Significance
p53 mutations are the most frequent genetic alteration in cancer and are often indicative of poor patient survival prognosis. The most prevalent missense mutations lead to a “gain of function” (GOF) that actively drives tumor progression, metastasis, and therapy resistance. Our study links the mutant p53 (mutp53) GOF to enhanced N-glycoprotein folding via ectonucleoside triphosphate diphosphohydrolase 5 (ENTPD5) in the calnexin/calreticulin cycle of the endoplasmic reticulum. Mutp53 thus increases expression of prometastatic cell surface proteins, such as receptors and integrins, not only quantitatively but also qualitatively, with respect to N-glycosylation state. Our study reveals N-glycoprotein quality control in the endoplasmic reticulum as an indispensable mechanism underlying the progression of tumors with GOF mutp53 that could provide new possibilities for treating prognostically challenging p53-mutated cancers.
Keywords: p53, tumor suppressor, ENTPD5, N-glycosylation, metastasis
Abstract
Mutations in the p53 tumor suppressor gene are the most frequent genetic alteration in cancer and are often associated with progression from benign to invasive stages with metastatic potential. Mutations inactivate tumor suppression by p53, and some endow the protein with novel gain of function (GOF) properties that actively promote tumor progression and metastasis. By comparative gene expression profiling of p53-mutated and p53-depleted cancer cells, we identified ectonucleoside triphosphate diphosphohydrolase 5 (ENTPD5) as a mutant p53 target gene, which functions as a uridine 5′-diphosphatase (UDPase) in the endoplasmic reticulum (ER) to promote the folding of N-glycosylated membrane proteins. A comprehensive pan-cancer analysis revealed a highly significant correlation between p53 GOF mutations and ENTPD5 expression. Mechanistically, mutp53 is recruited by Sp1 to the ENTPD5 core promoter to induce its expression. We show ENTPD5 to be a mediator of mutant p53 GOF activity in clonogenic growth, architectural tissue remodeling, migration, invasion, and lung colonization in an experimental metastasis mouse model. Our study reveals folding of N-glycosylated membrane proteins in the ER as a mechanism underlying the metastatic progression of tumors with mutp53 that could provide new possibilities for cancer treatment.
Mutations in the TP53 tumor suppressor gene are the most frequent genetic alterations in human cancer and commonly compromise the gene’s tumor suppressor activity. p53-knockout mice succumb to tumors very early in life, arguing that the loss of function associated with p53 mutations is sufficient on its own to explain the high mutation frequency observed in patients with cancer (1). However, in striking contrast to mutations in other tumor suppressor genes, the vast majority of TP53 gene alterations in patients with cancer neither ablate p53 expression nor produce unstable or truncated proteins. Instead, p53 mutations are mostly missense mutations resulting in expression of mutant p53 (mutp53) proteins with only single-amino acid substitutions that accumulate to steady-state levels greatly exceeding those of wild-type p53 (wtp53) in normal tissues. Immunohistochemical positivity for p53 is therefore considered a diagnostic marker for the presence of a TP53 mutation (2). The high prevalence of missense mutations suggests a selective advantage during cancer progression, so it was hypothesized early on in p53 research that p53 mutations are neomorphic and endow the mutp53 protein with novel oncogenic functions that actively promote cancer progression and therapy resistance (2). These oncogenic properties are generally referred to as the mutp53 gain of function (GOF).
Over the years, substantial experimental evidence for mutp53 GOF has accumulated (3–5). For example, mice expressing cancer-associated p53 hot spot mutations from the endogenous Trp53 gene locus are remarkably different from p53-deficient mice: tumorigenesis is accelerated, and the spectrum of tumors is shifted toward carcinomas and more metastatic tumors (6–8). Of note, the mutp53 GOF appears to be mutation-specific, as not all mutations engineered into the p53 gene show the same phenotype (8–10). Importantly, tumors arising in mice with mutp53 GOF are addicted to sustained mutp53 expression and undergo tumor regression or stagnation on mutp53 gene ablation, thereby providing proof-of-principle evidence for mutp53 GOF as an actionable cancer-specific drug target (11). Although previous research on drugging mutp53 was primarily focused on restoring wtp53-like functions to mutp53 (12), addiction to mutp53 implies that small compound inhibitors of the mutp53 GOF might suffice to induce therapeutic responses. Promising strategies include the promotion of mutp53 degradation (11), interference with mutp53 aggregation (13), and inhibition of mutp53-specific protein–protein interactions or downstream pathways (14, 15). A more detailed knowledge of the mutp53 GOF effector mechanisms is therefore instrumental for developing therapeutic targeting approaches.
Mechanistically, the mut53 GOF appears to involve a variety of different facets, including chemotherapy resistance, metabolic deregulation, and increased metastasis (2–5, 16). Although effects of wtp53 are primarily mediated by sequence-specific DNA binding to cognate p53 response elements located in regulatory regions of p53 target genes, this DNA binding is commonly prevented by cancer-associated missense mutations clustered in the DNA binding domain. Nevertheless, mutp53 has a broad effect on gene expression profiles by binding genes indirectly through interactions with other transcription factors; for example p63/p73, Sp1/Sp3, NF-Y, ETS2, vitamin D receptor, or SREBP2 (4), and by regulating chromatin-modifying enzymes such as the ATP-dependent nucleosome remodeling complex SWI/SNF and the histone H3 lysine 4 methyltransferases MLL1 and MLL2 (14, 16, 17). By increasing the expression of various receptor tyrosine kinases (RTKs), such as transforming growth factor β (TGFβ) receptor, epidermal growth factor receptor (EGFR), hepatocyte growth factor receptor (HGFR/c-MET), and platelet-derived growth factor (PDGF) receptor β, mutp53 enhances proinvasive signaling, which is further reinforced by stimulatory effects of mutp53 on integrin/RCP-driven receptor recycling (18–21).
Here, we identified ENTPD5 as a mutp53-specific target gene that promotes the calnexin/calreticulin-mediated folding of N-glycoproteins in the endoplasmic reticulum (ER) (22, 23) (SI Appendix, Fig. S1). The synthesis of N-linked glycans begins in the lumen of the ER with an en bloc transfer of an invariant presynthesized core oligosaccharide (24). Improperly folded glycoproteins are sensed by the UDP-glucose:glycoprotein glucosyltransferase (UGGT) and tagged by transfer of a glucose moiety from UDP-glucose to the core oligosaccharide. Such monoglucosylated glycoproteins are sequestered by the lectins calnexin and calreticulin, which serve as molecular chaperones, preventing aggregation and export of incompletely folded proteins from the ER. The single glucose residue can be removed by glucosidase II, which releases the bound protein from calnexin/calreticulin for export to the Golgi, unless recognized again as unfolded by UGGT and tagged with glucose for another round of chaperone-mediated folding. Completion of the calnexin/calreticulin cycle requires cleavage of UDP to UMP by the UDPase ENTPD5, so UMP can leave the ER through an antiporter in exchange for new UDP-glucose. Interestingly, RTKs that promote cell growth and proliferation, such as EGFR, are much more highly N-glycosylated than receptors that do not promote cell growth and proliferation (25), suggesting that high-level expression of ENTPD5 might be required to support the oncogenic functions of RTKs, and thereby prove critical for RTK-addicted tumor cells. In fact, ENTPD5 was shown to be essential for sustained tumor cell proliferation and in vivo tumor growth in xenograft mouse models of prostate cancer (22). Here, we show that mutp53 enhances ENTPD5 expression, thereby promoting N-glycoprotein maturation, tumor cell proliferation, invasion, and metastasis.
Results
Identification of ENTPD5 as a mutp53 Target Gene.
