Significance
Replacement of petrochemistry by bio-based processes requires microbes equipped with novel-to-nature capabilities. The efficiency of such engineered microbes strongly depends on their native metabolic networks, which, forged by eons of evolution, are complex and encoded by mosaic microbial genomes. Absence of a modular organization of genomes tremendously restricts genetic accessibility and presents a major hurdle for fundamental understanding and rational engineering of central metabolism. Using as a paradigm the nearly ubiquitous glycolytic pathway, we introduce a radical approach, enabling the “transplantation” of essential metabolic routes in the model and industrial yeast Saccharomyces cerevisiae. This achievement demonstrates that a modular design of synthetic genomes offers unprecedented possibilities for fast, combinatorial exploration, and optimization of the biological function of essential cellular processes.
Keywords: pathway swapping, glycolysis, Saccharomyces cerevisiae, modular genomes
Abstract
Recent developments in synthetic biology enable one-step implementation of entire metabolic pathways in industrial microorganisms. A similarly radical remodelling of central metabolism could greatly accelerate fundamental and applied research, but is impeded by the mosaic organization of microbial genomes. To eliminate this limitation, we propose and explore the concept of “pathway swapping,” using yeast glycolysis as the experimental model. Construction of a “single-locus glycolysis” Saccharomyces cerevisiae platform enabled quick and easy replacement of this yeast’s entire complement of 26 glycolytic isoenzymes by any alternative, functional glycolytic pathway configuration. The potential of this approach was demonstrated by the construction and characterization of S. cerevisiae strains whose growth depended on two nonnative glycolytic pathways: a complete glycolysis from the related yeast Saccharomyces kudriavzevii and a mosaic glycolysis consisting of yeast and human enzymes. This work demonstrates the feasibility and potential of modular, combinatorial approaches to engineering and analysis of core cellular processes.
Replacement of petrochemistry by bio-based processes is a key element for sustainable development and requires microbes equipped with novel-to-nature capabilities. Recent developments in synthetic biology enable introduction of entire metabolic pathways and, thereby, new functionalities for product formation and substrate consumption, into microbial cells (1). However, industrial relevance of the resulting strains critically depends on optimal interaction of the newly introduced pathways with the core metabolism of the host cell. Central metabolic pathways such as glycolysis, tricarboxylic acid cycle, and pentose phosphate pathways, are essential for synthesis of precursors, for providing free energy (ATP), and for redox-cofactor balancing. Optimization of productivity, product yield, and robustness therefore requires modifications in the configuration and/or regulation of these core metabolic functions.
Engineering of central metabolism is in some respects more challenging than the functional expression of heterologous product pathways. Millions of years of evolution of microorganisms have endowed their metabolic and regulatory networks with a level of complexity that cannot be efficiently reengineered by iterative, single-gene modifications. Enzymes of central metabolism are encoded by hundreds of genes that, especially in eukaryotes, are scattered across microbial genomes. Moreover, inactivation and subsequent replacement of genes involved in central metabolism is complicated by functional redundancy of isoenzymes (2, 3) as well as by the essential role of many of the corresponding biochemical reactions. Microbial platforms in which the configuration of key pathways can be remodelled in a swift, combinatorial manner would provide an invaluable asset for fundamental research and engineering of central metabolism.
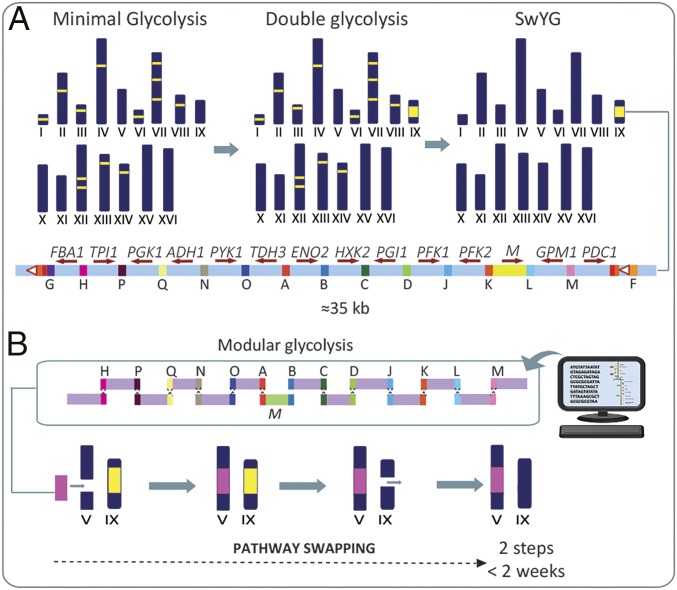
Whereas rapid, cost-effective assembly of entire synthetic genomes is becoming a realistic perspective for small bacterial genomes (4, 5), routine synthesis and expression of entire eukaryotic genomes is unlikely to be implemented in the next few years. Here, we propose and experimentally explore a modular approach to the engineering of central metabolism that involves versatile, synthetic microbial strain platforms in which entire metabolic pathways can, in a few simple steps, be replaced by any functional, newly designed configuration. As a proof of principle, we set out to construct a platform that enables swapping of the entire Embden–Meyerhoff–Parnas pathway of glycolysis, a strongly conserved metabolic highway for sugar utilization in the model eukaryote and industrial yeast Saccharomyces cerevisiae. Including the reactions leading to the formation of ethanol, the main fermentation product of S. cerevisiae, yeast glycolysis encompasses a set of 12 reactions, catalyzed by no fewer than 26 cytosolic isoenzymes. Several of these (e.g., Tpi1, Tdh3, and Adh1) are among the most abundant proteins in yeast cells. The genes encoding glycolytic enzymes are scattered over 12 of the 16 yeast chromosomes. Construction of a platform for glycolysis swapping involved a two-step approach (Fig. 1A). In the first step, described in a recent study by our group, the genetic complexity of yeast glycolysis was reduced by deleting the structural genes for 13 of the 26 glycolytic enzymes. Remarkably, a detailed systems analysis revealed that, under laboratory conditions, the phenotype of the resulting minimal glycolysis (MG) strain was virtually identical to that of the parental strain carrying a full complement of glycolytic genes (2). In a next step, the remaining 13 glycolytic genes in MG were expressed from a single chromosomal locus. Finally, the remaining scattered native genes were removed from their original loci, leading to switchable yeast glycolysis (SwYG), a yeast strain carrying a native, minimal single-locus glycolysis on chromosome IX. In SwYG, glycolysis can be swapped in two steps by integration of a new, heterologous, or synthetic glycolytic gene cluster, followed by removal of the minimal single-locus glycolysis that was initially integrated on chromosome IX (Fig. 1B).
