Abstract
Pulmonary disease caused by nontuberculous mycobacteria (NTM) is increasing worldwide. Mycobacterium avium is the most clinically significant NTM species in humans and animals, and comprises four subspecies: M. avium subsp. avium (MAA), M. avium subsp. silvaticum (MAS), M. avium subsp. paratuberculosis (MAP), and M. avium subsp. hominissuis (MAH). To improve our understanding of the genetic landscape and diversity of M. avium and its role in disease, we performed a comparative genome analysis of 79 M. avium strains. Our analysis demonstrated that MAH is an open pan-genome species. Phylogenetic analysis based on single nucleotide variants showed that MAH had the highest degree of sequence variability among the subspecies, and MAH strains isolated in Japan and those isolated abroad possessed distinct phylogenetic features. Furthermore, MAP strains, MAS and MAA strains isolated from birds, and many MAH strains that cause the progression of pulmonary disease were grouped in each specific cluster. Comparative genome analysis revealed the presence of genetic elements specific to each lineage, which are thought to be acquired via horizontal gene transfer during the evolutionary process, and identified potential genetic determinants accounting for the pathogenic and host range characteristics of M. avium.
Whole-genome sequencing and comparative genome analysis of a large number of related strains have recently emerged as a cost-effective and convenient approach for addressing many microbiological questions, such as evolution, outbreaks, antibiotic resistance, and pathogenicity. This approach has been applied to several pathogens, such as Staphylococcus aureus1, Streptococcus pneumonias2, Vibrio cholera3, and Propionibacterium acnes4,5.
Nontuberculous mycobacteria (NTM) are increasingly recognized as an important cause of morbidity in many countries, including the United States and Japan6,7,8,9,10. NTM infection is thought to be caused by NTM that reside in the environment, including in garden soil and bathrooms11,12,13. Pulmonary disease caused by NTM, which is both intractable and infectious, has variable clinical manifestations. Although some patients remain stable without treatment, others show signs of deterioration despite long-term multidrug therapy9,14,15. The causative NTM species of pulmonary disease vary by country. In Japan, the pulmonary disease-causing NTM with the highest incidence is Mycobacterium avium, followed by M. intracellulare, M. kansasii, and M. abscessus; today, the incidence per 100,000 population is estimated to be 14.78.
Among NTM species, M. avium is the most clinically significant species in humans and animals and comprises four subspecies that have specific pathogenic and host range characteristics as follows: M. avium subsp. avium (MAA) and M. avium subsp. silvaticum (MAS) are avian pathogens; M. avium subsp. paratuberculosis (MAP) causes John’s disease in ruminants; and M. avium subsp. hominissuis (MAH) infects mainly pigs and humans16,17,18. MAH is the causative pathogen of two main types of disease in humans: disseminated disease in immunocompromised hosts such as individuals infected with human immunodeficiency virus (HIV), and pulmonary disease in individuals without systemic immunosuppression9. MAA and MAH have also been isolated from other animals, such as deer, cattle, and horses19. MAP is transmitted into herds via the fecal-oral route through pastures or water contaminated with the feces of infected animals18. In contrast, MAS infects wood pigeons almost exclusively and is taxonomically very close to MAA17. However, the genetic differences among the four subspecies are still unknown.
The mechanisms involved in the development and exacerbation of pulmonary NTM disease have yet to be elucidated, but are possibly the result of both host and bacterial factors. Maekura et al. reported that pulmonary disease patients with serotype 4 MAH strains have significantly poorer prognoses than those with other serotypes20. In recent studies, variable number tandem repeats (VNTR) typing analysis of isolates from patients with pulmonary NTM disease revealed that isolates from patients with progressive pulmonary disease and those with stable pulmonary disease are clustered differently21,22,23, which suggests the involvement of bacterial factors in the progression of pulmonary NTM disease.
In our previous study, we determined the complete genome sequence of strain TH135 isolated from a serious case with worsening pulmonary MAH disease24, and further demonstrated the presence of a circular plasmid, pMAH13525. This novel plasmid consists of 194,711 nucleotides and has 164 coding sequences (CDSs), some of which encode proteins involved in the pathogenicity of mycobacteria and their resistance to antimicrobial agents. The screening of MAH isolates from humans and pigs for genes located on pMAH135 revealed that these genes are more commonly detected in isolates from patients with pulmonary MAH disease than in HIV-positive patients. However, the genes are almost entirely absent in isolates from pigs, suggesting that pMAH135 influences not only the pathological manifestations of MAH disease, but also the host specificity.
In this study, to improve our understanding of the genetic landscape and diversity of M. avium and its role in disease, we performed a comparative genome analysis of 79 M. avium strains including 46 novel MAH genomes.
Results
M. avium strains and general genomic features
To investigate the genomic features of MAH that cause the progression of pulmonary disease, we sequenced the genomes of 46 MAH isolates from 17 patients with progressive disease and 29 patients with stable disease, in addition to the previously determined complete genome sequence of MAH strain TH13524, which was used as a reference genome in the analysis. The average genome size of 46 novel MAH isolates was 5.351 Mb (ranging from 4.981 to 5.895 Mb), and the G + C content was 68.9% (ranging from 68.5 to 69.3%), with 370 contigs on average (ranging from 162 to 1,062) (Table 1). The relationship between genome size and G + C content was examined in these strains (Supplementary Fig. S1). Interestingly, a negative correlation (correlation coefficient −0.9491; p < 0.0001), wherein the G + C content became smaller as the genome size became larger. This indicates that MAH strains with a large genome might have evolved through acquisition of exogenous genes via phages, transposons, or other integrative and conjugative elements.
