Abstract
Cell death is a common and important feature of animal development, and cell death defects underlie many human disease states. The nematode Caenorhabditis elegans has proven fertile ground for uncovering molecular and cellular processes controlling programmed cell death. A core pathway consisting of the conserved proteins EGL-1/BH3-only, CED-9/BCL2, CED-4/APAF1, and CED-3/caspase promotes most cell death in the nematode, and a conserved set of proteins ensures the engulfment and degradation of dying cells. Multiple regulatory pathways control cell death onset in C. elegans, and many reveal similarities with tumor formation pathways in mammals, supporting the idea that cell death plays key roles in malignant progression. Nonetheless, a number of observations suggest that our understanding of developmental cell death in C. elegans is incomplete. The interaction between dying and engulfing cells seems to be more complex than originally appreciated, and it appears that key aspects of cell death initiation are not fully understood. It has also become apparent that the conserved apoptotic pathway is dispensable for the demise of the C. elegans linker cell, leading to the discovery of a previously unexplored gene program promoting cell death. Here, we review studies that formed the foundation of cell death research in C. elegans, and describe new observations that expand, and in some cases remodel, this edifice. We raise the possibility that, in some cells, more than one death program may be needed to ensure cell death fidelity.
Keywords: Cell death, C. elegans Apoptosis, Nonapoptotic cell death, Linker cell
Introduction
Cell death is a widespread process that is essential for life. Tissue sculpting, organ morphogenesis, and organ size control are but a few of the developmental events that integrally utilize programmed cell death to generate a functioning adult animal. It is therefore not surprising that many things go wrong when cell death goes awry (Fuchs, & Steller, 2011). Indeed, neurodegeneration and tumorigenesis, disease states against which armies of researchers have been amassed, result from too much or too little cell culling, respectively (Youle, & van der Bliek, 2012; Hanahan, & Weinberg, 2011). While the hypothesis that cell death is a regulated phenomenon in animal development was first experimentally addressed in vertebrates (Hamburger, & Levi-Montalcini, 1949) and insects (Lockshin, & Williams, 1965), the first systematic studies aimed at deciphering the molecular program promoting cell demise employed the free-living soil nematode Caenorhabditis elegans (Horvitz, 2003). Early observations of the cellular complement of adult C. elegans revealed little variation in the number and position of cells between individuals of similar ages, leading to the proposal that cell lineage in this animal may be invariant. This prediction was largely borne out by taking advantage of the transparent cuticle of the animal to observe cell divisions in vivo (Kimble, & Hirsh, 1979; Sulston, Albertson, & Thomson, 1980; Sulston, & Horvitz, 1977; Sulston, Schierenberg, White, & Thomson, 1983). This heroic effort culminated in a complete cell lineage tree documenting a generally predictable pattern of divisions that generate adult somatic tissue from the zygote. These studies demonstrated that precisely 1090 and 1178 somatic cells must be generated to produce a C. elegans hermaphrodite and male, respectively.
Among the generated cells, a small but substantial set (~12%) are eliminated. These cells become refractile under Differential Interference Contrast (DIC) optics (Fig. 1), acquire a rounded morphology, and eventually disappear. Ultrastructural studies reveal that these dying cells are engulfed by neighboring cells (Sulston, Schierenberg, White, & Thomson, 1983), and possess characteristics of apoptotic cell death, such as condensed nuclear chromatin, and reduced cytoplasmic volume (Shaham, & Horvitz, 1996b; Sulston, Schierenberg, White, & Thomson, 1983) (Fig. 1). Like the lineage itself, these cell death events are essentially invariant between individuals and target the same cells at the same time in development. In the hermaphrodite and male, 131 and 147 somatic cells are eliminated, respectively. Subsequent studies demonstrated that cell death is highly prevalent during germline development and maintenance, with roughly 50% of female meiosis products succumbing to apoptosis (Gumienny et al., 1999). Developmental death of germ cells in C. elegans differs from somatic cell death in that the identities of dying cells are not ascribed to their lineage (Gumienny et al., 1999; Sulston, Schierenberg, White, & Thomson, 1983). C. elegans therefore offers two arenas for understanding cell death control: one in which cell death and lineage are tightly coupled, and one in which stochastic processes apparently determine life and death. Studies of the former revealed a core pathway controlling apoptotic cell death from C. elegans to mammals.
Figure 1.
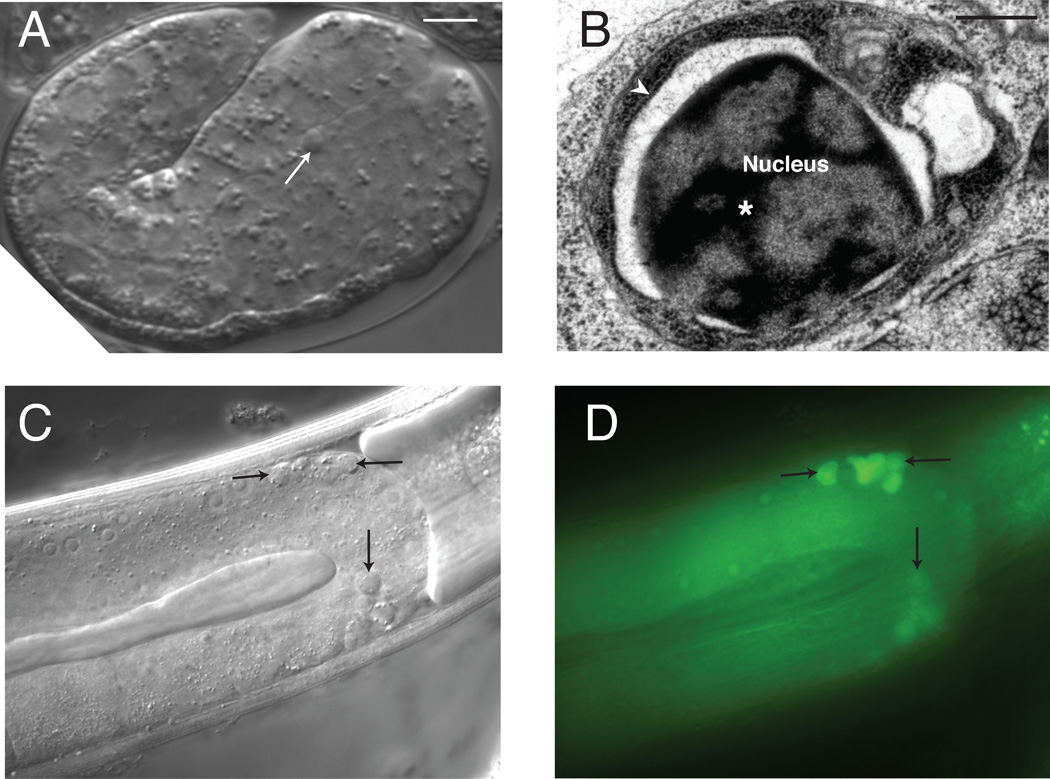
Apoptotic developmental cell death in C. elegans. (A) Differential Interference Contrast (DIC) image of a dying cell (arrow) in a developing embryo. Scale bar, 2.5 microns. (B) Electron micrograph of a dying embryonic cell. Note dark staining, condensed chromatin (asterisk) within the nucleus and shrunken cytoplasm. Arrowhead, nuclear envelope. Scale bar, 400 nm. (C) DIC image of dying cells in the developing gonad (arrows). (D) Fluorescence image of animal in (C) stained with the cell death marker SYTO12. Scale bar, 8 microns.
Here we describe these key components and their interactions and explore current understanding of the lineage-dependent mechanisms that trigger the activation of these killer genes and proteins. We also discuss a group of genes important for the clearance of dying cells and their relation to cell death execution, and delve into a number of mysteries that remain unanswered and which have the potential to expand and modify our understanding of why and how cells die. We end by describing a novel non-apoptotic C. elegans cell death program that promotes dismantling of the male-specific linker cell.
Core apoptosis regulators in C. elegans
Most cell death in C. elegans is controlled by the proteins CED-3, CED-4, CED-9, and EGL-1, whose functions and interactions have been worked out in some detail (Fig. 2). All four components of this canonical cell death pathway are conserved across disparate animal species, but are apparently absent from bacteria, fungi, and plants. Thus, it is likely that this pathway arose early on in the animal lineage.
Figure 2.
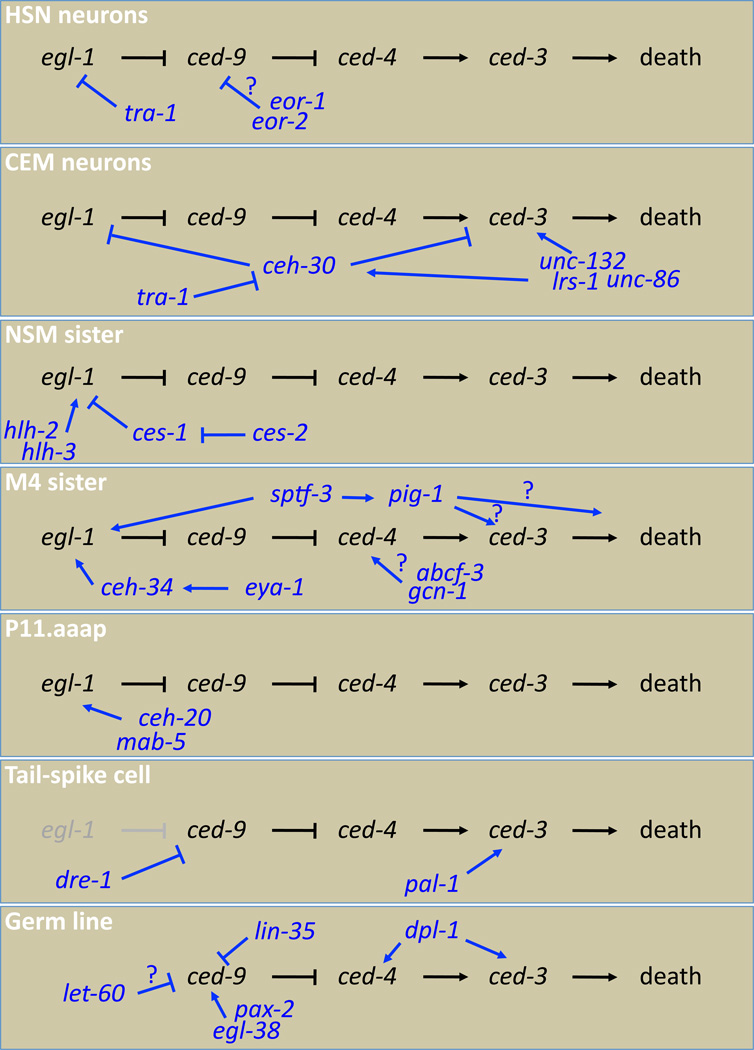
Apoptotic cell death control in C. elegans. Core cell death components (black) interact with a variety of regulators (blue) in different cells (white). Only cells for which direct control of the core apoptotic pathway is established are shown.
CED-3/caspase
The most downstream core component of the apoptotic cell death pathway is the protein CED-3, encoded by the ced-3 gene. The role of ced-3 in cell death was initially revealed from genetic studies. Animals mutant for the gene ced-1 (see below) accumulate unengulfed cell corpses during development that are easily detectable using DIC microscopy. A suppressor screen for animals lacking these corpses identified the recessive mutant ced-3(n717), in which most cells fated to die fail to do so. Characterization of the mutant revealed widespread inhibition of cell death, resulting in animals with extra cells (Ellis, & Horvitz, 1986). While lineage studies suggest that few, if any, of these extra cells divide (Ellis, & Horvitz, 1986; Hoeppner, Hengartner, & Schnabel, 2001), some are able to differentiate (Shaham, & Bargmann, 2002), incorporate into neural circuits (White, Southgate, & Thomson, 1991), and even substitute for their surviving sister cells, if those are experimentally ablated (Avery, & Horvitz, 1987). ced-3 mutant animals are alive, suggesting that at least under laboratory conditions cell death is not essential (Ellis, & Horvitz, 1986). However, some ced-3 animals exhibit defects in chemotaxis to attractive odors, and some exhibit pronounced developmental delay (Ellis, Yuan, & Horvitz, 1991), suggesting that in the wild, cell death likely confers a survival advantage.