Fig. 1.
Schematic overview of the glycolysis swapping approach. (A) Construction of SwYG that contains a single locus endogenous glycolysis platform for pathway swapping. (B) In silico design and in vivo assembly and integration of the glycolytic gene cluster on chromosome V, followed by the removal of the endogenous glycolysis on chromosome IX, leading to a strain with a redesigned glycolysis. M, selectable marker.
Results
Engineering of a Yeast Platform for Glycolysis Swapping.
A single-locus native glycolysis gene cluster was assembled from “glycoblocks” (Fig. 1 and SI Appendix, Fig. S1), 13 DNA cassettes each consisting of a S. cerevisiae glycolytic gene, including its native promoter and terminator, flanked by 60-bp synthetic homologous recombination (SHR) sequences (6). SHR sequences share no homology with the S. cerevisiae genome and can be used for efficient in vivo assembly and integration, by homologous recombination (HR), of the glycoblocks. Moreover, use of standardized SHR sequences enables flexible design and combinatorial assembly of different glycolytic pathway variants. The single-locus MG cluster was composed of 13 glycoblocks (corresponding to the 13 genes remaining in MG) and of a selectable marker (amdSYM, encoding Aspergillus nidulans acetamidase conferring the ability to grow on acetamide as sole nitrogen source) (7) flanked by SHR sequences (Fig. 1A). The single-locus native glycolytic cluster was integrated into the yeast genome to promote stable, single-copy expression, using combined assembly and targeted integration (CATI) (8) (SI Appendix, Fig. S1). To this end, recognition sequences for the I-SceI homing endonuclease were introduced at the SGA1 locus on chromosome IX of MG (SI Appendix, Fig. S1). The 13 glycoblocks and the amdSYM cassette were cotransformed to the modified MG strain, in which SCEI was induced by growth on galactose to introduce a double-strand DNA break at the SGA1 locus, thereby promoting integration of the glycoblocks (Fig. 2A and SI Appendix, Fig. S1). Four of five tested transformant colonies harbored the complete 35-kb single-locus native glycolysis (SinLoG-IX) integrated at the SGA1 locus. In a selected transformant (strain IMX382), next-generation (next-gen) sequencing showed that the in vivo assembled and integrated glycolytic gene cluster was virtually identical to its in silico blueprint (Fig. 2A). Six of the nine deviations in nucleotide sequence were found at the HR loci linking the glycoblocks, which may either reveal recombinase-based errors or simply errors in the primers used to construct the HR sequences. Of the glycolytic genes, only ADH1 was found to contain a mutation that was synonymous (A180A).
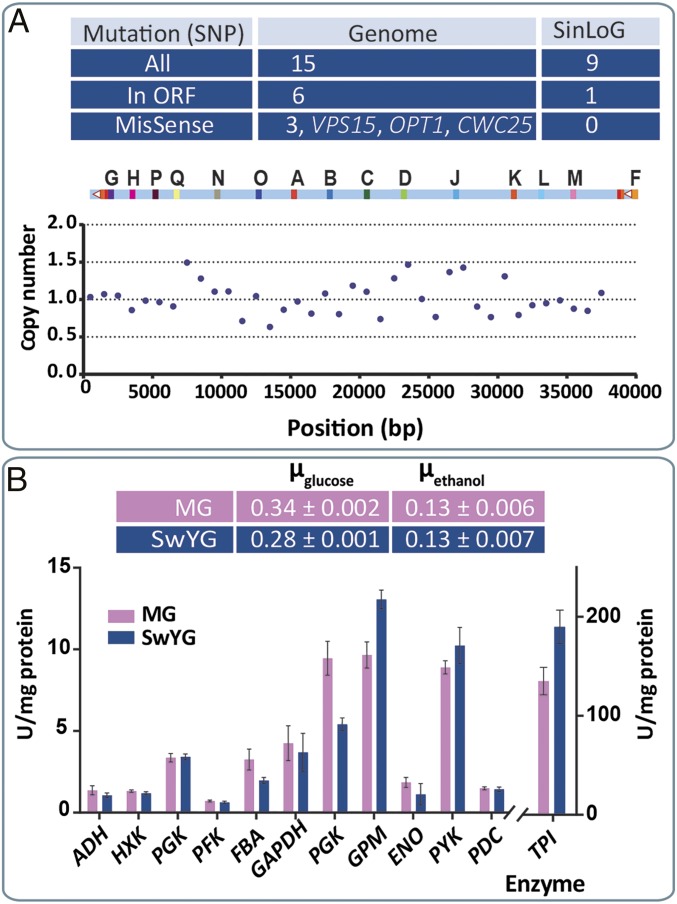
Fig. 2.