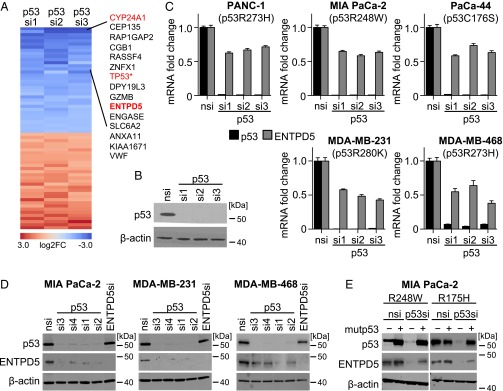
TP53 mutations are frequent mutational events in 75% of pancreatic adenocarcinoma that occur at the transition from benign pancreatic intraepithelial neoplasias to highly aggressive, invasive, and metastatic pancreatic ductal adenocarcinomas (PDAC) (20). Consistent with mutp53 driving invasion and metastatic progression, p53 accumulation in human PDAC significantly correlates with lymph node metastasis, and mice harboring pancreatic cancers driven by oncogenic Kras and the GOF mutant Trp53R172H allele show more metastases compared with mice harboring a Trp53 null allele (26). To better understand the underlying mechanism of the mutp53 GOF, we depleted the mutp53R273H protein in the human PDAC cell line PANC-1, using three independent siRNAs, and characterized the resulting gene expression changes by genome-wide expression profiling (Fig. 1 A and B). In total, 65 genes were found to be up- or down-regulated consistently by all three siRNAs, with an absolute log2FC of greater than 1 (Dataset S1). Among the top 15 down-regulated genes were TP53 itself and CYP24A1, a gene regulated by mutp53 in a vitamin D receptor-dependent manner (27), validating that the profiling strategy properly identifies mutp53 targets. Among the novel mutp53-regulated genes, we noted ENTPD5, a UDPase that promotes the calnexin/calreticulin-mediated folding of glycoproteins in the ER (22). As RTKs involved in cell proliferation, growth, and oncogenesis, including EGFR, MET, and PDGFRB, are the most heavily N-glycosylated receptors (25), and also decisive mediators of the prometastatic activity of mutp53 (18, 20, 21), we hypothesized that the ER-resident UDPase ENTPD5, which drives the calnexin/calreticulin cycle, might be a crucial downstream target of the mutp53 GOF.
Fig. 1.
Identification of ENTPD5 as a mutp53 target gene. (A) Heat map depicting differentially expressed genes on knockdown of mutp53 with three different siRNAs in PANC-1 cells. Shown are all protein coding genes with a fold change of >2 or <0.5 for all three p53 siRNAs vs. nontargeting siRNA control. *TP53 carries the R273H mutation. (B) Knockdown efficiency of mutp53 by the indicated p53 siRNAs in PANC-1 cells analyzed by Western blot. (C) ENTPD5 and p53 mRNA expression analyzed by quantitative reverse transcription PCR (RTqPCR) in indicated cell lines after transfection with siRNAs targeting p53 and an nsi. Shown are median ± SEM (n = 6) normalized to GAPDH. (D) Protein expression of p53 and ENTPD5 analyzed by Western blot after siRNA transfection. (E) MIA PaCa-2 cells were stably transduced with vectors for doxycycline-inducible expression of p53 R248W or R175H mutant. Doxycycline was added 24 h before transfection with an siRNA targeting the endogenous mutp53 3′UTR or a nontargeting siRNA control (nsi). p53 and ENTPD5 protein levels were determined by Western blot. In all Western blots, β-actin served as a loading control.
We therefore first validated regulation of ENTPD5 expression by mutp53 in other pancreatic cancer cell lines harboring distinct p53 missense mutations. Depletion of mutp53 by three independent siRNAs resulted in comparable down-regulation of ENTPD5 at the mRNA and protein level in PANC-1, MIA PaCa-2, and PaCa-44 cells containing the TP53 R273H, R248W, and C176S mutations, respectively (Fig. 1 C and D and SI Appendix, Fig. S2A). Similarly, stable knockdown of mutp53 with lentiviral shRNA vectors resulted in sustained down-regulation of ENTPD5 (SI Appendix, Fig. S2B). Furthermore, ENTPD5 down-regulation on mutp53 silencing was also observed in breast adenocarcinoma cell lines MDA-MB-231, MDA-MB-468, and T-47D with the TP53 R280K, R273H, and L194F mutations, respectively, indicating that the correlation between mutp53 and high-level ENTPD5 expression is not restricted to pancreatic cancer (Fig. 1 C and D and SI Appendix, Fig. S2A). In addition, targeting mutp53 with pharmacological inhibitors of the HSP90/HDAC6 chaperone machinery, which is a major determinant of mutp53 stability (11), selectively down-regulated ENTPD5 expression in mutp53-containing MIA PaCa-2, but not wtp53 or p53-null, cells (SI Appendix, Fig. S2C). Importantly, mutp53-dependent expression of ENTPD5 in a wide range of cell lines with different p53 missense mutations indicates that this regulation is not restricted to single missense mutations. In support, ENTPD5 expression in mutp53-depleted MIA PaCa-2 was not only restored by ectopic expression of p53R248W, the endogenous mutation in these cells, but similarly by p53R175H (Fig. 1E).
Mutp53 regulates some of the same target genes as described for wtp53; however, the outcome is often exactly reciprocal: Whereas mutp53 augments expression of, for example, vitamin D receptor target genes, wtp53 represses some of these target genes (4, 27). We therefore asked whether ENTPD5 is also regulated by wtp53. However, ectopic expression of wtp53 in p53-null Saos-2 cells, just like activation of endogenous wtp53 in MCF-7 or U2OS cells by DNA damage (etoposide) or nongenotoxic MDM2 inhibition (nutlin-3a), failed to alter ENTPD5 expression, despite strong activation of the bona fide wtp53 target gene p21CDKN1A (SI Appendix, Fig. S3). We conclude that ENTPD5 is a specific target gene of mutp53.
ENTPD5 Expression Levels Correlate with GOF p53 in Human Tumor Samples.
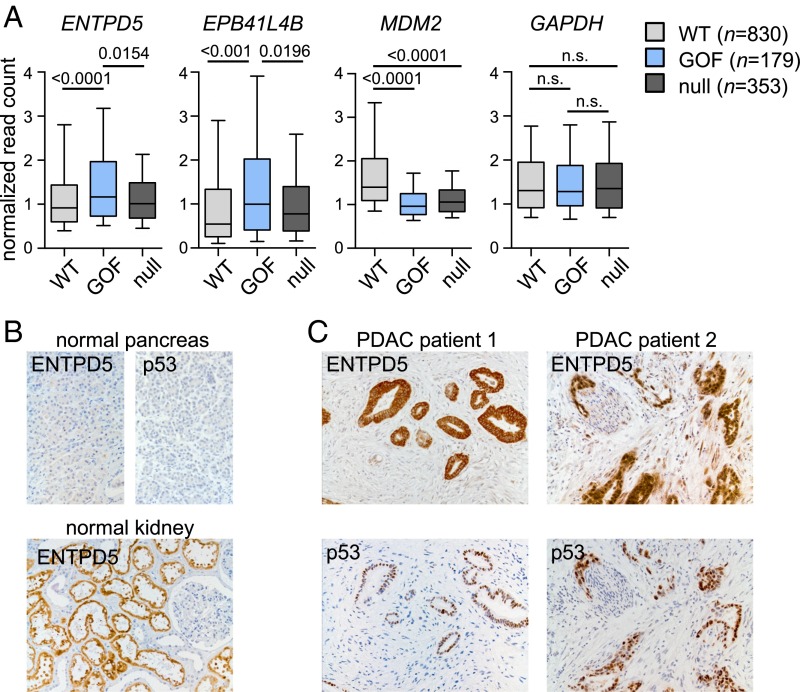
We examined the significance of our findings in the context of human tumor samples by an integrated analysis of exome and RNA sequencing data of The Cancer Genome Atlas (TCGA). To explore a correlation between TP53 GOF missense mutations and ENTPD5 expression, tumor samples were grouped according to their p53 mutation status as wild-type, GOF (missense mutation of R175H, R248Q, R248W, R249S, or R273H), or p53 null (p53 nonsense mutations or frameshift truncations), as previously described (14). Other p53 mutations (other missense mutations, in-frame insertions/deletions, or splicing mutations) were not included in further analysis because of an unpredictable effect on p53 function (14). We focused our further analysis on cancer types that include more than 5% of samples in the p53 GOF group. As expected, MDM2 as a canonical target gene of wtp53 was expressed at significantly higher levels in the group of tumors with wtp53, whereas expression of EPB41L4B, a previously reported prometastatic target gene of p53 GOF mutants (28), was elevated in the GOF group (Fig. 2A). Consistent with a mutp53-dependent expression, ENTPD5 RNA levels were also significantly higher in the group of tumors with GOF mutations compared with the group of wtp53- or p53-null tumors. Expression of GAPDH as a housekeeping gene showed no significant differences. Therefore, p53 GOF mutations correlate with high-level expression of ENTPD5 across a broad panel of patients with different tumor entities.
Fig. 2.
ENTPD5 expression levels correlate with GOF p53 in human tumor samples. (A) Box plots of TCGA RNA expression profiles in tumors with wtp53, p53 GOF, or p53 null. Statistical analysis was calculated using nonparametric Mann–Whitney U tests followed by Benjamini-Hochberg correction. (B and C) Immunohistochemistry of p53 and ENTPD5 in (B) normal human pancreas and kidney tissues (positive control) and (C) serial sections of human pancreatic adenocarcinoma samples.
