Significance
Most genome-editing nucleases introduce a double-strand break at a target site, leaving compatible DNA ends that are repaired by the nonhomologous end joining repair pathway to regenerate the target site. This leads to a cycle of cleavage, target site regeneration by ligation, and subsequent recleavage by the nuclease. Here, we bypass this cycle by developing and testing an RNA-guided dual active site nuclease that introduces two noncompatible DNA breaks at a target site, deleting the majority of the target site so that it cannot be regenerated. The dual nuclease (TevCas9) robustly functions in HEK293 and biases genome-editing outcomes toward defined length deletions at high frequencies. Potential applications include directional oligonucleotide ligation, deletion of DNA-binding sites, and in-frame protein deletions.
Keywords: CRISPR/Cas9, genome editing, NHEJ, I-TevI homing endonuclease
Abstract
The CRISPR/Cas9 nuclease is commonly used to make gene knockouts. The blunt DNA ends generated by cleavage can be efficiently ligated by the classical nonhomologous end-joining repair pathway (c-NHEJ), regenerating the target site. This repair creates a cycle of cleavage, ligation, and target site regeneration that persists until sufficient modification of the DNA break by alternative NHEJ prevents further Cas9 cutting, generating a heterogeneous population of insertions and deletions typical of gene knockouts. Here, we develop a strategy to escape this cycle and bias events toward defined length deletions by creating an RNA-guided dual active site nuclease that generates two noncompatible DNA breaks at a target site, effectively deleting the majority of the target site such that it cannot be regenerated. The TevCas9 nuclease, a fusion of the I-TevI nuclease domain to Cas9, functions robustly in HEK293 cells and generates 33- to 36-bp deletions at frequencies up to 40%. Deep sequencing revealed minimal processing of TevCas9 products, consistent with protection of the DNA ends from exonucleolytic degradation and repair by the c-NHEJ pathway. Directed evolution experiments identified I-TevI variants with broadened targeting range, making TevCas9 an easy-to-use reagent. Our results highlight how the sequence-tolerant cleavage properties of the I-TevI homing endonuclease can be harnessed to enhance Cas9 applications, circumventing the cleavage and ligation cycle and biasing genome-editing events toward defined length deletions.
Genome editing with engineered nucleases has revolutionized the targeted manipulation of the genomes of organisms ranging from bacteria to mammals (1). Zinc finger nucleases (ZFNs) (2), transcription-like effector nucleases (TALENs) (3), MegaTALs (fusion of a LAGLIDADG homing endonuclease and TALE domain) (4–6), and nucleases based on the CRISPR-associated protein 9 (Cas9) all represent programmable genome-editing nucleases that have successfully been used to introduce targeted changes in genomes (7–11). One of the most common applications of genome-editing nucleases is gene knockouts that are performed in the absence of an exogenously added repair template (12). In the case of Cas9, the blunt DNA ends introduced at DNA cleavage are substrates for error-free repair by the classical nonhomologous end-joining repair (c-NHEJ) pathway (13), regenerating the target site for recleavage by the nuclease. This cycle of cleavage, ligation, and target site regeneration is perturbed when the double-strand break (DSB) is sufficiently modified by exonucleolytic processing by c-NHEJ, or by the alternative NHEJ pathway (alt-NHEJ), to prevent cleavage by the nuclease (14–18). Imprecise repair by either of the NHEJ pathways generates the characteristic spectrum of heterogeneous length insertions or deletions (indels) centered around the break site (19, 20). The heterogeneous distribution of indels, and the fact that not all indels generate gene knockouts, means that downstream screening and confirmation of knockout genotypes is required. In addition, the chromatin context of the target site (21, 22), the cell cycle stage and cell type (23), and the nature of the DNA overhangs generated by genome-editing nucleases influence the types of indels and efficiency of gene knockouts (24). Coupling the expression of DNA end processing enzymes and genome-editing nucleases can bias gene knockouts by enhancing exonucleolytic end processing before ligation by NHEJ, with variable rates of success depending on the end-processing enzyme used (24, 25). Although more commonly used for homology-directed repair applications (26), paired Cas9 nickase variants can be used to generate gene knockouts but also generate heterogeneous length deletions.
Here, we provide a simple and robust solution to bypass the persistent cycle of cleavage and religation and shift genome-editing events toward deletions of defined length. We created a dual nuclease that introduces two noncompatible DNA breaks at a target site such that the majority of the target site is deleted, preventing regeneration of the target site and continued cleavage. We fused the monomeric nuclease and linker domains from the I-TevI homing endonuclease to Cas9 (27, 28), creating an RNA-guided TevCas9 dual nuclease that functions robustly in HEK293 cells at endogenous target sites. TevCas9 generates defined length deletions of 33–36 bp at high frequencies with minimal DNA end processing compared with Cas9. We envision that the noncompatible and directional nature of the TevCas9 overhangs will enhance applications such as site-directed mutagenesis by oligonucleotide ligation, deletion of binding sites, or epitope tagging.
Results
Fusion of I-TevI to Cas9 Creates an RNA-Guided Dual Nuclease.