Characterization of SwYG. Next-gen sequencing and copy number analysis of the auxotrophic SwYG (IMX589) containing a clustered set of glycolytic genes (SinLoG-IX) (A). Physiological characterization in shake-flask culture using chemically defined medium with glucose as carbon source of IMX606 (prototrophic SwYG strain) and the MG strain (IMX370) (B). Growth rates (hour−1) and enzyme activity data represent the average and SEM of at least two independent culture replicates.
S. cerevisiae IMX382 was further engineered by deleting the 13 remaining glycolytic genes from their native loci (Fig. 1A and SI Appendix, Fig. S2). The first five deletions, targeting PYK1, PGI1, TPI1, TDH3, and PGK1, were performed with standard deletion cassettes, using I-SceI–mediated marker removal to recycle multiple selection markers simultaneously without leaving scars in the genome (9) (SI Appendix, Fig. S3). For subsequent engineering, an expression cassette encoding the CRISPR endonuclease Cas9 was integrated at the PFK2 locus, thereby deleting PFK2 (SI Appendix, Fig. S4). The remaining glycolytic genes, PFK1, GPM1, HXK2, FBA1, ADH1, and PDC1, were deleted from their native loci with the CRISPR/Cas9 system (10). Except for ENO2, all glycolytic expression cassettes harbored by the SinLoG-IX cluster were able to complement a null mutation in the corresponding native gene. The native promoter of ENO2, designed to be 411 bp long to avoid expressing unwanted ORFs from the glycolytic gene cluster, proved to be too short to drive expression of ENO2 and was replaced by a longer promoter (1,012 bp) (SI Appendix, Table S9). This observation underlines the limited knowledge, even for glycolytic genes, on promotor structure and function in yeast and highlights the need for systematic design of synthetic promoter. This last genetic modification yielded SwYG (IMX589). Whole-genome sequencing of this strain confirmed: (i) the correct sequence of the single-locus native glycolysis (SI Appendix, Table S1) and its integration at the SGA1 locus, (ii) deletion of the native glycolytic genes from their original loci (SI Appendix, Fig. S5), and (iii) absence of duplicated glycolytic genes in the single-locus native glycolysis and in the genome (Fig. 2A and SI Appendix, Fig. S6). Relative to the ancestor MG strain, only six ORFs in the genome of SwYG contained a nucleotide difference. Three of these caused an amino acid substitution in the encoded protein (Fig. 2A and SI Appendix, Table S2). None affected glycolytic genes or genes that are known to be otherwise associated with glycolysis. The specific growth rate of the prototrophic SwYG (IMX606) measured in aerobic batch in shake-flask culture, on chemically defined medium using glucose as carbon source, was slightly lower than that of the parental MG strain (17% lower) (Fig. 2B). However, more accurate quantification of specific growth rates in tightly controlled bioreactors revealed a stronger impact of the relocalization of the glycolytic gene cluster. The specific growth rate of SwYG was decreased by 28% compared with its parent MG (Fig. 3). Concomitantly the major metabolic fluxes, i.e., the specific glucose uptake, and ethanol production rates, were decreased in SwYG. Remarkably, the biomass and product yields on glucose remained unaffected by the relocalization of the glycolytic genes to chromosome IX (Fig. 3), showing that the growth stoichiometry was conserved and that only the magnitude of fluxes was attenuated in SwYG. These decreased rates of SwYG are unlikely to result from differences in glycolytic capacity as the activities of glycolytic enzymes in cell extracts of these two strains were highly similar (Fig. 2B).
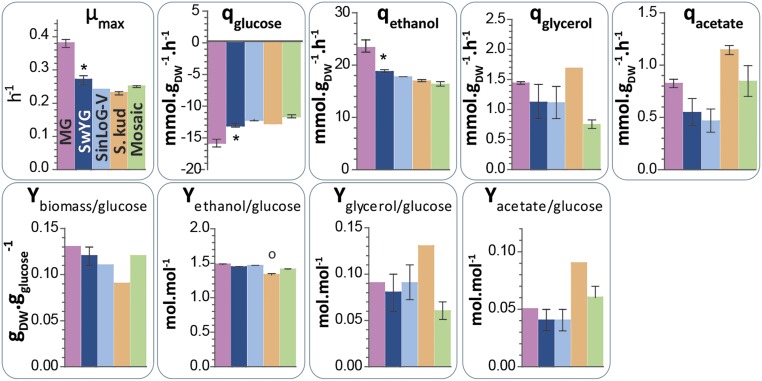
Fig. 3.
Physiological characterization during aerobic batches in bioreactors of MG (IMX370), SwYG (IMX606), SinLoG-V (IMX605), Sk-SinLoG-V (S. kud, IMX652), and mosaic SinLoG-V (Mosaic, IMX645). The strains were cultivated in chemically defined medium with glucose as carbon source. Bars and error bars represent the average and SEM of independent duplicate cultures. Stars indicate that the data from SwYG significantly differ from MG data; empty dots indicate significant differences between SwYG and the SwYG-based strains (P value <0.05, two-tailed t test, samples with equal variance). DW, biomass dry weight.
Chromosome Hopping of a Yeast Glycolysis Gene Cluster.