Table 1. General feature of 75 M. avium genomes.
| Genome no. | Strain | Subspeciesa | Source | Clusterb | Genome size (Mb) | GC (%) | No. of contigs |
|---|---|---|---|---|---|---|---|
| 1 | DH-6 | MAH | stable disease patientc | Ib | 5.895 | 68.5 | 537 |
| 2 | DH-7 | MAH | stable disease patient | Ib | 5.077 | 69.2 | 276 |
| 3 | HF-1 | MAH | stable disease patient | Ib | 5.252 | 68.9 | 313 |
| 4 | HF-3 | MAH | stable disease patient | Ib | 5.258 | 68.9 | 399 |
| 5 | IH-065 | MAH | stable disease patient | Ib | 5.122 | 69.1 | 484 |
| 6 | IH-068 | MAH | stable disease patient | IIa | 5.215 | 69.1 | 287 |
| 7 | IH-145 | MAH | stable disease patient | Ib | 5.218 | 69.0 | 342 |
| 8 | IH-149 | MAH | stable disease patient | Ib | 5.211 | 69.1 | 366 |
| 9 | IH-217 | MAH | stable disease patient | Ia | 5.331 | 69.1 | 162 |
| 10 | IH-483 | MAH | stable disease patient | Ia | 5.753 | 68.7 | 415 |
| 11 | IH-550 | MAH | stable disease patient | Ib | 5.192 | 69.1 | 419 |
| 12 | IH-532 | MAH | stable disease patient | Ib | 5.440 | 68.9 | 327 |
| 13 | IH-560 | MAH | stable disease patient | Ib | 5.384 | 68.9 | 328 |
| 14 | IH-660 | MAH | stable disease patient | Ib | 5.359 | 68.9 | 552 |
| 15 | IH-687 | MAH | stable disease patient | Ib | 5.090 | 69.1 | 272 |
| 16 | IH-788 | MAH | stable disease patient | Ib | 5.397 | 68.6 | 1,062 |
| 17 | IH-799 | MAH | stable disease patient | Ib | 5.106 | 69.1 | 402 |
| 18 | IH-801 | MAH | stable disease patient | Ia | 5.335 | 69.0 | 376 |
| 19 | IH-945 | MAH | stable disease patient | Ib | 5.090 | 69.1 | 510 |
| 20 | Kin-16 | MAH | stable disease patient | Ib | 5.217 | 69.0 | 268 |
| 21 | Kin-27 | MAH | stable disease patient | Ib | 5.127 | 69.1 | 347 |
| 22 | Kin-28 | MAH | stable disease patient | Ib | 5.390 | 68.9 | 327 |
| 23 | MK20-1 | MAH | stable disease patient | Ib | 5.453 | 68.8 | 486 |
| 24 | MK20-5 | MAH | stable disease patient | Ib | 5.340 | 69.0 | 295 |
| 25 | NN-108 | MAH | stable disease patient | Ib | 5.250 | 69.0 | 416 |
| 26 | NN-127 | MAH | stable disease patient | IIa | 5.209 | 69.1 | 242 |
| 27 | Tone-6 | MAH | stable disease patient | Ib | 5.812 | 68.6 | 434 |
| 28 | Tone-7 | MAH | stable disease patient | Ib | 5.463 | 68.9 | 377 |
| 29 | Tone-15 | MAH | stable disease patient | Ib | 5.498 | 68.8 | 511 |
| 30 | DH-2 | MAH | progressive disease patient | IIa | 4.981 | 69.3 | 308 |
| 31 | DH-5 | MAH | progressive disease patient | Ib | 5.449 | 68.9 | 326 |
| 32 | DH-8-4 | MAH | progressive disease patient | Ib | 5.130 | 69.1 | 389 |
| 33 | HF-4 | MAH | progressive disease patient | Ib | 5.222 | 69.0 | 304 |
| 34 | NB-36042 | MAH | progressive disease patient | Ib | 5.482 | 68.8 | 382 |
| 35 | NN-122 | MAH | progressive disease patient | Ib | 5.751 | 68.7 | 367 |
| 36 | Tone-1 | MAH | progressive disease patient | Ia | 5.659 | 68.7 | 371 |
| 37 | Tone-5 | MAH | progressive disease patient | Ib | 5.232 | 69.1 | 262 |
| 38 | Tone-12 | MAH | progressive disease patient | Ia | 5.398 | 68.9 | 354 |
| 39 | Tone-13 | MAH | progressive disease patient | Ia | 5.742 | 68.7 | 295 |
| 40 | Tone-16 | MAH | progressive disease patient | Ia | 5.354 | 69.0 | 226 |
| 41 | TR-M-1 | MAH | progressive disease patient | Ib | 5.442 | 68.8 | 336 |
| 42 | TR-M-2 | MAH | progressive disease patient | Ia | 5.515 | 68.8 | 299 |
| 43 | TR-M-3 | MAH | progressive disease patient | Ia | 5.280 | 69.0 | 352 |
| 44 | TR-M-4 | MAH | progressive disease patient | Ia | 5.588 | 68.8 | 294 |
| 45 | TR-M-5 | MAH | progressive disease patient | Ib | 5.264 | 69.0 | 286 |
| 46 | TR-M-6-4 | MAH | progressive disease patient | Ib | 5.156 | 69.1 | 322 |
| 47 | TH135d | MAH | progressive disease patient | Ia | 4.951 | 69.3 | 1 |
| 48 | MAH27-1 | MAH | household dust (Germany) | IIa | 4.794 | 69.0 | 977 |
| 49 | 12_062 | MAH | human (Belgium) | IIa | 5.095 | 69.0 | 175 |
| 50 | 12_067 | MAH | human (Belgium) | IIa | 5.108 | 69.0 | 216 |
| 51 | LYM122 | MAH | pig (Belgium) | IIa | 5.093 | 69.0 | 175 |
| 52 | LYM086 | MAH | pig (Belgium) | IIa | 5.308 | 69.0 | 199 |
| 53 | 10-5606 | MAH | pig (USA) | IIa | 5.529 | 68.7 | 1,690 |
| 54 | 10-4249 | MAH | deer (USA) | IIIa | 4.787 | 69.2 | 924 |
| 55 | 05-4293 | MAH | bird (USA) | IIa | 5.156 | 69.2 | 190 |
| 56 | 3388 | MAH | unknown (USA) | IIa | 5.052 | 69.2 | 128 |
| 57 | 100 | MAH | unknown (USA) | IIa | 5.499 | 68.8 | 340 |
| 58 | A5 | MAH | unknown (USA) | IIIa | 4.871 | 69.4 | 50 |
| 59 | 104 | MAH | AIDS patient (USA) | IIa | 5.475 | 69.0 | 1 |
| 60 | MAV_061107_1842 | MAH | lung disease patient (USA) | IIa | 5.321 | 69.1 | 16 |
| 61 | MAV_120809_2495 | MAH | lung disease patient (USA) | IIa | 5.712 | 68.9 | 97 |
| 62 | MAV_120709_2344 | MAH | lung disease patient (USA) | Ib | 5.629 | 68.8 | 62 |
| 63 | 10–5581 | unknown | elephant (USA) | Ia | 5.330 | 69.0 | 325 |
| 64 | 09–5983 | unknown | whitetail deer (USA) | IIa | 5.167 | 68.9 | 1,113 |
| 65 | XTB13-223 | unknown | human (USA) | IIa | 5.130 | 69.2 | 33 |
| 66 | 10-9275 | MAA | redtail hawk (USA) | IIb | 4.795 | 69.1 | 886 |
| 67 | 11–4751 | MAA | ruddy duck (USA) | IIb | 4.835 | 69.2 | 577 |
| 68 | 2285 R | MAA | human (USA) | IIa | 5.169 | 69.1 | 1 |
| 69 | DJO-44271 | MAA | human (USA) | IIa | 5.011 | 69.1 | 1 |
| 70 | DT78 | MAA | water buffalo’s ileum (USA) | IIa | 4.960 | 69.2 | 1,201 |
| 71 | ATCC25291 | MAA | hen (Canada) | IIb | 4.858 | 69.2 | 258 |
| 72 | ATCC49884 | MAS | unknown | IIb | 4.713 | 69.2 | 808 |
| 73 | K10 | MAP | Johne’s disease cow (USA) | IIIb | 4.830 | 69.3 | 1 |
| 74 | ATCC19698 | MAP | cow (USA) | IIIb | 4.736 | 69.3 | 1,092 |
| 75 | Pt146 | MAP | unknown (Australia) | IIIb | 4.583 | 69.0 | 955 |
| 76 | Pt154 | MAP | unknown (Australia) | IIIb | 4.500 | 68.9 | 1,077 |
| 77 | CLIJ623 | MAP | unknown (Australia) | IIIb | 4.523 | 69.0 | 915 |
| 78 | JQ5 | MAP | unknown (USA) | IIIb | 4.735 | 69.1 | 176 |
| 79 | JQ6 | MAP | unknown (USA) | IIIb | 4.697 | 69.0 | 176 |
aMAH; M. avium subsp. hominissuis, MAA; M. avium subsp. avium, MAS; M. avium subsp. silvaticum, MAP; M. avium subsp. paratuberculosis.
bCluster was classified by the phylogenetic analysis as shown in Fig. 1.
cPatients with different clinical courses in pulmonary diseases described in Material and Methods.
dStrain TH135 was used as the reference strain.
Our analysis included 32 additional M. avium genomes that are publicly available (Table 1). These genomes include all M. avium subspecies: fifteen MAH strains, six MAA strains, seven MAP strains, one MAS strain, and three M. avium strains of unknown subspecies.
