522 (cat. no. CP2191 RRID:AB_2094486; ECM Biosciences), and CRMP2 pTyr555 (cat. no. CP2251 RRID:AB_2094483; ECM Biosciences), SUMO1 (cat. no. S8070 RRID:AB_477543; Sigma-Aldrich), Numb (cat. no. ab4147 RRID:AB_304320; Abcam), Eps15 (cat. no. ab174291, RRID:AB_2620176; Abcam), and Nedd4-2 (cat. no. ab131167 RRID:AB_11157800; Abcam), Itch (cat. no. 611198 RRID:AB_398732; BD Biosciences), and Rab5 (cat. no. PA3-915 RRID:AB_2177668; Thermo Fisher Scientific), Rab11 (cat. no. 71–5300 RRID:AB_2533987; Thermo Fisher Scientific), and Actin (cat. no. A2228 RRID:AB_476697; Sigma-Aldrich).
Manipulations of protein expression in our experiments used the plasmids pCMV-Cdk5 (cat. no. 1872), pRK5-c-Fyn (cat. no. 16032), pRK5-DN-Fyn (cat. no. 16033) plasmids from Addgene. Detection of multiple bands when probing Fyn phosphorylation of CRMP2 is typical despite concrete evidence for a single modification of tyrosine 32 and no other CRMP2 residues (13). Mutations to pdsRed2-N1-CRMP2 (22) used in this study were introduced by QuikChange II XL mutagenesis kit (cat. no. 200521; Agilent) in the pdsRed2-CRMP2 plasmid. Plasmids were purified from DH5α Escherichia coli using the NucleoBond Xtra Maxi kit (cat. no. 740414; Macherey-Nagel). Introduced mutations were verified by DNA sequencing.
For RNA interference, siRNA CRMP2 (5′-GTAAACTCCTTCCTCGTGT-3′), siRNA Numb (5′-TAACTGGGAAGCTACACTTTCCAGT-3′), siRNA Eps15 (5′-CCCAGGCAATGATAGTCCCAAAGAA-3′), siRNA Nedd4-2 (5′-CATACTATGTCAATCATAATT-3′), siRNA Itch (5′-TCACAGTGGATGGAATTCCATATTT-3′), and siRNA control (cat. no. 12935300) were obtained from Thermo Fisher Scientific.
Culturing and Transfection of CAD and HEK293 Cell Lines.
Mouse neuron derived CAD (ECACC cat. no. 08100805, RRID:CVCL_0199) and human-derived HEK293 cells were grown in standard cell culture conditions, 37 °C in 5% (vol/vol) CO2, as previously described (20, 51, 52). All media were supplemented with 10% (vol/vol) FBS (HyClone) and 1% penicillin/streptomycin sulfate from 10,000 µg/mL stock. CAD cells were maintained in DMEM/F12 media, and HEK293 cells were maintained in DMEM media. HEK293 cell lines expressing various NaV1.X isoforms were obtained from Theodore R. Cummins (Indiana University School of Medicine, Indianapolis, IN). HEK293 cells stably expressing NaV1.X subtypes were generated by calcium phosphate precipitation transfection of hNaV1.1 in pTarget vector, rNaV1.3 or hNaV1.7 in pcDNA3.1-mod vector, or hNaV1.5 in pRcCMVII vector. Geneticin (cat. no. 10131035; Thermo Fisher Scientific) was used at 500 µg/mL to select for NaV1.X-expressing cells. CAD cells were chosen as a model neuron cell line as a result of ∼80% contribution by NaV1.7 to total sodium currents. This ∼80% contribution was determined by isoform-specific blockade of NaV1.7 by HWTX-IV (Alomone Laboratories) and ProTox-II (Sigma). Cells were transfected by using 1 µg/µL polyethylenimine (Sigma) (53) complexed with 2 µg/µL CRMP2 plasmid and/or 1 µg/µL of other indicated plasmids. Under these conditions, transfection efficiencies were ∼50%. To attain higher transfection efficiency required for protein quantification, several Western blots were performed on cells transfected with Lipofectamine 2000 (cat. no. 11668019; Thermo Fisher Scientific) according to the manufacturer’s instructions. In these cases, transfection efficiency was typically >95%. siRNAs were transfected by using Lipofectamine 2000 according to the manufacturer’s instructions at a concentration of 500 nM. All experiments were performed between 48 h and 72 h after transfection. Plasmid transfection was verified by dsRed fluorescence, and knockdown was verified by Western blot. A BLAST search of siRNA sequences against the rat transcriptome did not reveal potential off target effects. Knockdown-resistant plasmids were verified in Fig. S1 and also in previous studies (22).
Culturing and Transfection of Rat Primary DRG Neurons.
