Abstract
Adult stem cells or residential progenitor cells are critical to maintain the structure and function of adult tissues (homeostasis) throughout the lifetime of an individual. Mis-regulation of stem cell proliferation and differentiation often leads to diseases including cancer, however, how wildtype adult stem cells and cancer cells respond to cellular damages remains unclear. We find that in the adult Drosophila midgut, intestinal stem cells (ISCs), unlike tumor intestinal cells, are resistant to various cellular damages. Tumor intestinal cells, unlike wildtype ISCs, are easily eliminated by apoptosis. Further, their proliferation is inhibited upon autophagy induction, and autophagy-mediated tumor inhibition is independent of cas-pase-dependent apoptosis. Interestingly, inhibition of tumorigenesis by autophagy is likely through the sequestration and degradation of mitochondria, as compromising mitochondria activity in these tumor models mimics the induction of autophagy and increasing the production of mitochondria alleviates the tumor-suppression capacity of autophagy. Together, these data demonstrate that wildtype adult stem cells and tumor cells show dramatic differences in sensitivity to cellular damages, thus providing potential therapeutic implications targeting tumorigenesis.
Keywords: Stem cell, Apoptosis, Autophagy, Tumorigenesis, Drosophila, Raf, Mitochondria
1. Introduction
Adult stem cells or progenitors, by continuously supplying newly differentiated cells, sustain tissue homeostasis (Morrison and Spradling, 2008; Radtke and Clevers, 2005). Adult stem cells during their prolonged life time encounter various cellular damages, like accumulation of DNA damage and starvation (Liu et al., 2014; Potten and Loeffler, 1990) that they must counteract to maintain their robustness. Ineffective defense against these cellular damages impedes the delicate balance between stem cell self-renewal and differentiation, eventually leading to severe disease such as cancer (Biteau et al., 2008; Liu et al., 2014). However, how adult stem cells sense and respond to these cellular damages remains unclear.
Unbalanced stem cell proliferation toward progeny differentiation usually results in tumorigenesis (Clarke and Fuller, 2006; Simons and Clevers, 2011). Management of patients with advanced malignancies is a vexing problem (He et al., 2014). Tumors could be reduced or eliminated through surgical operation, radiotherapy or chemotherapy. However, tumors often become chemo- and radio-resistant, ultimately leading to cancer recurrence. Pre-existing or newly acquired drug resistance properties, like mutations preventing drug inhibition of cancer cells, additional mutations that activate multiple oncogenes, or the selection of cancer stem cells (CSCs) (Diehn et al., 2009; Matsui et al., 2008; Rexer et al., 2009), may underlie the mechanism of cancer recurrence. CSCs are defined as cells that possess stem cell-like properties and that are responsible for generating all the cell types in its cognate tumor (Magee et al., 2012).
The adult Drosophila intestine has proven to be an exquisite model system to study how adult stem cell proliferation and differentiation are regulated, especially as mammalian and Drosophila intestines share many similarities in terms of development, cellular make-up and genetic control (Casali and Batlle, 2009; Stainier, 2005; Wang, Hou, 2010). Previous studies have demonstrated that the Drosophila adult midgut contains intestinal stem cells (ISCs) that are located adjacent to the basement membrane of the midgut epithelium (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). ISCs undergo constant asymmetric self-renewing divisions and also produce non-dividing, undifferentiated ISC daughters, termed enteroblasts (EBs). In contrast to transit amplifying cells in mammalian intestinal crypts, Drosophila EBs do not proliferate, but directly differentiate into two conserved cell types, the absorptive enterocytes (ECs) and the secretory enteroendocrine cells (ee) (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). The differentiation of EB into either EC or ee is determined by the level of Notch signaling (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006; Perdigoto et al., 2011). In addition to specifying the fate of the progeny, Notch signaling also plays a critical role in ISC proliferation. In the absence of Notch signaling, ISC progenies continue to divide, rather than to exit the cell cycle or to differentiate into ECs, generating progenitor tumors (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006, 2007). Bidirectional Notch signaling is found to regulate ISC multipotency (Guo and Ohlstein, 2015). Additional evolutionarily conserved pathways, including the Wnt, JAK/STAT, EGFR, Hippo, and BMP pathways, regulate the proliferation and differentiation of ISCs (Amcheslavsky et al., 2009; Jiang et al., 2011; Jiang et al., 2009; Karpowicz et al., 2010; Li et al., 2013; Lin et al., 2008; Markstein et al., 2014; Ren et al., 2010; Staley and Irvine, 2010). Mis-regulation of these signaling pathways results in the formation of tumors in both human and fly (Jiang and Edgar, 2011; Radtke and Clevers, 2005; Takashima and Hartenstein, 2012; Yeung et al., 2011). Therefore, Drosophila ISC is an excellent model for its mammalian counterpart to study the mechanisms underlying tumor formation, in which the roles of various cellular damages during tumorigenesis can be studied in detail.
Here, we investigate the sensitivity of wildtype adult stem cells and tumor cells to various cellular damages in adult Drosophila. Interestingly, we find that wildtype adult stem cells are resistant to cellular damages, while tumor cells are very sensitive to these damages. Moreover, in contrast to its pro-survival function, induction of autophagy results in the suppression of tumorigenesis, and the suppression of tumorigenesis by autophagy is not mediated by caspase-dependent apoptosis. Importantly, we find that mitochondria, the energy source for cell proliferation and growth, play a key role in the mechanism of inhibition of tumorigenesis by autophagy.
2. Materials and methods
2.1. Fly lines and husbandry
Flies were maintained on standard media at 25 °C. 2–3 days old flies were selected and transferred to 29 °C, unless otherwise specified. Flies were transferred to new vials with fresh food every day and dissected at specific time points as indicated. In all experiments, only female posterior midguts were analyzed. Information about alleles and transgenes used in this study can be found either in FlyBase or as noted: UAS-Rafgof (Brand and Perrimon, 1994), UAS-Atg1 (gift from T. Neufeld) (Scott et al., 2007), UAS-Rpr (gift from Z. Wang), UAS-Hid (gift from Y. Cai), UAS-p53, UAS-GFP-Atg8 (gift from T. Neufeld) (Juhász et al., 2008), FRT19A-N 264-39 (Slizynska, 1938), FRT82B-DlRevF10, SerRX82 (Heitzler and Simpson, 1991), esgGal4, UAS-GFP, tubGal80ts (esgts) (Micchelli and Perrimon, 2006), esgGal4, UAS-RFP; tubGal80ts, UAS-p35 (BL5073), UAS-mito-GFP (BL8442), mito-YFP (BL7194), UAS-ND75RNAi (BL33911, HMS00854), UAS-wRNAi (BL33613, HMS00004) (from TRiP at Harvard Medical School)(Ni et al., 2011), UAS-Srl (gift from F. Christian)(Tiefenböck et al., 2009). Other stocks used include: hsFlp; Act-FRT>Y>FRT-Gal4 (Y: yellow), UAS-mRFP, hsFlp; Act-FRT>Y>FRT-Gal4, UAS-nlsGFP (Ito et al., 1997), UAS-Atg5RNAi (HMS01244, BL34899 and JF02703, BL27551).
