Abstract
The aim of this study is to characterize the factors related to peptidoglycan metabolism in isogenic hVISA/VISA ST100 strains. Recently, we reported the increase in IS256 transposition in invasive hVISA ST100 clinical strains isolated from the same patient (D1 and D2) before and after vancomycin treatment and two laboratory VISA mutants (D23C9 and D2P11) selected from D2 in independent experiments. High performance liquid chromatography-mass spectrometry (HPLC-MS) analysis of peptidoglycan muropeptides showed increased proportion of monomeric muropeptides and a concomitant decrease in the proportion of tetrameric muropeptide in D2 and derived mutants when compared to the original strain D1. In addition, strain D2 and its derived mutants showed an increase in cell wall thickness with increased pbp2 gene expression. The VISA phenotype was not stable in D2P11 and showed a reduced autolysis profile. On the other hand, the mutant D23C9 differentiates from D2 and D2P11 in the autolysis profile, and pbp4 transcription profile. D2-derived mutants exhibited differences in the susceptibility to other antimicrobials. Our results highlight the possibility of selection of different VISA phenotypes from a single hVISA-ST100 genetic background.
Keywords: : Staphylococcus aureus, vancomycin, hVISA, VISA, MRSA, ST100
Background
Staphylococcus aureus is an important opportunistic pathogen that has the ability to acquire resistance to a great number of antimicrobial agents. Currently, the main concern comes from methicillin-resistant S. aureus (MRSA), which generally harbor additional antibiotic resistance determinants.
Vancomycin (VAN) is the main alternative for the treatment of severe infections caused by MRSA, but from the late 90s, the increase in reports of vancomycin-intermediate S. aureus (VISA) and heterogeneous vancomycin-intermediate S. aureus (hVISA) has raised an alarm worldwide.1,2 Subsequently, increased reports of vancomycin failure started to appear and the efficacy of vancomycin in the treatment of many serious infections has been questioned.
To date, the underlying mechanism responsible for the hVISA/VISA phenotype is not yet fully understood. A thickened cell wall is a common feature described among most VISA isolates reported, possibly preventing vancomycin diffusion to its target in the division septum where peptidoglycan synthesis occurs. Notwithstanding, there is a large number of phenotypic and genotypic features associated with the resistance, and it has been proposed that VISA strains emerge by mutations from vancomycin-susceptible S. aureus by a step-wise process with hVISA as intermediary.3
Some hVISA/VISA strains were described to have abnormal growth rate, increased concentration of free D-Ala-D-Ala terminal peptidoglycan precursors, and reduced cell wall turnover and autolytic rates.2 It has been observed that after incubation in a drug-free medium or prolonged storage, resistance phenotype can revert to a more susceptible one.2,4 In addition, it has been described that the loss of functionality of the agr locus may confer an adaptive advantage under vancomycin selective pressure.5
Point mutations in regulatory genes such as vraSR, graSR, walKR, rpoB, and cmk6–10 were associated to VAN resistance. Furthermore, disruption of genes involved in cell wall synthesis (tca, walKR), by IS256 insertions, led to VISA phenotype.11–13
Recently, we reported the increase in IS256 transposition in an invasive hVISA ST100 clinical isolate and its derived VISA mutants.14 The aim of this study is to characterize the factors related to peptidoglycan metabolism in these isogenic ST100 strains.
Materials and Methods
Strains and culture conditions
S. aureus strains D1 and D2 were isolated from a patient suffering bone and joint infection, before and after 40 days of vancomycin treatment. Detailed clinical history of the patient was recently described.14 In vitro selection of D2-derived mutants (D23C9, D2P11) was performed in two independent experiments by serial passage with increasing concentrations of vancomycin (Sigma-Aldrich). All strains were grown on the BHI (Britania, Argentina) broth at 37°C with aeration. Mutant D23C9 and D2P11 were selected at 9 and 11 μg/ml of vancomycin, respectively.14 Relevant properties of these strains are listed in Table 1.
Table 1.