Phylogenetic relationships among M. avium genomes
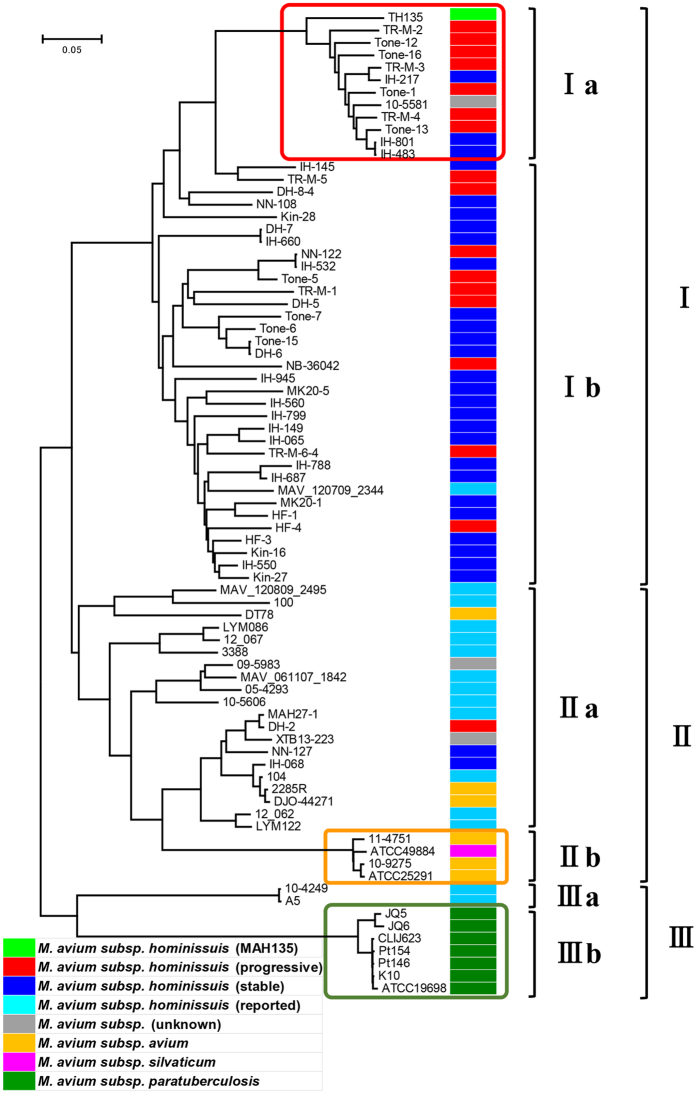
Comparative genome analysis of the 79 M. avium strains including four subspecies revealed that the total length of the core region, which was shared by all the 79 M. avium isolates, was 3,404,650 bp with 101,139 unique single nucleotide variants (SNVs). Phylogenetic analysis based on the SNVs in the core regions showed that the M. avium strains were roughly classified into three clusters: cluster I, cluster II, and cluster III (Fig. 1). Furthermore, it was shown that each cluster has a distinctive subcluster (cluster Ia, cluster IIb or cluster IIIb) comprised of strains with genetic distances that are clearly different from those of the others. Cluster I contained 93.5% (43/46) of the MAH genome sequenced in this study, MAH strain TH135 (reference strain), MAV_120709_2344, and strain 105581 of unknown subspecies. Cluster II contained 12 MAH strains isolated abroad, 2 strains of unknown subspecies, 3 MAH strains (IH-065, NN-127, and DH-2) sequenced in this study, all MAA strains, and MAS strain ATCC49884. Thus, the MAH strains isolated in Japan and those isolated in the United States, Belgium, and Germany formed different clusters. In cluster II, MAA strains were divided largely into two subclusters: cluster IIb containing 3 of the 6 MAA strains along with MAS strain ATCC49884 and cluster IIa containing the rest of the MAA strains along with MAH strains. Interestingly, all strains in cluster IIb were of avian origin. Among cluster III, cluster IIIb contained all MAP strains, and cluster IIIa contained two MAH strains.
Figure 1. Phylogenetic tree of 79 M. avium strains using 101,139 SNVs on 3,404,650 bases of core genome regions.
The evolutionary history was inferred using the neighbor-joining method60. The evolutionary distances were computed using the Maximum Composite Likelihood method61 and represent the number of base substitutions per site. Evolutionary analyses were conducted in MEGA658. The analysis included nucleotide sequences of genomes from 79 M. avium strains (red, MAH isolates from progressive disease patients; blue, MAH isolates from stable disease patients; light blue, MAH strains reported publicly; gray, other M. avium strains; orange, MAA strains; pink, MAS strain; green, MAP strains; and light green, MAH reference strain TH135).
MAH infects both pigs and humans, and several studies in Europe using molecular genotyping methods have shown high genetic similarity between MAH strains isolated from humans and pigs26,27,28,29, indicating the possibility of a common source of MAH infection among humans and pigs, as well as the possibility of pig–human zoonotic infections. In this study, three MAH isolates (10–5606, LYM122, and LYM086) from pigs in the United States and Belgium belonged to cluster IIa, which contains MAH human isolates, without the formation of a specific cluster. In addition, no major genetic differences between human and pig isolates were found. Specifically, high genetic similarity was found between human (12_062 and 12_067) and pig (LYM122 and LYM086) isolates in Belgium30, suggesting either a common source of infection or the zoonotic potential of MAH. However, this problem needs to be examined in detail using genome sequences of many pig isolates.
Next, we examined the phylogenetic relationships among 46 MAH isolates from patients with either progressive or stable disease. Of the 46 isolates, 43 (93.5%) were grouped in cluster I, while only 3 were in cluster II (Fig. 1). It is worth mentioning that 41.2% (7/17) of isolates from the patients with progressive disease and 10.3% (3/29) of those from the patients with stable disease were in cluster Ia, a subcluster of cluster I with a distinctively different genetic distance. The ratio of isolates from patients with progressive disease to those from patients from stable disease was significantly higher in cluster Ia than in other subclusters (Supplementary Table S1, p = 0.025 by Fisher’s exact test). It would be interesting to clarify the pathogenesis or evolutionary process of strain 105581, isolated in the United States, as it belongs in cluster Ia. These results indicate a specific genotype of MAH is associated with the progression of pulmonary MAH disease.
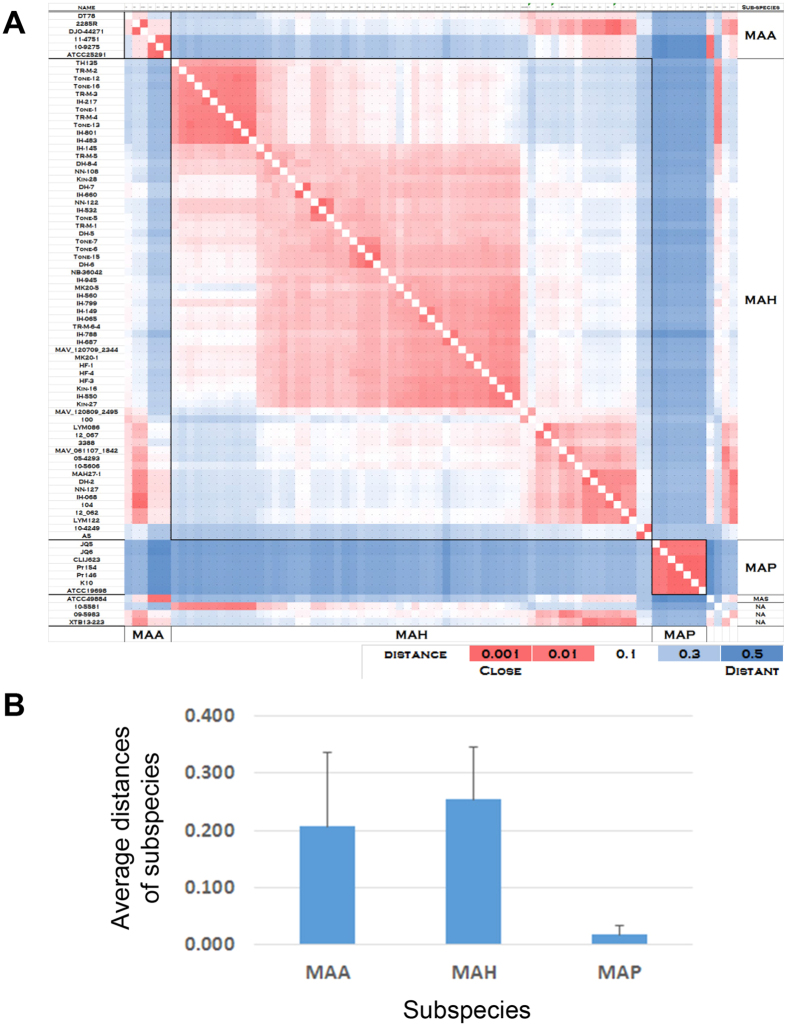
Furthermore, phylogenetic analysis currently shows that the degree of sequence diversity of M. avium genomes is different between subspecies. Therefore, we calculated the distance (substitution rate at the 101,139 SNV sites in the core regions) between each pair of M. avium subspecies (Fig. 2A). The average distance of MAH strains was 0.254, while those of MAA and MAP strains were 0.207 and 0.018, respectively (Fig. 2B). This result indicates that, among the M. avium subspecies, MAH exhibits the highest degree of sequence diversity, whereas the least diversity is observed in MAP.
Figure 2. Distance matrix among 79 M. avium isolates.
(A) Distances between 79 M. avium isolates were calculated as nucleotide substitution rates at all 101,139 SNV sites, and are colored according to the scale bar. (B) Average distance was calculated for MAAs, MAHs, and MAPs.
