Significance
Nutritional conditions during early development influence the plastic expression of adult phenotypes. Because heightened nutrition sensitivity often characterizes the development of exaggerated traits, there should be molecular mechanisms underlying trait-specific variability. This study reveals the molecular mechanisms underlying the expression of nutrition-sensitive mandibles in the beetle Gnatocerus cornutus. We found that epigenetic regulators, such as histone deacetylases (HDACs) and polycomb group (PcG) proteins, contribute specifically to the plastic expression of male mandibles, with little contribution to other body modules. In addition, HDAC1 and HDAC3 perturbation resulted in opposite phenotypic effects on mandible and wing modules. Our findings provide molecular evidence of a link between distinct epigenetic modifications and module-specific phenotypic plasticity of exaggerated traits.
Keywords: phenotypic plasticity, HDAC, Gnatocerus cornutus, exaggerated male weapon, sexual dimorphism
Abstract
Nutritional conditions during early development influence the plastic expression of adult phenotypes. Among several body modules of animals, the development of sexually selected exaggerated traits exhibits striking nutrition sensitivity, resulting in positive allometry and hypervariability distinct from other traits. Using de novo RNA sequencing and comprehensive RNA interference (RNAi) for epigenetic modifying factors, we found that histone deacetylases (HDACs) and polycomb group (PcG) proteins preferentially influence the size of mandibles (exaggerated male weapon) and demonstrate nutrition-dependent hypervariability in the broad-horned flour beetle, Gnatocerus cornutus. RNAi-mediated HDAC1 knockdown (KD) in G. cornutus larvae caused specific curtailment of mandibles in adults, whereas HDAC3 KD led to hypertrophy. Notably, these KDs conferred opposite effects on wing size, but little effect on the size of the core body and genital modules. PcG RNAi also reduced adult mandible size. These results suggest that the plastic development of exaggerated traits is controlled in a module-specific manner by HDACs.
Although individuals of the same species share similar genomic DNA, they often display different phenotypes depending upon external environmental conditions. This phenomenon, called “phenotypic plasticity,” generates adaptive phenotypes in a specific environment (1).
Sexually selected male ornaments, such as bird tails, and weapons, such as beetle horns, exhibit extraordinary phenotypic plasticity (2, 3). Such ornaments and weapons are often disproportionately enlarged in large individuals. This exaggerated feature of the ornaments and weapons exhibits “positive allometry,” which represents the enhanced growth of a specific body part relative to body size growth (4, 5). Males often adopt condition-dependent strategies to gain access to females, such as fighting, sneaking, or dispersal; therefore, sexual selection is thought to be a major ecological factor that leads to the evolution of striking plasticity in ornament and weapon phenotypes (6–8). In contrast, other body modules, such as invertebrate genitalia or the mammalian brain, generally develop to a constant size irrespective of nutritional conditions (i.e., canalization) (9–12). Such reduction in plasticity evolves when trait uniformity, rather than variability, is favored in selection (i.e., “stabilizing selection”) (11, 12).
Because heightened nutrition sensitivity often characterizes the development of exaggerated traits, there should be molecular mechanisms underlying trait-specific variability. A recent study on the rhinoceros beetle has shown that the development of horns is more sensitive to perturbation of pathways regulating nutrient intake (e.g., insulin/insulin-like growth factor signaling) than to the perturbation of other body modules (13, 14). This result provides molecular evidence that different body modules respond differently to nutritional signals. However, the mechanism underlying module-specific plasticity is still unclear (15).
When we focus on plasticity at the cellular level, epigenetic control by DNA methylation and histone modifications broadly regulates cell fate plasticity. During development, the epigenetic status allows cells to memorize cell-type or tissue-specific gene expression patterns (16–18). Consequently, cellular developmental fates are gradually restricted during cell differentiation. This fundamental cellular mechanism to regulate plasticity leads us to propose that epigenetic regulation may be involved in increased developmental plasticity of an exaggerated trait.
Among various roles, epigenetic systems have significant functions in memorizing the nutritional environments during early development so as to link the developmental conditions with adult phenotypes. In the differentiation of queens and workers in the honey bee, Apis mellifera, larvae fed with highly nutritious “royal jelly” develop into queens and larvae fed with poorly nutritious “worker jelly” develop into workers (19). This process is determined by the DNA methylation patterns of the larvae; thus, caste fate can be altered by knockdown (KD) of DNA methyltransferase 3 (DNMT3) (20). In mammals, poor nutritional environments during early embryogenesis can influence the epigenetic status of the individual, resulting in physiological variability among adults (21, 22).
On the basis of such findings, we hypothesized that epigenetic regulation contributes to the nutrition-sensitive plasticity of exaggerated traits. To test this notion, we used the broad-horned flour beetle, Gnatocerus cornutus, which has exaggerated and hypervariable male mandibles, with conditional expression as the developmental genetic model (23, 24) (Fig. 1A). Using epigenetic inhibitor drugs and RNA interference (RNAi)-mediated KD of genes for DNA methylation and histone modification, we showed that epigenetic perturbations in penultimate- and final-instar larvae specifically impact the development of mandibles, suggesting the epigenetic regulation of hypervariability in exaggerated traits.
