Significance
The ability of proteins to interact quickly and specifically is fundamental for most living activities. For a protein interaction to be formed, the corresponding surfaces need a high degree of chemical and structural complementarity. Here we investigated the level of plasticity of composition of a protein–protein interface without affecting the high-affinity binding. Using directed evolution and mild selection, we show that most interface residues can be exchanged without a loss in binding affinity. Conversely, many of the buried residues were highly conserved. Interestingly, the selection process resulted in sequences promoting faster binding kinetics. Selection of faster binders was further promoted by drastically shortening the incubation time, resulting in preequilibrium selection, which provides a path for evolution of fast-binding complexes.
Keywords: protein–protein interaction, in vitro evolution, association rate, interface plasticity
Abstract
Protein–protein interactions occur via well-defined interfaces on the protein surface. Whereas the location of homologous interfaces is conserved, their composition varies, suggesting that multiple solutions may support high-affinity binding. In this study, we examined the plasticity of the interface of TEM1 β-lactamase with its protein inhibitor BLIP by low-stringency selection of a random TEM1 library using yeast surface display. Our results show that most interfacial residues could be mutated without a loss in binding affinity, protein stability, or enzymatic activity, suggesting plasticity in the interface composition supporting high-affinity binding. Interestingly, many of the selected mutations promoted faster association. Further selection for faster binders was achieved by drastically decreasing the library–ligand incubation time to 30 s. Preequilibrium selection as suggested here is a novel methodology for specifically selecting faster-associating protein complexes.
Protein–protein interactions play important roles in most cellular processes. Proteins interact through chemical and structural complementarity of their mutual binding sites (1). Amino acids found in physical proximity form noncovalent interactions that stabilize the complex (2). Residues not directly involved in binding also play a role in complex formation by stabilizing the association encounter complex and transition state, which determine the interaction’s association rate constant (3). Protein interaction interfaces seem to have dual, seemingly contradicting, properties: On one hand, the interface is a unique area (4) on the protein surface that has evolved to bind specific partners (5). Although much is known about the molecular mechanism of binding, de novo design of protein interactions is still a complicated task, requiring in vitro evolution to achieve high-affinity binding (6–8). On the other hand, interfaces seem to be rather tolerant to mutagenesis (2, 9–11), implying that there are many ways to achieve high-affinity binding. The plastic nature of protein–protein interfaces becomes particularly clear when comparing the conservation levels of transient complex interfaces versus surface and core residues in homologous complexes (12, 13). Here it is shown that core residues are similarly conserved as permanent interaction interface residues but that transient interaction interface residues are hardly more conserved than other surface residues. The relatively low conservation levels of transient interface residues also make it difficult to predict the protein binding sites (contrary to permanent interfaces) (14, 15). In fact, it was suggested that the vast majority of interfacial residues are important only for conformational optimization of the few interacting residues, resulting in variability in their actual identity (16).
The interface formed between different β-lactamase proteins and their inhibitor protein BLIP is a good example of the use of different amino acid identities in homologous binding interfaces. Although the interface location is conserved, the interface residue identities are not, with different residues being used as hotspots for binding (5, 17). Despite this evolutionary lesson, in vitro evolution has seldom been used to specifically address this question (18). Weiss et al. (10) mutated the 19 interfacial residues of the human growth hormone with its receptor to alanine and found that only 7 substitutions affected binding. Thom et al. (19) emphasized the importance of allowing random mutagenesis all through the protein-coding sequence for interaction optimization. TEM1 and other β-lactamase enzymes were a subject for several directed evolution studies summarized in Yuan et al. (20), but these studies concentrated on β-lactamase enzymatic activity and substrate recognition whereas our study discusses the composition of protein–protein interaction interfaces.
Protein affinity maturation is usually done by multiple rounds of selection, from low to high stringency, and then combining the highest-contributing mutations to achieve tight binding. Our study was fundamentally different, as it was designed to explore the plasticity of the interaction between TEM1 β-lactamase and its inhibitor BLIP with no goal of optimizing the interaction affinity, which is already high: The TEM1–BLIP interaction possesses nanomolar affinity with an association rate constant of ∼105 M−1⋅s−1 and a dissociation rate constant of 10−4 s−1 (21). For the purpose of this study, we applied in vitro evolution to a randomly mutated TEM1 library, using yeast surface display for three rounds of mild selection, and sequenced the selected library assuming a relation between the occurrence of specific mutations (either above or below their occurrence in a nonselected library) and their contribution to binding (22). The subsequent use of deep sequencing has the potential to provide complete data on all point mutations at the binding surface extracted from a natural pool of binders. This allows for nonbiased mapping of mutational effects on binding and stability. The basic principle behind this method is that selection will enrich for amino acids that contribute to a specific function. Deep sequencing then provides the amino acid preference at any randomized position, which is related (with a Boltzmann-like distribution) to the contribution toward the selected trait (for example, binding) (22). In this study, the selected trait was binding to BLIP. Although co-occurring mutations may have also influenced the outcome of the analysis, they were not considered in the data analysis.
The results imply that most of the interface residues can be replaced by other amino acids without affecting the binding affinity, excluding three residues that apparently are hotspots for interaction (2, 4, 23, 24). Interestingly, the selection resulted in faster binders even when using the standard selection protocol. We were able to further increase the percentages of fast binders by drastically decreasing the protein–ligand incubation time (from 1 h to 30 s) and ligand concentration (from 1 µM to 50 nM). Here we present a selection method specifically resulting in faster binders.