The CED-3 protein is a founding member, together with mammalian caspase-1, of the caspase family of proteases (Yuan et al., 1993). Active CED-3 is derived from a precursor that is cleaved to generate three fragments. The N-terminal fragment, which has sequence homology to caspase recruitment domains (CARD), is not required for protease activity, while the middle and C-terminal fragments associate in pairs to form a tetramer, which is the active protease (Huang et al., 2013; Thornberry et al., 1992). Protease function is effected by an active-site cysteine residue that cleaves target proteins after the amino-acid aspartate. Aspartate residues also define the junctions between subunits in the precursor form, and biochemical studies have revealed a role for cross-catalysis in protease activation (Qi et al., 2010; Thornberry et al., 1992). Supporting this notion, the ability of the CED-3 precursor to trigger cell death in vivo is apparently positively correlated with CED-3 protein concentration (Shaham, & Horvitz, 1996b). Expression of middle and C-terminal fragments from separate mRNAs does not generally yield active protease, suggesting that cross-catalysis yields concerted structural changes that form the active moiety. However, for CED-3, and caspase-1, tagging each subunit with an N-terminal leucine zipper domain does result in formation of active protease (Chelur, & Chalfie, 2007), presumably because the leucine zipper facilitates co-translational interactions that promote folding into the active structure. As with caspase-3, a key mammalian effector caspase required for cell elimination in many natural and disease settings, CED-3 shows a preference for cleavage after the sequence DEVD (Xue, Shaham, & Horvitz, 1996).
Insertion of the gene encoding green fluorescent protein (GFP) into a genomic fragment containing the ced-3 locus reveals widespread expression in early embryos prior to the onset of most cell death events (Maurer, Chiorazzi, & Shaham, 2007). Expression then recedes and disappears in most cells, but turns on in the tail-spike cell (see below), which undergoes apoptosis just prior to hatching. Expression in larvae is generally undetectable, even though ced-3-dependent cell death takes place in early larval stages. Just before molting into the adult stage, GFP expression can be detected in the male tail (Maurer, Chiorazzi, & Shaham, 2007). As in the embryo, GFP here is expressed in many cells not fated to die. With few exceptions, therefore, expression of GFP is not detected in cells that die. This observation seemingly contradicts results that suggest a cell-autonomous role for CED-3 in cell death execution. Mosaic analysis demonstrates that ced-3 expression is required in lineages of dying cells (Yuan, & Horvitz, 1990), and ectopic expression studies suggest that only ced-3-expressing cells, but not their neighbors, can be induced to die (Shaham, & Horvitz, 1996b). While it is possible that the genomic fragment used to drive GFP expression in the studies of Maurer et al. is missing enhancer elements, and therefore generates expression below the threshold of detection, this seems unlikely as the fragment confers rescue in the absence of GFP, and GFP can be detected at high levels in many cells (Maurer, Chiorazzi, & Shaham, 2007). This puzzling observation may, therefore, suggest that turnover of GFP and the CED-3 precursor protein differs markedly, with CED-3 having a much longer half-life. If this is the case, then CED-3 protein expressed in early embryos must be maintained in most or all cells through early larval development.
While caspase enzymology and activity have been studied in C. elegans and many other organisms, the mechanism by which these proteases bring upon cellular demise is an enduring mystery. Are there many or few relevant targets? Are there many or few cellular processes that must be blocked or activated to ensure cell death? While a plethora of vertebrate caspase substrates have been described (reviewed in Poreba, Strozyk, Salvesen, & Drag, 2013), their relevance to cell death is generally not understood. One exception is the ICAD DNase inhibitory subunit, whose inactivation by caspase-mediated cleavage leads to DNA degradation in mammalian cells (Enari et al., 1998; Liu et al., 1998). Proteins related to ICAD or CAD, the active nuclease, are not encoded by the C. elegans genome. An RNAi screen for increased TUNEL staining isolated a number of nucleases (designated crn-1 to crn-6) that may be involved in DNA degradation following death. While RNAi against each gene weakly affects DNA end accumulation and cell death kinetics, no surviving cells are seen, suggesting that these nucleases are not essential for cell death (Parrish & Xue, 2003). The DNAse II homolog, NUC-1, however, does play a key role in C. elegans DNA degradation during apoptosis (Hevelone, & Hartman, 1988; Sulston, 1976; Wu, Stanfield, & Horvitz, 2000). C. elegans apoptotic DNA is cleaved into 10 bp fragments (Aruscavage, Hellwig, & Bass, 2010), and nuc-1 mutants lack these fragments and accumulate larger DNA in dying cells, as well as undegraded bacterial DNA in the gut (Aruscavage, Hellwig, & Bass, 2010; Sulston, 1976). nuc-1 mutant embryos stained using the TUNEL method (Gavrieli, Sherman, & Ben-Sasson, 1992) display an accumulation of DNA ends (Wu, Stanfield, & Horvitz, 2000), suggesting that NUC-1 degrades TUNEL-positive intermediates produced by another nuclease. One candidate may be the microRNA processing enzyme Dicer, which can be cleaved by CED-3 and may, surprisingly, itself promote DNA cleavage (Nakagawa et al., 2010). It is therefore possible that a DNA degradation system analogous to that in mammals is targeted by caspases in C. elegans.
Mutations in C. elegans ced-8, encoding an Xk-family protein, result in transient accumulation of cell corpses during embryogenesis (Ellis, Jacobson, & Horvitz, 1991; Stanfield, & Horvitz, 2000). Two recent studies suggest that CED-8 protein is also a caspase target and that its cleavage allows exposure of phosphatidyl serine (PS) on the surface of dying cells (Chen et al., 2013; Suzuki et al., 2013). PS then acts as one of multiple eat-me signals that promote the engulfment of dying cells by their neighbors (Venegas, & Zhou, 2007). A similar set of events takes place in mammalian cells, where the CED-8 related protein Xkr8 is a target of caspase cleavage (Suzuki et al., 2013).
A recent study also suggests that the fidelity of some cell death in C. elegans is enhanced by CED-3 cleavage of the GTPase activating protein CNT-1 (Nakagawa, Sullivan, & Xue, 2014). Cleaved CNT-1 can translocate to the cell membrane where it may control apoptotic signaling. Although transient effects on cell death are observed in the relevant mutants, nearly all cells fated to die eventually succumb. Thus, it remains unclear whether caspase cleavage of CNT-1 is an integral aspect of cell death execution.
C. elegans CED-11 may also function downstream of CED-3 caspase. Defects in this protein, a TRPM-related channel, do not block cell death, but result in a profound change in dying cell appearance under DIC optics (Strange, 2003). Instead of acquiring a refractile appearance, cell corpses appear swollen and vacuolated, a defect that is perhaps not unexpected from a membrane channel mutant. Whether CED-11 is activated by CED-3 cleavage is not known.
In addition to CED-3, the C. elegans genome encodes three other caspase-related proteins: CSP-1, −2, and −3 (Shaham, 1998). The csp-1 locus generates at least two functional transcripts, one encoding a zymogen with a long N-terminal domain (CSP-1A), and one with a short N-terminal region (CSP-1B). In vitro, CSP-1B can cleave itself and CED-3, presumably resulting in active enzymes. However, substrate specificities of CED-3 and CSP-1B appear to be different, as CED-3 cannot cleave the CSP-1B precursor. Furthermore, CED-3 can cleave the fluorescent substrate Ac-DEVD-AFC, but not Z-YVAD-AFC, and CSP-1B has the opposite substrate profile (Shaham, 1998). CSP-1 has been suggested to promote cell death during embryonic development (Denning, Hatch, & Horvitz, 2013). However, the effects of csp-1 mutants are subtle, raising the possibility that CSP-1 may not play a key cell death role.
The csp-2 locus also generates at least two transcripts, csp-2A, encoding a protein with a large N-terminal domain, and csp-2B, encoding a short domain. While the active-site cysteine of this protease is conserved, surrounding sequences are not, suggesting that this protease may be inactive. Indeed, in vitro activity of the protease could not be demonstrated (Shaham, 1998). While caspase inhibitor proteins are known in mammals and Drosophila (e.g. IAP proteins, (Goyal et al., 2000; Hay, Wassarman, & Rubin, 1995; Silke, & Vucic, 2014)), C. elegans proteins related to these appear not to have any role in cell death (Speliotes, Uren, Vaux, & Horvitz, 2000). CSP-2 has been suggested to inhibit CED-3 activity in the germ line (Geng et al., 2009), perhaps functioning analogously to IAP proteins.
The csp-3 gene encodes only a C-terminal caspase domain, and apparently arose from a partial duplication of the csp-2 locus (Shaham, 1998). CSP-3 does not contain an active site, suggesting it could act as a dominant-negative caspase inhibitor. It has been suggested that CSP-3 can inhibit CED-3 (Geng et al., 2008); however, this observation has been contested (Denning, Hatch, & Horvitz, 2013).
In contrast to mammals, therefore, C. elegans seems to employ only a single major caspase, CED-3, to control cell death, and while other caspase-related genes are expressed in this animal, they likely serve other functions. The csp-1, -2, and -3 caspase genes are not well conserved among nematode species, suggesting that whatever roles they have are likely to be tailored specifically to the biology of C. elegans. In this context, it is intriguing that the strongest ced-3 mutations block nearly all cell death in C. elegans, and most also have effects as heterozygotes. However, a deletion eliminating the protease domain fails to block nearly 20% of cell death events that occur in the animals pharynx (Shaham, Reddien, Davies, & Horvitz, 1999). This surprising observation suggests that CED-3 caspase activity may not account for all cell death in the animal. An alternative effector mechanism may be found through analysis of the strongest ced-3 alleles, which appear to interfere both with CED-3 protease activity and this hypothetical alternative effector pathway (Shaham, Reddien, Davies, & Horvitz, 1999).
CED-4
Recessive mutations in the ced-4 gene, like those in ced-3, promote the survival of somatic and germ cells slated to die during C. elegans development, suggesting a key role for CED-4 in promoting cell death (Ellis, & Horvitz, 1986). CED-4 protein is similar to the mammalian cell death gene Apaf-1, and is composed of four structural domains: an N-terminal CARD domain, a nucleotide-binding domain found in NTPases, a helical domain, and a C-terminal winged-helix domain (Yuan, & Horvitz, 1992; Zou et al., 1997, Qi et al., 2010). Structural studies suggest that CED-4 monomers can form an octameric complex with 4-fold symmetry (Qi et al., 2010) in which CARD domains seem to mediate much of the contact. This is in contrast to the proposed hepatmer formed by Apaf-1 (Acehan et al., 2002). Although explanations based on structural data for the different stoichiometries have been put forward (Qi et al., 2010), the difference remains an intriguing mystery. The nucleotide-binding domain of CED-4, together with its overall structure, bear resemblance to AAA+ ATPases (Qi et al., 2010), although significant deviations in multimer interfaces are observed compared with bona fide members of this family. Nonetheless, it is intriguing and perhaps functionally relevant that the C. elegans protein MAC-1, an AAA+ family ATPase, can bind CED-4 (as well as Apaf-1), and that overexpression of MAC-1 can prevent some natural cell deaths in the animal (Wu et al., 1999). The CED-4 nucleotide-binding domain binds ATP-Mg2+ more strongly than dATP, in contrast to Apaf-1 (Li et al., 1997; Seiffert, Vier, & Hacker, 2002). Whether ATP binding per se, or exchange of ADP for ATP is required for CED-4 function is still not clear.
Expression studies of CED-4, like those of CED-3, pose a conundrum. Polyclonal antibodies against CED-4 detect the protein in most cells in the early embryo, well before any cell death takes place (Chen et al., 2000). Staining gradually recedes and is absent from larvae, even though ced-4 is required for the handful of early larval cell death events that occur in C. elegans (Sulston, & Horvitz, 1977). CED-4 expression is also broadly detected in the germ line (Pourkarimi, Greiss, & Gartner, 2012). As with CED-3, several studies suggest that CED-4 functions within dying cells to promote their demise, and is also required for postembryonic induction of cell death in cells that usually survive (Shaham, & Horvitz, 1996b; Yuan, & Horvitz, 1990). Why CED-4 is broadly expressed in the embryo but not detectable in larvae is not at all clear. As with CED-3, technical concerns may resolve this paradox.
Staining of early embryos with a polyclonal CED-4 antibody suggests that CED-4 is associated with mitochondria (Chen et al., 2000). However, a different antibody detects perinuclear staining that does not overlap with mitochondria in later embryos and in the germline (Pourkarimi, Greiss, & Gartner, 2012). Consistent with the latter localization pattern, RNA interference directed against the inner nuclear membrane protein SUN-1 appears to block CED-4 accumulation at the nuclear periphery. Furthermore, CED-4 and SUN-1 proteins purified from bacteria can physically interact (Tzur, Margalit, Melamed-Book, & Gruenbaum, 2006). Whether CED-4 and SUN-1 bind in vivo is unclear, as they are normally segregated to different cellular compartments (cytoplasm vs. inner nuclear membrane). The antibody staining studies indicate a shift in CED-4 localization during development from mitochondria to perinuclear regions. However, it is also possible that the antibodies used are detecting different subpopulations of CED-4, or that technical issues, such as sample preparation, differ between experiments, providing only incomplete views of CED-4 localization.
