Significance
Synaptic AMPA-type and NMDA-type glutamate receptors (AMPARs and NMDARs) have different dynamic characteristics critical for synaptic plasticity. We find that the posttranslational modification, palmitoylation, changes the conformation of postsynaptic density protein 95 (PSD95), a major synaptic scaffold, promoting interactions with AMPARs and NMDARs. In synapses, we measured the conformation and orientation of palmitoylated PSD95 relative to the scaffold, synapse-associated protein 97 (SAP97), and found that changing PSD95 palmitoylation altered PSD95 and AMPAR levels, but not NMDAR levels. We conclude that palmitoylation regulates PSD95 conformation and retention of AMPAR and NMDARs at synapses. Differences in PSD95 palmitoylation appear to occur when AMPARs and NMDARs are in separate synaptic domains, likely contributing to differences in AMPAR and NMDAR dynamics in synapses.
Keywords: palmitoylation, PSD95, SAP97, NMDA receptor, AMPA receptor
Abstract
Postsynaptic density protein 95 (PSD95) and synapse-associated protein 97 (SAP97) are homologous scaffold proteins with different N-terminal domains, possessing either a palmitoylation site (PSD95) or an L27 domain (SAP97). Here, we measured PSD95 and SAP97 conformation in vitro and in postsynaptic densities (PSDs) using FRET and EM, and examined how conformation regulated interactions with AMPA-type and NMDA-type glutamate receptors (AMPARs/NMDARs). Palmitoylation of PSD95 changed its conformation from a compact to an extended configuration. PSD95 associated with AMPARs (via transmembrane AMPAR regulatory protein subunits) or NMDARs [via glutamate ionotropic receptor NMDA-type subunit 2B (GluN2B) subunits] only in its palmitoylated and extended conformation. In contrast, in its extended conformation, SAP97 associates with NMDARs, but not with AMPARs. Within PSDs, PSD95 and SAP97 were largely in the extended conformation, but had different orientations. PSD95 oriented perpendicular to the PSD membrane, with its palmitoylated, N-terminal domain at the membrane. SAP97 oriented parallel to the PSD membrane, likely as a dimer through interactions of its N-terminal L27 domain. Changing PSD95 palmitoylation in PSDs altered PSD95 and AMPAR levels but did not affect NMDAR levels. These results indicate that in PSDs, PSD95 palmitoylation, conformation, and its interactions are dynamic when associated with AMPARs and more stable when associated with NMDARs. Altogether, our results are consistent with differential regulation of PSD95 palmitoylation in PSDs resulting from the clustering of palmitoylating and depalmitoylating enzymes into AMPAR nanodomains segregated away from NMDAR nanodomains.
Postsynaptic densities (PSDs) at glutamatergic synapses organize and hold NMDA receptors (NMDARs), AMPA receptors (AMPARs), and other signaling molecules in place, apposed to sites of neurotransmitter release. Just below the PSD plasma membrane lies a latticework of vertical and parallel filaments that provides a structural scaffold to stabilize synaptic signaling molecules within PSDs (1, 2). Postsynaptic density protein 95 (PSD95) and synapse-associated protein 97 (SAP97) are members of a family of membrane-associated guanylate kinases (MAGUKs) (3). PSD95 is the most abundant scaffold protein in adult synapses, with ∼300 PSD95 molecules (2.3% of the mass of the PSD) in the average PSD, and is part of the lattice forming the core of the PSD (4). SAP97 is also a component of the PSD lattice. Estimates of its PSD copy numbers range from 90 SAP97 molecules per average PSD [0.9% of the mass of the PSD (4)] to lower values (5). As MAGUKs, PSD95 and SAP97 share a series of highly homologous protein-interacting domains but diverge at their N-terminal domains, which affects their trafficking into and out of the PSD, as well as interactions with AMPARs and NMDARs (3, 6, 7). The SAP97β-isoform, like almost all SAP97 molecules, contains an N-terminal L27 domain that interacts with other L27 domain-containing proteins, particularly with a different MAGUK, CASK (8). Most PSD95 molecules, like the PSD95α-isoform, contain, instead of an L27 domain, an N-terminal protein palmitoylation domain, which is required for PSD95 synaptic targeting and retention in the PSD (9–11).
The insoluble nature of isolated PSD fractions has prevented detailed biochemical characterization of interactions between MAGUKs (e.g., PSD95 and SAP97) and glutamate receptors (NMDARs and AMPARs) within PSDs, whereas in vitro binding analysis of these interactions has provided significant insights. The first two PDZ domains of PSD95 and SAP97 bind the C-terminal 5 to 7 aa of different AMPAR and NMDAR subunits (12, 13). PSD95 and SAP97 can both bind the NMDAR, GluN2 subunits, but also bind to different subunits. SAP97 can bind directly to AMPARs via the C terminus of GluA1 subunits (14). PSD95 does not bind to GluA1 subunits and, instead, interacts with the C terminus of AMPAR auxiliary subunits, transmembrane AMPAR regulatory proteins (TARPs), such as Stargazin (15). It has been proposed that when integrated into PSDs, PSD95 serves as a dynamic “slot” that binds AMPARs at the PSD periphery as they enter and exit PSDs (16–18).
Although it had been assumed that NMDARs and AMPARs are homogeneously distributed within PSDs, mounting evidence suggests that assumption is not true. Early immuno-EM studies showed a differential distribution of NMDARs and AMARs at the PSDs in hippocampal and cortical synapses (19, 20). New studies using superresolution light microscopy observed that AMPARs (21, 22) and palmitoylated PSD95 (22, 23) within PSDs were clustered in nanodomains. Using EM tomography, Chen et al. (1, 2) found NMDAR clusters were separate from AMPARs. The NMDAR nanodomain contained NMDAR–PSD95 complexes in a 1:2 stoichiometry, whereas other PSD regions with AMPARs contained AMPAR–PSD95 complexes in a 1:1 stoichiometry. PSD95-like vertical filaments interacted directly with both NMDAR and AMPAR complexes in PSDs (2), suggesting that there are differences in how PSD95 associates with the two glutamate receptor subtypes. When PSD95 was acutely knocked down, AMPAR nanodomains were lost, whereas NMDAR nanodomains were relatively preserved in PSDs (1). This result is consistent with NMDAR nanodomains being more stable than AMPAR nanodomains, and also consistent with the idea that there are important differences in how PSD95 associates with NMDARs and AMPARs at the PSD. Studies assaying the recycling of AMPARs and NMDARs at mature glutamatergic synapses have also found that, compared with AMPARs, NMDARs are more stable in PSDs (24–26). The presence of separate domains in PSDs, together with differences in how PSD95 interacts with AMPARs and NMDARs, suggests that PSD95 has an important role as a scaffold in forming and maintaining separate AMPAR and NMDAR nanodomains.
Previously, we found that SAP97 binding to AMPAR and NMDAR subunits was dependent on conformational change from a “compact” to “extended” conformation (27). Here, we assay PSD95 conformation and demonstrate that it changes from a compact to extended conformation regulated by its palmitoylation. Palmitoylation is a reversible posttranslational modification in which the fatty acid, palmitate, is attached to cysteine residues at the N terminus of PSD95 (28). It provides soluble proteins, such as PSD95, with an anchor to associate with membranes. PSD95 bound directly with NMDAR and AMPAR subunits only in its palmitoylated and extended conformation, and the binding was absent in the compact conformation. PSD95 and SAP97 were largely in the extended conformation at PSDs, and in the compact conformation outside of synapses. However, PSD95 was oriented perpendicular to the PSD plane, with its palmitoylated N terminus at the membrane, whereas SAP97 was oriented parallel to the PSD plane. Increasing PSD95 palmitoylation in PSDs increased PSD95 and AMPAR levels but did not affect NMDAR levels. Thus, in PSDs, PSD95 palmitoylation and conformation are more dynamic when associated with AMPARs and more stable when associated with NMDARs. Evidence of separate AMPAR and NMDAR nanodomains from other studies suggests partitioning of palmitoylating and depalmitoylating enzymes into AMPAR nanodomains separate from NMDAR nanodomains. We propose that regulation of PSD95 and SAP97 conformation changes, and their dynamics in PSDs, results in different interactions with NMDARs and AMPARs, thereby segregating and clustering the receptors in separate nanodomains more dynamic for AMPARs and more stable for NMDARs.
Results
Palmitoylation and Depalmitoylation Regulate PSD95 Conformation.
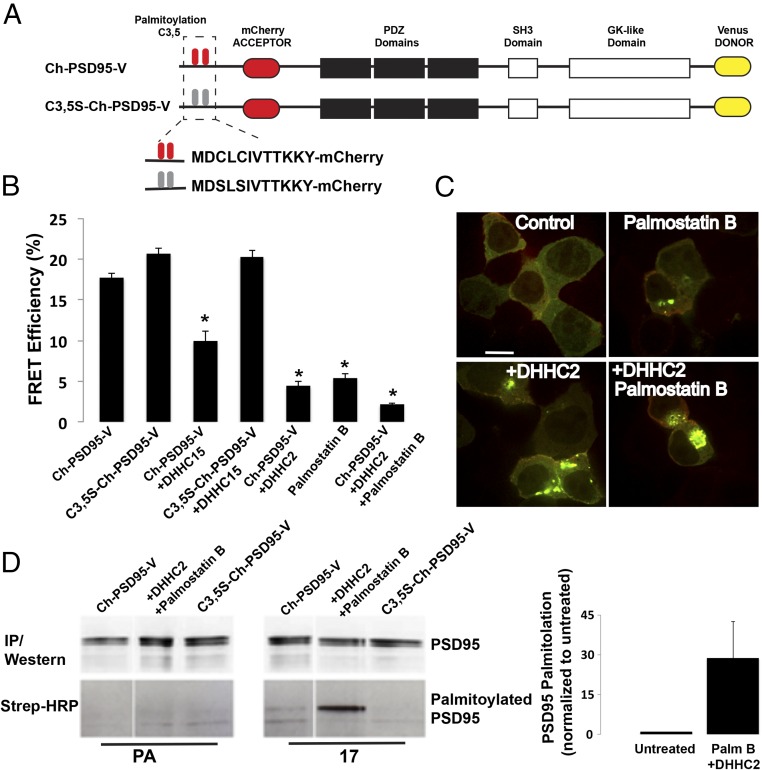
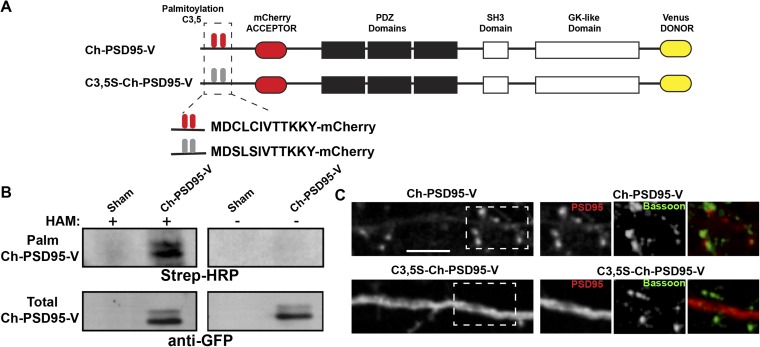
To assay for conformational changes in the predominant PSD95 splice variant, PSD95α (referred to here as PSD95), we generated a PSD95 FRET construct (Ch-PSD95-V; Fig. 1A). Ch-PSD95-V was created by adding the mCherry and Venus fluorescent proteins to the N and C termini of PSD95, respectively. The mCherry sequence was inserted downstream of the palmitoylation sites (3,5 Cys) (29) (Fig. 1A), which allowed palmitoylation of Ch-PSD95-V (Fig. S1). We also mutated the two palmitoylated cysteine residues at the N terminus to serines, as previously described (29), to make a palmitoylation-deficient version of the PSD95 FRET sensor (C3,5S-Ch-PSD95-V; Fig. 1A). The distribution of the PSD95 FRET construct in cultured rat hippocampal neurons was indistinguishable from the distribution of native PSD95 or PSD95 with GFP attached to the C terminus (e.g., see Fig. 3A and Fig. S1). In contrast to the intact Ch-PSD95-V, C3,5S-Ch-PSD95-V trafficking to spines in cultured hippocampal neurons was highly reduced (Fig. S1). These results are consistent with previous findings that PSD95 is palmitoylated to traffic into synapses (11).
Fig. 1.