As immunohistochemical detection of p53 in tumors usually indicates TP53 missense mutation (2), we analyzed p53 and ENTPD5 protein expression in human tissue samples (Fig. 2B and SI Appendix, Fig. S4). As expected, healthy pancreas tissue showed no expression of p53. ENTPD5 was only barely detectable in normal pancreas, whereas in human kidneys, used as positive controls (www.proteinatlas.org), proximal tubules were strongly positive (Fig. 2B). The majority of p53-immunopositive PDAC and breast cancer samples showed strong staining for ENTPD5, which in cases of heterogeneous p53 staining, correlated strikingly with the p53 expression pattern (Fig. 2C and SI Appendix, Fig. S4).
mutp53 Induces ENTPD5 Expression via Sp1.
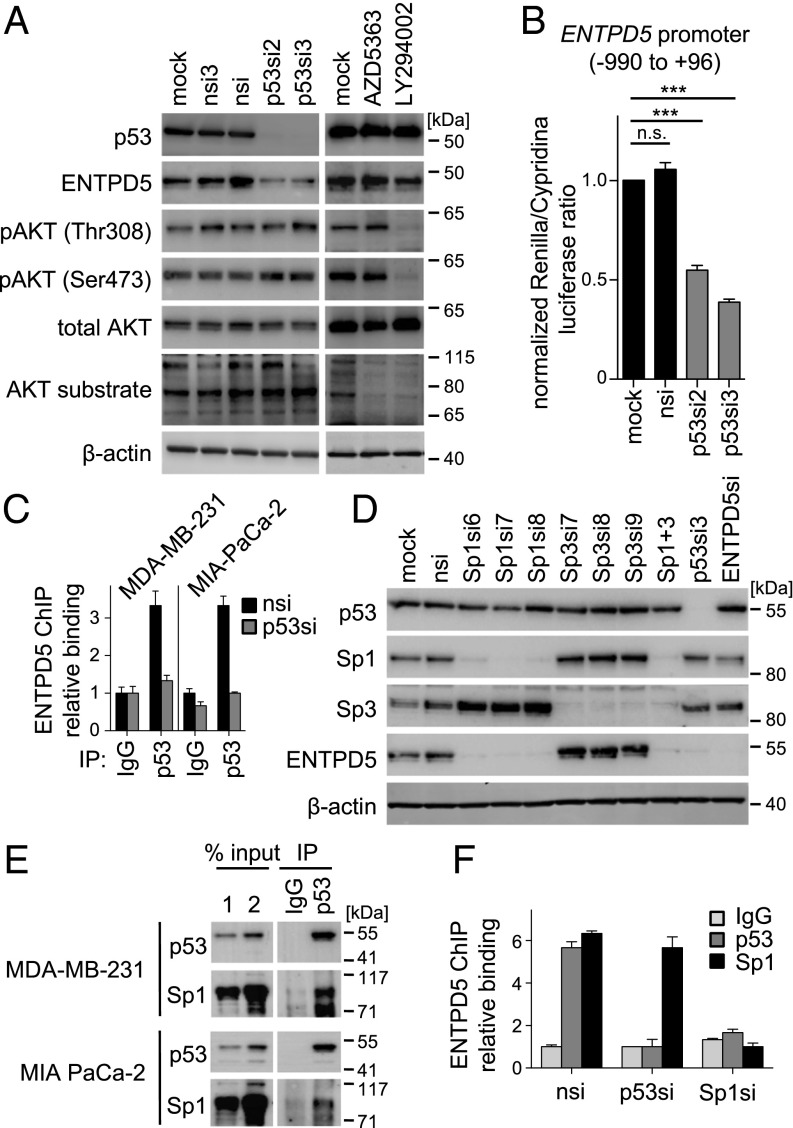
ENTPD5 was initially identified biochemically because of its highly elevated expression in phosphatase and tensin homolog (PTEN)-deficient mouse embryonic fibroblasts, in which elevated levels of phosphatidylinositol 3,4,5-trisphosphate relieve the ENTPD5 promoter from repression by FoxO transcription factors via the serine/threonine kinase AKT (22). We therefore asked whether ENTPD5 expression in mutp53 cancer cells is under control of AKT. First, knockdown of mutp53 in MIA PaCa-2 clearly reduced ENTPD5 expression, but did not affect the activating PI3K- and mammalian target of rapamycin complex 2 (mTORC2)-mediated phosphorylation of AKT at S473 and T308, and similarly did not reduce phosphorylation of AKT substrates, meaning that AKT signaling is not altered by mutp53 knockdown conditions sufficient to down-regulate ENTPD5 (Fig. 3A). Second, inhibition of AKT signaling in MIA PaCa-2 cells with pharmacological inhibitors of AKT (AZD5363) or PI3K (LY294002) did not result in ENTPD5 down-regulation (Fig. 3A). Together, this indicates that mutp53 does not induce ENTPD5 expression through AKT signaling.
Fig. 3.
mutp53 induces ENTPD5 expression via Sp1. (A) Western blot analysis of AKT signaling in MIA PaCa-2 cells after transfection with p53-targeting and nontargeting siRNAs or treatment with pharmacological inhibitors of AKT (AZD5363) and PI3K (LY294002). (B) ENTPD5 promoter activity (Renilla) in MIA PaCa-2 cells after depletion of mutp53 compared with controls (mock, nsi) normalized to cotransfected CMV promoter activity (Cypridina). Shown are mean ± SEM (n = 6). Statistical analysis was calculated using one-way analysis of variance and Bonferoni posttest (***P < 0.0001; n.s. not significant). (C) Chromatin immunoprecipitation in MDA-MB-231 and MIA PaCa-2 cells transfected with the indicated siRNAs. Binding of p53 to the ENTPD5 promoter was quantified by qPCR. Binding is shown as mean percentage input normalized to the IgG nsi-negative control sample ± SEM (n = 3). (D) Protein expression of p53, Sp1, Sp3, ENTPD5, and β-actin (control) in MIA PaCa-2 cells analyzed by Western blot after transfection with indicated siRNAs. (E) Protein extracts of MDA-MB-231 and MIA PaCa-2 cells were subjected to immunoprecipitation (IP) with IgG or p53 antibodies, followed by immunoblotting for p53 and Sp1. (F) Chromatin immunoprecipitation in MDA-MB-231 transfected with the indicated siRNAs. Binding of p53 and Sp1 to the ENTPD5 promoter was quantified by qPCR. Binding is shown as mean percentage input normalized to the IgG nsi negative control sample ± SEM (n = 3).
Mutp53 has been shown to regulate gene expression via a multitude of different mechanisms, both transcriptional and nontranscriptional (4). Low ENTPD5 mRNA levels in mutp53-depleted cells could, in principle, be explained by reduced mRNA production or increased degradation. To test for changes in mRNA production, we measured ENTPD5 promoter activity, using a luciferase reporter assay. Depletion of mutp53 reduced luciferase reporter activity to ∼50% (Fig. 3B), nicely recapitulating the effect on ENTPD5 mRNA steady-state levels (Fig. 1C). In addition, we detected specific binding of mutp53 to the ENTPD5 core promoter by chromatin immunoprecipitation in both MIA PaCa-2 and MDA-MB-231 cells (Fig. 3C). The signal was weaker than binding of wtp53 to its canonical target genes (29) suggesting that recruitment of mutp53 to the ENTPD5 promoter is indirect. Although there is no unifying hypothesis to explain the ability of mutp53 to regulate target gene promoter activity, one of the most commonly proposed mechanisms posits that mutp53 interacts with other sequence-specific transcription factors to modulate their transcriptional activity on respective target genes (4). In fact, the ENTPD5 core promoter contains a CpG island with multiple GC boxes, the characteristic binding motif for Sp-family members (SI Appendix, Fig. S5). Reduced ENTPD5 expression at the mRNA and protein level on depletion of Sp1 (not Sp3) in both MIA PaCa-2 and MDA-MB-231 cells indicates a specific requirement of Sp1 for sustained high-level ENTPD5 expression (Fig. 3D and SI Appendix, Fig. S6). Furthermore, endogenous coimmunoprecipitation experiments demonstrated interactions between mutp53 and Sp1 in both cell lines (Fig. 3E) which were stable in the presence of ethidium bromide, and therefore DNA-independent (SI Appendix, Fig. S7) (30). In chromatin immunoprecipitation experiments, mutp53 recruitment to ENTPD5 was dependent on Sp1, but not vice versa (Fig. 3F). Together, these data indicate that mutp53 docks onto Sp1 to increase ENTPD5 promoter activity.
mutp53 and ENTPD5 Promote N-Glycoprotein Folding and Maturation.
