Abstract
Lipopolysaccharide (LPS) O-antigen and enterobacterial common antigen (ECA) are two major polysaccharide structures on the surface of Salmonella enterica serovar Typhimurium. Previous studies have demonstrated that regulated truncation of LPS enhances the cross-reaction against conserved outer membrane proteins (OMPs) from enteric bacteria. We speculate that the regulation of both O-antigen and ECA may enhance the induction of immune responses against conserved OMPs from enteric bacteria. In this work we targeted rfbB and rffG genes which encode dTDP-glucose 4,6-dehydratases and share the same function in regulating O-antigen and ECA synthesis. We constructed a mutant, S496 (ΔrfbB6 ΔrffG7 ΔpagL73::TT araC PBAD rfbB-3), in which rfbB gene expression was dependent on exogenously supplied arabinose during in vitro growth and achieved the simultaneous tight regulation of both LPS and ECA synthesis, as demonstrated by the LPS profile and Western blotting using antisera against LPS and ECA. When administered orally, S. Typhimurium S496 was completely attenuated for virulence but still retained the capacity to colonize and disseminate in mice. In addition, we found that oral immunization with S496 resulted in increased immune responses against OMPs from enteric bacteria and enhanced survival compared with immunization with S492 possessing ΔrfbB6 ΔrffG8 mutations when challenged with lethal doses of Salmonella Choleraesuis or Salmonella Enteritidis. These results indicate that S. Typhimurium arabinose-regulated rfbB strain S496 is a good vaccine candidate, conferring cross-protection against lethal challenge with heterologous Salmonella.
Keywords: Salmonella Typhimurium, rfbB and rffG genes, Arabinose-regulated expression, Cross-protection
1. Introduction
Lipopolysaccharide (LPS) is one of the key virulence factors and a major surface component of Salmonella enterica serovar Typhimurium [1]. LPS is critical for successful Salmonella infection and survival in the host as the lack of LPS or modification of LPS will compromise many biological characteristics, including swimming and swarming motility [2], intestinal colonization [3], serum resistance [4], invasion/intracellular replication [5], and resistance to killing by macrophages [6]. Full-length LPS consists of three domains: lipid A, core oligosaccharide and O-antigen polysaccharide [1]. The O-antigen polysaccharide, which is composed of a polymer of repeating sugar units and is the primary surface antigen of Salmonella, contributes to defense against serum complement activation [7].
The enterobacterial common antigen (ECA) is the second major immunogenic antigen on the surfaces of Salmonella and other enteric bacteria [8]. ECA is composed of linear polysaccharide chains, consisting of a repetitive tri-saccharide composed of 4-acetamide-4,6-dideoxy-D-galactose (Fuc4NAc), N-acetyl-D-mannosaminuronic acid (ManNAcA) and N-acetyl-D-glucosamine (GlcNAc) in Salmonella [9]. Previous studies have demonstrated that ECA plays an important role in bacterial virulence [10] and that an ECA-deficient mutant was able to induce a protective immune response against lethal challenge of homologous and heterologous Salmonella when used as a live-attenuated vaccine in the mice model [11]. We and others have demonstrated that regulated O-antigen synthesis contributes to the induction of cross-reactive immune responses to conserved outer membrane proteins (OMPs), thereby enhancing cross protection against infection by multiple pathogens [12–14]. Regulated O-antigen synthesis can be achieved by the deletion of particular genes essential for O-antigen synthesis, such as pmi (manA) and galE, or by the replacement of the promoter of a gene of interest with a regulated promoter, such as the araC PBAD promoter [12,13]. The pmi and galE gene deletions resulted in mutants with a reversible rough LPS, in which the smooth LPS phenotype was restored when grown in the presence of mannose and galactose, respectively [15–17].
The dTDP-glucose 4,6-dehydratase, encoded by rfbB (rmlB) in the O-antigen gene cluster, is the second enzyme necessary for the synthesis of dTDP-rhamnose, which is the rhamnose donor for the S. Typhimurium O-antigen structure [18]. The rffG gene, which is located in the ECA gene cluster, is a functional homolog of rfbB and is responsible for synthesis of dTDP-Fuc4NAc, the precursor of Fuc4NAc in the ECA structure [9]. Previous results have demonstrated that these two genes are exchangeable [8,19]. We are planning to manipulate one gene to achieve the regulated synthesis of two major surface antigens, with the goal of maximal exposure of the conserved OMPs to the host immune system, thus inducing enhanced protective immunity.
In this study, we achieved regulated LPS and ECA synthesis via rfbB gene regulation by the arabinose-regulated araC PBAD promoter. In this system, LPS and ECA were synthesized in vitro, where arabinose was available, but not in vivo, where arabinose was absent. This vaccine strain displayed the wild-type phenotype at the time of immunization and became attenuated after colonization in the host tissue [13]. The resultant strain was evaluated for its capacity to induce immunity and cross-protection against S. Enteritidis and S. Choleraesuis.
2. Materials and methods
2.1. Bacterial strains, plasmids, media, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. The S. Typhimurium vaccine strains were derived from the highly virulent strain S100. S. Typhimurium cultures were grown at 37 °C in Luria–Bertani (LB) broth or on LB agar with or without 0.1% arabinose. Diaminopimelic acid (DAP) (50 µg/ml) was added for the growth of the Δasd strains [20]. LB agar containing 5% sucrose was used for sacB gene-based counterselection in the allelic exchange experiments [21].
Table 1.
Strains and plasmids used in this study.
| Strains and plasmids |
Description | Source or reference |
|---|---|---|
| Plasmids | ||
| pYA4278 | SacB mobRP4 R6K ori Cm+, pRE112-T-vector | [6] |
| pSS239 | ΔrfbB6 | pYA4278 |
| pSS240 | ΔrffG8 | pYA4278 |
| pYA4284 | ΔpagL7 | [35] |
| pSS242 | ctcgag AGGA gtcatt GTG | pSS241 |
| pSS243 | ctcgag AGGA gtcatt ATG | pSS241 |
| pSS244 | ctcgag GGAA gtcatt ATG | pSS241 |
| pSS245 | ctcgag GGAA gtcatt GTG | pSS241 |
| pYA4518 | GFP p15A ori Cm+ | [37] |
| pYA3700 | TT araC PBAD cassette plasmid; Apr | [39] |
| pSS250 | pYA4518-rfbB | |
| pSS251 | pYA4518-rffG | |
| Bacterial strains | ||
| S100 | Wild-type S. Typhimurium, isolated from infected duck |
[40] |
| S340 | S. Choleraesuis, wild-type virulent | [40] |
| S246 | S. Enteritidis, wild-type virulent | [40] |
| S490 | ΔrfbB6 | S100 |
| S491 | ΔrffG8 | S100 |
| S492 | ΔrfbB6 ΔrffG8 | S491 |
| S493 | ΔrfbB6 ΔrffG7 ΔpagL7 | S492 |
| S494 | ΔrfbB6 ΔrffG7 ΔpagL71:TT araC PBAD rfbB-1 | S493 |
| S495 | ΔrfbB6 ΔrffG7 ΔpagL72:TT araC PBAD rfbB-2 | S493 |
| S496 | ΔrfbB6 ΔrffG7 ΔpagL73:TT araC PBAD rfbB-3 | S493 |
| S497 | ΔrfbB6 ΔrffG7 ΔpagL74:TT araC PBAD rfbB-4 | S493 |
| E. coli | ||
| χ7232 |
endA1 hsdR17 () glnV44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 λpir deoR (Φ80dlac Δ (lacZ)M15) |
[41] |
| χ7213 |
thi-1 thr-1 leuB6 glnV44 fhuA21 lacY1 recA1 RP4- 2-Tc::Mu[λ pir] ΔasdA4 Δ(zhf-2:Tn10) |
[41] |
2.2. Plasmids and mutant strain construction
The primers used in this study are listed in Supplementary Table 1. DNA manipulations were performed as described elsewhere [22], and detailed procedures could be found in the supplementary materials and methods. Transformation of Escherichia coli and S. Typhimurium was accomplished by electroporation (2.5 kV, 5 ms). The transformants were selected on LB agar plates containing chloramphenicol (25 µg/ml). The construction of the ΔrfbB ΔrffG mutant and the arabinose-regulated mutant is described in the Supplementary Materials and Methods.