Relevant Characteristics of the Bacterial Strains Used in This Study
| D1 | D2 | D23C9 | D2P11 | Reference | |
|---|---|---|---|---|---|
| MIC (μg/ml) | |||||
| VAN | 0.5 | 1 | 4 | 8 | 15 |
| RIF | 8 | 4 | 1 | 2 | 15 |
| DAP | 0.094 | 0.38 | 0.5 | 1 | This study |
| DAP predifussion method (mm) | 30 | 30 | 22 | 18 | This study |
| VAN PAP-AUC/PAP-AUC Mu3 | 0.98 | 1.48 | 2.81 | 5.7 | 15 |
| Phenotype | hVISA | hVISA | VISA | VISA | 15 |
| Cell wall thickness (nm) (median; range) | 26.20; 23.57–28.74 | 33.41; 27.17–39.42 | 34.07; 27.23–45.15 | 35.54; 27.75–49.49 | This study |
DAP, daptomycin; hVISA, heterogeneous vancomycin-intermediate Staphylococcus aureus; RIF, rifampin; VAN, vancomycin.
Susceptibility testing
Stability of vancomycin resistance was assayed by determining the MIC (microdilution method) according to CLSI guidelines in three independent experiments for D2 and its derived mutants after 30 and 60 days of serial passage in BHI agar without antibiotic. Daptomycin susceptibility was analyzed by prediffusion method (Neosensitabs; Rosco Diagnostica) and Etest® (AB BioMerieux, Solna, Sweden) according to the manufacturer's instructions.
Transmission electron microscopy
Strains were grown in the BHI broth, until they reached an OD620nm = 0.5–0.7. Preparation and examination of S. aureus cells by transmission electron microscopy (TEM) were performed as described previously.15 Ultrathin sections of the samples were examined using a Zeiss 10C electron microscope and images analyzed with Image J 1.46r software (http://imagej.nih.gov/ij/). Cell wall thickness at nearly equatorially cut surfaces was measured (30 cells for each strain) using a magnification of 50,000 × , and the results for each strain were expressed as median and range.
Characterization of the peptidoglycan
Peptidoglycan was isolated as previously described16 and digested with mutanolysin (Sigma-Aldrich). Reaction mixture was incubated overnight at 37°C, and the soluble muropeptides were separated by reversed phase-high performance liquid Chromatography using a 3 μm ODS-Hypersil column (4.6–250 mm; Thermo Scientific), at 25°C. The compounds were detected at 210 nm. HPLC elution patterns are included in Supplementary Data 1; (Supplementary Data are available online at www.liebertpub.com/mdr) The areas of the peaks were added together, and individual peaks were expressed as a percentage of the total.
Relevant peaks were collected and lyophilized. Samples were hydrolyzed in 6 M HCl at 95°C for 16 hr. After evaporation, the dry residues were dissolved in 67 mM trisodium citrate-HCl (pH = 2.20) and injected into a Hitachi L8800 analyzer equipped with a 2620MSC-PS column (ScienceTec). The structure and purity of isolated PG fragments were confirmed by MALDI-TOF mass spectrometry (MS) on a PerSeptive Voyager-DE STR instrument (Applied Biosystems) equipped with a 337 nm laser.
Membrane purification and Penicillin Binding Proteins analysis
Membrane fractions were prepared by ultracentrifugation from cultures in the exponential phase as previously described.17,18 One hundred micrograms of membrane fraction was incubated with 25 μM of fluorescent ampicillin for 30 min to detect all PBPs.17 The labeling of PBP2A was performed by adding 200 μM ampicillin for 10 min at 37°C before incubation with the fluorescent antibiotic. Proteins were separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and then visualized with a fluorescence scanner (Molecular Imager FX PhosphoImager; BioRad).
Autolysis assay
Strains were grown in the BHI broth until they reached an OD620nm = 0.5–0.7, chilled on ice, harvested by centrifugation (3,000 g, for 10 min at 4°C), and washed with ice-cold water. Cells were then suspended to an OD620nm = 1.0 either in the lysis buffer (50 mM glycine buffer, pH = 8.00, containing 0.01% Triton X-100) as previously described19 or in 50 mM phosphate buffer, pH = 7.00, and incubated at 37°C with agitation. Autolysis was evaluated by following OD620nm.