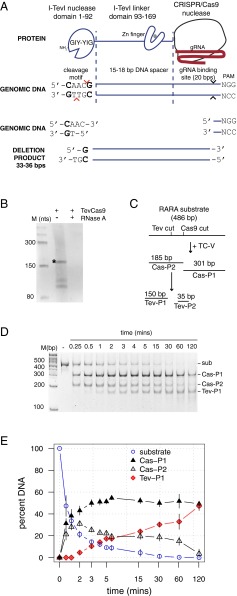
To create a TevCas9 RNA-guided dual nuclease, we fused the I-TevI nuclease domain (residues 1–92) and a portion of the linker domain (residues 92–169) to the N terminus of SpCas9 nuclease derived from the Streptococcus pyogenes type II CRISPR system (27, 28) (Fig. 1A). A TevCas9 target site includes the I-TevI 5′-CNNNG cleavage motif, a DNA spacer that interacts with the I-TevI linker and that separates the cleavage motif from the binding site, the DNA-binding site complementary to the gRNA, and the downstream NGG protospacer-associated motif (PAM) sequence. Previous studies revealed that the length of the DNA spacer is crucial for I-TevI cleavage function, with lengths of 14–19 bp supporting activity in various chimeric contexts (27, 29, 30). We coexpressed TevCas9 with a C-terminal histidine tag and a gRNA targeting a site in the retinoic acid receptor alpha gene (RARA.233, numbered according to start of target site in the RARA cDNA) from the same plasmid in Escherichia coli. The RARA.233 target site (Dataset S1) was predicted according to a binding model that used data from in vitro profiling of I-TevI linker–DNA spacer nucleotide preference (SI Appendix, Figs. S1–S3) and the activity of the I-TevI nuclease domain on all possible 64 CNNNG variants (30). The TevCas9 variant (TC-V) used in this experiment contains an activity-enhancing V117F substitution in the I-TevI linker.
Fig. 1.
Purification and characterization of a TevCas9 dual nuclease. (A) Schematic of TevCas9, organization of the DNA substrate, and cleavage products. (B) TevCas9 copurifies as a RNP with an RNA of the size predicted for the transcribed gRNA. Shown is a 12% (wt/vol) urea–polyacrylamide gel of TevCas9 protein samples treated with proteinase K and then with (+) or without (−) RNaseA. The marker is an RNA ladder with sizes in nucleotides. (C) Putative reaction scheme. TC-V, TevCas9-V117F; sub, substrate; Cas-P1 and Cas-P2, Cas9 cleavage products; and Tev-P1 and Tev-P2, I-TevI cleavage products. (D) Representative cleavage assay (in minutes) with TC-V and RARA.233 target site substrate. The substrate and cleavage products are indicated on the Right side of the gel. The gel image is inverted, and the 35-bp Tev-P2 is not shown. (E) Plot of reaction progress in minutes versus percent DNA for the RARA substrate. Data points are mean values of four independent experiments, with vertical bars representing SD.
We purified TC-V by metal-affinity chromatography, and showed that TC-V was complexed with an RNA species of the size predicted for the transcribed gRNA (Fig. 1B, *). To determine whether the TC-V/gRNA ribonucleoprotein (RNP) complex introduced two DSBs, we examined the cleavage profile on a PCR substrate that contained the RARA.233 target site. Cleavage was consistent with two sequential DSBs, first by the Cas9 nuclease and second by the I-TevI nuclease (Fig. 1 C and D). The predicted 33-bp product corresponding to the fragment excised between the I-TevI and Cas9 cleavage sites was difficult to reproducibly visualize and not used to quantitate reaction progress. Interestingly, cleavage by Cas9 was ∼30-fold faster than cleavage by I-TevI (Fig. 1E, as judged by time required for 50% product formation).
TevCas9 Functions Robustly in HEK293 Cells.