SNV distribution and conserved region of M. avium genomes
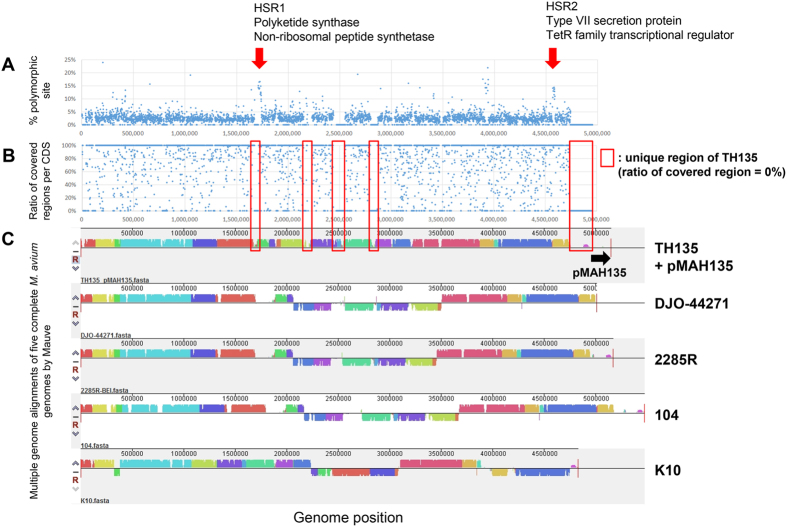
To understand whether there are “hot spot regions (HSRs)” for mutation and/or recombination in the M. avium genomes, we calculated the percentage of polymorphic sites for each CDS (Fig. 3A). We found several HSRs in M. avium genome, such as HSR1 (ranging from 1,712,045 to 1,737,933) encoding CDSs involved in mycobactin biosynthesis, including several nonribosomal peptide synthetases and polyketide synthases, and HSR2 (ranging from 4,565,477 to 4,587,364) encoding type VII secretion proteins and TetR family transcriptional regulators (Table 2). Mycobacteria synthesize siderophores, named mycobactin, to capture iron, which is an essential nutrient for almost all organisms31. HSR1 contains 7 CDSs with sequence homologies to the MbtB–MbtH proteins of other mycobacteria, and is involved in the synthesis of the siderophore core of mycobactin31. Pathogenic mycobacteria carry the type VII secretion systems, namely the ESX systems (ESX-1 to ESX-5), which are responsible for secreting 6-kDa early secreted antigenic target (ESAT-6) or mycobacteria-specific proteins with conserved N-terminal domains containing prolyl-glutamic acid (PE) and prolyl-prolyl glutamic acid (PPE) motifs32,33. Thus, CDSs associated with the pathogenicity of M. avium were present in HSR1 and HSR2. In addition, a lower ratio of covered regions per CDS corresponded to a lower similarity region analyzed by comparing five complete genomes, such as MAH strain TH135, MAA strain DJO-44271, MAA strain 2285 R, MAH strain 104, and MAP strain K10, using MAUVE. This result showed that the non-core regions identified in this analysis were unique or specific to each isolate (Fig. 3B,C).
Figure 3. SNV distribution in the core regions.
(A) SNV frequencies (percentages of polymorphic sites) of each coding sequence (CDS) in core regions. (B) Ratio of covered regions of each CDS. (C) Multiple genome alignments of five complete M. avium genomes (strains TH135 with pMAH135, DJO-44271, 2285 R, 104, and K10) analyzed using Mauve software.
Table 2. List of CDSs with higher percentage of polymorphic site.
| Regiona | Location (position) | Length | Covered length | Ratio of covered region per CDS | Polymorphic site (%) | Locus tag | Description |
|---|---|---|---|---|---|---|---|
| HSR1 | 1712045–1713700 | 1,656 | 1,012 | 61% | 14% | MAH_RS08025 | 2,3-dihydroxybenzoate-AMP ligase |
| HSR1 | 1713807–1717304 | 3,498 | 3,028 | 87% | 15% | MAH_RS08030 | non-ribosomal peptide synthetase |
| HSR1 | 1717313–1718077 | 765 | 765 | 100% | 14% | MAH_RS08035 | thioesterase |
| HSR1 | 1718083–1719402 | 1,320 | 1,320 | 100% | 14% | MAH_RS08040 | polyketide synthase |
| HSR1 | 1719402–1722443 | 3,042 | 1,275 | 42% | 16% | MAH_RS08045 | polyketide synthase |
| HSR1 | 1722445–1729017 | 6,573 | 6,419 | 98% | 14% | MAH_RS08050 | non-ribosomal peptide synthetase |
| HSR1 | 1729014–1733456 | 4,443 | 3,896 | 88% | 17% | MAH_RS08055 | non-ribosomal peptide synthetase |
| HSR1 | 1733453–1734739 | 1,287 | 1,287 | 100% | 12% | MAH_RS08060 | lysine 6-monooxygenase |
| HSR1 | 1734720–1734935 | 216 | 216 | 100% | 13% | MAH_RS08065 | protein mbtH |
| HSR1 | 1735111–1735647 | 537 | 537 | 100% | 12% | MAH_RS08070 | hypothetical protein |
| HSR1 | 1735941–1736456 | 516 | 516 | 100% | 12% | MAH_RS08075 | low molecular weight antigen MTB12 |
| HSR1 | 1736450–1737340 | 891 | 656 | 74% | 15% | MAH_RS08080 | RNA polymerase sigma-70 factor |
| HSR1 | 1737337–1737933 | 597 | 597 | 100% | 12% | MAH_RS08085 | 4-carboxymuconolactone decarboxylase |
| HSR2 | 4565477–4567111 | 1,635 | 1,635 | 100% | 14% | MAH_RS21020 | type VII secretion protein EccB |
| HSR2 | 4567108–4568961 | 1,854 | 1,714 | 92% | 10% | MAH_RS21025 | type VII secretion AAA-ATPase EccA |
| HSR2 | 4570937–4571842 | 906 | 906 | 100% | 13% | MAH_RS21035 | SAM-dependent methyltransferase |
| HSR2 | 4577124–4578050 | 927 | 927 | 100% | 12% | MAH_RS21070 | hypothetical protein |
| HSR2 | 4578152–4578814 | 663 | 647 | 98% | 14% | MAH_RS21075 | TetR family transcriptional regulator |
| HSR2 | 4580166–4581641 | 1,476 | 1,415 | 96% | 14% | MAH_RS21085 | acyltransferase |
| HSR2 | 4582027–4582575 | 549 | 549 | 100% | 14% | MAH_RS21090 | hypothetical protein |
| HSR2 | 4583277–4583933 | 657 | 657 | 100% | 13% | MAH_RS21100 | TetR family transcriptional regulator |
| HSR2 | 4583966–4585072 | 1,107 | 1,107 | 100% | 13% | MAH_RS21105 | alpha/beta hydrolase |
| HSR2 | 4585163–4587364 | 2,202 | 2,202 | 100% | 11% | MAH_RS21110 | acyl-CoA dehydrogenase |
aRegions HSR1 and HSR2 are shown in Fig. 3.
Noncore genome regions in cluster Ia
By comparing the genome sequences of the 79 M. avium strains, including the sequences of the chromosome and pMAH135 from strain TH135 as references, we identified noncore genome regions that were not shared by all strains (Fig. 4). The total length of the noncore regions was approximately 7.86 Mb, meaning each strain has on average about 100 kb unique sequences. It is noteworthy that the mean G + C content of 7.86 Mb noncore regions is 65.4%, which is much lower than that of the 79 M. avium genomes (68.9%). This result suggests that part of the specific noncore regions might have originated from other species via horizontal gene transfer. Clustering analysis of 1187 noncore regions with more than 1000 bp showed that MAH isolates from the progressive disease patients were significantly grouped in a specific cluster (cluster Ia), which was completely consistent with the results of clustering analysis based on SNVs in the core regions (Fig. 4). Among the noncore regions, we identified a genomic region (locus 1) that was mostly specific to cluster Ia.
Figure 4. Noncore genome comparison of 79 M. avium strains.
Rows represent 79 M. avium genomes and the sequence of the pMAH135 plasmid from strain TH135, and columns represent 1187 noncore regions longer than 1000 bp. Genomes and noncore regions were clustered based on similarity. The presence of a noncore region is shown in yellow, and its absence is shown in blue. Locus 1 is mostly unique to cluster Ia, which consists of many isolates from progressive disease patients. The presence of pMAH135 is also characteristic of cluster Ia. Loci 2 and 3 are mostly unique to cluster IIIb that includes MAP strains, and cluster IIb that includes MAA and MAS strains, respectively.