Results
Analyzing Interface Plasticity.
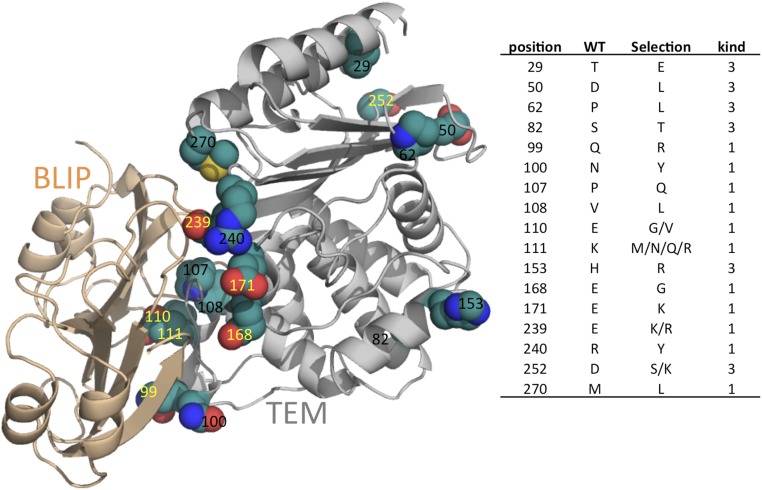
TEM1 was randomly mutated by mutazyme, resulting in an average of four mutations per clone. Because the mutations are randomly distributed throughout the gene, there is a strong bias toward single-nucleotide mutations of amino acid codons, which results in only a partial cover of the mutation space of the individual residues (see Table S1 for the probability of a given amino acid being mutated to each one of the 20 possible amino acids upon single-nucleotide exchange). However, because natural evolution also progresses by single mutations, there is a natural preference for single-nucleotide substitutions. The library of 108 clones underwent selection against binding to BLIP conjugated to the fluorescent protein YPET for FACS sorting. The library was selected three times against 1 µM Y–BLIP, and its gene composition was evaluated by deep sequencing. The sequencing data were analyzed using Enrich 2.0 software (25). The deep sequencing of TEM1 was done in four fragments covering 80% of the gene and 92% of the binding site (Fig. 1). Analysis of mutation enrichment (or reduction) was done by comparing the mutation frequency at each position with that of the original library or the previous selection round. This normalizes the inherent bias toward single-nucleotide mutations (Table S1). Our aim was to investigate the sequence space tolerated by this interaction without losing affinity, rather than its optimization. Mutations that were selected after the third round of the soft sorts are presented in Fig. 2. Most (11 out of 17) selected mutations are located at the interface. In addition, most of the mutations (11 out of 17) involved charged residues (either resulting in charge deletion or insertion or in replacement by the opposite charge). All of the selected mutations excluding T29E are located on the protein surface. Because charged residues around the interface are known to play an important role in dictating association rate constants (kons), the selected mutations were expressed and their kinetic rate constants were measured (Table 1).
Table S1.
Probability of a given WT amino acid being mutated to each of the 20 possible amino acids or to a stop codon
| A | C | D | E | F | G | H | I | K | L | M | N | P | Q | R | S | T | V | W | Y | * | |
| A | 33 | 6 | 6 | 11 | 11 | 11 | 11 | 11 | |||||||||||||
| C | 17 | 11 | 11 | 11 | 22 | 8 | 11 | 8 | |||||||||||||
| D | 11 | 17 | 17 | 11 | 11 | 11 | 11 | 11 | |||||||||||||
| E | 11 | 17 | 17 | 11 | 11 | 11 | 11 | 11 | |||||||||||||
| F | 11 | 17 | 11 | 28 | 11 | 11 | 11 | ||||||||||||||
| G | 11 | 6 | 6 | 6 | 33 | 17 | 6 | 11 | 3 | 3 | |||||||||||
| H | 11 | 17 | 11 | 11 | 11 | 17 | 11 | 11 | |||||||||||||
| I | 7 | 25 | 4 | 15 | 8 | 7 | 4 | 7 | 11 | 11 | |||||||||||
| K | 11 | 6 | 17 | 6 | 17 | 11 | 11 | 11 | 11 | ||||||||||||
| L | 6 | 4 | 8 | 39 | 3 | 7 | 4 | 7 | 4 | 11 | 2 | 6 | |||||||||
| M | 33 | 11 | 22 | 11 | 11 | 11 | |||||||||||||||
| N | 11 | 11 | 11 | 17 | 17 | 11 | 11 | 11 | |||||||||||||
| P | 11 | 6 | 11 | 33 | 6 | 11 | 11 | 11 | |||||||||||||
| Q | 11 | 17 | 11 | 11 | 11 | 17 | 11 | 11 | |||||||||||||
| R | 6 | 11 | 4 | 2 | 4 | 7 | 2 | 7 | 4 | 39 | 6 | 4 | 3 | 3 | |||||||
| S | 7 | 7 | 4 | 4 | 4 | 4 | 4 | 7 | 9 | 28 | 11 | 2 | 4 | 6 | |||||||
| T | 11 | 8 | 6 | 3 | 6 | 11 | 6 | 17 | 33 | ||||||||||||
| V | 11 | 6 | 6 | 6 | 11 | 8 | 17 | 3 | 33 | ||||||||||||
| W | 22 | 11 | 11 | 22 | 11 | 22 | |||||||||||||||
| Y | 11 | 11 | 11 | 11 | 11 | 11 | 17 | 17 |
Probability is given as a percentage (%). Rows, WT amino acids; columns, each of the 20 possible amino acids or a stop codon (*). The calculation is for a change in a single nucleotide. The calculation takes into account the amino acid sequence and assumes an equal probability for each possible DNA codon. Note that all entries in this table correspond to mutation probabilities in one direction.