PBP2 and PBP4 transcription analysis by RTqPCR
The transcription of PBP2 and PBP4 coding genes was evaluated by reverse transcription quantitative real-time polymerase chain reaction (RTqPCR) using primers described in Supplementary Data 2. The effect of vancomycin at subinhibitory concentration (¼ MIC) on the pbp2 and pbp4 transcription was further analyzed. RNA was isolated by triplicate, in three independent experiments, from bacteria grown in the BHI broth, in the presence of VAN (¼ MIC) or without antibiotic, until they reached an OD620nm = 0.5–0.7. Cells were collected by centrifugation and bacterial pellets were treated with lysozyme 15 mg/ml (Sigma-Aldrich) in 10 mM Tris-HCl (pH = 8.00), 0.1 mM EDTA, for 1 hr at 37°C and RNA was extracted using TRIZOL® Reagent (Invitrogen) with the Pure Link® RNA Mini Kit (AMBION) according to manufacturer's recommendations. RNA was quantified using NanoDrop™ 1000 spectrophotometer (Thermo Scientific) and treated with 3 U/μl DNAse for 1 hr at 37°C (RQ1 RNase free DNase; Promega). Reverse transcription was performed using 500 ng of RNA, 200 U of M-MLV™ Reverse Transcriptase (Invitrogen), and 50 μM Random primers (Invitrogen) according to manufacturer's recommendations. The qPCR reaction was carried out using a 1/100 dilution of cDNA, SYBR® Select Master Mix (Applied Biosystems) and the primers previously described in a 7500 Real-Time PCR System (Applied Biosystems).
The combination of gyrB and pta was used as reference genes. Cq values were converted into normalized relative quantity (NRQ) values using normalization to the geometrical average of the reference genes and the specific PCR efficiency for each gene.20–23
Statistical analysis
The NRQ values of pbp2 and pbp4 genes were converted into logarithmic values to obtain symmetrical data and compared by two-way analysis of variance (ANOVA). Multiple comparisons were performed with the Duncan post-test using Infostat Software.24 p Values of <0.05 were considered statistically significant.
Cell wall thickness values were compared with the nonparametrical Kruskal–Wallis test and Dunn post-test using Infostat Software. p Values of <0.05 were considered statistically significant.
Sequencing of candidate loci involved in VAN resistance
The complete vraTSR operon, walKR operon, graXRS operon, and the RRDR region of the rpoB gene were amplified by PCR.7,11,25–28 Purified amplicons were sequenced and compared with the S. aureus N315 genome sequence (NCBI Reference Sequence: NC_002745.2).
Results
Previously, we reported that strains isolated from the same patient (D1 and D2) and the laboratory-selected mutants (D23C9 and D2P11) displayed distinct antibiotic susceptibility patterns (Table 1). Daptomycin susceptibility was additionally explored in this study. Daptomycin prediffusion method was positive for D2P11, suggesting the potential reduction in daptomycin susceptibility for this strain. Daptomycin MIC values were higher for the derived mutants D2P11 and D23C9 compared to the parental strain, but all of them are classified as susceptible according to CLSI breakpoint (Table 1).
The stability of the hVISA/VISA phenotype was explored by the determination of vancomycin MIC after daily passages in the drug-free medium. While vancomycin MIC value remained unchanged in D2 and D23C9 mutant, the MIC of mutant D2P11 decreased twice (to 2 μg/ml) after 60 daily passages in the drug-free medium, suggesting some instability in its phenotype.
We compared D1 cell wall thickness with its derived mutants: the observed values were significantly different (p < 0.0001, Kruskal–Wallis). Increased cell wall thickness was detected by TEM in D2 and its derived mutants when compared to the D1 strain (p < 0.05, Dunn) (Table 1).
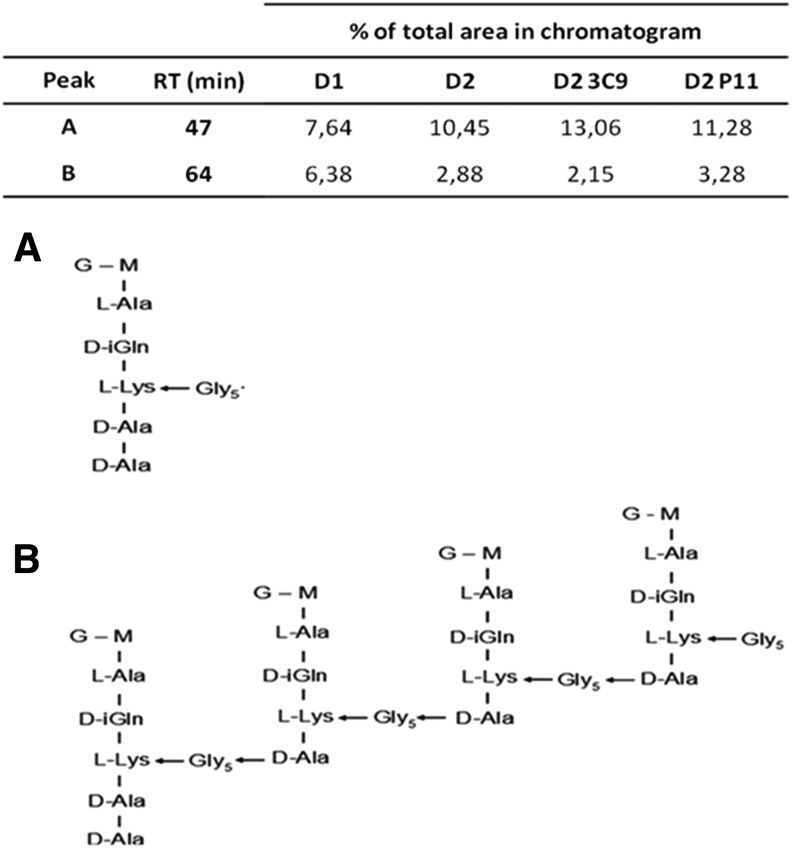
Purified peptidoglycan was prepared from the four strains, digested with M1 muramidase, and analyzed by reversed-phase HPLC. The muropeptide composition of D2, D23C9, and D2P11 showed an increased proportion in pentaglycine -disaccharide pentapeptide monomer together with a decreased proportion of the tetramer muropeptide when chromatograms were compared to D1 (Fig. 1). The difference was major in D23C9 mutant (Fig. 1).