Rat DRG neurons were isolated from 150–174-g Sprague–Dawley rats and then transfected by using previously developed procedures (15). In brief, removing dorsal skin and muscle and cutting the vertebral bone processes parallel to the dissection stage exposed DRGs. DRGs were then collected, trimmed at their roots, and digested in 3 mL bicarbonate-free, serum-free, sterile DMEM (cat. no. 11965; Thermo Fisher Scientific) solution containing neutral protease (3.125 mg⋅mL−1; cat. no.LS02104; Worthington) and collagenase Type I (5 mg⋅mL−1; cat. no. LS004194; Worthington) and incubated for 45 min at 37 °C under gentile agitation. Dissociated DRG neurons (∼1.5 × 106) were then gently centrifuged to collect cells and washed with DRG media DMEM containing 1% penicillin/streptomycin sulfate from 10,000 µg/mL stock, 30 ng⋅mL−1 nerve growth factor, and 10% (vol/vol) FBS (HyClone). Collected cells were resuspended in Nucleofector transfection reagent containing plasmids or siRNA at the working concentrations listed here earlier. Then, cells were subjected to electroporation protocol O-003 in an Amaxa Biosystem (Lonza) and plated onto 12- or 15-mm poly-d-lysine- and laminin-coated glass coverslips. Transfection efficiencies were routinely between 20% and 30%, with approximately ∼10% cell death. Small-diameter neurons were selected to target Aδ- and c-fiber nociceptive neurons. For rat DRG culture, small cells were considered to be approximately <30 µm as determined by an eyepiece micrometer within the objective lens. Successfully transfected cells were identified by fluorescence.
Patch-Clamp Electrophysiology.
Whole-cell voltage-clamp and current-clamp recordings were performed at room temperature by using an EPC 10 Amplifier-HEKA as previously described (8). The internal solution for voltage clamp CAD cell recordings contained (in mM): 110 CsCl, 5 MgSO4, 10 EGTA, 4 ATP Na2-ATP, and 25 Hepes (pH 7.3, 290–310 mOsm/L), and external solution contained (in mM): 100 NaCl, 10 tetraethylammonium chloride, 1 CaCl2, 1 CdCl2, 1 MgCl2, 10 d-glucose, 4 4-aminopyridine, 0.1 NiCl2, and 10 Hepes (pH 7.3, 310–315 mOsm/L). For DRG and HEK293 cells, the internal solution for voltage clamp contained (in mM): 140 CsF, 1.1 Cs-EGTA, 10 NaCl, and 15 Hepes (pH 7.3, 290–310 mOsm/L), and external solution contained (in mM): 140 NaCl, 3 KCl, 30 tetraethylammonium chloride, 1 CaCl2, 0.5 CdCl2, 1 MgCl2, 10 d-glucose, and 10 Hepes (pH 7.3, 310–315 mosM/L). For DRG current-clamp experiments, the internal solution contained (in mM): 140 KCl, 10 NaCl, 1 MgCl2, 1 EGTA, 10 Hepes (pH 7.2), and 1 ATP-Mg (pH 7.3, 285–295 mOsm/L), and external solution contained (in mM): 154 NaCl, 5.6 KCl, 2 CaCl2, 2.0 MgCl2, 1.0 glucose, and 10 Hepes (pH 7.4, 305–315 mOsm/L).
In experiments in which clathrin-mediated endocytosis was prevented with 20 µM Pitstop2 (cat. no. ab120687; Abcam), the compound was incubated in the tissue culture well for 30 min before the experiment. In experiments in which proteasome degradation was prevented with 20 µM Lactacystin (54) (cat. no. l7035; Sigma), the compound was incubated within the tissue culture well for 24 h before use in an experiment with replacement of media at 12 h with fresh Lactacystin. Pitstop2 and Lactacystin were included in the bath solution during experimentation.
Electrodes were pulled from standard wall borosilicate glass capillaries (Warner Instruments) with a P-97 electrode puller (Sutter Instruments) and heat-polished to final resistances of 1.5–3 MΩ when filled with internal solutions. Whole-cell capacitance and series resistance were compensated. Linear leak currents were digitally subtracted by P/4 method for voltage clamp experiments, and bridge balance was compensated in current-clamp experiments. Signals were filtered at 10 kHz and digitized at 10–20 kHz. Cells in which series resistance or bridge balance was more than 15 MΩ or fluctuated by more than 30% over the course of an experiment were omitted from datasets. Analysis was performed by using Fitmaster software (HEKA) and Origin 9.0 software (OriginLab).
Voltage-Clamp Protocols.
