Abstract
Variation of the HIV-1 subtype C reverse transcriptase region (RT) resulting in response to the selective pressures of drug therapy remains poorly characterized. Here, we compared the genetic variation resulting in the presence and absence of antiretroviral drug selective pressures on HIV-1 subtype C RT among nontreated and treated patients. The nucleotide variability, nonsynonymous and synonymous ratio, and the positively selected mutations were determined by comparing the RT sequences isolated at two time points among nontreated (baseline and follow-up) and treated patients (baseline and treatment failure). Compared to the nontreated patients, the intrapatient nucleotide variability, the number of nonsynonymous and synonymous substitutions was significantly higher among the treated patients. Among the mutations positively selected, the frequency of D121Y, I135R, and Q207E increased and the frequency of mutation S48T decreased significantly during treatment failure. Further studies are essential to discover the role of these mutations during treatment in HIV-1 subtype C.
The reverse transcriptase (RT) enzyme of the human immunodeficiency virus (HIV-1) converts viral genomic single-stranded RNA into double-stranded DNA. It is the essential step in the HIV-1 life cycle and therefore the RT region has been a target of antiretroviral therapy (ART). As an intrinsic property, HIV-1 RT lacks a proofreading function and this error-prone nature of RT together with the high rate of virus production sustained by HIV-1 infection in vivo contributes to the continuous generation of new viral variants.1,2 The variability is further increased by antiretroviral drugs, resulting in mutations that have a selective advantage during drug pressure.3–6 Studying the effect of drug treatment on HIV-1 variation is important in understanding the emergence of drug resistance and disease pathogenesis. Here we compared the genetic variation resulting from the presence and absence of ARV drug selective pressures on HIV-1 subtype C RT among nontreated and treated patients.
HIV-1-infected nontreated patients (n = 18) and patients treated (n = 16) for >6 months and failing the first line regimen were included. The demographic, clinical, and laboratory characteristics of the study groups are presented in Table 1. Peripheral blood samples from nontreated patients were collected during enrollment (baseline) and after a period of 8–12 months (follow-up). From the treated patients peripheral blood samples were collected at treatment failure and respective pretherapy (baseline) plasma samples that were collected before 12–16 months of therapy were retrospectively obtained from the archives (–70°C freezers). The study protocol was approved by the Institutional Review Board of the Y.R. Gaitonde Centre for AIDS Research and Education and the written informed consent was obtained from all the participants included in the study.
Table 1.
Demographic, Clinical, and Laboratory Characteristics of HIV-1-Infected ART-Treated and Nontreated Patientsa
| Characteristics | Treated (n = 16) | Nontreated (n = 18) |
|---|---|---|
| Gender | ||
| Male, n (%) | 10 (62.5) | 11 (61) |
| Female, n (%) | 6 (37.5) | 7 (39) |
| Age (years)/mean (±SD) | ||
| Male | 37 (3.9) | 33 (4) |
| Female | 33 (3) | 30 (2) |
| CD4+ T cell count (cells/μl)/median (IQR) | ||
| Baseline | 161(53–182) | 503 (366–672) |
| Follow-up | 318 (55–302) | 548 (315–885) |
| ART regimen | ||
| AZT + 3TC + NVP or EFV, n (%) | 6 (37.5) | None |
| d4T + 3TC + NVP or EFV, n (%) | 8 (50) | |
| ddI + 3TC + NVP or EFV, n (%) | 2 (12.5) |
AZT, zidovudine; ddI, didanosine; d4T, stavudine; EFV, efavirenz; NVP, nevirapine; 3TC, lamivudine; ART, antiretroviral therapy; HIV-1, human immunodeficiency virus type 1; IQR, interquartile range; SD, standard deviation.
HIV-1 RNA was isolated using the QIAamp viral RNA kit (QIAGEN, Inc., USA). HIV-1 RT (region 20–240) was amplified from cDNA using nested polymerase chain reaction (PCR) as described earlier7 with appropriate controls. Bidirectional population sequencing of purified products was done using an ABI 3100-Avant genetic analyzer (Applied Biosystems, USA). All the sequences were edited using the Seqscape software (Applied Biosystems, USA, version 2.5). The nucleotide variability and the Jukes–Cantor correction for multiple hits of the proportion of observed nonsynonymous (dN) and synonymous substitutions (dS) and its ratio (dN/dS) were determined by comparing the RT sequences isolated at two time points among nontreated (baseline and follow-up) and treated (baseline and treatment failure) patients using Syn-SCAN.8 Similarly the codon sites evolving under the influence of positive Darwinian selection were identified by comparing the RT sequences isolated at two time points among nontreated and treated patients using HyPhy with the codon substitution model MG94.9 Drug resistance mutations were identified using the Stanford HIV-1 drug resistance database (http://hivdb.stanford.edu/). The Mann–Whitney U test was used to compare the variables between the groups. The statistical analysis was performed using SPSS 13.0 statistical software (Chicago, IL). The GenBank accession numbers of the HIV-1 RT sequences described here are EU429988 through EU430023, EU545198 through EU545213, and EU545214 through EU545229.