Locus 1 in the specific noncore regions contains CDSs encoded by strain TH135 chromosome, the pMAH135 plasmid, and genomes of other mycobacteria except for strain TH135 (Fig. 4 and Supplementary Table S2). Specific regions (SR)-2, SR-4, SR-7, SR-8, and SR-9, which were previously identified on strain TH135 chromosome24, were present on this locus. These regions have low G + C content compared with the mean G + C content of strain TH135 chromosome and are flanked by genes that encode integrases of phage origin and/or transposases derived from transposons, which is an additional sign of foreign origin. Among these regions, SR-2 carries virulence-associated genes, namely, mce family genes and mmpL gene (Supplementary Table S2). Mycobacteria have several mce operons that comprise two yrbE and six mce genes (mceA to mceF), which are homologous to the permeases and substrate-binding proteins of ABC transporters, respectively34. MmpL and MmpS proteins are reported to mediate the transport of lipid metabolites for the biosynthesis of cell wall lipids in mycobacteria35,36,37. The high content of lipids, such as mycolic acids, in the cell walls plays a pivotal role in host survival38. Locus 1 also contains CDSs encoded by pMAH135. This plasmid encodes 164 CDSs, some of which encode proteins involved in mycobactin biosynthesis and the type VII secretion system, associated with pathogenicity of mycobacteria (Supplementary Table S2). Furthermore, locus 1 contains CDS with a 100% sequence identity to MmpL protein of M. avium 10–5581. These results suggest that virulence-associated CDSs in the noncore region specific to cluster Ia, probably acquired by horizontal gene transfer during evolution, play an important role in the pathogenicity of MAH isolates from patients with progressive disease.
Plasmid analysis of MAH isolates
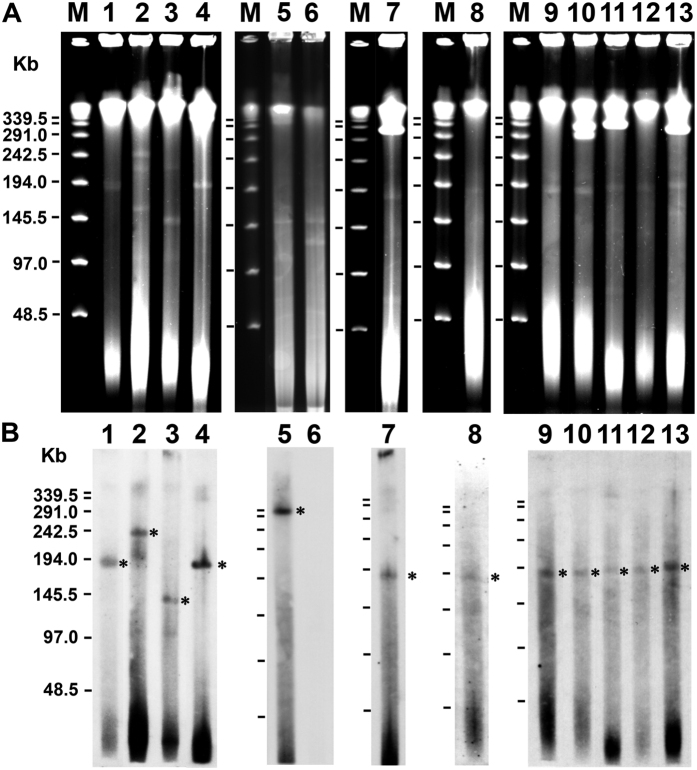
As shown in Fig. 4, CDSs encoded by pMAH135 were present in 12 strains (Tone-6, TR-M-1, IH-217, TR-M-3, IH-483, IH-801, Tone-12, Tone-13, Tone-1, Tone-16, TR-M-4, and TR-M-2) from 46 MAH isolates, suggesting the presence of pMAH135. Except for strains Tone-6 and TR-M-1 in cluster Ib, 10 strains were grouped in cluster Ia that consisted of many isolates from progressive disease patients (Fig. 1). We therefore conducted plasmid analysis of these strains with S1-PFGE and Southern hybridization, using a pMAH135-specific probe (MAH_p01). Among these 12 strains, 8 (IH-483, IH-801, Tone-12, Tone-13, Tone-1, Tone-16, TR-M-4, and TR-M-2) belonged to cluster Ia and carried a plasmid of approximately 194-kb, which was similar in size to pMAH135 (Fig. 5A) and MAH_p01 (Fig. 5B). The three remaining strains—Tone-6, IH-217, and TR-M-3—had MAH_p01 located on plasmids of approximately 388 kb, 242 kb, and 145 kb in size, respectively. Because MAH_p01 was absent, strain TR-M-1 was analyzed using MAH_p47 as another pMAH135-specific probe, which showed that MAH_p47 was located on the chromosome, not the plasmid.
Figure 5. Analysis of plasmids from MAH isolates.
PFGE of S1 nuclease-digested total DNA of MAH isolates (A) and Southern hybridization with a probe derived from MAH_p01 (B). Asterisks show plasmid bands hybridized with the probe. Lanes 1–13 represent, in order, strains TH135, IH-217, TR-M-3, TR-M-2, Tone-6, TR-M-1, IH-483, Tone-12, IH-801, Tone-13, Tone-1, Tone-16, and TR-M-4. The molecular size of the lambda ladder PFG marker (lane M) is shown in the left panel.
Noncore genome regions in clusters IIb and IIIb
As shown in Fig. 4, we found noncore regions specific to clusters IIb and IIIb. Locus 2 was present in cluster IIIb that contains MAP strains. Locus 3 was found in cluster IIb that contains MAS and MAA strains isolated from birds. These loci contain CDSs with sequence homologies to integrases and/or transposases, with lower G + C content compared with the mean G + C content of 79 M. avium genomes (Supplementary Table S2). Furthermore, locus 2 carries virulence-related CDSs with sequence homologies to genes encoding Mec, MmpL/MmpS, and PPE proteins, while CDS showing sequence homologies to genes encoding PPE protein is present in locus 3. PE/PPE family proteins are recognized as virulence factors that participate in antigenic variation and host immune evasion39. On the other hand, some regions were missing from the strains found in clusters IIb and IIIb. Locus 4 was absent from cluster IIIb strains, and locus 5 was absent from clusters IIb and IIIb strains. These loci contain several CDSs with sequence homologies to PPE proteins. Although the roles of many of the CDSs in the regions specific to clusters IIb and IIIb or in the missing regions are unclear, it is thought that these CDSs affects the specific pathogenic and host range characteristics of strains in clusters IIb and IIIb. Further study is necessary to elucidate the functions of those isolate-specific CDSs.
M. avium pan-genome
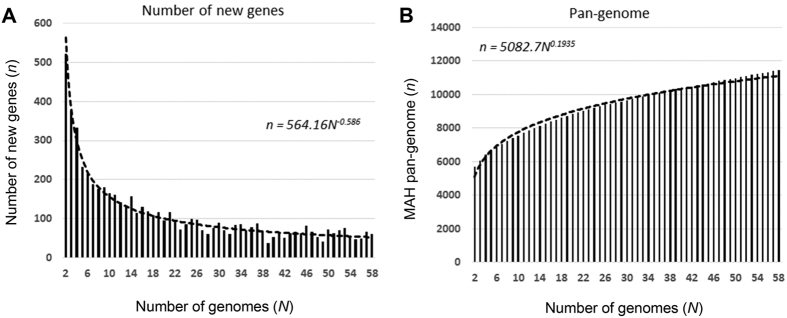
Based on the suggested diversity of MAH genome as described above, we estimated the pan-genome based on the 58 MAH genomes, including the reference strain TH135 genome and 11 additional publicly available MAH genomes. We first estimated the number of new genes that would be discovered by sequencing additional MAH genomes via power law regression analysis, n = κN–α 40 (Fig. 6A). Our analysis identified α as 0.586. When the 58th genome was added, the average number of new genes added by a novel genome was 52. We then estimated the number of MAH pan-genes that would be accumulated by sequencing additional MAH genomes using power law regression analysis, n = κNγ (Fig. 6B). The exponent γ as 0.194, and MAH had 11,151 pan-genes (n = 58). Based on these results, the pan-genome of MAH is defined as open because the exponent α was less than one and γ was greater than zero40. These results indicate that MAH has a high degree of genomic diversity.
Figure 6. M. avium pan-genome.
(A) Power law regression for new genes (n) discovered on the addition of new genome sequences (N). (B) Power law regression for total genes (n) accumulated with the addition of new genome sequences (N). Circles show the medians of n for 200 simulations. Error bars indicate the standard deviations for the 200 simulations.