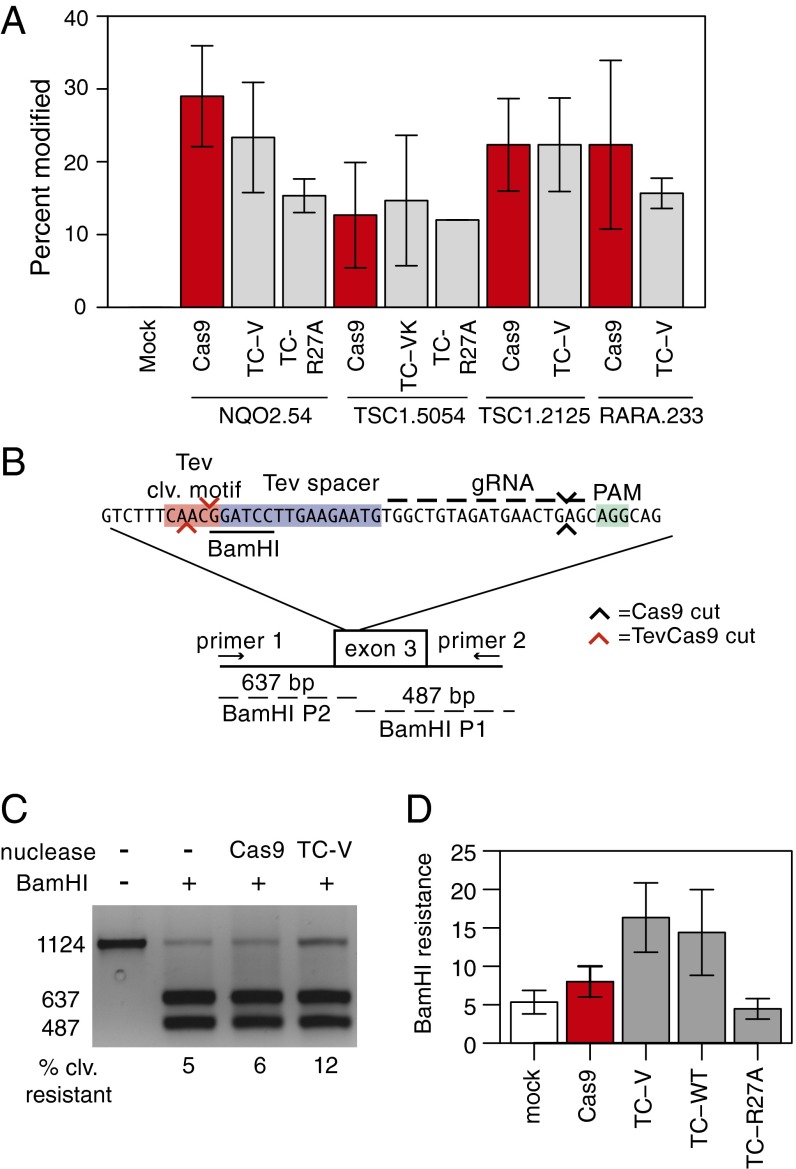
TevCas9 is targeted by the Cas9-associated gRNA, a property that allowed us to directly compare TevCas9 and Cas9 activity at the same sites in HEK293 cells. To do so, we used T7 endonuclease 1 (T7E1) mismatch cleavage assays to measure on-target modification at the RARA.232 site and at two other sites, one in the tuberous sclerosis 1 gene (TSC1.2125) and one in the quinone reductase 2 gene (NQO2.54) (Fig. 2A) (31, 32). With TC-V transfections, we observed modification rates ranging from 16 to 23% versus from 12 to 29% modification rates at the same sites when cells were transfected with Cas9. We also targeted TevCas9 to a second site in the TSC1 gene (TSC1.5054). The TevCas9 variant (TC-VK) used in this experiment contained V117F and K135N substitutions in the I-TevI linker domain that enhance activity on DNA spacers with a T-to-C substitution in position 6 of the DNA spacer (SI Appendix, Fig. S2). We observed very similar levels of modification with both TC-VK (mean = 15%) and Cas9 (mean = 13%) at the TSC1.5054 site. No activity at the NQO2.54, TSC1.2125, or TSC1.5054 sites was observed in cells transfected with TC-V programmed with the RARA.233 gRNA, showing that the I-TevI domains do not influence gRNA-mediated targeting of Cas9 (SI Appendix, Fig. S4). We also predicted off-target sites based on the NQO2.54 gRNA (33), and examined cleavage at these sites by T7E1 mismatch cleavage assays (SI Appendix, Fig. S4). No cleavage was observed at these sites, three of which had a correctly spaced CNNNG motif from the binding site for the gRNA. These experiments show that the I-TevI nuclease and variant linker domains can be directed by Cas9 to cleave target sites with diverse cleavage motifs and DNA spacers, suggesting that the I-TevI sequence requirements are not limiting for TevCas9 targeting.
Fig. 2.
TevCas9 activity in HEK293 cells. (A) T7E1 mismatch cleavage assays of PCR amplified target sites after transfection with Cas9 or TevCas9. TC-V, TevCas9-V117F; TC-VK, TevCas9-V117F/K135N; TC-R27A, TevR27ACas9 (R27A inactivates I-TevI cleavage activity). (B) TevCas9 target site in exon 3 of the human NQO2 gene, positions of PCR primers used for amplification, and sizes of BamHI cleavage products. The I-TevI cleavage motif and DNA spacers are highlighted by red and blue rectangles and the PAM motif by a green rectangle. I-TevI and Cas9 cleavage sites are represented by red and black arrows, respectively. (C) Agarose gel of BamHI cleavage assays on PCR products amplified from the NQO2 locus. Substrate (1,124 bp) and two BamHI cleavage products (673 bp and 487 bp) are indicated on the Left. The percent of substrate resistant to cleavage by BamHI is indicated below each lane. (D) Activity of TevCas9 variants at the NQO2 site measured by BamHI resistance. TevCas9 variants labeled as in A. In A and D, barplots are mean values of at least three independent experiments, with vertical bars representing SD.
TevCas9 Generates Defined Length Deletions in HEK293 Cells.