FIG. 1.
HPLC muropeptide profile analysis. Area under curve of relevant peaks (expressed as percentage of the total area), and proposed structures for muropeptides corresponding to peaks: (A) eluted at RT 47 min and (B) eluted at RT 64 min, after MS amino acid analysis. HPLC, high performance liquid chromatography; MS, mass spectrometry; RT, retention time.
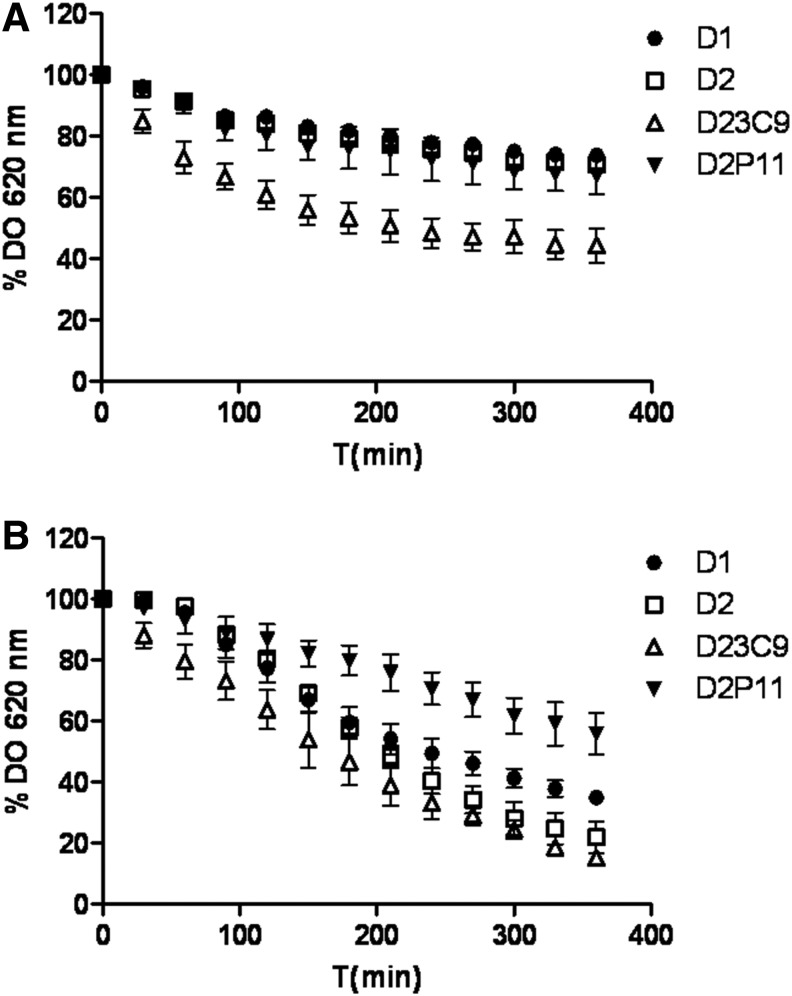
Moreover, mutant D23C9 also exhibited an enhanced autolysis rate both in the phosphate buffer and lysis buffer containing Triton X-100 (an inductor of autolysis), while a reduced autolytic activity in the lysis buffer was observed in D2P11 mutant (Fig. 2).
FIG. 2.