Fig. 1.
Hyperstable (HS)-TEM gene map. Fragments analyzed by deep sequencing are in bold and underlined; BLIP interface residues are in red.
Fig. 2.
(Left) Structural representation of the TEM–BLIP complex (gray and wheat, respectively) (PDB ID code 1JTG) and the mutations selected during the first three sorts of the TEM random library against BLIP. Selected mutations are presented as spheres (green, C; red, O; blue, N; yellow, S). Residues labeled in yellow are those that upon mutation increase kon by >1.6-fold (2× SE). (Right) A table indicating the specific selected mutations and their kind: 1, interface; 3, surface.
Table 1.
Characterization of the TEM mutations selected against BLIP
| Mutant | Kind | konmut/konwt* | koff† × 10−4 s−1 | KD‡ × 10−10 M | Tm,§ °C | Enzymatic activity¶ |
| WT | 1.0 | 0.85 | 2.8 | 55 | 0.0023 | |
| T29E | Buried | 0.6 | 47.6 | 0.0019 | ||
| K111Q | Interface | 0.7 | 57.6 | 0.0025 | ||
| K111M | Interface | 0.9 | 55.7 | 0.0016 | ||
| K111N | Interface | 0.9 | 1.5 | 3.8 | 54.3 | 0.0014 |
| S82T | Surface | 1.0 | 53.4 | 0.0017 | ||
| P62L | Surface | 1.0 | 53.2 | 0.0019 | ||
| H153R | Surface | 1.0 | 58 | 0.0017 | ||
| V108L | Interface | 1.0 | 54.4 | 0.0015 | ||
| R240Y | Interface | 1.1 | 1 | 2.2 | 55 | 0.0027 |
| M270L | Interface | 1.2 | 56.4 | 0.0023 | ||
| D50L | Surface | 1.2 | 55 | 0.0015 | ||
| N100Y | Interface | 1.3 | 52 | 0.0028 | ||
| P107Q | Interface | 1.3 | 3.3 | 6.1 | 54.3 | 0.0017 |
| D252K | Surface | 1.4 | 50.8 | 0.0025 | ||
| Q99R | Interface | 1.6 | 54.4 | 0.0019 | ||
| E168G | Interface | 1.6 | 1 | 2.1 | 55.8 | 0.0014 |
| D252S | Surface | 1.6 | 54.1 | 0.0012 | ||
| K111R | Interface | 1.9 | 55.8 | 0.0012 | ||
| E110V | Interface | 2.3 | 0.9 | 0.9 | 55.5 | 0.0016 |
| E110G | Interface | 3.3 | 0.5 | 0.6 | 56.4 | 0.0018 |
| E171K | Interface | 3.4 | 0.9 | 1.0 | 54 | 0.001 |
| E110G E168G | 3.6 | 1 | 1.0 | 54.6 | 0.0011 | |
| E110V E168G | 3.7 | 0.8 | 0.8 | 51 | 0.0015 | |
| E239K | Interface | 5.0 | 1.6 | 1.4 | 51.2 | 0.0019 |
| E239R | Interface | 5.2 | 56 | 0.0011 |
Stopped-flow measurements as described in Methods. The values are relative to WT (3 × 105 M−1⋅s−1). SE for kon values is 20%, and for konmut/konwt is 30% (see Methods for details).
From SPR measurements done at six different concentrations. SE is 25%.
KD = koff/kon, with koff values taken from SPR and kon from stopped-flow measurements. SE is 30%.
Protein thermal stability. SE is 1 °C.
Enzymatic activity was determined as the initial slope of CENTA catalysis using 5 nM enzyme and 375 µM substrate. SE is 30%.
With the exception of T29E, no mutation caused a significant reduction in kon (>2× SE), whereas about 40% of the mutations bound between 50% and fivefold faster than WT. For the mutations where koff was determined, a minor two- to fourfold increase was determined for three of them, whereas for the other seven mutations no significant change in koff was measured. The thermal stability shifted in either direction by up to 4 °C (which should not affect the protein at room temperature). We used the initial slope of substrate degradation with 5 nM enzyme as an indicator for β-lactamase activity. Overall, the effects of the mutations on enzymatic activity were small, with up to twofold increase or decrease being estimated by this measure.