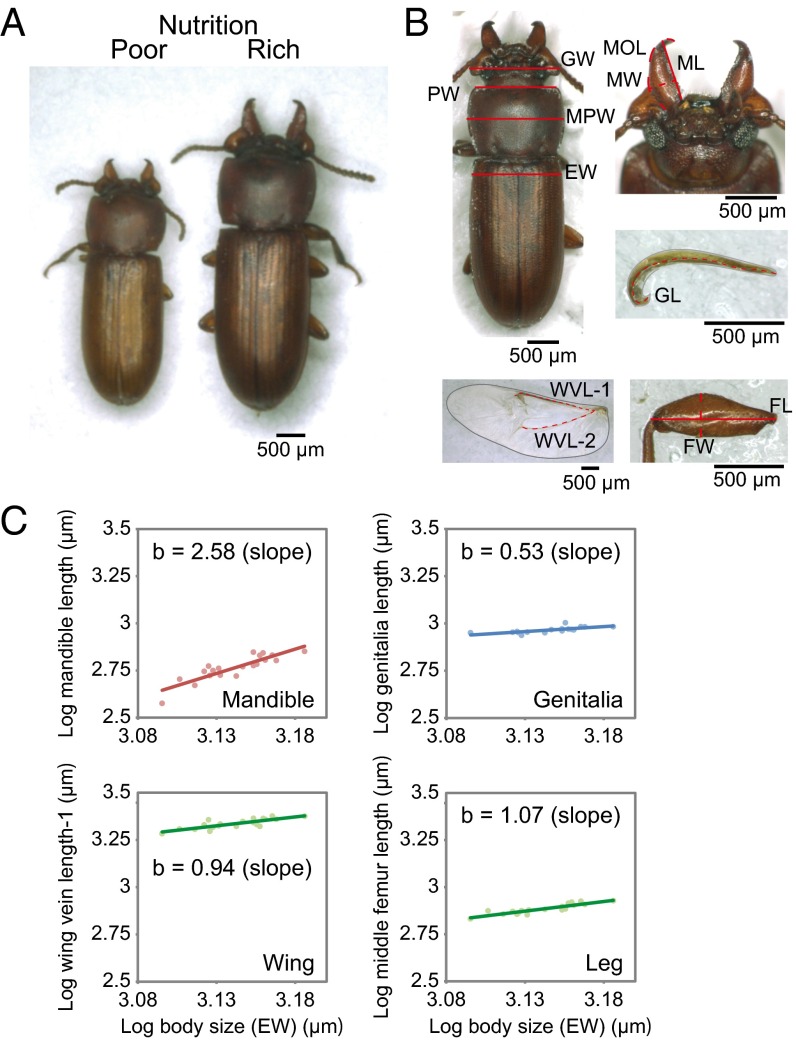
Fig. 1.
Trait-specific variabilities in weapon, somatic, and genital traits. (A) Male polymorphism under different nutritional conditions in the broad-horned flour beetle, G. cornutus. (B) Morphometric landmarks were used. EW, elytra width; FL, femur length; FW, femur width; GL, genitalia length; GW: gena width; MPW, maximum prothorax width; PW, prothorax width. FL and FW are landmarked for fore, middle, and hind legs as fore femur length (FFL), middle femur length (MFL), hind femur length (HFL), fore femur width (FFW), middle femur width (MFW), and hind femur width (HFW). (C) Allometric growth of each body module in G. cornutus males. Trait sizes are plotted against body size (EW) in the log–log scale. Y = a + bX, where b is a coefficient of regression. Note that the mandible exhibits hyperallometry, the genitalia show hypoallometry, and the wing and leg show isometry. The allometric relationships of other parts are indicated in SI Appendix, Table S1.
Results
First, we evaluated morphological variations in several body parts of G. cornutus (Fig. 1B) and estimated allometric relationships between each body part and elytra width (an index of whole-body size). When the slope of the regression line in the log–log plot is greater than 1, the trait is assumed to show hypervariability. Allometric analysis revealed that mandible traits showed steep positive allometries (i.e., hyperallometry), whereas genitalia size demonstrated hypoallometry (Fig. 1C and SI Appendix, Table S1 for all traits). Other body modules, such as legs and wings, were nearly isometric in relative growth to the elytra width. Such module-specific variability is likely linked with differential sensitivity to nutritional conditions in the larval stage (13, 25).
Before the gene KD experiments, inhibitors of DNMTs and histone deacetylases (HDACs) were applied to final-instar larvae to evaluate the involvement of DNA- and histone-modifying processes on mandible morphogenesis. Injection of a HDAC inhibitor, trichostatin A (TSA), caused 6.3% hypertrophy of the mandibles [SI Appendix, Fig. S1A and Table S2; treatment: P = 0.038, treatment × body size: P = 0.139, analysis of covariance (ANCOVA)]. In contrast, injection of the DNMT inhibitor 5-azacytidine (5-AzaC) caused no detectable changes in the adult mandible size but slightly affected the regression slope (SI Appendix, Fig. S1B and Table S2; treatment: P = 0.070, treatment × body size: P = 0.037, ANCOVA). These results suggest that histone acetylation may contribute to determining the size of the mandibles.
To elucidate the molecular basis for epigenetic control of phenotypic variability, we further identified the genes that encode five HDACs, 13 polycomb group (PcG) proteins, histone methyltransferases (HMTs), DNMTs, and other epigenetic factors by using de novo RNA sequencing (RNA-seq) (detailed information on RNA-seq and gene identification is provided in SI Appendix, Tables S3 and S4). Five different class I (HDAC1 and HDAC3), class II (HDAC4 and HDAC6), and class IV (HDAC11) HDACs of G. cornutus appear to have corresponding orthologs in the red flour beetle (Tribolium castaneum), fruit fly, and humans, supporting the structural conservation of these enzymes across wide taxa (SI Appendix, Fig. S2).
We then conducted comprehensive RNAi experiments (Fig. 2A) by injecting double-stranded RNA (dsRNA) of target genes into larvae at penultimate- and final-instar stages (i.e., early and late KDs) and estimated their influence on pupal and adult morphologies. First, we present the results of late KDs because early and late KDs yielded similar phenotypes. Late KDs reduced mRNA levels of HDAC1 (59.1%), HDAC3 (22.8%), HDAC6 (54%), and HDAC11 (69%) compared with the control treatment (GFP RNAi) in the heads [in each treatment, HDAC level was quantified by quantitative PCR (qPCR) and normalized to actin; details are provided in Materials and Methods and SI Appendix, Fig. S3 A–C]. The G. cornutus HDAC4 (GcHDAC4) KD was not effective (89.7%; P = 0.171, t test).