Autolysis assay performed using (A) 50 mM phosphate buffer pH = 7.00 and (B) lysis buffer (0.01% Triton X-100, 5 mM glycine pH = 8.00). Data were expressed as the percent loss of OD620nm at the indicated times compared to the zero time point. Each data point represents the mean and standard deviation from three independent experiments.
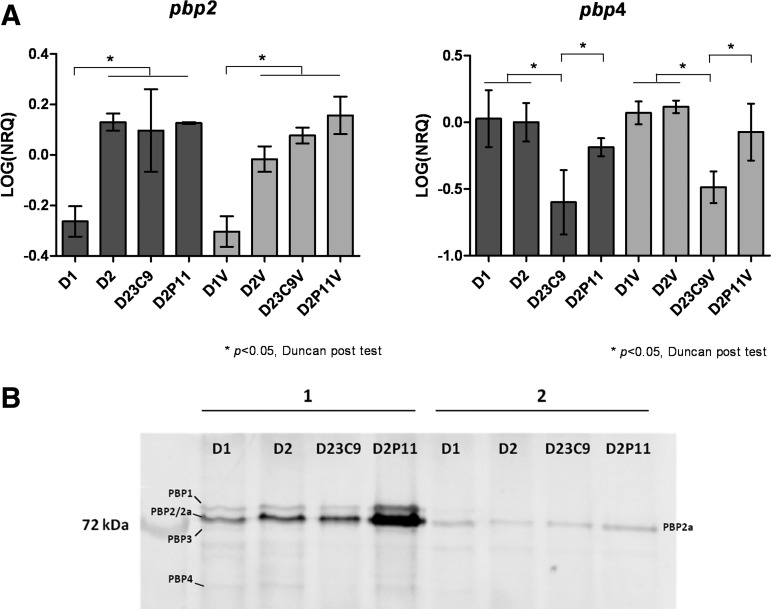
RTqPCR was performed to evaluate changes in the transcription of pbp2 and pbp4 genes, involved in peptidoglycan synthesis (Fig. 3A). The effect of subinhibitory concentrations of vancomycin (1/4 MIC) on the transcription of these genes was further analyzed. Transcription levels of the pbp2 and pbp4 genes were significantly different in the four strains (p = 0.013 and p = 0.040 respectively, two-way ANOVA). A higher pbp2 expression was detected in strains D2, D23C9, and D2P11 when compared to strain D1 (p < 0.05, Duncan post-test). This observation correlates with PBPs pattern analysis (Fig. 3B). In addition, mutant D23C9 showed a decreased pbp4 transcription level when compared with the other strains (p < 0.05, Duncan post-test). Nevertheless, variations in transcription profile were independent of the presence of subinhibitory concentrations of vancomycin (1/4 MIC) (p = 0.4026 and p = 0.3985, for pbp2 and pbp4, respectively, two-way ANOVA).
FIG. 3.
Expression of penicillin binding proteins. (A) Transcription levels of pbp2 and pbp4 genes by RTqPCR. Strains named with “V” were incubated with VAN (¼ MIC). NRQ, normalized relative quantity. Bars represent the media and the standard error of three determinations. A two-way analysis of variance was conducted to analyze the effect of two variables: strain and vancomycin treatment. Expression levels of pbp2 and pbp4 genes were significantly different (p = 0.0013 and p = 0.040) and were not affected by vancomycin treatment (p = 0.4026 and p = 0.3985, for pbp2 and pbp4, respectively). Horizontal bars show the differences detected by Duncan post-test. (B) PBPs pattern by 10% SDS-PAGE (100 μg of protein in each lane). (1) Proteins incubated 30 min at 37°C with 25 μM fluorescent ampicillin. (2) Proteins incubated 10 min at 37°C with 200 μM ampicillin followed by 30 min at 37°C with 25 μM fluorescent ampicillin. PBPs were visualized as described in Materials and Methods section. RTqPCR, reverse transcription quantitative real-time polymerase chain reaction; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
To further investigate genetic differences in candidate loci involved in hVISA/VISA phenotype, the complete vraTSR, walKR, and graXRS operons were amplified by PCR and sequenced. No mutations were found in these loci. In addition, the RRDR region of the rpoB gene was analyzed. The four strains carried a mutation leading to the amino acid substitution H481N, which was previously reported in clinical VISA strains.29
Discussion
HPLC-MS analysis of peptidoglycan muropeptides showed an increased proportion of monomeric muropeptides and a concomitant decrease in the proportion of tetrameric muropeptide in D2 and derived mutants when compared to the original strain D1. This fact has been described in hVISA and VISA strains, where a decrease in the peptidoglycan cross-linking degree has been shown.2 A much more pronounced decreased cross-linking was observed in mutant D23C9, as per the proportion of monomer muropeptides observed in the HPLC chromatogram. This observation could be related to a lower pbp4 gene transcription level observed by RTqPCR, and is in agreement with the reduced cross-linking associated with a loss of PBP4 previously reported in the strain MW2.30 In addition, strain D2 and its derived mutants showed an increase in cell wall thickness associated to an increased pbp2 gene expression. The changes observed in the transcription profile of pbp2 and pbp4 genes does not seem to be triggered by subinhibitory concentrations of vancomycin (1/4 MIC) as revealed by RTqPCR.
