Significance
Mutations in isocitrate dehydrogenase (IDH)1 and IDH2 contribute to malignant progression by producing the oncometabolite 2HG. In myeloid disorders, mutations at three positions in these genes are commonly observed, but in angioimmunoblastic T-cell lymphoma (AITL), IDH mutations are restricted to IDH2 arginine (R) 172. The complex microenvironment of AITL, where malignant T cells comprise a minority of the tumor, has made it difficult to evaluate the role of this mutation. Here, we provide clinical data showing that mutant IDH2 expression is restricted to malignant T cells and that 2HG may be a useful biomarker in AITL. In addition, using conditional knock-in mouse models, we find that only mutations at IDH2 R172 produce significant quantities of 2HG in lymphoid cells and alter lymphoid development.
Keywords: isocitrate dehydrogenase, AITL, lymphoma, 2-hydroxyglutarate, T cell
Abstract
Oncogenic isocitrate dehydrogenase (IDH)1 and IDH2 mutations at three hotspot arginine residues cause an enzymatic gain of function that leads to the production and accumulation of the metabolite 2-hydroxyglutarate (2HG), which contributes to the development of a number of malignancies. In the hematopoietic system, mutations in IDH1 at arginine (R) 132 and in IDH2 at R140 and R172 are commonly observed in acute myeloid leukemia, and elevated 2HG is observed in cells and serum. However, in angioimmunoblastic T-cell lymphoma (AITL), mutations are almost exclusively restricted to IDH2 R172, and levels of 2HG have not been comprehensively measured. In this study, we investigate the expression pattern of mutant IDH2 in the AITL tumor microenvironment and measure levels of 2HG in tissue and serum of AITL patients. We find that mutant IDH2 expression is restricted to the malignant T-cell component of AITL, and that 2HG is elevated in tumor tissue and serum of patients. We also investigate the differences between the three hotspot mutation sites in IDH1 and IDH2 using conditional knock-in mouse models. These studies show that in the lymphoid system, mutations in IDH2 at R172 produce high levels of 2HG compared with mutations at the other two sites and that lymphoid development is impaired in these animals. These data provide evidence that IDH2 R172 mutations may be the only variants present in AITL because of their capacity to produce significant amounts of the oncometabolite 2HG in the cell of origin of this disease.
Isocitrate dehydrogenase (IDH)1 and IDH2 are mutated in various malignancies, including gliomas, cholangiocarcinomas, chondrosarcomas, acute myeloid leukemias (AML) and other myeloid malignancies, and angioimmunoblastic T-cell lymphoma (1). Three hotspots are recurrently mutated via amino acid substitution at the IDH1R132, IDH2R140, and IDH2R172 arginine residues. These three mutations confer an enzymatic neoactivity to the IDH enzyme, which normally converts isocitrate to α-ketoglutarate (αKG), leading to the abnormal production of the D form of 2 hydroxyglutarate (2HG) (2, 3). 2HG is an oncometabolite that can competitively inhibit a large class of iron and αKG-dependent dioxygenases (4). It has been shown that 2HG can inhibit ten–eleven translocation (TET) proteins (5) and Jumonji (JMJ) family histone demethylases (6), resulting in epigenetic alterations to DNA and histone proteins that can impact cell differentiation in different systems. More widely, 2HG can inhibit other dioxygenases involved in a number of cellular processes including the hypoxia response (7–9) and collagen maturation (8).
Although these three IDH mutations all result in an abnormal increase in 2HG, differences in the distribution of these mutations among malignancies suggest that they may have different functional consequences. Indeed, IDH mutations in glioma are IDH1R132 in more than 90% of cases (10), whereas IDH mutations in AITL are present at IDH2R172 almost exclusively (11–14).
Angioimmunoblastic T-cell lymphoma (AITL) is one of the most frequent nodal peripheral T-cell lymphomas (PTCL) (15, 16). It preferentially affects the elderly and carries a poor prognosis, with a median survival of less than three years (15). Histologically, it is characterized by a unique tumor microenvironment consisting of reactive cells, hyperplastic postcapillary venules, and disruption of the follicular dendritic cell (FDC) network. These nonmalignant cells frequently account for up to 90% of the cell content. The small population of malignant cells display a T follicular helper (TFH) cell phenotype, and this population is thought to be the cell of origin for this disease (17). Recently, recurrent mutations in the epigenetic regulators TET2 (18, 19), IDH2 (11, 12), and DNA (cytosine-5)-methyltransferase 3A (DNMT3A) (20) as well as mutation of Ras homolog family member A (RHOA) at the G17V residue have been proposed as driver events in PTCL (13, 21, 22). Among these drivers, IDH2 mutations seem restricted to AITL, whereas TET2, DNMT3A, and RHOA mutations can be found in other PTCL entities as well, although they are enriched in PTCL of TFH origin (12, 13, 19, 23). These findings suggest that epigenetic alteration is a hallmark of malignant transformation in AITL.