The RT sequences from all the study subjects were subtype C and no intersubtype recombinants were observed. The intrapatient nucleotide variability [median (IQR)] of treated patients [5.3% (3.1–7.6)] was significantly higher (p < 0.03) compared to nontreated patients [1.9% (0.9–2.45)]. Similarly, the dN, dS significantly (p < 0.001) increased among treated [median (IQR): dN 0 (0–0); dS 0.02 (0–0.04)] compared to nontreated patients [median (IQR): dN 0.04 (0.02–0.04); dS 0.12 (0.05–0.15)]. Although previous studies have shown a reduction in the genetic variability of HIV during drug therapy,10–12 higher intrapatient gene variability observed among the treated patients in the present analysis could be attributed to viral escape from drug pressure, thereby enhanced replication efficiency, which in turn led to greater genetic variation.13 Several studies have found an inverse relationship between the rate of viral diversification and host disease progression,14–18 whereas others have not.19–21 The limitation of the present investigation is that the history of HIV seroconversion for the study population is not known and hence the consequence of drug treatment on the intrahost nucleotide variation could not be delineated from the duration of HIV infection. It should also be noted that earlier studies have shown that a higher selection pressure will be imposed by drug therapy22–24 and the strains resistant to nucleoside reverse transcriptase inhibitors (NRTIs) can increase HIV-1 mutation frequencies.25,26 However, in the present analysis, the ratio of dN/dS was <1 among the treated [median (IQR): 0.2 (0.08–0.25)] and nontreated patients [median (IQR): 0 (0–0)], which was similar to earlier reports.27–31 This implies that the RT region is highly conserved because of structural/functional constraints and consequently any mutations in this region will be deleterious to the virus.32,33
Among nontreated patients, two (11%) had transient drug resistance mutations associated with NRTIs and nonnucleoside reverse transcriptase inhibitors (NNRTIs) such as V108IV (6%) and Y181CY (6%), respectively, at baseline, which was observed to be wild type at follow-up. Among the NRTI mutations observed among the treated patients, M184V (62.5%) was predominant, followed by T215F/Y (19%). Mutations K219Q, M41L, D67N, and T69D was observed in 13% and K65R, K70R, V75I, V118F, and Q151M occurred in 6%. Among NNRTI mutations, Y181C (31%) was predominant followed by K103N/S, V106M, and G190A, which occurred in 25%. A98G/S and K101E were observed in 19% and 13%, respectively.
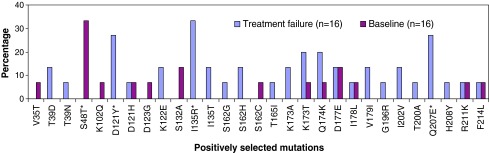
Positive selection of mutations D121Y, K122E, D123G, I135T, Q174R, I195L, I202V, Q207N, and R211K each at a frequency of 5.5% (1/18) was observed in the RT sequences amplified from nontreated patients. However, treated patients demonstrated positive selection (dN/dS > 1) of 29 mutations, among which the frequency of D121Y, I135R, and Q207E was observed to be significantly increased (p < 0.001) and the frequency of S48T significantly decreased (p < 0.001) during treatment failure compared to the baseline (Fig. 1). A study of B/C recombinants in China by Liao et al.34 has shown that both D121Y and I135R are the common subtype C and subtype B polymorphisms, respectively. Moreover, in the same study, position 207 was also found to be a polymorphic site. However, mutations at position 135 and 207 have been reported to be associated with reduced susceptibility to nevirapine and zidovudine, respectively, in subtype B and D viruses.35–39 Kantor et al.40 reported that treatment has a greater effect at position 121 in subtype C viruses. Previous investigations have revealed that the positive selection of beneficial mutations is an important mechanism in HIV evolution, both for drug resistance41,42 and immune escape.43–47 Even though mutations at position 135 and 207 in RT are known to be associated with a reduction in susceptibility to nevirapine and zidovudine in HIV-1 subtype B and D viruses,35–39 it is not clear if D121Y is related to drug resistance. Mutation S48T, observed to be negatively associated with treatment failure, is a common polymorphism that occurred at >50% of drug-naive patients in the present analysis similar to other studies of subtype C viruses.7,48–50 This finding shows that mutation S48T might be deleterious in terms of viral replication in the presence of resistance mutations, thus increasing the level of the genetic barrier to drug resistance.