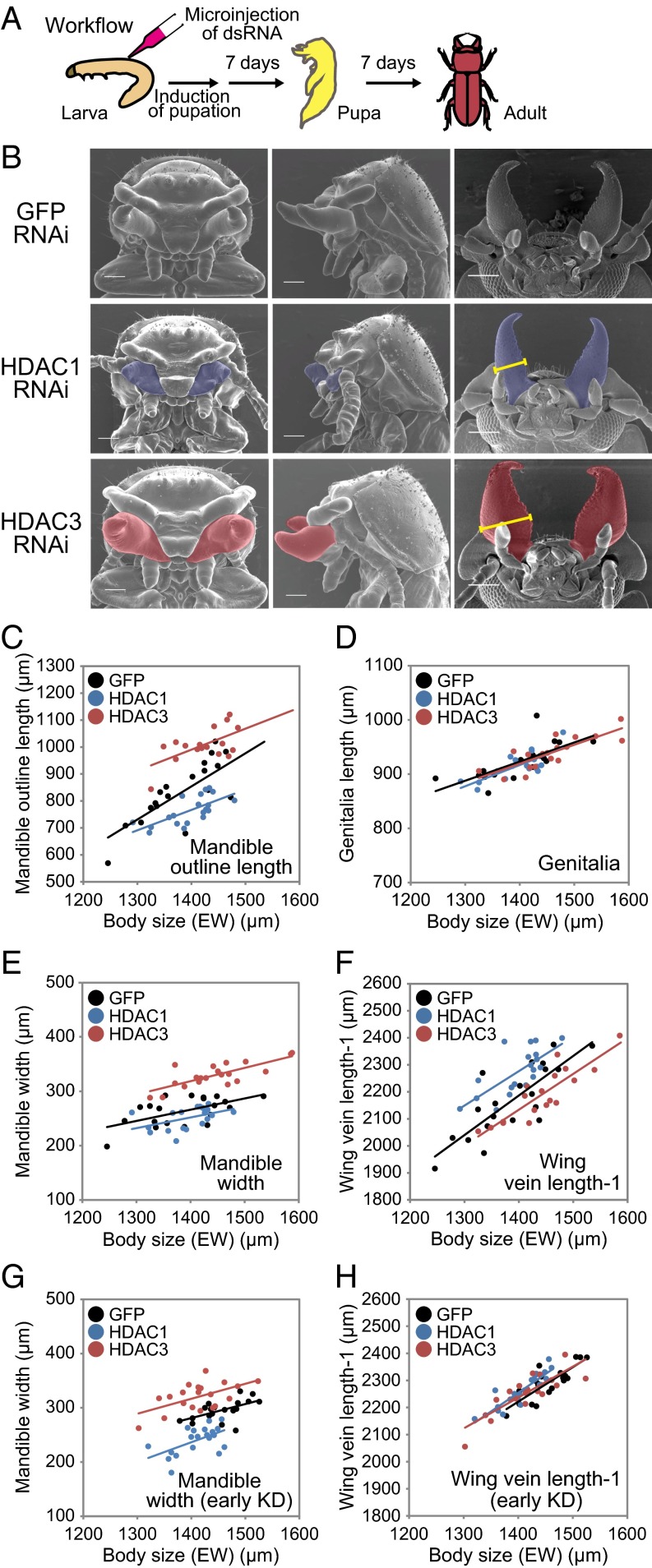
Fig. 2.
Effects of HDAC1 and HDAC3 KDs on pupal and adult morphologies. (A) Schematic workflow of the RNAi experiment. (B, Left and Middle) Pupal heads (front and side views). (B, Right) Adult heads (ventral views). Mandibles are colored blue (HDAC1) or red (HDAC3). (Scale bars: 200 μm.) (C–H) Effects of HDAC1 and HDAC3 KDs on adult male morphology. The stages of HDAC KDs are final-instar larvae (C–F) and penultimate-instar larvae (G and H). KDs of HDAC1 (blue dots and regression lines) and HDAC3 (red dots and lines) significantly increased and decreased the mandible sizes (C, D, and G) with respect to the GFP dsRNA-injected control (black dots and lines). ANCOVA results are shown in SI Appendix, Table S6 and S7. Effects of HDAC1 and HDAC3 KDs on other traits are shown in SI Appendix, Figs. S4 and S5.
KD of GcHDAC1 led to a clear curtailment of pupal mandible morphologies (Fig. 2B, Middle), whereas KD of GcHDAC3 resulted in mandible hypertrophy (Fig. 2B, Bottom). Microscopic inspection of pupal morphologies showed little morphogenetic effects on other traits (Fig. 2B, Left and Center), whereas gene silencing was effective throughout the body (SI Appendix, Fig. S3 A and B).
For morphological quantification of adult phenotypes, we carefully determined the appropriate strengths of gene KDs (conditions are discussed in Materials and Methods and SI Appendix, Table S5) that enabled successful development of adults, so as to avoid any systemic developmental failures due to the high dose of the gene KD. Using those RNAi conditions, we obtained adult males without notable developmental failures (Fig. 2B, Right). We found that the GcHDAC1 KD significantly reduced adult mandible outline length (MOL; Fig. 2C, blue dots and line; 9.8% reduction in intercept; treatment: P < 0.001, treatment × elytra width: P = 0.175, ANCOVA; detailed statistics are provided in SI Appendix, Table S6) and mandible width (MW; Fig. 2E, blue dots and line; 4.9% reduction in intercept; treatment: P < 0.001, treatment × elytra width: P = 0.934, ANCOVA; statistics are provided in SI Appendix, Table S6).
In contrast, the GcHDAC3 KD caused a marked increase in MOL (Fig. 2C, red dots and line; 12.8% increase in intercept; treatment: P < 0.001, treatment × body size: P = 0.118, ANCOVA; statistics are provided in SI Appendix, Table S6) and MW (Fig. 2E, red dots and line; 20.0% increase in intercept; treatment: P < 0.001, treatment × body size: P = 0.653, ANCOVA; statistics are provided in SI Appendix, Table S6). Thus, GcHDAC1 and GcHDAC3 KDs caused opposite effects on adult mandible morphogenesis.