The ced-4 genomic locus encodes at least two alternative transcripts, ced-4L and ced-4S, that differ by inclusion of an alternate splice site acceptor upstream of exon 4 of the gene, resulting in a 24 amino-acid insertion in CED-4L (Shaham, & Horvitz, 1996a). Overexpression of CED-4L potently inhibits cell death, suggesting that this protein may normally function as a dominant interfering protein, consistent with the predicted multimeric structure of CED-4S. A number of observations suggest that alternative splicing of ced-4 may have physiological importance. The alternative splice site and the sequence of the additional exon segment it introduces are highly conserved among nematodes, indicating functional significance. A mutation, ced-4(n2273), which removes the ced-4S-specific splice-acceptor site, appears to perturb not only the killer, but also a protective function encoded by the locus (Shaham, & Horvitz, 1996a). Finally, weak mutations in ced-4, which only partially block cell death, are suppressed by mutations in the spk-1 gene, which encodes an SR protein kinase (Galvin, Denning, & Horvitz, 2011). This family of kinases phosphorylates SR proteins that mediate splicing. Nonetheless, northern blot and PCR studies demonstrate that the inhibitory ced-4L transcript is expressed at one-tenth the level of ced-4S in whole animal assays (Shaham, & Horvitz, 1996a). It is therefore possible that ced-4S is expressed at high levels only in a subset of cells. The isoform-specific expression patterns of CED-4L and CED-4S are not known.
CED-4/Apaf-1-related proteins have been described in many animal species, but not outside the animal kingdom, suggesting the possibility that CED-3 and CED-4 evolved together. Nonetheless, loss of C. elegans ICD-1, a protein similar to the beta-subunit of the nascent polypeptide-associated complex, results in wide-spread cell death that appears, at least in part, to depend on CED-4, but not on CED-3 (Bloss, Witze, & Rothman, 2003). Also, mutations in the gene pvl-5, whose identity is still unknown, result in cell death that is inhibited by ced-3 mutations but not by ced-4 alleles (Joshi, & Eisenmann, 2004). Thus, under at least some circumstances, ced-3 and ced-4 may operate independently.
CED-9
Unlike ced-3 and ced-4, the ced-9 gene encodes a negative regulator of cell death. Increased CED-9 function, either through overexpression or by a mutation in a conserved domain, promotes survival of cells that normally die during C. elegans development (Hengartner, Ellis, & Horvitz, 1992; Hengartner, & Horvitz, 1994b). Loss-of-function mutations in ced-9 are generally lethal. Some alleles result in inappropriate and widespread death of embryonic cells not fated to die. However, strong loss-of-function mutations result in arrest very early in embryonic development with no evidence of ectopic cell death (Hengartner, Ellis, & Horvitz, 1992). While the phenotypic discrepancy between these allele classes is not understood, it may suggest that CED-9 has a vital cellular role independent of its role in cell death. Yeast two-hybrid studies identified the centrosome maturation and mitotic spindle assembly factor SPD-5 as a CED-9 binding protein, an interaction also validated by co-affinity purification from transfected human HEK293T cells (Dreze et al., 2009). spd-5 is required for early embryonic events, a spd-5 loss of function is also embryonic lethal. Thus, CED-9 may contribute to the essential functions of this gene. The ced-9 mRNA is generated from a longer RNA encoded by a splicing operon that also expresses the gene mev-1, encoding the mitochondrial succinate dehydrogenase b560 subunit (Ishii et al., 1998). A functional relationship between CED-9 and MEV-1 has not been established. However, RNAi against mev-1 produces embryonic lethality (Gonczy et al., 2000) and accumulation of cell corpses in the germ line (Sendoel et al., 2010), raising the untested possibility that strong loss-of-function lesions (such as early stop codon insertions) in ced-9 may interfere with mev-1 expression or mRNA processing, possibly through the nonsense mediated mRNA decay pathway.
CED-9 protein is similar in sequence to mammalian BCL2 and related proteins (Hengartner, & Horvitz, 1994b). The protein consists of seven alpha helices. Helix 1 corresponds to sequences that comprise BCL2 homology region 4 (BH4) in other BCL2 family members, but is not conserved in the primary sequence of CED-9. Helix 3 comprises the BH3 domain, helices 4 and 5 make up the BH1 domain, and helices 6 and 7 overlap the BH2 domain (Woo et al., 2003; Yan et al., 2004). Antisera against CED-9 reveal that the protein is localized to mitochondria in embryos and in the germ line (Chen et al., 2000; Pourkarimi, Greiss, & Gartner, 2012). It has been suggested that the C-terminal region of CED-9, which contains a transmembrane region like other BCL2 family members, contributes to this localization. Indeed, expression in C. elegans muscle cells of GFP fused to the transmembrane domain confers mitochondrial localization, and a GFP-CED-9 fusion protein lacking the transmembrane domain fails to localize to mitochondria (Tan, Fire, & Hill, 2007). However, ced-9 mutants can still be rescued by expression of cytosolic CED-9(ΔTM), or by expression of an ER-targeted CED-9 (Tan, Fire, & Hill, 2007). Thus, the transmembrane region appears not essential for function, but may facilitate mitochondrial localization.
BCL2 family members have been suggested to form membrane pores, and in vitro experiments demonstrate that they can assemble into channels of variable conductivity (Antonsson et al., 1997; Basanez et al., 2002). CED-9 can also associate with lipid membranes in vitro, inducing changes in membrane permeability, and this association appears independent of the C-terminal transmembrane domain (Tan, Zuckerman, Wells, & Hill, 2011). Whether pore formation is required for the in vivo function of CED-9, or any BCL-2 family member, is an important outstanding question.
Like CED-3 and CED-4, CED-9 is expressed ubiquitously early in the embryo; however, anti-CED-9 antibodies do not detect the protein in late embryos or in early larvae (Chen et al., 2000; Pourkarimi, Greiss, & Gartner, 2012). Nonetheless, a number of studies suggest that CED-9 acts cell autonomously in larvae to protect cells from cell death (Hengartner, Ellis, & Horvitz, 1992; Hengartner, & Horvitz, 1994b). Loss of ced-9 leads to abnormalities in cell death associated with male tail development in L4 larvae (Hengartner, Ellis, & Horvitz, 1992), and facilitates ectopic larval cell death induced by CED-3 overexpression (Shaham, & Horvitz, 1996b). Thus, even though protein expression has not been confirmed, CED-9 activity is detected postembryonically in both dying and living cells.
Although the death-inhibitory role of CED-9 has been extensively explored, genetic evidence suggests that the ced-9 gene also encodes a death-promoting activity. Overexpression of CED-9 can suppress inappropriate cell survival in animals carrying the ced-9(1950) gain-of-function mutation, and fewer cells survive inappropriately in ced-9(n1950) heterozygotes balanced by a wild-type copy of ced-9 compared with animals balanced with a deletion spanning the ced-9 locus. Also, double mutants harboring a weak ced-3/caspase mutation and a ced-9 loss-of-function lesion exhibit more ectopic cell survival than weak ced-3 mutants alone (Hengartner, & Horvitz, 1994a). The physical nature of this death-promoting activity is unknown, and could be due to a protein arising from alternative splicing or transcription/translation initiation, or a protein modification that competes with the activity of unmodified CED-9.
Finally, while CED-9, and perhaps CED-4L, are the best characterized cell death inhibitors in C. elegans, others may exist. DAD-1 is a core component of the multisubunit oligosaccharyltransferase that attaches mannose oligosaccharides to asparagine residues on proteins traveling through the endoplasmic reticulum (ER). Overexpression of DAD-1 can prevent normally-occurring cell death in C. elegans (Sugimoto et al., 1995), and its loss is embryonic lethal (Fraser et al., 2000). Whether DAD-1 is an integral component of the cell death machinery, or mediates modification of core cell death proteins, remains to be determined.
EGL-1
Genetic screens seeking mutants regulating vulval development in C. elegans identified a host of genes required for egg-laying, including the gene egl-1 (Trent, Tsung, & Horvitz, 1983). Dominant gain-of function mutations in egl-1 result in the inappropriate death of the two HSN neurons that regulate hermaphrodite egg-laying (Desai, Garriga, McIntire, & Horvitz, 1988). That these neurons normally die during male development suggested initially that egl-1 might be involved in sex determination; however, the mutant revealed no pleiotropies that might indicate sexual transformation (Desai, Garriga, McIntire, & Horvitz, 1988; Trent, Tsung, & Horvitz, 1983). While egl-1(gf)/egl-1(gf) mutants are 100% egg laying defective, about 30% of animals heterozygous for an egl-1(gf) allele and a deletion spanning the locus lay eggs normally (Conradt, & Horvitz, 1998). Using this observation, homozygous egl-1(gf) animals were mutagenized, and a loss-of-function mutation in the gene was obtained by screening for animals with normal egg laying. Remarkably, animals carrying such egl-1(lf) mutations are unable to execute the majority of developmental cell death in the animal, and contain many extra cells (Conradt, & Horvitz, 1998).
The egl-1 gene was cloned using complementation rescue and mapping of the loss-of-function allele of egl-1 (Conradt, & Horvitz, 1998). The predicted EGL-1 protein is only 106 amino acids long, and is homologous to the BH3 domain common to many mammalian cell death regulators (Kelekar, & Thompson, 1998). By sequence, EGL-1 is most similar to the BH3-only class of cell death regulators that promote cell death in a variety of settings. This similarity is also evident in structural studies of the protein, which has been shown to adopt an amphipathic alpha helix conformation (Yan et al., 2004).
EGL-1 expression differs markedly from that of the other core apoptotic regulators in C. elegans. During development, egl-1 expression is upregulated in cells that die (Conradt, & Horvitz, 1999; Thellmann, Hatzold, & Conradt, 2003), although another study suggests that the gene may also be functional elsewhere (Jagasia, Grote, Westermann, & Conradt, 2005). Furthermore, while CED-9, CED-4, and CED-3 all control germ cell death, EGL-1 appears to be dispensable for this stochastic form of cell death (Gumienny et al., 1999), perhaps indicating that EGL-1 can act only to guarantee deterministic activation of the apoptotic program. Consistent with its expression pattern, egl-1 gene regulatory sequences are extensive, and are found both upstream and downstream of the gene. The egl-1(gf) lesion that leads to the specific demise of the HSN neurons is a point mutation in a regulatory site 5.6 kb downstream of the egl-1 transcription unit, which prevents the binding of the inhibitory sex-determination factor TRA-1A (Conradt, & Horvitz, 1999). In the absence of TRA-1A binding, egl-1 is now expressed ectopically only in HSNs, leading to their removal.
Besides EGL-1, two other BH3-only proteins are made by C. elegans. DCT-1 is similar to mammalian BNIP3, and can cooperate with CED-3 caspase to induce cell death in cultured mammalian cells (Cizeau et al., 2000). However, a role in C. elegans cell death has not been established. CED-13, a protein more similar to EGL-1, is upregulated in the C. elegans germ line in response to radiation, and may participate in radiation-induced cell death, but does not appear to play a significant role in developmental cell death either in the soma or the germ line (Schumacher et al., 2005). Thus, unlike mammals, in which a host of BH3-only proteins controls cell death in a myriad of contexts and conditions, C. elegans appears to have a single member of this family dedicated to somatic developmental cell death.
The core apoptotic pathway of C. elegans
The current model for apoptotic cell death execution in C. elegans is shown in Fig. 2. The apoptotic cascade begins when EGL-1 inhibits the activity of CED-9. CED-9 inhibition of CED-4 is then relieved, allowing CED-4 to activate CED-3, promoting cell death. The model is supported by genetic, biochemical, and structural data, and predicts similarities and differences with pathway interactions described in mammals.
CED-4->CED-3
Genetic studies support the notion that CED-3 functions downstream of CED-4. Overexpression of CED-3 in neurons not fated to die can result in their demise, and this killing is reduced, but not eliminated, in animals harboring a ced-4(lf) mutation. Ectopic expression of CED-4 in the same neurons can also promote their death, however, this is nearly entirely abrogated in strong ced-3 mutants (Shaham, & Horvitz, 1996b). A parsimonious explanation for these genetic studies is that CED-4 facilitates the activity of CED-3. While direct binding of the two proteins in vivo has not yet been demonstrated, binding studies in 293T cells show that CED-4 and CED-3 can physically interact (Chinnaiyan et al., 1997; Chinnaiyan, ORourke, Lane, & Dixit, 1997). This interaction seems to occur between the CARD domain in CED-4 and the N-terminal domain of CED-3 (Irmler, Hofmann, Vaux, & Tschopp, 1997), although binding could also be achieved in the absence of the CED-3 N-terminal domain through the L2 loop of its C-terminal subunit (Huang et al., 2013).