2.3. LPS and ECA analysis
LPS was analyzed as previously described by Hitchcock and Brown [23]. The ECA phenotypes were characterized by immunoblotting. ECA was prepared as previously described [24]. The isolated ECA samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the resulting bands were transferred to nitrocellulose membranes. The membranes were probed with a 1:1000 dilution of the murine anti-ECA monoclonal antibody mAb898 [24].
2.4. Growth kinetics and motility and sensitivity assays
For determining the growth kinetics of the wild-type strain S100 and the mutant strains, bacterial density was monitored by optical density (OD) at 600 nm every hour. LB plates containing 0.3% agar with or without arabinose were inoculated with each strain for the motility assays. For the polymyxin B and sodium deoxycholate (DOC) sensitivity assays, 100 µl of 100-fold diluted bacterial cultures that had been grown to 0.8 of OD600 were inoculated with polymyxin B at a final concentration of 0.1 µg/ml or DOC at a final concentration of 10 mg/ml for 1 h at 37 °C. The bacteria were diluted to the appropriate concentration and plated on LB plates, and bacterial colony counts were used to calculate survival rates. The assays were repeated three times.
2.5. Attachment and invasion assays
The human intestinal cell line INT407 was obtained from the American Type Culture Collection (ATCC strain CCL-6). The attachment and invasion tests were performed three times as described previously [25].
2.6. Determinations of virulence and colonization in mice
All animal research was conducted in compliance with the Animal Welfare Act and regulations related to experiments involving animals following the principles stated in the Guide for the Care and Use of Laboratory Animals. All efforts were made to minimize animal suffering during the experiments.
Six-week-old female BALB/c mice were obtained from Dashuo Experimental Animal Ltd. (Chengdu; Sichuan, China). Determination of the 50% lethal dose (LD50) (n = 5/group) and colonization evaluation (n = 3/group) were conducted as previously described [12]. Peyer’s patches (PP), spleen and liver tissues were harvested on day 6 post-infection to evaluate colonization.
2.7. Immunization and lethal Salmonella challenge
The bacterial cultures were prepared as previously described [12]. Each mouse was orally inoculated with 20 µl of buffered saline gelatin (BSG) containing 1.0 × 109 colony-forming units (CFU) of each strain on day 0 and was boosted on day 28 with the same dose of the same strain. Blood was obtained from mice by eye venous plexus bleeding at biweekly intervals after the first immunization.
For the lethal challenge studies, the immunized mice were challenged by oral inoculation with 6.5 × 108 CFU, 2.4 × 107 CFU and 2.4 × 107 CFU of the wild-type S. Typhimurium, S. Choleraesuis and S. Enteritidis, respectively. The LD50 values of the wild-type S. Typhimurium, S. Choleraesuis and S. Enteritidis in BALB/c mice were 5.0 × 105, 1.0 × 105 and 1.0 × 105 by oral administration, respectively (data not shown). The challenged mice were monitored and deaths were recorded daily for 30 days.
2.8. Quantitative enzyme-linked immunosorbent assay (ELISA)
S. Typhimurium LPS was purchased from Sigma (St. Louis, MO, USA). Salmonella outer membrane proteins (SOMPs) were prepared as previously described in [26]. A quantitative ELISA was used for the analysis of antibodies in serum to S. Typhimurium LPS and SOMPs. The wells of polystyrene 96-well flat-bottom microtiter plates were coated with 100 ng/well LPS or SOMPs suspended in 100-µl volumes of sodium carbonate/bicarbonate coating buffer (50 mM Na2CO3, 50 mM NaHCO3, 0.1% sodium azide, pH 9.6). Goat anti-mouse Ig (H + L) (BD Pharmingen; San Diego, CA) in PBS was added to extra wells without coating with OMPs or LPS in triplicate to determine the standard curve. The coated plates were incubated overnight at 4 °C, followed by blocking with PBS containing 10% fetal bovine serum (FBS) for 1 h at room temperature. A 100-µl volume of 200-fold diluted sample was added to each corresponding well in triplicate, and 100-µl volume of the mouse IgG (BD Pharmingen; San Diego, CA, USA) with twofold dilutions in PBS from 0.5 mg/ml was successively added to the well coated by goat anti-mouse Ig (H + L) for the standard curve. The plates were incubated for 1 h at 37 °C and were then treated with biotinylated goat anti-mouse IgG (Southern Biotechnology Associates; Birmingham, AL, USA). The wells were developed with a streptavidin-alkaline phosphatase conjugate (Southern Biotechnology Associates; Birmingham, AL, USA), followed by p-nitrophenylphosphate substrate (Sigma-Aldrich; St. Louis, MO, USA) in diethanolamine buffer (pH 9.8). Color development (absorbance) was recorded at 405 nm using an iMark™ Microplate Reader (Bio-Rad; Hercules, CA, USA). The standard curve was drawn using Curve Expert (Hyams DG; Starkville, MS, USA), and the concentration of serum antibodies was calculated using the standard curve.
2.9. Statistical analysis
Antibody titers were expressed as means ± SEM. Statistical analyses were performed using the GraphPad Prism 5 software package (Graph Software; San Diego, CA, USA) [27]. One- or two-way ANOVA were performed to determine the statistical significance of the differences between or among mean values for the various experimental and control groups. The log-rank test was used for survival curves. P < 0.05 was considered a significant difference.