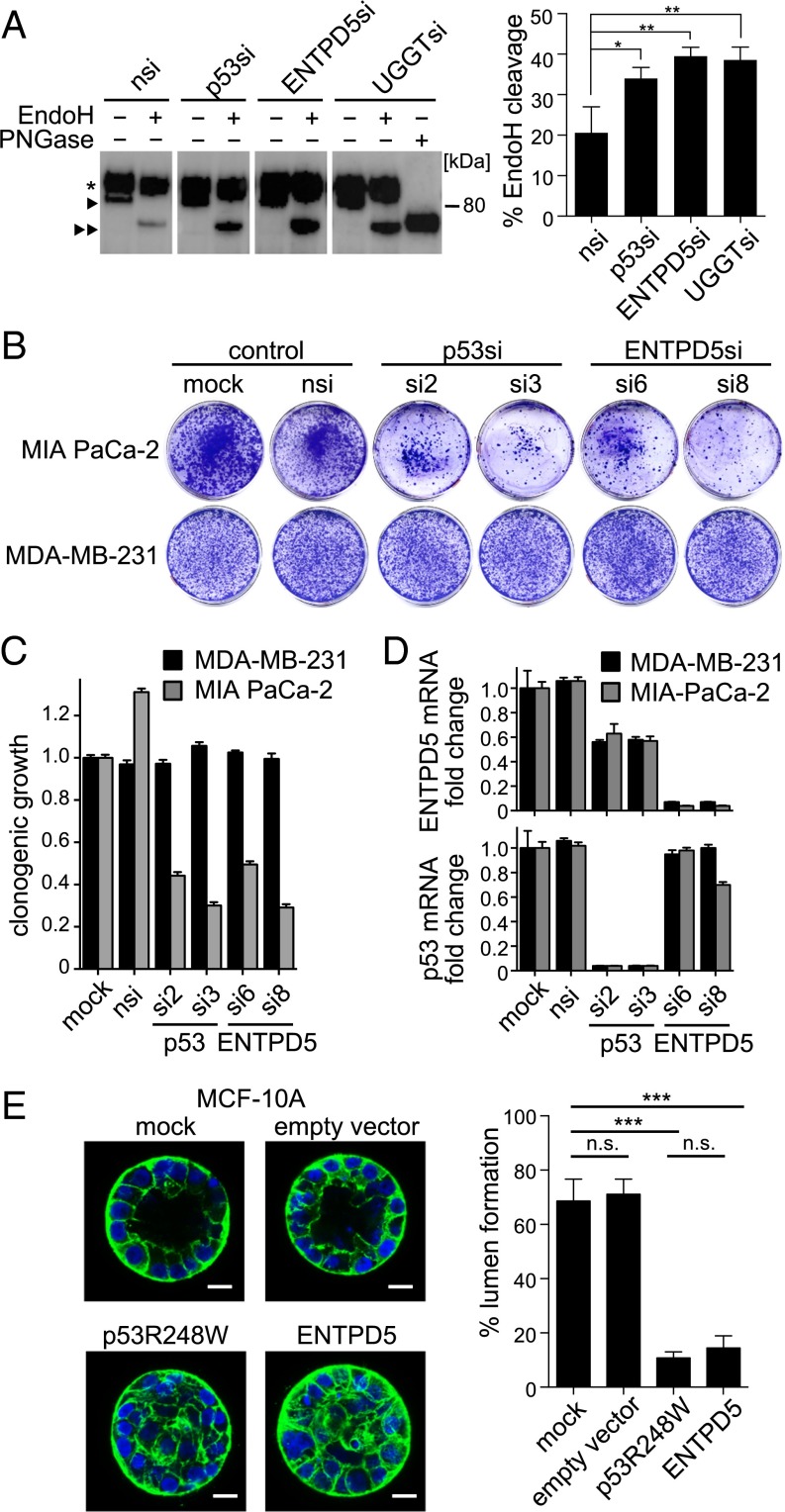
To explore whether mutp53 and ENTPD5 enhance folding and maturation of N-glycoproteins, we investigated endoglin/CD105, a coreceptor for TGFβ that is implicated in migration, invasion, and metastasis of breast and pancreatic cancer cells (19, 31, 32). Endoglin function relies exquisitely on protein folding in the calnexin/calreticulin cycle because endoglin mutants that colocalize with calnexin in the ER cause hereditary hemorrhagic telangiectasia (33, 34). Similar to the immature N-glycosylation pattern of hereditary hemorrhagic telangiectasia–associated endoglin mutants (33), we observed an increase in immature N-glycosylated wild-type endoglin in mutp53- and ENTPD5-depleted MDA-MB-231 cells (Fig. 4A). In detail, transfected wild-type endoglin appeared as two separate bands, both of which contained N-glycans that were susceptible to deglycosylation by peptide:N-glycosidase F (Fig. 4A). However, endoglycosidase H (EndoH) only cleaved the fast-migrating band that was enriched in mutp53- and ENTPD5-depleted cells (Fig. 4A). As EndoH specifically cleaves immature N-glycan side chains, but not the complex N-linked oligosaccharides acquired during processing in the Golgi, this indicates a defect in N-glycoprotein maturation induced by mutp53- or ENTPD5-depletion. UGGT-knockdown cells, which, as a positive control, cannot tag unfolded proteins with the single glucose moiety required for binding to calnexin and calreticulin, showed the same phenotype as mutp53- or ENTPD5-knockdown cells (Fig. 4A). Together, these results support that the mutp53-ENTPD5 axis promotes N-glycoprotein folding in the ER to enhance export to the Golgi, where N-glycan maturation generates the mature functional membrane proteins.
Fig. 4.
ENTPD5 promotes maturation of N-glycoproteins and phenocopies oncogenic effects of mutp53. (A) Protein extracts of MDA-MB-231 cells transfected with the indicated siRNAs and a plasmid encoding HA-tagged endoglin were immunoprecipitated with anti-HA antibody. Immunoprecipitates were treated with EndoH and PNGase F, as indicated, and subjected to immunoblotting with anti-HA antibody. *Mature endoglin; right-facing triangle, immature endoglin; double right-facing triangle, EndoH-cleaved endoglin. EndoH reaction products from at least four samples per siRNA-knockdown were quantified. Percentage EndoH cleavage was calculated as the band intensity ratio cleaved/(cleaved+uncleaved). Statistical analysis was performed using one-way analysis of variance corrected for multiple hypotheses testing via Benjamini-Hochberg correction (*P < 0.05; **P < 0.01). (B and C) Colony formation of MIA PaCa-2 and MDA-MB-231 cells transfected with indicated siRNAs. Colonies were visualized (B) and quantified (C), using crystal violet. Shown are mean ± SD (n = 3). (D) Knockdown efficiency of p53 and ENTPD5 determined by RTqPCR in MIA PaCa-2 and MDA-MB-231. Shown are mean ± SEM (n = 3) normalized to GAPDH. (E) Acini formation of MCF-10A cells transduced with vectors for doxycycline-inducible expression of either p53 R248W, ENTPD5, or controls (empty vector, mock) in 3D culture. (Scale bars, 20 µm.) The percentage of acini with a hollow lumen was quantified by fluorescence microscopy after staining F-actin (green) and nuclei (blue), mean ± SD (n = 3 experiments, >60 acini each). Statistical analysis was performed using one-way analysis of variance and Bonferoni posttest (***P < 0.0001; n.s. not significant).
ENTPD5 Phenocopies Oncogenic Effects of mutp53.
Given that ENTPD5 is a downstream target gene of the oncogenic mutp53, we asked whether mutp53 GOF activities are mediated by ENTPD5. Pro-oncogenic functions of mutp53 in proliferation, invasion, metastasis, and drug resistance are supported by numerous studies, but not all these activities are mutp53-dependent in every single tumor or cell line (5). For example, in vitro proliferation of MIA PaCa-2 cells under conventional 2D cell culture conditions is mutp53-dependent (35), whereas proliferation of MDA-MB-231 is not (19). Consistently, knockdown of mutp53 with 2 distinct siRNAs reduced clonogenic growth of MIA PaCa-2, but not MDA-MB-231, cells (Fig. 4B). As ENTPD5 is mechanistically implicated in folding of oncogenic RTKs involved in controlling cell proliferation, we examined the effect of ENTPD5 depletion in this system. As shown earlier, ENTPD5 is expressed in a mutp53-dependent manner in both cell lines (Fig. 1). Intriguingly, ENTPD5, similar to mutp53, is selectively essential for clonogenic growth of MIA PaCa-2 (Fig. 4 B–D), thereby phenocopying the clonogenic activity of mutp53 in a cell context-dependent manner.