2.2. Immunostainings and fluorescence microscopy
For standard immunostainings, intestines were dissected in 1 × PBS (10 mM NaH2PO4/Na2HPO4, 175 mM NaCl, pH7.4), and fixed in 4% paraformaldehyde for 25 min at room temperature. Samples were washed with 1 × PBT (0.1% Triton X-100 in 1 × PBS) and blocked in 3% BSA in 1 × PBT for 45 min. Primary antibodies were added to the samples and incubated at 4 °C overnight. The following primary antibodies were used: mouse mAb anti-Dl (C594.9B, 1:50, developed by S. Artavanis-Tsakonas, DSHB), mouse mAb anti-Prospero (MR1A, 1:100, developed by Chris Doe, DSHB), mouse anti-Cytochrome C (1:25, BD Biosciences), rabbit anti-pH3 (pSer10, Millipore, 1:2000), rabbit anti-GM130 (Abcam, 1:200), rabbit anti-Cova (this study, 1:100). Primary antibodies were detected using fluorescent-conjugated secondary antibodies from Jackson ImmunoResearch Laboratories. Secondary antibodies were incubated for 2 h at room temperature. DAPI (Sigma-Aldrich; 0.1 μg/ml) was added after secondary antibody stainings. The samples were mounted in mounting medium (70% glycerol containing 2.5% DABCO). All images were captured using a Zeiss inverted confocal microscope and were processed in Adobe Photo-shop and Illustrator.
2.3. Mosaic analyses
ISC clones were induced using MARCM system (Lee and Luo, 2001) by heat shocking 3–5 day-old adult flies at 37 °C for 60 min. Flies were maintained at 25 °C and transferred to new vials with fresh food every day. The sizes of the marked clones were assayed at 6 days after clone induction (6D ACI, clones from 5–10 midguts for each genotype were assayed). For AY-Gal4-mediated ectopic expression or RNAi knockdown (Act5C-FRT>Y>FRT-Gal4, UAS-mRFP or UAS-nlsGFP), adult flies with proper genotypes were heat-shocked for 1 h at 37 °C. Adult flies were reared at room-temperature and dissected at the time points indicated in the text after clone induction.
2.4. Starvation
Flies were cultured in vials with a disc of filter paper soaked with 5% sucrose at room temperature, and transferred to new vials with filter paper disc supplemented with 5% sucrose. Flies were starved for 3 days before analysis.
2.5. Oligomycin feeding
Oligomycin (Sigma-Aldrich) was dissolved in 100% DMSO at a concentration of 1 mg/ml and stored at −20 °C. Oligomycin was added at a final concentration of 10 μg/ml to the surface of the food. Flies were transferred to fresh vials with drugs supplemented every day. Flies were fed on the food with drug for 3 days before being analyzed.
2.6. Cell death detection (TUNEL assay)
Intestines were dissected in 1 × PBS and fixed in 4% paraformaldehyde for 25 min at room temperature. Samples were washed with 1 × PBT for 5 min. Cell death was detected by using In Situ Cell Death Detection Kit TMR Red (Roche) according to manufacturer’s instruction.
3. Data analysis
The number of intestines scored is indicated in the text. The number of esg+ cells was determined in at least five different guts. To determine the number of esg+ cells, confocal images from the posterior midgut were acquired using 40 × lens/1.0 zoom. The relative number of esg+ cells was determined using Image-Pro Plus software from each confocal image. Image-Pro-Plus software was used for the intensity of Mito-GFP fluorescence quantification. IOD (integrated optical density) value per esg+ cell (IOD/esg+) was used. At least 4 different images were analyzed for each sample. Statistical analysis was done using the Student’s t-test. PEMS 3.1 software was used for SD analyses and Sigma plot software for graph generation. The graphs were further modified using Adobe Photoshop and Illustrator.
4. Results
4.1. Wildtype progenitors, but not tumor cells, are resistant to apoptosis in the adult Drosophila midgut
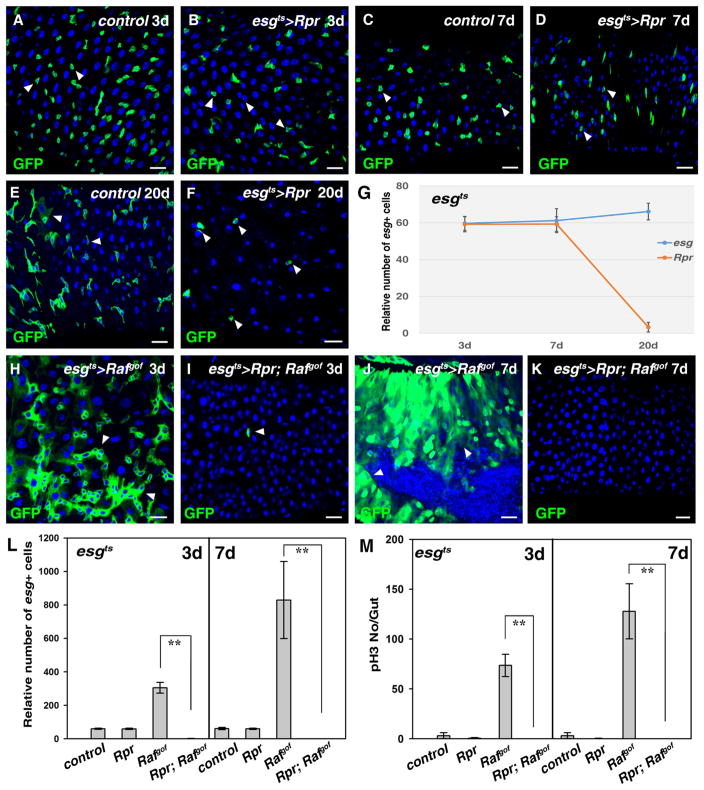
To examine the effect of cellular damage on wildtype stem cells, we induced cell death effectors in gut progenitors (ISCs and EBs), using esgGal4 and the temperature-sensitive Gal4 repressor, tubGal80ts (esgts)(Micchelli and Perrimon, 2006). Expression of the apoptotic factors, Reaper (Rpr), Hid (inhibitors of Drosophila Inhibitor of Apoptosis-1, DIAP-1), and Drosophila p53, in ISCs revealed that wildtype progenitors are resistant to apoptosis (Fig. 1A–G and data not shown). No obvious defects were observed in progenitors expressing the apoptotic factors for 3 and 7 days (Fig. 1A–D). Consistent with the role of Rpr in apoptosis, cell death (by TUNEL assay) could be observed in some esg+ cells after induction of Rpr for 7 days (Fig. S1). However, at 20 days of apoptosis induction, we observed a significant reduction in the number of esg+ progenitors, with the remaining cells showing aberrant cell morphology (Fig. 1E–G). These data are consistent with previous reports that wildtype progenitors are difficult to be ablated (Jiang et al., 2009; Lu and Li, 2015).