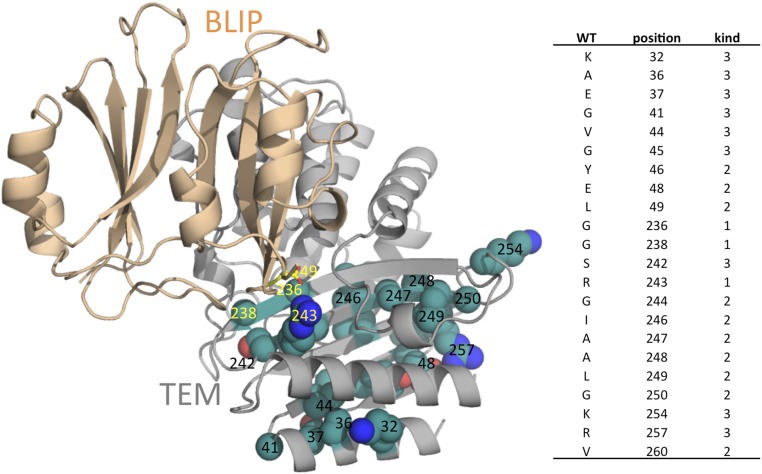
The deep-sequencing data also indicated specific positions that were underrepresented (i.e., strongly dominated by WT), and thus can be assumed to be particularly sensitive to mutagenesis. These positions are presented in Fig. 3. Positions with low mutagenesis rate are located at two sequence patches closely placed in the 3D structure. Almost all of the positions that did not tolerate mutagenesis are buried and probably relate to protein stability. Only three interfacial residues did not tolerate mutagenesis: G236, G238, and R243. Taking into account that single-nucleotide mutations cover only part of the amino acid repertoire, these results clearly demonstrate that the TEM1 interface is less conserved than its core and that the interface can readily be mutated without harming the high-affinity binding. These results are in line with the variation in β-lactamase sequences binding BLIP (5).
Fig. 3.
(Left) Structural representation of the TEM–BLIP complex (PDB ID code 1JTG). Positions that did not tolerate mutations are presented as spheres (green, C; red, O; blue, N). Residues important for binding are labeled in yellow. Arg243 on TEM forms a salt bridge with Asp49 on BLIP (represented as sticks), which is important for binding. (Right) A table indicating the conserved positions and their kind: 1, interface; 2, buried; 3, surface.
Selection for Faster Binders.
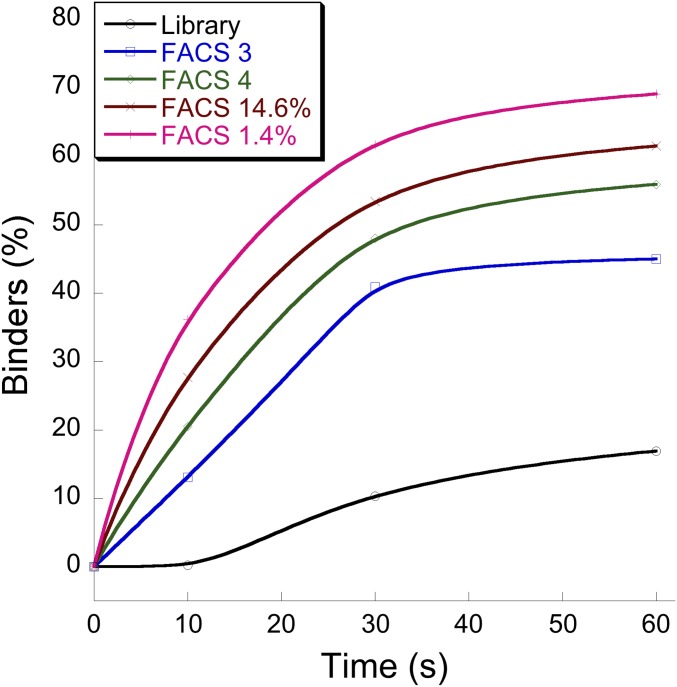
Because the TEM1 selection against BLIP seemed to specifically enhance the association rate constants, we now aimed to explicitly select for fast binders. Traditionally, selection using yeast display is done under equilibrium conditions, where one allows for sufficient time of binding before washing steps are initiated. Because we aimed to specifically select for fast binders, we significantly shortened the binding step before washing, as well as reduced the ligand concentration. Shorter binding time at lower protein concentration provides an advantage for faster binders to associate. Hence, in the kinetic selection, the library was incubated with a low ligand concentration (50 nM) for a very short time (30 s) before FACS sorting. Fig. 4 shows the percentage of bound cells after indicated incubation times (before ligand washing was commenced) of the given selected libraries. The sorted libraries displayed higher percentages of bound cells after very short incubation times. FACS 4, which was selected against 70 nM Y–BLIP, binds faster than FACS 3, which was selected against 1 µM Y-BLIP, despite these sorts being initiated after reaching equilibrium (1-h incubation with BLIP). The FACS 3 library was then incubated for only 30 s with 50 nM Y-BLIP before washing, collecting the top 14.6 and 1.4% of binders. This kinetically sorted library bound as a whole faster and with higher percentage (1.4% more than 14%) than BLIP. To verify the nature of the selected mutants, individual clones were sequenced from the 1.4% library. These clones had a very high representation of charge-altering mutations located within or in close proximity to the interface (Table 2). TransComp (26) is an algorithm that calculates kon values for protein complexes, taking into account, in addition to electrostatic forces, diffusion rates. We used TransComp to evaluate whether the selected mutations are predicted to increase the association rate constant and compared the results with experimental measurements of selected purified clones. Table 2 indicates that almost half of the selected clones are predicted by TransComp to improve the TEM–BLIP association rate constant by greater than twofold, while none is predicted to slow the interaction by greater than twofold. Seven of the clones were expressed and purified and their rates of association were determined. Most of the purified clones (five out of seven) showed significantly faster association rate constants than WT, in line with the calculated data. The four fastest selected clones increased kon by 2.6- to 5-fold, which equals a reduction in activation energy of 0.5 to 0.9 kcal/mol. These clones have mutations at positions E239K, H96R+D115G, E110V+T200I+M270K, and I47T+Q88K+E110V+A227T. Common to the four is an increase in positive net charge by +2.