In the light of these results, our findings point out that the development of the VISA phenotype is related to a modification of peptidoglycan metabolism involving at least PBP2 and PBP4, as described by other authors.2,15,19,31
It has been described that VISA phenotype might be achieved by accumulation of different mutations in other genes related to cell wall metabolism.3 Unlike results reported by other research groups, no mutations were found in the vraTSR, graXRS, or walKR operons (related to cell wall synthesis and homeostasis). Point mutations in several key genes that have been shown to contribute to vancomycin resistance, or even mutations in other loci implied in resistance to other antimicrobial families, could not be disregarded and perhaps explain the changes in antibiotic susceptibility profile of D2, D23C9, and D2P11.
Several authors have reported an increased resistance to daptomycin together with a decrease in vancomycin susceptibility.32–34 In this study, the VISA mutant D2P11 exhibited a twofold increase in daptomycin MIC value compared to the hVISA parental strain D2. This observation correlates with the reduction in the inhibition zone of D2P11 compared to D2 in the prediffusion assay. Nevertheless, all the strains remain daptomycin susceptible according to CLSI breakpoints.
It was described that vancomycin pressure (both in vitro and in vivo) is associated with changes in rifampin resistance.10,29 The VISA mutants described herein present a reduction in rifampin MIC value when compared to the hVISA clinical isolates D1 and D2 (rifampin resistant). However, the rpoB sequence was the same across all isolates. The mechanism by which rifampin resistance was reduced in these strains is yet to be determined.
Different features have been described in association to VISA strains such as reversion to a more susceptible phenotype, alteration in agr locus function, reduced autolysis rate, and lower expression of PBP4 leading to the development of reduced susceptibility to vancomycin.2,4 In this study, VISA phenotype of the mutant D23C9 was stable, displayed enhanced autolysis rate, decreased pbp4 gene expression, and a previously described loss of agr function14; on the other hand, the D2P11 mutant, selected by an independent experiment, showed a nonstable VISA phenotype, reduced autolysis rate (in lysis buffer containing Triton X-100), and showed no changes in pbp4 gene expression or agr function when compared to the parental strain D2.
In Argentina, MRSA strains belonging to CC5 and CC30 are within the most prevalent lineages both in hospital and community settings.35,36 It was described that some of the changes reported in VISA strains might be related/linked to specific MRSA genetic lineages.2 However, this study shows that a single genetic background (CC5-ST100) under vancomycin pressure could render VISA strains with differential features and a comparable level of vancomycin resistance. A similar observation was described by Vidaillac et al. in laboratory mutants belonging to ST105 (which also forms part of CC5).37 Therefore, it might be possible to select different VISA phenotypes from the same genetic background. This concept might explain the observed differences in D23C9 and D2P11 mutants.
The factors implied in the development of hVISA/VISA are probably diverse, and this perhaps might explain the diversity of mutations observed in strains reported from around the world, whose phenotypes are comparable, but not completely identical. Since mutations in a heterogeneous population (such as hVISA) might be randomly selected, different pathways could lead to the VISA phenotype. The pleiotropic effects of vancomycin selective pressure might lead to reduced susceptibility in association with changes in peptidoglycan metabolism.
Supplementary Material
Acknowledgments
This work was supported, in part, by grants from University of Buenos Aires, Argentina (UBACYT 2014–2017 20020130100381BA), Agencia Nacional de Promoción Científica y Tecnológica (PICT 2362), and Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) (PIP CONICET N°: 11220110100707CO) to M.M.; and BELSPO, IAP 7/44 iPROS to B.J. The bilateral cooperation was supported by scientific agreement between the Belgian Fund for Scientific Research and the Ministry of Science, Technology and Productive Innovation (MINCyT/FRS-FNRS, BE 0907). M.M. is member of “Carrera del Investigador” of CONICET. SDiG is a postdoctoral fellow of CONICET.