Fig. 1.
Tumor cells are more sensitive than wildtype progenitors to apoptosis in the adult Drosophila midgut. (A) Progenitors detected using esgGal4, UAS-GFP (green) in control midguts at 29 °C for 3 days (white arrowheads). (B) No obvious defects are observed in esgts>Rpr intestines at 29 °C for 3 days (white arrowheads). (C) Progenitors (green) in control midguts at 29 °C for 7 days (white arrowheads). (D) No obvious defects are observed in esgts>Rpr intestines at 29 °C for 7 days (white arrowheads). (E) Progenitors (green) in control midguts at 29 °C for 20 days (white arrowheads). Note that some large cells with GFP signal can be observed. (F) The number of esg+ cells is significantly reduced in esgts>Rpr intestines at 29 °C for 20 days (white arrowheads). Note that the morphology of the remaining esg+ cells is aberrant. (G) Quantification of the relative number of esg+ cells in the intestines of different genotypes at the indicated time points. n= 10–15 intestines. Mean ± SD is shown. **p<0.001. (H) Intestinal tumors in esgts>Rafgof intestines at 29 °C for 3 days (white arrowheads). (I) Tumor formation observed in esgts>Rafgof intestines is completely inhibited by co-expression of Rpr, and almost all esg+ cells are eliminated (white arrowhead). (J) Intestinal tumors in esgts>Rafgof intestines at 29 °C for 7 days (white arrowheads). Note that the intestines are deformed and tumor cells invade into the gut lumen. (K) All esg+ cells are eliminated in esgts>Rpr, Rafgof intestines at 29 °C for 7 days. (L) Quantification of relative number of esg+ cells in the intestines of different genotypes at 29 °C for 3 and 7 days, respectively. Note that because esgts>Rafgof intestines are highly deformed due to the formation of tumors, it is very difficult to accurately count the number of esg+ cells in these intestines. n=10–15 intestines. Mean ± SD is shown. **p<0.001. (M) Quantification of pH3 staining per gut in the intestines of different genotypes at 29 °C for 3 and 7 days, respectively. n=10–15 intestines. Mean ± SD is shown. **p<0.001. Blue indicates DAPI staining for DNA. Scale bars: 20 μm.
Next, we examined how tumor cells derived from ISCs respond to apoptosis. Ectopic activation of EGFR signaling in ISCs, following the expression of a constitutively active form of Raf (Rafgof), results in the formation of heterogeneous tumors (Fig. 1H) (Brand and Perrimon, 1994; Jiang et al., 2011; Markstein et al., 2014), reminiscent of the observation that ectopic activation of EGFR signaling is associated with human colorectal carcinoma (CRC) (Radtke and Clevers, 2005). Strikingly, when Rpr and Rafgof were co-expressed after 3 days of co-induction, not only the formation of Rafgof tumors was completely suppressed, but also, as no esg+ cells remained, Rafgof expressing ISC cells were almost completely ablated (Fig. 1H, I, and L). After 7 days of Rpr and Rafgof co-induction, esg+ cells were completely ablated from the whole intestines (Fig. 1J–L). Consistently, no pH3 positive cells were observed compared to esgts>Rafgof (Fig. 1M). A time course analysis from 0 to 3 days was performed to determine how apoptosis suppresses tumor growth. A transient increase of ISC number, together with an increase of pH3 positive cells, was detected in esgts>Rpr; Rafgof intestines at day 1. Most of the esg+ cells expressing Rpr and Rafgof were morphologically abnormal and some cell debris were observed at the 2nd day, while only some cell debris were observed at the 3rd day (Fig. S2). These data demonstrate that, unlike wildtype ISCs, Rafgof ISCs and their progenies are very sensitive to apoptosis.
4.2. Rafgof tumors are sensitive to autophagy induction in the Drosophila adult midgut
We further examined how wildtype progenitors and tumor cells respond to another kind of cellular damage, autophagy. Autophagy is a conserved catabolic cellular process in which bulk cytosol, protein aggregates, and organelles are sequestered and degraded in lysosomes (He and Klionsky, 2009; Mizushima et al., 2008). Autophagy is upregulated under stress conditions such as nutrient deprivation, hypoxia, heat, and drug treatment (Kondo et al., 2005; Levine, 2007). Previous studies have demonstrated that induction of the Ser/Thr kinase Atg1 can induce the formation of autophagosomes (He and Klionsky, 2009; Scott et al., 2007). Thus, we tested whether overexpression of Atg1 in the intestines could induce the formation of cytoplasmic vesicles containing GFP-Atg8/LC3, a hallmark of autophagosome (Juhász et al., 2008; Scott et al., 2007)(Fig. S3). GFP-Atg8-positive vesicles were barely observed when GFP-Atg8 was expressed alone, either using the FLP-out system (AY) or esgGal4 (Fig. S3A, D, and data not shown). However, GFP-Atg8-positive vesicles were readily detectable when Atg1 and GFP-Atg8 were co-expressed (Fig. S3B–D), demonstrating that overexpression of Atg1 in ISCs induces autophagy.