3. Results
3.1. Mutant construction
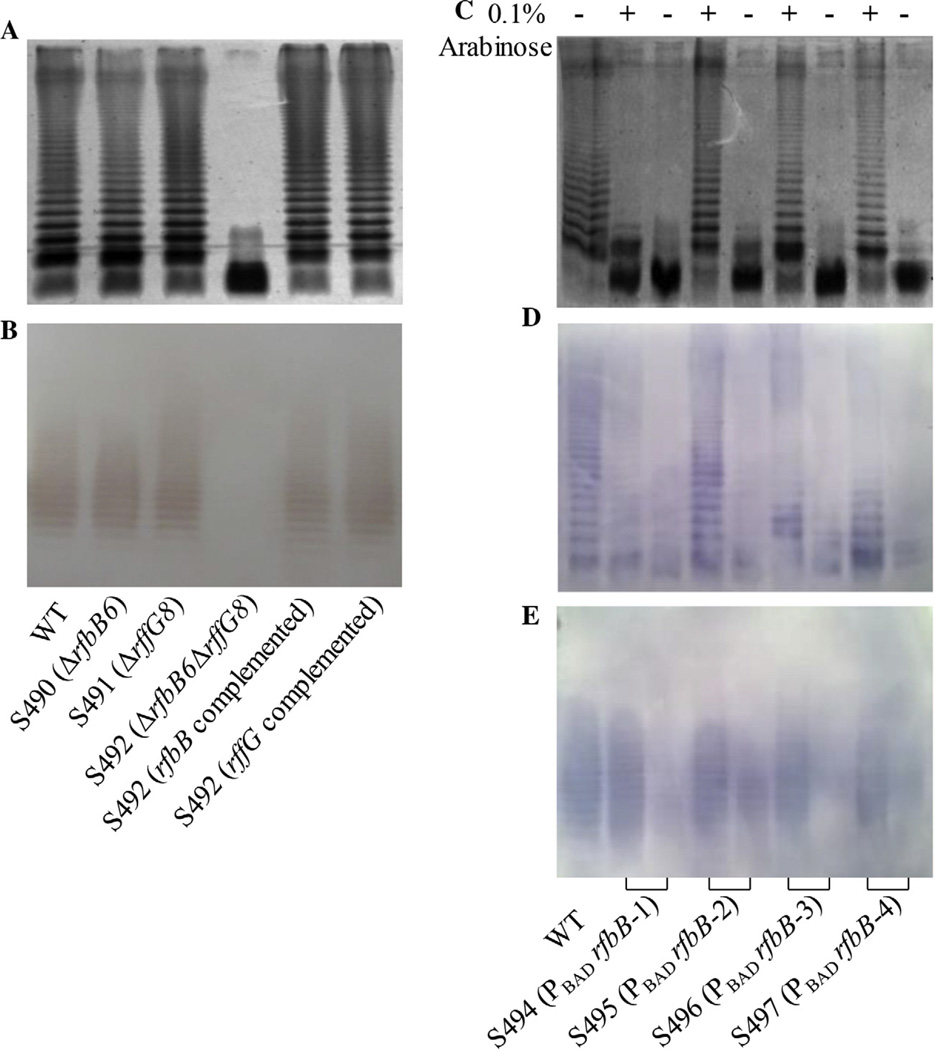
The single mutants S490 (ΔrfbB6) and S491 (ΔrffG8) had the same profiles of LPS and ECA as the parent wild-type strain, and double deletion of rfbB and rffG resulted in a mutant S492 with deficient LPS and ECA profiles (Fig. 1A and B), indicating that the proteins encoded by rfbB and rffG have the same function as dTDP-glucose 4,6-dehydratase and that the amount of dTDP-glucose 4,6-dehydratase encoded by either rfbB or rffG gene is sufficient for synthesizing the full LPS and ECA structures in LB media. To further elucidate this phenomenon, the two complement plasmids were constructed and transformed into the mutant S492 (ΔrfbB6 ΔrffG8). The LPS and ECA profiles indicated that each complement plasmid has capacity to restore truncated LPS and ECA to the wild-type state (Fig. 1A and B), which further demonstrated that the rfbB and rffG genes performed the same function in the LPS and ECA biosynthetic pathways [8,19].
Fig. 1.
Construction of Salmonella mutants. (A) LPS profiles. LPS from the wild-type S. Typhimurium S100 and the mutant strains S490 (ΔrfbB6), S491 (ΔrffG8), S492 (ΔrfbB6 ΔrffG8), S492 (rfbB complement plasmid) and S492 (rffG complement plasmid) were subjected to 12% SDS-PAGE and were silver-stained. (B) ECA profiles. ECA extracts from total membranes were separated by 12% SDS-PAGE and transferred to nitrocellulose membranes. The blots were probed with the ECA-specific mAb898. (C) LPS profiles of the wild-type S. Typhimurium S100 and the arabinose-regulated mutants. The LPS molecules from different mutant strains grown in nutrient broth with (+) or without (−) 0.1% arabinose were silver-stained after separation by 12% SDS-PAGE. (D) Western blots of LPS preparations from panel C. The blots were probed with anti-Salmonella group B antibodies. (E) ECA profiles of the wild-type S. Typhimurium S100 and the arabinose-regulated mutants. ECA from the wild-type S. Typhimurium S100 and the arabinose-regulated mutants grown in nutrient broth with (+) or without (−) 0.1% arabinose were separated by 12% SDS-PAGE and transferred to nitrocellulose membranes. The blots were probed with the ECA-specific mAb898.
To achieve regulated LPS and ECA synthesis, we applied the same strategy as we previously used for the regulation of wzy and rfaH expression in Salmonella to the rfbB gene [12,13]. We used the arabinose-regulated araC PBAD promoter to regulate rfbB expression to in turn regulate LPS and ECA synthesis. Four mutants with arabinose-regulated araC PBAD rfbB carrying variable Shine-Dalgarno (SD) sequences, ATG/GTG start codons or both were constructed (Table 1). The effects of arabinose-regulated rfbB on LPS and ECA synthesis were evaluated by silver staining and Western blotting. The mutants S496 (PBAD rfbB-3) and S497 (PBAD rfbB-4) produced an LPS and ECA pattern similar to that of the wild-type Salmonella when they were grown with arabinose and displayed the phenotypes of rough LPS and ECA-deficiency similar to that of the mutant S492 (ΔrfbB6 ΔrffG8) in the absence of arabinose, whereas the other two constructs did not show tight regulation by arabinose (Fig. 1C and E). Thus, S496 and S497 were selected for further phenotypic evaluation.