Apart from affecting tumor cell growth in 2D, mutp53 GOF has also been implicated in disruption of normal 3D tissue architectures, which is one of the hallmarks of cancer. For example, nontransformed MCF-10A mammary epithelial cells grown in a laminin-rich extracellular matrix undergo 3D morphogenesis and form spherical acinus-like structures of polarized cells surrounding a central hollow lumen. Transfection with p53 GOF mutants interferes with cell polarity and luminal clearance, reminiscent of the filled lumen phenotype observed in mammary ductal carcinoma in situ (15). Consistently, ∼70% of acini formed by mock or empty vector transduced MCF-10A cells showed a hollow lumen, whereas less than 20% of p53R248W transduced cells did (Fig. 4E). In support of ENTPD5 being a downstream target and mediator of the mutp53 GOF, ENTPD5 reduced lumen formation to a comparable extent (Fig. 4E). Enforced expression of either mutp53 or ENTPD5 therefore similarly disrupts normal 3D architectures. In summary, ENTPD5 phenocopies several oncogenic GOFs of mutp53 in tumor cell proliferation and architectural tissue remodeling.
ENTPD5 Mediates the Proinvasive mutp53 GOF.
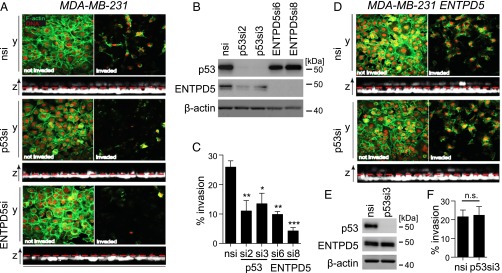
Numerous in vitro and mouse models have confirmed the ability of mutp53 to drive enhanced invasion and motility, likely by enhancing signaling through receptors such as TGFβ, EGFR, and MET (5). Given the link between ENTPD5 and folding of RTKs in the ER, we aimed to explore the contribution of ENTPD5 to mutp53-mediated extracellular matrix invasion. In light of previous reports on reduced invasive potential of MDA-MB-231 cells after mutp53 depletion (19), we depleted mutp53 or ENTPD5 in these cells with two independent siRNAs each and compared the effects on invasion of a matrigel matrix by confocal microscopy. Knockdown of either mutp53 or ENTPD5 reduced the number of invaded cells significantly, by more than 50% (Fig. 5 A–C). Of note, ENTPD5 protein levels were reduced more effectively by direct knock-down than by down-regulation with mutp53-targeted siRNAs (Fig. 5B). As invasion was also decreased more strongly in ENTPD5- than mutp53-depleted cells, this suggests a direct correlation between invasion and ENTPD5 expression.
Fig. 5.
ENTPD5 mediates the proinvasive mutp53 GOF. (A–C) Matrigel invasion of MDA-MB-231 cells after depletion of p53 or ENTPD5 by siRNA. (A) Cells were visualized by confocal imaging (nuclei, red; F-actin, green). Images of noninvading (Left) and invading cells (Right) from one representative experiment are shown. (B) Knockdown efficiency of ENTPD5 and p53 in MDA-MB-231 cells analyzed by Western blot. (C) Invasive cells were quantified by counting six randomly chosen sections of a transwell insert. Graph shows the percentage of invading cells compared with the total amount of plated cells. Error bars illustrate SEM (n = 3). Statistical analysis was performed using one-way analysis of variance and Bonferoni posttest (*P < 0.05; **P < 0.01; ***P < 0.001; n.s., not significant). (D and E) Matrigel invasion of MDA-MB-231 cells after depletion of p53 and simultaneous overexpression of ENTPD5. (E) Knockdown efficiency and (F) quantification of invading cells as described in A–C. In all Western blots, β-actin served as a loading control.
To test whether the invasion-promoting effects of mutp53 and ENTPD5 are epistatic, we repeated the mutp53 depletion in MDA-MB-231 cells stably transfected with ENTPD5 (Fig. 5 D–F). In these cells, mutp53-knockdown did not appreciably decrease ENTPD5 protein levels and, in turn, did not decrease matrigel invasion. We conclude that ENTPD5 is not only required for mutp53-driven invasion but was also sufficient to maintain invasion after depletion of mutp53.
Mutp53 Promotes Lung Colonization in Mice Through ENTPD5.
We next aimed to study the in vivo relevance of ENTPD5 for the mutp53 prometastatic GOF. Metastasis involves a cascade of events including invasion of host tissues adjacent to the primary tumor, entrance into the systemic vasculature, dissemination via the circulation, arrest in microvasculature, extravasation into the parenchyma of distant organs, and proliferation at these ectopic sites to form secondary colonies, also termed “colonization” (36, 37). As patients with cancer, and in particular patients with breast cancer, are often found to have hundreds, likely thousands, of disseminated tumor cells in their body, only some of which will ever develop metastatic relapse (38), the last steps in the cascade are likely rate-limiting. To specifically interrogate these late steps, tumor cells are commonly injected i.v. and examined for lung colonization as a readout. Importantly, endogenous p53R280K in MDA-MB-231 cells is known to be required for this process (19), which links the mutp53 GOF directly to these prognostically crucial steps in metastasis.
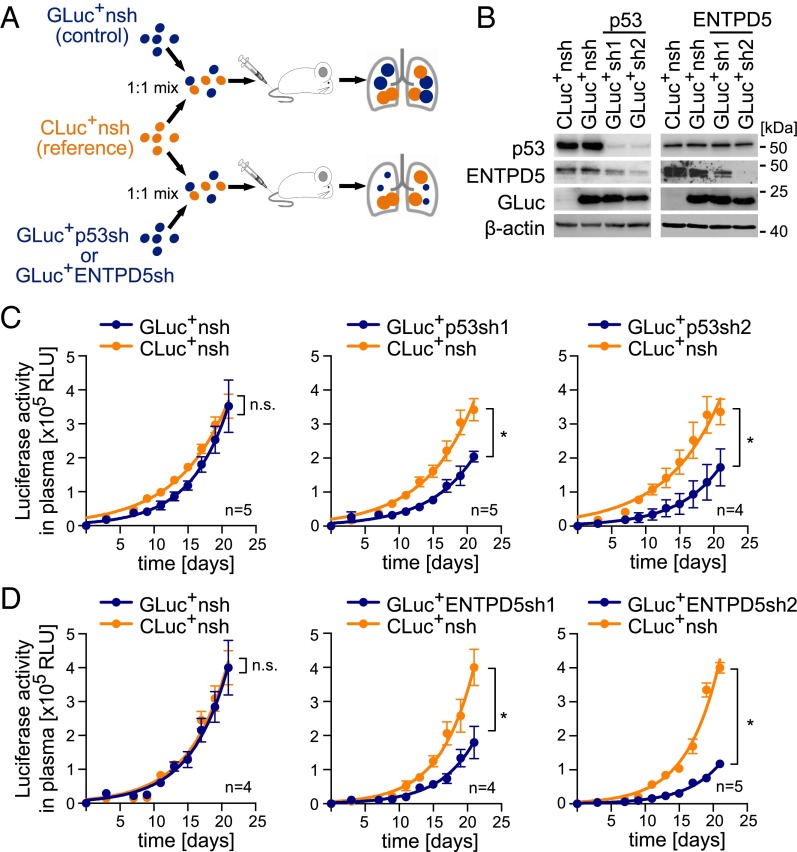
To compare the role of mutp53 and ENTPD5 in lung colonization, we stably transduced MDA-MB-231 cells with lentiviruses expressing either of two independent shRNAs against each target. To quantitatively track the proliferation of knock-down cells in vivo, we coupled shRNA expression to secreted luciferases that accumulate in the blood of tumor-bearing mice and can be used as an artificial tumor marker for longitudinal monitoring of tumor burden (39). To directly compare knockdown and control cells within the same animal, we used a dual luciferase labeling approach (39). First, MDA-MB-231 cells were labeled with Gaussia luciferase (GLuc) in conjunction with shRNAs targeting mutp53, ENTPD5 or a nontargeting (nsh) control (Fig. 6A). In addition, the nsh control was also coupled to Cypridina luciferase (CLuc), to be used as a reference in all experiments. Correct labeling and efficient knockdown were confirmed by Western blot (Fig. 6B) before groups of animals were injected i.v. with 1:1 mixtures of a single GLuc+ cell line (GLuc+nsh, GLuc+p53sh1/2, GLuc+ENTPD5sh1/2) and the CLuc+nsh reference (Fig. 6 A–D). In vivo proliferation of the different cell types was quantified for 3 wk by measuring the increase of GLuc and CLuc luciferase activities in blood samples. Although the CLuc activity (orange curves), which reflects the proliferation of the reference cells, increased similarly in all animal groups, the GLuc activities (blue curves) remained significantly lower when GLuc was coupled with mutp53- or ENTPD5-targeting shRNAs (Fig. 6 C and D). When GLuc was coupled to the control shRNA, no difference to CLuc is evident, indicating that the reduced GLuc increase observed on mutp53- or ENTPD5-knockdown is target-specific and not a result of different luciferase labels. ENTPD5 is therefore similarly essential for lung colonization by MDA-MB-231 cells as mutp53 itself.