Discussion
In our previous study, VNTR typing analysis using M. avium tandem repeats (MATR) of isolates from patients with pulmonary MAH disease demonstrated a relationship between VNTR genotype and disease progression. Furthermore, screening of these isolates for six genes located in pMAH135 indicated a relationship between disease progression and the presence of pMAH135 genes23. In this study, by comparing the genome sequences of 79 M. avium strains comprising four subspecies, we analyzed the phylogenetic relationships based on the SNVs among the M. avium subspecies, including MAH isolates from patients with different clinical courses, characterized the genetic diversity and features of SNVs in M. avium genomes, revealed the presence of genetic elements specific to each lineage phylogenetically classified into unique clusters, and identified potential genetic determinants associated with the host range characteristics of M. avium, as well as the progression of pulmonary MAH disease.
The phylogenetic analysis based on the SNVs in the core regions showed that the M. avium strains were roughly classified into three clusters. Furthermore, it was shown that each cluster has a distinctive subcluster (cluster Ia, cluster IIb or cluster IIIb), the constituent strains of which appear to have evolved from the common ancestor through unique evolutionary pathways (Fig. 1). MAH strains were present in clusters I through III, whereas all MAP strains belonged to cluster IIIb, and cluster IIb was formed specifically by MAA strains of avian origin and MAS strain. This suggests that strains in cluster IIIb have genomic feature associated with John’s disease and that MAA and MAS strains share genomic features that enable them to infect birds. Using VNTR analysis, Iwamoto et al. and Ichikawa et al. demonstrated a geographical difference in the genetic diversity of MAH41,42. In agreement with this, we found that MAH strains isolated in Japan formed a cluster (cluster I) that differs from the cluster (cluster II) containing MAH strains isolated in the United States or Germany, indicating that they have different genomic features. This may be one of the reasons for the high incidence of pulmonary MAH disease in Japan8.
The average distances of each M. avium subspecies calculated based on all SNV sites indicated that MAH strains have the highest degree of sequence diversity (Fig. 2). This is consistent with previous results of SNV-based multilocus sequencing analysis using 10 housekeeping genes43. Moreover, this analysis revealed that MAP strains exhibit the lowest sequence diversity. Our study further demonstrated that MAH is an open pan-genome species (Fig. 6), indicating that the acquisition and deletion of genetic elements occurred at high rates during the evolutionary process. Taken together, these results show that MAH is an M. avium species with high genetic diversity.
In our recent studies and that of Kikuchi et al., MATR-VNTR analysis of isolates from patients with pulmonary MAH disease demonstrated that isolates from progressive disease cases are grouped in a specific cluster22,23, and further revealed that many of the isolates from both groups are classified into the same cluster. These findings suggest that strains in this cluster are highly virulent. In this study, SNV-based phylogenetic analysis showed that isolates from progressive disease patients were notably grouped in cluster Ia (p = 0.025) (Fig. 1 and Supplementary Table S1). Interestingly, isolates in cluster Ia fully corresponded with those in the specific cluster described above obtained by MATR-VNTR analysis examining an identical set of isolates23. Although other clusters did not exhibit a complete match, these results indicate that genotypes based on SNVs overlap with VNTR genotypes. Taken together, these results suggest that the isolates in cluster Ia have unique genomic features associated with the progression of pulmonary MAH disease, and demonstrate that MATR-VNTR analysis can distinguish isolates from progressive disease patients simply. Therefore, this analysis is a clinically useful approach.
By analyzing the noncore regions, we identified genomic element (locus 1) specific to cluster Ia consisting of many MAH isolates from progressive disease patients (Fig. 4). This genomic element harbors virulence genes that account for the progression of pulmonary MAH disease. On locus 1, SR-2, which was previously identified as one of the specific regions on strain TH135 chromosome24, is present and carries virulence-associated mce family genes and mmpL gene. Although the precise mechanisms of Mce proteins remain unclear, they are thought to be mainly involved in the entry of mycobacteria into mammalian cells and their subsequent survival44,45. Furthermore, locus 1 contains CDSs that are encoded on pMAH135 and involved in mycobactin biosynthesis and the type VII secretion system. De Voss et al. reported that a M. tuberculosis mutant lacking the mbtB gene interrupts the biosynthesis of mycobactin and impairs the growth of macrophages46, suggesting that mycobactin plays a significant role in the pathogenicity of mycobacteria. ESX-5, which is similar to the ESX-related proteins encoded on pMAH135, mediates the secretion of ESAT-6-like proteins EsxN and EsxP, and is involved in inducing cell death in infected macrophages and modulating the immune response47. Thus, pMAH135 is thought to be involved in MAH pathogenicity. Interestingly, Ummels et al. reported that pMAH135 is a conjugative plasmid in slow-growing mycobacteria species, including M. avium48. Plasmid analysis by S1-PFGE revealed that eight isolates carry pMAH135 (Fig. 5), which probably originated from other mycobacteria by conjugation.
We previously reported that five IsMav6 genes, which is a novel insertion sequence, are coded by strain TH135 chromosome24,49. One was inserted into Shine–Dalgarno region of the cfp29 gene50, which is involved in the induction of interferon–γ production in hosts with mycobacterial infection, thus suggesting its influence on MAH pathogenicity. Interestingly, the frequency of isolates with IsMav6 inserted into the cfp29 gene determined by analyzing an identical set of isolates—but not those with IsMav6—was significantly higher in patients with progressive disease than in those with stable disease51. Comparisons of the detection rates between IsMav6 inserted into the cfp29 gene and four potential virulence factors specific to cluster Ia strains in isolates from patients with progressive disease showed that the former was higher than the latter (Supplementary Table S3). These results suggest that IsMav6 inserted into the cfp29 gene is one factor related to the progression of pulmonary MAH disease. Taken together, cluster Ia strains acquired genetic regions (e.g. SR-2 and pMAH135) encoding virulence genes via horizontal transfer during the evolutionary process, thereby acquiring pathogenicity resulting in disease progression. It will be intriguing in the future to discover how such virulence factors are involved in pathogenicity. Cluster Ia also contained three strains isolated from the stable disease patients. In addition, some isolates from the progressive disease patients did not belong to cluster Ia, but caused disease progression despite having identical genomic characteristics to isolates from the stable disease patients. Such cases could be explained by our hypothesis that suggests the influence of host factors, such as the host immune system, are stronger than bacterial factors on the clinical course of pulmonary MAH disease.
One of the clinical problems in the treatment of pulmonary NTM disease is the difficulty in judging the appropriate time to start therapy. The clinical course of patients with pulmonary NTM disease is diverse; some patients are stable without any treatment and others have worsening symptoms despite drug therapy, which leads to severe lung damage9,14. Clarithromycin-based multidrug therapy is recommended for pulmonary MAH disease; however, it requires a long treatment period (18–24 months) and is associated with the risk of adverse reactions from the use of multiple drugs, which places considerable financial, psychological, and physical burdens on patients52,53,54. Furthermore, the timing of treatment initiation influences the outcome and is therefore also important. Clear criteria for determining the timing of treatment are not currently available. The findings of this study demonstrated the potential of virulence genes encoded by SR-2 and pMAH135 specific to isolates from progressive disease patients as one indicator of the need to initiate therapy.
However, this study has some limitations. There were no objective standards used in judging patient status, either progressive or stable, and the classification of patients depended entirely on the decision made by each physician in charge at the individual participating hospitals. In addition, this was a retrospective study with a small number of subjects. Thus, this investigation can be regarded as a preliminary study. A future prospective study should evaluate our findings, and further investigate the way in which the virulence genes specific to isolates from progressive disease patients are involved in the pathogenicity.
In conclusion, the findings from our comparative genome analysis of 79 M. avium strains comprising four subspecies provided a perspective on the genetic diversity and evolution of M. avium strains, as well as genomic evidence that may explain the differences among M. avium subspecies. Of note, we revealed the presence of genetic elements specific to each lineage, which are thought to be acquired via horizontal gene transfer during the evolutionary process, and identified potential genetic determinants associated with not only the progression of pulmonary MAH disease but also the host range characteristics of M. avium. In the future, genome sequences of many NTM isolates should be investigated to elucidate in detail the various problems associated with NTM.