To test the feasibility of glycolysis swapping, we attempted to exchange the single-locus native glycolysis integrated on chromosome IX by a nearly identical copy integrated on chromosome V (SI Appendix, Fig. S7). SwYG was transformed with a complete set of glycoblocks (Fig. 4A), and with a CRISPR plasmid carrying a guide RNA (gRNA) designed to target Cas9 to the CAN1 locus on chromosome V, thus enabling in vivo assembly and integration of a second glycolytic gene cluster called SinLoG-V. Colony PCR showed that at least 2 of 12 G418-resistant transformants carried the complete set of genes in this SinLoG-V, correctly inserted at the CAN1 locus. After curing the URA3-carrying CRISPR plasmid, a selected clone was transformed with a 120-bp repair fragment and a new CRISPR plasmid carrying gRNAs targeting Cas9 to sequences positioned at each end of the SinLoG-IX, thereby excising the entire cluster from the genome. All three tested transformants were shown to lack the SinLoG-IX and to have retained the newly inserted single-locus native glycolysis on chromosome V. Whole-genome sequencing of one clone (IMX605) confirmed the successful relocalization of the entire glycolytic gene cluster and that no recombination had occurred between the glycoblocks or the excised single-locus native glycolysis gene cluster and the genome during glycolysis swapping (SI Appendix, Fig. S6). IMX605 grew as fast as SwYG in chemically defined medium (Fig. 4C) and displayed the same activity of the glycolytic enzymes in cell extracts (Fig. 4D). In IMX605, the position of the selectable marker and the ENO2 glycoblock were reversed compared with the SinLoG-IX cluster in SwYG. However, this different organization did not affect ENO2 expression (Fig. 4D). The similarity in specific growth rate between SwYG and IMX605 was also observed during growth in bioreactors (Fig. 3). These cultures in bioreactor also revealed identical metabolic rates and yields in the two strains (Fig. 3). This lack of locus-specific expression demonstrated that the CAN1 locus on chromosome V was a suitable “landing pad” for further testing of the pathway swapping concept.
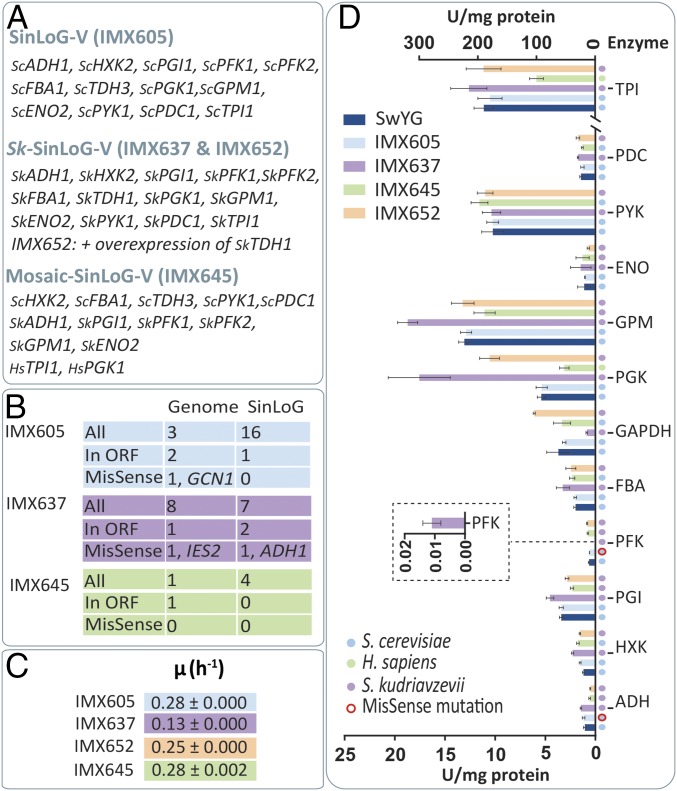
Fig. 4.
Characterization of yeast strains with a remodelled glycolysis. SinLoG-V (IMX605), Sk-SinLoG-V (IMX637), and mosaic SinLoG-V (IMX645) (A) were analyzed by next-gen sequencing to identify mutations compared with SwYG (B). The maximum specific growth rate on chemically defined medium with glucose as carbon source was measured (C), as well as in vitro enzymatic activity of the glycolytic enzymes from cell extracts (D). Data represent the average and SEM of at least two independent culture replicates.
S. cerevisiae Expressing a Heterologous Glycolytic Pathway.
Demonstration of the technical feasibility of pathway swapping opened up the way to test whether it is possible to integrally replace yeast glycolysis, an essential, tightly controlled metabolic pathway, by heterologous or synthetic variants. For this purpose, we selected a donor of glycolytic genes within the Saccharomyces genus. Saccharomyces kudriavzevii is a cold-tolerant close relative of S. cerevisiae, recently identified as an important contributor to wine making in cool climates (11, 12). Whereas glycolytic genes and enzymes of S. kudriavzevii have not been characterized in detail, its genome sequence is available (13). The complement of putative glycolytic genes in S. kudriavzevii and their sequences differ from those of the established S. cerevisiae glycolytic genes. However, putative S. kudriavzevii glycolytic genes with substantial identity (above 89% at the protein level) with their S. cerevisiae orthologs were easily identified by sequence comparison (SI Appendix, Table S3). Major glycolytic isoenzymes in S. kudriavzevii were selected based on high transcript levels of their structural genes (14). S. kudriavzevii does not contain a homolog of ScTDH3, the most highly expressed glycolytic gene in S. cerevisiae (15), but harbors two other putative TDH genes. SkTDH1 closely resembles ScTDH1, whereas SkTDH2 is more similar to ScTDH2 and ScTDH3. Based on its high expression level during wine fermentation, the gene homologous to ScTDH1 was selected for construction of a synthetic S. kudriavzevii glycolysis cluster. Because promoters within the Saccharomyces genus are functional in different species belonging to this group (16, 17), and promoters of S. kudriavzevii and S. cerevisiae are highly homologous (44–80% identity with an average of 72% for glycolytic promoters), S. kudriavzevii glycoblocks were constructed with their native promoters and terminators. Following the approach described above, the single-locus native glycolysis in SwYG was replaced by the S. kudriavzevii glycolysis integrated on chromosome V (Sk-SinLoG-V) (Fig. 4A). Transformants that only retained the S. kudriavzevii glycolysis, including a selected clone, IMX637, showed a strongly reduced specific growth rate on glucose, suggesting that the glycolytic function of the integrated set of S. kudriavzevii genes was suboptimal (Fig. 4C). Because SkTDH1 was not an ortholog of ScTDH3, we hypothesized that insufficient GAPDH activity caused the suboptimal specific growth rate. Indeed, the specific growth rate was almost completely restored to the level of SinLoG-V (IMX605) when SkTDH1 was overexpressed in the Sk-SinLoG-V strain IMX637 (strain IMX652) (Fig. 4C). Accordingly GAPDH activity, very low in the Sk-SinLoG-V strain, was boosted by overexpressing SkTDH1 (Fig. 4D). Enzyme activity assays also revealed a very low activity of phosphofructokinase in cell extracts of the Sk-SinLoG-V strain. Remarkably, this activity was fully restored upon overexpression of SkTDH1 (Fig. 4D). During growth in bioreactor, IMX652, carrying an overexpression of SkTDH1, grew nearly as fast as SwYG, but seemed to display a perturbation of its metabolic network (Fig. 3). Most of the observed increase in specific glycerol and acetate production rates and yields in IMX652, however, were not deemed statistically significant compared with SwYG (Fig. 3). Whereas these variations fell within measurement error, they would be consistent with a perturbation of flux distribution at the glyceraldehyde-3-phosphate branch point in favor of glycerol synthesis. Altogether these results illustrate how pathway swapping can identify interesting regulatory phenomena, and interesting leads for follow-up studies.