Other body parts, such as genitalia, were not affected by the gene KDs (Fig. 2D and SI Appendix, Table S6). Interestingly, however, GcHDAC1 KD led to a slight enlargement in hind wing size (Fig. 2F, blue dots and line; 4.4% increase in intercept; treatment: P < 0.001, treatment × body size, P = 0.175, ANCOVA; statistics are provided in SI Appendix, Table S6), whereas GcHDAC3 KD caused a reduction in hind wing size (Fig. 2F, red dots and line; 2.4% reduction in intercept; treatment: P < 0.001, treatment × body size: P = 0.175, ANCOVA; statistics are provided in SI Appendix, Table S6).
HDAC KDs were previously conducted at the end of the larval stage. Given that different organs may form at different times during development (26–29), mandible progenitor cells may retain the developmental flexibility to allow phenotypic plasticity until late stages, whereas other somatic/genital precursor cells may lose such flexibility at earlier stages of development. If such is the case, HDAC KDs at an early developmental stage may have non–organ-specific morphogenetic effects. We tested this notion by conducting HDAC KDs at earlier stages, particularly the penultimate-instar larvae stage, using a dose that enabled the larvae to develop into adults (SI Appendix, Table S5). Early-HDAC KDs had a similar direction of the effects in adult phenotypes to the effects observed in late KDs: HDAC1 KDs caused a reduction in mandible size but slight elongation of the wings, whereas HDAC3 KDs resulted in enlargement of the mandibles but had subtle effects on wing size (Figs. 2 G and H and 3A). However, upon careful inspection, the magnitude of effects was different for HDAC1 and HDAC3. Later HDAC1 KD resulted in much greater effects in the mandible compared with early KD, whereas the opposite was true for HDAC3: Early-HDAC3 KDs showed weaker effects in the mandible. In addition, early-HDAC1 KDs exhibited weaker effects in wings compared with late injections. Other traits, such as legs and genitalia, were unaffected by both the early- and late-HDAC KDs (Fig. 3 A and B and SI Appendix, Tables S6 and S7).
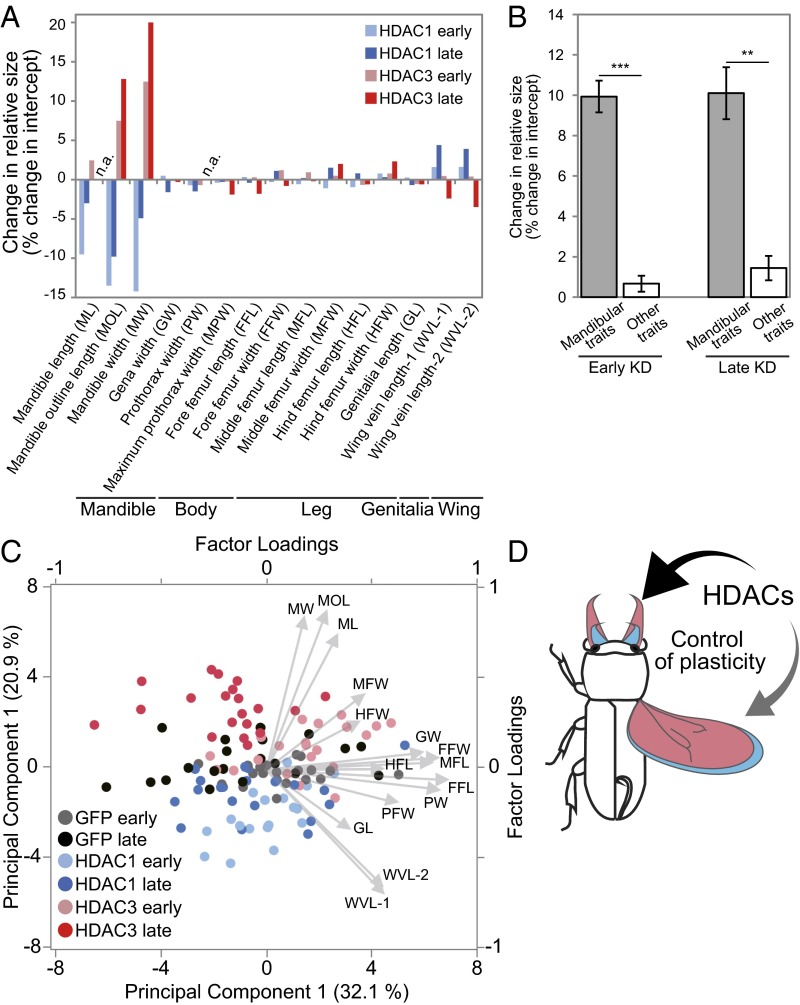
Fig. 3.
Overall phenotypic changes caused by HDAC1 and HDAC3 KDs. (A) Changes in adjusted trait sizes (i.e., changes of intercepts in ANCOVA with body size as covariates) (Fig. 2 C–H and SI Appendix, Figs. S4 and S5 and Tables S6 and S7). Light blue, HDAC1 early KD; blue, HDAC1 late KD; pink, HDAC3 early KD; red, HDAC3 late KD; n.a., not applicable because of significant changes in slopes. (B) Magnitudes of changes in mandibular traits (ML, MOL, and MW) and 12 other body traits under early- and late-HDAC KD conditions. Magnitudes of changes (changes of intercepts in absolute values) were significantly greater for mandibular traits than for other somatic and genital traits (early KD: P < 0.001, Z = 3.71; late KD: P = 0.0012, Z = 3.24; **P < 0.01, ***P < 0.001, U test). Mean ± SD values are shown. (C) PCA performed on the detrended data of 15 body traits [residuals of linear regression to body size (i.e., elytra width)]. PC1 indicates the relative sizes of organs, except the mandible and genitalia (factor loadings are provided in SI Appendix, Table S8). PC2 indicates an increase in relative mandible size and decrease in relative wing size. Arrows indicate factor loadings (also SI Appendix, Table S8). Note that the phenotypes of early- and late-HDAC1 KDs (light blue and blue dots) occupied distinct morphospaces to early- and late-HDAC3 KDs (pink and red dots) across the GFP control phenotype (black dots), suggesting that KD of HDAC1 and HDAC3 caused opposite phenotypic changes. (D) Schematic diagram of HDAC-mediated, module-specific plasticity. HDAC1 (blue) and HDAC3 (red) have antagonistic effects on mandible and wing sizes.