Steady state binding of CED-4 to CED-3 is correlated with activation of the procaspase. For example, in insect cells, co-expression of CED-4 and CED-3 leads to increased caspase activity dependent on the CED-3 N-terminal domain and the ATP binding domain of CED-4 (Seshagiri, Chang, & Miller, 1998; Seshagiri, & Miller, 1997). Conversely, a CED-4 mutation disrupting a hydrophobic binding surface between CED-4 and CED-3 reduces the ability of CED-4 to stimulate CED-3 activity (Huang et al., 2013). Mutations blocking CED-4 oligomerization appear to block processing of CED-3 in cultured cells, suggesting that oligomerization is likely important for activity (Yang, Chang, & Baltimore, 1998). CED-4 has also been suggested to promote the activity of the mature CED-3 caspase in vitro (Qi et al., 2010). However, the stoichiometry of CED-4:CED-3 here is 8:2 and not 8:8 as perhaps expected, raising the possibility that this complex may not be the only source of in vivo activity.
CED-9-|CED-4
The lethality observed in ced-9(lf) mutants is suppressed by loss-of-function mutations in either ced-3 or ced-4 (Hengartner, Ellis, & Horvitz, 1992), and cell death induced by CED-4 ectopic expression is reduced by CED-9 overexpression (Shaham, & Horvitz, 1996b). These observations, and the discovery that ced-9(gf) mutations block cell death, suggest that CED-4 and CED-3 function downstream of CED-9, and that CED-4 may be a direct target of CED-9. This hypothesis is bolstered by the demonstration that CED-9 and CED-4 physically interact (Spector, Desnoyers, Hoeppner, & Hengartner, 1997; Wu, Wallen, Inohara, & Nunez, 1997), and that mutations reducing CED-9 function in vivo also reduce CED-9-CED-4 binding (Spector, Desnoyers, Hoeppner, & Hengartner, 1997). Expression studies in the early embryo of wild-type and mutant C. elegans further corroborate this idea. While CED-4 protein, detected using anti-CED-4 sera, appears mitochondria-bound in wild-type animals, it is mainly localized to the nuclear periphery in ced-9(lf); ced-3(lf) double mutants in which the ced-3 mutation is used to keep ced-9(lf) animals alive (Chen et al., 2000). Thus, CED-9 appears to serve as a mitochondrial anchor for CED-4.
How CED-9 inhibits CED-4 from forming an active octamer is not understood; however, X-ray structures of CED-9 bound to CED-4 reveal a complex in which one CED-9 protein is bound to an asymmetric CED-4 dimer (Yan et al., 2005). Thus, it is possible that one mechanism by which CED-9 inhibits CED-4 activity is by preventing its oligomerization. While the contacts observed in this study are specific, they do conflict with solution studies suggesting that bacterially purified CED-9 and CED-4 form a 2:2 complex (Fairlie et al., 2006; Yan et al., 2005). Resolution of these differences is not yet at hand.
In vitro assays demonstrate that CED-9 protein can serve as a substrate for CED-3 caspase (Xue, & Horvitz, 1997), raising the possibility that CED-9 might also block cell death by competitively inhibiting CED-3 caspase activity. While global overexpression of CED-9 yields broad cell survival, mutations of the CED-9 aspartate residues targeting cleavage by CED-3 reduce the ability of CED-9 to inhibit death, consistent with this model. However, such lesions could also affect protein conformation and stability. It therefore remains unclear whether CED-9 cleavage by CED-3 has physiological significance.
EGL-1-|CED-9
Genetic studies demonstrate that EGL-1 functions upstream of both CED-4 and CED-3. Specifically, ectopic expression of EGL-1 in neurons that normally live can induce their death. This killing activity is entirely blocked in animals lacking either CED-3 or CED-4 function (Conradt, & Horvitz, 1998). Likewise, inappropriate death of the HSN neurons in animals carrying egl-1(gf) mutations is suppressed by loss-of-function mutations in ced-3 or ced-4 (Ellis, & Horvitz, 1986).
EGL-1 also appears to function upstream of CED-9. egl-1 (lf) mutations, which normally block programmed cell death, fail to do so in ced-9(lf); ced-3(weak lf) mutants that entirely lack CED-9 activity, suggesting that EGL-1 normally inhibits CED-9 activity (Conradt, & Horvitz, 1998). CED-9 tagged with Glutathione-S-Transferase (GST) can bind to in vitro translated EGL-1 (Conradt, & Horvitz, 1998), and CED-9 and EGL-1 can be co-crystallized to form a bound complex (Yan et al., 2004), raising the possibility that EGL-1 inhibition of CED-9 might occur through direct physical contact. The EGL-1-CED-9 crystal structure reveals that binding of EGL-1 results in large conformational changes to CED-9, introducing steric hindrance and misalignment of key residues involved in CED-9-CED-4 binding. Thus, EGL-1 likely acts by preventing the association of CED-9 with CED-4.
Consistent with this idea, overexpression of EGL-1 in early embryos leads to movement of CED-4 from mitochondria to the nuclear periphery as occurs in animals lacking CED-9 (Chen et al., 2000). Titration studies with purified proteins also demonstrate dissolution of the CED-4-CED-9 complex upon addition of EGL-1 (Yan et al., 2004) but not mammalian BH3-domain proteins (Fairlie et al., 2006), as do co-immunoprecipitation studies of the proteins from cultured mammalian cells (del Peso, Gonzalez, & Nunez, 1998). Further support for this model is provided by the CED-9 gain-of-function G169E mutation that results, in vivo, in cell survival (Hengartner, & Horvitz, 1994a). This mutation reduces the binding affinity of CED-9 for EGL-1 (del Peso et al., 2000; Parrish, Metters, Chen, & Xue, 2000), and introduces a bulky amino acid into the binding pocket for EGL-1, leaving the CED-4 binding interface intact. Thus, blocking EGL-1 binding can allow CED-9 to remain bound to CED-4.
The components of the core apoptosis pathway and their signs of interaction appear conserved from C. elegans to humans. Nonetheless, even within the core pathway, distinctions are obvious. In mammals, though the BCL2-related protein Diva/Boo can bind Apaf-1 (Inohara et al., 1998; Song, Kuang, Dixit, & Vincenz, 1999), this seems to be the exception. Most BCL2 family proteins appear to control Apaf-1 activity by regulating the release of cytochrome C, an Apaf-1 activating factor, from mitochondria. To date, no evidence implicating cytochrome C in C. elegans apoptosis has been revealed. Indeed, the cytochrome C binding domain in Apaf-1 appears not to be present in CED-4 (Li et al., 1997). Unlike CED-4, which is bound to mitochondria at least at some stages of C. elegans development, Apaf-1 is localized to the cytoplasm in a variety of cell types (Hausmann et al., 2000), consistent with a different mode of activation. Why the mammalian and C. elegans pathways appear to use similar proteins in different ways is not at all understood.
Whether mitochondria in general play a role in C. elegans cell death is also unclear. Some mitochondrial factors have been examined, including endonuclease G (CPS-6)(Parrish et al., 2001), Apoptosis Inducing Factor (WAH-1)(Wang et al., 2002), and the mitochondrial fission and fusion proteins DRP-1 and FZO-1 (Breckenridge et al., 2008; Jagasia, Grote, Westermann, & Conradt, 2005; Lu, Rolland, & Conradt, 2011). While animals carrying lesions in the genes encoding these factors have been reported in some cases to have cell death defects, the effects are rather weak. It is therefore unclear whether any of these proteins specifically interact with the cell death machinery or if the effects are due to more generic changes in cell state. Mutations in core apoptotic components do not obviously affect mitochondrial shape, fission, or fusion in homozygous early embryos (Breckenridge, Kang, & Xue, 2009). Perhaps more relevant, however, in dying cells, mitochondrial fragmentation is observed, is dependent on egl-1, and is blocked by a ced-9(gf) allele (Jagasia, Grote, Westermann, & Conradt, 2005). Importantly, fission is independent of ced-3 and ced-4. Thus, while mitochondrial fragmentation is not required for cell death, it does accompany the process.
Regulating apoptosis
Control of egl-1 expression
The induction of egl-1 expression in dying cells, and the relatively large and complex regulatory region required for this expression, suggests that the decision to live or die may be mediated by cell-specific transcription factors that bind to the egl-1 locus to promote and/or inhibit its transcription. As described above, this indeed appears to be the case in HSN neurons, where the sex-determination factor TRA-1A binds in the environs of egl-1 to block transcription in hermaphrodites (Conradt, & Horvitz, 1999) (Fig. 2). The CEM neurons are also sexually dimorphic cells. In this case, the cells survive in males but die in hermaphrodites (Sulston, Schierenberg, White, & Thomson, 1983), and sex-specific survival here is also mediated in part by control of egl-1 expression. The CEH-30 homeodomain transcription factor inhibits CEM death in males by acting with UNC-37/Groucho to block expression of egl-1 in these cells. In hermaphrodites, the TRA-1 protein promotes CEM neuron death by inhibiting transcription of the ceh-30 gene, thus allowing egl-1 expression (Nehme et al., 2010; Peden et al., 2007; Schwartz, & Horvitz, 2007)(Fig. 2).
Recessive mutations in the gene ces-2, encoding a bZIP transcription factor, and a dominant allele of the gene ces-1, encoding a Snail-like bHLH transcription factor, block the deaths of the NSM sister, and NSM sister and I2 sister cells, respectively (Ellis, & Horvitz, 1991; Metzstein et al., 1996; Metzstein, & Horvitz, 1999). In the NSM sisters, survival correlates with lack of induction of a Pegl-1::GFP reporter in the cells (Thellmann, Hatzold, & Conradt, 2003). CES-1 binds in vitro to Snail elements in an egl-1 genomic fragment that is required in vivo to mediate CES-1 activity. These elements overlap with E box sequences that can bind the HLH-2 and HLH-3 transcription factors, which promote NSM death and also promote egl-1 expression (Thellmann, Hatzold, & Conradt, 2003). Thus, CES-1 apparently inhibits egl-1 expression by blocking access of HLH-2/3 to the egl-1 promoter (Fig. 2).
Mutations in the ceh-34 and eya-1 genes, both encoding homeodomain transcription factors, block the deaths of the M4 and I3 sister cells (Fig. 2). CEH-34 and EYA-1 proteins physically interact, and CEH-34 binds to a regulatory site 5 of the egl-1 transcription unit. In both mutants, egl-1 expression in the M4 sister cell is abrogated (Hirose, Galvin, & Horvitz, 2010). The Sp1 transcription factor SPTF-3 is also required for expression of egl-1 in and death of the M4 sister cell (Hirose, & Horvitz, 2013). In a similar mode of regulation, a complex consisting of the CEH-20/Pbx1 and MAB-5/Hox transcription factors controls the death of the postembryonic cell P11.aaap (Fig. 2). In this case, a site approximately 6 kb downstream of the egl-1 start codon binds the CEH-20-MAB-5 complex, and mutations in that site or in its cognate binding factors eliminate egl-1 expression in the cell (Liu, Strauss, Potts, & Cameron, 2006).
Regulation of egl-1 expression may also govern the death of the embryonic cells ABprpppapp and ABplpppapp, whose demise appears to be weakly dependent on EGF signaling (Fig. 2). At least one of these cells survives inappropriately in about 30% of animals deficient in either the EGF ligand LIN-3 or its receptor, LET-23/EGFR, whose site of action is not firmly established. Regardless, a ~50% reduction in the levels of a Pegl-1::GFP reporter is seen in lin-3(lf) mutants (Jiang,& Wu, 2014).
egl-1 is not alone
While egl-1 is a hub for cell death decision-making, more is likely going on. In animals carrying both ced-9(strong lf) and ced-3(weak lf) mutations, cells destined to live do so, and only a few cells destined to die survive inappropriately (Hengartner, & Horvitz, 1994a). In these animals, many cells fated to die appear to do so on cue. Yet, since CED-9 activity is gone, egl-1 cannot be the relevant death effector in these cells, unless it also has CED-9-independent functions. A role for egl-1 in starvation-induced autophagy has been proposed, based on the observation that animals homozygous for an egl-1 (gf) mutation accumulate the autophagy marker LGG-1::DsRED even in the presence of food (Maiuri et al., 2007). However, autophagy genes are not required for cell death in C. elegans (Takacs-Vellai et al., 2005), making this an unlikely mechanism for cell death control. However, cell death regulators that function in place of EGL-1 have been uncovered.