Fig. 6.
Mutp53 and ENTPD5 are required for lung colonization in mice. (A) Overview of experimental procedure: MDA-MB-231 cells were transduced with vectors coexpressing GLuc or CLuc, together with shRNAs targeting p53, ENTPD5, or control (nsh). Mice were i.v. injected with 1:1 mixtures of CLuc+nsh/GLuc+nsh (control group), CLuc+nsh/GLuc+p53sh1/2, or CLuc+nsh/GLuc+ENTPD5sh1/2. (B) Knockdown efficiencies analyzed by Western blot. β-actin served as a loading control. (C and D) Tumor growth measured in terms of GLuc (blue line) and CLuc (orange line) luciferase activity in plasma. Error bars illustrate SEM for each mouse group. Statistics were performed by two-way analysis of variance (*P < 0.001); n, number of mice per group.
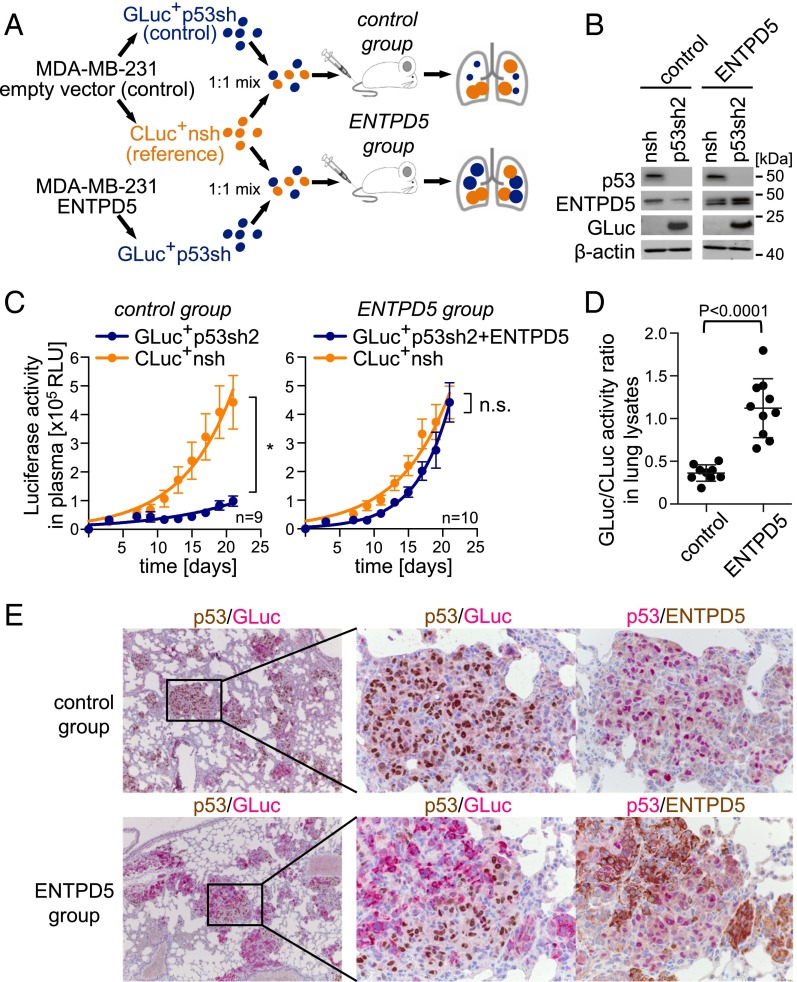
To further explore whether mutp53 and ENTPD5 are epistatic, we investigated the effect of mutp53 depletion on lung colonization of parental MDA-MB-231 (control) versus ENTPD5-overexpressing MDA-MB-231 cells (Fig. 7 A and B). Mutp53 was knocked-down in combination with GLuc labeling, yielding GLuc+p53sh and ENTPD5+GLuc+p53sh cells. A nontargeting shRNA coupled to CLuc served as a common reference (CLuc+nsh). The control group of mice received a 1:1 mixture of GLuc+p53sh and CLuc+nsh cells, and the ENTPD5 group a 1:1 mixture of ENTPD5+GLuc+p53sh and CLuc+nsh cells. As seen before (Fig. 6C), knockdown of mutp53 strongly dampened the increase of GLuc activity in the blood of control group mice (Fig. 7C), but not in the ENTPD5 group, despite equally efficient depletion of mutp53 (Fig. 7 B and E). This observation was confirmed by postmortem analysis of lungs from both groups. First, GLuc activity normalized to CLuc as a reference (GLuc/CLuc ratio) was significantly reduced in lungs of the control versus the ENTPD5 group (Fig. 7D). Second, consistent with their colonization defect, mutp53-depleted cells (red cytoplasmic GLuc staining) were strongly underrepresented in histological lung sections from the control group, and tumor nodules were mainly composed of mutp53-expressing (brown nuclear p53 staining) reference cells (Fig. 7E). In contrast, in the ENTPD5 group, we observed substantial colonization by mutp53-depleted tumor cells (red cytoplasmic GLuc staining, no brown nuclear p53 staining), with clear expression of exogenous ENTPD5, indicating that ENTPD5 can rescue the lung colonization defect on depletion of mutp53. ENTPD5 is therefore both required for lung colonization of mutp53 cells and sufficient to replace mutp53 in this function. In summary, these experiments demonstrate that ENTPD5 is a crucial mediator of the prometastatic mutp53 GOF in vivo.
Fig. 7.
Mutp53 promotes lung colonization in mice through ENTPD5. (A) Experimental procedure: MDA-MB-231 were transduced with ENTPD5 or empty vector (control) and vectors coexpressing GLuc or CLuc, together with shRNAs targeting p53 or control (nsh). Mice were i.v. injected with 1:1 mixtures of CLuc+nsh/GLuc+p53sh2 (control group) or CLuc+nsh/ENTPD5+GLuc+p53sh2 (ENTPD5 group). (B) Knockdown efficiencies analyzed by Western blot. β-actin served as a loading control. (C) Tumor growth measured in terms of GLuc (blue line) and CLuc (orange line) luciferase activity in plasma. Error bars illustrate SEM for each mouse group. Statistics were performed by two-way analysis of variance (*P < 0.001); n, number of mice per group. (D) GLuc and CLuc activity ratios measured in lung lysates of individual mice. Shown are mean ± SD. Statistical analysis was done using nonparametric Mann–Whitney U test. (E) Immunohistological double staining for p53 (brown)/GLuc (red) or p53 (red)/ENTPD5 (brown) of representative lungs from both experimental groups (control and ENTPD5).
Discussion
As TP53 is the most commonly mutated gene in cancer, mutp53 has always been considered a dream target for cancer therapy. However, wtp53 is a tumor suppressor that would need to be reactivated for cancer therapy, and as it is pharmacologically easier to inhibit a protein than to activate it, targeting mutp53 has proven challenging in practice. The realization that mutp53 cancer cells are not only addicted to the absence of wtp53 but also dependent on neomorphic GOF activities of the mutp53 protein (2, 4, 5, 11) has stimulated a new line of research aimed at inhibiting mutp53 directly or crucial downstream effectors, preferentially those that are considered druggable because of enzymatic activities.