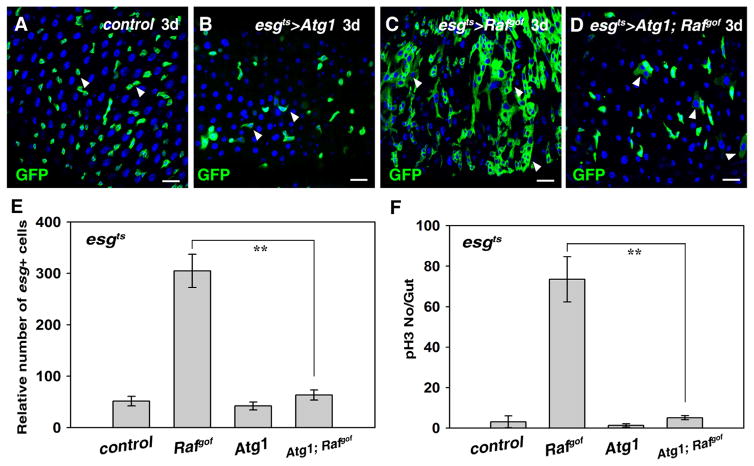
We did not detect any obvious defects in progenitor identity progeny differentiation when Atg1 was overexpressed in wildtype ISCs at 7 days, indicating that wildtype progenitors are also resistant to autophagy induction (Fig. 2A, B, Fig. S2, 4 and data not shown). Previous studies have found that both positive and negative effects of autophagy are implicated in tumorigenesis, making the contribution of autophagy to tumorigenesis controversial (Levine and Kroemer, 2008; Liang et al., 1999; Roy and Debnath, 2010). In contrast to its role as a tumor suppressor (Levine and Kroemer, 2008; Liang et al., 1999; Roy and Debnath, 2010), no obvious defects were found when autophagy was inhibited in the intestines (Fig. S6 and data not shown). Next, we examined the induction of autophagy during gut tumor formation. Surprisingly, the formation of Rafgof tumors was drastically suppressed when Atg1 was co-expressed in ISCs (Fig. 2A–D). The number of esg+ cells in esgts> Atg1, Rafgof intestines was dramatically reduced compared to that of esgts>Rafgof (Fig. 2D and E). Consistently, the number of pH3 positive cells was also significantly reduced compared to esgts>Rafgof intestines (Fig. 2F). A time course analysis from 0 to 3 days was performed to determine how autophagy suppresses tumor growth, revealing that Rafgof tumors were steadily suppressed by Atg1 co-induction (Fig. S2). Altogether, these data indicate that intestinal Rafgof tumors are sensitive to autophagy induction.
Fig. 2.
Tumorigenesis caused by Rafgof can be effectively inhibited by induction of autophagy in the Drosophila adult midgut. (A) Wildtype progenitors expressing esgGal4, UAS-GFP (green) in midguts at 29 °C for 3 days (white arrowheads). (B) No obvious defects are observed in esgts>Atg1 intestines (white arrowheads). (C) Intestinal tumors in esgts>Rafgof intestines at 29 °C for 3 days (white arrowheads). (D) Tumor formation in esgts>Rafgof intestines is significantly inhibited by co-expressing Atg1 (white arrowheads). (E) Quantification of the number of esg+ cells in the intestines of different genotypes. Note that because esgts>Rafgof intestines are highly deformed due to the formation of tumors, it is very difficult to accurately count the number of esg+ cells in these intestines. n=10–15 intestines. Mean ± SD is shown. **p<0.001. (F) Quantification of pH3 staining per gut in the intestines of different genotypes. n=10–15 intestines. Mean ± SD is shown. **p<0.001. Blue indicates DAPI staining for DNA. Scale bars: 20 μm.
4.3. Rafgof tumors are susceptible to autophagy induction in the Drosophila Malpighian tubules
To extend these observations to a different tissue, we expressed Rafgof in renal stem cells (RNSCs) of the Malpighian tubules, an organ analogous to mammalian kidney. As expression of Rafgof in RNSCs also leads to the formation of large heterogeneous tumors (Fig. S5) (Li et al., 2015), we co-expressed Rafgof and Atg1 in RNSCs using the FLP-out system (AY). Consistent with the results in the intestines, induction of Atg1 in these Rafgof renal tumors could almost completely suppress tumor formation (Fig. S5A–D).
Autophagy is negatively regulated by the nutrient sensor mammalian target of rapamycin (mTOR) (He and Klionsky, 2009). Thus, when mTOR is inhibited, autophagy is induced in response to nutrient deprivation. Consistent with the results observed with Atg1 overexpression, starvation could significantly inhibit the formation of renal tumors, albeit to a lesser extent (Fig. S5E). A similar result was observed in the intestines when flies were starved (data not shown). These results are also consistent with a previous study showing that feeding flies with rapamycin could significantly inhibit intestinal Rafgof tumors (Markstein et al., 2014). Collectively, these data indicate that Rafgof tumors are sensitive to autophagy induction.
To further demonstrate that the suppression of Rafgof tumors in the presence of Atg1 expression is mediated by autophagy, we depleted essential components required for the formation of autophagosomes. Depletion of essential components required for the formation of autophagosomes (such as Atg5) could significantly relieve the suppression of tumor formation associated with Atg1 expression (Fig. S6 and data not shown), which is consistent with autophagy induction suppressing tumorigenesis associated with Rafgof expression.
4.4. Notch-signaling-deficient progenitor tumors are also sensitive to autophagy induction in the adult Drosophila midgut
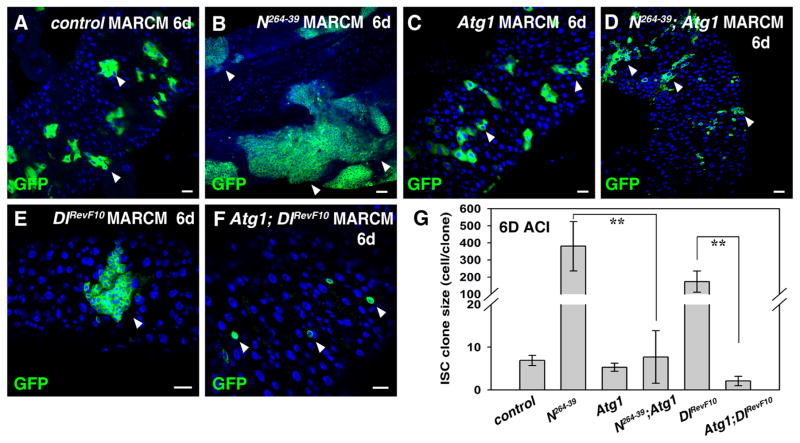
To determine whether the observed sensitivity of tumorigenesis to autophagy is tumor type specific or general, we generated tumors in the gut by perturbing the activity of the Notch signaling pathway. Notch signaling is not only required for ISC proliferation but also for progeny differentiation (Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). Loss of the Notch receptor leads to the formation of ISC-like tumors (Fig. 3A and B). When Atg1 was induced in Notch mutant clones, the formation of ISC-like tumors was dramatically suppressed (Fig. 3C, D, and G). Furthermore, loss of Delta (Dl) function, the ligand for Notch, also results in the formation of heterogeneous tumors (Fig. 3E), and the formation of these heterogeneous tumors was almost completely inhibited by Atg1 over-expression (Fig. 3F and G). These data demonstrate that tumors resulting from defective Notch signaling are also sensitive to autophagy induction, indicating that the tumor sensitivity to autophagy is not pathway specific.