FIG. 1.
Frequency of positively selected mutations in the RT sequences at baseline and treatment failure among the ART-treated patients. *p < 0.05 (Mann–Whitney U test). (Color image can be found at www.liebertonline.com/aid).
In conclusion, the present analysis reveals the higher selection pressure and genetic variability of HIV-1 subtype C RT during ART. Among the treated patients, a few positively selected mutations in RT not yet included as a candidate for drug resistance increased in frequency during treatment failure. A limitation of this study is the smaller sample size and population sequencing, which warrant further investigations that may provide new perspectives concerning the existence of polymorphisms that could influence the development of immune escape or drug resistance in HIV-1 subtype C viruses.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Bebenek K. Abbotts J. Roberts JD, et al. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989;264:16948–16956. [PubMed] [Google Scholar]
- 2.Mansky LM. Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Havlir DV. Eastman S. Gamst A, et al. Nevirapine- resistant human immunodeficiency virus: Kinetics of replication and estimated prevalence in untreated patients. J Virol. 1996;70:7894–7899. doi: 10.1128/jvi.70.11.7894-7899.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunthard HF. Wong JK. Ignacio CC, et al. Emergence of drug resistance in different tissue compartments in 10 patients using population-based sequencing after 1 year of potent antiretroviral therapy. International Workshop on HIV Drug Resistance, Treatment Strategies and Eradication, St. Petersburg, FL, 1997 (International Medical Press, London, 1997). Antiviral Ther. 1997;2:67. [Google Scholar]
- 5.Gonzales MJ. Wu TD. Taylor J, et al. Extended spectrum of HIV-1 reverse transcriptase mutations in patients receiving multiple nucleoside analog inhibitors. AIDS. 2003;17:91–99. doi: 10.1097/01.aids.0000050860.71999.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clavel F. Hance AJ. HIV drug resistance. N Engl J Med. 2004;350:1023–1035. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 7.Balakrishnan P. Kumarasamy N. Kantor R, et al. HIV type 1 genotypic variation in an antiretroviral treatment-naive population in southern India. AIDS Res Hum Retroviruses. 2005;21:301–305. doi: 10.1089/aid.2005.21.301. [DOI] [PubMed] [Google Scholar]
- 8.Gonzales MJ. Dugan JM. Shafer RW. Synonymous-non-synonymous mutation rates between sequences containing ambiguous nucleotides (Syn-SCAN) Bioinformatics. 2002;18(6):886–887. doi: 10.1093/bioinformatics/18.6.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pond SLK. Frost SDW. Muse SV. HyPhy: Hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 10.Ibanez A. Clotet B. Martinez MA. Human immunodeficiency virus type 1 population bottleneck during indinavir therapy causes a genetic drift in the env quasispecies. J Gen Virol. 2000;81:85–95. doi: 10.1099/0022-1317-81-1-85. [DOI] [PubMed] [Google Scholar]
- 11.Kristiansen TB. Pedersen AG. Eugen-Olsen J, et al. Genetic evolution of HIV in patients remaining on a stable HAART regimen despite insufficient viral suppression. Scand J Infect Dis. 2005;37:890–901. doi: 10.1080/00365540500333491. [DOI] [PubMed] [Google Scholar]
- 12.Shen L. Siliciano RF. Viral reservoirs, residual viremia, and the potential of highly active antiretroviral therapy to eradicate HIV infection. J Allergy Clin Immunol. 2008;22:22–28. doi: 10.1016/j.jaci.2008.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann D. Seebach J. Cosma A, et al. Therapeutic vaccination reduces HIV sequence variability. FASEB J. 2008;22:437–444. doi: 10.1096/fj.06-7975com. [DOI] [PubMed] [Google Scholar]
- 14.Wolinsky SM. Korber BTM. Neumann AU, et al. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;27:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- 15.Ganeshan S. Dickover RE. Korber BM, et al. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J Virol. 1997;71:663–677. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delwart EL. Pan H. Sheppard HW, et al. Slower evolution of human immunodeficiency virus type 1 quasispecies during progression to AIDS. J Virol. 1997;71:7498–7508. doi: 10.1128/jvi.71.10.7498-7508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williamson S. Perry SM. Bustamante CD, et al. A statistical characterization of consistent patterns of human immunodeficiency virus evolution within infected patients. Mol Biol Evol. 2005;22:456–468. doi: 10.1093/molbev/msi029. [DOI] [PubMed] [Google Scholar]