In HSN neurons, the EOR-1 protein, similar to the mammalian PLZF transcription factor, is required together with its novel cofactor, EOR-2, for death (Hoeppner et al., 2004; Howard, & Sundaram, 2002; Howell, Arur, Schedl, & Sundaram, 2010; Rocheleau et al., 2002). Neither gene promotes expression of egl-1. Furthermore, the effects of eor-1/2 mutations appear to require an intact ced-9 gene, suggesting that EOR-1/2 may work upstream of or in parallel to CED-9 and in parallel to EGL-1 (Fig. 2). In CEM neurons, three proteins, UNC-86, a POU homeodomain transcription factor, LRS-1, a leucyl t-RNA synthetase, and the kinase PIM-1 are required for cell death (Nehme et al., 2010; Peden et al., 2007). In hermaphrodites these genes promote expression of the ced-3 caspase gene but not egl-1. In males, they promote ceh-30 expression, and therefore indirectly inhibit egl-1 expression (Fig. 2).
In M4 sister cells, two proteins, GCN-1, which may be involved in controlling translation initiation factor levels, and the interacting protein ABCF-3, an AAA+ ATPase, appear to function together to promote cell death, and mutants in these genes have a weak survival defect. Here also, both proteins act independently of CED-9, and hence of EGL-1, to promote cell death (Hirose, & Horvitz, 2014). The mechanism of action of this protein complex is not known, but the similarity of ABCF-3 to AAA+ ATPases raises the possibility that they may interact with CED-4 (Fig. 2).
In addition to regulating egl-1 transcription in M4 sister cells, the SPTF-3/Sp1 protein also promotes expression of the gene pig-1 (Fig. 2). PIG-1, similar to MELK kinase, has been implicated in the control of a number of cell death events in C. elegans, and also appears to function in parallel to EGL-1 and CED-9. PIG-1 has been suggested to function in a ced-3-independent cell death pathway (Denning, Hatch, & Horvitz, 2012; Hirose, & Horvitz, 2013). However, the data are also consistent with a function within the canonical cell death pathway. Indeed, PIG-1 function in asymmetric cell division has been previously described (Chien, Brinkmann, Teuliere, & Garriga, 2013; Cordes, Frank, & Garriga, 2006), raising the possibility that PIG-1 may control the segregation of death effectors downstream of CED-9, such as CED-3 or CED-4. Such segregation could explain how an initially broad expression pattern of these genes leads to the death of specific cells. Supporting this notion, the polarity proteins PAR-4/LKB1 kinase, STRAD-1/STRAD, and MOP-25/MO25 appear to function together with PIG-1 in the death of the ABplpappap cell (Denning, Hatch, & Horvitz, 2012).
Parallel control of death is also seen in descendents of the Pn.p blast cells that normally die in the ventral cord of early larva and in pharyngeal cells. Here, components of the conserved retinoblastoma (Rb)/E2F pathway are required for cell death and also function independently of CED-9 and likely EGL-1 (Reddien, Andersen, Huang, & Horvitz, 2007).
egl-1 is not always required
While egl-1 control is essential for some cell death initiation, in some cases EGL-1 function is not required (Fig. 2). The tail-spike cell is a binucleate cell with a posterior process believed to function as a scaffold for developing tail epithelial cells. Five hours after it is born, the tail spike cell dies (Sulston, Schierenberg, White, & Thomson, 1983). While death requires ced-3 and ced-4, ced-9(gf) mutations do not block cell death, and egl-1(lf) animals only have a weak defect, with about 30% exhibiting tail-spike cell survival (Maurer, Chiorazzi, & Shaham, 2007), suggesting that EGL-1-CED-9 interactions are not important for the demise of this cell. In the tail-spike cell, transcription of the ced-3 caspase gene is temporally controlled. Expression of a Pced-3::GFP reporter is off until about 25 minutes before the cell rounds up and dies, suggesting that ced-3 transcription is an important regulatory node. The transcription factor PAL-1, similar to vertebrate Cdx2, is an important regulator of tail-spike cell death. PAL-1 is required for ced-3 expression, can bind to sequences upstream of the ced-3 start codon in vitro and probably in vivo, and mutations in pal-1 block tail-spike cell death (Maurer, Chiorazzi, & Shaham, 2007).
Besides PAL-1, the F-Box protein DRE-1 also regulates tail-spike cell death. dre-1 mutants have inappropriately surviving tail-spike cells, and like pal-1, dre-1 acts in parallel to egl-1. However, dre-1 requires a wild-type copy of ced-9 to carry out its function. Genetic and protein interaction studies suggest that DRE-1 physically interacts with SKR-1/SKP, which controls protein ubiquitylation, to promote cell death. DRE-1 also interacts with CED-9, which may, therefore, be its in vivo target (Chiorazzi et al., 2013).
egl-1(lf) and ced-9(gf) mutations block germ cell death in response to ionizing radiation (Gartner et al., 2000); however, as in the tail-spike cell, neither mutation affects normal physiological cell death in the germ line. Other regulators appear to take charge here (Fig. 2). Mutants in pax-2 and egl-38, encoding Pax2/5/8 family transcriptional regulators, have increased rates of germ cell death. Overexpression of these genes reduces germ line apoptosis (Park, Jia, Rajakumar, & Chamberlin, 2006). Mosaic analysis shows that egl-38 is required in the germline for germ cell survival, and RT-PCR, deletion, and chromatin immunoprecipitation studies suggest that both egl-38 and pax-2 act by activating ced-9 transcription (Park, Jia, Rajakumar, & Chamberlin, 2006). This function may also extend to embryonic cell death, as egl-38 and pax-2 single mutants also have increased cell corpses in embryos.
The Rb pathway also plays a role in developmental germ cell death control. LIN-35/Rb appears to inhibit ced-9 transcription in these cells, while the DP homolog DPL-1, and the E2F–like EFL-2 promote transcription of ced-3 and ced-4 (Schertel, & Conradt, 2007). Another tumor control pathway, the RAS/MAPK signaling pathway, also appears to be important. Mutations in the let-60/Ras, lin-45/Raf, and mek-2 and mpk-1 MAPK genes block germ cell death (Gumienny et al., 1999), and also lead to other developmental defects in the germ line, suggesting that they likely act early in cell fate determination (Fig. 2).
It is remarkable that so many of the regulators of the apoptotic pathway in C. elegans are related in sequence to mammalian oncogenes and tumor suppressors (Table 1). This supports the notion that manipulation of cell death pathways is an important aspect of tumor formation.
Table 1.
Some C. elegans apoptosis regulators are similar to mammalian tumor factors
|
C. elegans protein |
Function | Regulated cell (type) |
Mammalian homolog |
Tumor2 | References |
|---|---|---|---|---|---|
| ABCF-3 | AAA+ ATPase | M4 sister (neuron) |
ABCF3 | Liver cancer | (Hirose, & Horvitz, 2014) |
| CEH-20 | Transcription factor |
P11.aaap (neuron) |
PBX1 | Leukemia | (Liu, Strauss, Potts, & Cameron, 2006) |
| CEH-34 | Transcription factor |
M4 sister (neuron) |
SIX1 | Breast cancer, gastric cancer, others |
(Hirose, Galvin, & Horvitz, 2010) |
| CES-1 | Transcription factor |
NSM, I2 (neurons) |
SLUG | Acute lymphoblastic leukemia |
(Metzstein, & Horvitz, 1999) |
| CES-2 | Transcription factor |
NSM (neuron) |
HLF | Acute lymphoblastic leukemia |
(Metzstein et al., 1996) |
| EFL-2 | Transcription factor |
Germ cells, pharynx, others |
E2F | Colorectal cancer, pancreatic cancer, many others |
(Reddien, Andersen, Huang, & Horvitz, 2007; Schertel, & Conradt, 2007) |
| EGL-38 | Transcription factor |
Germ cells, embryonic cells(?) |
Pax2/5/8 | Renal cancer, colon cancer, many others |
(Park, Jia, Rajakumar, & Chamberlin, 2006) |
| EOR-1 | Transcription factor |
HSN (neuron) |
PLZF, BCL6 | Leukemia, lymphoma, others |
(Hoeppner et al., 2004; Howard, & Sundaram, 2002) |
| EYA-1 | Transcription factor |
M4 sister (neuron) |
EYA | Breast cancer, gastric cancer, others |
(Hirose, Galvin, & Horvitz, 2010) |
| DRE-1 | F-Box, ubiquitylation |
Tail-spike cell (epithelial) |
FBXO10/11 | B cell lymphoma |
(Chiorazzi et al., 2013) |
| LET-23 | Receptor tyrosine kinase |
Germ cells, ABpl/rpppap p (epithelial) |
EGFR | Lung cancer, medulloblasto ma, others |
(Jiang, & Wu, 2014) |
| LET-60 | GTPase | Germ cells, ABpl/rpppap p (epithelial) |
RAS | Many tumors | (Jiang, & Wu, 2014) |
| LIN-3 | Receptor tyrosine kinase ligand |
Germ cells, ABpl/rpppap p (epithelial) |
EGF | Lung cancer, medulloblasto ma, others |
(Jiang, & Wu, 2014) |
| LIN-35 | Transcription factor |
Germ cells, Pn.p descendents (neurons) |
Rb | Retinoblastom a, many tumors |
(Reddien, Andersen, Huang, & Horvitz, 2007; Schertel, & Conradt, 2007) |
| MAB-5 | Transcription factor |
P11.aaap (neuron) |
HOX | Breast cancer, many tumors |
(Liu, Strauss, Potts, & Cameron, 2006) |
| PAL-1 | Transcription factor |
Tail-spike cell (epithelial) |
Cdx2 | Intestinal cancer |
(Maurer, Chiorazzi, & Shaham, 2007) |
| PAR-4 | kinase | ABalapapaa, ABplpappap ABalppaaaa ABalppaapa ABaraaaapp ABplppaaap ABplpppapp ABalpapappa (epithelial, excretory cell) M4 sister (neuron) |
LKB1 | Liver cancer, leukemia, others |
(Chien, Brinkmann, Teuliere, & Garriga, 2013; Denning, Hatch, & Horvitz, 2012) |
| PAX-2 | Transcription factor |
Germ cells, embryonic cells (?) |
Pax2/5/8 | Renal cancer, colon cancer, many others |
(Park, Jia, Rajakumar, & Chamberlin, 2006) |
| PIG-1 | kinase | ABalapapaa, ABplpappap ABalppaaaa ABalppaapa ABaraaaapp ABplppaaap ABplpppapp ABalpapappa (epithelial, excretory cell) M4 sister (neuron) |
MELK | Glioblastoma, astocytoma, pediatric brain tumor, colon tumor, breast cancer, melanoma, rectal cancer |
(Chien, Brinkmann, Teuliere, & Garriga, 2013; Cordes, Frank, & Garriga, 2006; Denning, Hatch, & Horvitz, 2012; Hirose, & Horvitz, 2013) |
| UNC-86 | Transcription factor |
CEM (neurons) |
BRN3 | Neuroepitheli al cancer |
(Nehme et al., 2010; Peden et al., 2007) |
| UNC-132 | kinase | CEM (neuron) |
PIM-1 | Leukemia, lung cancer, pancreatic cancer, ovarian cancer |
(Nehme et al., 2010) |
| TRA-1 | Transcription factor |
HSN, CEM (neurons) |
Gli | Basal cell carcinoma, medulloblasto ma,breast cancer, glioma, leukemia, prostate cancer, GI cancers, others |
(Nehme et al., 2010; Peden et al., 2007; Schwartz, & Horvitz, 2007; Conradt, & Horvitz 1999) |
Tumors reporting associated changes in function of the indicated mammalian protein
The engulfment genes and their roles in cell death
Apoptosis in C. elegans is tightly coupled to engulfment. For many cells destined to die, engulfment by neighboring cells begins even before anaphase of the parent cell division is complete (Sulston, Schierenberg, White, & Thomson, 1983). Thus, cells destined to die often do not have an opportunity to function or interact with other cells. Only three C. elegans cells live for an extended period of time before death and engulfment ensue: the tail-spike cell (see above), the male-specific linker cell (see below), and the sister of Z1, a gonadal precursor whose function is not known (Sulston, Schierenberg, White, & Thomson, 1983). Genetic screens for mutants defective in the engulfment or degradation of cell corpses have identified a host of proteins required for these processes, and these have been extensively reviewed (Lu, & Zhou, 2012; Pinto, & Hengartner, 2012); we only briefly summarize the findings here.