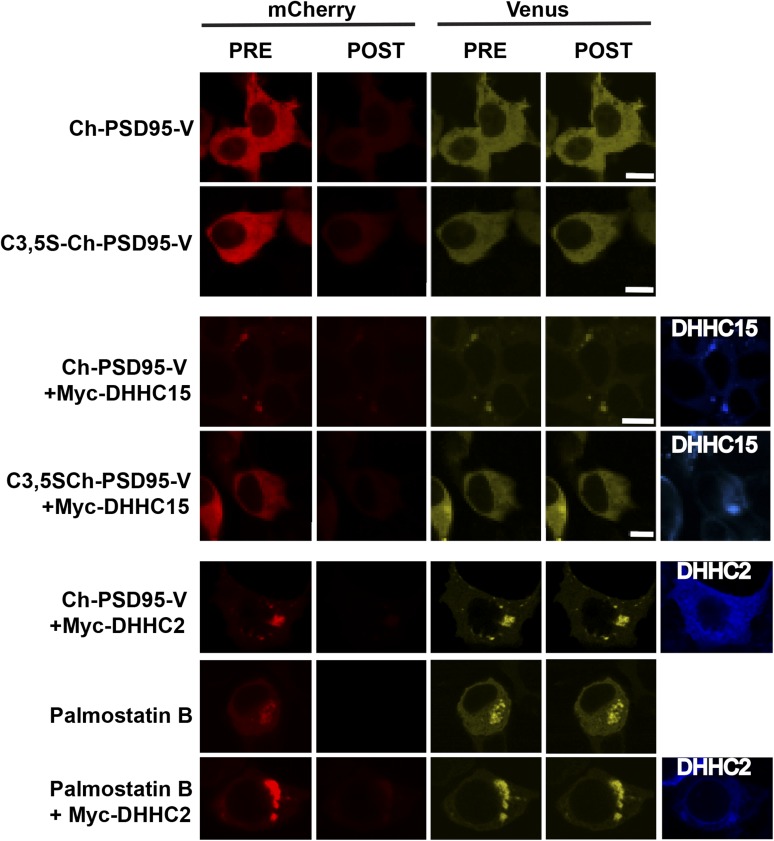
Palmitoylation of PSD95 triggers a change in PSD95 conformation. (A) Schematic of PSD95 (Ch-PSD95-V) and mutated PSD95 (C3,5S-Ch-PSD95-V) FRET sensor constructs. (B) FRET efficiency values for Ch-PSD95-V with changes in palmitoylation. FRET measurements were made in regions of interest in example cells shown in Fig. S3. Data are shown as mean ± SEM (n = 12–30 cells per condition; *P < 0.0001 relative to Ch-PSD95-V). (C) As an additional assay of the changes in PSD95 palmitoylation observed above, we used colocalization with PF11-GFP, a conformation-specific intrabody for palmitoylated PSD95. Representative images of HEK cells expressing PF11-GFP and PSD95 alone (Top Left), with DHHC2 (Bottom Left), with palmostatin B treatment (Top Right), or with palmostatin B treatment and DHHC2 expression (Bottom Right). Increased aggregation and colocalization with PF11-GFP is correlative with an increase in PSD95 palmitoylation. PSD95 alone is not highly palmitoylated (diffuse fluorescence and lack of colocalization with PF11-GFP) compared with its palmitoylation in the presence of DHHC2 and palmostatin B (coaggregation and colocalization with PF11-GFP). (Scale bar: 10 μm.) (D) Click chemistry palmitoylation assay was used to compare the level of Ch-PSD95-V palmitoylation under different conditions. HEK cells transiently expressing Ch-PSD95-V alone or with Myc-DHHC2 and palmostatin B treatment were labeled with 17-octadecynoic acid (17-ODYA). Ch-PSD95-V was immunoprecipitated and treated with biotin-azide under click chemistry reaction conditions, and subsequently analyzed by Western blotting with anti-PSD95 antibody (Top) and streptavidin (Bottom). The space after the first lane is due to cropping of lanes unrelated to the present analysis. PSD95 alone is not highly palmitoylated. In the bar graph, quantification of the normalized band intensities (ratio of streptavidin signal to PSD95 protein) indicates that Ch-PSD95-V palmitoylation increases 30-fold in the presence of DHHC2 and palmostatin B (n = 4 experiments). IP, immunoprecipitation.
Fig. S1.
Further characterization of PSD95 and mutated PSD95 FRET sensors. (A) Schematic of PSD95 (Ch-PSD95-V) and mutated PSD95 (C3,5S-Ch-PSD95-V) FRET sensor constructs as in Fig. 1A. (B) Ch-PSD95-V is palmitoylated. Ch-PSD95-V was expressed in HEK293 cells and assayed for palmitoylation by the acyl-biotin exchange (ABE) assay. (Top) ABE assay comparing sham-transected HEK293 cells (sham) and cells transfected with Ch-PSD95-V. Specific labeling was only observed with the hydroxylamine (HAM) treatment step, a step required in the ABE method to assay for palmitoylation. (Bottom) Total cellular expression of Ch-PSD95-V was assayed by Western blots. (C) Distribution of Ch-PSD95-V and C3,5S-Ch-PSD95-V in dendrites. (Left) Cultured hippocampal neurons were transfected with Ch-PSD95-V (Top) or C3,5S-Ch-PSD95-V (Bottom). Displayed are images of mCherry fluorescent signal in dendrites for each construct. Ch-PSD95-V concentrated in spines, consistent with wild-type PSD95 distribution. C3,5S-Ch-PSD95-V had a more diffuse distribution consistent with disrupted targeting and accumulation at spines. (Scale bar: 5 μm.) [Right; Insets of dashed boxes (Left)] Distribution of Ch-PSD95-V and C3,5S-Ch-PSD95-V at synapses. Neurons were fixed and immunolabeled with anti-Bassoon antibody to identify presynaptic sites. Ch-PSD95-V, but not C3,5S-Ch-PSD95-V, colocalized with anti-Bassoon staining.
Fig. 3.
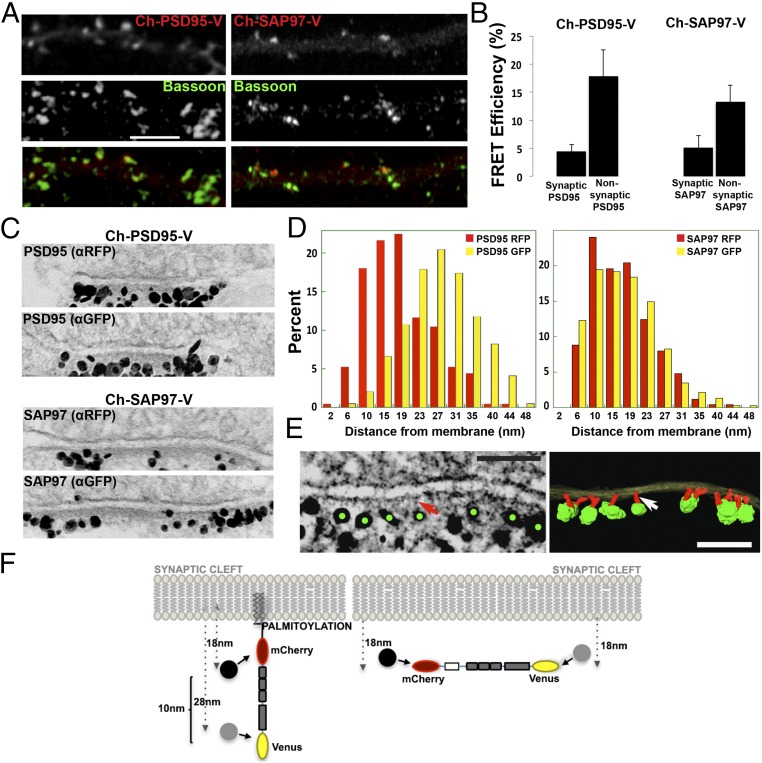
Conformation and orientation of PSD95 and SAP97 at synapses. (A) Immunolocalization of Ch-PSD95-V and Ch-SAP97-V at synapses. Ch-PSD95-V (Left) or Ch-SAP97-V (Right) was transfected into cultured hippocampal neurons at 14 days in vitro (DIV). Displayed are dendrites immunolabeled with anti-Bassoon, a presynaptic marker, with Ch-PSD95-V or Ch-SAP97-V expression [FRET construct fluorescence (Top), anti-Bassoon (Middle), merged image (Bottom)]. (Scale bar: 5 μm.) (B) FRET efficiency values for Ch-PSD95-V (Left) and Ch-SAP97-V (Right) at synaptic and nonsynaptic regions. At synaptic sites, the FRET efficiency of Ch-PSD95-V was 4.5 ± 1.2%, and at nonsynaptic sites, it was 17.9 ± 4.6% (mean ± SEM; n = 78 puncta, 10 cells, *P < 0.01). At synaptic sites, the FRET efficiency of Ch-SAP97-V was 5.2 ± 2.1%, and at nonsynaptic sites, it was 13.3 ± 2.9% (mean ± SEM; n = 58 puncta, 12 cells, *P < 0.02). (C) Immuno-EM experiment measuring the distance of N and C termini of Ch-PSD95-V (Top) and Ch-SAP97-V (Bottom) from the PSD membrane. Representative EM images of synapses labeled separately with silver-enhanced immunogold particles against RFP (mCherry; Top) or GFP (Venus, Bottom) of Ch-PSD95-V and Ch-SAP97-V at the PSD. (D) Quantification of immuno-EM labeling measuring the distance between anti-RFP–labeled gold particles (red) and the membrane or anti-GFP gold particles (yellow) and membrane. For Ch-PSD95-V, distances were 18 ± 8 nm (mean ± SD; n = 250, 49 spines) for anti-RFP particles and 28 ± 8 nm (n = 784, 37 spines, P < 0.0001 by Student’s t test) for anti-GFP particles. The measured distance between the N-terminal mCherry epitope and the C-terminal Venus epitope was therefore ∼10 nm as summarized in F. For CH-SAP97-V, distances were 17 ± 7 nm (mean ± SD; n = 250, 35 spines) for anti-RFP particles and 18 ± 8 nm (n = 377, 26 spines, P = 0.512) for anti-GFP particles. There was no difference in the measured distance between the N-terminal mCherry and C-terminal Venus epitopes on Ch-SAP97-V. (E) EM tomography on Venus immunolabeled Ch-PSD95-V at PSD. (Left) Tomogram of the immunolabeling of Ch-PSD95-V against the C-terminal Venus site showing silver-enhanced immunogold particles (green dots) at the distal ends of vertical filaments (red arrow). (Scale bar: 100 nm.) (Right) Surface-rendered structural model based on the tomograms. Postsynaptic membrane (translucent yellow), PSD95 molecules as vertical filaments (red; indicated by white arrow), and enhanced immunogold particles (green) are shown. (Scale bar: 100 nm.) (F) Model of PSD95 and SAP97 orientation at the PSD. (Left) For Ch-PSD95-V, the average distance for the N terminus was 18 nm, and it was 28 nm for the C terminus (D), consistent with an orientation perpendicular to the PSD membrane. (Right) In contrast, there was no difference in the distance to the membrane between the N-terminal and C-terminal domains of SAP97 (D), consistent with an orientation parallel to the PSD membrane. The PSD95 extended conformation and orientation perpendicular to the PSD membrane was confirmed by EM tomography in E for Ch-PSD95-V, with anti-GFP sliver-enhanced gold particles in green and PSD95 in red.
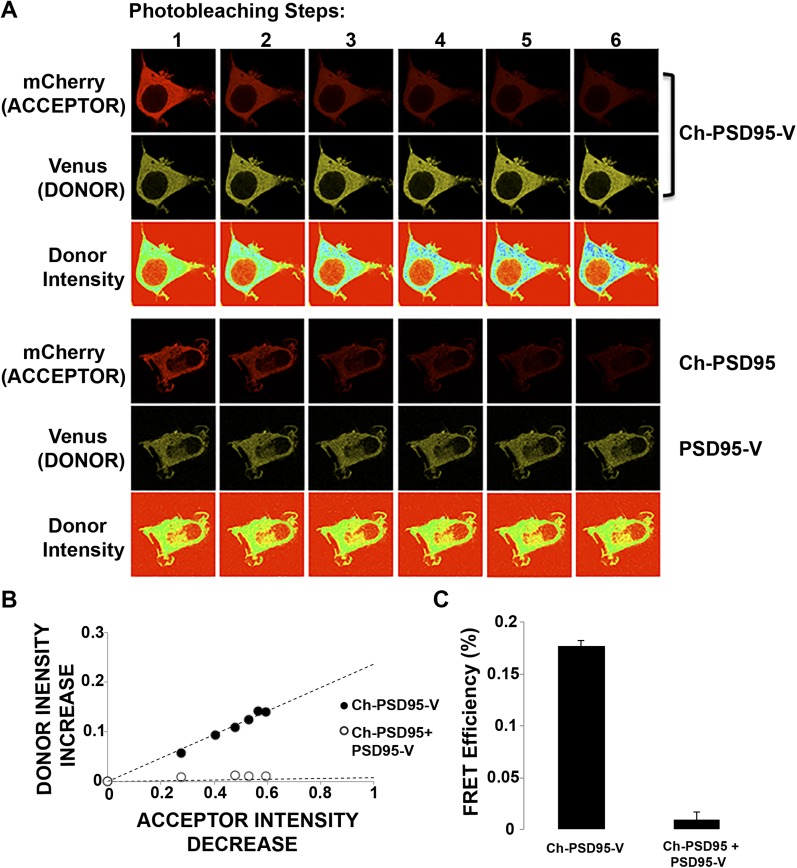
Ch-PSD95-V FRET efficiency (Fig. 1B) was determined using an acceptor photobleaching protocol (Fig. S2). The protocol measured increases in Venus fluorescence after successive photobleaching of mCherry. The FRET efficiency, 17.7%, was calculated from the fit to the values of the fluorescence changes with successive photobleaching. Importantly, no FRET was observed when the FRET pairs were attached separately to the N or C terminus of PSD95 (Ch-PSD95 and PSD95-Venus) and coexpressed in HEK293 cells (Fig. S2). The FRET measured using Ch-PSD95-V is therefore intramolecular and does not result from intermolecular FRET between two or more Ch-PSD95-V molecules. Using a FRET sensor of SAP97 conformation with the same FRET pair (Ch-SAP97-V), we previously found that SAP97 conformation changes from a compact conformation to an extended conformation when the protein CASK binds to its N-terminal domain (27). The relatively high FRET efficiency measurement of 17.7% is consistent with interacting FRET pairs of N and C termini. Therefore, PSD95 is predominantly in the compact conformation, as shown by EM of recombinant PSD95 molecules (30). If palmitoylation of PSD95 changes its conformation from a compact conformation to an extended conformation, similar to how CASK affects SAP97 conformation, we would expect a decrease in FRET efficiency of Ch-PSD95-V when it is palmitoylated. We would therefore expect the C3,5S-Ch-PSD95-V FRET sensor, which cannot be palmitoylated, to have the highest FRET efficiency. We observed a small, but not statistically significant, increase (17.7% vs. 20.6%) comparing FRET efficiencies of Ch-PSD95-V with C3,5S-Ch-PSD95-V (Fig. 1B, Top and Fig. S3, Top). The small change in FRET efficiency suggests that either the level of Ch-PSD95-V palmitoylation is relatively low or that palmitoylation has little effect on PSD95 conformation.