Interestingly, it has been shown that TET2 and DNMT3A mutations present in PTCL can be acquired in CD34-positive progenitor cells and can be observed not only in tumor cells, but also in other hematological lineages (13, 18). This finding is consistent with the relatively high frequency of these mutations in clonal hematopoiesis of undetermined significance, especially among elderly patients (24, 25). However, few data exist regarding the consequences of IDH2 mutation in AITL.
Here, using primary human samples and mouse models, we investigate the role of mutant IDH2 protein in T cells. In a series of human AITL samples, we detect mutant IDH2 in cells with a TFH phenotype, and not in other cells comprising the tumor microenvironment. We also show that IDH2-mutated AITL produces 2HG that is detectable in tumor tissue and in the serum of AITL patients, although at a lower level than is observed in IDH-mutated AML. Finally, using conditional knock-in (KI) mouse models bearing different Idh1 and Idh2 mutations, we find that the 2HG concentration is lower in lymphoid cells than in myeloid cells, and that 2HG is detectable in IDH2R172K KI T cells, but not in IDH1R132H KI T cells, which correlates with unique anomalies in the lymphoid lineages of IDH2R172K KI mice. These findings may explain the absence of IDH1R132 mutations in AITL and should provide a model for further investigation of this disease.
Results
IDH2R172 Mutant Protein Is Expressed in Inducible T-Cell Costimulator-Positive T Cells in AITL.
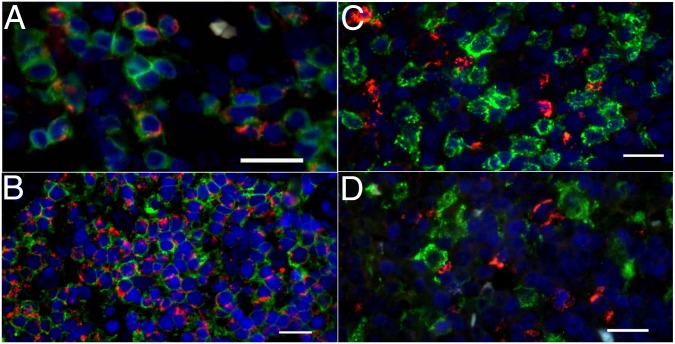
To determine which cells in the tumor microenvironment bear the IDH2 mutation and express the mutant protein, we developed immunohistochemical (IHC) and immunofluorescent (IF) assays by using a mutant-specific anti-IDH2R172K antibody. These assays were used to examine formalin fixed paraffin embedded (FFPE) tumor samples from nine AITL patients with IDH2R172K mutations, tumor cells being inducible T-cell costimulator (ICOS) positive in all cases (SI Appendix, Table S1). At low magnification, IDH2R172K staining overlapped with zones enriched in programmed cell death 1 (PD1)- and ICOS-positive tumor cells (SI Appendix, Fig. S1). This antibody stained slightly atypical small- to medium-sized cells with clear cytoplasm that were either scattered or grouped in variably sized aggregates, consistent with the AITL neoplastic component. The granular cytoplasmic staining of IDH2R172K and colocalization with cytochrome c oxidase subunit 4 (COX4) was compatible with the known mitochondrial localization of IDH2 (SI Appendix, Fig. S2). The number of IDH2R172K-positive cells was variable among the cases examined, and correlated with the estimate of tumor cell content obtained by using TFH-specific immunophenotypic markers (PD1, ICOS, and/or CXCL13) and with the IDH2 variant allele frequency (VAF) obtained by next-generation sequencing (SI Appendix, Table S1 and Fig. S3). Using double staining by both IHC and IF, we found that mutant IDH2 was present in CD3-positive T cells (Fig. 1A) expressing the TFH-associated antigen ICOS (Fig. 1B), a characteristic marker of reactive and neoplastic TFH cells. Interestingly, the presence of mutant IDH2 could not be detected in CD8+ T cells (Fig. 1C), CD163+ histiocytes (Fig. 1D), or PAX5+ B cells (SI Appendix, Fig. S1) in nine cases examined, which suggests that mutant IDH2 expression is restricted to malignant cells in AITL.
Fig. 1.
IDH2R172K mutant protein is present in malignant AITL T cells with a TFH phenotype. Immunofluorescence double staining for IDH2R172K (red) and CD3 (green) (A), IDH2R172K (red) and ICOS (green) (B), double staining IDH2R172K (red) and CD8 (green) (C), and IDH1R172K (red) and CD163 (green) (D). All images were acquired at 40× magnification. DAPI counterstaining for nuclei is shown in blue for all images. (Scale bars: 20 μm.)
IDH2-Mutated AITL Produces Detectable Levels of 2HG.