T7E1 mismatch cleavage assays report on the overall nuclease modification rate at a particular site, but cannot distinguish the relative contributions of the I-TevI and Cas9 active sites. To do so, we used a BamHI restriction site in the NQO2.54 target site that overlaps the predicted I-TevI CAACG cleavage motif (Fig. 2B). After transfection, the NQO2.54 target site (representing both modified and unmodified sites), was amplified from total cells and digested with BamHI. TevCas9 and Cas9 activity was estimated from the fraction of PCR products that are BamHI resistant (Fig. 2 C and D). In transfections with the TC-V and TC-WT (WT I-TevI) nucleases, we observed 14–16% BamHI cleavage resistance. This result implied that cleavage by the I-TevI nuclease domain at the CAACG motif knocked out the BamHI site. In contrast, transfections with Cas9 alone resulted in 5% BamHI cleavage resistance, implying that only a small fraction of Cas9-induced repair events destroy the BamHI site (Fig. 2D). A similar cleavage resistance assay was performed at the TSC1.5054 site and showed that TC-VK cleavage generated a higher level of PvuII resistance than in transfections with Cas9 alone (18% versus 6%) (SI Appendix, Fig. S5), consistent with deletion of the sequence between the I-TevI and Cas9 cleavage sites. We also took advantage of two adjacent NGG PAMs in the TSC1.5054 site to determine the effect of DNA spacer length on TevCas9 activity by simply changing the position of the gRNA (18_gRNA and 14_gRNA) (SI Appendix, Figs. S5 and S8A). We found that TC-VK programmed with the 18_gRNA produced ∼4.5-fold more PvuII resistance than with TC-VK programmed with the 14_gRNA (SI Appendix, Fig. S5), suggesting that the optimal DNA spacer length for the TevCas9 nuclease is between 15 and 18 bp.
Deep Sequencing Reveals Minimal Processing of TevCas9 Deletions.
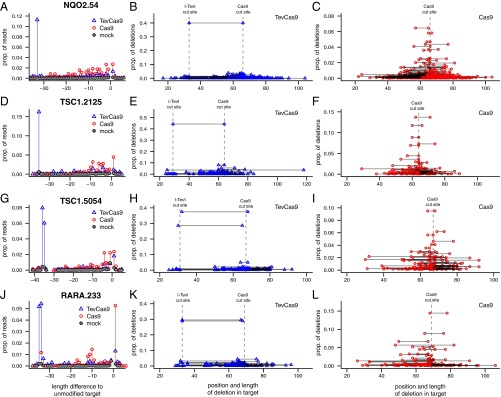
To more accurately assess indels introduced by TevCas9 and Cas9 in HEK293 cells, we used Illumina sequencing of PCR products amplified from the on-target sites to read out the type and length of indels. We first examined the reads for the length of deletions or insertions relative to the unmodified target site, and found that a high proportion of reads in the TevCas9 experiments were deletions that corresponded in length to the distance between the I-TevI and Cas9 cleavage sites (Fig. 3 A, D, G, and J) (SI Appendix, Fig. S6). For the Cas9 experiments, a range of insertion and deletion lengths were observed, including a high proportion of +1 insertions, particularly at the RARA.233 target site. The overall modification rates at each target site, estimated from indel-containing reads, was very similar for TevCas9 and Cas9, agreeing with the T7E1 mismatch cleavage assays (SI Appendix, Fig. S6). Strikingly, when we mapped the position of the deletions, the predominant deletions in the TevCas9 experiments were precise excisions of the DNA segment between the two cut sites (Fig. 3 B, E, H, and K). In contrast, we found that no single deletion product dominated the sequencing reads for the Cas9 experiment, that the deletion lengths were shorter and heterogeneous in length, and that the deletions were centered around the Cas9 cleavage site (Fig. 3 C, F, I, and L). These results demonstrate that TevCas9 cleavage generates defined length deletions at a high frequency in HEK293 cells, and that the two noncompatible DNA ends are repaired with minimal DNA processing (SI Appendix, Fig. S6).
Fig. 3.
TevCas9 generates deletions of precise lengths in HEK293 cells. Results from Illumina sequencing of PCR-amplified fragments for (A–C) NQO2, (D–F) TSC1.2125, (G–I) TSC1.5054, and (J–L) RARA target sites. (A, D, G, and J) Proportion of reads with length differences relative to the unmodified target, with blue triangles representing TevCas9, red open circles representing Cas9, and gray dots representing mock transfection. (B, D, F, and H) Proportion of TevCas9 reads with deletions mapped to the position in the target site. (C, F, I, and L) Proportion of Cas9 reads with deletions mapped to the position in the target site. Dotted vertical lines indicate I-TevI and Cas9 cleavage sites.
Positioning of the I-TevI Cleavage Motif and gRNA Biases In-Frame to Out-of-Frame Deletions.