Fig. 3.
Tumor formation in the absence of Notch signaling can be effectively inhibited by induction of autophagy in the adult Drosophila midgut. (A) ISC MARCM clones (green) in FRT control 6 days after clone induction (6D ACI) (white arrowheads). (B) Progenitor tumors (green) are formed in N264-39 ISC clones (6D ACI) (white arrowheads). (C). ISC MARCM clones (green) expressing Atg1 (6D ACI) (white arrowheads). (D) Progenitor tumors observed in N264-39 ISC clones are significantly inhibited by co-expression of Atg1 (white arrowheads). (E) Progenitor tumors (green) are formed in DlRevF10 ISC clones (6D ACI) (white arrowhead). (F) Progenitor tumors observed in DlRevF10 ISC clones are almost completely inhibited by co-expression of Atg1 (white arrowheads). (G) Quantification of relative size of ISC clones (cell/clone) in the intestines of different genotypes. Note that both N264-39 and DlRevF10 ISC clones are highly deformed, making it difficult to accurately count the number of mutant cells. n=5–10 intestines. Mean ± SD is shown. **p<0.001. Blue indicates DAPI staining for DNA. Scale bars: 20 μm.
4.5. The tumor suppression capacity of autophagy is not mediated by caspase-dependent apoptosis
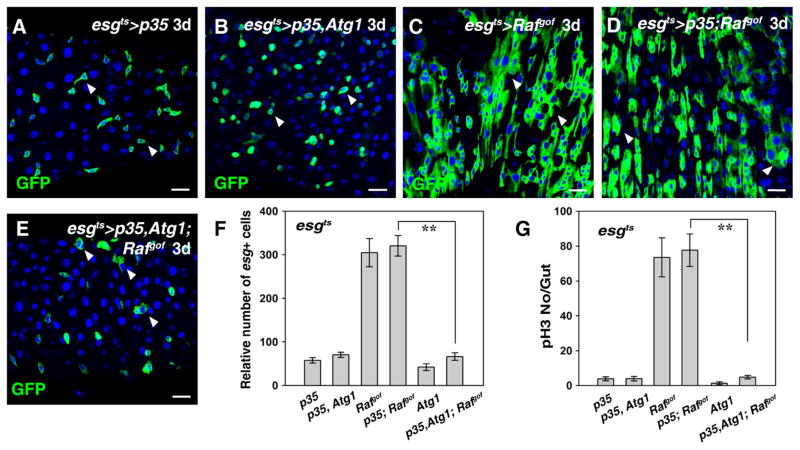
Previous studies have shown that induction of autophagy in certain tissues could induce apoptosis, thereby elimination of the cells (Berry and Baehrecke, 2007; Gorski et al., 2003; Lee and Baehrecke, 2001; Nezis et al., 2009; Scott et al., 2007). Thus, we examined whether apoptosis plays a role in autophagy-mediated suppression of tumorigenesis. Apoptosis is executed by caspases, which can be inhibited by the expression of anti-apoptotic p35 (Hay et al., 1994). To our surprise, no rescue of autophagy-mediated suppression of intestinal Rafgof tumors was observed in the presence of ectopic p35 expression, in terms of the number of esg+ cells and pH3-positive cells (Fig. 4). Consistently, no cell death (by TUNEL assay) was observed in Atg1 expressing cells (Fig. S7). These data indicate that caspase-dependent apoptosis is not involved in the suppression of autophagy-mediated suppression of tumorigenesis.
Fig. 4.
Atg1-mediated suppression of Rafgof tumor formation does not rely on caspase-dependent apoptosis. (A) Progenitors in esgts>p35 midgut at 29 °C for 3 days (white arrowheads). (B) No obvious effects are observed in intestines co-expressing Atg1 and p35 at 29 °C for 3 days (white arrowheads). (C) Intestinal tumors are formed in esgts>Rafgof intestines at 29 °C for 3 days (white arrowheads). (D) ISCs in esgts>p35; Rafgof midgut at 29 °C for 3 days (white arrowheads). (E) Atg1-mediated suppression of Rafgof tumor formation cannot be rescued by co-expression of p35 (white arrowheads). (F) Quantification of the number of esg+ cells in the intestines of different genotypes. Note that because esgts>Rafgof intestines are highly deformed due to the formation of tumors, it is very difficult to accurately count the number of esg+ cells in these intestines. n=10–15 intestines. Mean ± SD is shown. **p<0.001. (G) Quantification of pH3 staining per gut in the intestines of different genotypes. n=10–15 intestines. Mean ± SD is shown. **p<0.001. Blue indicates DAPI staining for DNA. Scale bars: 20 μm.
4.6. Autophagy-mediated suppression of tumorigenesis is associated with down-regulation of mitochondria
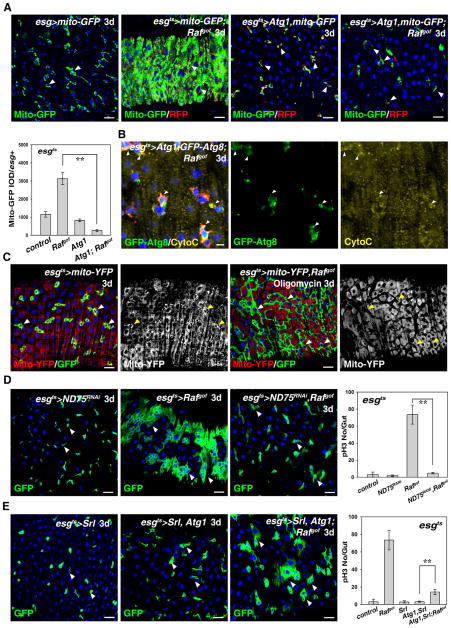
Autophagy is a conserved catabolic process in which bulk cytosol, protein aggregates, and organelles are sequestered and degraded in the lysosome (He and Klionsky, 2009; Mizushima et al., 2008). Interestingly, while autophagosomes were barely observed in Rafgof tumors, large autophagosomes were present in Atg1, Rafgof -expressing cells. These autophagosomes were larger than those in Atg1-expressing cells alone, indicating that an active autophagy process occurred in the tumor cells (Fig. S3E). Previous studies have demonstrated that mitochondria play important roles in energy production, cell proliferation/differentiation, and signaling transduction (Kabekkodu et al., 2015; McBride et al., 2006; Pinto et al., 2015; Wanet et al., 2015). Strikingly, mitochondria staining was dramatically increased in Rafgof tumors, indicating that increase of mitochondria in tumor cells may be involved in tumor cell proliferation (Fig. 5A), Interestingly, mitochondria staining in esgts>Atg1, Rafgof cells was drastically reduced compared with those of Rafgof tumors (Fig. 5A). Consistently, the intensity of Mito-GFP fluorescence (Mito-GFP IOD/esg+) was significantly reduced by induction of Atg1 expression compared with that of Rafgof tumors (Fig. 5A). These data suggest that the reduction of mitochondria in these tumors may be responsible for autophagy-mediated tumor suppression. Further examination of the content of these autophagosomes showed that these autophagosomes indeed contained mitochondria and Golgi apparatus as well (Fig. 5B and Fig. S8).