3.2. Phenotypic evaluation of the mutant strains
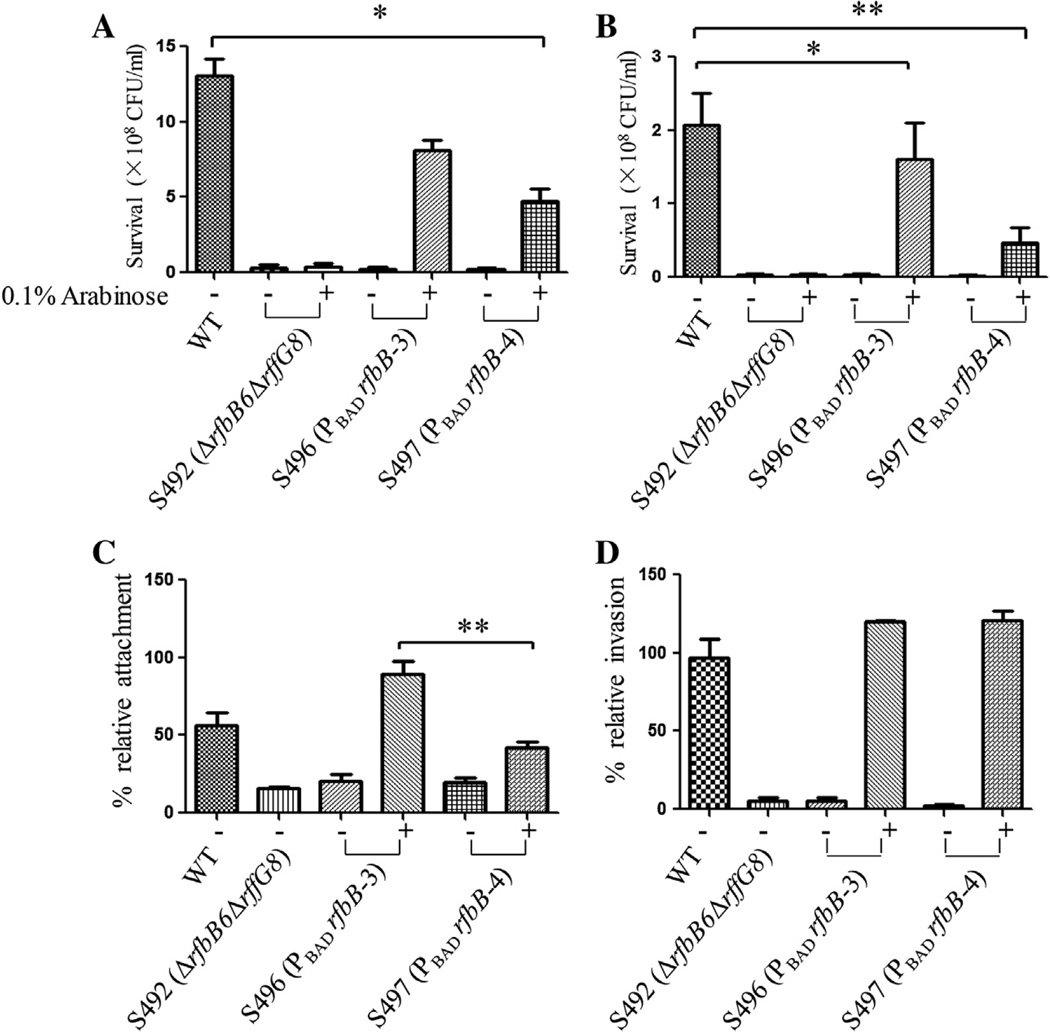
To investigate whether regulation of LPS and ECA synthesis affects Salmonella phenotypes, we performed several assays, including growth curve assays and assays of swarming capacity and susceptibility to environmental factors. The growth rates of the mutant strains were determined in LB broth with or without 0.1% arabinose. No differences were observed for growth rates of the mutants and the parent strain (Supplementary Fig. 1). As expected, the mutant S492 (ΔrfbB6 ΔrffG8) showed less ability to swarm in the absence of arabinose than the wild-type strain, and the arabinose-regulated mutants also had reduced motility capacity compared with the wild-type strain when grown in the absence of arabinose. The motility of the arabinose-regulated mutants was partially restored to wild-type levels when arabinose was available (Table 2). In assays examining susceptibility to environmental factors, the mutant S492 (ΔrfbB6 ΔrffG8) and the arabinose-regulated mutants also showed more sensitivity to polymyxin B and DOC than the wild-type strain in the absence of arabinose (P < 0.001) (Fig. 2B). When arabinose was supplied, the arabinose-regulated mutant S496 was restored to the wild-type level of resistance to polymyxin B but S497 still showed significant sensitivity to polymyxin B compared with the wild-type strain (P < 0.05) (Fig. 2A). Whereas both S496 and S497 exhibited significant sensitivity to DOC compared with the wild-type strain under arabinose availability (P < 0.05), S496 showed more resistance to DOC than S497.
Table 2.
Motility and virulence of S. Typhimurium strain S100 and mutant derivatives.
| Strains | 0.1% arabinose |
Motility (mm) on soft agar |
LD50 |
|---|---|---|---|
| S100 | − | 71 ± 2.00 | 5.0 × 105 |
| + | 75 ± 2.65 | ||
| S490 (ΔrfbB6) | − | 69 ± 2.60 | >2.5 × 109 |
| + | 70 ± 3.46 | ||
| S491 (ΔrffG8) | − | 72 ± 2.64 | 3.2 × 107 |
| + | 73 ± 3.00 | ||
| S492 (ΔrfbB6 ΔrffG8) |
− | 8 ± 1.00 | >2.3 × 109 |
| + | 8 ± 2.00 | ||
| S496 (ΔPBAD rfbB-3) | − | 8 ± 1.16 | >2 × 109 |
| + | 37 ± 2.00 | ||
| S497 (ΔPBAD rfbB-4) | − | 8 ± 0.00 | ND |
| + | 39 ± 3.00 |
ND: not determined.
Fig. 2.
Polymyxin B and DOC sensitivity and attachment/invasion assays for the mutant strains. (A) Polymyxin B sensitivity assay. Approximately 1 × 107 CFU/ml bacteria with 0.1 µg/ml polymyxin B were incubated for 60 min at 37 °C. The bacterial survival ratio was calculated after plating the bacteria on LB agar and counting colonies the following day. Values that are significantly different from those of the wild type as indicated by one-way ANOVA are indicated by asterisks (*P < 0.05). (B) DOC sensitivity assay. Approximately 1 × 107 CFU/ml bacteria with 10 mg/ml DOC were incubated for 60 min at 37 °C. The bacterial survival ratio was calculated after plating the bacteria on LB agar and counting colonies the following day. Values that are significantly different from those of the wild type as indicated by one-way ANOVA are indicated by asterisks (*P < 0.05, **P < 0.01). (C) Attachment efficiency of the Salmonella mutants to INT407 cells. The mutant strains were added to wells with INT407 cells to achieve an MOI of 5:1. A 1% Triton X-100 solution was added to lyse the INT407 cells after incubation for 60 min at 37 °C. After plating on petri dishes and overnight incubation, the colony counts were determined. Attachment was calculated as follows: percent attachment = 100 × (number of cell-associated bacteria/initial number of bacteria added). Values that are significantly different as determined by one-way ANOVA are indicated by asterisks (**P < 0.01). (D) Invasion efficiency of Salmonella mutants to INT407 cells. Prior to the lysis step, 100 µg/ml gentamicin was added to kill the extracellular bacteria, and the plate was incubated for an additional 60 min at 37 °C. Invasion was calculated as follows: percent invasion = 100 × (number of bacteria resistant to gentamicin/initial number of bacteria added).