Methods
Bacterial strains
MAH strain TH135 isolated from the sputum of a seriously ill patient with worsening pulmonary MAH disease at Higashinagoya National Hospital of the National Hospital Organization was used as the reference strain. As reported previously23, 46 MAH isolates used in genome analysis in this study were provided by nine National Hospital Organization hospitals across Japan. These clinical isolates were obtained from the sputa of 46 patients with distinct clinical courses (see below). Only one strain per patient was analyzed in this study. Of the patients diagnosed with pulmonary MAH disease (corresponding to the diagnostic criteria of the American Thoracic Society and the Infectious Diseases Society of America52) between July 2008 and September 2009, those who started clarithromycin-based multidrug treatment within 18 months, based on decisions made by the corresponding physician-in-charge because of deterioration in the patients’ condition, were classed as the progressive disease group (n = 17). Those who did not receive treatment because their condition was stable were classed as the stable disease group (n = 29). During the observation period, the condition of each patient was evaluated several times a year based on chest radiograph findings (including chest computed tomographic images), clinical symptoms, and/or microbiological findings. Parameters of age, sex, type of pulmonary disease, and the presence of underlying disease were not significantly different between the two groups23.
Identification of subspecies of M. avium, growth condition, and DNA isolation
The subspecies of M. avium clinical isolates was identified as MAH by sequence analysis of the 3′ fragment of the hsp65 gene55. The organism was grown in Middlebrook 7H9 liquid medium supplemented with 10% oleic acid/albumin/dextrose/catalase enrichment (Difco Laboratories, Detroit, MI) at 37 °C. DNA was extracted using the illustra bacterial genomicPrep Mini Spin Kit (GE Healthcare, Buckinghamshire, UK) according to the manufacturer’s instructions.
Whole genome sequencing and analysis of the core/noncore regions and pan-genome
All 46 MAH genomes were sequenced using Illumina MiSeq (150 bp, paired-end) with Nextera XT DNA Library Prep Kit (Illumina, CA) and assembled using CLC Genomics Workbench (Qiagen Inc., Valencia, CA) with the default settings. Core/noncore regions and pan-genome analysis were performed as previously described5. Briefly, the core regions were defined as genome sequences present in all 79 genomes, while the noncore regions were defined as those not present in all genomes. MAH strain TH135 was used as the reference genome. All other 78 genome sequences were mapped to the reference genome using Nucmer56. The unique regions from each genome were identified and added to the reference genome until all unique regions from all genomes were included in the pan-genome. Core regions were then subtracted from the pan-genome and the remaining regions were defined as noncore regions. Protein coding sequences were predicted by GeneMark57 using MAH strain 104 as a reference. The 79 concatenated sequences of the 101,139 single nucleotide variant (SNV) nucleotides in the core regions were used to construct a phylogenetic tree of the M. avium genomes. MEGA658 was used to calculate the distance based on the 101,139 SNVs in the core regions. Comparative genomic analysis was performed with five complete M. avium genomes (strains TH135 with pMAH135, DJO-44271, 2285 R, 104, and K10) using the Mauve multiple genome aligner59.
Plasmid analysis
Plasmid DNA analysis was performed using S1-pulsed-field gel electrophoresis (PFGE) and Southern hybridization using a specific probe, as described previously25. Bacteria in agarose gel plugs were treated with lysozyme and proteinase K, before digestion of the total DNA in the plugs by 10 U S1 nuclease (Takara Bio, Shiga, Japan) for 10 min at 37 °C. PFGE was performed using the Bio-Rad CHEF-DR III system at 14 °C and 6 V/cm2 for 24 h with a switch time of 1.6–21.3 s. After electrophoresis, Southern hybridization analysis including probe labeling was performed using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche, Mannheim, Germany) according to the manufacturer’s instructions. A pMAH135-specific probe was prepared by PCR with specific primers for MAH_p01, which encodes the repA gene (thought to be the origin of replication for pMAH135), and DNA from MAH strain TH135.
Statistical analysis
The correlation between genome size and G + C content was analyzed using Spearman’s rank correlation. Fisher’s exact test was used for categorical variables. All statistical analysis was performed using GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA). P values < 0.05 were considered significant.
Ethics Statement
This study was approved and carried out in accordance with guidelines and regulations by the Ethics Review Committee for Human Research of the Higashinagoya National Hospital, and written informed consent was obtained from all patients.
Nucleotide sequence accession numbers
Draft genome sequences reported here were deposited in DDBJ/EMBL/GenBank under accession no. PRJDB502.
Additional Information
How to cite this article: Uchiya, K. et al. Comparative genome analyses of Mycobacterium avium reveal genomic features of its subspecies and strains that cause progression of pulmonary disease. Sci. Rep. 7, 39750; doi: 10.1038/srep39750 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We are grateful to Drs Satoru Fujiuchi, Katsuhiro Kuwabara, Yuka Sasaki, Emiko Toyota, Ryoji Maekura, Kazunari Tsuyuguchi, Masahiro Shirai, Takefumi Saitoh, Seiji Kawabata, Yoshiaki Tao, Shuichi Takigawa, and Mitsunori Sakatani for kindly providing us with clinical isolates and clinical information. Library construction, sequencing and de novo assembly were supported by Okayama University Hospital Biobank. This work was supported by JSPS KAKENHI Grant Number 15K08049 (to K.U.).
Footnotes
Author Contributions K.U., S.T., T. Nakagawa, S.A., T. Nikai and K.O. contributed to the study design and data management. K.U., S.T. and S.A. participated in the study implementation and data analysis. K.U. and S.T. wrote the manuscript. All authors reviewed the manuscript.
References
- Harris S. R. et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327, 469–474 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher N. J. et al. Rapid pneumococcal evolution in response to clinical interventions. Science 331, 430–434 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. S. et al. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 364, 33–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitz-Gibbon S. et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J. Invest. Dermatol. 133, 2152–2160 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomida S. et al. Pan-genome and comparative genome analyses of propionibacterium acnes reveal its genomic diversity in the healthy and diseased human skin microbiome. mBio 4, e00003–00013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevots D. R. et al. Nontuberculous mycobacterial lung disease prevalence at four integrated health care delivery systems. Am. J. Respir. Crit. Care Med. 182, 970–976 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M. et al. Prognostic factors of 634 HIV-negative patients with Mycobacterium avium complex lung disease. Am. J. Respir. Crit. Care Med. 185, 575–583 (2012). [DOI] [PubMed] [Google Scholar]
- Namkoong H. et al. Epidemiology of pulmonary nontuberculous mycobacterial disease, Japan1. Emerg. Infect. Dis. 22, 1116–1117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout J. E. & Hamilton C. D. Mycobacterium avium complex disease. (ed. Schlossberg D.) 531–564 (ASM Press, 2011). [Google Scholar]
- Adjemian J., Olivier K. N., Seitz A. E., Holland S. M. & Prevots D. R. Prevalence of nontuberculous mycobacterial lung disease in U.S. Medicare beneficiaries. Am. J. Respir. Crit. Care Med. 185, 881–886 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa K. et al. Environmental risk factors for pulmonary Mycobacterium avium-intracellulare complex disease. Chest 140, 723–729 (2011). [DOI] [PubMed] [Google Scholar]
- von Reyn C. F., Maslow J. N., Barber T. W., Falkinham J. O. & Arbeit R. D. Persistent colonization of notable water as a source of Mycobacterium avium infection in AIDS. Lancet 343, 1137–1141 (1994). [DOI] [PubMed] [Google Scholar]
- Nishiuchi Y. et al. The recovery of Mycobacterium avium-intracellulare complex (MAC) from the residential bathrooms of patients with pulmonary MAC. Clin. Infect. Dis. 45, 347–351 (2007). [DOI] [PubMed] [Google Scholar]
- Weiss C. H. & Glassroth J. Pulmonary disease caused by nontuberculous mycobacteria. Expert Rev. Respir. Med. 6, 597–613 (2012). [DOI] [PubMed] [Google Scholar]
- Griffith D. E. et al. Clinical and molecular analysis of macrolide resistance in Mycobacterium avium complex lung disease. Am. J. Respir. Crit. Care Med. 174, 928–934 (2006). [DOI] [PubMed] [Google Scholar]