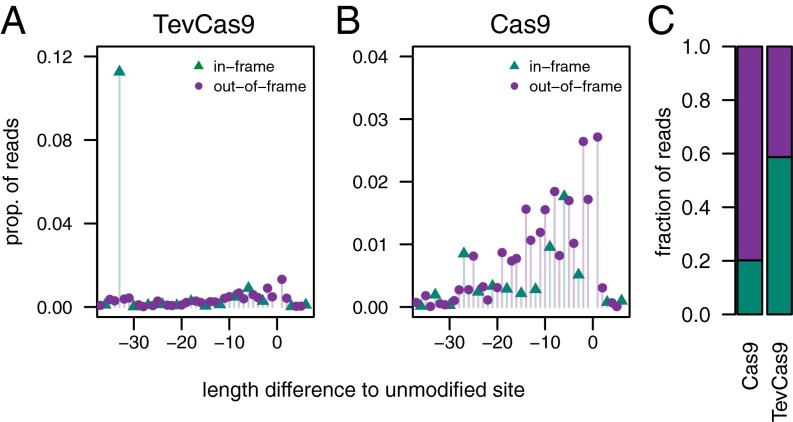
The relative position of the I-TevI and Cas9 cleavage sites at a given target site is determined by length of the DNA spacer that separates the I-TevI CNNNG cleavage motif from the gRNA binding site. At the NQO2.54 target, the distance between the I-TevI and Cas9 cleavage sites is 33 bp (measured from the I-TevI top strand cleavage site), which would generate an in-frame deletion of 11 aa. We examined the proportion of events at the NQO2.54 site and found that 59% of deletions are in frame versus 41% that are out of frame (Fig. 4). In contrast, 20% of events generated by Cas9 are in frame and 70% out of frame. Conversely, TevCas9 events can be biased toward out-of-frame events, as seen at the TSC1.2125 and RARA.233 sites that have an even number of bases between the I-TevI and Cas9 cut sites. In both cases, TevCas9 generates a higher percentage of out-of-frame deletions than does Cas9 (SI Appendix, Fig. S7).
Fig. 4.
TevCas9 can bias the proportion of in-frame to out-of-frame indels. Illumina read data for the NQO2 target site is plotted as the proportion of reads that are in frame (green) and out of frame (purple) for (A) TevCas9 (triangles) and (B) Cas9 (open circles). (C) Fraction of reads that are in frame or out of frame for TevCas9 and Cas9.
Discussion
Motivated by the observation that the majority of genome-editing nucleases generate a DSB with compatible cohesive ends, we hypothesized that a single-chain dual nuclease could bias genome-editing outcomes toward defined length deletions by generating two noncompatible DNA breaks at a target site. Subsequent repair would effectively delete the majority of the target site, preventing further cleavage. We previously created a dual nuclease termed MegaTev, a fusion of I-TevI to LAGLIDADG homing endonucleases (or meganuclease) (27, 34, 35). We showed that MegaTevs efficiently excises a 30-bp fragment from model DNA substrates in HEK293 cells (30). However, the utility of the MegaTev platform is complicated by reengineering of meganuclease specificity and by uncharacterized interactions of the I-TevI linker domain with the DNA spacer region that are critical for nuclease domain positioning at the 5′-CNNNG-3′ cleavage site (36, 37).
Here, we have improved on MegaTevs, in two ways. First, we profiled the nucleotides requirements of the I-TevI linker–DNA spacer interactions, identifying positions critical for activity and delineating a putative linker–DNA code that enabled a targeting model. Directed evolution experiments isolated I-TevI linker variants with activity on DNA spacers not cleaved by the WT linker. Second, we created an easier-to-target enzyme by fusing the I-TevI nuclease and linker domains to the N terminus of Cas9 (28), generating an RNA-guided TevCas9 nuclease with two active sites. TevCas9 can be purified as an RNP that could be used for direct transfection of cell lines (38).
The I-TevI nuclease and linker domains have successfully been fused to four different DNA-binding architectures that are used in genome editing: zinc fingers (27), TALEs (29), meganucleases (30), and, as reported here, Cas9. An on-going debate in the genome-editing field centers on the ease of use (targeting range) versus specificity (off-target effects) of the various reagents, particularly for common laboratory manipulations of cell lines or model organisms. Of the I-TevI–based nucleases, the TevCas9 fusions are the most user friendly, given the ease of programming Cas9–substrate interactions. Similarly, the identification and characterization of I-TevI linker variants makes the TevCas9 reagent easy to use in that targeting of a desired site does not require extensive cycles of engineering and optimization of I-TevI–DNA specificity. For TevCas9, we predicted a potential target site on average every 14 bp for the NQO2, TSC1, and RARA genes, with CG-rich regions having a higher number of potential sites due to the nature of the NGG PAM and I-TevI 5′-CNNNG-3′ cleavage motif. The targeting requirements of TevCas9 (two cleavage sites separated by a defined length spacer) are similar to the parameters for paired Cas9 nickases and FokI–dCas9 nucleases that function as dimers (two independent gRNAs separated by a defined length) (9–11, 26, 33). Whereas future refinements of the TevCas9 binding model will improve target site selection, especially for the I-TevI linker variants with preferences for substitutions in positions T6 or G8 of the DNA spacer, it is encouraging that the high density of predicted sites will not severely limit targeting of TevCas9. Moreover, our recent demonstration that I-TevI cleavage specificity can be modulated to include different cleavage motifs (notably 5′-NNNNG-3′) could also increase targeting potential by alleviating the requirement for a CNNNG motif (39). I-TevI fusions to Cas9 variants with altered PAM specificities (40, 41), or fusion to other Cas9 homologs (42), could similarly broaden the targeting range. It remains to be seen whether the I-TevI nuclease and linker domains mitigate Cas9 off-target effects, as observed with the chimeric ZF–Cas9 and FokI–dCas9 fusions (9, 11, 43).