3.3. Attachment and invasion assays
To evaluate the effects of the rfbB and rffG gene mutations on attachment and invasion, we examined the capacity of the mutant strains to attach to and invade INT407 human epithelial cells. The mutant S492 (ΔrfbB6 ΔrffG8) and the arabinose-regulated mutant strains in the absence of arabinose showed significant reductions in attachment (approximately 5-fold) compared with the parent strain (P < 0.05). No significant differences in attachment were observed between the arabinose-regulated mutant strains and the parent strain S100 in the presence of arabinose; similar results were observed for the invasion assays (Fig. 2C and D). The mutant S496 (PBAD rfbB-3) showed significantly higher attachment to the INT407 cells than the S497 mutant (PBAD rfbB-4) (89% vs 42%) (P < 0.01) (Fig. 2C), but no difference between these two mutants was observed for INT407 invasion (Fig. 2D). The mutant S496 (PBAD rfbB-3) possessed a more similar phenotype than S497 (PBAD rfbB-4) to the wild-type strain. S496 (PBAD rfbB-3) was selected for further evaluation in animal experiments.
3.4. Virulence and colonization of mutant strains in BALB/c mice
To assess the virulence of the mutants, the oral LD50 values for each strain were determined in BALB/c mice. Groups of female BALB/c mice received gradual doses of each strain orally and were monitored for 30 d after inoculation. The results showed that the LD50 of the wild-type S. Typhimurium S100 was 5.0 × 105 CFU, displaying the most virulence among all of the tested strains. The LD50 of S491 (ΔrffG8) of 3.2 × 107 CFU indicated that it was 64-fold less virulent than the wild-type S100. The mutant strains S490 (ΔrfbB6), S492 (ΔrfbB6 ΔrffG8) and S496 (PBAD rfbB-3) were avirulent while grown with arabinose prior to oral administration to mice (LD50 > 109 CFU) (Table 2).
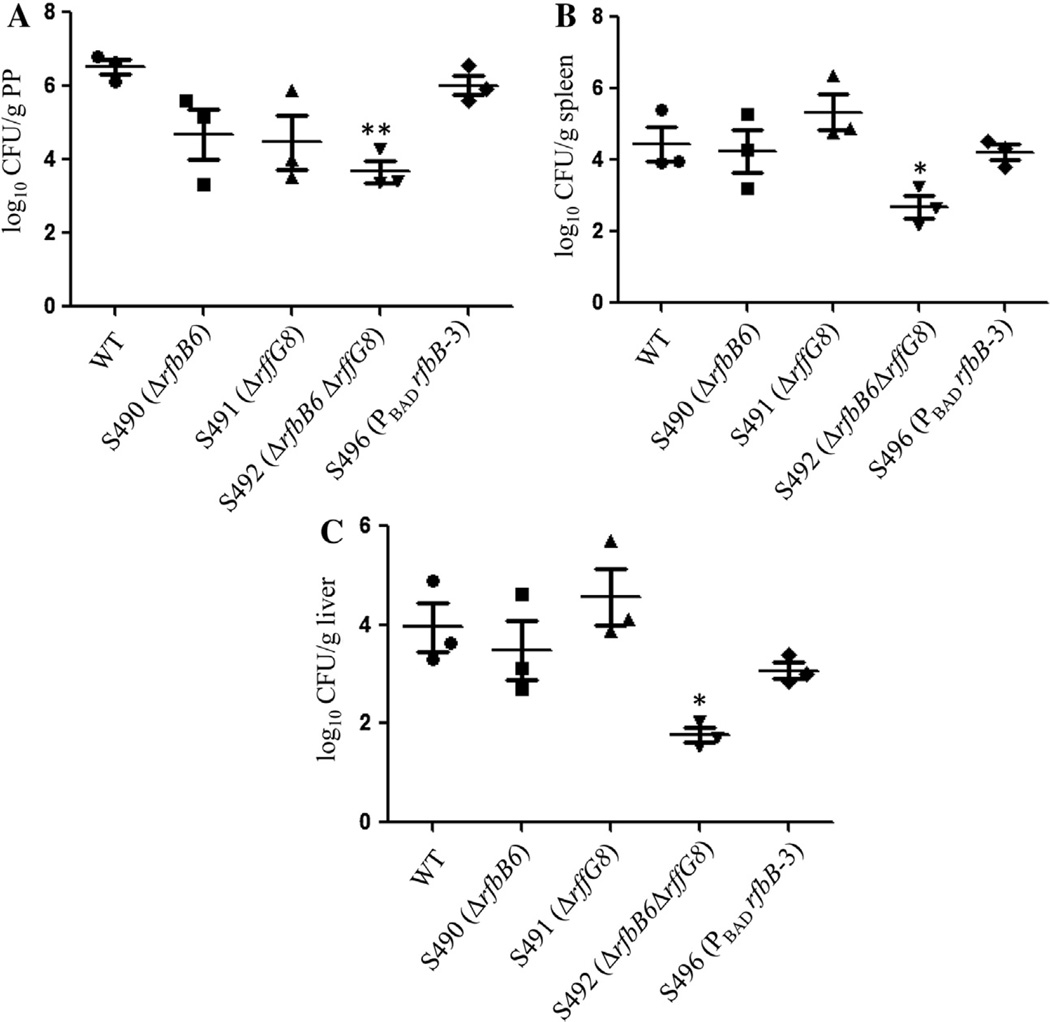
Due to the significant attenuation of these mutant strains, we also determined whether these mutants retained the capacity to efficiently colonize and disseminate in mouse tissues. The bacterial numbers in tissues from mice infected with the wild-type Salmonella strain recovered from PP (Fig. 3A), spleen (Fig. 3B), and liver (Fig. 3C) were approximately 106, 104, and 104, respectively. The colonization levels of the ΔrfbB6 and ΔrffG8 single mutants and the arabinose-regulated mutant S496 (PBAD rfbB-3) in the spleen and liver were similar to those of the parent strain S100. However, in all three different organs from infected mice including spleen, liver and PP, the mutant S492 (ΔrfbB6 ΔrffG8) showed significantly lower colonization levels than the wild-type S100 (P < 0.05), but no significant differences in the bacterial loads in the PP, spleen and liver were observed between the wild-type S100 and arabinose-regulated mutant S496 (PBAD rfbB-3) (P > 0.05) (Fig. 3).
Fig. 3.
Colonization by the wild-type Salmonella and mutants in mice organs. Groups of mice (n = 3/group) were orally inoculated with approximately 1 × 109 CFU of the indicated strains, and the viable bacteria were recovered from the PP, spleen, and liver of BALB/c mice 6 days after oral inoculation. The bacterial counts were determined and are displayed as log10 CFU/g of PP (A), spleen (B), and liver (C) tissue. Salmonella S100 is the wild-type strain that was used as a positive control. The horizontal lines represent the means and the error bars represent the SEM. Values that are significantly different from those of the wild type as indicated by one-way ANOVA are indicated by asterisks (*P < 0.05, **P < 0.01).