CAD and HEK293 cells were subjected to current-density (I-V) and fast-inactivation voltage protocols. In the I-V protocol, cells were held at a −80-mV holding potential before depolarization by 20-ms voltage steps from −70 mV to +60 mV in 5-mV increments. This allowed for collection of current density data to analyze activation of sodium channels as a function of current vs. voltage and also peak current density, which was typically observed near ∼0–10 mV and normalized to pF. Values of current in nA and cell size in pF are available in the figure legends. In the fast-inactivation protocol, cells were held at a −80-mV holding potential before hyperpolarizing and repolarizing pulses for 500 ms between −120 mV and −10 mV in 5-mV increments. This step conditioned various percentages of channels into fast-inactivated states so that a 0-mV test pulse for 20 ms could reveal relative fast inactivation normalized to maximum current. DRGs from rat and human were subjected to I-V protocol and H-infinity (prepulse inactivation) protocol. To estimate TTX-R contributions, I-V protocol was run after 10 m incubation with 500 nM TTX. Following holding at −100 mV, 200-ms voltage steps from −70 mV to +60 mV in 5-mV increments allowed for analysis of peak currents. The TTX-R peak current density was always measured at depolarizations near 0 mV and within the first 10 ms of the voltage step protocol. Given the previously identified properties of NaV1.8 and NaV1.9 TTX-R currents, this voltage-dependence and activation profile suggests our analysis of peak current density represents only NaV1.8 current (55). Thus, we analyzed sodium current present at 150 ms following a voltage pulse to −60 mV, an established method of isolating Nav1.9 current. In cells electroporated with CRMP2 plasmids, however, we observed no Nav1.9 current with this protocol. Analysis of NaV1.9 currents in response to changes of CRMP2 modification may therefore require extensive optimization of recording solutions and transfection protocols and cannot be inferred by our TTX-R current density data.
In the H-infinity protocol, cells were held at −100 mV and subjected to conditioning voltage steps for 1 s varying from −120 mV to 0 mV in 10-mV increments. This conditioning step was followed by a 0-mV test pulse for 200 ms to analyze current. The H-infinity protocol allowed subtraction of electrically isolated TTX-R (current available after −40 mV prepulse) from total current (current available after −120 mV prepulse) to estimate TTX-S current. This protocol is possible because of differential inactivation kinetics of TTX-R vs. TTX-S channels wherein only TTX-S current becomes activated and then fast-inactivated during the 1-s −40-mV pulse. For all protocols, a test pulse was performed before and after the voltage protocol to evaluate run-down or run-up of currents during the voltage protocols and to omit data from cells with currents that were altered as a function of time. Currents densities analyzed in voltage-clamp are represented as picoamps or nanoamps per picofarad (pA/pF, nA/pF).
Current Clamp Protocols.
To assess excitability, DRGs were held at their resting membrane potential and subjected to a 1-s depolarizing current step. The intensity of this current step was adjusted until the neuron fired an action potential to determine the rheobase of the cell. The stimulation amplitude was then set to 2× rheobase to collect data on relative DRG excitability analyzed by number of evoked action potentials during the 1-s current step. Spike height was determined by the potential difference between the action potential peak and the minimum voltage during after-hyperpolarization. Action potential (AP) half width was measured as the width of the action potential trace in milliseconds at half the AP spike height.
Human Sensory Neuron Cultures and Transfection.
Human sensory neurons cultures were purchased from Anabios. DRGs from consenting donors were dissected from thoracic vertebra T2 through T12 as described by Davidson et al. (56). For the DRGs recorded from in this manuscript, two donors were used. The first donor was a 27-y-old white man with no history of chronic pain who died of a motor vehicle accident. The second donor was a 40-y-old Hispanic man who died of cardiovascular complications. Connective tissue was removed before enzymatic digestion with AnaBios enzyme mixture at 37 °C for 2 h. Cells were centrifuged for 2 min at 200 × g, and solution was replaced three times with DMEM/F12 containing 1% horse serum to wash cells. Then, cells were mechanically disassociated by gentle trituration through fire-polished sterile Pasteur pipettes. The cells were plated onto poly-d-lysine–coated 12-mm glass coverslips and maintained at 37 °C with 5% (vol/vol) CO2 in DMEM/F12 (cat. no. 10565; Thermo Fisher Scientific) supplemented with 10% (vol/vol) horse serum (cat. no. SH30074.03; Thermo Fisher Scientific), 25 ng/mL human NGF (cat. no. 256-GF-100/CF, R&D Systems), 25 ng/mL human GDNF (cat. no. G1491, Promega), and penicillin/streptomycin. Half of the culture media was replaced with fresh media every 3 d. The cultures were mixed DRG/glial cells to facilitate the survival of the neurons. The size of the neurons was 30–120 µm, with an average size of 40–60 µm. At 10 d after the plating, cells were shipped in an isolated container to prevent any variation in temperature. The shipping time was usually 4 h, and no cell death was observed after shipping of DRG cultures. Lipofuscin pigment granules were evident in almost all neurons, but this did not preclude them for being amenable for patching.
For transfection of human DRGs, Lipofectamine 3000 (cat. no. L3000015; Thermo Fisher Scientific) was used. The plasmid DNAs were mixed with the P3000 reagent in OptiMEM (cat. no. 51985091; Thermo Fisher Scientific) and incubated for 5 min at room temperature. The plasmid/Lipofectamine 3000 complexes were added drop by drop on the DRGs. After 6 h at 37 °C, the entire media was carefully changed for a 1:1 mixture of conditioned human DRG media and fresh human DRG media. Although fluorescence was observable after only 24 h, the experiments were performed 48 h after transfection. Transfection efficiency was routinely greater than 60%.