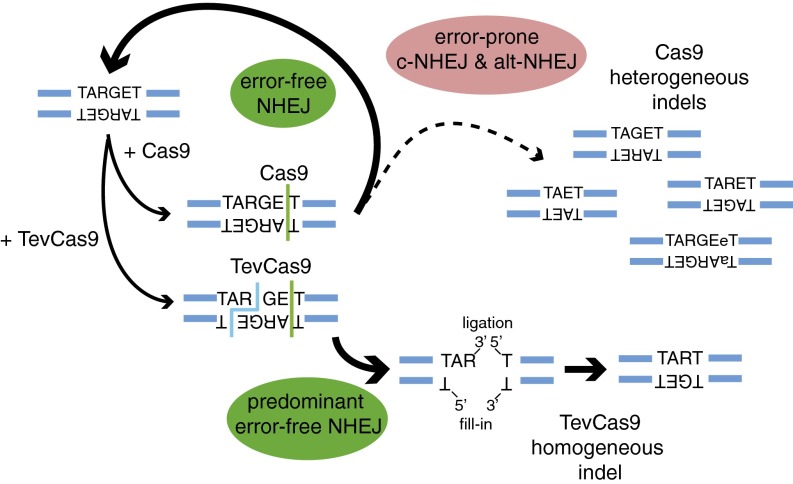
One striking result from our study was that TevCas9 generates defined length deletions of 33–36 bp in HEK293 with minimal DNA end processing. Sequences of the TevCas9 deletion products are consistent with repair by the c-NHEJ pathway whereby the protruding I-TevI 3′ overhang was directly ligated to the blunt end generated by Cas9, with subsequent fill-in of the 2-nt gap on the opposite strand (Fig. 5). The nature of the repair products must mean that TevCas9 sequesters the DNA ends from exonucleolytic processing associated with NHEJ pathways after cleavage, possibly because product release by I-TevI and Cas9 (at least in the context of the chimeric TevCas9) is slow (44, 45). Interestingly, our in vitro assays indicate that cleavage by I-TevI is ∼30-fold slower than Cas9, which is also supported by the deep sequencing data showing that approximately half of all TevCas9 indel events are consistent with cleavage only by Cas9. This observed difference in cleavage rates could reflect a suboptimal fusion point between I-TevI and Cas9 that hinders I-TevI activity, or inherently faster cleavage by Cas9. Improving the rate of I-TevI cleavage in the context of the chimeric TevCas9 nuclease by optimization of the fusion construct could conceivably bias events even more toward deletions of defined length than is observed now.
Fig. 5.
Model of how TevCas9 biases DNA repair outcomes. TevCas9 or Cas9 recognizes and cleaves a target site. The noncompatible DNA ends and 33- to 36-bp deletion generated by TevCas9 prevents regeneration of the target site. Compatible DNA ends generated by Cas9 are repaired by NHEJ, regenerating the target site and inducing a cycle of cleavage and ligation. At a lower rate (dashed line), some Cas9 events undergo repair by the alt-NHEJ pathway, getting heterogeneous length indels.
Cas9 is commonly used to create gene knockouts for downstream phenotypic studies (33). However, in-frame deletions could generate different phenotypes than knockouts if the deletions encompass functionally important regions. The TevCas9 nuclease can generate in-frame deletions of 11 aa efficiently, because the length of the deletion is determined by the distance between the I-TevI and Cas9 cleavage sites. This property of TevCas9, coupled with the directional nature of the TevCas9 deletion (a 3′ 2-nt overhang by I-TevI and a blunt DNA end by Cas9), could enhance ligation of oligonucleotides, enabling applications such as site-directed mutagenesis, swapping of functional domains, or epitope tagging (46). Indeed, preliminary data show that TevCas9 can promote both oligonucleotide insertion and homology-directed repair at the NQO2.54 site (SI Appendix, Fig. S8). Conversely, TevCas9 could be used to tile across promoter regions to delineate functional elements (47) or protein–DNA interaction sites in introns or other noncoding regions.
In summary, we have shown that the TevCas9 dual nuclease provides a strategy to bias genome-editing events toward deletions of defined lengths, escaping the persistent cycle of cleavage and ligation of compatible DSBs that results in heterogeneous length deletions observed with other genome-editing nucleases.
Materials and Methods
Bacterial Strains and Plasmid Construction.
E. coli DH5α and E. coli ER2566 [both from New England Biolabs (NEB)] were used for cloning and for protein expression and were grown in LB media. To create TevCas9 fusions, a human codon optimized version of I-TevI (amino acids 1–169) with a GGSGGS peptide at the C-terminal end was fused to the N terminus of SpCas9 using synthesis by overlap extension PCR and cloned into the AgeI and BglII sites of pX458 (Addgene). The TevCas9 gRNAs were cloned into the BbsI site of pX458. All constructs (Dataset S2) were confirmed by sequencing.
Protein Purification.