3.5. Evaluation of immunogenicity and protective efficacy
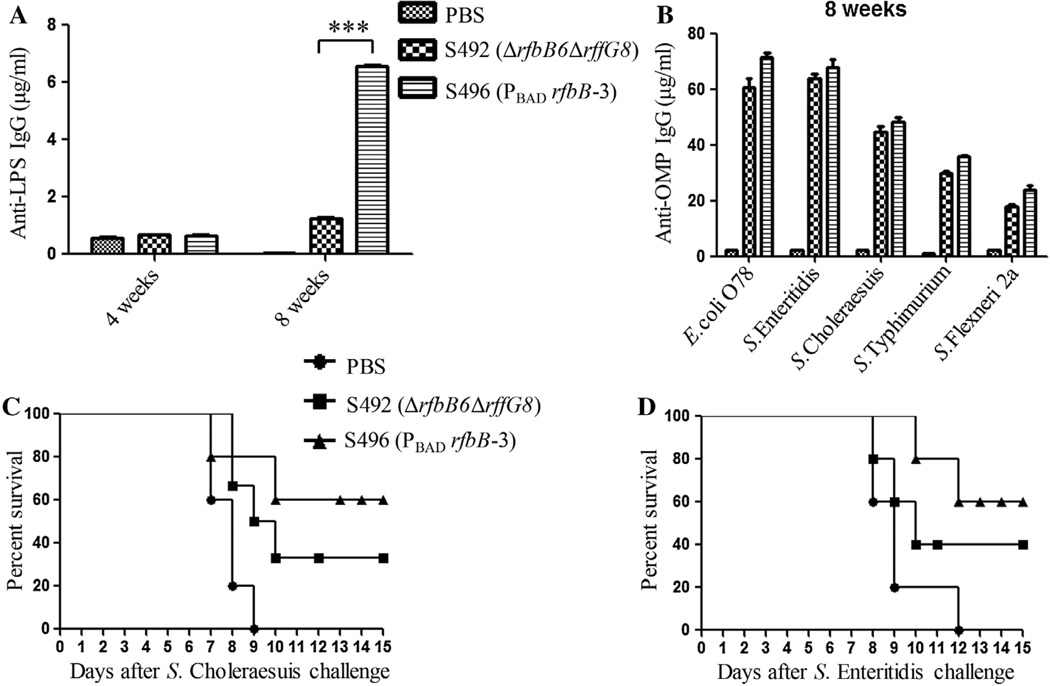
As the mutant S496 (PBAD rfbB-3) with the arabinose-regulated synthesis of LPS and ECA was significantly attenuated but nonetheless efficiently colonized and disseminated to mouse tissues (Table 2 and Fig. 3), we then determined whether immunization with this mutant induces protective immunity and enhances protection efficacy. The mutant S496 (PBAD rfbB-3) strain induced the highest titer of anti-LPS IgG, which was significantly higher than that of the mutant S492 (ΔrfbB6 ΔrffG8) at week 8 (P < 0.001) (Fig. 4A). The anti-OMP IgG level from mice immunized with the mutant S496 (PBAD rfbB-3) was higher than that from mice immunized with S492 (ΔrfbB6 ΔrffG8) with no significant difference (Fig. 4B). The immunized mice were challenged orally with 6.5 × 108 CFU of the wild-type S. Typhimurium 4 wks after the second immunization. The mice immunized with either S492 (ΔrfbB6 ΔrffG8) or S496 (ΔPrfbB-3) were fully protected against a subsequent lethal challenge of 6.5 × 108 CFU of the wild-type Salmonella S100, and the mice in the control group succumbed after a challenge of 15 days.
Fig. 4.
Serum immune responses and protection efficacy in orally immunized mice. Total serum IgG specific for S. Typhimurium LPS (A) and serum IgG against OMPs from S. Typhimurium and other enteric bacteria (B) were measured via quantitative ELISA. Each group consisted of 12 mice. The data represent antibody concentrations in pooled sera from orally immunized mice at the indicated number of weeks post-immunization (***P < 0.001, S492 (ΔrfbB6 ΔrffG8) versus S496 (PBAD rfbB-3)). The protection rates against S. Choleraesuis (C) (n = 6/group) and S. Enteritidis (D) (n = 6/group) challenge are shown. The immunized mice were challenged by oral inoculation with 200 × LD50 S. Choleraesuis and 200 × LD50 S. Enteritidis. The mice were monitored daily and deaths were recorded for 30 days to calculate the protection rates after challenge. Statistical analyses were performed using the log-rank test.
3.6. Evaluation of cross-reactive immunity and cross-protection against heterologous Salmonella challenge
The arabinose-regulated mutant S496 (PBAD rfbB-3) induced a strong immune response to OMPs from S. Typhimurium (Fig. 4B). Therefore, it was worthwhile to investigate whether S496 (PBAD rfbB-3) elicits cross-reactive immunity and confers cross-protection against other Salmonella infections. The serum IgG responses to OMPs from other enteric bacteria were determined in the blood of mice immunized with S496 (PBAD rfbB-3) or S492 (ΔrfbB6 ΔrffG8) at week 8. Although S496 (PBAD rfbB-3) was able to induce a higher IgG immune response to OMPs than S492 (ΔrfbB6 ΔrffG8) from all of the tested bacteria, no significant differences were observed in the immune responses (Fig. 4B).
To evaluate the cross protective efficacy conferred by S496 (PBAD rfbB-3), the clinically relevant serogroups S. Choleraesuis and S. Enteritidis were selected to challenge the immunized mice with doses of 2.4 × 107 (~200 × LD50) and 2.6 × 107 CFU (~200 × LD50) by oral administration 4 wks following the second immunization, respectively. For the challenge with S. Choleraesuis, the survival rate of the mice immunized with S496 (PBAD rfbB-3) was 60%, which was higher than the protection level afforded by S492 (ΔrfbB6 ΔrffG8) (33.3%) and the mock-immunized controls (0%) (Fig. 4C). There were significant increases in survival after S492 (ΔrfbB6 ΔrffG8) or S496 (PBAD rfbB-3) immunization following S. Enteritidis challenge compared with the oral mock-immunized animals (Fig. 4D) (P < 0.01). Moreover, S496 (PBAD rfbB-3) afforded better cross-protection than S492 (ΔrfbB6 ΔrffG8) (Fig. 4C and D).
4. Discussion
The presence of multiple O-antigen serotypes in pathogenic Salmonella poses a major challenge for designing and developing a universal Salmonella vaccine to induce cross-protective immunity against numerous Salmonella serotypes, such as S. Typhimurium, S. Choleraesuis and S. Enteritidis [28]. O-antigen polysaccharide and ECA are two major immunogenic antigens on the surface of Salmonella [8,9]. Previous studies have indicated that O-antigen-deficient strains are typically less immunogenic and fail to effectively colonize in the host organ due to an inability to resist host pressures [6].