To compare the magnitudes of trait size changes caused by gene KDs, changes in the heights of intercepts (calculated as the percentage of changes in the adjusted mean by using elytra width as a covariate; SI Appendix, Table S6) were calculated (Fig. 3A). Mandibular traits [mandible length (ML), MOL, and MW] exhibited drastic size changes, whereas other body parts showed few changes (Fig. 3A). The magnitudes of the changes were significantly greater for mandibular traits than for other traits (Fig. 3B; late KD: P = 0.0012, U test). Therefore, two HDACs appear to regulate size plasticity of mandibles and wings antagonistically, irrespective of which stage was perturbed (Fig. 3 B–D).
To summarize the overall morphological changes caused by RNAi, we performed principal component analysis (PCA) using all body traits. To remove multicollinearity, the residuals of the linear regressions on body size (i.e., elytra width) were used for PCA. Principal component 1 (PC1; 32.1% contribution) was highly positively loaded by all traits except the mandibular traits and genitalia, both of which showed small factor loadings (Fig. 3C and SI Appendix, Table S8). Given that the residuals of the linear regressions on elytra width were used in PCA, PC1 was considered to reflect the relative sizes of organs except the mandible and genitalia (Fig. 3C and SI Appendix, Table S8). PC2 (20.9% contribution) was strongly positively loaded by ML, MOL, and MW, and it was strongly negatively loaded by wing vein length (WVL-1 and WVL-2; SI Appendix, Table S8). This result indicates that PC2 reflected a relative increase in the mandibular size and decrease in wing size; thus, PC2 likely reflects the morphological changes caused by RNAi (Fig. 3C; other PCs are shown in SI Appendix, Table S8). The PC1/PC2 plot shows that the KD phenotypes of GcHDAC1 (Fig. 3C, blue and light blue dots represent late and early KDs, respectively) and GcHDAC3 (Fig. 3C, red and pink dots represent late and early KDs, respectively) were mostly located above and below the control phenotype (Fig. 3C, black dots; GFP RNAi), confirming that the two HDACs have distinct and antagonistic effects on the mandible and wing, irrespective of KD timing. Importantly, PC2 factor loadings for traits other than mandibular and wing traits were relatively small (Fig. 3C and SI Appendix, Table S8), indicating that traits other than mandible and wing traits are less affected by HDAC perturbations.
In addition to HDACs, we examined KD effects of other epigenetic regulators, such as PcG proteins (components of polycomb-repressive complexes PRC1 and PRC2), DNMTs, and HMTs, because they have been demonstrated to modulate the epigenetic status of developing cells (30–33) (SI Appendix, Table S4). KDs of the two PcG proteins led to the reduction of mandible size (SI Appendix, Fig. S6 A–C and Table S9), suggesting that PRC1 and PRC2 complexes are indeed involved in the determination of mandible size. However, other DNA methylation factors and histone modification factors, including DNMTs (GcDNMT1 and GcDNMT2) and HMTs (GcSETMAR and GcPRMT5), did not affect mandible development significantly (a list of examined genes is provided in SI Appendix, Table S4). These results suggest that the plasticity of exaggerated organs is selectively controlled by a subset of HDACs and PcG complexes, which are mainly involved in chromatin silencing (34).
Discussion
In this study, we used we used the sexually dimorphic horned flour beetle, G. cornutus, to investigate how epigenetic regulation influences the expression of nutrition-sensitive exaggerated traits (i.e., mandibles). We found that epigenetic regulators, such as HDACs and PcG proteins, contribute to the plastic expression of male mandibles, but there is little contribution to other body modules. In addition, HDAC1 and HDAC3 perturbation resulted in opposite phenotypic effects on mandible and wing modules. These findings provide molecular evidence that indicates a link between the module-specific phenotypic plasticity of exaggerated traits and distinct epigenetic modifications.
Hypervariability of exaggerated traits is widely found in animal taxa, and it is thought to be the result of heightened nutrition sensitivity (13, 14, 35, 36). Module-specific response to insulin signaling is a plausible mechanism that regulates trait variability (13, 15), but the mechanism underlying module specificity needs to be elucidated (15).
Experimental perturbation of development by HDAC KDs revealed that phenotypic modifications are most striking in exaggerated traits (i.e., mandibular traits) (Figs. 2 and 3). Our finding proposed a mechanistic explanation that the more plastic development of the mandible is characterized by its relative sensitivity to epigenetic perturbations. Epigenetic regulation and nutrient-sensing mechanisms have been suggested to be tightly linked (37–41). It is thus likely that nutrient signals, such as the insulin-like growth factor, may differentially affect downstream target genes in an organ-specific manner via epigenetic modifications. In this study, we found that variability in the weapon size was particularly influenced by HDAC KD, although the regression slope of the mandible size was not affected in the HDAC KDs. The detailed mechanism determining the slope is under investigation; however, PcG proteins (PCGF3 and EHZ2; SI Appendix, Fig. S6) whose KDs had clear reductions in slope are the hopeful candidates of relative growth regulation in future studies.