TevCas9 or Cas9 variants with C-terminal 6× histidine tags and gRNA were cloned in pACYC–Duet1 expression plasmids under control of separate T7-regulated promoters, and expression was induced overnight at 16 °C with 1 mM isopropyl β-d-thiogalactopyranoside (IPTG). Clairified lysate was loaded onto a 1-mL Ni-NTA affinity column (GE Healthcare), and protein/gRNA complexes were eluted in steps of increasing imidazole. Fractions were dialyzed into storage buffer [20 mM Tris⋅HCl, pH 8, 500 mM NaCl, 1 mM DTT, 5% (vol/vol) glycerol] and frozen at −80 °C. To confirm gRNA copurification, purified protein (4.4 μg) was treated with 20 μg of proteinase K in NEBuffer 3 at 37 °C for 30 min, and half was then treated with RNase A for 30 min at 37 °C, heated to 95 °C for 5 min, and run on a 12% (wt/vol) urea–polyacrylamide gel.
TevCas9 Endonuclease Assays.
Cleavage assays were performed with a 90:1 molar ratio of TevCas9:DNA substrate in a pooled reaction (20 nM DNA substrate, 50 mM Tris⋅HCl, pH 8, 125 mM NaCl, 10 mM MgCl2, 1 mM DTT). Aliquots were taken at the appropriate time points, stopped [200 mM EDTA, 30% (vol/vol) glycerol, 1% SDS, and xylene cyanol], and treated with proteinase K and RNase A before electrophoresis on an 8% (vol/vol) polyacrylamide gel in Tris-borate EDTA buffer.
HEK293 Transfections.
HEK293 cells were cultured in DMEM supplemented with 10% (vol/vol) FBS and grown at 37 °C with 5% (vol/vol) CO2. Approximately 1 × 105 cells were seeded into 24-well plates and transfected with 500 ng of pX458 plasmid using JetPRIME (Polyplus Transfection). Transfection reagent was replaced with fresh media after 4 h and incubated for another 48 h at 37 °C with 5% (vol/vol) CO2. We did not select for transfected cells before isolating genomic DNA.
T7 Endonuclease I Mismatch Repair Assays and Restriction Endonuclease Assays.
For T7E1 mismatch assays (31, 32) PCR products were boiled at 95 °C for 5 min and cooled to 50 °C before flash freezing at −20 °C for 2 min. PCR product (250 ng) was incubated with 2U of T7E1 (NEB) with NEBuffer 2 for 15 min at 37 °C. For restriction endonuclease assays, 250 ng of PCR product was digested with BamHI or PvuII for 1 h at 37 °C.
Next Generation DNA Sequencing.
Genomic DNA from three independent transfections of HEK293 cells with TevCas9, Cas9, and one mock transfection, was used to PCR amplify target sites with barcoded primers (SI Appendix, Table S2) and then pooled. A single-end 250-bp run was performed on an Illumina Mi-Seq platform at the London Regional Genomics Centre. Reads were trimmed to 200 nt and parsed for barcodes using custom Perl scripts. Reads were analyzed for length differences (insertions and deletions) relative to the unmodified site by extracting sequence between anchor sites that flanked the TevCas9 and Cas9 target sites. The proportion of insertions or deletions was the ratio of the number of reads with length N to the total number of extracted reads, excluding reads with fewer than 1,000 counts. To map deletions, we used the UBLAST function of USEARCH (48), with the following parameters: -evalue 0.01, -mismatch −5, -gapopen 20IQ/*LQ/*RQ, -gapext 0IQ, -strand plus, and -maxhits 2. Searches were parsed for deletion start and deletion end point and length, and each deletion was plotted as a proportion relative to all mapped deletions.
Supplementary Material
Acknowledgments
We thank Ben Kleinstiver and Thomas McMurrough for helpful suggestions on the design and testing of the TevCas9 variants and Jean Macklaim for assistance with bioinformatics. This work was supported by the Natural Sciences and Engineering Research Council of Canada Discovery Grants RGPIN-2015-04800 (to D.R.E.), RGPIN-2015-03878 (to G.B.G.), and RGPIN-2013-355799 (to C.S-P.); Schulich Faculty of Medicine Gap Funding (to D.R.E.); and a Doctoral Postgraduate Scholarship from the Natural Sciences and Engineering Research Council of Canada (to J.M.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616343114/-/DCSupplemental.