Two parallel and partially-redundant pathways control recognition of dying cells (Ellis, Jacobson, & Horvitz, 1991). One consists of the membrane receptor CED-1, which appears to recognize the lipid phosphatidyl serine on the surface of dying cells (Zhou, Hartwieg, & Horvitz, 2001), CED-6, an adapter protein that may function in signal transduction (Liu, & Hengartner, 1998; Liu, & Hengartner, 1999), and CED-7, a membrane ABC transporter that may be involved in phosphatidyl serine presentation (Wu, & Horvitz, 1998). CED-1 and CED-6 function in the engulfing cell, and CED-7 is required in both dying and engulfing cells. Unlike CED-1 and CED-6, CED-7 is required only in the soma, whereas the phospholipid scramblase PLSC-1 exposes phosphatidyl serine in dying germ cells (Venegas, & Zhou, 2007). The other pathway consists of the proteins CED-2/CrkII, CED-5/Dock180, CED-10/Rac, and CED-12/Elmo (Gumienny et al., 2001; Reddien, & Horvitz, 2000; Wu et al., 2001; Zhou et al., 2001). CED-2, −5, and −12, appear to function together to regulate CED-10 activity. CED-10 likely acts to control assembly of the actin cytoskeleton during the extension of engulfing cell protrusions around the dying cell.
It has been suggested that CED-1 and CED-6 also function through CED-10 (Kinchen et al., 2005), although this has been debated (Yu et al., 2006). Nonetheless, recent studies show that the dynamin protein DYN-1 mediates signaling downstream of CED-1 in a pathway involving clathrin and epsin, which promote actin assembly at the phagocytic cup (Shen et al., 2013). Thus, CED-1 may indirectly impact actin remodeling.
Once an apoptotic cell has been incorporated into a phagosome within the engulfing cell, a series of events alter the phagosomal surface, culminating in lysosomal targeting. Phosphatidylinositol 3-phosphate (PtdIns(3)P) coats the phagosomal membrane, allowing binding of BAR-domain sorting nexins to recruit lysosomes (Lu et al., 2011). In parallel, RAB-5, which binds initially to the phagosomal surface, is replaced by RAB-7, through activity of the SAND-1 and CCZ-1 complex (Kinchen, & Ravichandran, 2010; Nieto et al., 2010), and phagolysosome formation ensues through activities of the conserved HOPs complex (Kinchen et al., 2008) and the proteins UNC-108/RAB2 and RAB-14, which promote acidification of the compartment (Guo et al., 2010; Mangahas, Yu, Miller, & Zhou, 2008).
While engulfment has generally been viewed as independent of cell death execution, some observations suggest a more intimate connection. In animals carrying weak ced-3 caspase mutations, some cells destined to die in the developing ventral cord of C. elegans larvae succumb and are removed, while others survive inappropriately. Occasionally, cells that by DIC microscopy appear far gone recover and survive. Thus, in these mutants, cells slated to die teeter on the edge between life and death. Remarkably, in animals also homozygous for mutations in genes controlling engulfment, significantly more cells survive and develop (Reddien, Cameron, & Horvitz, 2001). These results demonstrate that at least under some conditions, engulfment may promote cell death. Similarly, in C. elegans males, the cells B.alapaav and B.arapaav have been reported to survive in ced-1 and ced-2 mutants, presumably because they fail to be engulfed by the P12.pa cell (Hedgecock, Sulston, & Thomson, 1983).
While the mechanism by which engulfing cells promote cell death of their neighbors is not clear, some molecular insight may be gleaned from studies of DNA degradation accompanying cell death. As described above, mutations in the DNase II gene nuc-1 result in accumulation of TUNEL-staining DNA in developing embryos, revealing an activity for an early-acting DNAse. Mutations in the engulfment genes ced-1 and ced-7, but not ced-2, -5, -6, or -10, prevent accumulation of TUNEL-reactive fragments (Wu, Stanfield, & Horvitz, 2000). Thus, CED-1 and CED-7 are required for initiating the degradation of DNA in dying cells. While this could be taken to mean that DNAse activity resides in the engulfing cells, this interpretation is unlikely, since in ced-2, -5, -6, and -10, mutants, engulfment also fails to take place, but DNase activation still occurs. Rather, barring technical issues with TUNEL staining in these mutants, it seems that a signal unrelated to engulfment per se, but dependent on ced-1 and ced-7, promotes DNase activation in dying cells. Although it is likely that ced-1 and ced-7 function in the engulfing cell to activate DNA degradation, this has not been directly tested, and would be a prerequisite for defining the functions of these genes in DNA degradation.
Linker cell death
The linker cell is a male-specific leader cell born in the ventral midbody in the second larval stage (L2). The cell guides the elongation of the gonad, and upon reaching the cloacal region in the late L4 stage, it dies. Death may allow fusion of the gonad with the cloaca, resulting in an open germ system competent for sperm transfer during fertilization. Anecdotal reports suggested that linker cell death may require the neighboring engulfing cells, U.l/rp (Hedgecock, Sulston, & Thomson, 1983); however, later studies demonstrated that the cell can die following ablation of these dedicated engulfing cells, or in animals in which the linker cell fails to migrate properly (Abraham, Lu, & Shaham, 2007). Anecdotal reports also suggested initially that linker cell death is partially dependent on ced-3 and ced-4 (Ellis, & Horvitz, 1986). However, further studies of the genetic requirements for death, coupled with extensive morphological and ultrastructural observations revealed that linker cell death does not require ced-3 or ced-4. Indeed the cell death process could proceed in its entirety in animals carrying mutations in any of the core apoptotic pathway genes (Abraham, Lu, & Shaham, 2007). Thus, linker cell death proceeds by a previously unexplored mode of cell death. Consistent with these novel genetic requirements, the ultrastructural changes accompanying the cell death process are very different from those seen in apoptotic cells. While apoptosis is characterized by nuclear condensation, cytoplasmic shrinkage, and morphologically intact organelles (until the very end) (Fig. 1), the dying linker cell exhibits none of these features. Instead, the cell displays pronounced nuclear envelope invagination (crenellation), as well as swelling of endoplasmic reticulum and mitochondria (Abraham, Lu, & Shaham, 2007)(Fig. 3). These changes are often observed in cells dying during vertebrate development, and are prevalent in degenerating cells of patients with polyQ diseases.
Figure 3.
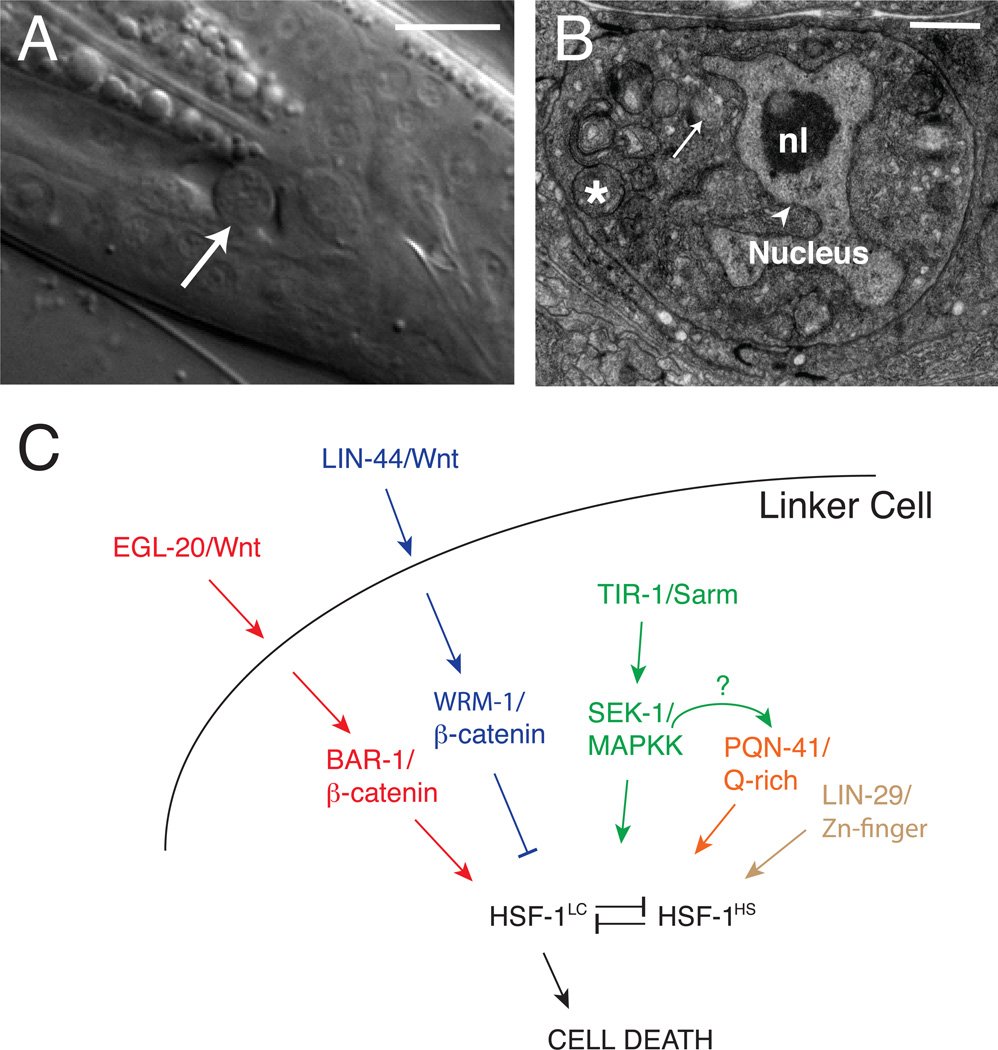
Linker cell death in C. elegans. (A) DIC image of a dying linker cell (arrow). Scale bar, 5 microns. (B) Electron micrograph of a dying linker cell. Note crenellated nucleus (arrowhead), intact nucleolus (nl), and swollen organelles (asterisk and arrow). Scale bar, 700 nm. (C) A model for linker cell death. Proteins in the same regulatory pathway are color-matched. How regulators impinge on HSF-1 function is not known.
Genetic and molecular screens have uncovered genes involved in linker cell death. Initiation of the death process appears to be dependent on both temporal and positional cues. The LIN-29 Zn-finger transcription factor, a downstream component of the C. elegans heterochronic pathway controlling developmental timing, promotes linker cell demise (Abraham, Lu, & Shaham, 2007). Two opposing Wnt pathways mediate the spatial signal (M. Kinet and S. Shaham, submitted). The Wnt LIN-44 is secreted by male tail cells and acts through the MIG-1 and CFZ-2 Frizzled-like receptors, LIT-1/Nemo-like kinase, and WRM-1/beta-catenin—all of which function in the linker cell—to inhibit linker cell death. This protective pathway is antagonized by the Wnt EGL-20, also expressed in neighboring cells, and its linker-cell effectors LIN-17/Frizzled, MOM-5/Frizzled, MIG-5/Dishevelled, and BAR-1/beta-catenin (Fig. 3).
The Wnt responsive transcription factor POP-1/TCF/LEF does not appear to play a role in linker cell death (M. Kinetand S.Shaham, submitted), suggesting other possible targets. One candidate is the Q-rich protein PQN-41. A C-terminal polypeptide encoded by the locus, and consisting of 35% glutamine residues, is sufficient to rescue linker cell survival exhibited by pqn-41 mutants (Blum et al., 2012). At least some PQN-41 isoforms appear to be localized to nuclei, suggesting that they could function to mediate a transcriptional output of Wnt signaling. Another candidate is the transcription factor HSF-1, which in other contexts transduces heat and stress signals to promote a protective cellular response. In the linker cell, however, hsf-1 (lf) mutations block linker cell death, suggesting that it, instead, promotes cell dismantling (M. Kinetand S. Shaham, submitted). Supporting this observation, stress targets of HSF-1 are not induced in the linker cell. Importantly, an HSF-1 gain-of-function mutation can rescue inappropriate linker cell survival in Wnt pathway mutants, as well as all other known mutants that block linker cell death (M. Kinet and S. Shaham, submitted). HSF-1, therefore, may be a key downstream regulatory node, and determining its targets is an important next step.
The kinase SEK-1 and its adapter protein TIR-1 also promote linker cell death, and their loss can also be compensated for by the hsf-1 (gf) allele. Genetic interaction studies suggest that these proteins might function in the same pathway as PQN-41 (Blum et al., 2012). Drosophila and murine homolgs of TIR-1, dSarm and Sarm, respectively, have been implicated in axon distal segment degeneration following axotomy (Osterloh et al., 2012), raising the possibility that the dismantling program of the linker cell may share conserved features with non-apoptotic degenerative programs in other systems. Supporting this idea, mutations in ubiquitin-dependent protein degradation system components block linker cell death (J. Malin and S. Shaham, in preparation), and have likewise been implicated in axon degeneration (Korhonen, & Lindholm, 2004).
Linker cell engulfment also has novel genetic requirements. Mutations in apoptotic corpse engulfment genes do not affect engulfment of the linker cell (Abraham, Lu, & Shaham, 2007), suggesting that alternative pathways must exist. Genetic screens have identified a number of mutants in which engulfment and/or corpse degradation appear blocked (L. Kutscher and S. Shaham, unpublished data). Characterization of these mutants will likely reveal aspects of this new machinery.