Because GcHDAC KDs reduced target GcHDAC expression throughout the body, mandible-specific effects show that the mandibular progenitor cells have a specific sensitivity to the fluctuation in HDAC activities. There are two possibilities regarding how mandibles are specifically affected by epigenetic perturbations. First, the developmental timing for size determination is relatively late in the mandibles, and thus HDAC KDs had mandible-specific effects. Second, however, early-HDAC KDs also influenced mandible sizes without largely affecting other morphological traits. Therefore, it is unlikely that the first possibility explains the results (Fig. 3 A and B). Thus, we considered another scenario to explain the module-specific effect of epigenetic perturbation, which is that epigenetic fixation is more flexible in the mandible traits than in other traits. Despite a generally similar direction of effects of early- and late-HDAC KDs, there were small but detectable differences. Early-HDAC1 KD yielded stronger effects on mandibles than late-HDAC1 KD, whereas early-HDAC3 KD exhibited milder effects than late-HDAC3 KD (Fig. 3A). Additionally, effects of early-HDAC KDs on wings were generally small. These results suggest stage-dependent distinct involvements of two HDACs in mandibles and wings, which needs to be elucidated in future studies.
The GcHDAC1 and GcHDAC3 KD experiments implicate that these HDACs may have different roles in size determination of the mandible and wing. HDAC1 and HDAC3 belong to the same HDAC group (class I), which exhibits relatively broad substrate specificities (42). The diversification of class I HDACs has been observed in basal eukaryotes, such as fungi (42), and is thought to have originated in early metazoans or earlier (43). In multicellular organisms, class I HDACs are very similar proteins, but they often have distinct functions (44, 45). In chicken B cells, HDAC1 and HDAC2 showed different effects on the diversification of Ig hypervariable regions (45). It has been suggested that class I HDACs can function distinctly by recruiting different transcriptional factors (46, 47). Therefore, GcHDAC1 and GcHDAC3 may distinctly contribute to the fine-tuning of the mandible size by recruiting different target genes. The functional diversification of HDACs observed in beetle weapon development implicates a widespread evolutionary mechanism of HDACs in generating morphological modularity during evolution.
The trade-off relationship between the weapon and flight apparatus is often observed as a characteristic of weaponed insects, reflecting the diverse reproductive strategy of males (48–50). A distinct type of HDAC (e.g., HDAC1, HDAC3) that causes antagonistic responses to the mandible and wing may be associated with the trade-off relationship, but further investigations of the epigenetic status of the genes involved in mandible and wing development are required to confirm this possibility.
Considering the generality of the epigenetic status to define cellular plasticity, we propose that the evolution of module-specific plasticity is associated with the evolution of epigenetic regulation. In the case of sexually selected exaggerated traits, condition-dependent selections for fighting or sneaking/dispersal are thought to generate the evolution of heightened plasticity (6–8). In contrast, other body modules, such as genitalia, are under stabilizing selection that favors the production of uniform organ size irrespective of developmental conditions (i.e., canalization) (10–12). In response to selections that favor heightened plasticity or reduced plasticity, acquisition of epigenetic flexibility or stability may evolve in the development of target organs. The similar process of differential canalization may be important in stem cell functions (51–53). Weak canalization is assumed to be critical in genes responsible for the maintenance of pluripotency in mammalian somatic stem cells, which must flexibly and dynamically respond to the environmental cues for differentiation. Although the detailed mechanism is open to debate, our finding provides the insight that module-specific epigenetic regulation may underlie hypervariability and weak canalization of a class of environment-sensitive traits.
Materials and Methods
Insect Husbandry.
Our stock population of G. cornutus originated from adults collected in Miyazaki City (31°54′, 131°25′), Japan, and was maintained at the National Food Research Institute and Okayama University (24). G. cornutus was reared as described previously (54).
Synthesis of dsRNA.
The cDNA was prepared from the prepupal whole body of G. cornutus. The dsRNA was synthesized using an Ambion MEGAscript T7 Transcription Kit (Invitrogen). The DNA template for in vitro transcription was produced with PCR using gene-specific primers with the T7 polymerase promoter at their 5′ ends (SI Appendix, Table S5). The DNA template of GFP was produced using primers GFPiF2 and GFPiR5 (55). The dsRNA was quantified and diluted to 1 μg/μL and stored at −80 °C.
Microinjection of Epigenetic Drugs and dsRNA.
Final-instar and penultimate-instar larvae were randomly selected from the stock culture. Stock solutions of TSA (with 2 μg/μL dimethyl sulfoxide) and 5-AzaC (with 10 μg/μL water) were prepared. Dosage was optimized to produce maximal nonlethal effects by the stated criteria (TSA: 46 ng per larva, 5AzaC: 3.48 μg per larva), and drugs were injected into the larvae by using Nanoject II (Drummond Scientific) under CO2 anesthesia. The injected larvae were individually isolated in 24-well culture plates (Thermo Scientific), so that they could develop into prepupae (the adult morphogenetic stage) within 3–4 d (36, 54). No food was provided after the treatment. The developmental stage of the injected larvae was observed once a day. Individuals that successfully became adults were used for measurements.
The dsRNA was also injected in the penultimate- and final-instar larvae (i.e., early and late KDs). Stock solutions of dsRNA (1 μg/μL water) for each target gene were prepared. To avoid any systemic developmental failures due to high doses of RNAi, dsRNA dose adjustment was performed as described in SI Appendix, Table S5. For morphological observations and measurements of adults subjected to HDAC1 and HDAC3 RNAi, moderate doses were used to avoid severe defects during pupal maturation and adult eclosion.
Morphological Observations and Measurements.
Morphological observations of pupae and adults were performed using a scanning electron microscope (VE-8800; KEYENCE). To evaluate the morphogenetic effects of epigenetic perturbations, 16 body parts of adult males were quantified (Fig. 1B) using mandible sizes (ML, MOL, and MW) as indices of exaggerated traits. Elytra width was considered as an index of body size and was used as a covariate to examine the effect of the treatments on trait sizes (25, 56).
Body parts sizes were measured using a microscope monitoring system (VHX-200; KEYENCE) and ImageJ (NIH) (57). All analyses were performed using R 3.1.1 and JMP 11.
Sequencing of Genes for HDACs and PcGs in G. cornutus.