Truncation of LPS in Salmonella may result from blocking the synthesis of dNDP-sugars that are the precursors of core or O-antigen chains, from the inactivity of glycosyltransferases that are responsible for the formation of sugar linkages, or from the interruption of O-antigen processing genes [6,12,13,29]. Deletion of the gylcosyltransferase genes or O-antigen processing genes leads to a nonreversible incomplete core or O-antigen of LPS [6]; however, deletion of genes such as galE or pmi, which are responsible for dNDP-sugar synthesis, results in mutants with reversible LPS [15,17,30]. It is obvious that an LPS- and ECA-negative strain would have a decreased capacity to withstand acidic, osmotic, bile and other stresses from the host, resulting in a decreased colonization capacity and impacting the induction of immune responses [6,11]. Thus, an optimal live oral Salmonella vaccine would retain its capacity to colonize and invade host lymphoid tissues while remaining completely avirulent [15,31–33]. In this study, we achieved construction of a mutant in which both O-antigen and ECA synthesis were simultaneously regulated by arabinose availability, resulting in enhanced cross-reactive immunity and cross-protection against challenge with multiple Salmonella serotypes compared with the double mutant.
Previous studies have demonstrated that the rfbB gene in the O-antigen gene cluster has functional homology with the rffG gene located in the ECA gene cluster [8,19]. Our results showed that the rfbB and rffG genes were exchangeable in terms of their impact on LPS and ECA synthesis (Fig. 1). Therefore, we targeted the rfbB gene to regulate O-antigen and ECA synthesis by arabinose availability via the araC PBAD promoter, which has been widely used to regulate expression of essential genes [12,13,34]. As the rfbB and rffG genes are located within operons, our strategy was to insert araC PBAD rfbB with a variable SD sequence and a start codon at a different position to maintain operon integrity. In this case, the araC PBAD rfbB cassette was inserted into the pagL position as we have demonstrated that deletion of pagL does not alter Salmonella virulence [35]. We obtained an ideal arabinose-regulated rfbB expression strain, S496, by assaying a series of biological characteristics and demonstrating wild-type characteristics when the strain was grown with arabinose (Figs. 1–3).
The animal experiments showed that the mutant S496 (PBAD rfbB-3) was fully attenuated (LD50 > 109 CFU) but retained the capacity to disseminate in host organs (Fig. 3). Previous studies have shown that immunization with attenuated Salmonella strains with regulated LPS synthesis can elicit cross-reactive antibodies reacting with OMPs from different Salmonella serovars [6,29,36]. In addition, an ECA-deficient mutant induced a protective immune response against lethal challenge of homologous and heterologous Salmonella when used as a live-attenuated vaccine [11,37]. We observed that mutant S496 (PBAD rfbB-3) induced higher levels of cross-reactive antibodies against OMPs from multiple enteric bacteria than the double mutant S492 (ΔrfbB6 ΔrffG8) lacking O-antigen and ECA polysaccharides, indicating that regulation of both O-antigen and ECA would be beneficial for inducing cross-reactive immune responses against conserved OMPs from multiple Salmonella infections. The PBAD rfbB-3 mutant strain was able to provide higher cross-protection levels against S. Choleraesuis and S. Enteritidis than the mutant S492 (ΔrfbB6 ΔrffG8) (Fig. 4), but no significant difference was observed for the immune responses or protection efficacy (P > 0.05) (Fig. 4). We did not observe a significant advantage of S496 (PBAD rfbB-3) over S492 (ΔrfbB6 ΔrffG8) for inducing cross-reactive immune responses in this study, which may be due to the fact that the double mutant (ΔrfbB6 ΔrffG8) still colonized and persisted well in the host organs (Fig. 3). As more than one means of attenuating Salmonella is necessary for live attenuated vaccine development [33,38], we will introduce other genetically unlinked mutations into this mutant to ensure the safety of the attenuated Salmonella strain and to evaluate cross-reactive immune responses and protection efficacy in future works.
In summary, we constructed a Salmonella mutant with arabinose-regulated O-antigen and ECA synthesis, and we demonstrated that immunization with this mutant induces cross-protection against heterologous Salmonella infection. Our results indicate that this mutation is a good choice to combine with other mutations to maximally unmask conserved immunogenic epitopes on the surface of Salmonella to develop a universal live vaccine for preventing infection by enteric bacteria.
Supplementary Material
Acknowledgments
We thank Dr. Susanne Haussler (The Hannover Medical School, The Hannover University, Germany) for kindly providing the anti-ECA mAb. This work was supported by grants 31270981 and 31472179 from the National Natural Science Foundation of China, and by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (Grant NIH R01 AI112680 to Q.K.).
Footnotes
Appendix A. Supplementary material
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vaccine.2016.07.010.
References
- 1.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toguchi A, Siano M, Burkart M, Harshey RM. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J Bacteriol. 2000;182:6308–6321. doi: 10.1128/jb.182.22.6308-6321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nevola JJ, Laux DC, Cohen PS. In vivo colonization of the mouse large intestine and in vitro penetration of intestinal mucus by an avirulent smooth strain of Salmonella typhimurium and its lipopolysaccharide-deficient mutant. Infect Immun. 1987;55:2884–2890. doi: 10.1128/iai.55.12.2884-2890.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaio MF, Rowland H. Bactericidal and opsonizing effects of normal serum on mutant strains of Salmonella typhimurium. Infect Immun. 1985;49:647–653. doi: 10.1128/iai.49.3.647-653.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagy G, Danino V, Dobrindt U, Pallen M, Chaudhuri R, Emödy L, et al. Down-regulation of key virulence factors makes the Salmonella enterica serovar Typhimurium rfaH mutant a promising live-attenuated vaccine candidate. Infect Immun. 2006;74:5914–5925. doi: 10.1128/IAI.00619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kong Q, Yang J, Liu Q, Alamuri P, Roland KL, Curtiss R. Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar Typhimurium. Infect Immun. 2011;79:4227–4239. doi: 10.1128/IAI.05398-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray GL, Attridge SR, Morona R. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J Bacteriol. 2006;188:2735–2739. doi: 10.1128/JB.188.7.2735-2739.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rick P, Silver R. Enterobacterial common antigen and capsular polysaccharides. In: Neidhardt, editor. Escherichia coli and Salmonella: cellular and molecular biology. Washington, DC: ASM Press; 1996. pp. 104–122. [Google Scholar]