Fig. S2.
Ch-PSD95-V FRET in HEK293 cells. (A) Representative images of the photobleaching method performed on Ch-PSD95-V in HEK293 cells to obtain Ch-PSD95-V intramolecular FRET efficiency (Top). Venus and mCherry fluorescence images were acquired before and after six acceptor photobleaching steps. Venus (donor) pseudocolor intensity is displayed in the third row. (Bottom) No rise in donor intensity is observed when Ch-PSD95 is coexpressed with PSD95-V. (B) Ch-PSD95-V FRET efficiency determined from the linear relationship between increases in donor (mCherry) fluorescence with decreases in acceptor (Venus) with photobleaching. The photobleaching steps for ChPSD95-V in A are plotted as the percent decrease in acceptor intensity versus the percent increase in donor intensity (●). A corresponding rise in donor intensity is not observed when Ch-PSD95 is coexpressed with PSD95-V (○). (C) FRET efficiency value, ET, was calculated from the data above using the equation: ET = (1 − Ida/Id), where Ida and Id are the fluorescence intensities of the donor in the presence and absence of acceptor, respectively. FRET efficiency for Ch-PSD95-V is 17.7 ± 0.4% (mean ± SEM; n = 27 cells, *P < 0.00001) compared with 0.9 ± 0.7% (mean ± SEM; n = 19 cells) in cells coexpressing Ch-PSD95 and PSD95-V.
Fig. S3.
Palmitoylation conditions for Ch-PSD95-V FRET measured in HEK293 cells. Representative images of cells used for the Ch-PSD95-V FRET analysis in Fig. 1B to test for changes in conformation and palmitoylation. Images are displayed from HEK293 cells expressing Ch-PSD95-V or C3,5S-Ch-PSD95-V (first and second rows), with Myc-DHHC15 (third and fourth rows), Myc-DHHC2 (fifth row), palmostatin B treatment (sixth row), or palmostatin B treatment and Myc-DHHC2 expression (seventh row). Shown are the initial and final (PRE and POST) Venus and mCherry fluorescence signals acquired over the course of acceptor photobleaching. Epitope-tagged DHHC proteins were detected with anti-Myc antibodies (blue). (Scale bars: 10 μm.)
To assess further how palmitoylation affects PSD95 conformation, we altered conditions in the HEK cells to increase PSD95 palmitoylation. First, we coexpressed zinc finger DHHC-type containing 15 (DHHC15), a palmitoyl-acyl transferase (PAT), which increased its palmitoylation and redistributed PSD95 into perinuclear clusters with DHHC15 when expressed in heterologous cells with PSD95 (31). When coexpressed, perinuclear clusters of Ch-PSD95-V that colocalized with DHHC15 were observed (Fig. S3). The clustering was not observed when C3,5S-Ch-PSD95-V replaced Ch-PSD95-V (Fig. S3). Thus, the clusters are consistent with an interaction between DHHC15 and Ch-PSD95-V that increases Ch-PSD95-V palmitoylation. Ch-PSD95-V FRET efficiency with DHHC15 coexpression was significantly reduced from 18.0% to 9.9%, whereas the FRET efficiency of C3,5S-Ch-PSD95-V coexpressed with DHHC15 did not change (Fig. 1B).
Next, we coexpressed Ch-PSD95-V with a different PAT, DHHC2. DHHC2 differs from other PATs that can palmitoylate PSD95, such as DHHC15 and DHHC3, in that it is trafficked to the neuronal plasma membrane at PSDs, and appears to be the native PAT that palmitoylates PSD95 at PSDs (9, 23). Ch-PSD95-V redistributed into even larger perinuclear clusters (Fig. S3) when coexpressed with DHHC2. The Ch-PSD95-V FRET efficiency in the clusters with DHHC2 coexpression was further reduced, from 18.0% to 4%, compared with the reduction to 9.9% observed with DHHC15 coexpression (Fig. 1C). As an alternative to coexpression with DHHC proteins, we used palmostatin B to inhibit the palmitoyl-thioesterase (PPT) that depalmitoylates PSD95, and thereby to increase its palmitoylation. Treatment of cells expressing Ch-PSD95-V with palmostatin B again caused Ch-PSD95-V redistribution into perinuclear clusters similar in size to the clusters observed with DHHC2 coexpression (Fig. S3), consistent with Ch-PSD95-V clustering caused by its palmitoylation. Ch-PSD95-V FRET efficiency decreased from 18.0% to 5% under this condition (Fig. 1B). For a final condition, we coexpressed DHHC2 with Ch-PSD95-V, and the cells were also treated with palmostatin B. The largest Ch-PSD95-V perinuclear clusters were observed under these conditions, suggesting that Ch-PSD95-V palmitoylation is highest under these conditions (Fig. S3), and the FRET efficiency decreased from 18.0% to 2% (Fig. 1B). Altogether, our results are consistent with palmitoylation of Ch-PSD95-V causing a change in its conformation from a compact to extended conformation.
To test directly whether the PSD95 in the perinuclear clusters was palmitoylated, we obtained an “intrabody,” PF11, a recombinant IgG fragment that specifically recognizes and binds to palmitoylated PSD95 but not to depalmitoylated PSD95 (23). PF11, tagged with GFP (PF11-GFP), was coexpressed with PSD95. In the PSD95-alone condition, PF11 and PSD95 were almost always evenly distributed throughout the cell cytoplasm whether expressed together or alone. When the PSD95 perinuclear clusters formed with coexpression of DHHC2 and/or palmostatin B treatment (Fig. 1C), PF11-GFP colocalized predominantly in the clusters, demonstrating the presence of palmitoylated PSD95. The conditions that increased PSD95 clustering, and presumably its palmitoylation (coexpression of DHHC2 and/or palmostatin B), also increased PF11-GFP clustering and colocalization. No clustering of PF11-GFP was observed without coexpression of PSD95. Together, our data demonstrate that the clustering of PSD95 is caused by its palmitoylation and that the size and extent of the clustering correlate with the degree to which PSD95 is palmitoylated.
Using click chemistry methods (32) to quantitate changes in Ch-PSD95-V palmitoylation, we estimated the change in PSD95 palmitoylation with the addition of DHHC2 and palmostatin B treatment in the cells. The results were somewhat variable, but indicated that the DHHC2 enzyme, together with palmostatin B treatment, resulted in an increase in PSD95 palmitoylation of ∼30-fold (Fig. 1D).
PSD95 Palmitoylation Is Required for Its Associations with AMPARs and NMDARs.
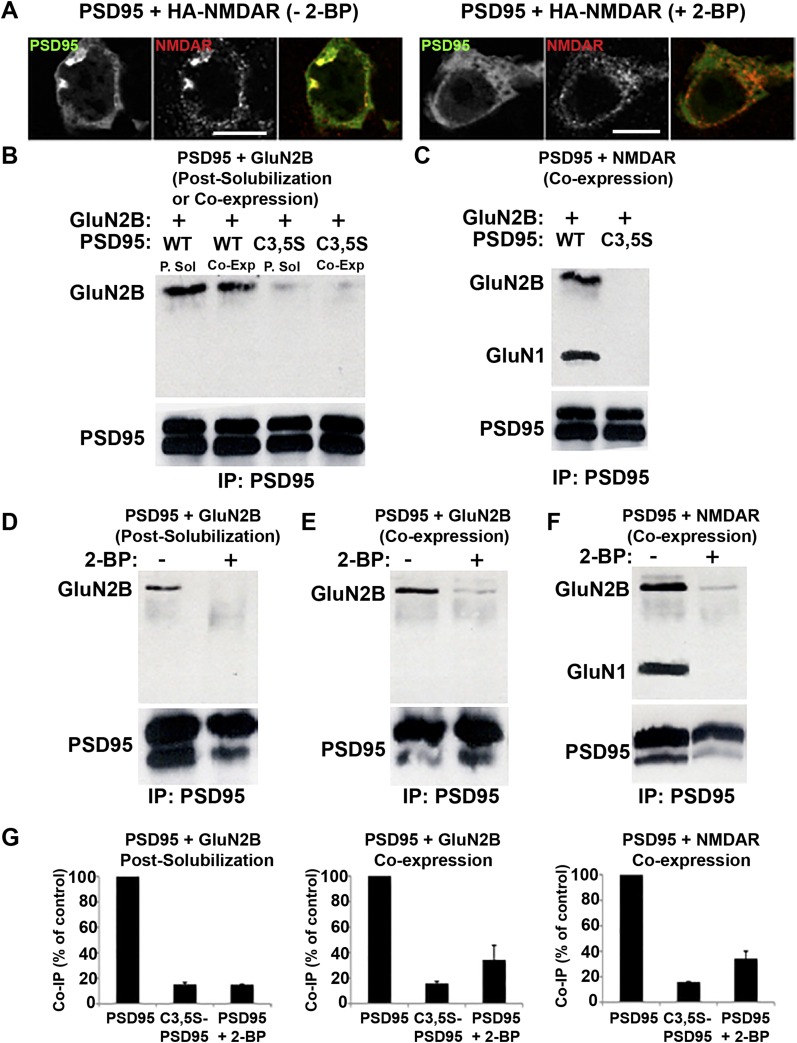
We performed additional experiments in HEK cells to examine how palmitoylation and changes in PSD95 conformation might regulate its interactions with AMPARs and NMDARs. PSD95 does not associate directly with AMPAR subunits. Instead, PSD95 associates indirectly with AMPARs through a direct interaction with their auxiliary TARP subunits. PSD95–TARP interactions depend on the TARP PDZ-binding domain at its C terminus (33). In HEK cells, we examined whether palmitoylation regulates the interaction between PSD95 and the TARP Stargazin. When Ch-PSD95-V was expressed with AMPAR GluA1 subunits and Stargazin, we observed it in clusters with GluA1 and Stargazin at the cell surface and at intracellular sites (Fig. 2A). We did not find any PSD95-GluA1-Stargazin clusters when PSD95 was replaced by C3,5S-PSD95 (Fig. 2A). PSD95 also binds directly to NMDAR, GluN2 subunits. Consistent with this property, we observed strong colocalization between GluN2B-containing NMDARs and PSD95 when coexpressed in HEK293 cells in small clusters at the cell surface and at intracellular sites as previously reported (34) (Fig. 2B). Again, we did not find any PSD95-NMDAR clusters when PSD95 was replaced by C3,5S-PSD95 (Fig. 2B). Furthermore, PSD95-NMDAR clusters disappeared when HEK293 cells were treated with 2-bromopalmitate (2-BP), an inhibitor of palmitoylation (Fig. S4A).
Fig. 2.