Transcript sequences of the G. cornutus orthologs for HDACs, PcG genes, and DNA methylation factors and histone modification factors were identified using de novo RNA-seq. Total RNA was isolated from the heads of final-instar larvae and prepupae (24 and 48 h after attaining the prepupal stage). The cDNA was synthesized using the template-switching method (58). Library samples of cDNA were run using an Illumina HiSeq 2000 platform at the Beijing Genomics Institute for 100 cycles with paired-end reads. All postprocessed reads were pooled together and assembled de novo with Trinity (version: Trinityrnaseq_r20131110) (59). We obtained 412,824 contigs by de novo assembly with Trinity. G. cornutus orthologs of histone-modifying genes were identified using the Kyoto Encyclopedia of Genes and Genomes Automatic Annotation Server (60), with manual checks of sequence similarities to T. castaneum orthologs. The DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank accession numbers for Gc-HDACs and Gc-PcGs are listed in SI Appendix, Table S4.
In addition, cross-species gene orthologies (G. cornutus, T. castaneum, Drosophila melanogaster, and Homo sapiens) were confirmed by phylogenetic relationships of proteins.
Quantitative RT-PCR for HDAC Expression Analysis.
The cDNA was prepared from the head, thorax, abdomen, and whole body of dsRNA-injected larvae and prepupae. Quantitative RT-PCR was performed using the KAPA SYBR Fast qPCR Kit (Kapa Biosystems) with Applied Biosystems StepOnePlus. Actin gene Gc-ACT was used as the reference for comparative threshold cycle quantification. Primer sequences for the actin gene, HDACs, and PcG genes are listed in SI Appendix, Table S10.
Detailed materials and methods are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Naoki Irie, Dr. Alexander S. Mikheyev, and Dr. Kazuto Kugo for technical advice on RNA-seq. This study was supported by grants from the Japan Society for the Promotion of Science (Grant 26870121 to Y.O. and Grants 23114003, 26291018, and 15K14443 to K. Ohta); the Platform for Dynamic Approaches to Living System from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan (to K. Ohta, M.S., and Y.O.); and the Yamada Science Foundation (to Y.O.), and by Grant-in-Aid for Scientific Research 25128706 from MEXT (to T.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank databases (accession nos. LC096257–LC096261, LC100104, and LC100109).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615688114/-/DCSupplemental.
References
- 1.West-Eberhard MJ. Phenotypic plasticity and the origins of diversity. Annu Rev Ecol Syst. 1989;20:249–278. [Google Scholar]
- 2.West-Eberhard MJ. Sexual selection, social competition, and speciation. Q Rev Biol. 1983;58(2):155–183. [Google Scholar]
- 3.Andersson MB. Sexual Selection. Princeton Univ Press; Princeton: 1994. [Google Scholar]
- 4.Huxley JS. 1932. Problems of Relative Growth (Methuen & Co. LTD, London)
- 5.Shingleton AW, Frankino WA. New perspectives on the evolution of exaggerated traits. BioEssays. 2013;35(2):100–107. doi: 10.1002/bies.201200139. [DOI] [PubMed] [Google Scholar]
- 6.Gross MR. Disruptive selection for alternative life histories in salmon. Nature. 1985;313(5997):47–48. [Google Scholar]
- 7.Emlen DJ. Alternative reproductive tactics and male-dimorphism in the horned beetle Onthophagus acuminatus (Coleoptera: Scarabaeidae) Behav Ecol Sociobiol. 1997;41(5):335–341. [Google Scholar]
- 8.Okada K, Miyatake T, Nomura Y, Kuroda K. Fighting, dispersing, and sneaking: body‐size dependent mating tactics by male Librodor japonicus beetles. Ecol Entomol. 2008;33(2):269–275. [Google Scholar]
- 9.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150(3811):563–565. [Google Scholar]
- 10.Guthrie HA, Brown ML. Effect of severe undernutrition in early life on growth, brain size and composition in adult rats. J Nutr. 1968;94(4):419–426. doi: 10.1093/jn/94.4.419. [DOI] [PubMed] [Google Scholar]
- 11.Eberhard WG, et al. One size fits all? Relationships between the size and degree of variation in genitalia and other body parts in twenty species of insects and spiders. Evolution. 1998;52(2):415–431. doi: 10.1111/j.1558-5646.1998.tb01642.x. [DOI] [PubMed] [Google Scholar]
- 12.House CM, Simmons LW. No evidence for condition-dependent expression of male genitalia in the dung beetle Onthophagus taurus. J Evol Biol. 2007;20(4):1322–1332. doi: 10.1111/j.1420-9101.2007.01346.x. [DOI] [PubMed] [Google Scholar]
- 13.Emlen DJ, Warren IA, Johns A, Dworkin I, Lavine LC. A mechanism of extreme growth and reliable signaling in sexually selected ornaments and weapons. Science. 2012;337(6096):860–864. doi: 10.1126/science.1224286. [DOI] [PubMed] [Google Scholar]
- 14.House CM, et al. Macronutrient balance mediates the growth of sexually selected weapons but not genitalia in male broad‐horned beetles. Funct Ecol. 2015;30:769–779. [Google Scholar]
- 15.Tang HY, Smith-Caldas MS, Driscoll MV, Salhadar S, Shingleton AW. FOXO regulates organ-specific phenotypic plasticity in Drosophila. PLoS Genet. 2011;7(11):e1002373. doi: 10.1371/journal.pgen.1002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 17.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76(1):75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 18.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 19.Weaver N. Rearing of honeybee larvae on royal jelly in the laboratory. Science. 1955;121(3145):509–510. doi: 10.1126/science.121.3145.509. [DOI] [PubMed] [Google Scholar]
- 20.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319(5871):1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 21.Barker DJP, Osmond C. Infant mortality, childhood nutrition, and ischaemic heart disease in England and Wales. Lancet. 1986;1(8489):1077–1081. doi: 10.1016/s0140-6736(86)91340-1. [DOI] [PubMed] [Google Scholar]
- 22.Gluckman PD, Hanson MA. Living with the past: Evolution, development, and patterns of disease. Science. 2004;305(5691):1733–1736. doi: 10.1126/science.1095292. [DOI] [PubMed] [Google Scholar]
- 23.Okada K, Miyanoshita A, Miyatake T. Intra-sexual dimorphism in male mandibles and male aggressive behavior in the broad-horned flour beetle Gnatocerus cornutus (Coleoptera: Tenebrionidae) J Insect Behav. 2006;19(4):457–467. [Google Scholar]