- 18.Lemey P. Pond SLK. Drummond AJ, et al. Synonymous substitution rates predict HIV disease progression as a result of underlying replication dynamics. PLoS Comput Biol. 2007;3:e29. doi: 10.1371/journal.pcbi.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markham RB. Wang WC. Weisstein AE, et al. Patterns of HIV-1 evolution in individuals with differing rates of CD4 T cell decline. Proc Natl Acad Sci USA. 1998;95:12568–12573. doi: 10.1073/pnas.95.21.12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McNearney T. Hornickova Z. Markham R, et al. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc Natl Acad Sci USA. 1992;89:10247–10251. doi: 10.1073/pnas.89.21.10247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HY. Perelson AS. Park SC, et al. Dynamic correlation between intrahost HIV-1 quasispecies evolution and disease progression. PLoS Comput Biol. 2008;4(12):e1000240. doi: 10.1371/journal.pcbi.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finzi D. Blankson J. Siliciano JD, et al. Latent infection of CD4 (+) T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5(5):512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 23.Ribeiro RM. Bonhoeffer S. Nowak MA. The frequency of resistant mutant virus before antiviral therapy. AIDS. 1998;12(5):461–465. doi: 10.1097/00002030-199805000-00006. [DOI] [PubMed] [Google Scholar]
- 24.Rouzine IM. Coffin JM. Evolution of human immunodeficiency virus under selection and weak recombination. Genetics. 2005;170(1):7–18. doi: 10.1534/genetics.104.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mansky LM. Bernard LC. 3'-Azido-3'-deoxythymidine (AZT) and AZT-resistant reverse transcriptase can increase the in vivo mutation rate of human immunodeficiency virus type 1. J Virol. 2000;74:9532–9539. doi: 10.1128/jvi.74.20.9532-9539.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansky LM. Pearl DK. Gajary LC. Combination of drugs and drug-resistant reverse transcriptase results in a multiplicative increase of human immunodeficiency virus type 1 mutant frequencies. J Virol. 2002;76:9253–9259. doi: 10.1128/JVI.76.18.9253-9259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cornelissen M. van den Burg R. Zorgdrager F, et al. Pol gene diversity of five human immunodeficiency virus type 1 subtypes: Evidence for naturally occurring mutations that contribute to drug resistance, limited recombination patterns, and common ancestry for subtypes B and D. J Virol. 1997;71:6348–6358. doi: 10.1128/jvi.71.9.6348-6358.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunthard HF. Leigh-Brown AJ. D'Aquila RT, et al. Higher selection pressure from antiretroviral drugs in vivo results in increased evolutionary distance in HIV-1 pol. Virology. 1999;259:154–165. doi: 10.1006/viro.1999.9774. [DOI] [PubMed] [Google Scholar]
- 29.De S Leal E. Holmes EC. Zanotto PM. Distinct patterns of natural selection in the reverse transcriptase gene of HIV-1 in the presence and absence of antiretroviral therapy. Virology. 2004;325:181–191. doi: 10.1016/j.virol.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Ceccherini-Silberstein F. Gago F. Santoro M, et al. High sequence conservation of human immunodeficiency virus type 1 reverse transcriptase under drug pressure despite the continuous appearance of mutations. J Virol. 2005;79:10718–10729. doi: 10.1128/JVI.79.16.10718-10729.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ceccherini-Silberstein F. Svicher V. Sing T, et al. Characterization and structural analysis of novel mutations in human immunodeficiency virus type 1 reverse transcriptase involved in the regulation of resistance to nonnucleoside inhibitors. J Virol. 2007;81:11507–11519. doi: 10.1128/JVI.00303-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Overbaugh J. Bangham CR. Selection forces and constraints on retroviral sequence variation. Science. 2001;292:1106–1109. doi: 10.1126/science.1059128. [DOI] [PubMed] [Google Scholar]
- 33.Holmes EC. 2003 Error thresholds, the constraints to RNA virus evolution. Trends Microbiol. 11:543–6. doi: 10.1016/j.tim.2003.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao L. Xing H. Li X, et al. Genotypic analysis of the protease and reverse transcriptase of HIV type 1 isolates from recently infected injecting drug users in western China. AIDS Res Hum Retroviruses. 2007;23(8):1062–1065. doi: 10.1089/aid.2007.0050. [DOI] [PubMed] [Google Scholar]