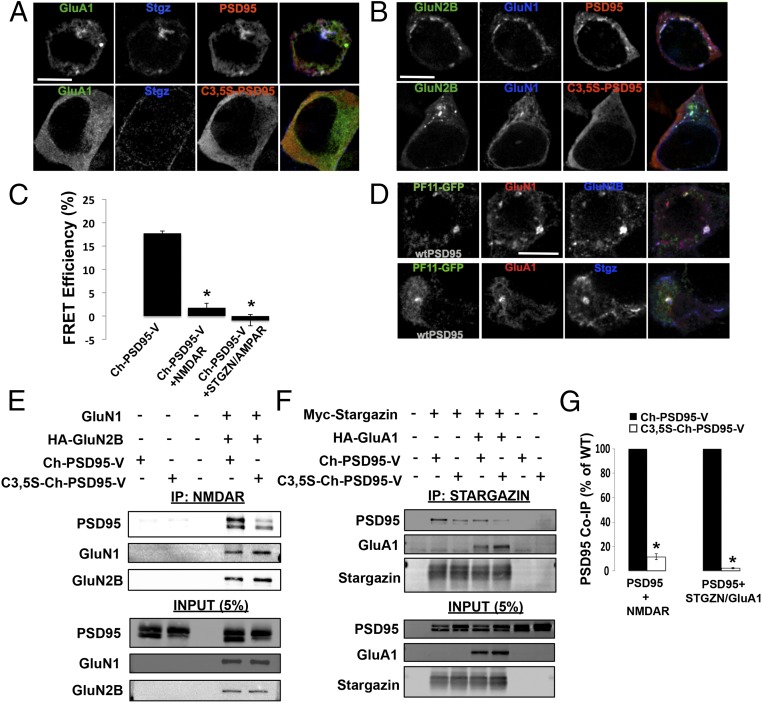
Palmitoylation of PSD95 regulates interactions with AMPARs and NMDARs. (A) Immunolocalization of HA-Stargazin, GFP-GluA1, and untagged wild-type (wt) PSD95 or C3,5S-PSD95 in HEK293 cells. (Top) PSD95, HA-Stargazin, and GFP-GluA1 AMPAR subunits transfected in HEK293 cells. (Bottom) C3,5S-PSD95 replaced PSD95. In both sets of images, PSD95 or C3,5S-PSD95 was detected with anti-PSD95 antibodies, and with anti-HA antibodies for Stargazin. (Scale bar: 10 μm.) (B) Immunolocalization of NMDARs (GFP-GluN2B and Flag-GluN1) and untagged PSD95 or C3,5S-PSD95 in HEK293 cells. (Top) PSD95, GFP-GluN2B, and Flag-GluN1 NMDAR subunits transfected in HEK293 cells. (Bottom) C3,5S-PSD95 replaced PSD95. In both sets of images, PSD95 or C3,5S-PSD95 was detected with anti-PSD95 antibodies, and with anti-Flag antibodies for GluN1. (Scale bar: 10 μm.) (C) Ch-PSD95-V FRET analysis to test for conformational changes that occur with interactions with NMDARs and AMPARs in HEK293 cells. Cells were transfected with Ch-PSD95-V and NMDARs, consisting of HA-GluN2B/Flag-GluN1, or with AMPARs, consisting of Myc-Stargazin/HA-GluA1. (D) Palmitoylated PSD95 specifically colocalized in puncta with NMDARs and AMPARs in HEK293 cells. Cells were transfected with PF11-GFP, the conformation-specific intrabody for palmitoylated PSD95, untagged wtPSD95, and either Flag-GluN1 plus HA-GluN2B (NMDARs, Top) or mCherry-GluA1 plus HA-Stargazin (AMPARs, Bottom). Flag-GluN1, HA-GluN2B, and HA-Stargazin were detected with anti-Flag and anti-HA antibodies. (Scale bar: 10 μm.) (E) Loss of PSD95 palmitoylation sites disrupts interactions between PSD95 and NMDARs. PSD95–NMDAR interactions were assayed by co-IP and Western blots. A representative Western blot shows the levels of Ch-PSD95-V that coimmunoprecipitated with GluN2B subunits. Cell lysates prepared from HEK293 cells expressing NMDARs (Flag-GluN1 and HA-GluN2B) were mixed with separate lysates from cells expressing Ch-PSD95-V or C3,5S-Ch-PSD95-V. IPs were performed with anti-HA antibodies specific for GluN2B, and anti-PSD95 and anti-HA antibodies were used to blot. (F) Loss of PSD95 palmitoylation sites disrupts interactions between PSD95 and AMPARs containing Stargazin. A representative Western blot displays the levels of Ch-PSD95-V that coimmunoprecipitated with Stargazin subunits alone, or with Stargazin and GluA1 subunits. Cell lysates prepared from HEK293 cells expressing Stargazin-containing AMPARs (Myc-Stargazin and HA-GluA1) or Stargazin alone (Myc-Stargazin) were mixed with separate lysates from cells expressing Ch-PSD95-V or C3,5S-Ch-PSD95-V. IPs were performed with anti-Myc antibodies specific for Stargazin, and anti-PSD95 and anti-HA antibodies were used to blot. (G, Left) Quantification of band intensities for Ch-PSD95-V and C3,5S-Ch-PSD95-V that coprecipitated with NMDAR subunits from four separate Western blots. Band intensities are plotted as the percentage of wild-type (WT) PSD95, with Ch-PSD95-V set at 100%. The mean value for C3,5S-Ch-PSD95-V band intensities was 11.6 ± 2.3% (SEM), with *P < 0.001. (G, Right) Quantification of band intensities for Ch-PSD95-V and C3,5S-Ch-PSD95-V that coprecipitated with AMPAR subunits from four separate Western blots. Band intensities are plotted as the percent of WT PSD95, with Ch-PSD95-V set at 100%. The mean value for C3,5S-Ch-PSD95-V band intensities was 2.23 ± 0.5% (SEM), with *P < 0.001.
Fig. S4.
Loss of PSD95 palmitoylation alters coprecipiatation and colocalization with NMDAR subunits. (A) Treatment with 2-BP prevents colocalization between NMDAR subunits and PSD95. HEK293 cells expressing HA-NMDAR and PSD95-GFP were untreated (Left) or treated with 2-BP (Right). PSD95 was visualized via its GFP tag, and immunostaining with anti-HA antibody was used to visualize NMDARs. (Scale bars: 10 μm.) (B) PSD95 C3,5S mutation reduces PSD95 interactions with GluN2B. To assay PSD95-NMDAR interactions in a cell system, PSD95-GFP, wild-type (WT), or C3,5S mutant was coexpressed (Co-Exp) with the NMDAR subunit, GluN2B, in COS7 cells. To assay in vitro interactions, PSD95-GFP and GluN2B were expressed separately in two sets of HEK293 cells and their lysates were mixed postsolubilzation (P.Sol). PSD95 constructs in cell lysates were immunoprecipitated with anti-GFP antibody and analyzed on Western blots with anti-GluN2B and anti-PSD95 antibodies. Displayed are representative Western blots. IP, immunoprecipitation. (C) PSD95 C3,5S mutation inhibits PSD95 interactions with NMDARs. PSD95-GFP (WT or C3,5S mutant) and NMDAR subunits, GluN2B and HA-GluN1, were coexpressed in HEK293 cells. Lysates were prepared, and PSD95 constructs were immunoprecipitated with anti-GFP antibody and probed with anti-GluN2B, anti-HA, and anti-PSD95 antibodies. Displayed are representative Western blots. (D and E) Palmitoylation inhibitor, 2-BP, blocks PSD95 interactions with GluN2B subunits. (D) PSD95-GFP was expressed in HEK293 cells and treated with 2-BP (100 μM), and lysates were prepared and mixed with lysates from cells expressing GluN2B. (E) Alternatively, PSD95-GFP and GluN2B were coexpressed, and cells were treated with 2-BP. Lysates were processed as described in B. Displayed are representative Western blots. (F) PSD95 and NMDAR interactions are inhibited by 2-BP. PSD95-GFP and NMDARs were coexpressed in HEK293 cells and treated with 2-BP (100 μM), and lysates were prepared as described in B. Displayed are representative Western blots. (G) Quantification of band intensity changes for coprecipitation of PSD95 or C3,5S-PSD95 with NMDAR subunits. Quantification of effects of C3,5S mutation or 2-BP treatment (100 μM) on interaction between PSD95 and GluN2B/NMDARs. The C3,5S mutation caused GluN2B and PSD95 coimmunoprecipitation (Co-IP) to decrease to 15.2 ± 1.4% when the interaction occurs in vitro and to 15.8 ± 1.5% when the interaction occurs in cells. The 2-BP treatment decreased the co-IP to 15.0 ± 0.4% (in vitro) and to 34.3% ± 11.4% (in cells). For NMDAR–PSD95 interactions, the C3,5S mutation decreased co-IP to 15.8 ± 0.3% and 2-BP treatment decreased co-IP to 34.2 ± 5.8%. Data are expressed as mean ± SEM for four experiments per condition.
To examine the conformation of PSD95 where interactions with the different receptor subunits occur, we measured Ch-PSD95-V FRET efficiency in the PSD95 clusters colocalized with glutamate receptor subunits (Fig. 2C). In puncta where Ch-PSD95-V and GluN2B-containing NMDARs in HEK293 cells colocalized, the Ch-PSD95-V FRET efficiency was reduced from 18.0% to 1.8% (Fig. 2C), significantly lower than when coexpressed with DHHC15 or DHHC2, or when treated with palmostatin B (Fig. 1). These conditions were similar to conditions where PSD95 appears to be most highly palmitoylated, that is, when in puncta coexpressed with DHHC2 and treated with palmostatin B. Similarly, when in puncta or clusters where Ch-PSD95-V and Stargazin-containing AMPARs colocalized, the Ch-PSD95-V FRET efficiency was reduced from 18.0% to −0.9% (Fig. 2C). At such low FRET efficiencies, we found that Ch-PSD95-V is both in an extended conformation and highly palmitoylated (Fig. 1). The low FRET efficiencies at clusters where PSD95 colocalizes with AMPARs or NMDARs thus suggest that the interactions between PSD95 and AMPARs and NMDARs are regulated by PSD95 palmitoylation, parallel to its changes in conformation.
To determine if the PSD95 localized to clusters containing AMPARs or NMDARs represents a palmitoylated pool, we used colocalization with the PF11 intrabody as a visual assay (Fig. 2D). PF11-GFP, which specifically recognizes palmitoylated PSD95, but not depalmitoylated PSD95 (23), was coexpressed with wild-type PSD95 and either GluN2B-containing NMDARs or Stargazin-containing AMPARs in HEK293 cells. PF11-GFP colocalized with NMDAR subunits, demonstrating the presence of palmitoylated PSD95 in PSD95-NMDAR clusters (Fig. 2D). Similarly, we observed colocalization in intracellular clusters between PF11-GFP and Stargazin-containing AMPARs (Fig. 2D). These data provide further evidence that PSD95 associated with either receptor subtype is specifically palmitoylated.
To test directly whether palmitoylation of PSD95 regulates the associations between PSD95 and NMDARs or AMPARs, we performed a series of coimmunoprecipitation experiments. Different combinations of PSD95 and NMDAR or AMPAR subunits expressed in HEK293 cells were used to compare results for PSD95 and C3,5S-PSD95, which cannot be palmitoylated. Detailed coimmunoprecipitation experiments were first performed for PSD95 or C3,5S-PSD95, and for GluN1 and/or GluN2B subunits. Surprisingly, we observed the same levels of coimmunoprecipitation between PSD95 and NMDAR subunits whether PSD95 was coexpressed with the NMDAR subunits in the same cells before solubilization or if the two components were expressed in a separate set of cells and the lysates were mixed before coimmunoprecipitations were performed (Fig. S4 B–G). Similar results with coimmunoprecipitations were obtained with PSD95 when it was expressed alone in HEK cells and the solute mixed with either the solute of cells expressing GluN2B alone or together with GluN1.
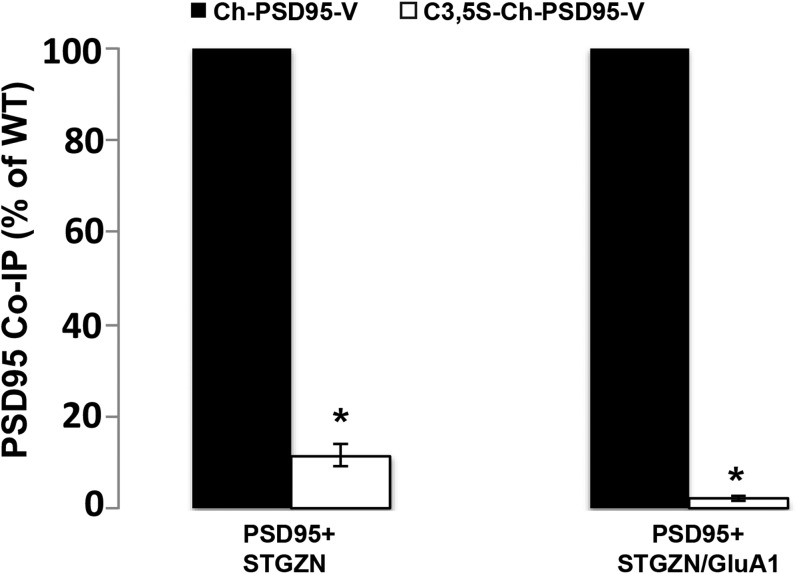
When palmitoylation-deficient C3,5S-PSD95 was substituted for PSD95, we no longer observed colocalization between PSD95 and NMDARs in HEK293 cells (Fig. 2B) nor could we coimmunoprecipitate C3,5S-PSD95 with GluN2B or with GluN2B-containing NMDARs (Fig. 2 E and G and Fig. S4 B and C). Similar results were obtained in a different study using GluN2A substituted for GluN2B (35). Thus, the association between PSD95 and NMDARs requires PSD95 N-terminal palmitoylation. We further tested this property using the palmitoylation inhibitor, 2-BP, which disrupted PSD95 coimmunoprecipitation with NMDAR subunits (Fig. S4) and prevented GluN2B-containing NMDARs from colocalizing with PSD95 (Fig. S4). We found a similar requirement for PSD95 N-terminal palmitoylation and its association with the TARP, Stargazin, associated with the AMPAR subunit GluA1 (Fig. 2 F and G). We observed a 33% decrease in the amount of PSD95 that immunoprecipitated with Stargazin alone versus Stargazin complexed with GluA1 (Fig. S5). This discrepancy is likely due to differences in the stoichiometry of PSD95 in the complexes formed with Stargazin alone versus Stargazin and GluA1. Importantly, both the Stargazin and Stargazin–GluA1 complexes coprecipitate much more with PSD95 possessing intact palmitoylation sites than with C3,5S-PSD95 lacking its palmitoylation sites. We conclude that PSD95 must be palmitoylated to stabilize interactions with NMDARs through its GluN2B subunit and with AMPARs through TARPs.