S. cerevisiae Expressing a Mosaic Glycolysis.
To further test the pathway swapping concept, we constructed a mosaic SinLoG composed of a combination of five S. cerevisiae, six S. kudriavzevii, and two Homo sapiens genes. HsTPI1 and HsPGK1 can complement null mutations in their S. cerevisiae orthologs (18, 19). The most abundant splicing variant of HsTPI1 (20) and the single splicing variant of HsPGK1 from muscle were codon optimized for expression in S. cerevisiae (SI Appendix, Table S4) and each stitched to the promoter and terminator of their respective yeast orthologs. The resulting human glycoblocks were pooled with HXK2, TDH3, PYK1, FBA1, and PDC1 glycoblocks from S. cerevisiae and PGI1, PFK1, PFK2, ENO2, GPM1, and ADH1 from S. kudriavzevii and transformed to SwYG, resulting in the integration of a mosaic glycolysis gene cluster in chromosome V (mosaic SinLoG-V gene cluster). Subsequent removal of the native SinLoG-IX gene cluster yielded strain IMX645, carrying a mosaic single-locus glycolytic gene cluster encoding a set of enzymes capable of supporting the entire glycolytic flux. Whole-genome sequencing confirmed the absence of the SinLoG-IX and the presence of the complete mosaic SinLoG-V. Just a single, silent nucleotide variation was detected within an ORF of the mosaic SinLoG IMX645 strain and its mosaic SinLoG-V cluster was identical to the in silico design (Fig. 4B and SI Appendix, Fig. S6 and Table S1). Although HsTPI1 and HsPGK1 expression was driven by the ScTPI1 and ScPGK1 promoters, in vitro activity of both encoded enzymes was approximately 50% lower in the mosaic glycolysis strain than in the native SinLoG-V IMX605 strain (Fig. 4D, t test, P value <0.01). Also the activity of SkADH was approximately 50% lower in IMX645 than in strain IMX605 (Fig. 4D, t test, P value <0.01). Despite these lower enzyme activities, the strain carrying the mosaic SinLoG-V grew as fast as SinLoG-V IMX605 both in shake flask and bioreactor, and its metabolic fluxes were undisturbed (Figs. 3 and 4), consistent with the notion that most glycolytic enzymes in S. cerevisiae have an overcapacity under standard laboratory growth conditions (15).
Discussion
This study demonstrates how modern genome-editing techniques can make essential biological processes, the partially redundant genetic information for which is scattered over an entire eukaryotic genome, accessible to fast, combinatorial analysis and optimization.
The high efficiency of the pathway swapping approach exceeded our expectations. Pathway swapping involves the transient, simultaneous presence in the yeast nucleus of two SinLoGs sharing high, and in one of the experiments even near-complete, sequence identity. One of these is integrated in chromosome V, whereas a second, 35-kb SinLoG, is excised from chromosome IX. DNA ends are highly recombinogenic and can interact directly with homologous sequences (21) and homologous recombination is the main double-strand break repair mechanism in growing S. cerevisiae (22, 23). The excised SinLoG-IX, or fragments generated by unspecific nuclease activity, might recombine with the newly integrated SinLoG-V. In practice, unintended genome rearrangements caused by HR, either between the engineered genetic elements themselves or between engineered genetic elements and the native yeast genome, were not observed. These results highlight the amazing efficiency and versatility of in vivo assembly and CRISPR/Cas9-facilitated genome editing in S. cerevisiae (6, 24).
Whereas genetic reduction of the glycolytic pathway did not lead to a detectable phenotype (2), clustering of the entire glycolytic gene set on a single locus resulted in decreased growth rate and metabolic fluxes. A logical explanation for this phenotype could be a decreased glycolytic capacity, resulting from reduced expression of the clustered and relocalized glycolytic genes; however, the activity of the glycolytic enzymes measured in vitro remained remarkably similar between the SwYG and MG strains. Relocalization of the entire glycolytic gene cluster from chromosome IX to V further substantiated the insensitivity of gene expression to large-scale targeted genome remodelling in S. cerevisiae. Whereas it is well documented that the genetic context can strongly influence gene expression, current knowledge does not allow for predictions on how the genomic site at which large synthetic gene clusters are integrated will affect transcription of genes harbored by such clusters. Using single-gene reporter systems, several studies have shown that localization in the vicinity of centromeres and telomeres leads to gene silencing, whereas proximity to autonomously replicating sequences (ARSs) tends to enhance transcription (25, 26). Although not completely understood at a mechanistic level, nucleosome positioning also modulates transcription (27). In the present study, glycolytic genes were concatenated and integrated at two different loci, without noticeable impact on expression. Whereas these rearrangements appear drastic, our design aimed at limiting epigenetic effects by integrating the glycolytic gene clusters in regions distant (>30 kb) from telomeres and centromeres and from active ARSs (>3 kb). Furthermore, nucleosome positioning in promoter regions is important for transcription and is strongly influenced by promoter sequences (28). In the SinLoGs, expression of the glycolytic genes was driven by S. cerevisiae promoters or by highly homologous S. kudriavzevii promoters. The genetic design of the SinLoGs may therefore explain the remarkable insensitivity of gene expression to relocalization.