For voltage-clamp recordings, the internal solution contained (in mM): 140 CsF, 1 EGTA, 10 NaCl, 1 MgCl2, 30 glucose, and 10 Hepes (pH 7.3, 290–310 mOsm/L), and external solution contained (in mM): 50 NaCl, 100 tetraethylammonium chloride, 1.8 CaCl2, 0.1 CdCl2, 1 MgCl2, 10 d-glucose, and 10 Hepes (pH 7.3, 310–315 mOsm/L). Extensive neurite arborization prevented the level of series resistance compensation typically achieved in rat DRG culture and produced space-clamp errors that limited collection of channel biophysical data (Fig. S6).
Confocal Imaging.
Immunocytochemistry was performed on DRGs that were transfected as described earlier. Briefly, cells were fixed by using 4% (vol/vol) paraformaldehyde and 4% (vol/vol) sucrose for 20 min at room temperature. Permeabilization was achieved by a 30-min incubation in PBS solution, 0.1% Triton X-100, following which nonspecific binding sites were saturated by PBS solution containing 3% (vol/vol) BSA for 30 min. Cell staining was performed with anti-NaV1.7, anti-Rab5, or anti-Rab11 primary antibodies in PBS solution with 3% (vol/vol) BSA overnight at 4 °C. Cells were then washed three times in PBS solution and incubated with PBS solution containing 3% (vol/vol) BSA and secondary antibodies (Alexa 488 goat anti-mouse and Alexa 594 goat anti-rabbit; Life Technologies) for 1 h at room temperature. Immunofluorescent micrographs were acquired on a Nikon C1si scanning confocal microscope by using CFI Plan APO VC 60× oil-immersion objective with a 1.4 n.a. Camera gain and other relevant settings were kept constant. Colocalization analysis preformed with NiS Elements-Nikon (version 4.20; Nikon Instruments).
Cell-Surface Biotinylation.
Biotinylation was performed as described previously (15, 57, 58). CAD cells were processed 48 h after transfection. Briefly, live cells were incubated with 0.5 mg⋅mL−1 sulfosuccinimidyl 2-(biotinamido)ethyl-1,3′dithiopropionate (EZ-link Sulfo NHS-SS-biotin; cat. no. 21335; Thermo Fisher Scientific) for 30 min at 4 °C in cold PBS solution, pH 8.0. Excess biotin was quenched by three washes with ice-cold PBS solution containing 100 mM glycine then washed three times with ice-cold PBS solution. The cells were lysed in RIPA lysis buffer [20 mM Tris⋅HCl, pH 7.4, 50 mM NaCl, 2 mM MgCl2, 1% (vol/vol) Nonidet P-40, 0.5% (mass/vol) sodium deoxycholate with protease inhibitors (cat. no. B14002; Biotool), phosphatase inhibitors (cat. no. B15002; Biotool), and BitNuclease (cat. no. B16002; Biotool)]. The biotinylated proteins were separated by adsorption onto Dynabeads M-280 Streptavidin (cat. no. 11205D; Thermo Fisher Scientific) for 3 h at 4 °C. Beads were washed three times with RIPA buffer, resuspended in Laemmli buffer, and heated at 95 °C for 5 min before immunoblotting.
IP of Endogenously SUMOylated Proteins and CRMP2 from Cell Lysate.
CAD cells or DRG tissues were lysed into the IP buffer as previously described (59). Buffer composition was 20 mM Tris⋅HCl, pH 7.4, 50 mM NaCl, 2 mM MgCl2, 10 mM N-Ethylmaleimide, 1% (vol/vol) Nonidet P-40, 0.5% (mass/vol) sodium deoxycholate, 0.1% (mass/vol) SDS with Protease inhibitors (cat. no. B14002; Biotool), phosphatase inhibitors (cat. no. B15002, Biotool), and BitNuclease (cat. no. B16002; Biotool). Total protein concentration was determined by BCA protein assay (cat. no. PI23225; Thermo Fisher Scientific), and then 500 μg of total protein were incubated with 2 μg of CRMP2 antibody overnight at 4 °C under gentle agitation. For IP of endogenously SUMOylated proteins, 0.5% SDS was added to the lysates at 0.5% (mass/vol) final concentration, before boiling for 5 min at 95 °C. Then, 500 µg of total proteins were incubated with 5 μg of SUMO1 antibody overnight at 4 °C under gentle agitation. Protein G magnetic beads (cat. no. 10009D; Thermo Fisher Scientific), preequilibrated with the IP buffer, were then added to the lysates and incubated for 1 h at 4 °C to capture immunocomplexes. Beads were washed four times with IP buffer to remove nonspecific binding of proteins before resuspension in Laemmli buffer and boiling at 95 °C for 5 min before immunoblotting.
In Vitro SUMOylation of CRMP2.