Fig. S5.
Further analysis of the role of PSD95 palmitoylation in regulating interactions with AMPARs. Additional quantification for the experiments displayed in Fig. 2F, which demonstrated that loss of PSD95 palmitoylation sites disrupted interactions between PSD95 and AMPARs containing Stargazin. Band intensities from four separate Western blots were used in the analysis and are plotted as the percentage of WT PSD95, with Ch-PSD95-V set at 100% when immunoprecipitated with Stargazin alone. The mean value for C3,5S-Ch-PSD95-V band intensities was 22.8 ± 4.5% (SEM) with Stargazin alone, and the mean values for Ch-PSD95-V and C3,5S-Ch-PSD95-V were 67.3 ± 10.3% and 1.5 ± 0.5%, respectively (*P < 0.001), when immunoprecipitated with Stargazin and GluA1.
PSD95 and SAP97 Have Similar Conformations but Different Orientations in PSDs.
PSD95 is highly palmitoylated in PSDs (11) and appears to be primarily oriented in an extended conformation perpendicular to the plane of the postsynaptic membrane (2). When expressed in cultured hippocampal neurons, Ch-PSD95-V, but not C3,5S-Ch-PSD95-V, was observed at PSDs as assayed by costaining with presynaptic Bassoon (Fig. 3A and Fig. S1). When FRET measurements were performed on Ch-PSD95-V outside of synapses, the FRET efficiency was high, 17.9% (Fig. 3B), identical to what was measured in HEK293 cells (Fig. 1B). For Ch-PSD95-V puncta at synapses, the FRET efficiency was low, 4.5% (Fig. 3B), very similar to the values obtained for Ch-PSD95-V in HEK293 cells in puncta formed when coexpressed with DHHC2 or when treated with palmostatin B (Fig. 1B). However, the FRET efficiency of the Ch-PSD95-V synaptic puncta was significantly higher than the values obtained when Ch-PSD95-V colocalized with Stargazin-containing AMPARs or GluN2B-containing NMDARs in HEK cells (Fig. 2C). The Ch-PSD95-V FRET efficiency outside of synapses indicates that nonsynaptic PSD95α is mostly in the compact conformation, and is likely not palmitoylated. At synapses, largely integrated in PSDs, the Ch-PSD95-V FRET efficiency indicates that most, but not all, of PSD95 is in the extended conformation and palmitoylated.
Using a FRET sensor of SAP97 conformation with the same FRET pair (Ch-SAP97-V), measurements of synaptic and extrasynaptic SAP97 conformations were obtained (Fig. 3B). When expressed in cultured hippocampal neurons, Ch-SAP97-V at PSDs was marked by costaining with presynaptic Bassoon (Fig. 3A). When FRET measurements were performed on Ch-SAP97-V outside of synapses, the FRET efficiency was high, 13%, similar to what we had measured when expressed alone in HEK293 cells (27). For Ch-SAP97-V puncta at synapses, the FRET efficiency was low, 5%, very similar to the values obtained for Ch-SAP97-V in HEK293 cells in puncta formed when coexpressed with CASK or NMDARs (27). The Ch-SAP97-V FRET efficiency outside of synapses indicates that a sizable pool of nonsynaptic SAP97 is mostly in the compact conformation, and is likely not associated with CASK or NMDARs, which associated with SAP97 in the extended conformation (27). At synapses, the Ch-SAP97-V FRET efficiency indicates that most, but not all, of SAP97 is in the extended conformation.
As a more direct measurement of PSD95 conformation, we used antibodies specific to mCherry (RFP) or Venus (YFP) to label the tags on Ch-PSD95-V expressed in cultured rat hippocampal neurons (Fig. 3C). We then used immuno-EM to measure the distance between the mCherry and Venus tags of Ch-PSD95-V in PSDs at spines, calculated from their individual distances to the postsynaptic membrane, and to visualize Ch-PSD95-V with EM tomography (Fig. 3 C–F). We found that immunogold labels with antibodies specific for N-terminal mCherry on PSD95 were located, on average, 18 nm from the postsynaptic membrane, whereas labels with antibodies specific for C-terminal Venus were located, on average, 28 nm away from the membrane (Fig. 3 D and F). The averaged ∼10-nm separation between the PSD95 mCherry and Venus sites confirms the distance between the two fluorophores based on the FRET estimates for Ch-PSD95-V at synapses (Eq. S2). It is also consistent with PSD95 being oriented as a vertical filament, perpendicular to the plane of the postsynaptic membrane, with the palmitoylated N-terminal tag at the membrane and the C-terminal tag further away. This orientation was directly visualized when we performed EM tomography using immunogold labeling to the C-terminal Venus tag of Ch-PSD95-V in PSDs (Fig. 3E). Ch-PSD95-V appeared as a vertical filament perpendicular to the plane of the postsynaptic membrane with immunogold particles at the distal ends of the vertical filaments (1, 2).
We performed immunogold EM on tagged SAP97 to estimate the distance between the mCherry and Venus tags of Ch-SAP97-V in PSDs (Fig. 3 C and D). In contrast to what we observed with Ch-PSD95-V, there was no significant difference between the distance of the immunogold-labeled N-terminal mCherry tag and the C-terminal Venus tag from the postsynaptic membrane; both immunogold-labeled termini were, on average, 18 nm away from the membrane (Fig. 3 C and D) even though the FRET efficiency was essentially the same as the FRET efficiency of Ch-PSD95-V. This apparent inconsistency between the PSD95 and SAP97 can only be explained if SAP97 is in the extended conformation but oriented parallel to the plane of the postsynaptic membrane at the PSD (Fig. 3F).
Increasing PSD95 Palmitoylation in PSDs Correlates with Increases in AMPAR but Not NMDAR Levels.
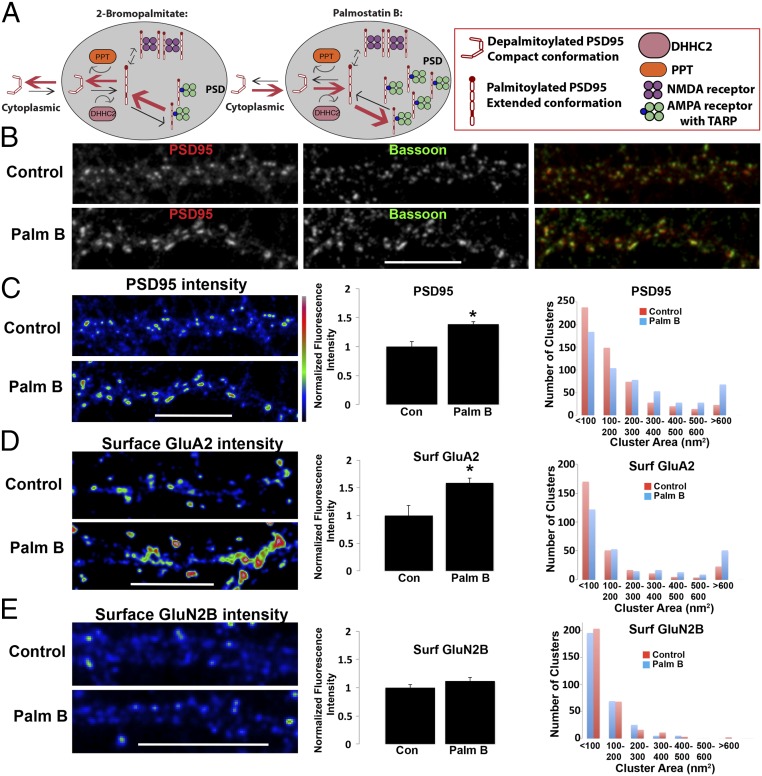
Because of our findings with HEK cells that palmitoylation regulates interactions with AMPARs and NMDARs (Fig. 2), we examined whether PSD95 palmitoylation regulates interactions with these receptors at cultured hippocampal neuronal synapses. The model in Fig. 4A is based on previous studies of PSD95 palmitoylation in neuronal cultures, which established that blockade of PSD95 palmitoylation with the inhibitor 2-BP (11, 23) reduced PSD95 and AMPAR levels but did not alter NMDAR levels at PSDs. Similar results were obtained by overexpressing C3,5S-PSD95, which cannot be palmitoylated (36). For PSD95 to be palmitoylated, it must interact with the PATs or DHHC proteins. DHHC2 is found primarily in dendrites or within PSDs (9, 23, 31). It palmitoylates PSD95 while in PSDs, and 2-BP acts largely by blocking DHHC2 in PSDs (23), allowing the unidentified PPT in PSDs to dominate and depalmitoylate PSD95 (23). As shown in the model, these findings also suggested that PSD95 depalmitoylation occurs at PSDs, and depalmitoylated PSD95 lacking its membrane anchor remains for some time in the PSD before exiting.
Fig. 4.
Synaptic PSD95 and surface AMPARs increase with PSD95 palmitoylation. (A, Left) Summary model of previous results with 2-BP treatment of rat hippocampal neuronal cultures and how PSD95, AMPAR, and NMDAR levels were affected. (A, Right) Prediction of how palmostatin B blockade of PPT will affect PSD95, AMPAR, and NMDAR levels based on the model. (B) Control (Top) and palmostatin B-treated (Bottom) cultures (4 h) at DIV17 were coimmunolabeled with anti-PSD95 (Left) and anti-Bassoon, a presynaptic marker (Center) antibodies. Merged channel images are displayed (Right). (C, Left) Intensity profile of PSD95 fluorescence signal in B. (C, Center) Quantification of endogenous PSD95 pixel intensities on dendrites of untreated control and palmostatin B-treated cultures in B. Data are shown as mean ± SEM; control cells, 100 ± 8%; palmostatin B cells, 139 ± 5% (n = 5–8 fields per group, *P < 0.02 relative to control). (C, Right) Histogram showing the distribution of PSD95 punctal size (area) on control and palmostatin B-treated dendrites (n = ∼545 puncta per group). (D, Left) Intact (nonpermeabilized) neurons (DIV17) were stained with an anti-GluA2 antibody to visualize surface AMPARs on control (Top) and palmostatin B-treated (Bottom) neurons. Displayed are intensity profiles of surface GluA2 fluorescence signal. (D, Center) Quantification of endogenous surface GluA2 staining on dendrites of untreated control and palmostatin B-treated cultures is shown. Data are shown as mean ± SEM; control cells, 100 ± 18%; palmostatin B cells, 159 ± 8% (n = 6–10 fields per group, *P < 0.01 relative to control). (D, Right) Histogram showing the distribution of surface GluA2 punctal size (area) on control and palmostatin B-treated dendrites (n = ∼280 puncta per group). (E, Left) Intact (nonpermeabilized) neurons (DIV17) were stained with an anti-GluN2B antibody to visualize surface GluN2B-containing NMDARs on control (Top) and palmostatin B-treated (Bottom) neurons. Displayed are intensity profiles of surface GluN2B fluorescence signal. (E, Center) Quantification of endogenous surface GluN2B staining on dendrites of untreated control and palmostatin B-treated cultures. Data are shown as mean ± SEM; control cells, 100 ± 5%; palmostatin B cells, 112 ± 6% [n = 8–10 fields per group, P = 0.08 (nonsignificant relative to control)]. (E, Right) Histogram showing the distribution of surface GluN2B punctal size (area) on control and palmostatin B-treated dendrites (n = ∼300 puncta per group).
Based on our results using palmostatin B in HEK cells and increasing PSD95 palmitoylation (Fig. 1), we further tested the model (Fig. 4A) by treating hippocampal cultures with palmostatin B to increase PSD95 palmitoylation in PSDs. We would expect, as shown in Fig. 4A, that palmostatin B should have the opposite effect of 2-BP with respect to PSD95 and receptor levels in PSDs. The levels of PSD95 (Fig. 4 B and C), cell-surface GluA2-containing AMPARs (Fig. 4D), and cell-surface GluN2B-containing NMDARs (Fig. 4E) were measured by immunostaining at spiny synapses. The staining intensities for each were measured for palmostatin B-treated and untreated conditions, as were the puncta size for each for the two conditions. As predicted in the model (Fig. 4A), the intensity and size of the puncta were significantly increased for PSD95 and AMPARs, but not for NMDARs. The trend toward a larger increase in levels observed for AMPARs compared with PSD95, although not significant, is predicted by the model because a portion of the PSD95 pool is bound to the NMDARs, and therefore is not changed with its palmitoylation. Thus, the results are consistent with the idea that when in PSDs, PSD95 is readily palmitoylated and depalmitoylated if associated with AMPARs, but in a more stable palmitoylated state if associated with NMDARs.
Discussion
Because of the different roles AMPARs and NMDARs play in synaptic plasticity, a major question is how AMPAR and NMDAR dynamics are regulated by PSD scaffolding molecules. Here, we address this question by examining how palmitoylation regulates the conformation of a major PSD scaffolding molecule, PSD95, as well as its interactions with AMPARs and NMDARs. We previously found that SAP97 association with AMPARs and NMDARs is dictated by its conformation. An intramolecular SAP97 FRET sensor was used to demonstrate that SAP97 is in either an extended or compact conformation in vivo, and that SAP97 binds GluA1-containing AMPARs in the compact conformation and GluN2B-containing NMDARs in the extended conformation (27). SAP97 is normally in the compact conformation, but binding of the CASK L27 domain to the SAP97 N-terminal L27 domain, or forming dimers of SAP97 through L27-L27 domain binding, changes SAP97 to an extended conformation. Thus, modification of the SAP97 N-terminal domain determines its conformation and whether it binds to AMPARs or NMDARs (27).