Fig. 4.
FACS measurements of binders as a function of incubation time with the ligand (Y–BLIP) of the different HS-TEM libraries: library, random nonselected original library; FACS 3, three nonstringent sorts against BLIP; FACS 4, FACS 3 followed by a stringent sort (low BLIP concentration); FACS 14.6%, the best 14.6% binders of FACS 3 after a 30-s incubation with BLIP; FACS 1.4%, the best 1.4% binders of FACS 3 after a 30-s incubation with BLIP. The x axis shows time of incubation of the library before washing was initiated before FACS measurements.
Table 2.
Sequences and relative kon values of individual clones resulting from stringent kinetic selection
| Clone | Sequence* | TransComp† | konmut/konwt‡ |
| 1 | I95L A126T | 1.3 | |
| 2 | H96R D115G | 2.5 | 3.2 |
| 3 | K111R I208V T269N | 1.4 | |
| 4 | K192N | 0.9 | |
| 5 | Y262N | 1.0 | |
| 6 | L51F K111N E121D A201T D214N S223P | 0.7 | |
| 7 | I56V V80A T141A E171V | 1.2 | |
| 8 | E110V T200I M270K | 8.6 | 3.6 |
| 9 | G91S G116S L162H | 0.9 | 1.1 |
| 10 | E239K | 6.1 | 5.0 |
| 11 | E171G E279V | 1.4 | |
| 12 | L102R | 1.2 | |
| 13 | Q90H K111R A135T V159E T182R | 9.7 | |
| 14 | R94H S124G | 0.7 | |
| 15 | E63G D115G | 2.1 | 1.2 |
| 16 | I47T Q88K E110V A227T | 3.9 | 2.6 |
| 17 | T133A T188A K215R | 1.8 | |
| 18 | K111R S223P | 2.6 | |
| 19 | Q99R T128I L137S | 1.1 | |
| 20 | T188M P219L S266G | 1.0 | |
| 21 | L75P E110V M186V I246V | 3.8 | |
| 22 | F60L | 1.0 | |
| 23 | K111R | 3.4 | 1.9 |
| 24 | L113R E239K | 9.5 | |
| 25 | E110V P226Q | 3.8 | |
| 26 | R240A | 0.6 |
Interface residues are underlined.
TransComp predicted kon rates at an ionic strength of 0.15 M, taking into account the influence of all occurring mutations, relative to WT.
Stopped-flow measurements as described in Methods, relative to WT (3 × 105 M-1⋅s−1). The SE for konmut/konwt is 30%.
Discussion
Our study focused on the selection of a randomly mutated TEM1 library against its tight binder BLIP. Deep-sequencing analysis of the selected clones revealed an interesting property of the interface: Most of the interface seems to be plastic, tolerating substitution with other amino acids, with some of them even improving on binding affinity (Fig. 2). One should, however, note that the selected interface substitutions were restricted to one to two other amino acids (except position 111, where four substitutions were selected) out of the possible allowed substitutions by single-nucleotide mutations (Table S1). This shows that potentially this interface has many solutions to achieving binding at a similarly high affinity (nanomolar). The results are also in line with the sequence alignment of different natural TEM1 genes binding BLIP, which also showed a high degree of plasticity of the interface (5, 27). Nevertheless, in the natural environment, negative selection against nonspecific binders may limit the number of interface solutions obtained by a specific complex (28).
Three positions were essential for binding and were not replaced by other amino acids in the library (Fig. 3). G236 and G238 were found to be critical for tight BLIP binding and seem to be important for substrate specificity (29); R243 was shown to contribute to binding by forming a salt bridge with D49 on BLIP, with its mutation decreasing the TEM–BLIP affinity by 10-fold (30). It should also be noted that most positively selected mutations are located on the surface of TEM1, whereas negatively selected positions are mostly located in the core of the protein. This result is in agreement with other studies involving TEM1 libraries (31, 32). One can assume that many of these core mutations have detrimental effects on protein stability, and thus were not selected. Indeed, the Tm of the positively selected clones is close to that of the WT protein. It has previously been shown that poorly folded proteins are reduced in yeast surface display, which could be the case also here (33).
It is interesting to note that the selection was acting to specifically increase the association rate constant, whereas not having much effect on the dissociation rate constant, TEM1 enzymatic activity, or its thermal stability (Table 1). In accordance, none of the stabilizing mutations reported by previous studies was strongly selected here, implying that the selection was specifically acting upon TEM1–BLIP binding (34), especially when taking into account the opposite influence of mutations on function versus thermal stability (35). Usually, selection for binding specifically slows the dissociation rate constant, as the procedure involves multiple washing steps, which result in dissociation of the complex (36). However, apparently due to the slow dissociation rate constant of the WT complex, the bound ligands in our study remained bound throughout the washing steps, providing an advantage for faster binders. Previous studies have shown that increasing association rate constants for the TEM1–BLIP complex is feasible by introducing charge mutations either within or in the proximity of the interface (3, 37–39). The potential to enhance association is shown also in the selections presented here (Table 1). To further promote faster binders, the library was incubated with a low ligand concentration for a very short time period (30 s), not reaching equilibrium. Computational analysis using TransComp, a predictor of kon, showed that many of the selected clones indeed bind faster. The computational results were validated by experimental measurements for seven of the selected clones (Table 2). It should be noted that the selection shown in Table 1 results from deep-sequencing analysis, with a read of 200 bp, which was analyzed by selecting for frequently occurring mutations. Conversely, in the subsequent, fast library selection (Table 2), selected clones were directly sequenced, with one to six mutations per clone. Kinetic selection might be useful for any process where fast-binding variants are important. Faster binding allows for binding to occur at lower ligand concentrations and after shorter periods of time, which would have a clear advantage in protein–drug screening, with faster association improving drug functionality.