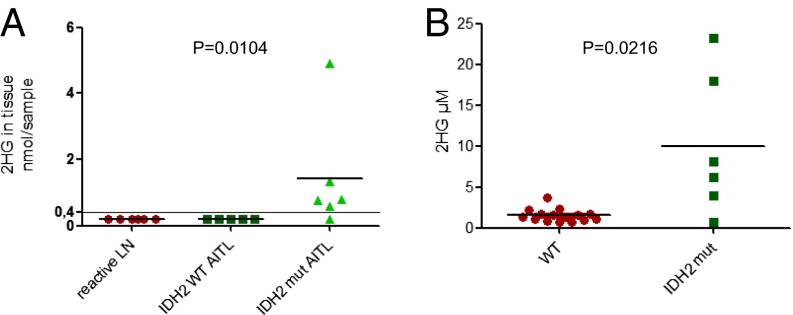
We measured the level of 2HG in frozen human tissues, including six reactive lymph nodes, five IDH2 wild-type (WT), and six IDH2- (three IDH2R172K, one R172G, one R172T, and one R140Q) mutated AITL samples. Within the limit of the technique and the quantity of tissue, 2HG was undetectable in all reactive tissues and in IDH2 WT AITL, but was detected in all but one IDH2 mutant AITL (Fig. 2A), at a median level of 0.775 nmol per sample (<0.05–4.9 nmol). The only IDH2 mutant sample with undetectable levels of 2HG bore an IDH2R140Q mutation detected with a low VAF of 1.5%.
Fig. 2.
IDH2-mutated AITL tumors produce 2HG that is detectable in tissue and serum. (A) 2HG concentration in tissue obtained from reactive lymph nodes (LN) and tissue from IDH2 WT and IDH2 mutant AITL tumors. (B) 2HG in the serum of AITL patients of the indicated mutational status. The Mann–Whitney test was used to compare WT AITL versus mutated AITL.
We also assessed the level of 2HG in the serum of a separate cohort of 21 AITL patients, including 6 IDH2-mutated patients (2 IDH2R172K, 2 IDH2R172G, 1 R172S, and 1 R172M) and found that the median level of 2HG was higher in IDH2-mutated AITL than in WT AITL patients (7.14 versus 1.37 µM, P = 0.001; Fig. 2B). Because of a cutaneous rash, which is a frequent symptom of this disease, steroids were introduced 2 wk before serum collection in the IDH2R172G mutant patient displaying a low level of 2HG. It is possible that steroid treatment contributed to a 2HG decrease in this patient, but this effect remains to be confirmed in larger patient series.
It is noteworthy that this level of 2HG is significantly lower than the level previously reported in IDH1- or IDH2-mutated AML (median 21 µM, range 2.4–305.6 µM) (26). This discrepancy may be due to a higher tumor burden and proliferation rate in AML compared with AITL. In AITL, the involved lymph nodes are rarely bulky, and tumor cells bearing the mutation are usually scattered among a prominent population of reactive cells which, as shown above, are not mutated. However, this difference could also be due to differences in the expression level of the mutant enzyme in different cell types (mature T cells in AITL and immature myeloid cells in AML).
2HG Production Depends on the IDH Mutation Type and Cell Context.
To explore the difference in impact of common IDH mutations in the lymphoid system, we generated three different conditional KI mice representing the most frequent amino acid substitutions for each commonly mutated residue: IDH1R132H, IDH2R140Q, and IDH2R172K. We crossed these three mouse models with Vav-cre to generate heterozygous Vav-cre IDH1R132H, Vav-cre IDH2R140Q, and Vav-cre IDH2R172K mice, which express the mutant protein in the entire hematopoietic system, including lymphoid cells. These experimental animals are designated IDH1R132H, IDH2R140Q, and IDH2R172K in all experiments.
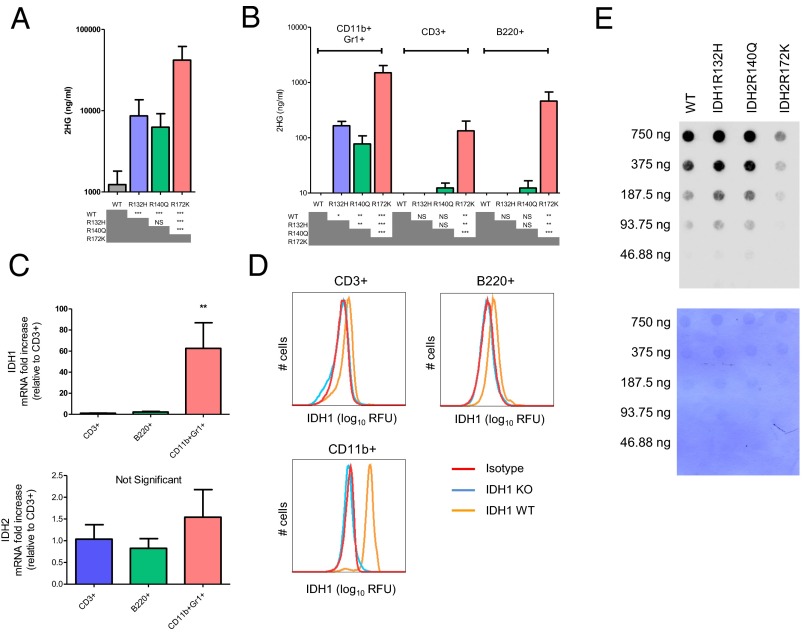
All three mutants had higher levels of 2HG in serum compared with WT littermate controls, although there were differences between the different lines. 2HG was significantly higher in IDH2R172K mice than in IDH1R132H or IDH2R140Q (Fig. 3A).