In vitro SUMOylation of CRMP2 was performed by first preparing a bacteria lysate containing the proteins necessary to achieve SUMOylation. The pSUMO1 plasmid (cat. no. 52258; Addgene) (60) was transformed into BL21 E. coli and grown in Luria-Bertani media until reaching an optical density of 0.6–0.8, and then protein synthesis was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) during ∼19 h at 16 °C. Bacteria cells were resuspended in 20 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Triton X-100, and 20% (vol/vol) glycerol, supplemented with Complete EDTA-free protease inhibitors (Roche). Disruption of the bacteria was performed by two rounds of high-pressure homogenization at 10,000 PSI with a LM10 Microfluidizer (Microfluidics), and the lysate was centrifuged for 1 h at 130,000 × g at 4 °C. Supernatant was divided into aliquots, flash-frozen, and kept at –80 °C until use. His-CRMP2, Cdk5, and p25 were purified as previously described (22, 61). Cdk5 was activated by incubation with its cofactor p25 (1 μM each) for 2 h at room temperature. The Cdk5/p25 complex (200 nM final) was then added to 5 μM WT CRMP2 along with pSUMO1 bacterial lysate. In vitro SUMOylation reaction was performed for 1 h at 37 °C with 10 µM ATP. The product of the reaction was then analyzed by Western blotting with a CRMP2 antibody. The products were also analyzed for expression of the E2 SUMO-conjugating enzyme Ubc9.
Immunoblot Preparation and Analysis.
Indicated samples were loaded on 4–20% Novex gels (cat. no. EC60285BOX; Thermo Fisher Scientific). Proteins were transferred for 1 h at 120 V using TGS [25 mM Tris, pH 8.5, 192 mM glycine, 0.1% (mass/vol) SDS], 20% (vol/vol) methanol as transfer buffer to PVDF membranes (0.45 μm; cat. no. IPVH00010; Millipore), preactivated in pure methanol. After transfer, the membranes were blocked at room temperature for 1 h with TBST (50 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 0.1% Tween 20) with 5% (mass/vol) nonfat dry milk, and then incubated separately in indicated primary antibodies in TBST, 5% (mass/vol) BSA, overnight at 4 °C. Following incubation in HRP-conjugated secondary antibodies from Jackson ImmunoResearch, blots were revealed by enhanced luminescence (WBKLS0500; Millipore) before exposure to photographic film or fluorescence [goat anti-rabbit DyLight 800, cat no. 355571, Thermofisher (Figs. 5D and 6 A and E)], or 3,3'-Diaminobenzidine [DAB (PI34002), Thermofisher; Fig. 7 D and E and Fig. S4 A and D]. The fluorescent images (whole) were converted to grayscale and inverted prior to use. Films were scanned, digitized, and quantified by using Un-Scan-It gel scanning software (version 6.1; Silk Scientific).
Data Analysis.
All data columns are shown as mean ± SEM. Individual data points are displayed on top of columns as diamond symbols for patching data. In Western blots, n is presented as the number of separate experiments (minimum of three). Western blots were quantified by using Un-Scan-It gel (version 6.1; Silk Scientific). Steady-state activation was analyzed by using the I-V protocol data for channel conductance G at each voltage step as a function of maximum conductance (G/Gmax). Within the equation G = I(Vpulse − Erev), I is the maximum current in response to Vpulse in mV, and Erev is the reversal potential plotted by Fitmaster software (HEKA). Steady-state fast inactivation (FI) was analyzed by using FI protocol data for channel current I, where maximum current to each voltage step is divided by the maximal observed current within the protocol. The steady-state activation and inactivation curves for individual cells were fitted by using the Boltzmann equation in Origin 9.0 (Origin Lab) and averaged. In the Boltzmann equation, y = 1/(1 + exp[(V1/2 − V)/k], where V1/2, V, and k represent midpoint voltage of kinetics, test potential, and slope factor, respectively.
No data were excluded from the analyses. Unless otherwise stated, statistical differences between control and experimental conditions were determined by using Kruskal–Wallis nonparametric test followed by Dunnett’s post hoc test or a Mann–Whitney nonparametric test when comparing only two conditions with R software (R Foundation for Statistical Computing). P values < 0.05 were judged to be statistically significant.
Acknowledgments
We thank Dr. Yoshio Goshima (Yokohama City University Graduate School of Medicine, Yokohama, Japan) for providing CRMP2-pY32 antibody and Dr. Pascale Giraudon (Lyon Neuroscience Research Center, Lyon, France) for providing CRMP2-pY479 antibody; members of the R. Khanna laboratory for technical assistance regarding this project, and Isabel N. Angeles for assistance with plasmid cloning. This work was supported by a Career Development Award from the Arizona Health Science Center (to M.K.), National Scientist Development Grant SDG5280023 from the American Heart Association (to R.K.), Neurofibromatosis New Investigator Award NF1000099 from the Department of Defense Congressionally Directed Military Medical Research and Development Program (to R.K.), and a Young Investigator Award from the Children’s Tumor Foundation (to A.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1610531113/-/DCSupplemental.