Fig. 5.
Mitochondria affect Atg1-mediated inhibition of Rafgof tumors. (A) Mitochondria (green, mito-GFP) stainings in Rafgof tumors (esgts>mRFP) are greatly increased, but are significantly reduced by induction of autophagy (compare the 2nd and 4th panels) (white arrowheads). The intensity of Mito-GFP fluorescence (Mito-GFP IOD/esg+) is also greatly reduced by induction of autophagy. (B) Mitochondria (Cyto-C, yellow) are present within autophagosomes (green, labeled with GFP-Atg8) (white arrowheads). (C) Rafgof tumors (green) can be inhibited by feeding oligomycin (white arrowheads). Note that, for an unknown reason, mitochondria staining (by mito-YFP, red) is greatly reduced in cells expressing Rafgof after oligomycin feeding (compare the 2nd panel and 4th panels, yellow arrowheads). (D) Knockdown of ND75 greatly inhibits Rafgof tumor formation (white arrowheads). Quantification of pH3 numbers per gut in the intestines with different genotypes. n=10–15 intestines. Mean ± SD is shown. **p<0.001. (E) Increasing mitochondrial biogenesis and activity can rescue Atg1-mediated inhibition of Rafgof tumors (white arrowheads, compare with the 2nd panel in D). Quantification of pH3 staining per gut in the intestines of different genotypes. n=10–15 intestines. Mean ± SD is shown. **p<0.001. Blue indicates DAPI staining for DNA. Scale bars: 5 μm (A) and 10 μm (B–E).
As mitochondria produce ATP, the energy critical for many essential cellular activities, including proliferation and differentiation, we tested whether inhibition of ATP production could mimic autophagy-mediated tumor suppression. Thus, we fed flies with the ATP synthesis inhibitor, oligomycin, which strikingly inhibited tumorigenesis (Fig. 5C). Of note for unknown reason, mitochondria signals in esg+ cells in the oligomycin-fed Rafgof flies were reduced (Fig. 5A and C). To further confirm the role of mitochondria activity in tumorigenesis, we inhibited the ATP synthesis electron transport by depleting ND75, a subunit of the complex I. Consistent with the oligomycin result, inhibition of ND75 suppressed tumorigenesis associated with Rafgof expression. The number of esg+ cells and ISCs undergoing mitosis were dramatically reduced by ND75 depletion (Fig. 5D). Altogether, these data suggest that reduction of mitochondria, and the reduction of ATP production as a consequence, may be responsible for autophagy-mediated tumor suppression.
Finally, we tested whether an increase in the number and activity of mitochondria in these tumors could alleviate the tumor suppression capacity of autophagy. Previous studies have shown that ectopic expression of the Drosophila PGC-1 homolog Spargel (Srl) increases mitochondria biogenesis and activity (Rera et al., 2011; Tiefenböck et al., 2009). Strikingly, expression of Srl could significantly alleviate the suppression of tumorigenesis by autophagy, as the number of esg+ cells and ISCs undergoing mitosis were significantly increased in the presence of Srl (Fig. 5E). Collectively, these data demonstrate that autophagy-mediated suppression of tumorigenesis is likely achieved, in part, through down-regulation of mitochondria.
5. Discussion
Adult stem cells play an essential role in the maintenance of tissue homeostasis. Environmental and cellular insults leading to cellular injuries, like DNA damage, dramatically impact stem cell functions and can lead to organ failure or cancer development (Morrison and Spradling, 2008; Radtke and Clevers, 2005). Yet little is known about the mechanisms by which adult stem cells respond to such cellular damages and resume normal cellular functions. Our data demonstrate that wildtype progenitors, unlike tumor cells, are resistant to cellular damages, like apoptosis and autophagy. It will be interesting to study the mechanism (s) underlying this difference in sensitivity to damage.
We find that tumor cells clearly show different sensitivities to apoptosis and autophagy: apoptosis induction can completely eliminate all tumor cells in a short period of time, while inducing autophagy in tumors can only significantly inhibit tumorigenesis, but cannot completely ablate tumor cells (Figs. 1–3). Previous studies have shown that high levels of autophagy in certain dying cells during metamorphosis and oogenesis in Drosophila act in concert with the apoptotic machinery to promote cell elimination, and ectopic induction of autophagy has been found to lead to apoptotic cell death (Berry and Baehrecke, 2007; Gorski et al., 2003; Lee and Baehrecke, 2001; Nezis et al., 2009; Scott et al., 2007). However, we find that autophagy-mediated tumor suppression in Rafgof gut tumors does not act through caspase-dependent apoptosis: (1) apoptosis induction can rapidly eliminate all tumor cells, while autophagy induction only suppresses tumor formation, but not elimination of esg+ cells, and significant amount of esg+ cells can still be observed in esgts>Atg1, Rafgof intestines after 7 days (Figs. 1, 2, Fig. S2, and data not shown); and (2) no cell death (by TUNEL assay) was observed when Atg1 was induced, and ectopic expression of the caspase inhibitor p35 could not rescue autophagy-mediated suppression of tumorigenesis (Fig. S7 and Fig. 4). These data indicate that different downstream effectors are used in apoptosis- and autophagy-mediated tumor-igenesis suppression, respectively.