Promoters of glycolytic genes are among the strongest in yeast and, consistent with the essentiality of the encoded proteins, glycolytic genes are constitutively expressed (29). Colocalization, in a single locus, of 13 genes that are heavily loaded with RNA polymerase II, might affect the conformation of DNA and thereby locally affect transcription. Moreover, Pol II disruption of chromatin has been proposed to increase sensitivity to several stresses (30) and to affect binding of proteins such as cohesin that play an important role in genetic stability (31). This colocalization of glycolytic genes into a 35-kb transcriptional hotspot had remarkably little impact on the expression of the glycolytic enzymes. Similar to the recently developed “telomerator” (26), SwYG offers an attractive experimental model to systematically explore the impact of broader genomic context and thereby guide the de novo design of synthetic yeast chromosomes.
The present data suggest that the reduced growth phenotype of the SwYG strain lineage is unlikely to result from a reduced glycolytic activity. Several other mechanisms, directly or indirectly related to the pathway swapping concept, could contribute to this slower growth phenotype. A potential epigenetic factor is DNA replication and the requirement for regularly spaced ARSs along chromosomes. Whereas ARSs are typically spaced by 30–40 kb in S. cerevisiae (32), insertion of the 35-kb sequence carrying the glycolytic genes resulted in a spacing between adjacent confirmed ARSs (ARS504.2 and ARS507, and ARS912 and ARS913) of 82 and 74 kb for chromosomes V and IX, respectively (33). In their design for a synthetic chromosome III, Annaluru et al. kept a conservative approach by maintaining 12 of the 19 native ARSs, with a maximum spacing between ARSs of ∼50 kb (34). Although it has been proposed that 120–300 kb of chromosomal DNA could be replicated from a single replication origin (35), a quantitative evaluation of the impact on the physiology of S. cerevisiae of increasing spacing between ARSs would facilitate the design of large chromosomal constructs and synthetic chromosomes. Another factor potentially involved in the slow growth phenotype of SwYG is the secondary function, unrelated to their catalytic role in glycolysis, of three glycolytic enzymes (36), which may be affected by the genetic relocalization. However, involvement of these secondary functions in the slow growth phenotype of SwYG does not appear very likely as the vacuolar role of Fba1 and Eno2 is not expected to lead to visible growth defects under acidic environments (the pH used was ≤6) (37, 38) and the physiological characterization of SwYG did not suggest an altered regulatory activity of Hxk2 (39). Alternatively, we cannot rule out that factors external to the clustering and relocalization of glycolytic genes are responsible for the slower growth of SwYG. For instance three genes, VPS15, CWC25, and OPT1, of which the first two are essential, have a missense mutation in SwYG compared with its parent MG. These mutations may be deleterious for the growth of S. cerevisiae. Further research is ongoing to evaluate contribution of these factors in the slow growth phenotype of the SwYG and SwYG-derived strains.
Functional replacement of the entire S. cerevisiae glycolysis by that of its close relative, the cold-tolerant yeast S. kudriavzevii, provided a proof of principle that pathway swapping can be used to rapidly express and study entire metabolic pathways in a heterologous context. S. kudriavzevii and S. cerevisiae are sympatric and both show fast, fermentative sugar dissimilation in glucose-rich media (40). Pathway swapping demonstrated that a set of S. kudriavzevii glycolytic enzymes can support glycolysis and growth of S. cerevisiae. Coevolution in the same ecological niches may have led to similar optima in terms of expression level and transcriptional regulation and explain the highly similar activities observed for most glycolytic enzymes upon replacement of all S. cerevisiae glycolytic genes by their S. kudriavzevii counterparts, controlled by their native promoters. Functional replacement of the full complement of 26 glycolytic genes in S. cerevisiae, deletion of a significant number of which is lethal in wild-type strains, by a set of 13 heterologous variants, demonstrates the potential of the pathway swapping concept for studying essential metabolic pathways. Moreover, it paves the way for modifications and, indeed, complete redesign of other multigene, essential cellular processes.
Approximately 60% of the S. cerevisiae genes share significant homology with human genes (41) and, moreover, an estimated 30% (42) of human genes connected with specific diseases have a yeast ortholog. The popularity of S. cerevisiae as a model eukaryote is further boosted by its experimental tractability and by the availability of a wide range of tools and technologies, that make this yeast particularly well suited for high throughput studies (43). “Humanized” yeast strains provide powerful models to explore effects of therapeutics, gene dosage, and wild-type or disease-causing variants of human genes on protein function (43). Recent examples include breakthroughs in research on cell-autonomous mechanisms of neurodegeneration and identification of drug candidates against neurodegenerative diseases (44, 45). Recent large-scale studies on the ability of human genes to complement native genes in S. cerevisiae demonstrated that complementation of haploid yeast gene knockouts is a reliable approach for functional characterization of human gene variants (43). Although elegant, many such studies are limited to single-gene complementation and require the generation of multiple yeast strains that each contain only a single ortholog of a studied heterologous gene. Pathway swapping enables the systematic analysis of heterologous complementation of entire pathways and should enable humanization of, for example, the complete glycolytic pathway. Availability of strains containing a fully or partially humanized glycolytic pathway will enable testing of the impact of mutations or drugs on human proteins in their natural glycolytic context and thereby identify potential synergetic effects between native human proteins.