Fig. 3.
2HG production depends on the specific Idh mutation and the cell context. (A) 2HG level measured by mass spectrometry in serum of IDH KI mice aged 3–6 mo of the indicated genotypes. (B) 2HG level in 500,000 CD11b+Gr1+ myeloid, CD3+ T, and B220+ B cells. (C) Quantitative RT-PCR measurement of IDH1 and IDH2 expression in CD11b+ Gr1+ myeloid cells, CD3+ T cells, and B220+ B cells. RPS9 is used as a housekeeping gene, and results are shown relative to T cells. Results from three independent experiments are shown. In A–C, bars represent the mean and error bars represent SD. Two-tailed Student's t tests were performed to determine statistical significance by using the Bonferroni correction for multiple comparisons. (D) FACS analysis of IDH1 expression in the indicated cell types derived from bone marrow and spleen of IDH1 KO or IDH1 WT mice compared with isotype control. ANOVA with Tukey's multiple comparison correction was used to determine statistical significance. (E) Dot blot measurement of the level of 5hmC in T cells from IDH KI mice of the indicated genotypes. DNA loading control is shown in Lower. *P < 0.05, **P < 0.01, and ***P < 0.001. NS, not significant.
We next examined the level of 2HG in different cell subsets: CD3+-positive T cells, B220-positive B cells and CD11b+Gr1+ myeloid cells. Similar to the observations in serum, the intracellular level of 2HG in myeloid cells was highest in IDH2R172K followed by IDH1R132H and IDH2R140Q (Fig. 3B). Interestingly, in all mice, 2HG levels were higher in myeloid cells than in the lymphoid populations (B cells and T cells). In fact, 2HG was undetectable in IDH1R132H KI lymphoid cells. There was a small but not statistically significant increase in 2HG in IDH2R140Q mice and a significant increase in IDH2R172K KI lymphoid cells (Fig. 3B).
We investigated whether the expression level of IDH1 and IDH2 could partially explain the differences in 2HG production in these different cell types. The baseline expression of IDH1 was much lower in WT lymphoid cells than in CD11b myeloid cells at both the mRNA level (Fig. 3C) and the protein level (Fig. 3D). However, IDH2 expression was similar in these different cell types where no statistically significant differences were observed (Fig. 3C).
One well-characterized effect of 2-HG is the inhibition of the TET family of enzymes, which results in decreased production of 5-hydroxymethylcytosine (5hmC) in DNA. It is thought that these epigenetic changes may contribute to malignant transformation in the hematopoietic system. Therefore, we analyzed 5hmC levels in CD3+ T cells bearing different IDH mutations. We observed a dramatic decrease in 5hmC in DNA of IDH2R172K KI T cells, whereas 5hmC levels were unchanged in IDH1R132H or IDH2R140Q KI T cells compared with WT controls (Fig. 3E).
Contrary to the IDH1R132H or IDH2R140Q Mutations, the IDH2R172K Mutation Impairs Lymphoid Development.
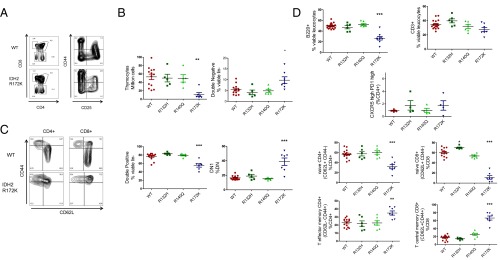
Given the differences observed between the IDH mutants in lymphoid cells, we examined the lymphoid populations in the three different KI mice between the ages of 3 and 7 mo. Thymic development was significantly impaired in the IDH2R172K animals where we observed an important decrease in the total number of thymocytes, a decrease in CD4+CD8+ double positive thymocytes, and an increase in the proportion of double negative (DN) cells, predominantly DN1, defined as lin− CD4− CD8− CD44+ CD25−. These results suggest that double positive cells may be more sensitive to apoptosis caused by introduction of this mutation (Fig. 4 A and B). No differences were observed in cells derived from the thymus of IDH1R132H or IDH2R140Q mice. Similarly, in the spleen, we observed that only IDH2R172K KI mice displayed anomalies in mature lymphoid populations. There was a decrease in the percentage of B cells, and although the proportion of CD3 + T cells was similar among the genotypes, we observed changes in T-cell differentiation. CD4+ and CD8+ naive T cells (CD62L+ CD44−) were decreased, and CD8+ central memory cells (CD44+ CD62L+) were increased (Fig. 4 C and D). No significant change in the TFH cell proportion, defined as CD4-positive PD1 high CXCR5 high cells was observed in these mice. Thus, in vivo, anomalies in lymphoid populations were restricted to IDH2R172 KI mice.