In summary, in this work, we have shown that the natural interface between TEM1 and BLIP is not unique, and that it can be altered without loss in binding affinity. Moreover, we show that the relaxed selection conditions selected for faster-binding variants, which could be enhanced by changing the selection protocol to a novel one where selection was done under preequilibrium conditions.
Methods
Yeast Library Formation.
The yeast (Eby100) library was created following the procedure described by Benatuil et al. (40). DNA mutagenesis was performed with mutazyme (GeneMorph II Random Mutagenesis Kit; 200550; Agilent), aiming for four mutations per gene. DNA amplification was done with Taq DNA polymerase as described by Chao et al. (41). The library was prepared based on a stabilized variant of the TEM1 β-lactamase previously described (42). The library size was estimated to be ∼108 by plating serial dilutions on selection plates lacking tryptophan. The library was further examined for the correct gene insertion and for the average number of mutations introduced per gene by sequencing 20 single clones.
Library Sorting.
The first three sorts of the library were not stringent, using 1 μM ligand BLIP for selection and collecting the 4 to 50% best binders (the FACS gate remained constant and the percentage of gated clones rose through the selection). BLIP was fused to the fluorescent protein YPET, which allowed it to be detected by FACS. After the three sorts, the selected libraries were analyzed by deep sequencing. An additional two stringent sorts were performed using 70 nM Y-BLIP, collecting the best 4% of binders. These sorts were done after a 1-h incubation on ice. An additional, stringent kinetic sort was done following the third nonstringent selection, using 50 nM Y-BLIP and allowing only 30 s of library–ligand incubation at room temperature. The highest 14 and 1.4% of binders were collected, and are referred to as “kinetic selection.”
Deep-Sequencing Analysis.
The original random library and the first and third mild sorts were analyzed by deep sequencing to follow the selection progression. Deep sequencing was done on an Illumina in a HiSeq rapid with cBot mode with a read length of 200 nt. The DNA amplifications were done on the libraries DNA-extracted from yeast by the Zymoprep Yeast Plasmid Miniprep I Kit (Zymo Research; D2001). The TEM1 gene (792 bp) was sequenced as a mixture of four ∼200-bp fragments. The deep-sequencing results were analyzed using Enrich 2.0 software (25). The analysis of mutation enrichment (or decline) was done by comparing the mutation frequency at each position of each amino acid with the frequency of occurrence in the unselected library and with the previous selection rounds. Mutations were individually expressed, purified, and analyzed if they stood up to one of the following criteria. (i) A specific mutation had a frequency of greater than fourfold that in the original library after sort 3. (ii) The mutation frequency after the first sort was greater than three times that of the original library, and the mutation frequency after sort 3 was higher compared with sort 1. (iii) The mutation frequency after sort 1 was greater than 1.8-fold higher than the original library and increased by at least 1.4-fold after sort 3. A position was defined to be not tolerant to mutagenesis when its total mutation frequency was decreased by a factor of 5 after sort 1 relative to the random library, and further declined at sort 3.
Protein Production.
The proteins were produced and purified using Ni beads binding a 6×His tag as detailed (43).
Definition of Buried, Surface, and Interface Residues.
Swiss-PdbViewer (spdbv.vital-it.ch/) was used to define surface and buried residues, with buried residues having <10% accessibility. As input, we used Protein Data Bank (PDB) ID code 1BTL. Interface residues were defined by their proximity to a heavy atom on the second chain, with 4 Å taken as cutoff.
Measuring Binding Constants.
Protein binding affinities were estimated by surface plasmon resonance (SPR) using the ProteOn XPR36 Protein Interaction Array System (Bio-Rad). All SPR measurements were made in PBS/0.005% Tween 20 running buffer. The ligands were bound to Bio-Rad GLC SPR chips using the high-affinity biotin–avidin interaction. Surface regeneration was done by two short cycles of 8 M urea that enabled complex dissociation. The analyte was injected at six different concentrations, and the affinity and kinetic binding constants were determined by ProteOn Manager Version 3 (Bio-Rad) using the Langmuir reaction model. Association rate constants were determined also using a stopped-flow spectrometer (Applied Photophysics), as it was shown to be a more reliable method for this purpose. The measurements were done as described (44) using Hepes buffer (pH 7.2).
β-Lactamase Activity Assay.
β-Lactamase activity was estimated with the commercially available yellow chromogenic substrate CENTA (Calbiochem; 219475), whose absorbance at 405 nm rises upon catalysis by β-lactamase. As a measure of relative activity, the initial slope at the linear phase of the reaction (steady state) was determined using 5 nM enzyme and 375 µM substrate.
Protein Stability Measurements.