Self-digestion of subcellular components through autophagy provides an energy and nutrient source allowing temporary survival of starvation. Autophagy has been implicated in many developmental and disease contexts in multicellular organisms, including cancer, although the contribution of autophagy to tumor-igenesis remains controversial (Chang and Neufeld, 2010; He and Klionsky, 2009; Klionsky and Emr, 2000; Levine, 2007; Meijer and Codogno, 2006; Mizushima et al., 2008; Roy and Debnath, 2010; Zirin and Perrimon, 2010). We examined the response of tumor cells to autophagy in vivo, and provide evidence that autophagy suppresses tumorigenesis likely through the sequestration and degradation of mitochondria. As we found that not only mitochondria but also some Golgi material is engulfed when autophagy is induced, mitochondria are only parts of the subcellular components engulfed by autophagosomes (Fig. 5 and Fig. S8), indicating that the elimination of mitochondria we observe does not correspond to the specialized process of eliminating dysfunctional mitochondria through autophagy, termed mitophagy (He and Klionsky, 2009; Kim et al., 2007; Mizushima et al., 2008).
Previously, JNK signaling has been shown to protect flies during bacterial infection-induced ROS/oxidative stress by stimulating the expression of several Atg genes (Tang et al., 2013; Wu et al., 2009). However, the induction of Atg genes was found to be transient, reaching peak levels at 4–6 h after ROS induction, followed by a dramatic reduction 8 h after bacterial infection (Tang et al., 2013; Wu et al., 2009). Consistent with our observations that autophagy suppresses tumorigenesis, Atg1 overexpression effectively blocked JNK activation and ISC proliferation under oxidative stress conditions (Tang et al., 2013).
In human, CRC is the second leading cause of cancer mortality in the western world (Radtke and Clevers, 2005). Oncological studies of a genetic model for CRC have established that CRC development involves multiple steps and that activation of receptor tyrosine kinases, particularly EGFR signaling, is an early event in the development of colon adenomas (Calcagno et al., 2008; Walther et al., 2009). Similar to the observations in mammals, ectopic activation of EGFR signaling in Drosophila ISCs by expressing Rafgof results in the formation of heterogeneous tumors, which provide an excellent model to study the role of cellular damages, like apoptosis and autophagy, in tumorigenesis in vivo (Markstein et al., 2014). Our genetic analyses demonstrate that tumor cells, but not wildtype progenitors, are very sensitive to apoptosis and autophagy. Based on the conservation of Drosophila and mammalian ISCs, our data suggest that induction of apoptosis and autophagy may provide a promising avenue for the clinical treatment of patients with tumors, like CRC.
In summary, our data demonstrate that unlike wildtype progenitors, tumor cells are very sensitive to apoptosis and autophagy induction in vivo, and that autophagy-mediated tumorigenesis suppression involves the engulfment of mitochondria. Therefore, based on the conservation of Drosophila and mammalian intestines, manipulation of apoptosis, autophagy levels and mitochondria activity may have significant outcomes on cancer therapy.
Supplementary Material
Acknowledgments
We are grateful to Thomas Neufeld, Yu Cai, Zhaohui Wang, Frei Christian, Lei Liu, Quan Chen, Developmental Studies Hybridoma Bank (DSHB), Bloomington Stock Center, and the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for stocks and reagents. We thank Jonathan Zirin and Hong-Wen Tang for comments. This work is supported by grants from the National Natural Science Foundation of China (Nos. 31271582 and 31471384), and Beijing Municipal Commission of Education (Nos. 010135336400 and 010155310500). NP is an investigator of the Howard Hughes Medical Institute.
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2016.01.040.
References
- Amcheslavsky A, Jiang J, Ip YT. Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell. 2009;4:49–61. doi: 10.1016/j.stem.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteau B, Hochmuth CE, Jasper H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell. 2008;3:442–455. doi: 10.1016/j.stem.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Raf acts downstream of the EGF receptor to determine dorsoventral polarity during Drosophila oogenesis. Genes Dev. 1994;8:629–639. doi: 10.1101/gad.8.5.629. [DOI] [PubMed] [Google Scholar]
- Calcagno SR, Li S, Colon M, Kreinest PA, Thompson EA, Fields AP, Murray NR. Oncogenic K-ras promotes early carcinogenesis in the mouse proximal colon. Int J Cancer. 2008;122:2462–2470. doi: 10.1002/ijc.23383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali A, Batlle E. Intestinal stem cells in mammals and Drosophila. Cell Stem Cell. 2009;4:124–127. doi: 10.1016/j.stem.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Chang YY, Neufeld TP. Autophagy takes flight in Drosophila. FEBS Lett. 2010;584:1342–1349. doi: 10.1016/j.febslet.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124:1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Diehn M, Cho RW, Clarke MF. Therapeutic implications of the cancer stem cell hypothesis. Semin Radiat Oncol. 2009;19:78–86. doi: 10.1016/j.semradonc.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski SM, Chittaranjan S, Pleasance ED, Freeman JD, Anderson CL, Varhol RJ, Coughlin SM, Zuyderduyn SD, Jones SJM, Marra MA. A SAGE approach to discovery of genes involved in autophagic cell death. Curr Biol. 2003;13:358–363. doi: 10.1016/s0960-9822(03)00082-4. [DOI] [PubMed] [Google Scholar]
- Guo Z, Ohlstein B. Bidirectional Notch signaling regulates Drosophila intestinal stem cell multipotency. Science. 2015:350. doi: 10.1126/science.aab0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YC, Zhou FL, Shen Y, Liao DF, Cao D. Apoptotic death of cancer stem cells for cancer therapy. Int J Mol Sci. 2014;15:8335. doi: 10.3390/ijms15058335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzler P, Simpson P. The choice of cell fate in the epidermis of Drosophila. Cell. 1991;64:1083–1092. doi: 10.1016/0092-8674(91)90263-x. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Jiang H, Edgar BA. Intestinal stem cells in the adult Drosophila midgut. Exp Cell Res. 2011;317:2780–2788. doi: 10.1016/j.yexcr.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Grenley MO, Bravo MJ, Blumhagen RZ, Edgar BA. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell. 2011;8:84–95. doi: 10.1016/j.stem.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, Edgar BA. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhász G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabekkodu SP, Chakrabarty S, Shukla V, Varghese VK, Singh KK, Thangaraj K, Satyamoorthy K. Mitochondrial biology: from molecules to diseases. Mitochondrion. 2015;24:93–98. doi: 10.1016/j.mito.2015.07.008. [DOI] [PubMed] [Google Scholar]