Fig. 4.
IDH2 R172K mutant impairs lymphoid development and T-cell differentiation. (A) Representative FACS profiles of WT and IDH2R172 thymocytes. (B) Quantification of thymocyte number, Lin− CD4+ CD8+ double positive proportion, Lin− CD4− CD8− double negative proportion, and type I double negative (DN1) proportion, defined as Lin− CD4− CD8− CD44+ CD25− thymocytes. The Lin− population was CD11b−, Ter119−, Gr1−, B220−, NK1.1− CD11c−. (C) Representative FACS profile of WT and IDH2R172K CD4+ and CD8+ T-cell differentiation in spleen. (D) Quantification of lymphoid population in the spleen: B220+ B cells; CD3+ T cells; TFH cells defined as CD4+ PD1 high CXCR5 high, naive CD4+ and CD8+ T cells defined as CD62L+ and CD44−; CD4+ T-effector memory cells defined as CD62L−, CD44+, and CD4+; CD8+ central memory T cells, defined as CD62L+ CD44+ and CD8+. ANOVA test with Dunnett's multiple comparison test (each KI was compared with WT) were performed, and *P < 0.05, **P < 0.01, and ***P < 0.001.
Together, these results indicate that the IDH mutation encoding IDH2R172K, contrary to mutations encoding IDH2R140Q or IDH1R132H, when present in a T cell, has the ability to increase 2HG concentrations enough to inhibit the TET enzymes, leading to 5hmC decrease. Furthermore, only the IDH2R172K mutant affects lymphoid development in this accurate in vivo model. These findings are consistent with the fact that in AITL, almost all IDH mutations are at IDH2R172, unlike in other malignancies. The low potential of IDH1R132H or IDH2R140Q to produce 2HG in T cells may explain why these mutations are not observed in AITL.
Discussion
The importance of IDH2 mutations in human AITL has been somewhat unclear, because the level of 2HG produced in clinical samples from these malignancies has not been well-characterized. A recent report described an increase in 2HG production after IDH2R172K mutant protein was overexpressed in the Jurkat lymphoblastic T-cell leukemia cell line (12) or primary CD4+ T cells. However, these in vitro systems do not accurately model human AITL. Furthermore, a single case report has described a detectable increase in 2HG in a tumor biopsy, but not in the serum of an IDH2 mutated AITL patient (27). These findings illustrate the importance of investigating the significance and consequences of IDH2 mutations in AITL. Here, using a set of patient samples collected before the initiation of therapy, we have demonstrated that 2HG was significantly increased and detectable in the majority of patients. Although we cannot completely exclude, in rare cases, that the IDH2 mutation occurs in a hematopoietic progenitor cell, as has been described for TET2 and DNMT3A mutations in PTCL (13, 18) our data showing that the IDH2 mutant protein is present only in the ICOS-positive T-cell compartment in 9/9 cases suggests that this mutation occurs most often in more mature T cells. These findings also suggest that 2HG production persists during malignant progression, and that it could be a useful diagnostic marker. The usefulness of 2HG to monitor minimal residual disease, as it is beginning to be used in AML (26), remains to be determined in further longitudinal studies.
Given that all hotspot IDH1 and IDH2 mutations cause acquisition of the same neoenzymatic activity and production of the 2HG oncometabolite, the reasons for the different distributions of IDH mutations in different malignancies are still unclear. Specifically, the reason that IDH mutations in AITL occur almost exclusively at IDH2R172 is unknown. One possibility is that specific mutations result in more efficient production of 2HG, leading to a broader spectrum of downstream effects that are critical for the malignant transformation of specific cell populations. Previous in vitro studies have shown that overexpression of IDH2R140Q and IDH2R172K in cell lines resulted in different levels of 2HG production, with IDH2R172K being more efficient than IDH2R140Q. This work also showed that the subcellular localization of the mutant enzyme can affect 2HG production, because relocalization of mutant IDH1 from the cytosol to the mitochondria dramatically increased 2HG production (28). However, studying the quantitative effects of IDH mutant proteins using overexpression systems has drawbacks, because 2HG production is influenced by the level of protein expression, which is often not physiological in these model systems. Furthermore, because IDH mutations are always heterozygous, and the IDH enzyme acts as a dimer, the balance of expression between the wild-type and mutant alleles may also influence enzymatic activity. This effect has been observed in previous in vitro work where 2HG production by overexpressed IDH1R132H depended on the expression level of both WT and mutant proteins (28). Given this biological complexity, more accurate in vivo models should be more informative in determining the critical differences in the effects of specific IDH mutations.