References
- 1.Dib-Hajj SD, Yang Y, Waxman SG. Genetics and molecular pathophysiology of Na(v)1.7-related pain syndromes. Adv Genet. 2008;63:85–110. doi: 10.1016/S0065-2660(08)01004-3. [DOI] [PubMed] [Google Scholar]
- 2.Jarecki BW, Sheets PL, Jackson JO, 2nd, Cummins TR. Paroxysmal extreme pain disorder mutations within the D3/S4-S5 linker of Nav1.7 cause moderate destabilization of fast inactivation. J Physiol. 2008;586(17):4137–4153. doi: 10.1113/jphysiol.2008.154906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waxman SG. Nav1.7, its mutations, and the syndromes that they cause. Neurology. 2007;69(6):505–507. doi: 10.1212/01.wnl.0000268068.02343.37. [DOI] [PubMed] [Google Scholar]
- 4.Bennett DL, Woods CG. Painful and painless channelopathies. Lancet Neurol. 2014;13(6):587–599. doi: 10.1016/S1474-4422(14)70024-9. [DOI] [PubMed] [Google Scholar]
- 5.Hong S, Morrow TJ, Paulson PE, Isom LL, Wiley JW. Early painful diabetic neuropathy is associated with differential changes in tetrodotoxin-sensitive and -resistant sodium channels in dorsal root ganglion neurons in the rat. J Biol Chem. 2004;279(28):29341–29350. doi: 10.1074/jbc.M404167200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laedermann CJ, et al. Dysregulation of voltage-gated sodium channels by ubiquitin ligase NEDD4-2 in neuropathic pain. J Clin Invest. 2013;123(7):3002–3013. doi: 10.1172/JCI68996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Dougherty PM. Enhanced excitability of primary sensory neurons and altered gene expression of neuronal ion channels in dorsal root ganglion in paclitaxel-induced peripheral neuropathy. Anesthesiology. 2014;120(6):1463–1475. doi: 10.1097/ALN.0000000000000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dustrude ET, Wilson SM, Ju W, Xiao Y, Khanna R. CRMP2 protein SUMOylation modulates NaV1.7 channel trafficking. J Biol Chem. 2013;288(34):24316–24331. doi: 10.1074/jbc.M113.474924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukata Y, et al. CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol. 2002;4(8):583–591. doi: 10.1038/ncb825. [DOI] [PubMed] [Google Scholar]
- 10.Yoshimura T, et al. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell. 2005;120(1):137–149. doi: 10.1016/j.cell.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 11.Cole AR, et al. Distinct priming kinases contribute to differential regulation of collapsin response mediator proteins by glycogen synthase kinase-3 in vivo. J Biol Chem. 2006;281(24):16591–16598. doi: 10.1074/jbc.M513344200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arimura N, et al. Phosphorylation of collapsin response mediator protein-2 by Rho-kinase. Evidence for two separate signaling pathways for growth cone collapse. J Biol Chem. 2000;275(31):23973–23980. doi: 10.1074/jbc.M001032200. [DOI] [PubMed] [Google Scholar]
- 13.Uchida Y, et al. Semaphorin3A signaling mediated by Fyn-dependent tyrosine phosphorylation of collapsin response mediator protein 2 at tyrosine 32. J Biol Chem. 2009;284(40):27393–27401. doi: 10.1074/jbc.M109.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varrin-Doyer M, et al. Phosphorylation of collapsin response mediator protein 2 on Tyr-479 regulates CXCL12-induced T lymphocyte migration. J Biol Chem. 2009;284(19):13265–13276. doi: 10.1074/jbc.M807664200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brittain JM, et al. Suppression of inflammatory and neuropathic pain by uncoupling CRMP-2 from the presynaptic Ca2+ channel complex. Nat Med. 2011;17(7):822–829. doi: 10.1038/nm.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moutal A, François-Moutal L, Brittain JM, Khanna M, Khanna R. Differential neuroprotective potential of CRMP2 peptide aptamers conjugated to cationic, hydrophobic, and amphipathic cell penetrating peptides. Front Cell Neurosci. 2015;8:471. doi: 10.3389/fncel.2014.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brustovetsky T, Pellman JJ, Yang XF, Khanna R, Brustovetsky N. Collapsin response mediator protein 2 (CRMP2) interacts with N-methyl-D-aspartate (NMDA) receptor and Na+/Ca2+ exchanger and regulates their functional activity. J Biol Chem. 2014;289(11):7470–7482. doi: 10.1074/jbc.M113.518472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimura T, et al. CRMP-2 regulates polarized Numb-mediated endocytosis for axon growth. Nat Cell Biol. 2003;5(9):819–826. doi: 10.1038/ncb1039. [DOI] [PubMed] [Google Scholar]
- 19.Santolini E, et al. Numb is an endocytic protein. J Cell Biol. 2000;151(6):1345–1352. doi: 10.1083/jcb.151.6.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, et al. Development and characterization of novel derivatives of the antiepileptic drug lacosamide that exhibit far greater enhancement in slow inactivation of voltage-gated sodium channels. ACS Chem Neurosci. 2011;2(2):90–106. doi: 10.1021/cn100089b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dib-Hajj SD, Yang Y, Black JA, Waxman SG. The Na(V)1.7 sodium channel: from molecule to man. Nat Rev Neurosci. 2013;14(1):49–62. doi: 10.1038/nrn3404. [DOI] [PubMed] [Google Scholar]