Looking ahead
In C. elegans, mutations in ced-3 or ced-4 block most developmental cell death that takes place in the animal. While the mammalian counterparts of these genes clearly participate in apoptotic events, mutations in these genes have surprisingly weak effects on developmental cell death. caspase-3 and caspase-9 mutants are viable, as are Apaf-1 knockout mice (Honarpour et al., 2000). In some specific instances, the mammalian mutants exhibit a slight delay in cell death timing; however, the process eventually gains traction, and cells give up the ghost. Why are CED-3 and CED-4, therefore, essential for most cell death in nematodes but not in mammals? Why do mammals apparently have parallel programs ensuring cellular demise even if apoptosis fails? While the answers remain unknown, one explanation invokes the inherent developmental differences between C. elegans and mammals. The nematode lineage is essentially invariant, and cell identity and survival are nearly always determined cell-autonomously by lineal history. Honing of cell differentiation programs therefore leads to reproducible development and cell death that is not very error prone. In mammals, however, cell identity and cell death are very often regulated by cell-cell communication. While cell-autonomous processes are confined by the cell membrane, cell-cell signaling is, by its nature, less spatially contained. Thus, to ensure robust death, alternative death processes may have evolved. In this light, it is intriguing that the only C. elegans cell in which cell-cell signaling has been unambiguously demonstrated to control survival, the linker cell, also employs an alternative cell death form. If this hypothesis is correct, two predictions may be made. First, ced-3 and ced-4 may have a role in linker cell death, but this may only be elicited in sensitized genetic backgrounds. Second, the non-apoptotic death program governing linker cell death may very well be conserved in mammals. Experiments testing both predictions are currently underway, and may eventually reveal an even deeper similarity in cell death processes among animals.
Acknowledgments
We are grateful to members of our laboratory for comments and for sharing unpublished results. Every attempt to include all relevant citations was made, and we apologize to those authors whose work was not cited either because of space constraints or because of our oversight. This study was supported by NIH grants HD078703 and NS081490 to S. S.
References
- Abraham MC, Lu Y, Shaham S. Dev. Cell. 2007 doi: 10.1016/j.devcel.2006.11.012. In press. [DOI] [PubMed] [Google Scholar]
- Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW. Mol Cell. 2002;9:423–432. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- Antonsson B, Conti F, Ciavatta A, Montessuit S, Lewis S, Martinou I, et al. Science. 1997;277:370–372. doi: 10.1126/science.277.5324.370. [DOI] [PubMed] [Google Scholar]
- Aruscavage PJ, Hellwig S, Bass BL. PLoS One. 2010;5:e11217. doi: 10.1371/journal.pone.0011217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L, Horvitz HR. Cell. 1987;51:1071–1078. doi: 10.1016/0092-8674(87)90593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basanez G, Sharpe JC, Galanis J, Brandt TB, Hardwick JM, Zimmerberg J. J Biol Chem. 2002;277:49360–49365. doi: 10.1074/jbc.M206069200. [DOI] [PubMed] [Google Scholar]
- Bloss TA, Witze ES, Rothman JH. Nature. 2003;424:1066–1071. doi: 10.1038/nature01920. [DOI] [PubMed] [Google Scholar]
- Blum ES, Abraham MC, Yoshimura S, Lu Y, Shaham S. Science. 2012;335:970–973. doi: 10.1126/science.1215156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckenridge DG, Kang BH, Kokel D, Mitani S, Staehelin LA, Xue D. Mol Cell. 2008;31:586–597. doi: 10.1016/j.molcel.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckenridge DG, Kang BH, Xue D. Curr Biol. 2009;19:768–773. doi: 10.1016/j.cub.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelur DS, Chalfie M. Proc Natl Acad Sci U S A. 2007;104:2283–2288. doi: 10.1073/pnas.0610877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Hersh BM, Conradt B, Zhou Z, Riemer D, Gruenbaum Y, et al. Science. 2000;287:1485–1489. doi: 10.1126/science.287.5457.1485. [DOI] [PubMed] [Google Scholar]
- Chen YZ, Mapes J, Lee ES, Skeen-Gaar RR, Xue D. Nat Commun. 2013;4:2726. doi: 10.1038/ncomms3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien SC, Brinkmann EM, Teuliere J, Garriga G. Genetics. 2013;193:897–909. doi: 10.1534/genetics.112.148106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnaiyan AM, Chaudhary D, O’Rourke K, Koonin EV, Dixit VM. Nature. 1997;388:728–729. doi: 10.1038/41913. [DOI] [PubMed] [Google Scholar]
- Chinnaiyan AM, ORourke K, Lane BR, Dixit VM. Science. 1997;275:1122–1126. doi: 10.1126/science.275.5303.1122. [DOI] [PubMed] [Google Scholar]
- Chiorazzi M, Rui L, Y Y, Ceribelli M, ishbi N, Maurer CW, et al. Proc Natl Acad Sci U S A. 2013;110:3943–3948. doi: 10.1073/pnas.1217271110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizeau J, Ray R, Chen G, Gietz RD, Greenberg AH. Oncogene. 2000;19:5453–5463. doi: 10.1038/sj.onc.1203929. [DOI] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR. Cell. 1999;98:317–327. doi: 10.1016/s0092-8674(00)81961-3. [DOI] [PubMed] [Google Scholar]
- Cordes S, Frank CA, Garriga G. Development. 2006;133:2747–2756. doi: 10.1242/dev.02447. [DOI] [PubMed] [Google Scholar]
- del Peso L, Gonzalez VM, Inohara N, Ellis RE, Nunez G. J Biol Chem. 2000;275:27205–27211. doi: 10.1074/jbc.M000858200. [DOI] [PubMed] [Google Scholar]
- del Peso L, Gonzalez VM, Nunez G. J Biol Chem. 1998;273:33495–33500. doi: 10.1074/jbc.273.50.33495. [DOI] [PubMed] [Google Scholar]
- Denning DP, Hatch V, Horvitz HR. Nature. 2012;488:226–230. doi: 10.1038/nature11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning DP, Hatch V, Horvitz HR. PLoS Genet. 2013;9:e1003341. doi: 10.1371/journal.pgen.1003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai C, Garriga G, McIntire SL, Horvitz HR. Nature. 1988;336:638–646. doi: 10.1038/336638a0. [DOI] [PubMed] [Google Scholar]
- Dreze M, Charloteaux B, Milstein S, Vidalain PO, Yildirim MA, Zhong Q, et al. Nat Methods. 2009;6:843–849. doi: 10.1038/nmeth.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis HM, Horvitz HR. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- Ellis RE, Horvitz HR. Development. 1991;112:591–603. doi: 10.1242/dev.112.2.591. [DOI] [PubMed] [Google Scholar]
- Ellis RE, Jacobson DM, Horvitz HR. Genetics. 1991;129:79–94. doi: 10.1093/genetics/129.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RE, Yuan JY, Horvitz HR. Annu Rev Cell Biol. 1991;7:663–698. doi: 10.1146/annurev.cb.07.110191.003311. [DOI] [PubMed] [Google Scholar]
- Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- Fairlie WD, Perugini MA, Kvansakul M, Chen L, Huang DC, Colman PM. Cell Death Differ. 2006;13:426–434. doi: 10.1038/sj.cdd.4401762. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Fuchs Y, Steller H. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvin BD, Denning DP, Horvitz HR. Proc Natl Acad Sci U S A. 2011;108:1998–2003. doi: 10.1073/pnas.1018805108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO. Mol Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- Gavrieli Y, Sherman Y, Ben-Sasson SA. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Shi Y, Nakagawa A, Yoshina S, Mitani S, Xue D. Nat Struct Mol Biol. 2008;15:1094–1101. doi: 10.1038/nsmb.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng X, Zhou QH, Kage-Nakadai E, Shi Y, Yan N, Mitani S, et al. Cell Death Differ. 2009;16:1385–1394. doi: 10.1038/cdd.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, et al. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Embo J. 2000;19:589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, et al. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- Guo P, Hu T, Zhang J, Jiang S, Wang X. Proc Natl Acad Sci U S A. 2010;107:18016–18021. doi: 10.1073/pnas.1008946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Levi-Montalcini R. J Exp Zool. 1949;111:457–501. doi: 10.1002/jez.1401110308. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hausmann G, OReilly LA, van Driel R, Beaumont JG, Strasser A, Adams JM, et al. J Cell Biol. 2000;149:623–634. doi: 10.1083/jcb.149.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wassarman DA, Rubin GM. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Sulston JE, Thomson JN. Science. 1983;220:1277–1279. doi: 10.1126/science.6857247. [DOI] [PubMed] [Google Scholar]
- Hengartner MO, Ellis RE, Horvitz HR. Nature. 1992;356:494–499. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- Hengartner MO, Horvitz HR. Nature. 1994a;369:318–320. doi: 10.1038/369318a0. [DOI] [PubMed] [Google Scholar]
- Hengartner MO, Horvitz HR. Cell. 1994b;76:665–676. doi: 10.1016/0092-8674(94)90506-1. [DOI] [PubMed] [Google Scholar]
- Hevelone J, Hartman PS. Biochem Genet. 1988;26:447–461. doi: 10.1007/BF02399412. [DOI] [PubMed] [Google Scholar]
- Hirose T, Galvin BD, Horvitz HR. Proc Natl Acad Sci U S A. 2010;107:1547–1584. doi: 10.1073/pnas.1010023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Horvitz HR. Nature. 2013;500:354–358. doi: 10.1038/nature12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Horvitz HR. PLoS Genet. 2014;10:e1004512. doi: 10.1371/journal.pgen.1004512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeppner DJ, Hengartner MO, Schnabel R. Nature. 2001;412:202–206. doi: 10.1038/35084103. [DOI] [PubMed] [Google Scholar]
- Hoeppner DJ, Spector MS, Ratliff TM, Kinchen JM, Granat S, Lin SC, et al. Dev Biol. 2004;274:125–138. doi: 10.1016/j.ydbio.2004.06.022. [DOI] [PubMed] [Google Scholar]
- Honarpour N, Du C, Richardson JA, Hammer RE, Wang X, Herz J. Dev Biol. 2000;218:248–258. doi: 10.1006/dbio.1999.9585. [DOI] [PubMed] [Google Scholar]
- Horvitz HR. Biosci Rep. 2003;23:239–303. doi: 10.1023/b:bire.0000019187.19019.e6. [DOI] [PubMed] [Google Scholar]
- Howard RM, Sundaram MV. Genes Dev. 2002;16:1815–1827. doi: 10.1101/gad.998402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell K, Arur S, Schedl T, Sundaram MV. Genetics. 2010;184:899–913. doi: 10.1534/genetics.109.111591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Jiang T, Choi W, Qi S, Pang Y, Hu Q, et al. Genes Dev. 2013;27:2039–2048. doi: 10.1101/gad.224428.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Gourley TS, Carrio R, Muniz M, Merino J, Garcia I, et al. J Biol Chem. 1998;273:32479–32486. doi: 10.1074/jbc.273.49.32479. [DOI] [PubMed] [Google Scholar]
- Irmler M, Hofmann K, Vaux D, Tschopp J. FEBS Lett. 1997;406:189–190. doi: 10.1016/s0014-5793(97)00271-8. [DOI] [PubMed] [Google Scholar]
- Ishii N, Fujii M, Hartman PS, Tsuda M, Yasuda K, Senoo-Matsuda N, et al. Nature. 1998;394:694–697. doi: 10.1038/29331. [DOI] [PubMed] [Google Scholar]