References
- 1.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346(6213):1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 2.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 3.Bogdanove AJ, Voytas DF. TAL effectors: Customizable proteins for DNA targeting. Science. 2011;333(6051):1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 4.Boissel S, et al. megaTALs: A rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 2014;42(4):2591–2601. doi: 10.1093/nar/gkt1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sather BD, et al. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci Transl Med. 2015;7(307):307ra156. doi: 10.1126/scitranslmed.aac5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoddard BL. Homing endonucleases from mobile group I introns: Discovery to genome engineering. Mob DNA. 2014;5(1):7. doi: 10.1186/1759-8753-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sternberg SH, Doudna JA. Expanding the biologist’s toolkit with CRISPR-Cas9. Mol Cell. 2015;58(4):568–574. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 8.Carroll D. Genome engineering with targetable nucleases. Annu Rev Biochem. 2014;83:409–439. doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- 9.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32(6):577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aouida M, et al. Efficient fdCas9 synthetic endonuclease with improved specificity for precise genome engineering. PLoS One. 2015;10(7):e0133373. doi: 10.1371/journal.pone.0133373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32(6):569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161(3):1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: The increasing spectrum of non-homologous end joining pathways. Mutat Res. 2011;711(1-2):61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McVey M, Lee SE. MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 2008;24(11):529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18(1):134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 17.Deriano L, Roth DB. Modernizing the nonhomologous end-joining repertoire: Alternative and classical NHEJ share the stage. Annu Rev Genet. 2013;47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- 18.Richardson CD, Ray GJ, Bray NL, Corn JE. Non-homologous DNA increases gene disruption efficiency by altering DNA repair outcomes. Nat Commun. 2016;7:12463. doi: 10.1038/ncomms12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beumer KJ, et al. Comparing zinc finger nucleases and transcription activator-like effector nucleases for gene targeting in Drosophila. G3 (Bethesda) 2013;3(10):1717–1725. doi: 10.1534/g3.113.007260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S, et al. A large-scale in vivo analysis reveals that TALENs are significantly more mutagenic than ZFNs generated using context-dependent assembly. Nucleic Acids Res. 2013;41(4):2769–2778. doi: 10.1093/nar/gks1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, et al. Probing the impact of chromatin conformation on genome editing tools. Nucleic Acids Res. 2016;44(13):6482–6492. doi: 10.1093/nar/gkw524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daboussi F, et al. Chromosomal context and epigenetic mechanisms control the efficacy of genome editing by rare-cutting designer endonucleases. Nucleic Acids Res. 2012;40(13):6367–6379. doi: 10.1093/nar/gks268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin S, Staahl BT, Alla RK, Doudna JA. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Certo MT, et al. Coupling endonucleases with DNA end-processing enzymes to drive gene disruption. Nat Methods. 2012;9(10):973–975. doi: 10.1038/nmeth.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delacôte F, et al. High frequency targeted mutagenesis using engineered endonucleases and DNA-end processing enzymes. PLoS One. 2013;8(1):e53217. doi: 10.1371/journal.pone.0053217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleinstiver BP, et al. Monomeric site-specific nucleases for genome editing. Proc Natl Acad Sci USA. 2012;109(21):8061–8066. doi: 10.1073/pnas.1117984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinstiver BP, et al. The I-TevI nuclease and linker domains contribute to the specificity of monomeric TALENs. G3 (Bethesda) 2014;4(6):1155–1165. doi: 10.1534/g3.114.011445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wolfs JM, et al. MegaTevs: Single-chain dual nucleases for efficient gene disruption. Nucleic Acids Res. 2014;42(13):8816–8829. doi: 10.1093/nar/gku573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu P, et al. Mutation detection using Surveyor nuclease. Biotechniques. 2004;36(4):702–707. doi: 10.2144/04364PF01. [DOI] [PubMed] [Google Scholar]
- 32.Guschin DY, et al. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- 33.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sethuraman J, Majer A, Friedrich NC, Edgell DR, Hausner G. Genes within genes: Multiple LAGLIDADG homing endonucleases target the ribosomal protein S3 gene encoded within an rnl group I intron of Ophiostoma and related taxa. Mol Biol Evol. 2009;26(10):2299–2315. doi: 10.1093/molbev/msp145. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi R, et al. Tapping natural reservoirs of homing endonucleases for targeted gene modification. Proc Natl Acad Sci USA. 2011;108(32):13077–13082. doi: 10.1073/pnas.1107719108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dean AB, et al. Zinc finger as distance determinant in the flexible linker of intron endonuclease I-TevI. Proc Natl Acad Sci USA. 2002;99(13):8554–8561. doi: 10.1073/pnas.082253699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Q, et al. Role of the interdomain linker in distance determination for remote cleavage by homing endonuclease I-TevI. J Mol Biol. 2008;379(5):1094–1106. doi: 10.1016/j.jmb.2008.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24(6):1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy AC, Wilson GG, Edgell DR. Perpetuating the homing endonuclease life cycle: Identification of mutations that modulate and change I-TevI cleavage preference. Nucleic Acids Res. 2016;44(15):7350–7359. doi: 10.1093/nar/gkw614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleinstiver BP, et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015;33(12):1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523(7561):481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10(11):1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolukbasi MF, et al. DNA-binding-domain fusions enhance the targeting range and precision of Cas9. Nat Methods. 2015;12(12):1150–1156. doi: 10.1038/nmeth.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mueller JE, Smith D, Belfort M. Exon coconversion biases accompanying intron homing: Battle of the nucleases. Genes Dev. 1996;10(17):2158–2166. doi: 10.1101/gad.10.17.2158. [DOI] [PubMed] [Google Scholar]
- 45.Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34(3):339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 46.Lackner DH, et al. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat Commun. 2015;6:10237. doi: 10.1038/ncomms10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanjana NE, et al. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016;353(6307):1545–1549. doi: 10.1126/science.aaf7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.