- 24.Okada K, Miyatake T. Effect of losing on male fights of broad-horned flour beetle, Gnatocerus cornutus. Behav Ecol Sociobiol. 2010;64(3):361–369. [Google Scholar]
- 25.Okada K, Miyatake T. Plasticity of size and allometry in multiple sexually selected traits in an armed beetle Gnatocerus cornutus. Evol Ecol. 2010;24(6):1339–1351. [Google Scholar]
- 26.Miyazaki S, et al. Ergatoid queen development in the ant Myrmecina nipponica: Modular and heterochronic regulation of caste differentiation. Proc Biol Sci. 2010;277(1690):1953–1961. doi: 10.1098/rspb.2010.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moczek AP, Nagy LM. Diverse developmental mechanisms contribute to different levels of diversity in horned beetles. Evol Dev. 2005;7(3):175–185. doi: 10.1111/j.1525-142X.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 28.Moczek AP, Nijhout HF. A method for sexing final instar larvae of the genus Onthophagus Latreille (Coleoptera: Scarabaeidae) Coleopt Bull. 2002;56(2):279–284. [Google Scholar]
- 29.Svácha P. What are and what are not imaginal discs: Reevaluation of some basic concepts (Insecta, Holometabola) Dev Biol. 1992;154(1):101–117. doi: 10.1016/0012-1606(92)90052-i. [DOI] [PubMed] [Google Scholar]
- 30.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 31.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 32.Greer EL, Shi Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13(5):343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20(10):1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 34.van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23(4):474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 35.Gotoh H, et al. Juvenile hormone regulates extreme mandible growth in male stag beetles. PLoS One. 2011;6(6):e21139. doi: 10.1371/journal.pone.0021139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okada Y, Gotoh H, Miura T, Miyatake T, Okada K. Juvenile hormone mediates developmental integration between exaggerated traits and supportive traits in the horned flour beetle Gnatocerus cornutus. Evol Dev. 2012;14(4):363–371. doi: 10.1111/j.1525-142X.2012.00554.x. [DOI] [PubMed] [Google Scholar]
- 37.Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16(1):9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153(1):56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr. 2005;135(6):1382–1386. doi: 10.1093/jn/135.6.1382. [DOI] [PubMed] [Google Scholar]
- 40.Jang H, Serra C. Nutrition, epigenetics, and diseases. Clin Nutr Res. 2014;3(1):1–8. doi: 10.7762/cnr.2014.3.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinclair KD, et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc Natl Acad Sci USA. 2007;104(49):19351–19356. doi: 10.1073/pnas.0707258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gallinari P, Di Marco S, Jones P, Pallaoro M, Steinkühler C. HDACs, histone deacetylation and gene transcription: From molecular biology to cancer therapeutics. Cell Res. 2007;17(3):195–211. doi: 10.1038/sj.cr.7310149. [DOI] [PubMed] [Google Scholar]
- 43.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: Functional implications of phylogenetic analysis. J Mol Biol. 2004;338(1):17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 44.Bates EA, Victor M, Jones AK, Shi Y, Hart AC. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J Neurosci. 2006;26(10):2830–2838. doi: 10.1523/JNEUROSCI.3344-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurosawa K, Lin W, Ohta K. Distinct roles of HDAC1 and HDAC2 in transcription and recombination at the immunoglobulin loci in the chicken B cell line DT40. J Biochem. 2010;148(2):201–207. doi: 10.1093/jb/mvq054. [DOI] [PubMed] [Google Scholar]
- 46.Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21(18):6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guenther MG, Yu J, Kao GD, Yen TJ, Lazar MA. Assembly of the SMRT-histone deacetylase 3 repression complex requires the TCP-1 ring complex. Genes Dev. 2002;16(24):3130–3135. doi: 10.1101/gad.1037502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomkins JL, Kotiaho JS, Lebas NR. Phenotypic plasticity in the developmental integration of morphological trade-offs and secondary sexual trait compensation. Proc Biol Sci. 2005;272(1562):543–551. doi: 10.1098/rspb.2004.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okada K, Nomura Y, Miyatake T. Relations between allometry, male–male interactions and dispersal in a sap beetle, Librodor japonicus. Anim Behav. 2007;74(4):749–755. [Google Scholar]
- 50.Yamane T, Okada K, Nakayama S, Miyatake T. Dispersal and ejaculatory strategies associated with exaggeration of weapon in an armed beetle. Proc Biol Sci. 2010;277(1688):1705–10. doi: 10.1098/rspb.2009.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gifford CA, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153(5):1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu J, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152(3):642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hawkins RD, et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6(5):479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ozawa T, Ohta K, Shimada M, Okada K, Okada Y. Environmental factors affecting pupation decision in the horned flour beetle Gnatocerus cornutus. Zoolog Sci. 2015;32(2):183–187. doi: 10.2108/zs140203. [DOI] [PubMed] [Google Scholar]
- 55.Tomoyasu Y, Denell RE. Larval RNAi in Tribolium (Coleoptera) for analyzing adult development. Dev Genes Evol. 2004;214(11):575–578. doi: 10.1007/s00427-004-0434-0. [DOI] [PubMed] [Google Scholar]
- 56.Okada K, Miyatake T. Genetic correlations between weapons, body shape and fighting behaviour in the horned beetle Gnatocerus cornutus. Anim Behav. 2009;77(5):1057–1065. [Google Scholar]
- 57.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aird SD, et al. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis) BMC Genomics. 2013;14(1):790. doi: 10.1186/1471-2164-14-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35(Web Server issue) Suppl 2:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.