PSD95 is homologous to SAP97 except for its N-terminal domain, which has a palmitoylation site instead of an L27 domain. Using an intramolecular PSD95 FRET sensor, we demonstrate that, similar to SAP97, PSD95 is in either an extended or compact conformation in HEK cells, where PSD95 conformation is regulated by palmitoylation of the N-terminal domain. When palmitoylated, PSD95 is in the extended conformation; when depalmitoylated, it is in the compact conformation. Palmitoylated PSD95 binds GluN2B-containing NMDARs (Fig. 2E) like SAP97, but it also binds AMPARs through interactions with TARP subunits (Fig. 2F). These findings are consistent with a previous structural model based on EM tomography (1, 2, 37), where PSD95, as an extended filament, interacts with structures corresponding to NMDARs and AMPARs within PSDs.
Our results are relevant to understanding why there are different PSD-MAGUK isoforms and how they can compensate for each other in transgenic mouse knockouts (38–40). The nature of palmitoylation as a posttranslational modification explains, in part, the different orientations for PSD95 and SAP97 in PSDs. The PSD-MAGUK α-isoform splice variants of PSD95, SAP97, and PSD-93 all have N-terminal palmitoylation domains, and when palmitoylated, all would be anchored in the PSD membrane. Furthermore, palmitoylation would be expected to change their conformation from the compact to extended conformation as with PSD95 (Fig. 5B), resulting in the extended conformation perpendicular to the PSD membrane (Fig. 3F). The PSD-MAGUK β-isoform splice variants of PSD95, SAP97, and PSD-93 have an N-terminal L27 domain that replaces the palmitoylation domain. Lacking the ability to be palmitoylated, the β-isoform splice variants would not be expected to anchor their N termini to the plasma membrane. FRET data from the β-isoform of SAP97 in dissociated hippocampal neurons indicated that SAP97 exists as N termini-interacting dimers within PSDs (30). Intramolecular FRET data indicate that L27-L27–mediated SAP97 dimers are in the extended conformation, as opposed to a closed compact conformation (27). Altogether, it appears that SAP97 and the other PSD-MAGUK β-isoforms in PSDs are predominantly in the extended conformation as L27-L27–mediated, head-to-head dimers parallel to the plane of the membrane and that the L27-L27 interaction drives the conformational change from the compact to extended conformation. In this extended conformation, SAP97 interacts with NMDARs but not with AMPARs (27). Thus, in PSDs, we would expect that SAP97, and perhaps the other PSD-MAGUK β-isoforms, associate with NMDARs but not with AMPARs.
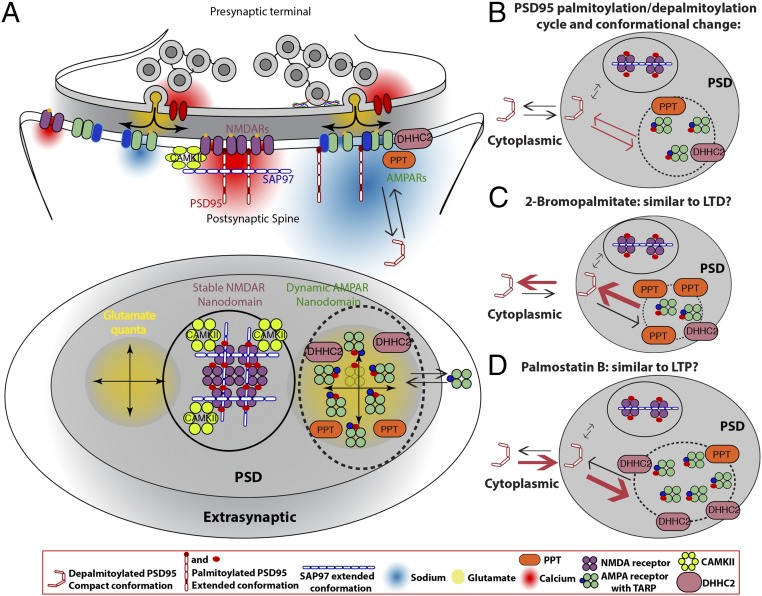
Fig. 5.
Model of PSD95 and SAP97 scaffolding separate AMPAR and NMDAR nanodomains and how the PSD95 palmitoylation cycle regulates AMPAR dynamics in PSDs. Based on our results and the work of others, we propose that AMPARs and NMDARs in PSDs are differentially scaffolded into separate nanodomains (NMDAR nanodomain, circle; AMPAR nanodomain, dotted circle). In the NMDAR nanodomains, stronger PSD95–SAP97 interactions stabilize NMDARs and prevent access of palmitoylation/depalmitoylation enzymes, DHHC2 and PPT. The more dynamic AMPAR nanodomain results from weaker interactions between PSD95 and TARP-containing AMPARs, and allows access of palmitoylation/depalmitoylation enzymes. (A) Lateral (Top) and en face (Bottom) schematic views of the presynaptic domain synapsing on the PSD) of a spine head. In the model, we propose functional rationales for why AMPARs and NMDARs cluster in separate nanodomains in PSDs. First, separate nanodomains increase receptor packing density, and thereby increase the sodium current density for AMPARs (red cloud) and calcium current density for NMDARs (yellow). Second, separate nanodomains allow strategic positioning of the nanodomains. As shown, positioning of the AMPAR nanodomain with respect to the presynaptic vesicle release site will regulate the size of the AMPAR response, whereas positioning of the NMDAR nanodomain with respect to downstream calcium response proteins will regulate intracellular calcium signaling at the PSD. Finally, separate domains allow other proteins to segregate in either AMPAR or NMDAR nanodomains. An example is shown in B–D for the palmitoylation of PSD95. (B–D) Details of how the palmitoylation cycle changes PSD95 conformation and its association with AMPARs and NMDARs. (B) PSD95 in the compact conformation attaches to the PSD and is stabilized there in the extended conformation when palmitoylated. In this conformation, PSD95 associates mainly with AMPAR subunits containing TARP subunits, such as Stargazin, by entering AMPAR nanodomains or, occasionally, with GluN2B NMDAR subunits in NMDAR nanodomains. The subunits dissociate from PSD95 when depalmitoylation reverses PSD95 conformation, changing back to the compact conformation. In this way, the PSD95 palmitoylation/depalmitoylation cycle regulates the number of AMPAR slots in AMPAR nanodomains. (C) We propose that the number of AMPAR slots, and thus the levels of AMPARs and PSD95, are decreased by decreasing PSD95 palmitoylation by DHHC2 blockade with 2-BP incubation. (D) Number of AMPAR slots, as well as the levels of AMPARs and PSD95, is increased by increasing PSD95 palmitoylation by PPT blockade with palmostatin B incubation.
PSD95 palmitoylation is more dynamic when PSD95 is associated with AMPARs and more stable when it is associated with NMDARs (Fig. 4). What accounts for the changes in PSD95 palmitoylation when it interacts with AMPARs but not with NMDARs? Observations by superresolution light microscopy show palmitoylated PSD95 and AMPARs clustered in nanodomains (23). EM tomography suggested that the NMDAR nanodomains contained NMDAR–PSD95 complexes in 1:2 stoichiometry and that other PSD regions with AMPARs, but without NMDARs, contained extended PSD95–AMPAR complexes in a 1:1 stoichiometry (2). When PSD95 was acutely knocked down, AMPAR nanodomains were lost, whereas NMDAR nanodomains in PSDs were relatively preserved, consistent with the NMDAR nanodomains being more stable (1, 41). The presence of separate AMPAR and NMDAR domains in PSDs provides a simple explanation for why PSD95 palmitoylation and depalmitoylation are different at AMPARs and NMDARs. By segregating the palmitoyltransferase, DHHC2, and PPT very close to or within AMPAR nanodomains, and away from NMDAR nanodomains (Fig. 5), the PSD95 palmitoylation cycle would be dynamically driven in the AMPAR nanodomain but slower in the NMDAR nanodomain. As proposed (Fig. 5), palmitoylated and extended PSD95 in AMPAR nanodomains interacts with AMPARs through the AMPAR TARP PDZ-binding domain with a stoichiometry of 1:1. This stoichiometry could contribute to more dynamic AMPAR scaffolding in the AMPAR nanodomain. In NMDAR nanodomains, palmitoylated and extended PSD95 interacts with NMDARs through their GluN2B subunit PDZ-binding domain with a 2:1 stoichiometry. We propose that the higher stoichiometry of NMDAR–PSD95 interactions allows the dimeric, extended SAP97 to bind to NMDARs via interactions between SAP97 and PSD95, contributing to a lattice that serves to stabilize NMDAR within nanodomains separate from AMPAR nanodomains.
We propose that changes in PSD95 palmitoylation, depalmitoylation, and conformation occur independent of interaction with AMPARs or NMDARs (Fig. 5B). The alternative, that these processes occur while PSD95 is associated with the receptors, is also possible, but is not shown in the model. Supporting the notion that PSD95 and SAP97 both interact with NMDARs are the findings that PSD95 and SAP97 in brain homogenate can interact in a domain-specific manner (42) and that SAP97 is complexed with NMDARs (43). However, simultaneous PSD95, PSD93, and SAP102 knockdown in a SAP97 knockout mouse line did not further decrease NMDAR currents (44), suggesting that SAP97 associations with NMDARs in PSDs are not essential. Alternatively, without PSD95, PSD93, and SAP102, loss of SAP97 may have no effect because PSD95, PSD93, and/or SAP102 is required for SAP97–NMDAR interactions.
A number of functional advantages for clustering of AMPARs and NMDARs in separate nanodomains are apparent in the model displayed in Fig. 5. First, separate nanodomains increase the packing density of the receptors, and thereby increase the density of local sodium current at AMPARs and calcium current density at NMDARs. Second, separate nanodomains allow strategic positioning of receptors with respect to other synaptic domains, for instance, positioning clusters of NMDARs closer to or further away from intracellular, downstream, calcium-dependent effectors to regulate the effects of the intracellular calcium influx (45). Because of the lower affinity of AMPARs for glutamate (46), the positioning of AMPAR nanodomains close to or away from the glutamate release sites could regulate the level of AMPAR activation (21, 45). Increased AMPAR packing density, aligned with the presynaptic release sites, could significantly increase or decrease AMPAR synaptic currents without adding or subtracting AMPARs from the PSD. Third, different signaling molecules, as well as posttranslational modifying enzymes, could segregate with either AMPAR or NMDAR nanodomains, as shown for the palmitoylation enzymes in the AMPAR nanodomains and for CAMKII in the NMDAR nanodomains. Separation of modifying and signaling proteins could play a critical role in regulating trafficking in the PSD, so that the rate of AMPAR/NMDAR entry and exit, and other proteins into the domains, can be separately regulated, especially during plasticity events, such as long-term potentiation (LTP) and long-term depression (LTD). Evidence that separate PSD domains may have a role in synaptic plasticity includes the finding that treatment of cultures with 2-BP has an effect similar to LTD, whereas palmostatin B treatment has an effect similar to LTP (Fig. 5 C and D). Effects include the change in the size of the PSD with LTP and LTD, which we assume reflects the PSD95 cluster size (Fig. 4C), and changes in the ratio of the levels of AMPARs to NMDARs (Fig. 4 D and E).
Experimental Procedures
Culture of HEK293 Cells and Primary Hippocampal Neurons.
HEK293 cells were maintained in DMEM supplemented with 10% (vol/vol) calf serum (HyClone). Cells were transiently transfected with cDNA using a calcium phosphate protocol (47). Hippocampal cultures were prepared using neurobasal media, 2% (vol/vol) B27, and 2 mM l-glutamine. Briefly, hippocampi from embryonic day (E) 18 to E19 Sprague–Dawley rats were dissected and dissociated in 0.05% trypsin (vol/vol; Invitrogen), and cells were plated at a density of ∼4 × 105 cells per millimeter on polyethylenimine-coated, 12-mm coverslips. A detailed description of cell culture protocol and reagents is provided in SI Experimental Procedures.
Electron Microscopy.