- Karpowicz P, Perez J, Perrimon N. The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development. 2010;137:4135–4145. doi: 10.1242/dev.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- Lee CY, Baehrecke EH. Steroid regulation of autophagic programmed cell death during development. Development. 2001;128:1443–1455. doi: 10.1242/dev.128.8.1443. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 2001;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Levine B. Cell biology: autophagy and cancer. Nature. 2007;446:745–747. doi: 10.1038/446745a. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Liu S, Cai Y. EGFR/MAPK signaling regulates the proliferation of drosophila renal and nephric stem cells. J Genet Genom. 2015;42:9–20. doi: 10.1016/j.jgg.2014.11.007. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Han L, Shi L, Lin X. Trachea-derived dpp controls adult midgut homeostasis in Drosophila. Dev Cell. 2013;24:133–143. doi: 10.1016/j.devcel.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Lin G, Xu N, Xi R. Paracrine wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature. 2008;455:1119–1123. doi: 10.1038/nature07329. [DOI] [PubMed] [Google Scholar]
- Liu JC, Lerou PH, Lahav G. Stem cells: balancing resistance and sensitivity to DNA damage. Trends Cell Biol. 2014;24:268–274. doi: 10.1016/j.tcb.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Li Z. No intestinal stem cell regeneration after complete progenitor ablation in Drosophila adult midgut. J Genet Genom. 2015;42:83–86. doi: 10.1016/j.jgg.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Magee Jeffrey A, Piskounova E, Morrison, Sean J. Cancer stem cells: impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283–296. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Dettorre S, Cho J, Neumüller RA, Craig-Müller S, Perrimon N. Systematic screen of chemotherapeutics in Drosophila stem cell tumors. Proc Natl Acad Sci USA. 2014;111:4530–4535. doi: 10.1073/pnas.1401160111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui W, Wang Q, Barber JP, Brennan S, Smith BD, Borrello I, McNiece I, Lin L, Ambinder RF, Peacock C, Watkins DN, Huff CA, Jones RJ. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008;68:190–197. doi: 10.1158/0008-5472.CAN-07-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Meijer AJ, Codogno P. Signalling and autophagy regulation in health, aging and disease. Mol Asp Med. 2006;27:411–425. doi: 10.1016/j.mam.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy flghts disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis IP, Lamark T, Velentzas AD, Rusten TE, Bjørkøy G, Johansen T, Papassideri IS, Stravopodis DJ, Margaritis LH, Stenmark H, Brech A. Cell death during Drosophila melanogaster early oogenesis is mediated through autophagy. Autophagy. 2009;5:298–302. doi: 10.4161/auto.5.3.7454. [DOI] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, Binari R, Booker M, Brennecke J, Perkins LA, Hannon GJ, Perrimon N. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- Perdigoto CN, Schweisguth F, Bardin AJ. Distinct levels of Notch activity for commitment and terminal differentiation of stem cells in the adult fly intestine. Development. 2011;138:4585–4595. doi: 10.1242/dev.065292. [DOI] [PubMed] [Google Scholar]
- Pinto MCX, Kihara AH, Goulart VAM, Tonelli FMP, Gomes KN, Ulrich H, Resende RR. Calcium signaling and cell proliferation. Cell Signal. 2015;27:2139–2149. doi: 10.1016/j.cellsig.2015.08.006. [DOI] [PubMed] [Google Scholar]
- Potten CS, Loeffler M. Stem cells: attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the crypt. Development. 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- Radtke F, Clevers H. Self-renewal and cancer of the gut: two sides of a coin. Science. 2005;307:1904–1909. doi: 10.1126/science.1104815. [DOI] [PubMed] [Google Scholar]
- Ren F, Wang B, Yue T, Yun EY, Ip YT, Jiang J. Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc Natl Acad Sci USA. 2010;107:21064–21069. doi: 10.1073/pnas.1012759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Bahadorani S, Cho J, Koehler, Christopher L, Ulgherait M, Hur, Jae H, Ansari, William S, Lo T, Jr, Jones DL, Walker, David W. Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 Homolog. Cell metabolism. 2011;14:623–634. doi: 10.1016/j.cmet.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexer BN, Engelman JA, Arteaga CL. Overcoming resistance to tyrosine kinase inhibitors: Lessons learned from cancer cells treated with EGFR antagonists. Cell Cycle. 2009;8:18–22. doi: 10.4161/cc.8.1.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Debnath J. Autophagy and tumorigenesis. Semin Immunopathol. 2010;32:383–396. doi: 10.1007/s00281-010-0213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RC, Juhász G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons Benjamin D, Clevers H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 2011;145:851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- Slizynska H. Salivary chromosome analysis of the white-facet region of Drosophila melanogaster. Genetics. 1938;23:291–299. doi: 10.1093/genetics/23.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier DYR. No organ left behind: tales of gut development and evolution. Science. 2005;307:1902–1904. doi: 10.1126/science.1108709. [DOI] [PubMed] [Google Scholar]
- Staley BK, Irvine KD. Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr Biol. 2010;20:1580–1587. doi: 10.1016/j.cub.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima S, Hartenstein V. Genetic control of intestinal stem cell specification and development: a comparative view. Stem Cell Rev. 2012;8:597–608. doi: 10.1007/s12015-012-9351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang HW, Liao HM, Peng WH, Lin HR, Chen CH, Chen GC. Atg9 interacts with dTRAF2/TRAF6 to regulate oxidative stress-induced JNK activation and autophagy induction. Dev Cell. 2013;27:489–503. doi: 10.1016/j.devcel.2013.10.017. [DOI] [PubMed] [Google Scholar]
- Tiefenböck SK, Baltzer C, Egli NA, Frei C. The Drosophila PGC31 homologue Spargel coordinates mitochondrial activity to insulin signalling. EMBO J. 2009;29:171–183. doi: 10.1038/emboj.2009.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 2009;9:489–499. doi: 10.1038/nrc2645. [DOI] [PubMed] [Google Scholar]
- Wanet A, Arnould T, Najimi M, Renard P. Connecting mitochondria, metabolism, and stem Cell Fate. Stem Cells Dev. 2015;24:1957–1971. doi: 10.1089/scd.2015.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Hou SX. Regulation of intestinal stem cells in mammals and Drosophila. J Cell Physiol. 2010;222:33–37. doi: 10.1002/jcp.21928. [DOI] [PubMed] [Google Scholar]
- Wu H, Wang MC, Bohmann D. JNK protects Drosophila from oxidative stress by trancriptionally activating autophagy. Mech Dev. 2009;126:624–637. doi: 10.1016/j.mod.2009.06.1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung TM, Chia LA, Kosinski CM, Kuo CJ. Regulation of self-renewal and differentiation by the intestinal stem cell niche. Cell Mol Life Sci. 2011;68:2513–2523. doi: 10.1007/s00018-011-0687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirin J, Perrimon N. Drosophila as a model system to study autophagy. Semin Immunopathol. 2010;32:363–372. doi: 10.1007/s00281-010-0223-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.