Here, using conditional KI mouse models in which IDH1 and IDH2 are expressed at physiological levels, we show that IDH2R172K produces significantly greater amounts of 2HG than IDH2R140Q in the hematopoietic system, with this effect being particularly prominent in lymphoid cells. 2HG production by IDH1R132H was similar to IDH2R140Q in the serum, and was intermediate between IDH2R172K and IDH2R140Q in myeloid cells. Furthermore, this model revealed that the potential of a specific IDH mutant protein to produce 2HG depends on cell context within the hematopoietic system, as we observed consistently higher 2HG concentrations in myeloid cells than in lymphoid cells. This difference in 2HG production could be explained by a number of factors, including differences in gene expression between cell types, mitochondrial vs. cytoplasmic localization of mutant IDH enzymes, or metabolic status of disparate cells types. Here, we present data suggesting that that the expression of IDH1 vs. IDH2 may account for some of the differences in the potential of these mutations to differentially affect the myeloid vs. the lymphoid system.
One question that remains is the significance of different amino acid substitutions at the same residue in either IDH1 or IDH2. In this report, we have compared the effects of IDH1R132H, IDH2R140Q, and IDH2R172K mutations in terms of their potential to produce 2HG in vivo and their effects on lymphoid development. However, the effects of other amino acid substitutions affecting residue IDH1R132, IDH2R140, or IDH2R172 in the hematopoietic system is unknown. Clinical differences exist in the prevalence of different amino acid substitutions, depending on the malignancy in question, suggesting that additional work to investigate these differences is also warranted.
This point is of particular interest for AITL, which presents a spectrum of amino acid substitutions at IDH2R172, although mutations at the IDH1R132 and IDH2R140 hotspot positions are exceedingly rare. In AITL, IDH2R172K mutations represent ∼40% of IDH2R172 mutations, in contrast to AML where IDH2R172K substitutions represent 98% of IDH2R172 mutations according to the COSMIC database. Analysis of 2HG levels in a larger series of IDH2-mutated AITL may clarify whether different mutations at R172 produce significantly different levels of 2HG. However, as in AML, differences in 2HG levels in clinical samples may also be affected by tumor burden and proliferation status.
One remaining question is to determine how IDH2 mutations drive AITL oncogenesis, given that they frequently coexist with TET2 mutations. Indeed, these two mutations are virtually mutually exclusive in AML where it is thought that TET2 inhibition is a major oncogenic consequence of IDH mutations (5). However, beyond TET2 inhibition, 2HG can inhibit multiple α-KG–dependent dioxygenases involved in various cell functions, including histone methylation, the hypoxia response, and collagen maturation (1) that all could contribute to the oncogenic process. The recent description of TET2 independent effects of mutant IDH on the DNA damage response in the hematopoietic system supports this view (29). Furthermore, the frequent coexistence of TET2 and IDH2 mutations within the same tumor suggests that these two mutations may synergize to drive the transformation in this cell type. Although our results do not provide a comprehensive mechanistic understanding of the oncogenic effects of IDH2 mutations in AITL lymphomagenesis, they suggest that ongoing production of 2HG by malignant AITL cells may be important for the maintenance of this disease. They also suggest that mutations at R172 in IDH2 have a greater capacity to produce 2HG in lymphoid cells, and are capable of impairing lymphoid cell development, which may explain the predominance of this mutation in AITL. This observation is an important first step. Further study of the effects of the combination of TET2, IDH2, and RHOA mutations should bring a better understanding of how these mutations cooperate to drive disease and how they could be targeted for effective treatment.
Materials and Methods
Patient Samples.
Patients samples (frozen and FFPE tissues and pretreatment serum) were collected within the framework of a multicentric T-cell lymphoma consortium (Tenomic). The study was approved by the local ethics committee (Comité de Protection des Personnes Ile de France IX 08–009) and included informed consent by patients.
IDH2 Genotyping.
Because of the lack of sensitivity of Sanger sequencing, a next-generation sequencing method was used to determine mutational status of AITL samples. Briefly, 20 ng of DNA was extracted from FFPE samples and was amplified by using an Ampliseq custom panel. Amplicons were then digested, barcoded, and amplified by using the Ion Ampliseq Library kit 2.0 and Ion Xpress barcode adapter kit (ThermoFisher) according to the manufacturer’s instructions. After quantification of DNA, 8 pM of each library was multiplexed and clonally amplified on Ion sphere particles (ISP) by emulsion PCR performed on the Ion One Touch 2 instrument with the Ion PGM template OT2 200 kit (ThermoFisher) according to the manufacturer’s instructions. After quality control, the ISP templates were enriched, loaded on an Ion 316 chip, and sequenced on a PGM sequencer with the Ion PGM Hi-Q Sequencing Kit according to the manufacturer’s instructions. Mutations were detected by using the Variant Caller plug-in version 5.0.0.7 with low stringency settings (ThermoFisher).
Immunohistochemistry and Immunofluorescence.
Deparaffinized tissue sections were stained with a mouse monoclonal IDH2R172K antibody (New East, 26163) after appropriate antigen retrieval with EDTA buffer pH 9 (DiaPath Martinengo). For double staining of IDH2R172K by immunofluorescence or immunochemistry, CD3 (DAKO), ICOS (Spring Bioscience/Roche), CD8 (DAKO), PAX55 (DAKO), CD163 (DAKO), or COX4 (Diagomic) antibodies were used, using a described method (30).