Disclosure Statement
No competing financial interests exist.
References
- 1.Hiramatsu K. 1998. The emergence of Staphylococcus aureus with reduced susceptibility to vancomycin in Japan. Am. J. Med. 104:7S–10S [DOI] [PubMed] [Google Scholar]
- 2.Howden B.P., Davies J.K., Johnson P.D., Stinear T.P., and Grayson M.L. 2010. Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 23:99–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howden B.P., Peleg A.Y., and Stinear T.P. 2014. The evolution of vancomycin intermediate Staphylococcus aureus (VISA) and heterogenous-VISA. Infect. Genet. Evol. 21:575–582 [DOI] [PubMed] [Google Scholar]
- 4.Boyle-Vavra S., Berke S.K., Lee J.C., and Daum R.S. 2000. Reversion of the glycopeptide resistance phenotype in Staphylococcus aureus clinical isolates. Antimicrob. Agents Chemother. 44:272–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakoulas G., Moellering R.C., Jr., and Eliopoulos G.M. 2006. Adaptation of methicillin-resistant Staphylococcus aureus in the face of vancomycin therapy. Clin. Infect. Dis. 42 Suppl. 1:S40–S50 [DOI] [PubMed] [Google Scholar]
- 6.Galbusera E., Renzoni A., Andrey D.O., Monod A., Barras C., Tortora P., Polissi A., and Kelley W.L. 2011. Site-specific mutation of Staphylococcus aureus VraS reveals a crucial role for the VraR-VraS sensor in the emergence of glycopeptide resistance. Antimicrob. Agents Chemother. 55:1008–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howden B.P., Stinear T.P., Allen D.L., Johnson P.D., Ward P.B., and Davies J.K. 2008. Genomic analysis reveals a point mutation in the two-component sensor gene graS that leads to intermediate vancomycin resistance in clinical Staphylococcus aureus. Antimicrob. Agents Chemother. 52:3755–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuo M., Cui L., Kim J., and Hiramatsu K. 2013. Comprehensive identification of mutations responsible for heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA)-to-VISA conversion in laboratory-generated VISA strains derived from hVISA clinical strain Mu3. Antimicrob. Agents Chemother. 57:5843–5853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shoji M., Cui L., Iizuka R., Komoto A., Neoh H.M., Watanabe Y., Hishinuma T., and Hiramatsu K. 2011. walK and clpP mutations confer reduced vancomycin susceptibility in Staphylococcus aureus. Antimicrob. Agents Chemother. 55:3870–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watanabe Y., Cui L., Katayama Y., Kozue K., and Hiramatsu K. 2011. Impact of rpoB mutations on reduced vancomycin susceptibility in Staphylococcus aureus. J. Clin. Microbiol. 49:2680–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jansen A., Turck M., Szekat C., Nagel M., Clever I., and Bierbaum G. 2007. Role of insertion elements and yycFG in the development of decreased susceptibility to vancomycin in Staphylococcus aureus. Int. J. Med. Microbiol. 297:205–215 [DOI] [PubMed] [Google Scholar]
- 12.Maki H., McCallum N., Bischoff M., Wada A., and Berger-Bachi B. 2004. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:1953–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McEvoy C.R., Tsuji B., Gao W., Seemann T., Porter J.L., Doig K., Ngo D., Howden B.P., and Stinear T.P. 2013. Decreased vancomycin susceptibility in Staphylococcus aureus caused by IS256 tempering of WalKR expression. Antimicrob. Agents Chemother. 57:3240–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Gregorio S., Fernandez S., Perazzi B., Bello N., Famiglietti A., and Mollerach M. 2016. Increase in IS256 transposition in invasive vancomycin heteroresistant Staphylococcus aureus isolate belonging to ST100 and its derived VISA mutants. Infect. Genet. Evol. 43:197–202 [DOI] [PubMed] [Google Scholar]
- 15.Hanaki H., Kuwahara-Arai K., Boyle-Vavra S., Daum R.S., Labischinski H., and Hiramatsu K. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199–209 [DOI] [PubMed] [Google Scholar]
- 16.de Jonge B.L., Chang Y.S., Gage D., and Tomasz A. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 267:11248–11254 [PubMed] [Google Scholar]
- 17.Galleni M., Lakaye B., Lepage S., Jamin M., Thamm I., Joris B., and Frere J.M. 1993. A new, highly sensitive method for the detection and quantification of penicillin-binding proteins. Biochem. J. 291(Pt 1):19–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malachowa N., and DeLeo F.R. 2010. Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 67:3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sieradzki K., and Tomasz A. 2003. Alterations of cell wall structure and metabolism accompany reduced susceptibility to vancomycin in an isogenic series of clinical isolates of Staphylococcus aureus. J. Bacteriol. 185:7103–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., and Wittwer C.T. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622 [DOI] [PubMed] [Google Scholar]