- 22.Brittain JM, Wang Y, Eruvwetere O, Khanna R. Cdk5-mediated phosphorylation of CRMP-2 enhances its interaction with CaV2.2. FEBS Lett. 2012;586(21):3813–3818. doi: 10.1016/j.febslet.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 23.von Kleist L, et al. Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition. Cell. 2011;146(3):471–484. doi: 10.1016/j.cell.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 24.Laedermann CJ, Decosterd I, Abriel H. Ubiquitylation of voltage-gated sodium channels. Handb Exp Pharmacol. 2014;221:231–250. doi: 10.1007/978-3-642-41588-3_11. [DOI] [PubMed] [Google Scholar]
- 25.Kawahashi K, Sakata T, Hayashi S. Modulation of Notch signal transduction by endocytotic regulators Numb and the Nedd4 family of ubiquitin ligases. A Dros Res Conf. 2007;48:444C. [Google Scholar]
- 26.Woelk T, et al. Molecular mechanisms of coupled monoubiquitination. Nat Cell Biol. 2006;8(11):1246–1254. doi: 10.1038/ncb1484. [DOI] [PubMed] [Google Scholar]
- 27.Horvath CA, Vanden Broeck D, Boulet GA, Bogers J, De Wolf MJ. Epsin: inducing membrane curvature. Int J Biochem Cell Biol. 2007;39(10):1765–1770. doi: 10.1016/j.biocel.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 28.Polo S, et al. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature. 2002;416(6879):451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 29.Di Marcotullio L, et al. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol. 2006;8(12):1415–1423. doi: 10.1038/ncb1510. [DOI] [PubMed] [Google Scholar]
- 30.Osterhout DJ, Wolven A, Wolf RM, Resh MD, Chao MV. Morphological differentiation of oligodendrocytes requires activation of Fyn tyrosine kinase. J Cell Biol. 1999;145(6):1209–1218. doi: 10.1083/jcb.145.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bramham CR, et al. The Arc of synaptic memory. Exp Brain Res. 2010;200(2):125–140. doi: 10.1007/s00221-009-1959-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Craig TJ, et al. Homeostatic synaptic scaling is regulated by protein SUMOylation. J Biol Chem. 2012;287(27):22781–22788. doi: 10.1074/jbc.M112.356337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chowdhury S, et al. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52(3):445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dulubova I, et al. A Munc13/RIM/Rab3 tripartite complex: from priming to plasticity? EMBO J. 2005;24(16):2839–2850. doi: 10.1038/sj.emboj.7600753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castillo PE, Schoch S, Schmitz F, Südhof TC, Malenka RC. RIM1alpha is required for presynaptic long-term potentiation. Nature. 2002;415(6869):327–330. doi: 10.1038/415327a. [DOI] [PubMed] [Google Scholar]
- 36.Girach F, Craig TJ, Rocca DL, Henley JM. RIM1α SUMOylation is required for fast synaptic vesicle exocytosis. Cell Reports. 2013;5(5):1294–1301. doi: 10.1016/j.celrep.2013.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fotia AB, et al. Regulation of neuronal voltage-gated sodium channels by the ubiquitin-protein ligases Nedd4 and Nedd4-2. J Biol Chem. 2004;279(28):28930–28935. doi: 10.1074/jbc.M402820200. [DOI] [PubMed] [Google Scholar]
- 38.Jang SY, et al. Injury-induced CRMP4 expression in adult sensory neurons; A possible target gene for ciliary neurotrophic factor. Neurosci Lett. 2010;485(1):37–42. doi: 10.1016/j.neulet.2010.08.058. [DOI] [PubMed] [Google Scholar]
- 39.Tsutiya A, Nishihara M, Goshima Y, Ohtani-Kaneko R. Mouse pups lacking collapsin response mediator protein 4 manifest impaired olfactory function and hyperactivity in the olfactory bulb. Eur J Neurosci. 2015;42(6):2335–2345. doi: 10.1111/ejn.12999. [DOI] [PubMed] [Google Scholar]
- 40.Weiss J, et al. Loss-of-function mutations in sodium channel Nav1.7 cause anosmia. Nature. 2011;472(7342):186–190. doi: 10.1038/nature09975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang YR, et al. Activation of cyclin-dependent kinase 5 (Cdk5) in primary sensory and dorsal horn neurons by peripheral inflammation contributes to heat hyperalgesia. Pain. 2007;127(1-2):109–120. doi: 10.1016/j.pain.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 42.Yang L, Gu X, Zhang W, Zhang J, Ma Z. Cdk5 inhibitor roscovitine alleviates neuropathic pain in the dorsal root ganglia by downregulating N-methyl-D-aspartate receptor subunit 2A. Neurol Sci. 2014;35(9):1365–1371. doi: 10.1007/s10072-014-1713-9. [DOI] [PubMed] [Google Scholar]