2HG Detection.
2HG in patient serum was quantified by using a recently published liquid chromatography tandem mass spectrometry method allowing a rapid, accurate, and precise quantification (31). Briefly, deuterated (R,S)-2-hydroxyglutaric acid, disodium salt is used as internal standard and added to samples before a solid phase extraction on Phenomenex STRATA-XL-A (200 mg, 3 mL) 33-μm cartridges. Samples are separated on a C18 column and analyzed on a Xevo TQ-MS Waters mass spectrometer with an electrospray ionization source. 2HG in human tissues was measured in 1 cm2 and 30-µm-thick slides of frozen tissue. Tissues were lysed by using the TissueLyser technology, according to manufacturer’s instructions (Qiagen), before 2HG quantification. 2HG mouse whole blood and in 500,000 CD3+, B220+, or CD11b+Gr1+ FACS sorted KI mouse cells was measured after metabolite extraction in 80% (vol/vol) aqueous methanol, as described. Extracts were subjected to ion-paired reverse-phase LC coupled to negative mode electrospray triple-quadrupole mass spectrometry, using multiple reaction monitoring. Integrated elution peaks were compared with metabolite standard curves for absolute quantification (32).
Mice.
Conditional IDH1R132H-KI, IDH2R140Q, and IDH2R172K mice were created by using an equivalent strategy, placing a lox-stop-lox cassette in the intron upstream of the exon bearing a targeted mutation, as described (33). IDH2KI mice were developed by Agios Pharmaceuticals and Wuxi Apptec. IDH1KI and IDH2KI mice were bred with Vav-cre mice (Jackson Laboratories; catalog no. 008610) to produce Vav-IDH1 KI, and Vav-IDH2 KI animals. All animal experiments were approved by the University Health Network (UHN) Animal Care Committee.
Mice were euthanized between 4 and 7 mo, and the different genotypes were age matched for each experiment. Single-cell suspension for further applications were obtained by a gentle mechanical disruption with a syringe plunger then filtered with a cell strainer (100 µm and 40 µm) and were counted with a Vi-Cell XR (Beckman Coulter).
5hmC Dot Blot.
Dot blotting to detect 5hmC was performed as described (28). Briefly, genomic DNA was prepared from FACS-sorted CD3+ T cells isolated from IDH1R132H, IDH2R140Q, IDH2R172K, and WT mice by using the Nucleospin Tissue kit (Macherey-Nagel). Genomic DNA was denatured in 0.1 M NaOH at 90 °C for 10 min, followed by neutralization with chilled 0.2 M Tris⋅HCl (pH 8). Twofold serial dilutions of denatured genomic DNA were blotted onto Hybond N+ membranes (Amersham) in an assembled BIO-DOT apparatus (Bio-Rad). After blotting, membranes were air-dried, cross-linked (1200 µJ/cm2) by using a UV Stratalinker 2400 (Stratagene), blocked with 5% (wt/vol) skim milk, and incubated overnight at 4 °C with rabbit anti-5hmC Ab (Active Motif, catalog no. 39769).
Flow Cytometry.
Single-cell suspensions were prepared from spleen, thymus, or bone marrow preparations and treated to lyse red blood cells. Cells (1–2 × 106) were immunostained with antibodies recognizing the following: CD3 (145-2C-11), CD4 (GK1.5), CD5 (53-7.3), CD8 (53-6.7), CD25 (PC61), CD44 (IM7), CD62L (MEL-14), CD11b (M1-70), Gr1 (RB6-8C5) B220 (RA3-6B2), CXCR5 (2G8), and PD1 (RMP1-30) all from BioLegend; BD Biosciences; or eBioscience. IDH1 intracellular staining was performed by using the antibody manufacturer’s instructions. After blocking and extracellular staining, cells were fixed with 1.6% (wt/vol) paraformaldehyde and incubated for 15 min at room temperature. After wash, cells were resuspended in 900 µL of prechilled methanol and incubated 30 min on ice. Cells were then washed and incubated with anti-IDH1 antibody (D2H1; Cell Signaling) or an isotype control antibody at 1/200 dilution for 60 min at room temperature. Alexa Fluor 488 at 1/500 dilution was used after wash. Data were acquired by using a BD FCM CantoII flow cytometer and analyzed with the FlowJo analysis program (Tree Star).
RT-PCR.
Real-time RT-PCR was performed as described (34). Each sample was assayed in triplicate, and data were normalized to the housekeeping gene RPS9, which was determined to have a constant expression among these different cell populations. Results were calculated by using the comparative threshold cycle method (2-ΔΔCt). The primers used were the following: Idh1 (forward 5′-CAGGCTCATAGATGACATGGTGG-3′, reverse 5′-CACTGGTCATCATGCCAAGGGA-3′), Idh2 (forward 5′-GGCTGTCAAGTGTGCCACAATC-3′, reverse 5′-TTGGCTCTCTGAAGACGGTTCC-3′).