The modular pathway swapping approach opens up unprecedented possibilities. Applications ranging from functional analysis of heterologous proteins, testing of kinetic models (now hindered by the multiplicity of paralogs) (46) or screening drugs, to more technical aspects such as exploring the effect of genomic location of highly expressed native pathways, are now within reach. Continued improvements in CRISPR-mediated removal of scattered genes (24) should even further facilitate functional clustering and fast, modular swapping of key pathways/processes.
A worldwide research effort has already led to the first synthetic yeast chromosome and is progressing toward the synthesis of an entire yeast genome (34). The present study demonstrates that a modular design of such synthetic yeast genomes, in which the genetic information for key processes is functionally clustered, offers unprecedented possibilities for fast, combinatorial exploration and optimization of the biological function of multigene, essential cellular functions.
Materials and Methods
SI Appendix, Figs. S1–S12 and Tables S1–S12 provides detailed information about the following: strains and media, molecular biology techniques, construction of glycoblocks and marker cassettes, construction of deletion cassettes and CRISPR-Cas9 plasmids, construction of the SwYG strain, construction of glycolytic gene clusters in the CAN1 locus, excision of the native SinLoG cassette, SkTDH1 overexpression, sequencing, determination of specific growth rates and in vitro enzyme activities in shake-flask cultures, and cultivation in bioreactors.
Supplementary Material
Acknowledgments
We thank M. Niemeijer, R. Brinkman, and M. Zaidi for their valuable contribution to the construction and confirmation of the S. kudriavzevii and H. sapiens glycoblocks and Prof. Amparo Querol and Prof. Eladio Barrio for kindly providing S. kudriavzevii CR85 and for advice on selecting the genes to design S. kudriavzevii minimal glycolysis. This work was supported by the Technology Foundation Stichting voor Technische Wetenschappen (STW) (Vidi Grant 10776) and by the European Research Council (Grant CoG-648141-AdLibYeast).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) database, www.ncbi.nlm.nih.gov (BioProject accession no. PRJNA317665).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606701113/-/DCSupplemental.
References
- 1.Meadows AL, et al. Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature. 2016;537(7622):694–697. doi: 10.1038/nature19769. [DOI] [PubMed] [Google Scholar]
- 2.Solis-Escalante D, et al. A minimal set of glycolytic genes reveals strong redundancies in Saccharomyces cerevisiae central metabolism. Eukaryot Cell. 2015;14(8):804–816. doi: 10.1128/EC.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wieczorke R, et al. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999;464(3):123–128. doi: 10.1016/s0014-5793(99)01698-1. [DOI] [PubMed] [Google Scholar]
- 4.Serrano L. Synthetic biology: Promises and challenges. Mol Syst Biol. 2007;3:158. doi: 10.1038/msb4100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson DG, et al. One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc Natl Acad Sci USA. 2008;105(51):20404–20409. doi: 10.1073/pnas.0811011106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuijpers NG, et al. A versatile, efficient strategy for assembly of multi-fragment expression vectors in Saccharomyces cerevisiae using 60 bp synthetic recombination sequences. Microb Cell Fact. 2013;12:47. doi: 10.1186/1475-2859-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solis-Escalante D, et al. amdSYM, a new dominant recyclable marker cassette for Saccharomyces cerevisiae. FEMS Yeast Res. 2013;13(1):126–139. doi: 10.1111/1567-1364.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuijpers NG, et al. One-step assembly and targeted integration of multigene constructs assisted by the I-SceI meganuclease in Saccharomyces cerevisiae. FEMS Yeast Res. 2013;13(8):769–781. doi: 10.1111/1567-1364.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solis-Escalante D, et al. Efficient simultaneous excision of multiple selectable marker cassettes using I-SceI-induced double-strand DNA breaks in Saccharomyces cerevisiae. FEMS Yeast Res. 2014;14(5):741–754. doi: 10.1111/1567-1364.12162. [DOI] [PubMed] [Google Scholar]
- 10.DiCarlo JE, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41(7):4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lopes CA, Barrio E, Querol A. Natural hybrids of S. cerevisiae × S. kudriavzevii share alleles with European wild populations of Saccharomyces kudriavzevii. FEMS Yeast Res. 2010;10(4):412–421. doi: 10.1111/j.1567-1364.2010.00614.x. [DOI] [PubMed] [Google Scholar]
- 12.Arroyo-López FN, Orlić S, Querol A, Barrio E. Effects of temperature, pH and sugar concentration on the growth parameters of Saccharomyces cerevisiae, S. kudriavzevii and their interspecific hybrid. Int J Food Microbiol. 2009;131(2–3):120–127. doi: 10.1016/j.ijfoodmicro.2009.01.035. [DOI] [PubMed] [Google Scholar]
- 13.Scannell DR, et al. The awesome power of yeast evolutionary genetics: New genome sequences and strain resources for the Saccharomyces sensu stricto genus. G3 (Bethesda) 2011;1(1):11–25. doi: 10.1534/g3.111.000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tronchoni J, Medina V, Guillamón JM, Querol A, Pérez-Torrado R. Transcriptomics of cryophilic Saccharomyces kudriavzevii reveals the key role of gene translation efficiency in cold stress adaptations. BMC Genomics. 2014;15:432. doi: 10.1186/1471-2164-15-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daran-Lapujade P, et al. The fluxes through glycolytic enzymes in Saccharomyces cerevisiae are predominantly regulated at posttranscriptional levels. Proc Natl Acad Sci USA. 2007;104(40):15753–15758. doi: 10.1073/pnas.0707476104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliveira BM, Barrio E, Querol A, Pérez-Torrado R. Enhanced enzymatic activity of glycerol-3-phosphate dehydrogenase from the cryophilic Saccharomyces kudriavzevii. PLoS One. 2014;9(1):e87290. doi: 10.1371/journal.pone.0087290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young ET, et al. Evolution of a glucose-regulated ADH gene in the genus Saccharomyces. Gene. 2000;245(2):299–309. doi: 10.1016/s0378-1119(00)00035-4. [DOI] [PubMed] [Google Scholar]