The broad spectrum of reported mutp53 neomorphic activities (5) shows a remarkable focus on proinvasive and prometastatic functions. This correlates strikingly with the preferential occurrence of p53 mutations at the transition from benign to invasive stages of cancer, probably studied best in the context of colorectal and pancreatic cancer progression. The investigation of molecular mechanisms underlying mutp53-dependent stimulation of invasion and metastasis therefore promises to identify the targets of mutp53 that account for its high mutation frequency in aggressive cancers. Previous research has delineated a key role of mutp53 in enhancing proinvasive signaling via membrane receptors such as RTKs and integrins. In part, this is achieved by mutp53 transcriptionally up-regulating the expression of receptors (20), by suppressing receptor targeting miRNAs (40), or by enhancing receptor recycling (18, 21). Our study adds to this a GOF activity of mutp53, which is mediated by the mutp53 target gene ENTPD5, operates in the ER, and promotes glycoprotein folding via the calnexin/calreticulin cycle (22). If the calnexin/calreticulin cycle is inhibited, the folding efficiency of glycoproteins is often decreased, resulting in quality control breakdown, with the consequence of nonfunctional misfolded proteins exiting the ER (24). Cell surface receptors have varying numbers of N-glycan sites, with growth-promoting receptors and integrins being more heavily N-glycosylated than others (25), which as a consequence renders signaling through highly N-glycosylated receptors more susceptible to changes in quality control and folding efficiency. Stimulation of the calnexin/calreticulin cycle through increased ENTPD5 expression might therefore provide support for previously described mutp53-mediated effects on receptor expression and recycling by ensuring an optimal receptor quality with respect to their folding and N-glycosylation. In fact, it seems this support is critically essential, as knockdown of ENTPD5 impairs clonogenic growth of cells that depend on mutp53 (Fig. 4 B–D), reduces matrigel invasion (Fig. 5 A–C), and inhibits lung colonization by circulating tumor cells (Fig. 6). Amazingly, enforced expression of ENTPD5 rescues matrigel invasion (Fig. 5 D–F) and lung colonization (Fig. 7) in mutp53-depleted cells, indicating it can even compensate for the loss of some other GOF activities that contribute to these prometastatic activities. Stimulating calnexin/calreticulin-driven quality control would lay the grounds for other mechanisms, such as increased expression or recycling, to become effective. Obviously, a simple quantitative increase in receptor expression is only of limited effect if the receptors are misfolded. Similarly, increased recycling can only augment signaling if the recycled receptors are functional. It therefore seems intuitive that mutp53 not only increases receptor quantity but, in parallel, also activates mechanisms that ensure optimal receptor quality.
Mutp53 affects target gene expression by a multitude of different mechanisms (4). The interaction of mutp53 with various transcription factors has been studied in the most detail. In particular, the p53 family transcription factors p63 and p73 have been extensively explored in the context of mutp53-driven invasion and metastasis, as, in contrast to wtp53, mutp53 interacts directly with both p63 and p73, and thereby inhibits their transactivating functions (41). The inhibitory interaction with p63/p73 likely contributes to the prometastatic activity of mutp53 in vivo, as p53/p63 and p53/p73 double-heterozygous mice show a higher incidence of metastatic tumors than p53 single-heterozygotes (42). Mechanistically, the interaction of mutp53 with p63 enhances tumor cell invasion by stimulating TGFβ signaling (19); RCP-driven recycling of EGFR, MET, and integrins (18, 21); and down-regulation of Dicer-dependent processing of antimetastatic miRNAs (43, 44). In contrast, our study shows that ENTPD5 is regulated by mutp53 in an Sp1-dependent manner (Fig. 3 C–F). A role for Sp1 in transcriptional regulation by both wtp53 and mutp53 has been previously reported for other genes (45). For example, transfected mutp53 interacts with Sp1 and stimulates Sp1 binding and histone acetyltransferase recruitment to the EGFR promoter (46). Although we cannot formally exclude additional recruitment of mutp53 via other transcription factors, Sp1 is likely the dominant recruiting factor, as depletion of Sp1 prevented mutp53 from binding the ENTPD5 promoter.
p53 mutations on one allele are commonly followed by inactivation of the remaining wtp53 allele via loss of heterozygosity (LOH). In the absence of LOH, mutp53 can suppress wtp53 function in a dominant-negative manner (2, 4). In contrast, wtp53 suppression by mutp53 is not always efficient. Patients with Li-Fraumeni syndrome carry germ-line heterozygous p53 mutations, and yet exhibit normal development and develop tumors only later in adult life (47). Likewise, mice heterozygous for the Trp53R172H mutation have the same lifespan as mice heterozygous for the null-allele (6). Even in tumor cells wtp53 can be triggered by DNA damage to induce senescence in the presence of mutp53 (48). GOF experiments performed in the presence of wtp53 must therefore be interpreted with extreme caution (4). The mutp53 cell lines in our experiments did not express wtp53, excluding a dominant-negative effect as an underlying cause of ENTPD5 regulation. In the case of clinical tumor samples, tissue heterogeneity limits the ability to accurately infer LOH status, as a true heterozygous (non-LOH) state is difficult to distinguish from a contamination by wtp53 stromal cells or the coexistence of wild-type and p53-mutated tumor subclones. It therefore remains to be seen whether LOH affects the regulation of ENTPD5 by mutp53.
Mutp53 depletion caused a two- to fourfold decrease in tumor cell invasion and lung colonization, in many cases correlating with the degree of mutp53/ENTPD5 inhibition (Figs. 5–7). Translation into clinically meaningful antitumor activity will therefore rely on the development of effective ENTPD5 targeting approaches. Compared with the experimental RNAi approach used in our study, small molecules are often more effective inhibitors, raising hope that the antitumor effects can be further enhanced by pharmacological inhibitors of the ENTPD5 UDPase activity. Entpd5-knockout mice are viable and show hepatopathy and aspermia only after 1 y of age, promising a sufficiently broad therapeutic window for ENTPD5 inhibitors, despite interference with a central step in protein biosynthesis (49).
In summary, our study has identified ENTPD5 as a specific target of mutp53 that operates in the calnexin/calreticulin cycle of the ER. High-level ENTPD5 expression correlates with p53 GOF mutations across a broad panel of tumor entities and requires mutp53 docking to Sp1 bound to the ENTPD5 promoter. ENTPD5 promotes N-glycoprotein folding via the calnexin/calreticulin cycle and is essential for mutp53-mediated tumor cell proliferation, architectural tissue remodelling, extracellular matrix invasion, and lung colonization. As ENTPD5 mediates key protumorigenic effector functions of mutp53, it might represent a promising target for the treatment of tumors with p53 GOF mutations.
Materials and Methods
Additional experimental details are provided in SI Appendix.
N-Glycosylation Analysis.
For analysis of N-glycosylated proteins, MDA-MB-231 cells were transfected with siRNAs targeting p53, ENTPD5, UGGT, or a nontargeting control (nsi). Forty-eight hours after siRNA transfection, cells were transfected with a pCMV-HA-endoglin plasmid (33), using Lipofectamine 2000 (Thermo Fisher Scientific). Twenty-four hours after plasmid transfection, cells were harvested in NET buffer [150 mM NaCl, 50 mM Tris⋅HCl at pH 7.4, 0.5% Nonidet P-40, 10% (vol/vol) glycerol, 5 mM EDTA], and HA-endoglin was immunoprecipitated using anti-HA antibody (HA.11, Covance). Immunoprecipitated endoglin was eluted and denatured in 1× Glycoprotein Denaturation Buffer (0.5% SDS, 40 mM DTT; New England Biolabs, #P0702L) for 10 min at 100 °C. Samples were then digested in the manufacturer’s buffer with EndoH (New England Biolabs, P0702L) or PNGase F (New England Biolabs, P0708S) for 2.5 h at 37 °C. Immunoblotting was performed using anti-HA antibody (Cell signaling, 3724). Detected bands were quantified using ImageLab 5.0 software (Bio-Rad Laboratories).
Matrigel Invasion Assay.
Matrigel invasion assays were performed as described (50). Transwell inserts (Greiner Bio-One) were coated with 50 µL growth-factor-reduced Matrigel at 5 mg/mL (Corning). Fifteen thousand cells were seeded to the inverted transwell inserts and allowed to become adherent. ThinCerts were inverted, and medium was added to the top [10% (vol/vol) FBS] and the bottom (0.5% FBS). After 24 h, cells were fixed with 8% (wt/vol) formaldehyde and stained with rhodamine-phalloidin and sytox green (Invitrogen). Invasion assays were analyzed by laser-scanning microscopy, using a LSM 700 confocal laser scanning microscope (Zeiss). Numbers of noninvaded versus invaded cells in each optical section from six randomly chosen fields were counted using Photoshop CS6 (Adobe).
Experimental in Vivo Metastasis Model.