- Mijs W. et al. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp avium for bird-type isolates and ‘M. avium subsp hominissuis’ for the human/porcine type of M. avium. Int. J. Syst. Evol. Microbiol. 52, 1505–1518 (2002). [DOI] [PubMed] [Google Scholar]
- Thorel M. F., Krichevsky M. & Levy-Frebault V. V. Numerical taxonomy of mycobactin-dependent mycobacteria, emended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov., Mycobacterium avium subsp. paratuberculosis subsp. nov., and Mycobacterium avium subsp. silvaticum subsp. nov. Int. J. Syst. Bacteriol. 40, 254–260 (1990). [DOI] [PubMed] [Google Scholar]
- Cocito C. et al. Paratuberculosis. Clin. Microbiol. Rev. 7, 328–345 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorel M. F., Huchzermeyer H., Weiss R. & Fontaine J. J. Mycobacterium avium infections in animals. Literature review. Vet. Res. 28, 439–447 (1997). [PubMed] [Google Scholar]
- Maekura R. et al. Clinical and prognostic importance of serotyping Mycobacterium avium-Mycobacterium intracellulare complex isolates in human immunodeficiency virus-negative patients. J. Clin. Microbiol. 43, 3150–3158 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S. J. et al. Mycobacterial genotypes are associated with clinical manifestation and progression of lung disease caused by Mycobacterium abscessus and Mycobacterium massiliense. Clin. Infect. Dis. 57, 32–39 (2013). [DOI] [PubMed] [Google Scholar]
- Kikuchi T. et al. Association between mycobacterial genotypes and disease progression in Mycobacterium avium pulmonary infection. Thorax 64, 901–907 (2009). [DOI] [PubMed] [Google Scholar]
- Moriyama M., Ogawa K., Nakagawa T., Nikai T. & Uchiya K. Association between a pMAH135 plasmid and the progression of pulmonary disease caused by Mycobacterium avium. Kekkaku 91, 9–15 (2016). [PubMed] [Google Scholar]
- Uchiya K. et al. Comparative genome analysis of Mycobacterium avium revealed genetic diversity in strains that cause pulmonary and disseminated disease. PLoS One 8, e71831 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiya K. et al. Characterization of a novel plasmid, pMAH135, from Mycobacterium avium subsp. hominissuis. PLoS One 10, e0117797 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasoota P., Chansiripornchai N., Kallenius G., Hoffner S. E. & Svenson S. B. Comparison of Mycobacterium avium complex (MAC) strains from pigs and humans in Sweden by random amplified polymorphic DNA (RAPD) using standardized reagents. Vet. Microbiol. 78, 251–259 (2001). [DOI] [PubMed] [Google Scholar]
- Komijn R. E. et al. Prevalence of Mycobacterium avium in slaughter pigs in The Netherlands and comparison of IS1245 restriction fragment length polymorphism patterns of porcine and human isolates. J. Clin. Microbiol. 37, 1254–1259 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirkkonen T. et al. Comparison of variable-number tandem-repeat markers typing and IS1245 restriction fragment length polymorphism fingerprinting of Mycobacterium avium subsp. hominissuis from human and porcine origins. Acta Vet. Scand. 52, 21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radomski N. et al. Determination of genotypic diversity of Mycobacterium avium subspecies from human and animal origins by mycobacterial interspersed repetitive-unit-variable-number tandem-repeat and IS1311 restriction fragment length polymorphism typing methods. J. Clin. Microbiol. 48, 1026–1034 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruffaerts N. et al. Genome sequences of four strains of Mycobacterium avium subsp. hominissuis, isolated from swine and humans, differing in virulence in a murine intranasal infection model. Genome announcements 4, e00533–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez G. M. Control of iron metabolism in Mycobacterium tuberculosis. Trends Microbiol. 14, 320–327 (2006). [DOI] [PubMed] [Google Scholar]
- Abdallah A. M. et al. Type VII secretion-mycobacteria show the way. Nat. Rev. Microbiol. 5, 883–891 (2007). [DOI] [PubMed] [Google Scholar]
- Yu X. & Xie J. Roles and underlying mechanisms of ESAT-6 in the context of Mycobacterium tuberculosis-host interaction from a systems biology perspective. Cell. Signal. 24, 1841–1846 (2012). [DOI] [PubMed] [Google Scholar]
- Casali N. & Riley L. W. A phylogenomic analysis of the Actinomycetales mce operons. BMC Genomics 8, 60 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. S., Chen B., McNeil M. & Jacobs W. R. Complex lipid determine tissue specific replication of Mycobacterium tuberculosis in mice. Nature 402, 79–83 (1999). [DOI] [PubMed] [Google Scholar]
- Converse S. E. et al. MmpL8 is required for sulfolipid-1 biosynthesis and Mycobacterium tuberculosis virulence. Proc. Natl. Acad. Sci. USA 100, 6121–6126 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshayes C. et al. MmpS4 promotes glycopeptidolipids biosynthesis and export in Mycobacterium smegmatis. Mol. Microbiol. 78, 989–1003 (2010). [DOI] [PubMed] [Google Scholar]
- Daffé M. & Draper P. The envelope layers of mycobacteria with reference to their pathogenicity. Adv. Microbial. Physiol. 39, 131–203 (1998). [DOI] [PubMed] [Google Scholar]
- Akhter Y., Ehebauer M. T., Mukhopadhyay S. & Hasnain S. E. The PE/PPE multigene family codes for virulence factors and is a possible source of mycobacterial antigenic variation: perhaps more? Biochimie 94, 110–116 (2012). [DOI] [PubMed] [Google Scholar]
- Tettelin H., Riley D., Cattuto C. & Medini D. Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 11, 472–477 (2008). [DOI] [PubMed] [Google Scholar]
- Iwamoto T. et al. Genetic diversity of Mycobacterium avium subsp. hominissuis strains isolated from humans, pigs, and human living environment. Infect. Genet. Evol. 12, 846–852 (2012). [DOI] [PubMed] [Google Scholar]
- Ichikawa K. et al. Genetic diversity of clinical Mycobacterium avium subsp. hominissuis and Mycobacterium intracellulare isolates causing pulmonary diseases recovered from different geographical regions. Infect. Genet. Evol. 36, 250–255 (2015). [DOI] [PubMed] [Google Scholar]
- Turenne C. Y., Collins D. M., Alexander D. C. & Behr M. A. Mycobacterium avium subsp. paratuberculosis and M. avium subsp. avium are independently evolved pathogenic clones of a much broader group of M. avium organisms. J. Bacteriol. 190, 2479–2487 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda S., Bomfim G., Knights R., Huimabyron T. & Riley L. W. Cloning of an Mycobacterium tuberculosis DNA fragment associated with entry and survival inside cells. Science 261, 1454–1457 (1993). [DOI] [PubMed] [Google Scholar]
- Chitale S. et al. Recombinant Mycobacterium tuberculosis protein associated with mammalian cell entry. Cell. Microbiol. 3, 247–254 (2001). [DOI] [PubMed] [Google Scholar]
- De Voss J. J. et al. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA 97, 1252–1257 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdallah A. M. et al. The ESX-5 secretion system of Mycobacterium marinum modulates the macrophage response. J. Immunol. 181, 7166–7175 (2008). [DOI] [PubMed] [Google Scholar]
- Ummels R. et al. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. mBio 5, e01744–01714 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa K. et al. Characterization of Mycobacterium avium clinical isolates in Japan using subspecies-specific insertion sequences, and identification of a new insertion sequence, ISMav6. J. Med. Microbiol. 58, 945–950 (2009). [DOI] [PubMed] [Google Scholar]
- Rosenkrands I. et al. Identification and characterization of a 29-kilodalton protein from Mycobacterium tuberculosis culture filtrate recognized by mouse memory effector cells. Infect. Immun. 66, 2728–2735 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T. et al. Multicenter study on clinical features and genetic characteristics of Mycobacterium avium strains from patients in Japan with lung disease caused by M. avium. Kekkaku 87, 687–695 (2012). [PubMed] [Google Scholar]
- Griffith D. E. et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 175, 367–416 (2007). [DOI] [PubMed] [Google Scholar]
- Glassroth J. Pulmonary disease due to nontuberculous mycobacteria. Chest 133, 243–251 (2008). [DOI] [PubMed] [Google Scholar]
- Thomson R. M. & Yew W. W. When and how to treat pulmonary non-tuberculous mycobacterial diseases. Respirology 14, 12–26 (2009). [DOI] [PubMed] [Google Scholar]
- Turenne C. Y., Semret M., Cousins D. V., Collins D. M. & Behr M. A. Sequencing of hsp65 distinguishes among subsets of the Mycobacterium avium complex. J. Clin. Microbiol. 44, 433–440 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S. et al. Versatile and open software for comparing large genomes. Genome Biol. 5, R12 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodovsky M. & McIninch J. Recognition of genes in DNA sequence with ambiguities. Biosystems 30, 161–171 (1993). [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. C., Mau B., Blattner F. R. & Perna N. T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N. & Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987). [DOI] [PubMed] [Google Scholar]
- Tamura K., Nei M. & Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 101, 11030–11035 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.