Transfection of 3-wk-old rat hippocampal cultures with FRET constructs using the Clontech CalPhos Mammalian Transfection Kit was followed by 40–44 h of incubation at 35 °C. For immuno-EM, these transfected neurons were fixed in 4% (vol/vol) paraformaldehyde in 0.1 M phosphate buffer at pH 7.4 for 45 min, Cultures were washed with buffer, permeabilized with 0.1% saponin, and blocked with 5% (vol/vol) normal goat serum in PBS for 1 h. They were then incubated with the primary antibody for 1 h, washed, incubated with the secondary antibody conjugated to 1.4-nm gold (Nanogold; Nanoprobes) for 1 h, washed again, and fixed with 2% (vol/vol) glutaraldehyde in PBS. Samples were silver-enhanced for 5–10 min (HQ Silver Enhancement Kit; Nanoprobes), treated with 0.2% osmium tetroxide in buffer for 30 min and then with 0.25% uranyl acetate overnight, washed, dehydrated in ethanol, and finally embedded in Epon. No specific labeling was present at PSDs when primary antibody was omitted from the protocol. Antibodies used were against mCherry, an RFP (rabbit polyclonal, 1:500; MBL), and against Venus, an YFP (mouse monoclonal, 1:500; Invitrogen). For immuno-EM, conventional thin EM sections were cut, grids were unselectively sampled, and images were collected of all synapses encountered in a JEOL-200CX transmission electron microscope with a bottom-mounted CCD camera (Advanced Microscopy Techniques Corp.). Immunogold particles in the immediate proximity of PSDs were analyzed, and the distance from the center of each particle to the postsynaptic membrane was measured with ImageJ (NIH). For EM tomography, sections 80 nm thick were mounted on Formvar-coated, 200-mesh, copper/nickel grids, and ∼3 nm of carbon was evaporated to stabilize the grid. Gold particles (∼10 nm) were applied to both sides of the grid as fiducial markers. Sections were scanned to identify spines for collecting dual-axis EM tomography series on an FEI Tecnai 300 kV transmission electron microscope with a field emission gun and bottom-mounted CCD camera at a dose of ∼2,000 electrons per square nanometer per series. After the first series was acquired, the grid was rotated 90° and a second series was taken. Tilt increments were 2°, extending from +70° to −70°, and pixel sizes were 0.7 nm (2,048 × 2,048 image). Dual-axis image series were reconstructed by back-projection, and the 3D volume data were merged with IMOD (48). Fine alignment error was typically less than 0.2 pixel. The 3D volume data (tomogram) were analyzed and interpreted with EM3D (49), and segmented and surface-rendered with Amira (FEI). Segmentation, measurements, and surface rendering were performed as previously described (2, 37).
A detailed description of additional methods, reagents, imaging protocols, and calculation of FRET efficiencies is provided in SI Experimental Procedures.
SI Experimental Procedures
Culture of HEK293 Cells and Primary Hippocampal Neurons.
HEK293 cells were maintained in DMEM supplemented with 10% (vol/vol) calf serum (HyClone). Cells were transiently transfected with cDNA using a calcium phosphate protocol (47). Cells transfected with HA-GluR1 or HA-NMDAR were maintained in media containing 1 mM kynurenic acid (Sigma) or 100 M D(–)-2-Amino-5-phosphonopentanoic acid (Sigma-Aldrich) and 10 M MK-801 (RBI), respectively, to prevent excitotoxicity. Hippocampal cultures were prepared using neurobasal media (NBM), 2% (vol/vol) B27, and 2 mM l-glutamine. Briefly, hippocampi from E18–E19 Sprague–Dawley rats were dissected and dissociated in 0.05% trypsin (Invitrogen), and cells were plated at a density of ∼4 × 105 cells per millimeter on polyethylenimine-coated, 12-mm coverslips. Coverslips were maintained in NBM containing B27 and GlutaMAX (all from Invitrogen). Neuronal cultures were transfected at 12–14 d in vitro with the Lipofectamine 2000 transfection reagent (Invitrogen), according to the manufacturer’s recommendations, with the exception that 1–2.5 g of each cDNA in 62.5 μL of NBM and 2.5 μL of Lipofectamine 2000 in 62.5 μL of NBM were mixed and added to coverslips in 12-well plates.
Antibodies and Reagents.
The following antibodies were used: anti-Bassoon antibody (Synaptic Systems), anti-PSD95 antibodies (Stressgen or Neuromab), anti-NR2B antibody (BD Biosciences), anti-GluA1 antibody (Calbiochem), anti-GluA2 antibody (EMD Millipore), anti-HA (rabbit polyclonal) antibody (Bethyl Laboratories), anti-Myc 9E10 antibody (Santa Cruz Biotechnology), and anti-GFP antibody (Sigma-Aldrich). Alexa Fluor anti-rat 488/568, Alexa Fluor anti-rabbit 647, Alexa Fluor anti-rabbit 750, Alexa Fluor anti-mouse 647, and Alexa Fluor anti-mouse 750 were purchased from Molecular Probes.
Plasmids and Transfections.
Rat GluN1 and GluN2B were obtained from J. Boulter, University of California, Los Angeles). GluN1 and GluN2B subunits were tagged at the NH2 terminus with the FLAG (DYKDDDDK) and HA (YPYDVPDYA) epitopes, respectively, using the extension overlap PCR method (50), and subcloned into a pCB6 mammalian expression vector. Construction of GFP-SAP97β (splice variants I1b, I3, and I5) has been previously described (51, 52). To generate Ch-SAP97-V, EcoRI restriction enzyme sites were placed on the N and C termini of mCherry and KpnI sites were placed on the N and C termini of Venus using PCR. Using EcoR1 and KpnI, mCherry and/or Venus was subcloned into SAP97β in pEGFP-N1 and screened for correct directional insertion. To generate Ch-PSD95-V, Cherry-PSD95, and PSD95-Venus, EcoR1 sites were place on either end of mVenus (in pCS2) and subcloned in-frame by restriction digest into the EcoR1 site in PSD95-GFP (in pGW1) to generate PSD95-Venus. A KpnI site was introduced within the N terminus of PSD95α by a C-to-G point mutation to base pair 39 using Quikchange (Invitrogen). PCR was used to attach KpnI sites to the ends of mCherry, and mCherry was subcloned into PSD95-V or PSD95α to generate Ch-PSD95 or Ch-PSD95-V, respectively. C3,5S mutations were introduced by G-to-C point mutations in residues 3 and 5 using Quikchange. The cDNA encoding PF11-GFP (PSD95 intrabody) was obtained from M. Fukata, National Institute for Physiological Sciences, Okazaki, Japan, and F. Perez, Institut Curie Centre de Recherche, Paris.
Immunostaining and Image Analysis.
HEK293 cells and neuronal cultures (24 h posttransfection) were washed twice in PBS at 22–25 °C, fixed in 4% paraformaldehyde (PFA)/sucrose (vol/vol, room temperature for 15 min) and washed three times in PBS (5–10 min). For permeabilization, cells were incubated in 0.1% Triton-X in PBS (10 min), incubated in blocking solution [2% (wt/vol) glycine, 2% (wt/vol) BSA, 0.2% gelatin, and 50 mM NH4Cl in 1 PBS; 10 min], and then incubated with the indicated primary antibody diluted in blocking solution (1 h). For surface labeling, the permeabilization step was omitted and primary antibody incubation was overnight at 4 °C. Following primary antibody incubation, cells were washed three times in blocking solution (5–10 min) and overlaid with an appropriate secondary antibody diluted in blocking solution (1 h). Cells were then washed three times in PBS (5–10 min), and the slips were mounted in 80% glycerol or Prolong Gold (Invitrogen). Fluorescence images were acquired using a Leica SP5 Tandem Scanner Spectral 2-Photon scanning confocal microscope (Leica Microsystems) or Marianas Yokogawa type spinning disk confocal microscope with a back-thinned EMCCD camera. Images were processed using ImageJ (NIH) and Adobe Photoshop software. Surface expression of endogenous GluA2 and GluN2B and synaptic PSD95 was quantified using “sum of slice” z projections of images. Each field was background-subtracted, and mean intensities were calculated. Thresholded punctal size was analyzed using the Analyze Particle function in ImageJ.
Calculation of FRET Efficiency.
Images were acquired using a Leica SP5 laser scanning confocal microscope operating with an argon laser tuned to 514 nm, a diode-pumped solid state (DPSS) laser tuned to 561 nm, and a HeNe laser line tuned to 633 nm. Cells were examined with a 63× 1.4-N.A. (Leica) oil immersion objective and 4× zoom. FRET was measured using the acceptor photobleaching method (53). Cells were bleached in the mCherry channel by scanning a region of interest (ROI) five to 10 times using the 561-nm DPSS laser line at 100% intensity. The time of bleach ranged from 3 to 10 s depending on the size of the ROI for bleaching. For HEK293 cells, a scanning speed of 800 Hz was used to maximize the degree of acceptor bleaching at each step. For neurons, a scanning speed of 8,000 Hz was used. Before and after each bleach step, Venus images were collected to assess changes in donor fluorescence. Because increases in Venus fluorescence caused by bleaching of the mCherry acceptor could potentially be masked by bleaching of Venus related simply to the imaging process itself, the effect of photobleaching due to imaging was minimized by collecting Venus images at 1% of the laser intensity. To ensure that bleaching due to imaging was minimal, we monitored the level of bleaching in each experiment by collecting five images of the donor (Venus) before bleaching. The gain of the photomultiplier tubes was adjusted so as to eliminate cross-talk and to achieve the best possible dynamic range.
To calculate the FRET energy transfer efficiency (ET), we used the formula
| [S1] |
where Ida and Id are the fluorescence intensities of the donor in the presence and absence of acceptor, respectively. To obtain these values, fluorescence intensities were analyzed using ImageJ software and donor fluorescence variations were plotted against acceptor depletion over successive bleaching steps. The slope of the linear relationship was used to determine Ida and Id. As a reference, fluorescence of analogous peptides with only a donor, only an acceptor, or both were measured to control for both pseudo-FRET artifacts and intermolecular FRET. The average distance (r) between donor and acceptor was calculated using the formula
| [S2] |
where R0 is the Förster distance corresponding to 57 Å for our Venus and mCherry sensors. This calculation assumes a random orientation factor (κ2 = 2/3). To prepare HEK293 cells for FRET analysis, cells were fixed in 4% (vol/vol) PFA/(wt/vol) sucrose, washed in PBS, and mounted in 80% (vol/vol) glycerol. To define ROIs with cell markers (i.e., AMPARs, NMDARs, presynaptic markers), immunolabeling was performed with primary antibody, washed three times, and labeled with an Alexa 647- or Alexa 750-conjugated secondary antibody, which was imaged on the Far Red or Far Far Red channel at the end of a photobleaching protocol. Images of these markers were used to create a mask to isolate regions for FRET analysis. Argons lasers (514 nm and 568 nm) were used to excite Venus and mCherry, respectively (spectral channel settings: donor, 535 nm ± 10 nm; acceptor, 610 nm ± 25 nm). Low resonant scanning at 800 Hz was used in HEK293 cell experiments, whereas high resonant scanning at 8,000 Hz was used for neuron experiments.
Immunoprecipitation and Immunoblot Analysis.
Cells were pelleted by brief centrifugation, resuspended, washed once with PBS, and solubilized in lysis buffer [150 mM NaCl, 5 mM EDTA (pH 7.4), 50 mM Tris (pH 7.4), 0.02% NaN3] containing 1% Triton X-100 plus NEM (2 mM), phenylmethanesulphonylfluoride (2 mM), leupeptin (10 g⋅mL−1), Nα-Tosyl-L-lysine chloromethyl ketone hydrochloride (10 g⋅mL−1), chymotrypsin (10 g⋅mL−1), and pepstatin (g⋅mL−1). Following a 1-h solubilization (4 °C), samples were centrifuged at 14,000 × g for 30 min at 4 °C. Subsequent analyses were performed using the Triton X-100–soluble fraction. Immunoprecipitations were performed by overnight antibody incubation at 4 °C. Protein–antibody complexes were isolated by incubation with Protein G-Sepharose for 3 h at 4 °C. For immunoblotting, proteins separated by SDS/PAGE were transferred to PVDF membranes. After transfer, the PVDF membrane was blocked with 3% (wt/vol) milk in wash buffer [10 mM Tris (pH 7.4), 0.05% Tween 20, and 150 mM NaCl]. Membranes were washed briefly in wash buffer and then incubated for 1 h with primary antibodies. The blots were washed and incubated with secondary antibody (goat antibody to mouse, rabbit, or chicken Alexa 488, 568, or 647) at the appropriate dilution for 1 h. After washing, membranes were imaged on a Bio-Rad Molecular Imager Pharos-FX (Biorad). Quantification was done using ImageJ.
PSD95 Palmitoylation.
The acyl-biotin exchange method was performed as previously described (54). Click chemistry was performed with the alkyne-containing palmitate analog, 17-octadecynoic acid (17-ODYA), which was reacted with biotin-azide as previously described (32, 55). Cells were transfected with C3,5S-Ch-PSD95-V or Ch-PSD95-V alone, or with Ch-PSD95-V and Myc-DHHC2 (and palmostatin B treatment). At 24 h following transfection, cells were deprived of fatty acids by incubation in media supplemented with charcoal-treated FBS (Sigma-Aldrich) for 1 h before labeling. The 17-ODYA and palmitic acid were dissolved in DMSO to generate 100 mM stock solutions. To facilitate cellular uptake of fatty acids, before labeling, fatty acids were saponified by incubation with a 20% (wt/vol) molar excess of potassium hydroxide at 65 °C for 15 min. The saponified fatty acids were then dissolved in prewarmed, serum-free culture media containing 20% (wt/vol) fatty acid-free BSA at 37 °C, followed by an additional 30-min incubation at 37 °C. After deprivation of fatty acids, cells were washed with warm PBS and incubated in fresh media without supplement. BSA-conjugated fatty acids were added to cells in serum-free media (300 μL of fatty acid-BSA to 3 mL of media). Cells were incubated with fatty acids for 5 h at 37 °C to allow for metabolic labeling of proteins that are palmitoylated during the labeling period. Following labeling, cells were washed three times with PBS and then lysed in EDTA-free radioimmunoprecipitation assay (RIPA) buffer. PSD95 was subsequently immunoprecipitated using anti-PSD95 antibody (Neuromab). Next, beads were reacted with biotin azide under standard click chemistry conditions, where EDTA-free RIPA buffer was mixed with 0.1 mM biotin azide, 0.1 mM TBTA (Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine), 1 mM TCEP (Tris(2-carboxyethyl)phosphine hydrochloride), and 1 mM CuSO4. The click reaction mix was incubated with the beads in the dark at room temperature for 30 min. Proteins were eluted and processed for SDS/PAGE and immunoblot analysis.