- 43.Liu YN, Yang X, Suo ZW, Xu YM, Hu XD. Fyn kinase-regulated NMDA receptor- and AMPA receptor-dependent pain sensitization in spinal dorsal horn of mice. Eur J Pain. 2014;18(8):1120–1128. doi: 10.1002/j.1532-2149.2014.00455.x. [DOI] [PubMed] [Google Scholar]
- 44.Valdes AM, et al. Role of the Nav1.7 R1150W amino acid change in susceptibility to symptomatic knee osteoarthritis and multiple regional pain. Arthritis Care Res (Hoboken) 2011;63(3):440–444. doi: 10.1002/acr.20375. [DOI] [PubMed] [Google Scholar]
- 45.Meinecke I, et al. Modification of nuclear PML protein by SUMO-1 regulates Fas-induced apoptosis in rheumatoid arthritis synovial fibroblasts. Proc Natl Acad Sci USA. 2007;104(12):5073–5078. doi: 10.1073/pnas.0608773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Theile JW, Cummins TR. Recent developments regarding voltage-gated sodium channel blockers for the treatment of inherited and acquired neuropathic pain syndromes. Front Pharmacol. 2011;2:54. doi: 10.3389/fphar.2011.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmalhofer WA, et al. ProTx-II, a selective inhibitor of NaV1.7 sodium channels, blocks action potential propagation in nociceptors. Mol Pharmacol. 2008;74(5):1476–1484. doi: 10.1124/mol.108.047670. [DOI] [PubMed] [Google Scholar]
- 48.Yang S, et al. Discovery of a selective NaV1.7 inhibitor from centipede venom with analgesic efficacy exceeding morphine in rodent pain models. Proc Natl Acad Sci USA. 2013;110(43):17534–17539. doi: 10.1073/pnas.1306285110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JH, et al. A monoclonal antibody that targets a NaV1.7 channel voltage sensor for pain and itch relief. Cell. 2014;157(6):1393–1404. doi: 10.1016/j.cell.2014.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Clare JJ. Targeting voltage-gated sodium channels for pain therapy. Expert Opin Investig Drugs. 2010;19(1):45–62. doi: 10.1517/13543780903435340. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y, et al. Merging structural motifs of functionalized amino acids and α-aminoamides results in novel anticonvulsant compounds with significant effects on slow and fast inactivation of voltage-gated sodium channels and in the treatment of neuropathic pain. ACS Chem Neurosci. 2011;2(6):317–322. doi: 10.1021/cn200024z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou L, et al. First evidence of overlaps between HIV-Associated Dementia (HAD) and non-viral neurodegenerative diseases: Proteomic analysis of the frontal cortex from HIV+ patients with and without dementia. Mol Neurodegener. 2010;5:27. doi: 10.1186/1750-1326-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson SM, et al. Inhibition of transmitter release and attenuation of anti-retroviral-associated and tibial nerve injury-related painful peripheral neuropathy by novel synthetic Ca2+ channel peptides. J Biol Chem. 2012;287(42):35065–35077. doi: 10.1074/jbc.M112.378695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fenteany G, et al. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268(5211):726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 55.Maruyama H, et al. Electrophysiological characterization of the tetrodotoxin-resistant Na+ channel, Na(v)1.9, in mouse dorsal root ganglion neurons. Pflugers Arch. 2004;449(1):76–87. doi: 10.1007/s00424-004-1315-0. [DOI] [PubMed] [Google Scholar]
- 56.Davidson S, et al. Human sensory neurons: Membrane properties and sensitization by inflammatory mediators. Pain. 2014;155(9):1861–1870. doi: 10.1016/j.pain.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joiner WJ, Khanna R, Schlichter LC, Kaczmarek LK. Calmodulin regulates assembly and trafficking of SK4/IK1 Ca2+-activated K+ channels. J Biol Chem. 2001;276(41):37980–37985. doi: 10.1074/jbc.M104965200. [DOI] [PubMed] [Google Scholar]
- 58.Brittain JM, et al. Neuroprotection against traumatic brain injury by a peptide derived from the collapsin response mediator protein 2 (CRMP2) J Biol Chem. 2011;286(43):37778–37792. doi: 10.1074/jbc.M111.255455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chi XX, et al. Regulation of N-type voltage-gated calcium channels (Cav2.2) and transmitter release by collapsin response mediator protein-2 (CRMP-2) in sensory neurons. J Cell Sci. 2009;122(pt 23):4351–4362. doi: 10.1242/jcs.053280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weber AR, Schuermann D, Schär P. Versatile recombinant SUMOylation system for the production of SUMO-modified protein. PLoS One. 2014;9(7):e102157. doi: 10.1371/journal.pone.0102157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moutal A, et al. (S)-Lacosamide binding to Collapsin Response Mediator Protein 2 (CRMP2) regulates CaV2.2 activity by subverting its phosphorylation by Cdk5. Mol Neurobiol. 2016;53(3):1959–1976. doi: 10.1007/s12035-015-9141-2. [DOI] [PubMed] [Google Scholar]