Protein thermal stability was determined by the method described (45). The assay is based on the tendency of SYPRO Orange (Sigma; S5692) to bind hydrophobic areas, which are exposed upon protein unfolding due to a gradual increase in temperature (0.05 °C/s from 25 to 99 °C) controlled by an Applied Biosystems ViiA 7 RT-PCR instrument. The assay was optimized for β-lactamase, with the measurements done in a 20-μL volume using 10 μM protein in PBS buffer with the dye diluted 500×.
Error Calculations.
Reported values for individual measurements were calculated from two to four repeats of the different experiments. The errors for each type of experiment were calculated from all of the available data (and not for each mutant separately), as the different mutants were measured using the same method and thus their respective SD is expected to be similar.
Acknowledgments
We acknowledge Prof. Huan-Xian Zhou and Sanbo Qin for help in running the TransComp server, and Dr. Douglas M. Fowler for guiding us in using the Enrich software. This work was supported by a grant of the Israel Science Foundation (1549/14).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1613122113/-/DCSupplemental.
References
- 1.Reichmann D, et al. The modular architecture of protein-protein binding interfaces. Proc Natl Acad Sci USA. 2005;102(1):57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moreira IS, Fernandes PA, Ramos MJ. Hot spots—A review of the protein-protein interface determinant amino-acid residues. Proteins. 2007;68(4):803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 3.Selzer T, Albeck S, Schreiber G. Rational design of faster associating and tighter binding protein complexes. Nat Struct Biol. 2000;7(7):537–541. doi: 10.1038/76744. [DOI] [PubMed] [Google Scholar]
- 4.DeLano WL, Ultsch MH, de Vos AM, Wells JA. Convergent solutions to binding at a protein-protein interface. Science. 2000;287(5456):1279–1283. doi: 10.1126/science.287.5456.1279. [DOI] [PubMed] [Google Scholar]
- 5.Schreiber G, Keating AE. Protein binding specificity versus promiscuity. Curr Opin Struct Biol. 2011;21(1):50–61. doi: 10.1016/j.sbi.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joachimiak LA, Kortemme T, Stoddard BL, Baker D. Computational design of a new hydrogen bond network and at least a 300-fold specificity switch at a protein-protein interface. J Mol Biol. 2006;361(1):195–208. doi: 10.1016/j.jmb.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 7.Fleishman SJ, et al. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332(6031):816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karanicolas J, et al. A de novo protein binding pair by computational design and directed evolution. Mol Cell. 2011;42(2):250–260. doi: 10.1016/j.molcel.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowie JU, Reidhaar-Olson JF, Lim WA, Sauer RT. Deciphering the message in protein sequences: Tolerance to amino acid substitutions. Science. 1990;247(4948):1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- 10.Weiss GA, Watanabe CK, Zhong A, Goddard A, Sidhu SS. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci USA. 2000;97(16):8950–8954. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pál G, Kouadio J-LK, Artis DR, Kossiakoff AA, Sidhu SS. Comprehensive and quantitative mapping of energy landscapes for protein-protein interactions by rapid combinatorial scanning. J Biol Chem. 2006;281(31):22378–22385. doi: 10.1074/jbc.M603826200. [DOI] [PubMed] [Google Scholar]
- 12.Choi YS, Yang JS, Choi Y, Ryu SH, Kim S. Evolutionary conservation in multiple faces of protein interaction. Proteins. 2009;77(1):14–25. doi: 10.1002/prot.22410. [DOI] [PubMed] [Google Scholar]
- 13.Mintseris J, Weng Z. Structure, function, and evolution of transient and obligate protein-protein interactions. Proc Natl Acad Sci USA. 2005;102(31):10930–10935. doi: 10.1073/pnas.0502667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neuvirth H, Raz R, Schreiber G. ProMate: A structure based prediction program to identify the location of protein-protein binding sites. J Mol Biol. 2004;338(1):181–199. doi: 10.1016/j.jmb.2004.02.040. [DOI] [PubMed] [Google Scholar]
- 15.Zhang QC, et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490(7421):556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbreth RN, Esaki K, Koide A, Sidhu SS, Koide S. A dominant conformational role for amino acid diversity in minimalist protein-protein interfaces. J Mol Biol. 2008;381(2):407–418. doi: 10.1016/j.jmb.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Palzkill T. Dissecting the protein-protein interface between β-lactamase inhibitory protein and class A β-lactamases. J Biol Chem. 2004;279(41):42860–42866. doi: 10.1074/jbc.M406157200. [DOI] [PubMed] [Google Scholar]
- 18.Clackson T, Wells JA. In vitro selection from protein and peptide libraries. Trends Biotechnol. 1994;12(5):173–184. doi: 10.1016/0167-7799(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 19.Thom G, et al. Probing a protein-protein interaction by in vitro evolution. Proc Natl Acad Sci USA. 2006;103(20):7619–7624. doi: 10.1073/pnas.0602341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan L, Kurek I, English J, Keenan R. Laboratory-directed protein evolution. Microbiol Mol Biol Rev. 2005;69(3):373–392. doi: 10.1128/MMBR.69.3.373-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albeck S, Schreiber G. Biophysical characterization of the interaction of the β-lactamase TEM-1 with its protein inhibitor BLIP. Biochemistry. 1999;38(1):11–21. doi: 10.1021/bi981772z. [DOI] [PubMed] [Google Scholar]