- 9.Rick PD, Mayer H, Neumeyer BA, Wolski S, Bitter-Suermann D. Biosynthesis of enterobacterial common antigen. J Bacteriol. 1985;162:494–503. doi: 10.1128/jb.162.2.494-503.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valtonen MV, Larinkari UM, Plosila M, Valtonen VV, Mäkelä PH. Effect of enterobacterial common antigen on mouse virulence of Salmonella typhimurium. Infect Immun. 1976;13:1601–1605. doi: 10.1128/iai.13.6.1601-1605.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbreath JJ, Colvocoresses Dodds J, Rick PD, Soloski MJ, Merrell DS, Metcalf ES. Enterobacterial common antigen mutants of Salmonella enterica serovar Typhimurium establish a persistent infection and provide protection against subsequent lethal challenge. Infect Immun. 2012;80:441–450. doi: 10.1128/IAI.05559-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong Q, Liu Q, Roland KL, Curtiss R. Regulated delayed expression of rfaH in an attenuated Salmonella enterica serovar Typhimurium vaccine enhances immunogenicity of outer membrane proteins and a heterologous antigen. Infect Immun. 2009;77:5572–5582. doi: 10.1128/IAI.00831-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong Q, Liu Q, Jansen AM, Curtiss R. Regulated delayed expression of rfc enhances the immunogenicity and protective efficacy of a heterologous antigen delivered by live attenuated Salmonella enterica vaccines. Vaccine. 2010;28:6094–6103. doi: 10.1016/j.vaccine.2010.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germanier R, Fürer E. Immunity in experimental salmonellosis II. Basis for the avirulence and protective capacity of galE mutants of Salmonella Typhimurium. Infect Immun. 1971;4:663–673. doi: 10.1128/iai.4.6.663-673.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Wang S, Scarpellini G, Gunn B, Xin W, Wanda S-Y, et al. Evaluation of new generation Salmonella enterica serovar Typhimurium vaccines with regulated delayed attenuation to induce immune responses against PspA. Proc Natl Acad Sci USA. 2009;106:593–598. doi: 10.1073/pnas.0811697106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hone D, Morona R, Attridge S, Hackett J. Construction of defined galE mutants of Salmonella for use as vaccines. J Infect Dis. 1987;156:167–174. doi: 10.1093/infdis/156.1.167. [DOI] [PubMed] [Google Scholar]
- 17.Collins LV, Attridge S, Hackett J. Mutations at rfc or pmi attenuate Salmonella typhimurium virulence for mice. Infect Immun. 1991;59:1079–1085. doi: 10.1128/iai.59.3.1079-1085.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allard ST, Giraud M-F, Whitfield C, Graninger M, Messner P, Naismith JH. The crystal structure of dTDP-d-glucose 4, 6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium, the second enzyme in the dTDP-L-rhamnose pathway. J Mol Biol. 2001;307:283–295. doi: 10.1006/jmbi.2000.4470. [DOI] [PubMed] [Google Scholar]
- 19.Marolda CL, Valvano MA. Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7: K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J Bacteriol. 1995;177:5539–5546. doi: 10.1128/jb.177.19.5539-5546.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayama K, Kelly SM, Curtiss R. Construction of an Asd+ expression-cloning vector: stable maintenance and high level expression of cloned genes in a Salmonella vaccine strain. Nat Biotechnol. 1988;6:693–697. [Google Scholar]
- 21.Kang HY, Dozois CM, Tinge SA, Lee TH, Curtiss R. Transduction-mediated transfer of unmarked deletion and point mutations through use of counterselectable suicide vectors. J Bacteriol. 2002;184:307–312. doi: 10.1128/JB.184.1.307-312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Hitchcock PJ, Brown TM. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peters H, Jürs M, Jann B, Jann K, Timmis KN, Bitter-Suermann D. Monoclonal antibodies to enterobacterial common antigen and to Escherichia coli lipopolysaccharide outer core: demonstration of an antigenic determinant shared by enterobacterial common antigen and E. coli K5 capsular polysaccharide. Infect Immun. 1985;50:459–466. doi: 10.1128/iai.50.2.459-466.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gahring LC, Heffron F, Finlay BB, Falkow S. Invasion and replication of Salmonella Typhimurium in animal cells. Infect Immun. 1990;58:443–448. doi: 10.1128/iai.58.2.443-448.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang HY, Srinivasan J, Curtiss R. Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar Typhimurium vaccine. Infect Immun. 2002;70:1739–1749. doi: 10.1128/IAI.70.4.1739-1749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motulsky HJ. Prism 5 statistics guide. San Diego: GraphPad Software Inc; 2007. [Google Scholar]
- 28.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive nontyphoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet. 2012;379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy G, Palkovics T, Otto A, Kusch H, Kocsis B, Dobrindt U, et al. “Gently rough”: the vaccine potential of a Salmonella enterica regulatory lipopolysaccharide mutant. J Infect Dis. 2008;198:1699–1706. doi: 10.1086/593069. [DOI] [PubMed] [Google Scholar]
- 30.Hone DM, Attridge SR, Forrest B, Morona R, Daniels D, LaBrooy JT, et al. A galE via (Vi antigen-negative) mutant of Salmonella Typhi Ty2 retains virulence in humans. Infect Immun. 1988;56:1326–1333. doi: 10.1128/iai.56.5.1326-1333.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galen JE, Curtiss R. The delicate balance in genetically engineering live vaccine. Vaccine. 2014;32:4376–4385. doi: 10.1016/j.vaccine.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S, Kong Q, Curtiss R. New technologies in developing recombinant attenuated Salmonella vaccine vectors. Microb Pathogenesis. 2013;58:17–28. doi: 10.1016/j.micpath.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curtiss R. Bacterial infectious disease control by vaccine development. J Clin Invest. 2002;110:1061–1066. doi: 10.1172/JCI16941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Curtiss R, Wanda S-Y, Gunn BM, Zhang X, Tinge SA, Ananthnarayan V, et al. Salmonella enterica serovar Typhimurium strains with regulated delayed attenuation in vivo. Infect Immun. 2009;77:1071–1082. doi: 10.1128/IAI.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong Q, Six DA, Roland KL, Liu Q, Gu L, Reynolds CM, et al. Salmonella synthesizing 1-dephosphorylated lipopolysaccharide exhibits low endotoxic activity while retaining its immunogenicity. J Immunol. 2011;187:412–423. doi: 10.4049/jimmunol.1100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beal RK, Wigley P, Powers C, Barrow PA, Smith AL. Cross-reactive cellular and humoral immune responses to Salmonella enterica serovars Typhimurium and enteritidis are associated with protection to heterologous re-challenge. Vet Immunol Immunopathol. 2006;114:84–93. doi: 10.1016/j.vetimm.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Curtiss R. Efficient generation of influenza virus with a mouse RNA polymerase I-driven all-in-one plasmid. Virol J. 2015;12:95. doi: 10.1186/s12985-015-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curtiss R, Xin W, Li Y, Kong W, Wanda SY, Gunn B. New technologies in using recombinant attenuated Salmonella vaccine vectors. Crit Rev Immunol. 2010;30:257–270. doi: 10.1615/critrevimmunol.v30.i3.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun W, Wang S, Curtiss R. Highly efficient method for introducing successive multiple scarless gene deletions and markerless gene insertions into the Yersinia pestis chromosome. Appl Environ Microbiol. 2008;74:4241–4245. doi: 10.1128/AEM.00940-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Q, Liu Q, Zhao X, Liu T, Yi J, Liang K, et al. Immunogenicity and cross-protective efficacy induced by outer membrane proteins from Salmonella Typhimurium mutants with truncated LPS in mice. Int J Mol Sci. 2016;17(3) doi: 10.3390/ijms17030416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roland K, Curtiss R, Sizemore D. Construction and evaluation of a Δcya Δcrp Salmonella typhimurium strain expressing avian pathogenic Escherichia coli O78 LPS as a vaccine to prevent airsacculitis in chickens. Avian Dis. 1999;43:429–441. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.