- 21.Hellemans J., Mortier G., De Paepe A., Speleman F., and Vandesompele J. 2007. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valihrach L., and Demnerova K. 2012. Impact of normalization method on experimental outcome using RT-qPCR in Staphylococcus aureus. J. Microbiol. Methods 90:214–216 [DOI] [PubMed] [Google Scholar]
- 23.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., and Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Rienzo J.A., Casanoves F., Balzarini M.G., Gonzalez L., Tablada M., and Robledo C.W. 2013. InfoStat versión 2013. Available at www.infostat.com.ar (accessed July23, 2013)
- 25.Aubry-Damon H., Soussy C.J., and Courvalin P. 1998. Characterization of mutations in the rpoB gene that confer rifampin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 42:2590–2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato Y., Suzuki T., Ida T., and Maebashi K. 2010. Genetic changes associated with glycopeptide resistance in Staphylococcus aureus: predominance of amino acid substitutions in YvqF/VraSR. J. Antimicrob. Chemother. 65:37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen H.Y., Chen C.C., Fang C.S., Hsieh Y.T., Lin M.H., and Shu J.C. 2011. Vancomycin activates sigma(B) in vancomycin-resistant Staphylococcus aureus resulting in the enhancement of cytotoxicity. PLoS One 6:e24472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schreiber F., Szekat C., Josten M., Sahl H.G., and Bierbaum G. 2013. Antibiotic-induced autoactivation of IS256 in Staphylococcus aureus. Antimicrob. Agents Chemother. 57:6381–6384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hiramatsu K., Kayayama Y., Matsuo M., Aiba Y., Saito M., Hishinuma T., and Iwamoto A. 2014. Vancomycin-intermediate resistance in Staphylococcus aureus. J. Glob. Antimicrob. Resist. 2:12. [DOI] [PubMed] [Google Scholar]
- 30.Memmi G., Filipe S.R., Pinho M.G., Fu Z., and Cheung A. 2008. Staphylococcus aureus PBP4 is essential for beta-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob. Agents Chemother. 52:3955–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sieradzki K., and Tomasz A. 1999. Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus. J. Bacteriol. 181:7566–7570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cafiso V., Bertuccio T., Spina D., Purrello S., Campanile F., Di Pietro C., Purrello M., and Stefani S. Modulating activity of vancomycin and daptomycin on the expression of autolysis cell-wall turnover and membrane charge genes in hVISA and VISA strains. PLoS One 7:e29573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui L., Tominaga E., Neoh H.M., and Hiramatsu K. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob. Agents Chemother. 50:1079–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rincon S., Panesso D., Diaz L., Carvajal L.P., Reyes J., Munita J.M., and Arias C.A. 2014. Resistance to “last resort” antibiotics in Gram-positive cocci: the post-vancomycin era. Biomedica 34 Suppl. 1:191–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez Furst M.J., de Vedia L., Fernandez S., Gardella N., Ganaha M.C., Prieto S., Carbone E., Lista N., Rotryng F., Morera G.I., Mollerach M., and Stryjewski M.E. 2013. Prospective multicenter study of community-associated skin and skin structure infections due to methicillin-resistant Staphylococcus aureus in Buenos Aires, Argentina. PLoS One 8:e78303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gardella N., Picasso R., Predari S.C., Lasala M., Foccoli M., Benchetrit G., Famiglietti A., Catalano M., Mollerach M., and Gutkind G. 2005. Methicillin-resistant Staphylococcus aureus strains in Buenos Aires teaching hospitals: replacement of the multidrug resistant South American clone by another susceptible to rifampin, minocycline and trimethoprim-sulfamethoxazole. Rev. Argent. Microbiol. 37:156–160 [PubMed] [Google Scholar]
- 37.Vidaillac C., Gardete S., Tewhey R., Sakoulas G., Kaatz G.W., Rose W.E., Tomasz A., and Rybak M.J. 2013. Alternative mutational pathways to intermediate resistance to vancomycin in methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 208:67–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.