- 35.Brown A. Precious H. Whitcomb JM, et al. Reduced susceptibility of human immunodeficiency virus type 1 (HIV-1) from patients with primary HIV infection to nonnucleoside reverse transcriptase inhibitors is associated with variation at novel amino acid sites. J Virol. 2000;74:10269–10273. doi: 10.1128/jvi.74.22.10269-10273.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoeckli T. MaWhinney S. Uy J, et al. Phenotypic, genotypic analysis of biologically cloned human immunodeficiency virus type 1 isolates from patients treated with zidovudine, lamivudine. Antimicrob Agents Chemother. 2002;46:4000–4003. doi: 10.1128/AAC.46.12.4000-4003.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vavro C. Florance A. Irlbeck D, et al. GlaxoSmithKline, Research Triangle Park, NC, USA; Mutations at codon 135 at baseline are associated with the accumulation of NNRTI-resistance mutations while on EFV-containing regimens; San Francisco, CA. 2004. Presented at 11th CROI. [Google Scholar]
- 38.Gao Y. Paxinos E. Galovich J, et al. Characterization of a subtype D human immunodeficiency virus type 1 isolate that was obtained from an untreated individual and that is highly resistant to nonnucleoside reverse transcriptase inhibitors. J Virol. 2004;78(10):5390–5401. doi: 10.1128/JVI.78.10.5390-5401.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu J. Whitcomb J. Kuritzkes D. Effect of the Q207D mutation in HIV type 1 reverse transcriptase on zidovudine susceptibility and replicative fitness. J Acquir Immune Defic Syndr. 2005;40:20–23. doi: 10.1097/01.qai.0000172369.82456.36. [DOI] [PubMed] [Google Scholar]
- 40.Kantor R. Katzenstein DA. Efron B, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: Results of a global collaboration. PLoS Med. 2005;2:e112. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frost SD. Günthard HF. Wong JK, et al. Evidence for positive selection driving the evolution of HIV-1 env under potent antiviral therapy. Virology. 2001;284:250–258. doi: 10.1006/viro.2000.0887. [DOI] [PubMed] [Google Scholar]
- 42.Chen L. Lee C. Distinguishing HIV-1 drug resistance, accessory, and viral fitness mutations using conditional selection pressure analysis of treated versus untreated patient samples. Biol Direct. 2006;1:14. doi: 10.1186/1745-6150-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z. Nielsen R. Goldman N, et al. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross HA. Rodrigo A. Immune mediated positive selection drives human immunodeficiency virus type 1 molecular variation and predicts disease duration. J Virol. 2002;76:11715–11719. doi: 10.1128/JVI.76.22.11715-11720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore CB. John M. James IR, et al. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439–1443. doi: 10.1126/science.1069660. [DOI] [PubMed] [Google Scholar]
- 46.Yusim K. Kesmir C. Gaschen B, et al. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J Virol. 2002;76:8757–8768. doi: 10.1128/JVI.76.17.8757-8768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williamson S. Adaptation in the env gene of HIV-1 and evolutionary theories of disease progression. Mol Biol Evol. 2003;20:1318–1325. doi: 10.1093/molbev/msg144. [DOI] [PubMed] [Google Scholar]
- 48.Handema R. Terunuma H. Kasolo F, et al. Prevalence of drug-resistance-associated mutations in antiretroviral drug-naive Zambians infected with subtype C HIV-1. AIDS Res Hum Retroviruses. 2003;19:151–160. doi: 10.1089/088922203762688667. [DOI] [PubMed] [Google Scholar]
- 49.Deshpande A. Recordon-Pinson P. Deshmukh R, et al. Molecular characterization of HIV type 1 isolates from untreated patients of Mumbai (Bombay), India, and detection of rare resistance mutations. AIDS Res Hum Retroviruses. 2004;20:1032–1035. doi: 10.1089/aid.2004.20.1032. [DOI] [PubMed] [Google Scholar]
- 50.Sen S. Tripathy SP. Chimanpure VM, et al. Human immunodeficiency virus type 1 drug resistance mutations in peripheral blood mononuclear cell proviral DNA among antiretroviral treatment-naive and treatment-experienced patients from Pune, India. AIDS Res Hum Retroviruses. 2007;23:489–497. doi: 10.1089/aid.2006.0221. [DOI] [PubMed] [Google Scholar]