Supplementary Material
Acknowledgments
We thank all members of the Tenomic consortium. We are grateful for the administrative assistance of Ms. Irene Ng and for technical assistance from the Princess Margaret Cancer Center flow cytometry facility, genotyping facility, and animal resource center, and the UHN Genomics Centre, and we thank Lionel Mercier for his help in 2HG dosage. This work was supported by a grant from the Canadian Institutes of Health Research (to T.W.M. and R.A.C.); grants from the Association pour la Recherche contre le Cancer, the Institut National du Cancer and la Fondation pour la Recherche Médicale (to P.G.); and by a grant from the Leukemia and Lymphoma Society (to T.W.M.). F.L. received a grant from the Institut National du Cancer.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617929114/-/DCSupplemental.
References
- 1.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: Mechanisms, models, and clinical opportunities. Cancer Discov. 2013;3(7):730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 2.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowdhury R, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koivunen P, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483(7390):484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasaki M, et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 2012;26(18):2038–2049. doi: 10.1101/gad.198200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairns RA, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012;119(8):1901–1903. doi: 10.1182/blood-2011-11-391748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C, et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood. 2015;126(15):1741–1752. doi: 10.1182/blood-2015-05-644591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakata-Yanagimoto M, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46(2):171–175. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- 14.Odejide O, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123(9):1293–1296. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Leval L, Gisselbrecht C, Gaulard P. Advances in the understanding and management of angioimmunoblastic T-cell lymphoma. Br J Haematol. 2010;148(5):673–689. doi: 10.1111/j.1365-2141.2009.08003.x. [DOI] [PubMed] [Google Scholar]
- 16.de Leval L, et al. Angioimmunoblastic T-cell lymphoma is the most common T-cell lymphoma in two distinct French information data sets. Haematologica. 2015;100(9):e361–e364. doi: 10.3324/haematol.2015.126300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Leval L, et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood. 2007;109(11):4952–4963. doi: 10.1182/blood-2006-10-055145. [DOI] [PubMed] [Google Scholar]
- 18.Quivoron C, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20(1):25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Lemonnier F, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood. 2012;120(7):1466–1469. doi: 10.1182/blood-2012-02-408542. [DOI] [PubMed] [Google Scholar]
- 20.Couronné L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366(1):95–96. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- 21.Palomero T, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014;46(2):166–170. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo HY, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46(4):371–375. doi: 10.1038/ng.2916. [DOI] [PubMed] [Google Scholar]
- 23.Vallois D, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood. 2016;128(11):1490–1502. doi: 10.1182/blood-2016-02-698977. [DOI] [PubMed] [Google Scholar]
- 24.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janin M, et al. Serum 2-hydroxyglutarate production in IDH1- and IDH2-mutated de novo acute myeloid leukemia: A study by the Acute Leukemia French Association group. J Clin Oncol. 2014;32(4):297–305. doi: 10.1200/JCO.2013.50.2047. [DOI] [PubMed] [Google Scholar]
- 27.Churchill H, Naina H, Boriack R, Rakheja D, Chen W. Discordant intracellular and plasma D-2-hydroxyglutarate levels in a patient with IDH2 mutated angioimmunoblastic T-cell lymphoma. Int J Clin Exp Pathol. 2015;8(9):11753–11759. [PMC free article] [PubMed] [Google Scholar]
- 28.Ward PS, et al. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem. 2013;288(6):3804–3815. doi: 10.1074/jbc.M112.435495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inoue S, et al. Mutant IDH1 downregulates ATM and alters DNA repair and sensitivity to DNA damage independent of TET2. Cancer Cell. 2016;30(2):337–348. doi: 10.1016/j.ccell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amé-Thomas P, et al. CD10 delineates a subset of human IL-4 producing follicular helper T cells involved in the survival of follicular lymphoma B cells. Blood. 2015;125(15):2381–2385. doi: 10.1182/blood-2015-02-625152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poinsignon V, et al. Quantitation of isocitrate dehydrogenase (IDH)-induced D and L enantiomers of 2-hydroxyglutaric acid in biological fluids by a fully validated liquid tandem mass spectrometry method, suitable for clinical applications. J Chromatogr B Analyt Technol Biomed Life Sci. 2016;1022:290–297. doi: 10.1016/j.jchromb.2016.04.030. [DOI] [PubMed] [Google Scholar]
- 32.Hao Z, et al. Idh1 mutations contribute to the development of T-cell malignancies in genetically engineered mice. Proc Natl Acad Sci USA. 2016;113(5):1387–1392. doi: 10.1073/pnas.1525354113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki M, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488(7413):656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inoue S, et al. Mule/Huwe1/Arf-BP1 suppresses Ras-driven tumorigenesis by preventing c-Myc/Miz1-mediated down-regulation of p21 and p15. Genes Dev. 2013;27(10):1101–1114. doi: 10.1101/gad.214577.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.