- 18.Grüning NM, Du D, Keller MA, Luisi BF, Ralser M. Inhibition of triosephosphate isomerase by phosphoenolpyruvate in the feedback-regulation of glycolysis. Open Biol. 2014;4:130232. doi: 10.1098/rsob.130232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kachroo AH, et al. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science. 2015;348(6237):921–925. doi: 10.1126/science.aaa0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maquat LE, Chilcote R, Ryan PM. Human triosephosphate isomerase cDNA and protein structure. Studies of triosephosphate isomerase deficiency in man. J Biol Chem. 1985;260(6):3748–3753. [PubMed] [Google Scholar]
- 21.Orr-Weaver TL, Szostak JW, Rothstein RJ. Yeast transformation: A model system for the study of recombination. Proc Natl Acad Sci USA. 1981;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 23.Daley JM, Sung P. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol Cell Biol. 2014;34(8):1380–1388. doi: 10.1128/MCB.01639-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mans R, et al. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 2015;15(2):15. doi: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flagfeldt DB, Siewers V, Huang L, Nielsen J. Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast. 2009;26(10):545–551. doi: 10.1002/yea.1705. [DOI] [PubMed] [Google Scholar]
- 26.Mitchell LA, Boeke JD. Circular permutation of a synthetic eukaryotic chromosome with the telomerator. Proc Natl Acad Sci USA. 2014;111(48):17003–17010. doi: 10.1073/pnas.1414399111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wyrick JJ, et al. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402(6760):418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 28.Iyer VR. Nucleosome positioning: Bringing order to the eukaryotic genome. Trends Cell Biol. 2012;22(5):250–256. doi: 10.1016/j.tcb.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinisch JJ, Rodicio R. Fructose-1,6-bisphophate aldolase, triosephosphate isomerase, glyceraldhyde-3-phosphate dehydrogenases, and phosphoglycerate mutase. In: Zimmermann FK, Entian K-D, editors. Yeast Sugar Metabolism. CRC; Boca Raton, FL: 1997. pp. 119–140. [Google Scholar]
- 30.Ide S, Miyazaki T, Maki H, Kobayashi T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science. 2010;327(5966):693–696. doi: 10.1126/science.1179044. [DOI] [PubMed] [Google Scholar]
- 31.Bausch C, et al. Transcription alters chromosomal locations of cohesin in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27(24):8522–8532. doi: 10.1128/MCB.01007-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dhar MK, Sehgal S, Kaul S. Structure, replication efficiency and fragility of yeast ARS elements. Res Microbiol. 2012;163(4):243–253. doi: 10.1016/j.resmic.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Siow CC, Nieduszynska SR, Müller CA, Nieduszynski CA. OriDB, the DNA replication origin database updated and extended. Nucleic Acids Res. 2012;40(Database issue):D682–D686. doi: 10.1093/nar/gkr1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Annaluru N, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344(6179):55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williamson DH. The timing of deoxyribonucleic acid synthesis in the cell cycle of Saccharomyces cerevisiae. J Cell Biol. 1965;25(3):517–528. doi: 10.1083/jcb.25.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gancedo C, Flores CL. Moonlighting proteins in yeasts. Microbiol Mol Biol Rev. 2008;72(1):197–210. doi: 10.1128/MMBR.00036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu M, Sautin YY, Holliday LS, Gluck SL. The glycolytic enzyme aldolase mediates assembly, expression, and activity of vacuolar H+-ATPase. J Biol Chem. 2004;279(10):8732–8739. doi: 10.1074/jbc.M303871200. [DOI] [PubMed] [Google Scholar]
- 38.Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev. 2006;70(1):177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diderich JA, Raamsdonk LM, Kruckeberg AL, Berden JA, Van Dam K. Physiological properties of Saccharomyces cerevisiae from which hexokinase II has been deleted. Appl Environ Microbiol. 2001;67(4):1587–1593. doi: 10.1128/AEM.67.4.1587-1593.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonçalves P, Valério E, Correia C, de Almeida JM, Sampaio JP. Evidence for divergent evolution of growth temperature preference in sympatric Saccharomyces species. PLoS One. 2011;6(6):e20739. doi: 10.1371/journal.pone.0020739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Botstein D, Chervitz SA, Cherry JM. Yeast as a model organism. Science. 1997;277(5330):1259–1260. doi: 10.1126/science.277.5330.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Foury F. Human genetic diseases: A cross-talk between man and yeast. Gene. 1997;195(1):1–10. doi: 10.1016/s0378-1119(97)00140-6. [DOI] [PubMed] [Google Scholar]
- 43.Laurent JM, Young JH, Kachroo AH, Marcotte EM. Efforts to make and apply humanized yeast. Brief Funct Genomics. 2016;15(2):155–163. doi: 10.1093/bfgp/elv041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 45.Park SK, et al. Development and validation of a yeast high-throughput screen for inhibitors of Aβ42 oligomerization. Dis Model Mech. 2011;4(6):822–831. doi: 10.1242/dmm.007963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vavouri T, Semple JI, Lehner B. Widespread conservation of genetic redundancy during a billion years of eukaryotic evolution. Trends Genet. 2008;24(10):485–488. doi: 10.1016/j.tig.2008.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.