Statistical Analysis.
Results are expressed as mean ± SEM of n samples unless stated otherwise. Statistical significance was assessed by a two-tailed Student’s t test or ANOVA/Tukey post hoc test as appropriate.
Acknowledgments
We thank Drs. Simon Alford and Nicolas Brunel for extensive discussions and help with the model proposed in Fig. 5 and Dr. Anitha Govind for her critical reading and helpful comments on the manuscript. We thank Drs. Vytas Bindokas and Paul Selvin for their technical support with FRET experiments. We also thank Drs. Christine Winters, Rita Azzam, Susan Cheng, and Virginia Crocker for help on immuno-EM and Drs. Alioscka Sousa and Richard Leapman for help on EM tomography data acquisition. This work was supported by the NIH under Grants NS043782, MH081251, and DA019695 (to W.N.G.) and by National Institute of Neurological Disorders and Stroke intramural funds (to T.S.R.). This project was also supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences of the NIH through Grant UL1 RR024999 (to O.J.). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612963113/-/DCSupplemental.
References
- 1.Chen X, et al. PSD-95 is required to sustain the molecular organization of the postsynaptic density. J Neurosci. 2011;31(17):6329–6338. doi: 10.1523/JNEUROSCI.5968-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X, et al. Organization of the core structure of the postsynaptic density. Proc Natl Acad Sci USA. 2008;105(11):4453–4458. doi: 10.1073/pnas.0800897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu W. PSD-95-like membrane associated guanylate kinases (PSD-MAGUKs) and synaptic plasticity. Curr Opin Neurobiol. 2011;21(2):306–312. doi: 10.1016/j.conb.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X, et al. Mass of the postsynaptic density and enumeration of three key molecules. Proc Natl Acad Sci USA. 2005;102(32):11551–11556. doi: 10.1073/pnas.0505359102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lowenthal MS, Markey SP, Dosemeci A. Quantitative mass spectrometry measurements reveal stoichiometry of principal postsynaptic density proteins. J Proteome Res. 2015;14(6):2528–2538. doi: 10.1021/acs.jproteome.5b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chetkovich DM, et al. Postsynaptic targeting of alternative postsynaptic density-95 isoforms by distinct mechanisms. J Neurosci. 2002;22(15):6415–6425. doi: 10.1523/JNEUROSCI.22-15-06415.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elias GM, Nicoll RA. Synaptic trafficking of glutamate receptors by MAGUK scaffolding proteins. Trends Cell Biol. 2007;17(7):343–352. doi: 10.1016/j.tcb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 8.Lee S, Fan S, Makarova O, Straight S, Margolis B. A novel and conserved protein-protein interaction domain of mammalian Lin-2/CASK binds and recruits SAP97 to the lateral surface of epithelia. Mol Cell Biol. 2002;22(6):1778–1791. doi: 10.1128/MCB.22.6.1778-1791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noritake J, et al. Mobile DHHC palmitoylating enzyme mediates activity-sensitive synaptic targeting of PSD-95. J Cell Biol. 2009;186(1):147–160. doi: 10.1083/jcb.200903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craven SE, Bredt DS. Synaptic targeting of the postsynaptic density protein PSD-95 mediated by a tyrosine-based trafficking signal. J Biol Chem. 2000;275(26):20045–20051. doi: 10.1074/jbc.M910153199. [DOI] [PubMed] [Google Scholar]
- 11.El-Husseini Ael-D, et al. Synaptic strength regulated by palmitate cycling on PSD-95. Cell. 2002;108(6):849–863. doi: 10.1016/s0092-8674(02)00683-9. [DOI] [PubMed] [Google Scholar]
- 12.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269(5231):1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 13.Niethammer M, Kim E, Sheng M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J Neurosci. 1996;16(7):2157–2163. doi: 10.1523/JNEUROSCI.16-07-02157.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leonard AS, Davare MA, Horne MC, Garner CC, Hell JW. SAP97 is associated with the alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR1 subunit. J Biol Chem. 1998;273(31):19518–19524. doi: 10.1074/jbc.273.31.19518. [DOI] [PubMed] [Google Scholar]
- 15.Dakoji S, Tomita S, Karimzadegan S, Nicoll RA, Bredt DS. Interaction of transmembrane AMPA receptor regulatory proteins with multiple membrane associated guanylate kinases. Neuropharmacology. 2003;45(6):849–856. doi: 10.1016/s0028-3908(03)00267-3. [DOI] [PubMed] [Google Scholar]
- 16.Opazo P, Sainlos M, Choquet D. Regulation of AMPA receptor surface diffusion by PSD-95 slots. Curr Opin Neurobiol. 2012;22(3):453–460. doi: 10.1016/j.conb.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 17.Schnell E, et al. Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proc Natl Acad Sci USA. 2002;99(21):13902–13907. doi: 10.1073/pnas.172511199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bats C, Groc L, Choquet D. The interaction between Stargazin and PSD-95 regulates AMPA receptor surface trafficking. Neuron. 2007;53(5):719–734. doi: 10.1016/j.neuron.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 19.Kharazia VN, Weinberg RJ. Tangential synaptic distribution of NMDA and AMPA receptors in rat neocortex. Neurosci Lett. 1997;238(1-2):41–44. doi: 10.1016/s0304-3940(97)00846-x. [DOI] [PubMed] [Google Scholar]
- 20.Takumi Y, Ramírez-León V, Laake P, Rinvik E, Ottersen OP. Different modes of expression of AMPA and NMDA receptors in hippocampal synapses. Nat Neurosci. 1999;2(7):618–624. doi: 10.1038/10172. [DOI] [PubMed] [Google Scholar]
- 21.MacGillavry HD, Song Y, Raghavachari S, Blanpied TA. Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron. 2013;78(4):615–622. doi: 10.1016/j.neuron.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nair D, et al. Super-resolution imaging reveals that AMPA receptors inside synapses are dynamically organized in nanodomains regulated by PSD95. J Neurosci. 2013;33(32):13204–13224. doi: 10.1523/JNEUROSCI.2381-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukata Y, et al. Local palmitoylation cycles define activity-regulated postsynaptic subdomains. J Cell Biol. 2013;202(1):145–161. doi: 10.1083/jcb.201302071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lüscher C, et al. Role of AMPA receptor cycling in synaptic transmission and plasticity. Neuron. 1999;24(3):649–658. doi: 10.1016/s0896-6273(00)81119-8. [DOI] [PubMed] [Google Scholar]
- 25.Ehlers MD. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 2000;28(2):511–525. doi: 10.1016/s0896-6273(00)00129-x. [DOI] [PubMed] [Google Scholar]
- 26.Lin JW, et al. Distinct molecular mechanisms and divergent endocytotic pathways of AMPA receptor internalization. Nat Neurosci. 2000;3(12):1282–1290. doi: 10.1038/81814. [DOI] [PubMed] [Google Scholar]
- 27.Lin EI, Jeyifous O, Green WN. CASK regulates SAP97 conformation and its interactions with AMPA and NMDA receptors. J Neurosci. 2013;33(29):12067–12076. doi: 10.1523/JNEUROSCI.0816-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Craven SE, El-Husseini AE, Bredt DS. Synaptic targeting of the postsynaptic density protein PSD-95 mediated by lipid and protein motifs. Neuron. 1999;22(3):497–509. doi: 10.1016/s0896-6273(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 29.Topinka JR, Bredt DS. N-terminal palmitoylation of PSD-95 regulates association with cell membranes and interaction with K+ channel Kv1.4. Neuron. 1998;20(1):125–134. doi: 10.1016/s0896-6273(00)80440-7. [DOI] [PubMed] [Google Scholar]
- 30.Nakagawa T, et al. Quaternary structure, protein dynamics, and synaptic function of SAP97 controlled by L27 domain interactions. Neuron. 2004;44(3):453–467. doi: 10.1016/j.neuron.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 31.Fukata M, Fukata Y, Adesnik H, Nicoll RA, Bredt DS. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44(6):987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 32.Martin BR, Cravatt BF. Large-scale profiling of protein palmitoylation in mammalian cells. Nat Methods. 2009;6(2):135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, et al. Stargazin regulates synaptic targeting of AMPA receptors by two distinct mechanisms. Nature. 2000;408(6815):936–943. doi: 10.1038/35050030. [DOI] [PubMed] [Google Scholar]
- 34.Kim E, Sheng M. Differential K+ channel clustering activity of PSD-95 and SAP97, two related membrane-associated putative guanylate kinases. Neuropharmacology. 1996;35(7):993–1000. doi: 10.1016/0028-3908(96)00093-7. [DOI] [PubMed] [Google Scholar]
- 35.Dong YN, Waxman EA, Lynch DR. Interactions of postsynaptic density-95 and the NMDA receptor 2 subunit control calpain-mediated cleavage of the NMDA receptor. J Neurosci. 2004;24(49):11035–11045. doi: 10.1523/JNEUROSCI.3722-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. PSD-95 involvement in maturation of excitatory synapses. Science. 2000;290(5495):1364–1368. [PubMed] [Google Scholar]
- 37.Chen X, et al. Identifying individual scaffolding molecules in the postsynaptic density. Microsc Microanal. 2008;14(Suppl 2):1068–1069. doi: 10.1017/S1431927608085449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlüter OM, Xu W, Malenka RC. Alternative N-terminal domains of PSD-95 and SAP97 govern activity-dependent regulation of synaptic AMPA receptor function. Neuron. 2006;51(1):99–111. doi: 10.1016/j.neuron.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 39.Elias GM, Elias LA, Apostolides PF, Kriegstein AR, Nicoll RA. Differential trafficking of AMPA and NMDA receptors by SAP102 and PSD-95 underlies synapse development. Proc Natl Acad Sci USA. 2008;105(52):20953–20958. doi: 10.1073/pnas.0811025106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elias GM, et al. Synapse-specific and developmentally regulated targeting of AMPA receptors by a family of MAGUK scaffolding proteins. Neuron. 2006;52(2):307–320. doi: 10.1016/j.neuron.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 41.Chen X, et al. PSD-95 family MAGUKs are essential for anchoring AMPA and NMDA receptor complexes at the postsynaptic density. Proc Natl Acad Sci USA. 2015;112(50):E6983–E6992. doi: 10.1073/pnas.1517045112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai C, Li H, Rivera C, Keinänen K. Interaction between SAP97 and PSD-95, two Maguk proteins involved in synaptic trafficking of AMPA receptors. J Biol Chem. 2006;281(7):4267–4273. doi: 10.1074/jbc.M505886200. [DOI] [PubMed] [Google Scholar]
- 43.Frank RA, et al. NMDA receptors are selectively partitioned into complexes and supercomplexes during synapse maturation. Nat Commun. 2016;7:11264. doi: 10.1038/ncomms11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levy JM, Chen X, Reese TS, Nicoll RA. Synaptic consolidation normalizes AMPAR quantal size following MAGUK loss. Neuron. 2015;87(3):534–548. doi: 10.1016/j.neuron.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lisman J, Yasuda R, Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat Rev Neurosci. 2012;13(3):169–182. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clements JD, Lester RA, Tong G, Jahr CE, Westbrook GL. The time course of glutamate in the synaptic cleft. Science. 1992;258(5087):1498–1501. doi: 10.1126/science.1359647. [DOI] [PubMed] [Google Scholar]
- 47.Claudio T. Stable expression of heterologous multisubunit protein complexes established by calcium phosphate- or lipid-mediated cotransfection. Methods Enzymol. 1992;207:391–408. doi: 10.1016/0076-6879(92)07028-m. [DOI] [PubMed] [Google Scholar]
- 48.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 49.Harlow ML, Ress D, Stoschek A, Marshall RM, McMahan UJ. The architecture of active zone material at the frog's neuromuscular junction. Nature. 2001;409(6819):479–484. doi: 10.1038/35054000. [DOI] [PubMed] [Google Scholar]
- 50.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77(1):51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 51.Bresler T, et al. Postsynaptic density assembly is fundamentally different from presynaptic active zone assembly. J Neurosci. 2004;24(6):1507–1520. doi: 10.1523/JNEUROSCI.3819-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu H, Reuver SM, Kuhlendahl S, Chung WJ, Garner CC. Subcellular targeting and cytoskeletal attachment of SAP97 to the epithelial lateral membrane. J Cell Sci. 1998;111(Pt 16):2365–2376. doi: 10.1242/jcs.111.16.2365. [DOI] [PubMed] [Google Scholar]
- 53.Kenworthy AK. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods. 2001;24(3):289–296. doi: 10.1006/meth.2001.1189. [DOI] [PubMed] [Google Scholar]
- 54.Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 2004;36(2):276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- 55.Yap MC, et al. Rapid and selective detection of fatty acylated proteins using omega-alkynyl-fatty acids and click chemistry. J Lipid Res. 2010;51(6):1566–1580. doi: 10.1194/jlr.D002790. [DOI] [PMC free article] [PubMed] [Google Scholar]