- Jagasia R, Grote P, Westermann B, Conradt B. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- Jiang HS, Wu YC. PLoS Genet. 2014;10:e1004513. doi: 10.1371/journal.pgen.1004513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi P, Eisenmann DM. Genetics. 2004;167:673–685. doi: 10.1534/genetics.103.020503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelekar A, Thompson CB. Trends Cell Biol. 1998;8:324–330. doi: 10.1016/s0962-8924(98)01321-x. [DOI] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. Dev Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Cabello J, Klingele D, Wong K, Feichtinger R, Schnabel H, et al. Nature. 2005;434:93–99. doi: 10.1038/nature03263. [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Doukoumetzidis K, Almendinger J, Stergiou L, Tosello-Trampont A, Sifri CD, et al. Nat Cell Biol. 2008;10:556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen JM, Ravichandran KS. Nature. 2010;464:778–782. doi: 10.1038/nature08853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen L, Lindholm D. J Cell Biol. 2004;165:27–30. doi: 10.1083/jcb.200311091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, et al. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Liu H, Strauss TJ, Potts MB, Cameron S. Development. 2006;133:641–650. doi: 10.1242/dev.02234. [DOI] [PubMed] [Google Scholar]
- Liu QA, Hengartner MO. Cell. 1998;93:961–972. doi: 10.1016/s0092-8674(00)81202-7. [DOI] [PubMed] [Google Scholar]
- Liu QA, Hengartner MO. Curr Biol. 1999;9:1347–1350. doi: 10.1016/s0960-9822(00)80061-5. [DOI] [PubMed] [Google Scholar]
- Liu X, Li P, Widlak P, Zou H, Luo X, Garrard WT, et al. Proc Natl Acad Sci U S A. 1998;95:8461–8466. doi: 10.1073/pnas.95.15.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshin RA, Williams CM. J Insect Physiol. 1965;11:123–133. doi: 10.1016/0022-1910(65)90099-5. [DOI] [PubMed] [Google Scholar]
- Lu N, Shen Q, Mahoney TR, Liu X, Zhou Z. Mol Biol Cell. 2011;22:354–374. doi: 10.1091/mbc.e10-09-0756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N, Zhou Z. Int Rev Cell Mol Biol. 2012;293:269–309. doi: 10.1016/B978-0-12-394304-0.00013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Rolland SG, Conradt B. Proc Natl Acad Sci U S A. 2011;108:E813–E822. doi: 10.1073/pnas.1103218108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, et al. Embo J. 2007;26:2527–2539. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangahas PM, Yu X, Miller KG, Zhou Z. J Cell Biol. 2008;180:357–373. doi: 10.1083/jcb.200708130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer CW, Chiorazzi M, Shaham S. Development. 2007;134:1357–1368. doi: 10.1242/dev.02818. [DOI] [PubMed] [Google Scholar]
- Metzstein MM, Hengartner MO, Tsung N, Ellis RE, Horvitz HR. Nature. 1996;382:545–547. doi: 10.1038/382545a0. [DOI] [PubMed] [Google Scholar]
- Metzstein MM, Horvitz HR. Mol Cell. 1999;4:309–319. doi: 10.1016/s1097-2765(00)80333-0. [DOI] [PubMed] [Google Scholar]
- Nakagawa A, Shi Y, Kage-Nakadai E, Mitani S, Xue D. Science. 2010;328:32–34. doi: 10.1126/science.1182374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa A, Sullivan KD, Xue D. Nat Struct Mol Biol. 2014;21:1082–1090. doi: 10.1038/nsmb.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehme R, Grote P, Tomasi T, Loser S, Holzkamp H, Schnabel R, et al. Cell Death Differ. 2010;17:1266–1276. doi: 10.1038/cdd.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C, Almendinger J, Gysi S, Gomez-Orte E, Kaech A, Hengartner MO, et al. J Cell Sci. 2010;123:2001–2007. doi: 10.1242/jcs.062331. [DOI] [PubMed] [Google Scholar]
- Osterloh JM, Yang J, Rooney TM, Fox AN, Adalbert R, Powell EH, et al. Science. 2012;337:481–484. doi: 10.1126/science.1223899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Jia H, Rajakumar V, Chamberlin HM. Development. 2006;133:4193–4202. doi: 10.1242/dev.02614. [DOI] [PubMed] [Google Scholar]
- Parrish J, Li L, Klotz K, Ledwich D, Wang X, Xue D. Nature. 2001;412:90–94. doi: 10.1038/35083608. [DOI] [PubMed] [Google Scholar]
- Parrish J, Metters H, Chen L, Xue D. Proc Natl Acad Sci U S A. 2000;97:11916–11921. doi: 10.1073/pnas.210391597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden E, Kimberly E, Gengyo-Ando K, Mitani S, Xue D. Genes Dev. 2007;21:3195–3207. doi: 10.1101/gad.1607807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto SM, Hengartner MO. Curr Opin Cell Biol. 2012;24:881–888. doi: 10.1016/j.ceb.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Poreba M, Strozyk A, Salvesen GS, Drag M. Cold Spring Harb Perspect Biol. 2013;5:a008680. doi: 10.1101/cshperspect.a008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourkarimi E, Greiss S, Gartner A. Cell Death Differ. 2012;19:406–415. doi: 10.1038/cdd.2011.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S, Pang Y, Hu Q, Liu Q, Li H, Zhou Y, et al. Cell. 2010;141:446–457. doi: 10.1016/j.cell.2010.03.017. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Andersen EC, Huang MC, Horvitz HR. Genetics. 2007;175:1719–1733. doi: 10.1534/genetics.106.068148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Cameron S, Horvitz HR. Nature. 2001;412:198–202. doi: 10.1038/35084096. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR. Nat Cell Biol. 2000;2:131–136. doi: 10.1038/35004000. [DOI] [PubMed] [Google Scholar]
- Rocheleau CE, Howard RM, Goldman AP, Volk ML, Girard LJ, Sundaram MV. Genetics. 2002;161:121–131. doi: 10.1093/genetics/161.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertel C, Conradt B. Development. 2007;134:3691–3701. doi: 10.1242/dev.004606. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Schertel C, Wittenburg N, Tuck S, Mitani S, Gartner A, et al. Cell Death Differ. 2005;12:153–161. doi: 10.1038/sj.cdd.4401539. [DOI] [PubMed] [Google Scholar]
- Schwartz HT, Horvitz HR. Genes Dev. 2007;21:3181–3194. doi: 10.1101/gad.1607007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiffert BM, Vier J, Hacker G. Biochem Biophys Res Commun. 2002;290:359–365. doi: 10.1006/bbrc.2001.6211. [DOI] [PubMed] [Google Scholar]
- Sendoel A, Kohler I, Fellmann C, Lowe SW, Hengartner MO. Nature. 2010;465:577–583. doi: 10.1038/nature09141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshagiri S, Chang WT, Miller LK. FEBS Lett. 1998;428:71–74. doi: 10.1016/s0014-5793(98)00493-1. [DOI] [PubMed] [Google Scholar]
- Seshagiri S, Miller LK. Curr Biol. 1997;7:455–460. doi: 10.1016/s0960-9822(06)00216-8. [DOI] [PubMed] [Google Scholar]
- Shaham S. J Biol Chem. 1998;273:35109–35117. doi: 10.1074/jbc.273.52.35109. [DOI] [PubMed] [Google Scholar]
- Shaham S, Bargmann CI. Genes Dev. 2002;16:972–983. doi: 10.1101/gad.976002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham S, Horvitz HR. Cell. 1996a;86:201–208. doi: 10.1016/s0092-8674(00)80092-6. [DOI] [PubMed] [Google Scholar]
- Shaham S, Horvitz HR. Genes Dev. 1996b;10:578–591. doi: 10.1101/gad.10.5.578. [DOI] [PubMed] [Google Scholar]
- Shaham S, Reddien PW, Davies B, Horvitz HR. Genetics. 1999;153:1655–1671. doi: 10.1093/genetics/153.4.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, He B, Lu N, Conradt B, Grant BD, Zhou Z. Development. 2013;140:3230–3243. doi: 10.1242/dev.093732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silke J, Vucic D. Methods Enzymol. 2014;545:35–65. doi: 10.1016/B978-0-12-801430-1.00002-0. [DOI] [PubMed] [Google Scholar]
- Song Q, Kuang Y, Dixit VM, Vincenz C. Embo J. 1999;18:167–178. doi: 10.1093/emboj/18.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector MS, Desnoyers S, Hoeppner DJ, Hengartner MO. Nature. 1997;385:653–656. doi: 10.1038/385653a0. [DOI] [PubMed] [Google Scholar]
- Speliotes EK, Uren A, Vaux D, Horvitz HR. Mol Cell. 2000;6:211–223. doi: 10.1016/s1097-2765(00)00023-x. [DOI] [PubMed] [Google Scholar]
- Stanfield GM, Horvitz HR. Mol Cell. 2000;5:423–433. doi: 10.1016/s1097-2765(00)80437-2. [DOI] [PubMed] [Google Scholar]
- Strange K. Physiol Rev. 2003;83:377–415. doi: 10.1152/physrev.00025.2002. [DOI] [PubMed] [Google Scholar]
- Sugimoto A, Hozak RR, Nakashima T, Nishimoto T, Rothman JH. Embo J. 1995;14:4434–4441. doi: 10.1002/j.1460-2075.1995.tb00122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE. Philos Trans R Soc Lond B Biol Sci. 1976;275:287–297. doi: 10.1098/rstb.1976.0084. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Albertson DG, Thomson JN. Dev Biol. 1980;78:542–576. doi: 10.1016/0012-1606(80)90352-8. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Suzuki J, Denning DP, Imanishi E, Horvitz HR, Nagata S. Science. 2013;341:403–406. doi: 10.1126/science.1236758. [DOI] [PubMed] [Google Scholar]
- Takacs-Vellai K, Vellai T, Puoti A, Passannante M, Wicky C, Streit A, et al. Curr Biol. 2005;15:1513–1517. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]
- Tan FJ, Fire AZ, Hill RB. Cell Death Differ. 2007;14:1925–1935. doi: 10.1038/sj.cdd.4402215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan FJ, Zuckerman JE, Wells RC, Hill RB. Protein Sci. 2011;20:62–74. doi: 10.1002/pro.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thellmann M, Hatzold J, Conradt B. Development. 2003;130:4057–4071. doi: 10.1242/dev.00597. [DOI] [PubMed] [Google Scholar]
- Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, et al. Nature. 1992;356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- Trent C, Tsung N, Horvitz HR. Genetics. 1983;104:619–647. doi: 10.1093/genetics/104.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzur YB, Margalit A, Melamed-Book N, Gruenbaum Y. Proc Natl Acad Sci U S A. 2006;103:13397–13402. doi: 10.1073/pnas.0604224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venegas V, Zhou Z. Mol Biol Cell. 2007;18:3180–3192. doi: 10.1091/mbc.E07-02-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Yang C, Chai J, Shi Y, Xue D. Science. 2002;298:1587–1592. doi: 10.1126/science.1076194. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1991;331:263–271. [Google Scholar]
- Woo JS, Jung JS, Ha NC, Shin J, Kim KH, Lee W, et al. Cell Death Differ. 2003;10:1310–1319. doi: 10.1038/sj.cdd.4401303. [DOI] [PubMed] [Google Scholar]
- Wu D, Chen PJ, Chen S, Hu Y, Nunez G, Ellis RE. Development. 1999;126:2021–2031. doi: 10.1242/dev.126.9.2021. [DOI] [PubMed] [Google Scholar]
- Wu D, Wallen HD, Inohara N, Nunez G. J Biol Chem. 1997;272:21449–21454. doi: 10.1074/jbc.272.34.21449. [DOI] [PubMed] [Google Scholar]
- Wu YC, Horvitz HR. Cell. 1998;93:951–960. doi: 10.1016/s0092-8674(00)81201-5. [DOI] [PubMed] [Google Scholar]
- Wu YC, Stanfield GM, Horvitz HR. Genes Dev. 2000;14:536–548. [PMC free article] [PubMed] [Google Scholar]
- Wu YC, Tsai MC, Cheng LC, Chou CJ, Weng NY. Dev Cell. 2001;1:491–502. doi: 10.1016/s1534-5807(01)00056-9. [DOI] [PubMed] [Google Scholar]
- Xue D, Horvitz HR. Nature. 1997;390:305–308. doi: 10.1038/36889. [DOI] [PubMed] [Google Scholar]
- Xue D, Shaham S, Horvitz HR. Genes Dev. 1996;10:1073–1083. doi: 10.1101/gad.10.9.1073. [DOI] [PubMed] [Google Scholar]
- Yan N, Chai J, Lee ES, Gu L, Liu Q, He J, et al. Nature. 2005;437:831–837. doi: 10.1038/nature04002. [DOI] [PubMed] [Google Scholar]
- Yan N, Gu L, Kokel D, Chai J, Li W, Han A, et al. Mol Cell. 2004;15:999–1006. doi: 10.1016/j.molcel.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Yang X, Chang HY, Baltimore D. Science. 1998;281:1355–1357. doi: 10.1126/science.281.5381.1355. [DOI] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Odera S, Chuang CH, Lu N, Zhou Z. Dev Cell. 2006;10:743–757. doi: 10.1016/j.devcel.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Yuan J, Horvitz HR. Development. 1992;116:309–320. doi: 10.1242/dev.116.2.309. [DOI] [PubMed] [Google Scholar]
- Yuan J, Shaham S, Ledoux S, Ellis HM, Horvitz HR. Cell. 1993;75:641–652. doi: 10.1016/0092-8674(93)90485-9. [DOI] [PubMed] [Google Scholar]
- Yuan JY, Horvitz HR. Dev Biol. 1990;138:33–41. doi: 10.1016/0012-1606(90)90174-h. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Caron E, Hartwieg E, Hall A, Horvitz HR. Dev Cell. 2001;1:477–489. doi: 10.1016/s1534-5807(01)00058-2. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hartwieg E, Horvitz HR. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]