- 22.Tripathi A, Varadarajan R. Residue specific contributions to stability and activity inferred from saturation mutagenesis and deep sequencing. Curr Opin Struct Biol. 2014;24:63–71. doi: 10.1016/j.sbi.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 23.DeLano WL. Unraveling hot spots in binding interfaces: Progress and challenges. Curr Opin Struct Biol. 2002;12(1):14–20. doi: 10.1016/s0959-440x(02)00283-x. [DOI] [PubMed] [Google Scholar]
- 24.Hu Z, Ma B, Wolfson H, Nussinov R. Conservation of polar residues as hot spots at protein interfaces. Proteins. 2000;39(4):331–342. [PubMed] [Google Scholar]
- 25.Fowler DM, Araya CL, Gerard W, Fields S. Enrich: Software for analysis of protein function by enrichment and depletion of variants. Bioinformatics. 2011;27(24):3430–3431. doi: 10.1093/bioinformatics/btr577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin S, Pang X, Zhou H-X. Automated prediction of protein association rate constants. Structure. 2011;19(12):1744–1751. doi: 10.1016/j.str.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schreiber G. Kinetic studies of protein-protein interactions. Curr Opin Struct Biol. 2002;12(1):41–47. doi: 10.1016/s0959-440x(02)00287-7. [DOI] [PubMed] [Google Scholar]
- 28.Zarrinpar A, Park S-H, Lim WA. Optimization of specificity in a cellular protein interaction network by negative selection. Nature. 2003;426(6967):676–680. doi: 10.1038/nature02178. [DOI] [PubMed] [Google Scholar]
- 29.Rudgers GW, Palzkill T. Identification of residues in β-lactamase critical for binding β-lactamase inhibitory protein. J Biol Chem. 1999;274(11):6963–6971. doi: 10.1074/jbc.274.11.6963. [DOI] [PubMed] [Google Scholar]
- 30.Gretes M, et al. Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the β-lactamase inhibitory proteins BLIP, BLIP-I, and BLP. J Mol Biol. 2009;389(2):289–305. doi: 10.1016/j.jmb.2009.03.058. [DOI] [PubMed] [Google Scholar]
- 31.Jacquier H, et al. Capturing the mutational landscape of the beta-lactamase TEM-1. Proc Natl Acad Sci USA. 2013;110(32):13067–13072. doi: 10.1073/pnas.1215206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng Z, et al. Deep sequencing of systematic combinatorial libraries reveals β-lactamase sequence constraints at high resolution. J Mol Biol. 2012;424(3–4):150–167. doi: 10.1016/j.jmb.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shusta EV, Kieke MC, Parke E, Kranz DM, Wittrup KD. Yeast polypeptide fusion surface display levels predict thermal stability and soluble secretion efficiency. J Mol Biol. 1999;292(5):949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- 34.Kather I, Jakob RP, Dobbek H, Schmid FX. Increased folding stability of TEM-1 β-lactamase by in vitro selection. J Mol Biol. 2008;383(1):238–251. doi: 10.1016/j.jmb.2008.07.082. [DOI] [PubMed] [Google Scholar]
- 35.Shoichet BK, Baase WA, Kuroki R, Matthews BW. A relationship between protein stability and protein function. Proc Natl Acad Sci USA. 1995;92(2):452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levin KB, et al. Following evolutionary paths to protein-protein interactions with high affinity and selectivity. Nat Struct Mol Biol. 2009;16(10):1049–1055. doi: 10.1038/nsmb.1670. [DOI] [PubMed] [Google Scholar]
- 37.Selzer T, Schreiber G. New insights into the mechanism of protein-protein association. Proteins. 2001;45(3):190–198. doi: 10.1002/prot.1139. [DOI] [PubMed] [Google Scholar]
- 38.Shaul Y, Schreiber G. Exploring the charge space of protein-protein association: A proteomic study. Proteins. 2005;60(3):341–352. doi: 10.1002/prot.20489. [DOI] [PubMed] [Google Scholar]
- 39.Schreiber G, Haran G, Zhou H-X. Fundamental aspects of protein-protein association kinetics. Chem Rev. 2009;109(3):839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benatuil L, Perez JM, Belk J, Hsieh C-M. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng Des Sel. 2010;23(4):155–159. doi: 10.1093/protein/gzq002. [DOI] [PubMed] [Google Scholar]
- 41.Chao G, et al. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1(2):755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 42.Dellus-Gur E, Toth-Petroczy A, Elias M, Tawfik DS. What makes a protein fold amenable to functional innovation? Fold polarity and stability trade-offs. J Mol Biol. 2013;425(14):2609–2621. doi: 10.1016/j.jmb.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 43.Khait R, Schreiber G. FRETex: A FRET-based, high-throughput technique to analyze protein-protein interactions. Protein Eng Des Sel. 2012;25(11):681–687. doi: 10.1093/protein/gzs067. [DOI] [PubMed] [Google Scholar]
- 44.Phillip Y, et al. Contrasting factors on the kinetic path to protein complex formation diminish the effects of crowding agents. Biophys J. 2012;103(5):1011–1019. doi: 10.1016/j.bpj.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lavinder JJ, Hari SB, Sullivan BJ, Magliery TJ. High-throughput thermal scanning: A general, rapid dye-binding thermal shift screen for protein engineering. J Am Chem Soc. 2009;131(11):3794–3795. doi: 10.1021/ja8049063. [DOI] [PMC free article] [PubMed] [Google Scholar]