Animal experiments were approved by the regional board (RP Giessen), in accordance with the German animal welfare law and the European legislation for the protection of animals used for scientific purposes (2010/63/EU). Lung colonization after i.v. tail vein injection of tumor cells was performed as previously described (39). In brief, MDA-MB-231 cells were labeled ex vivo with Gaussia or Cypridina luciferases by transduction with lentiviral vectors coexpressing shRNAs targeting p53 or ENTPD5. Nontargeting shRNAs were used as a control. After successful transduction and puromycin selection, different GLuc- and CLuc-labeled tumor cells were mixed in a 1:1 ratio. A total of 1 × 106 cells of these mixtures were injected i.v. into the tail vein of immunocompromised 6–12-wk-old Rag2tm1.1Flv;Il2rgtm1.1Flv male and female mice kept under SPF conditions. Required sample sizes were calculated by an a priori power analysis. For induction of ENTPD5 expression, doxycycline was freshly prepared and administered via the drinking water in darkened bottles [1 mg/mL doxycycline; 2% (wt/vol) sucrose]. Drinking water was changed every second to third day.
Supplementary Material
Acknowledgments
We thank Bassam Ali for providing endoglin plasmid, Michael Krause and Sigrid Bischofsberger for performing microarray experiments and excellent technical assistance in immunohistochemistry, and Claudia Wickenhauser for critical reading of the manuscript, as well as other members of the participating laboratories for helpful discussions. This work was supported by grants from Deutsche Forschungsgemeinschaft (DFG TRR81, KFO210, STI 182/7-1), European Research Council, Bundesministerium für Bildung und Forschung, Rhön Klinikum AG, German Center for Lung Research (to T.S.); by Deutsche José Carreras Leukämie-Stiftung, Deutsche Krebshilfe (111250, 111444), and Universities of Giessen and Marburg Lung Center (to T.S. and O.T.); by Von-Behring-Röntgen-Stiftung (to J.P.C.); and by Universitätsklinikum Giessen und Marburg (to M.M. and A.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the EBI ArrayExpress database (accession no. E-MTAB-4672).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612711114/-/DCSupplemental.
References
- 1.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 2.Brosh R, Rotter V. When mutants gain new powers: News from the mutant p53 field. Nat Rev Cancer. 2009;9(10):701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 3.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2(2):a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freed-Pastor WA, Prives C. Mutant p53: One name, many proteins. Genes Dev. 2012;26(12):1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller PAJ, Vousden KH. Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell. 2014;25(3):304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lang GA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119(6):861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119(6):847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Hanel W, et al. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ. 2013;20(7):898–909. doi: 10.1038/cdd.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol. 2007;9(5):573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- 10.Lee MK, et al. Cell-type, dose, and mutation-type specificity dictate mutant p53 functions in vivo. Cancer Cell. 2012;22(6):751–764. doi: 10.1016/j.ccr.2012.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Alexandrova EM, et al. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature. 2015;523(7560):352–356. doi: 10.1038/nature14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheok CF, Verma CS, Baselga J, Lane DP. Translating p53 into the clinic. Nat Rev Clin Oncol. 2011;8(1):25–37. doi: 10.1038/nrclinonc.2010.174. [DOI] [PubMed] [Google Scholar]
- 13.Soragni A, et al. A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell. 2016;29(1):90–103. doi: 10.1016/j.ccell.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu J, et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525(7568):206–211. doi: 10.1038/nature15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freed-Pastor WA, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148(1-2):244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prives C, Lowe SW. Cancer: Mutant p53 and chromatin regulation. Nature. 2015;525(7568):199–200. doi: 10.1038/nature15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfister NT, et al. Mutant p53 cooperates with the SWI/SNF chromatin remodeling complex to regulate VEGFR2 in breast cancer cells. Genes Dev. 2015;29(12):1298–1315. doi: 10.1101/gad.263202.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller PAJ, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139(7):1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 19.Adorno M, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137(1):87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 20.Weissmueller S, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor β signaling. Cell. 2014;157(2):382–394. doi: 10.1016/j.cell.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller PAJ, et al. Mutant p53 enhances MET trafficking and signalling to drive cell scattering and invasion. Oncogene. 2013;32(10):1252–1265. doi: 10.1038/onc.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang M, et al. The ER UDPase ENTPD5 promotes protein N-glycosylation, the Warburg effect, and proliferation in the PTEN pathway. Cell. 2010;143(5):711–724. doi: 10.1016/j.cell.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Israelsen WJ, Vander Heiden MG. ATP consumption promotes cancer metabolism. Cell. 2010;143(5):669–671. doi: 10.1016/j.cell.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 24.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 25.Lau KS, et al. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129(1):123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 26.Morton JP, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci USA. 2010;107(1):246–251. doi: 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stambolsky P, et al. Modulation of the vitamin D3 response by cancer-associated mutant p53. Cancer Cell. 2010;17(3):273–285. doi: 10.1016/j.ccr.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Girardini JE, et al. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell. 2011;20(1):79–91. doi: 10.1016/j.ccr.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 29.Schlereth K, et al. Characterization of the p53 cistrome--DNA binding cooperativity dissects p53's tumor suppressor functions. PLoS Genet. 2013;9(8):e1003726. doi: 10.1371/journal.pgen.1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai JS, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89(15):6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oxmann D, et al. Endoglin expression in metastatic breast cancer cells enhances their invasive phenotype. Oncogene. 2008;27(25):3567–3575. doi: 10.1038/sj.onc.1211025. [DOI] [PubMed] [Google Scholar]
- 32.Fujiwara K, et al. Migratory activity of CD105+ pancreatic cancer cells is strongly enhanced by pancreatic stellate cells. Pancreas. 2013;42(8):1283–1290. doi: 10.1097/mpa.0b013e318293e7bd. [DOI] [PubMed] [Google Scholar]
- 33.Ali BR, et al. Endoplasmic reticulum quality control is involved in the mechanism of endoglin-mediated hereditary haemorrhagic telangiectasia. PLoS One. 2011;6(10):e26206. doi: 10.1371/journal.pone.0026206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mallet C, et al. Functional analysis of endoglin mutations from hereditary hemorrhagic telangiectasia type 1 patients reveals different mechanisms for endoglin loss of function. Hum Mol Genet. 2015;24(4):1142–1154. doi: 10.1093/hmg/ddu531. [DOI] [PubMed] [Google Scholar]
- 35.Yan W, Chen X. Identification of GRO1 as a critical determinant for mutant p53 gain of function. J Biol Chem. 2009;284(18):12178–12187. doi: 10.1074/jbc.M900994200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fidler IJ. The pathogenesis of cancer metastasis: The ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 37.Shibue T, Weinberg RA. Integrin beta1-focal adhesion kinase signaling directs the proliferation of metastatic cancer cells disseminated in the lungs. Proc Natl Acad Sci USA. 2009;106(25):10290–10295. doi: 10.1073/pnas.0904227106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braun S, et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. 2005;353(8):793–802. doi: 10.1056/NEJMoa050434. [DOI] [PubMed] [Google Scholar]
- 39.Charles JP, et al. Monitoring the dynamics of clonal tumour evolution in vivo using secreted luciferases. Nat Commun. 2014;5:3981. doi: 10.1038/ncomms4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, Cheng B, Miao L, Mei Y, Wu M. Mutant p53-R273H gains new function in sustained activation of EGFR signaling via suppressing miR-27a expression. Cell Death Dis. 2013;4:e574. doi: 10.1038/cddis.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21(5):1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flores ER, et al. Tumor predisposition in mice mutant for p63 and p73: Evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7(4):363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 43.Su X, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467(7318):986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller PAJ, Trinidad AG, Caswell PT, Norman JC, Vousden KH. Mutant p53 regulates Dicer through p63-dependent and -independent mechanisms to promote an invasive phenotype. J Biol Chem. 2014;289(1):122–132. doi: 10.1074/jbc.M113.502138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2(8):a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaughan CA, et al. Addiction of lung cancer cells to GOF p53 is promoted by up-regulation of epidermal growth factor receptor through multiple contacts with p53 transactivation domain and promoter. Oncotarget. 2016;7(11):12426–12446. doi: 10.18632/oncotarget.6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malkin D, et al. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250(4985):1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 48.Jackson JG, et al. p53-mediated senescence impairs the apoptotic response to chemotherapy and clinical outcome in breast cancer. Cancer Cell. 2012;21(6):793–806. doi: 10.1016/j.ccr.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Read R, et al. Ectonucleoside triphosphate diphosphohydrolase type 5 (Entpd5)-deficient mice develop progressive hepatopathy, hepatocellular tumors, and spermatogenic arrest. Vet Pathol. 2009;46(3):491–504. doi: 10.1354/vp.08-VP-0201-R-AM. [DOI] [PubMed] [Google Scholar]
- 50.Kitzing TM, et al. Positive feedback between Dia1, LARG, and RhoA regulates cell morphology and invasion. Genes Dev. 2007;21(12):1478–1483. doi: 10.1101/gad.424807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.